Summary
Plants have evolved a sophisticated immunity system for specific detection of pathogens and rapid induction of measured defences. Over‐ or constitutive activation of defences would negatively affect plant growth and development. Hence, the plant immune system is under tight positive and negative regulation. MAP kinase phosphatase1 (MKP1) has been identified as a negative regulator of plant immunity in model plant Arabidopsis. However, the molecular mechanisms by which MKP1 regulates immune signalling in wheat (Triticum aestivum) are poorly understood. In this study, we investigated the role of TaMKP1 in wheat defence against two devastating fungal pathogens and determined its subcellular localization. We demonstrated that knock‐down of TaMKP1 by CRISPR/Cas9 in wheat resulted in enhanced resistance to rust caused by Puccinia striiformis f. sp. tritici (Pst) and powdery mildew caused by Blumeria graminis f. sp. tritici (Bgt), indicating that TaMKP1 negatively regulates disease resistance in wheat. Unexpectedly, while Tamkp1 mutant plants showed increased resistance to the two tested fungal pathogens they also had higher yield compared with wild‐type control plants without infection. Our results suggested that TaMKP1 interacts directly with dephosphorylated and activated TaMPK3/4/6, and TaMPK4 interacts directly with TaPAL. Taken together, we demonstrated TaMKP1 exert negative modulating roles in the activation of TaMPK3/4/6, which are required for MAPK‐mediated defence signalling. This facilitates our understanding of the important roles of MAP kinase phosphatases and MAPK cascades in plant immunity and production, and provides germplasm resources for breeding for high resistance and high yield.
Keywords: MAP kinase phosphatase 1, wheat defence responses, MPK3, MPK4, MPK6, CRISPR/Cas9
Introduction
Plants have evolved complex mechanisms to recognize invading pathogens and mount appropriate defence responses to fight off infection (Dodds and Rathjen, 2010; Schwessinger and Ronald, 2012). They detect the presence of pathogen/microbe‐associated molecular patterns (PAMPs/MAMPs) and pathogen effector proteins, and subsequently initiate a diverse array of defence responses, via highly conserved signalling mechanisms (Teixeira et al., 2019; Zhou and Zhang, 2020). The recognition of PAMPs by plasma membrane‐localized pattern recognition receptors (PRRs) ensues PAMP‐triggered immunity (PTI). Adapted pathogens employ secreted effectors to subvert PTI. In response, plant intracellular immune receptors detect specific pathogen effectors, resulting in effector‐triggered immunity (ETI) (Li et al., 2020; Monaghan and Zipfel, 2012). There are several early signalling events and defence responses during PTI and ETI. These include intracellular calcium flux, activation of the mitogen‐activated protein kinase (MAPK) cascades, production of extracellular reactive oxygen species (ROS), and increased biosynthesis of salicylic acid (SA). ETI is often collimated with hypersensitive response (HR), which is rapid and localized cell death at the site of infection that plays an important role in containing invading pathogens, especially biotrophic fungi (Wang et al., 2020).
MAPK activation is one of the earliest signalling events that function downstream of immune receptors and transduce phytopathogenic stimuli into cellular defence responses (Zhang et al., 2018). MAPK cascades are three‐kinase modules consisting of MAPK kinase kinases (MKKKs/MEKKs), MAPK kinases (MKKs/MEKs), and MAPKs (Ichimura et al., 2002). In MAPK cascades, MAPKs are phosphorylated and activated by MAPKKs, which are in turn activated by their upstream kinases MAPKKKs (Zhang and Klessig, 2001). Numerous members of MAPK cascades have been reported to be involved in defence signalling. Notably, most research in the past has been focused on two hallmark parallel MAPK cascades (Devendrakumar et al., 2018). In Arabidopsis, the MKKK3/5‐MKK4/5‐MPK3/6 cascade downstream of multiple plant receptor kinases positively regulates both PTI and ETI responses (Sun et al., 2018; Zhang and Klessig, 2001). The Arabidopsis MEKK1‐MKK1/2‐MPK4 cascade is another best studied MAPK cascade involved in flg22‐induced immune signalling and mechanical wounding responses linked to ROS signalling (Takahashi et al., 2011).
MAPKs are activated through sequential phosphorylation and deactivated upon dephosphorylation by phosphatases. Three types of protein phosphatases are known to deactivate MAPK. These include tyrosine phosphatases, serine/threonine phosphatases, and dual specificity phosphatases (DSPs) that are capable of removing the phosphate groups from phosphoserine, phosphothreonine, and phosphotyrosine residues (Keyse, 2000). MAPK phosphatase 1 (MKP1), belonging to the group of DSPs, has been previously described as a broad negative regulator of plant stress responses through its regulation of MAPK signalling. In Arabidopsis, AtMKP1 (At3g55270) has been shown to be a crucial regulator of AtMPK6 (At2g43790) activity, determining the outcome of the cellular reaction and the level of genotoxic resistance (Ulm et al., 2001, 2002). In rice, OsMKP1 negatively regulates MAPK‐mediated phosphorylation of the transcription factor MYB4 which in turn negatively regulates vascular lignification through inhibiting lignin biosynthesis and vascular resistance to Xanthomonas oryzae pv. Oryzicola (Xoo) (Lin et al., 2022). OsMKP1 also negatively regulates SA‐ and ROS‐dependent mesophyll resistance to Xoo via dephosphorylation of MAPK (Lin et al., 2022; Zhang et al., 2022), which is consistent with earlier studies that AtMKP1 interacts with its targets MPK3 (At3g45640) and MPK6, thereby negatively regulating SA biosynthesis and ROS to maintain the best balance between growth and pathogen resistance (Anderson et al., 2011; Bartels et al., 2009). Phosphorylation of AtMKP1 was also shown to be required for PAMP responses and bacteria resistance (Escudero et al., 2019; Jiang et al., 2017), and increased bacterial resistance in Atmkp1 was found to be attributable to a decrease in the extracellular accumulation of bioactive metabolites that induce the bacteria's type III secretion system critical for bacterial pathogenicity (Anderson et al., 2014). Notably, AtMKP1 was also reported to be phosphorylated and activated by its physiological substrate, MPK6 (Park et al., 2011). In addition, AtMKP1 directly regulates RBOHD, an NADPH oxidase for ROS production during PTI process (Escudero et al., 2019). A more recent study showed that AtMKP1 promotes osmotolerance by suppressing the PAD4‐independent immune response activated by MPK3/6 (Uchida et al., 2022).
Heterologous expression of TdMKP1 isolated from durum wheat (Triticum durum) in Arabidopsis was shown to increase salt stress tolerance of the transgenic plants (Zaidi et al., 2016). TdMKP1 contains a canonical 14–3‐3 binding motif and was shown to interact with and regulated by 14–3‐3 proteins, a family of highly conserved acidic proteins that act as dynamic coordinators of many biological processes (Ghorbel et al., 2017). In gel assays showed that TdMKP1 could be regulated by calmodulin, bivalent cations and possibly TdMPK3 but its phosphorylation status seemed dispensable for an optimal phosphatase activity (Ghorbel et al., 2019). To date, whether MKP1 in wheat especially the allohexaploid bread wheat (Triticum aestivum) also plays an important role in negative regulation of immune signalling similar as AtMKP1 in Arabidopsis has yet to be determined.
In this study, we identified three TaMKP1 homoeologous genes in the genome of bread wheat and examined their expression patterns in response to pathogen infection. More importantly, we obtained definitive evidence to indicate that TaMKP1 plays a role in negative regulation of defence responses via inhibiting the activation of three key components of MPK3, MPK4 and MPK6 that function in two parallel MAPK cascades. We also generated wheat mkp1 plants by clustered regularly interspaced short palindromic repeats/CRISPR‐associated 9 (CRISPR/Cas9) technology and found that while Tamkp1 plants showed increased resistance to biotrophic rust and powdery mildew fungi, surprisingly, they also displayed improved wheat agronomic traits (plant height, ears and grain weight). The research findings presented in this study suggest novel molecular mechanisms that underlie the interplay between growth and defence in wheat and hint new strategies for improving both disease resistance and yield characteristics through genetic manipulation of TaMKP1.
Results
TaMKP1 expression is rapidly induced upon fungal infection
The genome of allohexaploid bread wheat contains three MKP1 homoeologs (TraesCS1A02G045300, TraesCS1B02G058600 and TraesCS1D02G046000) with ~97% nucleotide sequence identity between each other (Figure S1). The three deduced TaMPK1 proteins show ~54% sequence identity to AtMKP1, and were designated TaMKP1a, TaMKP1b and TaMKP1d, respectively, according to their chromosomal (Figure 1a). Phylogenetic analysis with the three TaMKP1 homoeologs and 16 representative MPK1s from other plant species showed that TaMKP1d is most closely related to predicted MKP1 proteins from Aegilops tauschii (which contributes to the wheat D genome) while TaMKP1a is most closely related to MPK1 form from Durum wheat Triticum turgidum (Figure 1b).
Figure 1.
