Abstract
Most of the seven flavivirus nonstructural proteins (NS1 to NS5) encoded in the distal two-thirds of the RNA positive-sense genome are believed to be essential components of RNA replication complexes. To explore the functional relationships of these components in RNA replication, we used trans-complementation analysis of full-length infectious RNAs of Kunjin (KUN) virus with a range of lethal in-frame deletions in the nonstructural coding region, using as helper a repBHK cell line stably producing functional replication complexes from KUN replicon RNA. Recently we showed that replication of KUN RNAs with large carboxy-terminal deletions including the entire RNA polymerase region in the NS5 gene, representing 34 to 75% of the NS5 coding content, could be complemented after transfection into repBHK cells. In this study we have demonstrated that KUN RNAs with deletions of 84 to 97% of the NS1 gene, or of 13 to 63% of the NS3 gene including the entire helicase region, were also complemented in repBHK cells with variable efficiencies. In contrast, KUN RNAs with deletions in any of the other four nonstructural genes NS2A, NS2B, NS4A, and NS4B were not complemented. We have also demonstrated successful trans complementation of KUN RNAs containing either combined double deletions in the NS1 and NS5 genes or triple deletions in the NS1, NS3, and NS5 genes comprising as much as 38% of the entire nonstructural coding content. Based on these and our previous complementation results, we have generated a map of cis- and trans-acting elements in RNA replication for the nonstructural coding region of the flavivirus genome. These results are discussed in the context of our model on formation and composition of the flavivirus replication complex, and we suggest molecular mechanisms by which functions of some defective components of the replication complex can be complemented by their wild-type counterparts expressed from another (helper) RNA molecule.
Kunjin virus (KUN) is an Australian flavivirus closely related to other members of the Japanese encephalitis (JE) virus subgroup (32). The KUN genome consists of single-stranded RNA of positive polarity comprising 11,022 nucleotides (20) with one long open reading frame coding for 3433 amino acids in three structural proteins (C, prM, and E) and seven nonstructural (NS) proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) (11). NS proteins are assumed to be involved primarily in the replication of viral RNA as a part of a replication complex (RC). Our previous studies on characterization and partial purification of the functionally active KUN RC (8–10), coprecipitations of NS proteins and double-stranded RNA (dsRNA) in a radioimmunoprecipitation reaction, and colocalizations of NS proteins and dsRNA defined by electron microscopy using immunogold labeling of cryosections (31, 44, 45) demonstrated that the RC consists of dsRNA as a template for RNA synthesis closely associated with most of the NS proteins in virus-induced membrane structures. This group of experiments indicated a consensus protein composition for the RC of NS1, NS3, NS5, NS2A, and NS4A colocalized with dsRNA in vesicle packets. These induced membranes were usually adjacent to the putative site of proteolytic cleavage located in convoluted membranes and paracrystalline arrays immunogold labeled in cryosections with antibodies to NS2B, NS3, and NS4A. The four small NS proteins (NS2A, NS2B, NS4A, and NS4B) are hydrophobic and relatively nonconserved in amino acid sequence. NS4B is membrane associated in cytoplasm, but a role in replication has not yet been shown (44). We proposed that the collections of induced membranes function as virus factories visible as discrete foci by immunofluorescence (IF) (45) and in which nascent RNA pulse-labeled by bromo-substituted uridine was readily detected (46). The KUN RC was stable to detergent treatment and continued to synthesize nascent RNA after several hours of inhibition of protein synthesis by cycloheximide late in infection (46). NS5 protein of flaviviruses contains motifs for methyltransferase (MT) and RNA-dependent RNA polymerase (RdRp) (25, 26, 34), and NS3 protein contains motifs for serine protease, nucleoside triphosphatase, and helicase (16, 17, 43). Recombinant NS5 of dengue type 1 virus was shown to possess RdRP activity in vitro (41), and we have recently shown an in vitro RdRp activity for recombinant KUN NS5 protein (K. J. Guyatt, E. G. Westaway, and A. A. Khromykh, unpublished data). Nucleoside triphosphatase and/or helicase activities were demonstrated for NS3 proteins of West Nile and dengue type 2 viruses (27, 43; for a review on protease activity of the NS2B-NS3 complex, see reference 37). A number of studies from other groups and our own results showed possible involvement of NS1 in flavivirus RNA replication (24, 28–30, 33, 45). The functions of other NS proteins in RNA replication have not been defined.