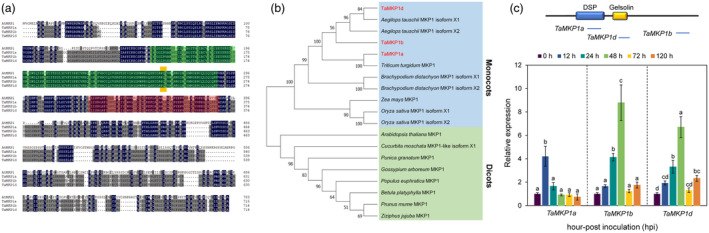
Identification of bread wheat MKP1 (TaMKP1) homologues and expression patterns. (a) Multiple alignment of the amino acid sequences of TaMKP1a, TaMKP1b, TaMKP1d, and AtMKP1. The green box indicates the DSP (dual‐specificity phosphatase) domain, the red box represents the Gelsolin repeat region, and the yellow region represents the FXF motif. (b) Phylogenetic analysis between TaMKP1 and other plants. (c) The expression level of TaMKP1 induced by Pst CYR34. TaMKP1 contains DSP and Gelsolin domains, and the horizontal line under the domain indicates the TaMKP1 RT‐qPCR amplified fragment.
To assess the role of TaMKP1 in the regulation of disease resistance, wheat plants were challenged with a virulent Puccinia striiformis f. sp. tritici (Pst) race (CYR34). The expression level of TaMKP1a in the inoculated leaves rapidly increased by more than 4 times at 12 h‐post inoculation (hpi), and then gradually dropped back to the level prior to infection. TaMKP1b and TaMKP1d expression was upregulated at 24 hpi and reached a maximum at 48 hpi, with 8.7‐ and 6.7‐fold increases, respectively (Figure 1c). This result implies that TaMKP1 may be involved in early responses of wheat to fungal infection. The total transcripts of TaMKP1 were found to be most abundant in mature leaves, which is over 2‐fold higher (P < 0.05) than those of root tissue. In grains and culms, the total transcripts of TaMKP1 were slightly less abundant compared to those in roots (Figure S2).
Knocking down TaMKP1 results in enhanced resistance in wheat
To determine the role of TaMKP1 in regulating defence responses in wheat, a well‐established virus‐induced gene silencing (VIGS) system was employed. Specifically, two artificial microRNA (amiRNA) genes that aim to target all of the three TaMKP1 homoeologs were made and expressed in wheat using the BSMV‐mediated gene‐silencing system. At 12 dpi with BSMV, leaf photo‐bleaching was observed in plants inoculated with the virus harbouring the amiRNA construct targeting the wheat phytoene desaturase gene (TaPDS), and mild chlorotic mosaic phenotypes were apparent on leaves inoculated with the virus carrying no insert or one of the two amiRNA constructs targeting TaMKP1 (Figure 2a), indicating that the BSMV‐based VIGS system was working. Subsequently, the fourth leaves were inoculated with fresh urediniospores of Pst‐CYR34. As expected, numerous uredia were produced on mock (BSMV‐Y) leaves at 14 dpi. By contrast, noticeably less Pst uredia were observed on leaves infected with BSMV:TaMKP1‐V1 or BSMV:TaMKP1‐V2 (Figure 2a). QRT‐PCR showed that the transcription levels of TaMKP1 in the fourth leaves of plants inoculated with either BSMV:TaMKP1‐V1 or BSMV:TaMKP1‐V2 were suppressed below 35% of that of the mock plants at 24, 48, 72, and 120 hpi with Pst‐CYR34 (Figure 2b). When TaMKP1 was silenced, the proportion of surface spores in leaves of BSMV‐TaMKP1 plants decreased significantly compared with control plants (Figure 2c). In addition, the proportion of spore heaps and the activity of the TaMKP1 enzyme in TaMKP1‐silenced plants showed a significant decrease compared to those in the mock plants (Figure 2d,e).
Figure 2.
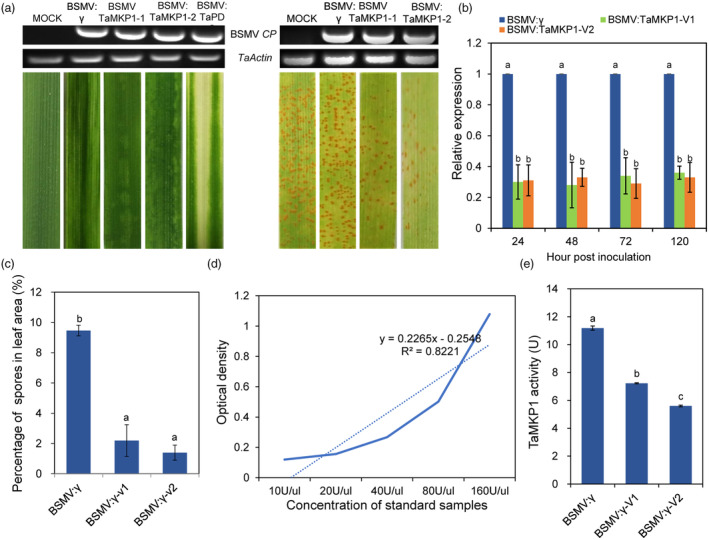
The silencing of TaMKP1 enhances the resistance of wheat to pathogens. (a) Wheat leaf symptoms after virus inoculation and phenotype of wheat leaves silenced for TaMKP1 gene infected by Pst. (b) Relative transcript levels of TaMKP1 in control and TaMKP1‐silenced plants. (c) The percentage of Pst urediniospores on wheat leaves. (d) The standard curve for MKP1 enzyme activity. (e) Changes in MKP1 enzyme activity in silenced plants.
Knocking out TaMKP1 by CRISPR/Cas9 results in clear enhanced resistance
To more definitively assess the role of TaMKP1 in regulation of defence signalling in wheat, CRISPR/Cas9 gene‐editing was used to knock out TaMKP1 in wheat. To achieve this, a single guide RNA was designed to target an exonic region where the nucleotide sequence is identical among the three TaMKP1 genes (Figure 3a). Successful transformation of Fielder immature embryos was achieved through Agrobacterium mediated transformation, resulting in T0 events. Mutant lines were identified using the bar gene test strip (Figure S3). To identify indel mutations in the target genes, specific primers were designed to amplify and sequence the target region of the three TaMPK1 homeologs. Among eight transgenic T1 lines analysed by targeted sequencing, a frameshift indel resulting in an earlier stop codon in each of the three TaMPK1 homeologs were found in two lines (#8 and #9), while one frameshift indel in one of the three TaMKP1 homeologs was identified in the remaining six lines (Figures 3a and S4). T2 progenies of the 8 T1 lines with the respective homozygous mutations were obtained and subjected to infection tests with Pst CYR34. For simplicity, aa, bb and dd are used to denote homozygous knockout mutations in TaMKP1 in the A, B and D genome, respectively. Not surprisingly, while aabbdd plants of line #8 and #9 showed the highest levels of resistance, which was reflected in lower fungal biomass at 14 dpi, which decreased by 86.5% and 86%, respectively. AABBdd plants from line #10 and #19 exhibited the second lowest incidence and fungal biomass, which decreased by 83.9% and 83.7%, respectively (Figure 3b). AAbbDD plants from line #1 and #12 and aaBBDD plants from line #17 and #21 were also significantly less susceptible compared to the WT Fielder plants, even though they were not as resistant as aabbdd and AABBdd plants (Figure 3b). DAB and Trypan Blue staining were used to respectively assess the in‐situ accumulation of hydrogen peroxide (H2O2) and HR cell death in infected leaves at 24 hpi. While no obvious H2O2 was detected in WT plants, plants of the aabbdd genotype developed massive H2O2 with the highest positive area, followed by plants of the AABBdd, AAbbDD and aaBBDD genotypes (Figure 3c). Results of HR cell death was consistent with the development of macro‐lesions in Pst‐infected leaves (Figure 3b) and were also highly similar to patterns of H2O2 production and accumulation (Figure 3d).
Figure 3.