To investigate the mechanisms of formation and operation of the flavivirus RC, we have been using our recently developed trans-complementation system consisting of (i) defective full-length KUN RNAs with in-frame deletions and point mutations in the NS genes and (ii) repBHK cells with persistently replicating KUN replicon RNA as a helper for provision of functional NS proteins (22–24). Our recent results on complementation of full-length RNAs containing progressive carboxy-terminal deletions in the NS5 gene demonstrated that translation of its N-terminal half and not the corresponding RNA sequence per se was essential for the replication of defective (NS5-deleted) RNA in repBHK cells (23). We proposed that the N-terminal half of NS5 protein is able to form defective but complementable RC via interactions with other NS proteins. We also showed recently that efficient complementation of the KUN NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase (24), indicating a significant difference in the mechanism of complementation between these two proteins. Our complementation results with KUN NS1 and those with yellow fever virus (YF) NS1 protein (28, 29), combined with our previous results on colocalization and coprecipitation of NS proteins with each other and with dsRNA (31, 42, 45) and on binding of NS2A and NS5 to the KUN 3′ untranslated region (3′UTR) (31; A. Khromykh, unpublished data), as well as results of others on NS3-NS5 interactions and binding to the 3′UTR (7, 19), allowed us to propose a model of formation of the flavivirus RC (23). According to the proposed model, the RC begins to form via binding of NS3 and possibly NS2A to defined conserved sequences in the N-terminal half of NS5 during their translation. On completion of translation, the partially assembled complex attaches to the adjacent 3′UTR via binding of NS2A probably to the 3′-terminal stem-loop at which the NS3 and NS5 components also bind. The complex attached to the RNA is then transported to the membrane site of replication by affinity of the hydrophobic regions of NS2A interacting with those of membrane-bound NS4A, which in turn is bound to the lumenal NS1 via hydrophilic extensions of NS4A through the membrane. This completes the assembly of RC. Based on this model and the observed difference in complementation requirements for NS1 and NS5 proteins, we proposed different mechanisms for trans complementation depending on their relationship to the membrane site and their location in the RC (24). Thus, defective NS1 can be efficiently complemented by an exchange in the lumen with individually expressed wild-type helper NS1. In contrast, defective NS5 and wild-type helper NS5 can be efficiently exchanged only if they are both expressed as components of partially assembled defective and wild-type RCs, respectively.
The aim of this study was to complete our trans-complementation experiments with the entire nonstructural coding region of the KUN genome. We present here the results of successful trans complementation of RNAs with large in-frame deletions in the KUN NS1 and NS3 genes, as well as of RNAs with multiple in-frame deletions in the NS1, NS3, and NS5 genes in the same RNA molecule. No trans complementation was detected for RNAs with in-frame deletions in the NS2A, NS2B, NS4A, and NS4B genes. A comprehensive map of cis- and trans-acting elements in the nonstructural coding region is presented based on these and our previous complementation results, and it is discussed in relation to our proposed model for formation of the flavivirus RC and suggested mechanisms of trans complementation.
MATERIALS AND METHODS
Cells.
BHK-21 cells were maintained in Dulbecco's modification of minimal essential medium supplemented with 10% fetal bovine serum. repBHK cells containing stably replicating KUN replicon RNA (22) were maintained in the same medium supplemented with 1 mg of G418 (Geneticin, Gibco BRL) per ml.
Construction of plasmids.
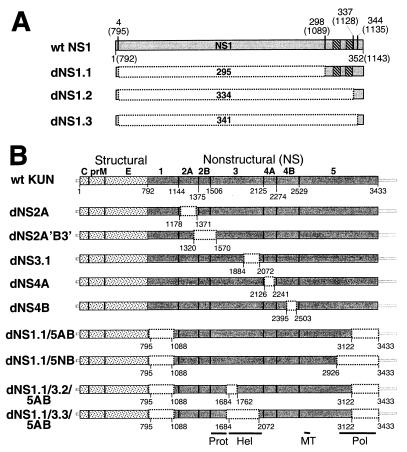
All deletion constructs were prepared from our recently described, highly efficient KUN full-length cDNA clone FLSDX (22) either by digestion with appropriate restriction enzymes, fill-in of ends with Klenow DNA polymerase, and subsequent religation of purified fragments (constructs dNS2A'B3', dNS3.1, dNS1.1/3.2/5AB, and dNS1.1/3.3/5AB [Fig. 1B]) or by PCR amplification of small cDNA fragments with high-fidelity Pfu DNA polymerase (Stratagene), using primers containing appropriate restriction sites, followed by assembly of intermediate plasmids (constructs dNS1.1, dNS1.2, and dNS1.3 [Fig. 1A] and dNS2A, dNS4A, and dNS4B [Fig. 1B]). Constructs dNS1.1/5AB and dNS1.1/5NB (Fig. 1B) were prepared by transferring fragments with deletions of the NS1 gene from NS1.1 into ns5dAB and ns5dNB, respectively (23). Deletions were designed to maintain an open reading frame and were confirmed by restriction digests with appropriate restrictases and/or by sequencing analysis. The details of the plasmid constructions can be obtained from A. Khromykh on request.
FIG. 1.
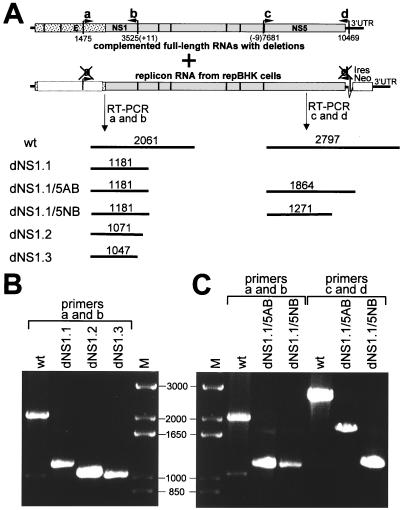
Schematic representation of deletion constructs. (A) Deletions in the NS1 protein. Filled boxes represent KUN NS1 protein, with numbers indicating NS1 amino acid positions. Numbers in brackets show corresponding amino acid positions in the KUN polyprotein (11). Open boxes with dotted borders show in-frame deletions, with the numbers in them indicating the total number of deleted amino acids. Striped boxes represent two amino acid sequences (KUN polyprotein amino acids 1103 to 1113 and 1119 to 1128 [11]) conserved among flaviviruses. wt, wild type. (B) Single, double, and triple deletions in the KUN nonstructural proteins. Dotted boxes and filled boxes represent KUN structural and nonstructural regions, respectively, with the name of the proteins shown above. Thick lines show KUN 5′ and 3′ untranslated regions. Numbers below the boxes in construct wt KUN indicate the first amino acid of the following protein in the KUN polyprotein (11). Open boxes with dotted borders show in-frame deletions with the positions of deleted amino acids indicated below. Apostrophes in dNS2A'B3' construct indicate that only very small proportions of the genes (NS2A and NS3) were deleted, in contrast to the deletion of an entire NS2B gene (indicated as B without apostrophe). Bars under the bottom construct indicate the position of domains for protease (Prot), helicase (Hel), methyltransferase (MT), and RNA polymerase (Pol).
RNA transcription, transfection, IF, and Northern blotting.
All full-length RNA transcripts were prepared with SP6 RNA polymerase from XhoI-linearized plasmid DNAs and electroporated into BHK-21 or repBHK cells as described previously (21, 22). All transcripts with deleted sequences were noninfectious. Detection of replication of complemented KUN full-length RNA in transfected repBHK cells was performed by indirect IF analysis of acetone-fixed cells with KUN anti-E antibodies and by Northern blot hybridization of total cell RNA with a 32P-labeled AatII-ClaI cDNA fragment representing 568 nucleotides of the KUN virus prM-E region (KUN nucleotides 522 to 1089 [11, 20]) as described previously (22, 23). Rehybridization of Northern blots with a 32P-labeled cDNA fragment representing cDNA sequence of the encephalomyocarditis virus RNA internal ribosomal entry site (EMCV IRES probe [Fig. 3C and 5C]) for detection of replicon RNA C20DXrepNeo in repBHK cells (22) was performed as above, using the same membranes from which the previously used prM-E probe was removed by boiling for 10 min in 0.5% sodium dodecyl sulfate solution.