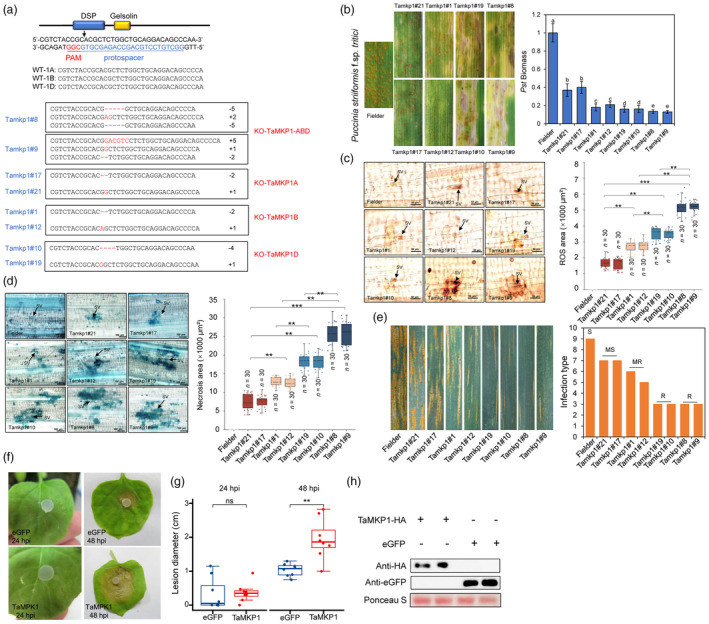
TaMKP1 plays a negative regulatory role in the interaction between plants and pathogens. (a) Selection of TaMKP1 mutation targets and identification of mutation types in TaMKP1 homologues. Deleted nucleotides are represented by “‐”. Inserted nucleotides are highlighted in red. The numbers on the right indicate the number of nucleotides involved in the indel events, represented by “+” or “−”. (b) Leaf phenotypes of aaBBDD, AAbbDD, AABBdd and aabbddd compared with Fielder at 14 dpi with Pst CYR34, and Pst biomass statistics. (c) ROS production was observed in Pst‐infested leaves at 24 hpi. (d) Histological observation of host cell death on the Tamkp1 leaves at 120 hpi with CYR34. Student's t‐tests were used to assess the differences between knockdown plants and control plants. Significance levels were denoted as * for P < 0.05, ** for P < 0.01, *** for P < 0.001 and ****for P < 0.001. SV refers to substomatal vesicle. (e) Characterization of Tamkp1 plants for disease resistance at the adult plant stage. S represents susceptible, MS represents moderately susceptible, MR represents moderately resistant, and R represents resistant. (f) Observations on the infection phenotype in overexpressed TaMKP1 Nicotiana benthamiana inoculated with Sclerotinia sclerotiorum. (g) Statistical analysis of lesion diameter following inoculation with S. sclerotiorum. ** indicate a significant difference (P < 0.01) according to Student's t‐test. (h) Western blot was performed to confirm the expression of TaMKP1 in N. benthamiana.
To confirm the enhanced resistance to virulent Pst race CYR34 in the TaMKP1 knockout lines is indeed due to PTI de‐repression, WT and mutant plants of the above‐described genotypes were treated with chitin or flg22. Our results showed that both chitin and flg22 more strongly induced H2O2 production in the mutant plants of different genotypes than WT plants (Figure S5). This result is consistent with more pronounced expression of TaCERK1 and TaRLCK185 (which is indicative of hyperactive chitin‐induced immune signalling) upon chitin treatment and TaFLS2 and TaMEKK1 (which is indicative of hyperactive flg22‐induced immune signalling) upon flg22 treatment in the mutant plants compared with WT plant (Figure S5).
T2 plants of the TaMKP1 knockout lines were subjected to mixed infection tests at the adult stage with four prevalent and severe Pst strains (CYR31, CYR32, CYR33 and CYR34). T2 plants of the aabbdd genotype and AABBdd genotypes supported least Pst urediniospores, while T2 plants of the AAbbDD and AABBdd genotypes also showed significantly reduced susceptibility to prevalent Pst races when compared the WT plants (Figure 3e). These results further demonstrated that TaMKP1 homeologs are negative regulators of resistance against Pst in wheat and suggested that TaMKP1d play the largest role among the three homeologs. To see if the enzymatic function of MKP1 is indeed conserved across plant species, we ectopically expressed TaMKP1 (which is identical to TaMKP1b and TaMKP1d at the protein level) in Nicotiana benthamiana leaves via Agrobacterium‐mediated transient expression. The TaMKP1 gene was translationally in‐frame fused with a C‐terminal hemagglutinin (HA) tag and TaMKP1‐HA fusion gene construct was transiently expressed in N. benthamiana leaves along with the eGFP construct (as control). Twenty‐four hours after Agroinfiltration, a Sclerotinia sclerotiorum strain virulent on N. benthamiana was inoculated on the infiltrated leaves. Larger lesions were observed on the leaves expressing TaMKP1 compared with those expressing eGFP (Figure 3f,g). Western blot analysis showed that both TaMKP1‐HA and eGFP were expressed in the infiltrated leaves (Figure 3h). This result suggested that TaMKP1 from wheat is capable of dephosphorylating relevant N. benthamiana proteins and consequently downregulating immune responses in distantly dicot plant species.
Next, plants of the Tamkp1 mutants inoculated with a virulent powdery mildew Blumeria graminis f. sp. tritici (Bgt) to evaluate the scope and effectiveness of their enhanced resistance. The results indicated that Tamkp1 plants have significantly reduced susceptibility to compared to the WT as shown by visual scoring of the infected leaves and Bgt biomass (Figure 4a). Similar to the infection phenotypes with Pst‐CYR34, the number and size of micro‐colonies of Bgt mycelium on leaves of the aabbdd mutant plants were most significantly reduced, followed by those of AABBdd, and then AAbbDD and aaBBDD genotypes compared to the WT (Figure 4b,c), while Bgt‐induced HR cell death in the infected leaves exhibited a reverse tendency based on visualization of cell death stained by trypan blue (Figure 4d). These results further substantiated our conclusion that knocking out TaMKP1 genes in wheat results in enhanced resistance to biotrophic fungal pathogens.
Figure 4.
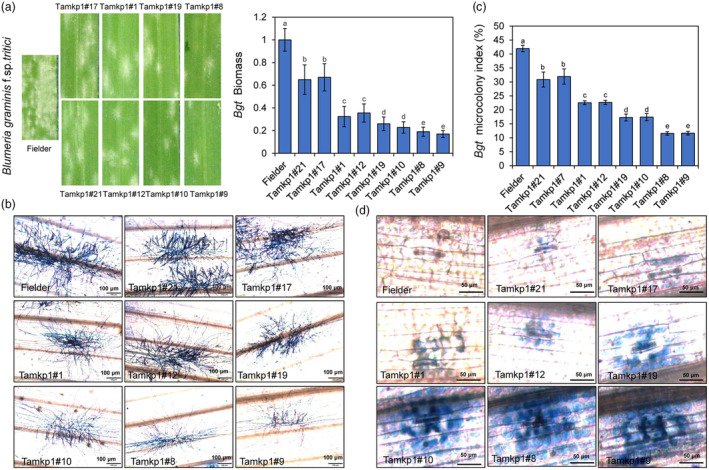
Tamkp1 plants demonstrate broad‐spectrum disease resistance. (a) Leaf phenotypes of Tamkp1 plants compared with Fielder at 7 dpi with Bgt, and Bgt biomass statistics. (b) The leaves of T2 generation of Tamkp1 plants were infected, and 7 dpi, they were stained with trypan blue to make fungal structures visible. (c) Statistical analysis of Bgt microcolony formation on Fielder and Tamkp1 leaves. (d) Host cell death on the Tamkp1 leaves with Bgt.
Nucleus‐localized TaMKP1 represses immune signalling
The N‐terminal domain of human MKP‐1 is responsible for both nuclear localization and for binding of its MAPK substrates (Kim and Asmis, 2017). Durum wheat MKP1 was localized in the nucleus and interacts with MPK3/6 (Zaidi et al., 2016). To investigate whether nuclear localization of TaMKP1 is required for repressing defence signalling, a nuclear localization signal (NLS) or a nuclear export signal (NES) was added to C‐terminus of TaMKP1‐GFP to make TaMKP1‐GFP‐NLS (or NES) constructs. Each of the two constructs were co‐expressed with the nuclear marker H2B‐mCherry (Long et al., 2019) in N. benthamiana leaves. Fluorescence microscopy showed that as expected, TaMKP1‐GFP‐NLS was localized in the nucleus, whereas TaMKP1‐GFP‐NES was found in the cytoplasm with little GFP signal seen in the nucleus (Figure 5a). Then, the infiltrated leaves were inoculated with S. sclerotiorum. Interestingly, leaves transiently expressing TaMKP1‐GFP‐NLS displayed significantly larger infection lesions compared with those expressing the TaMKP1‐GFP‐NES (Figure 5b). Gel blot analysis showed that TaMKP1‐GFP‐NES was expressed and distributed in both the cytoplasm and nucleus, mainly in the cytoplasm, while TaMKP1‐GFP‐NLS was only in the nucleus (Figure 5c). These observations suggest that TaMKP1 proteins mainly act to repress defence signalling in the nucleus.
Figure 5.
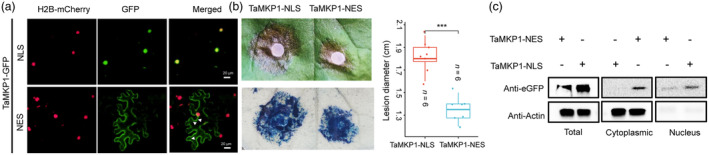
Nucleus‐localized TaMKP1 contributes to plant susceptibility against pathogens. (a) Observation of TaMKP1 nuclear localisation and nuclear export signals. Scale bar: 20 μm. (b) TaMKP1‐nuclear localisation and nuclear export signal region of the lesion phenotype and statistical analysis of lesion diameter **** indicate a significant difference (P < 0.0001) according to Student's t‐test. (c) Western blot analysis to verify the expression of TaMKP1‐NES and TaMKP1‐NLS in plants.
TaMKP1 interacts with, dephosphorylates and activates TaMPK3, TaMPK4 and TaMPK6
Previous studies showed that AtMKP1 interacts with its dominant substrate, AtMPK6, to repress defence/stress signalling in Arabidopsis (Anderson et al., 2011; Ulm et al., 2002). To identify wheat MAPKs targeted by and interacting with TaMKP1, wheat genes encoding core members of the MAPK cascades (i.e. TaMPK3, TaMPK4 and TaMPK6) were identified in silico (Figure S6) and cloned into the prey vector pGBKT7 for directed yeast two‐hybrid (Y2H) assays with TaMKP1a cloned into the bait vector pGADT7. Interestingly, as shown in Figure 6a, TaMKP1 was able to interact with TaMPK3, TaMPK4 and TaMPK6 in yeast cells (Figure 6a).
Figure 6.
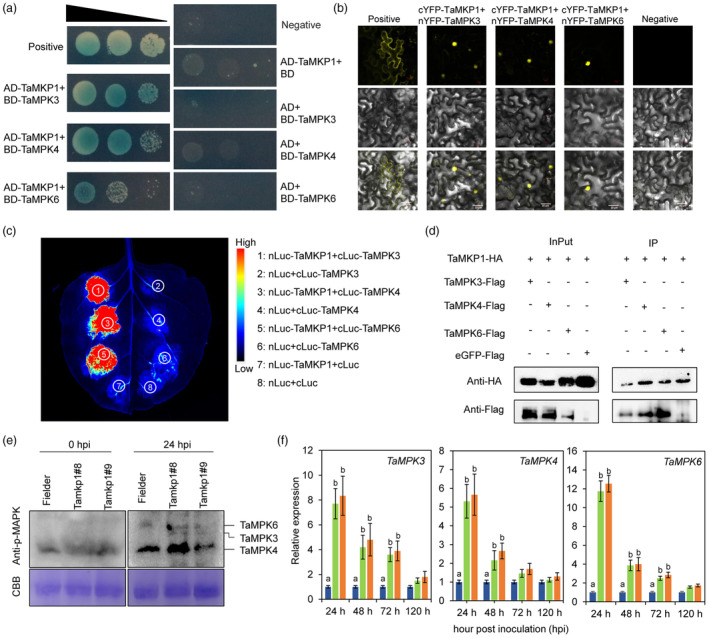
TaMKP1 interacts with, dephosphorylates, and activates TaMPK3/4/6. (a) Yeast Two‐Hybrid (Y2H) assay was used to validate the interaction between TaMKP1 and TaMPK3/4/6 in yeast cells. (b) In vivo BiFC analysis of TaMKP1 interaction with TaMPK3/4/6. (c) The LUC assay confirmed the interaction between TaMKP1 and TaMPK3/4/6. (d) In vivo co‐immunoprecipitation of TaMKP1 with TaMPK3, TaMPK4 or MPK6. (e) TaMKP1 affects phosphorylation levels of TaMPK3/4/6 in vivo. Immunoblot assay for MAPK phosphorylation using anti‐phospho‐MAPK antibodies. The Tamkp1 plants and Filder were treated with Pst CYR34. Proteins were extracted from Tamkp1 mutant and Fielder leaves at 0, and 24 h. CBB staining is shown as a protein loading control. (f) The relative transcript levels of TaMPK3, TaMPK4, and TaMPK6 in Tamkp1 plants.
To confirm whether TaMKP1 interacts with TaMPK3, TaMPK4, and TaMPK6 in planta, bimolecular fluorescence complementation (BiFC) assay was performed. To this end, TaMKP1a was translationally fused with the C‐terminal fragment of YFP, and TaMPK3, TaMPK4 and TaMPK6 were translationally fused with the N‐terminal fragment of YFP. The cYFP‐TaMKP1a fusion gene was co‐expressed with nYFP‐TaMPK3 or nYFP‐TaMPK4, or nYFP‐TaMPK6 in N. benthamiana leaves. At 48 h after agroinfiltration, strong YFP signal was predominantly detected in the nuclei of leaf epidermal cells co‐expressing any of the three above combinations (Figure 6b), indicating that TaMKP1 interacts with each of the three TaMPKs and the interaction mainly occurs in the nucleus. Similar results supporting the in planta direct interaction between TaMKP1 and each of the three TaMPKs were obtained using Luciferase complementation (LUC) assays (Figure 6c). To further validate the interaction of TaMKP1 and TaMPK3/4/6 in vivo, co‐immunoprecipitation (Co‐IP) assays were performed using TaMKP1 tagged with hemagglutinin (HA) and TaMPK3/4/6 tagged with FLAG via co‐transient expression in leaves of N. benthamiana. As expected, Co‐IP results showed that TaMKP1 was able to pull down the three TaMPKs, further demonstrating that TaMKP1 interacts with TaMPK3/4/6 (Figure 6d).
The physical interaction of TaMKP1 with TaMPK3/4/6 suggests that TaMPK3/4/6 are authentic substrates of TaMKP1. To test whether TaMKP1 dephosphorylates TaMPK3/4/6, we analysed the phosphorylation levels of TaMPKs in Tamkp1 plants. The results showed that Pst actively induced MPK3, MPK4, and MPK6 phosphorylation at higher levels in Tamkp1 plants than that in Fielder (Figure 6e). The transcripts of TaMPK3/4/6 genes in Tamkp1 plants were detected using qRT‐PCR. Compared to the control plants, the expression levels of TaMPK3/4/6 significantly increased at 24 hpi and 48 hpi with Pst (Figure 6f).
TaMPK4 interacts with TaPAL
In recent years, a few downstream target proteins of MPK3/6 have been identified. These include transcription factors ERF3 and WRKY33 (Wang et al., 2018b, 2023; Zhang et al., 2021). In this study, we utilized Y2H technology to screen for interacting targets of TaMPK4 in the wheat nucleus system. As a result of this screening, we identified TaPAL. Y2H, BiFC and LUC assays were used to determine whether TaMPK3/4/6 interacts with TaPAL. Y2H assays finding that TaPAL interact with TaMPK4 but not TaMPK3/6 (Figure 7a). LUC assays supported the finding that TaMPK4 interact with TaPAL (Figure 7b). The BiFC results showed that YFP signal was distributed in the nucleus of N. benthamiana co‐expressing nYFP‐TaMPK4 with cYFP‐TaPAL (Figure 7c).
Figure 7.
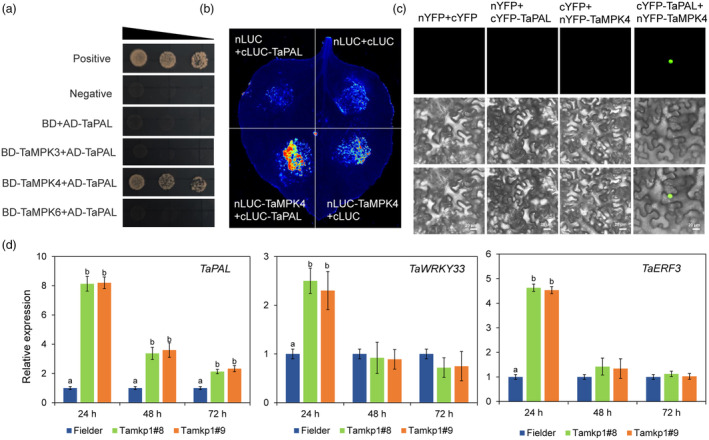
TaMPK4 interacts with TaPAL. (a) Y2H assay was used to validate the interaction between TaPAL and TaMPK4. (b) The LUC assay confirmed the interaction between TaPAL and TaMPK4. (c) BiFC analysis of TaPAL interaction with TaMPK4. Scale bar: 20 μm. (d) The relative transcript levels of TaPAL, TaWRKY33, and TaERF3 in Tamkp1 plants.
Moreover, to determine whether TaMPK4 regulates TaPAL transcription, we used qRT‐PCR to analyse TaPAL transcript levels in Tamkp1 plants and Fielder. Compared to the corresponding controls, the TaPAL transcript level was up‐regulated in the Tamkp1 plants, suggesting that TaMPK4 overexpression might activate TaPAL expression. In addition, transcript levels of TaWRKY33 and TaERF3 were up‐regulated in Tamkp1 plants (Figure 7d).
Wheat TaMkp1 mutants exhibit better performance in growth and reproduction
Defence activation is costly, hence there must be a defence‐growth trade‐off in plants (Huot et al., 2014). To test if loss of TaMKP1 genes indeed incurs a fitness cost in wheat, a field evaluation of different TaMKP1 mutant lines was conducted along with the WT plants. During the early growth stage, all mutant lines were phenotypically similar to the WT. However, to our surprise, during the later growth stage, all mutant lines showed a slight yet significant increase in plant height. Among them, plants of the aabbdd lines exhibited the greatest height, followed by the AABBdd lines, and then the aaBBDD and AAbbDD lines (Figure 8a,d).
Figure 8.