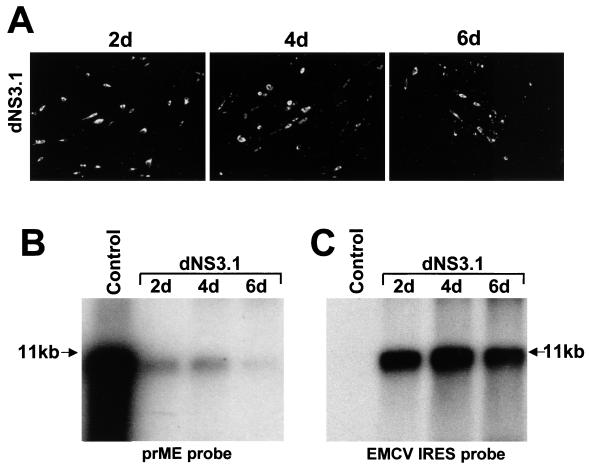
FIG. 3.
Complementation of KUN RNA with a single deletion in the NS3 gene. dNS3.1 RNA has a deletion of 189 codons in the C-terminal region of the helicase region in the NS3 gene (KUN polyprotein amino acids 1884 to 2072 inclusive [Fig. 1B]). (A) Selected fields of repBHK cells transfected with dNS3.1 RNA and stained with KUN anti-E antibodies at days 2, 4, and 6 (2d, 4d, and 6d) after transfection. (B and C) Northern blot analysis with radioactive prM-E (B) and EMCV IRES (C) cDNA probes of the same blot containing samples of total RNA (∼5 μg) isolated from repBHK cells transfected with dNS3.1 RNA. Northern blot hybridization was performed first with the prME probe and then rehybridized with the EMCV IRES probe as described in Materials and Methods. The arrows in panels B and C indicate the position in the gel of RNA of about 11 kb, determined as in Fig. 1B. Control lanes in panels B and C contain ∼10 ng of in vitro-transcribed full-length KUN RNA. The gels were exposed to X-ray film for 2.5 days (B) and for 16 h (C).
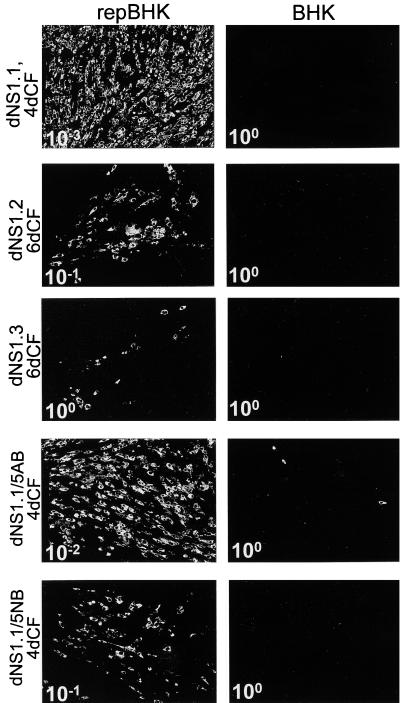
FIG. 5.
Characterization of secreted complemented viruses by IF analysis. Panels show selected fields of repBHK and BHK cells infected with diluted (10−1, 10−2, or 10−3) or undiluted (100) CF collected at 4 or 6 days (4dCF and 6dCF) after transfection of repBHK cells with corresponding RNAs (as shown on the left) and stained with anti-E antibodies at 2 days after infection.
Determination of the infectious titers of secreted complemented viruses.
The titers of complemented virus in infectious units (IU) per milliliter were detected by infection of repBHK cells with serial dilutions of the culture fluids (CFs) from transfected repBHK cells and counting E-positive foci at day 2 after infection.
RESULTS
Complementation of defective KUN RNAs with large deletions in the NS1 gene.