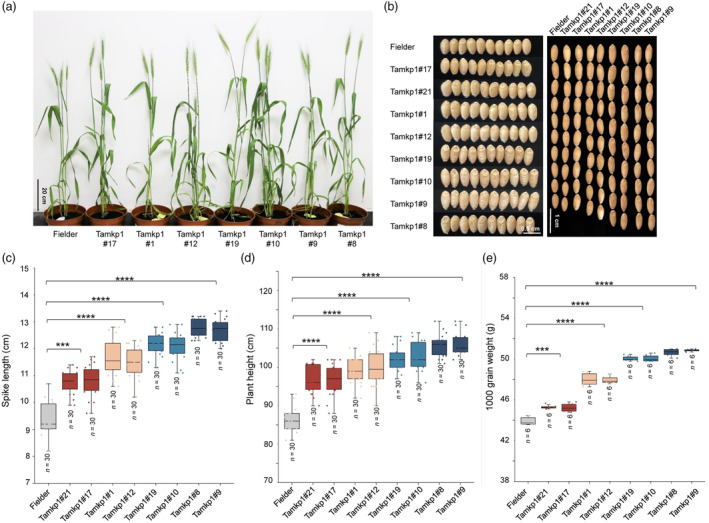
Field performances of the different Tamkp1 plants. (a) The growth conditions of Tamkp1 plants and Fielder in the field. (b) Mature paddy wheat grains of Fielder and Tamkp1. (c) Spike length statistics of Tamkp1 plants and Fielder. (d) plant length statistics of Tamkp1 plants and Fielder. (e) 1000 grain weight statistics of Tamkp1 plants and Fielder.
Consistent with the performance in plant height, all TaMKP1 mutant lines exhibited a significant increase in spike length (Figure 8c), and in grain length, grain width, and grain yield (Figure 8b,e) compared to the WT. Interestingly, the degree of increase in those traits remained more or less the same order among the mutant lines, that is aabbdd (most increase), AABBdd, aaBBDD, and then AAbbDD (least increase). The unexpected yet exciting results from our field experiments strongly suggest that the defence‐growth trade‐off is not a constant and that these two contrasting traits may reconcile under certain context such as when TaMKP1 genes in wheat are ablated.
TaMKP1 gene regulatory network and global germplasm relationships
To understand the gene regulation of TaMKP1, a gene regulatory network (GRN) was constructed based on wGRN (http://wheat.cau.edu.cn/wGRN/) (Chen et al., 2023). GRN analysis revealed that TaMKP1 was regulated by multiple transcription factors, with the top 6 ranked factors being calmodulin‐binding transcription activator 2‐like, auxin response factor 4‐like, transcription factor MYC2‐like, MYB family transcription factor PHL7, NAC domain‐containing protein 78, and ethylene‐responsive transcription factor RAP2‐13 (Figure S7; Table S1). These results indicated that TaMKP1 not only participates in the response of plants to biotic stress, but also plays an important role in other physiological processes of plants.
Wheat is a globally important staple crop. To enhance our knowledge of the genetic diversity of wheat varieties and the significance of the TaMKP1 gene, a genomic‐based germplasm network (GGNet) was constructed based on WheatComp Database (http://wheat.cau.edu.cn/WheatCompDB/) (Yang et al., 2022b). Fielder is one of the American cultivars (AMC). This variety has a close genetic relationship with African landrace (ASL), Asian cultivar (ASC), European cultivar (EUC), Chinese cultivar (CNC), Chinese landrace (CNL) and other germplasm resources (Figure S7). The research findings suggested that potential application prospects exist for enhancing disease resistance and increasing yield in global wheat through the modification of the TaMKP1 gene.
Discussion
Crop diseases are one of the major factors that significantly affect the yield and quality of wheat. The exact role of TaMKP1 in the interaction between wheat and fungus remains unknown. In this study, we cloned and functionally characterized MKP1 from wheat. We also proved that TaMKP1 interacts with TaMPK3/4/6. Importantly, we successfully generated Tamkp1 plants by CRISPR‐Cas9 technology and confirmed their resistance to Pst and Bgt. Our results demonstrated that simultaneous knock‐out of all three homologues of TaMKP1 confers resistance to stripe rust, TaMKP1d plays a greater role than other homologues.
In rice, OsMAPKKKε promotes the OsMKK4–OsMAPK3/6 cascade, and thus, the ability to perceive chitin and resistance to rice blast fungus was improved (Wang et al., 2017). OsILA1, indentified as a negative regulator of the OsMAPKK4–OsMAPK6 cascade and is involved in rice–Xoo interactions (Chen et al., 2021). OsMAPK6‐OsLIC‐OsWRKY30 module is an immune signalling pathway in response to the bacterial pathogens (Wang et al., 2022a). Interestingly, SCRE6 interacts with and dephosphorylates the OsMPK6 in rice, thereby suppressing plant immunity (Zheng et al., 2022). This study suggested that pathogens can exploit the tyrosine phosphatase family to inhibit the MAPK pathway, facilitating their infection. The MAPK pathway is also associated with plant yield. OsMKKK10, OsMKK4, and OsMAPK6 function in a common pathway to regulate grain size (Xu et al., 2018a). OsMAPK6 plays a key role in rice grain size through cell proliferation, brassinosteroids (BR) signalling, and homeostasis (Liu et al., 2015). Loss of function mutations in OsMKP1 results in larger grains, OsMKP1 determines grain size by restricting cell proliferation in grain hulls. OsMKP1 is a key factor influencing grain size by affecting OsMAPK6 (Xu et al., 2018b). In Arabidopsis, AtMKP1 negatively regulates MPK6‐mediated PAMP responses and resistance against bacteria, AtMPK6, but not AtMPK3, is necessary for the mkp1‐dependent increase in resistance to Pseudomonas syringae pv. tomato DC3000 (Anderson et al., 2011), and AtMKP1 was phosphorylated by AtMPK6 (Park et al., 2011). Activation of the MPK3 and MPK6 is sufficient to trigger pipecolic acid production and mount systemic acquired resistance (SAR) (Wang et al., 2018b). Pathogen‐responsive MPK3/MPK6 cascade is an essential signalling pathway that control, respectively, the organic acid metabolism, a main branch of osmotic regulation in guard cells that function interdependently to control stomatal opening/closure, whichis is essential for stomatal immunity (Su et al., 2017). A study pointed out that MPK3/MPK6 cascade is a key hub in orchestrating the trade‐off between growth and defence (Su et al., 2018). NtMKP1 negatively regulates wound response and resistance against both necrotrophic pathogens and herbivorous insects by suppression of jasmonic acid (JA) or ethylene (ET) pathways via inactivation of MAPK (Oka et al., 2013). In contrast, silencing of NbMKP1 increased the accumulation of Bamboo mosaic virus and Foxtail mosaic virus (Lee et al., 2018). Other studies have found that substrates for MPK3/6 play important roles downstream of the MPK3/MPK6 cascade in regulating plant immunity and yield. However, these mechanisms have yet to be fully understood in wheat. In general, these studies demonstrated that MAPK kinases play important roles between plant immunity and yield. Understanding the regulatory mechanism of MAPKs will undoubtedly benefit the development of future crop varieties by providing durable resistance against pathogens and other agronomic traits (Zhang et al., 2023).
In our study, TaMKP1 was found to negatively regulate the disease resistance of wheat, and TaMKP1 directly interacts with and dephosphorylates TaMPK3/4/6 (Figures 6 and S8). Interestingly, Tamkp1 plants showed corresponding improvements in plant height and yield, the relative expression of TaMPK6 was upregulated in TaMKP1 knockout plants (Figure 6f), suggesting that TaMKP1 regulates production by affecting TaMPK6 expression. In the previous view, plants should effectively balance their immune response with growth and development. Excessive and intense immune activity can have deleterious effects on plant production, leading to defects and reduced yields. (Lozano‐Duran and Zipfel, 2015; Ning et al., 2017). However, some studies have also shown that while improving plant resistance, it does not change growth traits and even increases yield. IPA1 promotes both yield and disease resistance by sustaining a balance between growth and immunity (Wang et al., 2018a). Disruption of ROD1 results in resistance to a variety of pathogens, whereas the natural ROD1 allele with agroecology‐specific distribution prevalent in indica rice enhances resistance without affecting yield (Gao et al., 2021). The knockout of TaPsIPK1 enhances resistance to stripe rust in wheat but does not result in any significant developmental defects (Wang et al., 2022b). Overexpression of TaPIP2;10 plants improved wheat resistance and grain yield (Lu et al., 2022). Our study demonstrated that Tampk1 effectively enhanced disease resistance and led to a significant increase in wheat yield.
Hexaploid wheat (T. aestivum, genome AABBDD) is derived from a cross between tetraploid T. turgidum (genome AABB) and A. tauschii (genome DD) (Marcussen et al., 2014). Through genome mapping, it was found that the wheat D genome exhibits unique characteristics in terms of disease resistance, stress resistance, adaptability and quality, disease resistance and quality‐related genes were significantly expanded (Luo et al., 2017). The close affinities of TaMKP1 in various wheat varieties worldwide suggested that there is strong potential for the application of TaMKP1 in promoting increased resistance and yield in wheat.
Experimental procedures
Plant materials and pathogens
Wheat cultivar Fielder, Pst races (CYR31, CYR32, CYR33, CYR34), and Bgt were used in this study. All plant materials were grown in a greenhouse under a 16‐h/8‐h light cycle at a constant 12 ± 2 °C. The fresh Pst urediniospores were rubbed using a smear method onto the first leaves. Inoculated seedlings were placed in a humidifier at 100% humidity closed and treated in the dark for 24 h and then recovered. Bgt maintained on wheat in a growth chamber with a 16‐h/8‐h light cycle, at a constant 22 ± 2 °C.
RNA extraction and gene expression analysis
Total RNA was extracted from the wheat leaves infected by Pst., high‐quality first‐strand cDNA was generated to clone TaMKP1 homoeologs. For gene expression analysis, quantitative reverse transcription PCR (qRT‐PCR) was performed on a Analytik qTOWER3G. Specific primers for each gene were designed using SnapGene, and synthesized by Sangon Biotech (https://www.sangon.com/) (Table S2). SYBR Premix Ex Taq (TaKaRa, http://www.takara‐bio.com/) was used for qRT‐PCR assays. The expression levels of target genes were normalized to TaEF‐1a. The statistical significance was evaluated by Student's t‐test (P < 0.05).
Virus‐Induced gene silencing
The Barley stripe mosaic virus (BSMV)‐based virus‐induced gene silencing (VIGS) assay was performed, as described previously (Liu et al., 2023). Briefly, the specific fragment of TaMKP1 were selected and connected it with BSMV:γ to form a recombinant vector. Then, BSMV:α, BSMV:β, BSMV:γ and the recombinant vector were linearized. In vitro transcription was performed with the linearized plasmid as the template. The BSMV mixture was applied to fully expanded Suwon11 wheat plants by rubbing. Twelve days after inoculation with BSMV, CYR34 was inoculated in the heart leaves of wheat. The feasibility was tested using the TaPDS as a positive control.
Yeast two‐hybrid (Y2H)
The coding fragments of TaMKP1, TaMPK3, TaMPK4 and TaMPK6 were amplified through using the primers listed in Table SX and inserted into the vectors pGADT7(AD) and pGBKT7(BD) to generate AD‐TaMKP1, BD‐TaMPK3, BD‐TaMPK4 and BD‐TaMPK6. The constructed vectors were then transformed into the yeast strains Y2H gold cultured in SD‐His‐Leu medium. Then, the monoclonal colonies were selected and diluted with sterile ddH2O. Then, 5 μL of each colony were coated in SD‐His‐Leu‐Trp‐Ade + X‐α‐gal medium for identification. pGBKT7‐53 and pGADT7‐T were used as positive control, and pGBKT7‐Lam and pGADT7‐T were used as negative control. The interaction protein pairs were confirmed according to the growth of the colony.
Bimolecular fluorescence complementation (BiFC)
The ORF of TaMKP1, TaMPK3, TaMPK4 and TaMPK6 were inserted into the vectors pCV‐nYFP and pCV‐cYFP. The recombinant vectors were transformed into Agrobacterium tumefaciens strain GV3101, and then co‐infiltrated into Nicotiana benthamiana leaves in different combinations. The positive control plants were agro‐infiltrated with A. tumefaciens carrying pCV‐nYFP‐AV2 and pCV‐cYFP‐AC1 vectors. The negative control plants were agro‐infiltrated with A. tumefaciens carrying empty pCV‐nYFP and pCV‐cYFP vectors. At 2 days after infiltration, injected areas from the leaves were observed by fluorescence microscopy using a Zeiss LSM780 microscope.
Split‐luciferase complementation assay
The method of LUC assay was described as previous studies (Yang et al., 2022a). GV3101 containing 35S:LUCN‐TaMKP1 constructs mixed with 35S:LUCC‐TaMPK3, 35S:LUCC‐TaMPK4 or 35S:LUCC‐TaMPK6 constructs at 1:1 ratio was infiltrated into N. benthamiana t leaves and incubated in the growth room for 48 h. 1 mM luciferin was sprayed onto the leaves for CCD imaging.
Co‐immunoprecipitation (Co‐IP) assays
The coding fragments of TaMKP1, TaMPK3, TaMPK4 and TaMPK6 were amplified through using the primers listed and inserted into the vectors pCNF3 to generate pCNF3‐TaMKP1‐HA, pCNF3‐TaMKP3‐Flag, pCNF3‐TaMKP4‐Flag and pCNF3‐TaMKP6‐Flag. Leaves infiltrated with the Agrobacterium GV3101 harbouring pCNF3‐TaMKP1‐HA, pCNF3‐TaMKP3‐Flag, pCNF3‐TaMKP4‐Flag and pCNF3‐TaMKP6‐Flag were harvested and ground into powder under liquid nitrogen at 48 hpi. Co‐IP assays were conducted as described before (Wang et al., 2016).
Immunoblotting
In the experiments, protein extracts from wheat leaves were fractionated through electrophoresis using a regular 12.5% w/v SDS‐PAGE gel. The resulting protein blots were then subjected to hybridization with commercially obtained α‐pMPK antibodies in order to ascertain the phosphorylation status of MPK3, MPK4 and MPK6 (Lu et al., 2022).
Construction of the CRISPR/Cas9‐related vectors
To investigate off‐target effects, three sgRNA targets for TaMKP1 were designed on the conserved domains of all three genomes of wheat based on the prediction of the WheatCrispr (https://crispr.bioinfo.nrc.ca/WheatCrispr/).
The delivered of the constructed vector into wheat immature embryos were performed as previously described (Li et al., 2021). The vector contains the bar gene, the wheat leaves were ground into juice, and 200 μL ddH2O was added, the bar gene test strips were used to detect whether there were positive plants.
Molecular characterization of different mutant lines
Wheat genomic DNA from leaf tissue was extracted using cetyl trimethyl ammonium bromide (CTAB) and PCR amplification was performed using PrimeSTAR® Max DNA Polymerase (https://www.takarabiomed.com.cn/) and 50–100 ng of genomic DNA as a template. To screen for mutations in the TaMKP1 gene, suitable primers were designed for PCR amplification and sequencing based on the indicated target loci and previous research methods (Liu et al., 2019). Any plant carrying not causing coding frame shift were excluded from further experiments.
3,3‐diaminabenzadine (DAB) and trypan blue staining
The staining method is based on the previous method with some modifications (Melichar et al., 2008). The plant tissues were stained with 0.1% DAB for 8 h. The leaves were then placed in 95% ethanol in a boiling water bath for 5 min. The leaves were placed in a saturated chloral hydrate solution for decolorisation and then photographed under the microscope when the chlorophyll had completely faded.
Plant tissues were soaked in 0.4% trypan blue staining solution and vacuum‐treated for 5 min, followed by a boiling water bath for 5 min. The leaves were decolored in saturated chloral hydrate solution and photographed when the chlorophyll had completely faded.
Statistical analysis
All experiments were performed using three replicates of the biological samples. The data is presented as means ± standard deviation, and data were analysed using SPSS. Differences between those in knockdown/overexpression plants and control plants were assessed using Student's t‐tests. Statistical significance was indicated as follows: * for P < 0.05, ** for P < 0.01, *** for P < 0.001, and **** for P < 0.0001. Inter‐group variations in the bar graph depicting letter identification were examined using analysis of ANOVA and LSD Duncan. If the P‐value was greater than 0.05, the letters were considered the same, indicating no significant difference. If the P‐value was less than or equal to 0.05, the letters were considered different, indicating a significant difference.
Funding
This work was supported by National Natural Science Foundation of China grant (31801719), National Key R&D Program of China (2022YFD1901402), Chongqing Technology Innovation and Application Development Special Project (CSTB2022TIAD‐LUX0004), and Fundamental Research Funds for the Central Universities (SWU‐XDJH202318).
Conflict of interest
The authors have no conflict of interest to declare.
Supporting information
Table S1 TaMKP1 gene regulatory relationship.
Table S2 Primers used in this study.
Figure S1 Alignment and analysis of nucleotide sequences of three homologous of TaMKP1 and AtMKP1.
Figure S2 Expression levels of TaMKP1 in different tissues.
Figure S3 The Bar gene test strip is used to screen for positive mutants.
Figure S4 Sequence information for the Tamkp1 mutant.
Figure S5 Effects of chitin and flg22 on stimulating ROS production in Tamkp1 plants.
Figure S6 Analysis of wheat MAPKs‐like MAPK cascade pathway genes.
Figure S7 TaMKP1 gene regulatory network and global germplasm relationships.
Figure S8 TaMKP1 negatively regulates wheat disease resistance by inhibiting TaMPK3/4/6.
Acknowledgements
We express our sincere gratitude to Professor Genying Li from the Shandong Academy of Agriculture Sciences for their invaluable assistance during the creation process of the Tamkp1 mutants. At the same time, we are grateful for the assistance provided by the China National Rice Research Institute during the sequencing process of the Tamkp1 mutants.
Contributor Information
Shunyuan Xiao, Email: xiao@umd.edu.
Yuheng Yang, Email: yyh023@swu.edu.cn.
Data availability statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
References
- Anderson, J.C. , Bartels, S. , Besteiro, M.A.G. , Shahollari, B. , Ulm, R. and Peck, S.C. (2011) Arabidopsis MAP Kinase Phosphatase 1 (AtMKP1) negatively regulates MPK6‐mediated PAMP responses and resistance against bacteria. Plant J. 67, 258–268. [DOI] [PubMed] [Google Scholar]
- Anderson, J.C. , Wan, Y. , Kim, Y.M. , Pasa‐Tolic, L. , Metz, T.O. and Peck, S.C. (2014) Decreased abundance of type III secretion system‐inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae . Proc. Natl. Acad. Sci. U.S.A. 111, 6846–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels, S. , Anderson, J.C. , Gonzalez Besteiro, M.A. , Carreri, A. , Hirt, H. , Buchala, A. , Metraux, J.P. et al. (2009) MAP kinase phosphatase1 and protein tyrosine phosphatase1 are repressors of salicylic acid synthesis and SNC1‐mediated responses in Arabidopsis . Plant Cell, 21, 2884–2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J. , Wang, L.H. , Yang, Z.Y. , Liu, H.B. , Chu, C.L. , Zhang, Z.Z. , Zhang, Q.L. et al. (2021) The rice Raf‐like MAPKKK OsILA1 confers broad‐spectrum resistance to bacterial blight by suppressing the OsMAPKK4‐OsMAPK6 cascade. J. Integr. Plant Biol. 63, 1815–1832. [DOI] [PubMed] [Google Scholar]
- Chen, Y.M. , Guo, Y.W. , Guan, P.F. , Wang, Y.F. , Wang, X.B. , Wang, Z.H. , Qin, Z. et al. (2023) A wheat integrative regulatory network from large‐ scale complementary functional datasets enables trait‐associated gene discovery for crop improvement. Mol. Plant 16, 393–414. [DOI] [PubMed] [Google Scholar]
- Devendrakumar, K.T. , Li, X. and Zhang, Y.L. (2018) MAP kinase signalling: interplays between plant PAMP‐ and effector‐triggered immunity. Cell. Mol. Life Sci. 75, 2981–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N. and Rathjen, J.P. (2010) Plant immunity: towards an integrated view of plant‐pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Escudero, V. , Torres, M.A. , Delgado, M. , Sopena‐Torres, S. , Swami, S. , Morales, J. , Munoz‐Barrios, A. et al. (2019) Mitogen‐activated protein kinase phosphatase 1 (MKP1) negatively regulates the production of reactive oxygen species during Arabidopsis immune responses. Mol. Plant Microbe. In. 32, 464–478. [DOI] [PubMed] [Google Scholar]
- Gao, M. , He, Y. , Yin, X. , Zhong, X. , Yan, B. , Wu, Y. , Chen, J. et al. (2021) Ca2+ sensor‐mediated ROS scavenging suppresses rice immunity and is exploited by a fungal effector. Cell, 184, 5391–5404.e5317. [DOI] [PubMed] [Google Scholar]
- Ghorbel, M. , Cotelle, V. , Ebel, C. , Zaidi, I. , Ormancey, M. , Galaud, J.P. and Hanin, M. (2017) Regulation of the wheat MAP kinase phosphatase 1 by 14‐3‐3 proteins. Plant Sci. 257, 37–47. [DOI] [PubMed] [Google Scholar]
- Ghorbel, M. , Zaidi, I. , Ebel, C. and Hanin, M. (2019) Differential regulation of the durum wheat MAPK phosphatase 1 by calmodulin, bivalent cations and possibly mitogen activated protein kinase 3. Plant Physiol. Biochem. 135, 242–252. [DOI] [PubMed] [Google Scholar]
- Huot, B. , Yao, J. , Montgomery, B.L. and He, S.Y. (2014) Growth–defense tradeoffs in plants: a balancing act to optimize fitness. Mol. Plant, 7, 1267–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura, K. , Shinozaki, K. , Tena, G. , Sheen, J. , Henry, Y. , Champion, A. , Kreis, M. et al. (2002) Mitogen‐activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7, 301–308. [DOI] [PubMed] [Google Scholar]
- Jiang, L.Y. , Anderson, J.C. , Besteiro, M.A.G. and Peck, S.C. (2017) Phosphorylation of Arabidopsis MAP kinase phosphatase 1 (MKP1) is required for PAMP responses and resistance against bacteria. Plant Physiol. 175, 1839–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keyse, S.M. (2000) Protein phosphatases and the regulation of mitogen‐activated protein kinase signalling. Curr. Opin. Cell Biol. 12, 186–192. [DOI] [PubMed] [Google Scholar]
- Kim, H.S. and Asmis, R. (2017) Mitogen‐activated protein kinase phosphatase 1 (MKP‐1) in macrophage biology and cardiovascular disease. A redox‐regulated master controller of monocyte function and macrophage phenotype. Free Radic. Biol. Med. 109, 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, C.C. , Wu, Y.J. , Hsueh, C.H. , Huang, Y.T. , Hsu, Y.H. and Meng, M.H. (2018) Mitogen‐activated protein kinase phosphatase 1 reduces the replication efficiency of Bamboo mosaic virus in Nicotiana benthamiana . Mol. Plant Pathol. 19, 2319–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, P. , Lu, Y.J. , Chen, H. and Day, B. (2020) The lifecycle of the plant immune system. Crit. Rev. Plant Sci. 39, 72–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J.H. , Zhang, S.J. , Zhang, R.Z. , Gao, J. , Qi, Y.P. , Song, G.Q. , Li, W. et al. (2021) Efficient multiplex genome editing by CRISPR/Cas9 in common wheat. Plant Biotechnol. J. 19, 427–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, H. , Wang, M. , Chen, Y. , Nomura, K. , Hui, S. , Gui, J. , Zhang, X. et al. (2022) An MKP‐MAPK protein phosphorylation cascade controls vascular immunity in plants. Sci. Adv. 8, eabg8723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, S.Y. , Hua, L. , Dong, S.J. , Chen, H.Q. , Zhu, X.D. , Jiang, J.E. , Zhang, F. et al. (2015) OsMAPK6, a mitogen‐activated protein kinase, influences rice grain size and biomass production. Plant J. 84, 672–681. [DOI] [PubMed] [Google Scholar]
- Liu, Q. , Wang, C. , Jiao, X.Z. , Zhang, H.W. , Song, L.L. , Li, Y.X. , Gao, C.X. et al. (2019) Hi‐TOM: a platform for high‐throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 62, 1–7. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Xiao, M. , Fang, A. , Tian, B. , Yu, Y. , Bi, C. , Ma, D. et al. (2023) LysM proteins TaCEBiP and TaLYK5 are involved in immune responses mediated by chitin coreceptor TaCERK1 in wheat. J. Agric. Food Chem. 71, 13535–13545. [DOI] [PubMed] [Google Scholar]
- Long, Y.P. , Xie, D.J. , Zhao, Y.Y. , Shi, D.Q. and Yang, W.C. (2019) BICELLULAR POLLEN 1 is a modulator of DNA replication and pollen development in Arabidopsis . New Phytol. 222, 588–603. [DOI] [PubMed] [Google Scholar]
- Lozano‐Duran, R. and Zipfel, C. (2015) Trade‐off between growth and immunity: role of brassinosteroids. Trends Plant Sci. 20, 12–19. [DOI] [PubMed] [Google Scholar]
- Lu, K. , Chen, X.C. , Yao, X.H. , An, Y.Y. , Wang, X. , Qin, L.N. , Li, X.X. et al. (2022) Phosphorylation of a wheat aquaporin at two sites enhances both plant growth and defense. Mol. Plant, 15, 1772–1789. [DOI] [PubMed] [Google Scholar]
- Luo, M.‐C. , Gu, Y.Q. , Puiu, D. , Wang, H. , Twardziok, S.O. , Deal, K.R. , Huo, N. et al. (2017) Genome sequence of the progenitor of the wheat D genome Aegilops tauschii . Nature, 551, 498–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcussen, T. , Sandve, S.R. , Heier, L. , Spannagl, M. , Pfeifer, M. , International Wheat Genome Sequencing Consortium , Jakobsen, K.S. et al. (2014) Ancient hybridizations among the ancestral genomes of bread wheat. Science, 345, 1250092. [DOI] [PubMed] [Google Scholar]
- Melichar, J.P.E. , Berry, S. , Newell, C. , MacCormack, R. and Boyd, L.A. (2008) QTL identification and microphenotype characterisation of the developmentally regulated yellow rust resistance in the UK wheat cultivar Guardian. Theor. Appl. Genet. 117, 391–399. [DOI] [PubMed] [Google Scholar]
- Monaghan, J. and Zipfel, C. (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 15, 349–357. [DOI] [PubMed] [Google Scholar]
- Ning, Y.S. , Liu, W.D. and Wang, G.L. (2017) Balancing immunity and yield in crop plants. Trends Plant Sci. 22, 1069–1079. [DOI] [PubMed] [Google Scholar]
- Oka, K. , Amano, Y. , Katou, S. , Seo, S. , Kawazu, K. , Mochizuki, A. , Kuchitsu, K. et al. (2013) Tobacco MAP kinase phosphatase (NtMKP1) negatively regulates wound response and induced resistance against Necrotrophic Pathogens and Lepidopteran Herbivores. Mol Plant Microbe In 26, 668–675. [DOI] [PubMed] [Google Scholar]
- Park, H.C. , Song, E.H. , Nguyen, X.C. , Lee, K. , Kim, K.E. , Kim, H.S. , Lee, S.M. et al. (2011) Arabidopsis MAP kinase phosphatase 1 is phosphorylated and activated by its substrate AtMPK6. Plant Cell Rep. 30, 1523–1531. [DOI] [PubMed] [Google Scholar]
- Schwessinger, B. and Ronald, P.C. (2012) Plant innate immunity: perception of conserved microbial signatures. Annu. Rev. Plant Biol. 63, 451–482. [DOI] [PubMed] [Google Scholar]
- Su, J.B. , Zhang, M.M. , Zhang, L. , Sun, T.F. , Liu, Y.D. , Lukowitz, W. , Xu, J. et al. (2017) Regulation of stomatal immunity by interdependent functions of a pathogen‐responsive MPK3/MPK6 cascade and abscisic Acid. Plant Cell 29, 526–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, J. , Yang, L. , Zhu, Q. , Wu, H. , He, Y. , Liu, Y. , Xu, J. et al. (2018) Active photosynthetic inhibition mediated by MPK3/MPK6 is critical to effector‐triggered immunity. PLoS Biol. 16, e2004122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, T. , Nitta, Y. , Zhang, Q. , Wu, D. , Tian, H. , Lee, J.S. and Zhang, Y. (2018) Antagonistic interactions between two MAP kinase cascades in plant development and immune signaling. EMBO Rep. 19, e45324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi, F. , Mizoguchi, T. , Yoshida, R. , Ichimura, K. and Shinozaki, K. (2011) Calmodulin‐dependent activation of MAP kinase for ROS homeostasis in Arabidopsis . Mol. Cell, 41, 649–660. [DOI] [PubMed] [Google Scholar]
- Teixeira, P.J.P. , Colaianni, N.R. , Fitzpatrick, C.R. and Dangl, J.L. (2019) Beyond pathogens: microbiota interactions with the plant immune system. Curr. Opin. Microbiol. 49, 7–17. [DOI] [PubMed] [Google Scholar]
- Uchida, K. , Yamaguchi, M. , Kanamori, K. , Ariga, H. , Isono, K. , Kajino, T. , Tanaka, K. et al. (2022) MAP KINASE PHOSPHATASE1 promotes osmotolerance by suppressing PHYTOALEXIN DEFICIENT4‐independent immunity. Plant Physiol. 189, 1128–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, R. , Revenkova, E. , di Sansebastiano, G.P. , Bechtold, N. and Paszkowski, J. (2001) Mitogen‐activated protein kinase phosphatase is required for genotoxic stress relief in. Genes Dev. 15, 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulm, R. , Ichimura, K. , Mizoguchi, T. , Peck, S.C. , Zhu, T. , Wang, X. , Shinozaki, K. et al. (2002) Distinct regulation of salinity and genotoxic stress responses by Arabidopsis MAP kinase phosphatase 1. EMBO J. 21, 6483–6493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z. , Li, N. , Jiang, S. , Gonzalez, N. , Huang, X. , Wang, Y. , Inzé, D. et al. (2016) SCFSAP controls organ size by targeting PPD proteins for degradation in Arabidopsis thaliana . Nat. Commun. 7, 11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, C. , Wang, G. , Zhang, C. , Zhu, P.K. , Dai, H.L. , Yu, N. , He, Z.H. et al. (2017) OsCERK1‐mediated chitin perception and immune signaling requires receptor‐like cytoplasmic kinase 185 to activate an MAPK cascade in rice. Mol. Plant 10, 619–633. [DOI] [PubMed] [Google Scholar]
- Wang, J. , Zhou, L. , Shi, H. , Chern, M. , Yu, H. , Yi, H. , He, M. et al. (2018a) A single transcription factor promotes both yield and immunity in rice. Science, 361, 1026–1028. [DOI] [PubMed] [Google Scholar]
- Wang, Y.M. , Schuck, S. , Wu, J.N. , Yang, P. , Doring, A.C. , Zeier, J. and Tsuda, K. (2018b) A MPK3/6‐WRKY33‐ALD1‐pipecolic acid regulatory loop contributes to systemic acquired resistance. Plant Cell, 30, 2480–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R.Y. , He, F. , Ning, Y.S. and Wang, G.L. (2020) Fine‐Tuning of RBOH‐Mediated ROS Signaling in Plant Immunity. Trends Plant Sci. 25, 1060–1062. [DOI] [PubMed] [Google Scholar]
- Wang, L.H. , Chen, J. , Zhao, Y.Q. , Wang, S.P. and Yuan, M. (2022a) OsMAPK6 phosphorylates a zinc finger protein OsLIC to promote downstream OsWRKY30 for rice resistance to bacterial blight and leaf streak. J. Integr. Plant Biol. 64, 1116–1130. [DOI] [PubMed] [Google Scholar]
- Wang, N. , Tang, C. , Fan, X. , He, M. , Gan, P. , Zhang, S. , Hu, Z. et al. (2022b) Inactivation of a wheat protein kinase gene confers broad‐spectrum resistance to rust fungi. Cell 185, 2961–2974.e2919. [DOI] [PubMed] [Google Scholar]
- Wang, Z. , Li, X. , Yao, X. , Ma, J. , Lu, K. , An, Y. , Sun, Z. et al. (2023) MYB44 regulates PTI by promoting the expression of EIN2 and MPK3/6 in Arabidopsis. Plant Commun. 4, 100628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, R. , Duan, P.G. , Yu, H.Y. , Zhou, Z.K. , Zhang, B.L. , Wang, R.C. , Li, J. et al. (2018a) Control of grain size and weight by the OsMKKK10‐OsMKK4‐OsMAPK6 signaling pathway in Rice. Mol. Plant 11, 860–873. [DOI] [PubMed] [Google Scholar]
- Xu, R. , Yu, H.Y. , Wang, J.M. , Duan, P.G. , Zhang, B.L. , Li, J. , Li, Y. et al. (2018b) A mitogen‐activated protein kinase phosphatase influences grain size and weight in rice. Plant J. 95, 937–946. [DOI] [PubMed] [Google Scholar]
- Yang, S. , Cai, W.W. , Shen, L. , Cao, J.S. , Liu, C.L. , Hu, J. , Guan, D.Y. et al. (2022a) A CaCDPK29‐CaWRKY27b module promotes CaWRKY40‐mediated thermotolerance and immunity to Ralstonia solanacearum in pepper. New Phytol. 233, 1843–1863. [DOI] [PubMed] [Google Scholar]
- Yang, Z.Z. , Wang, Z.H. , Wang, W.X. , Xie, X.M. , Chai, L.L. , Wang, X.B. , Feng, X.B. et al. (2022b) ggComp enables dissection of germplasm resources and construction of a multiscale germplasm network in wheat. Plant Physiol. 188, 1950–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi, I. , Ebel, C. , Belgaroui, N. , Ghorbel, M. , Amara, I. and Hanin, M. (2016) The wheat MAP kinase phosphatase 1 alleviates salt stress and increases antioxidant activities in Arabidopsis. J. Plant Physiol. 193, 12–21. [DOI] [PubMed] [Google Scholar]
- Zhang, S.Q. and Klessig, D.F. (2001) MAPK cascades in plant defense signaling. Trends Plant Sci. 6, 520–527. [DOI] [PubMed] [Google Scholar]
- Zhang, M.M. , Su, J.B. , Zhang, Y. , Xu, J. and Zhang, S.Q. (2018) Conveying endogenous and exogenous signals: MAPK cascades in plant growth and defense. Curr. Opin. Plant Biol. 45, 1–10. [DOI] [PubMed] [Google Scholar]
- Zhang, T.Y. , Li, Z.Q. , Zhao, Y.D. , Shen, W.J. , Chen, M.S. , Gao, H.Q. , Ge, X.M. et al. (2021) Ethylene‐induced stomatal closure is mediated via MKK1/3‐MPK3/6 cascade to EIN2 and EIN3. J. Integr. Plant Biol. 63, 1324–1340. [DOI] [PubMed] [Google Scholar]
- Zhang, D.D. , Lv, B. and Qiu, J.L. (2022) Being tough: The secret weapon of plants against vascular pathogens. Mol. Plant, 15, 934–936. [DOI] [PubMed] [Google Scholar]
- Zhang, H. , Liu, Y. , Zhang, X. , Ji, W. and Kang, Z. (2023) A necessary considering factor for breeding: growth‐defense tradeoff in plants. Stress Biol. 3, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X.H. , Fang, A.F. , Qiu, S.S. , Zhao, G.S. , Wang, J.Y. , Wang, S.Z. , Wei, J.J. et al. (2022) Ustilaginoidea virens secretes a family of phosphatases that stabilize the negative immune regulator OsMPK6 and suppress plant immunity. Plant Cell, 34, 3088–3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou, J.M. and Zhang, Y.L. (2020) Plant immunity: danger perception and signaling. Cell, 181, 978–989. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 TaMKP1 gene regulatory relationship.
Table S2 Primers used in this study.
Figure S1 Alignment and analysis of nucleotide sequences of three homologous of TaMKP1 and AtMKP1.
Figure S2 Expression levels of TaMKP1 in different tissues.
Figure S3 The Bar gene test strip is used to screen for positive mutants.
Figure S4 Sequence information for the Tamkp1 mutant.
Figure S5 Effects of chitin and flg22 on stimulating ROS production in Tamkp1 plants.
Figure S6 Analysis of wheat MAPKs‐like MAPK cascade pathway genes.
Figure S7 TaMKP1 gene regulatory network and global germplasm relationships.
Figure S8 TaMKP1 negatively regulates wheat disease resistance by inhibiting TaMPK3/4/6.
Data Availability Statement
The data that support the findings of this study are available on request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.