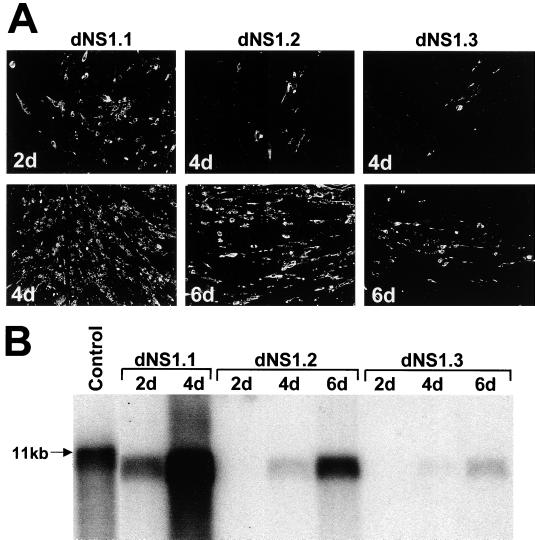
Encouraged by the results on efficient trans complementation of YF RNA YFΔSK with a deletion of 260 codons in the YF NS1 gene (28) and by our recent success in trans complementation of KUN RNAs ns1/C10A and ns1/C11A with lethal cysteine-to-alanine mutations in the KUN NS1 gene (24), we decided to define the maximum extent of deletions in the NS1 gene which can be complemented in trans. We prepared the three full-length KUN cDNA constructs dNS1.1, dNS1.2, and dNS1.3, containing progressive deletions of 295, 334, and 341 amino acids, respectively, which represented 84, 95, and 97% of the NS1 gene (Fig. 1A). In all three deletions, the first three amino acids of NS1 were retained to ensure proper cleavage by cellular signal peptidase. Resulting RNAs were transfected into repBHK cells which provided helper wild-type NS1 protein, and complementation of their replication was monitored by IF analysis with anti-E antibodies and by Northern blot analysis with prME-specific probes as described previously (22–24). KUN dNS1.1 RNA was complemented efficiently and resulted in the secretion of complemented virus which rapidly spread in cell monolayers from days 2 to 4 after transfection, as judged by the IF results with anti-E antibodies (Fig. 2A) and Northern blotting with the prME-specific probe (Fig. 2B). The titer of the complemented virus in 4 day CF, determined as described in Materials and Methods, was ∼6 × 106 IU/ml (Table 1). Further extension of the deletion into the C-terminal part of the NS1 gene resulted in dramatic decreases in the efficiency of complementation. dNS1.2 RNA contained a deletion of an additional 40 codons including two conserved amino acid motifs (Fig. 1A) (11). Complementation of dNS1.2 RNA was first detected by IF at day 4 after transfection in ∼2 to 5% of repBHK cells (Fig. 2A, 4d) and as a weak band in Northern blot (Fig. 2B, lane 4d), but by day 6 after transfection ∼50 to 70% of cells were E-positive (Fig. 2A, 6d) and a relatively strong radiolabeled band was detected in Northern blot (Fig. 2B, lane 6d). Further deletion of another eight codons, leaving only the last eight codons of the KUN NS1 gene (Fig. 1A, dNS1.3) previously shown for other flaviviruses to be essential for proper cleavage of the following NS2A protein (13, 18, 35), resulted in an even further decrease in the complementation efficiency of the corresponding RNA and slower release and spread of the complemented virus in repBHK cells, producing only ∼10% E-positive cells by IF and only a weak band in Northern blot by day 6 after transfection (Fig. 2). Corresponding titers of the secreted complemented dNS1.2 and dNS1.3 viruses in day 6 CFs were ∼3 × 104 and ∼3 × 102 IU/ml, respectively (Table 1). In separate experiments we confirmed proper cleavage at the N terminus of NS2A protein during translation of complemented dNS1.2 and dNS1.3 RNAs (data not shown). No E-positive cells were ever detected after transfection of any of the NS1-deleted RNAs into normal BHK cells (results not shown). We concluded from these results that although deletion of almost the entire NS1 gene (97%) could be complemented in trans, efficient trans complementation depended on the presence of NS1 codons 299 to 344, containing two amino acid motifs strongly conserved amongst all flaviviruses.
FIG. 2.
Complementation of KUN RNAs with large deletions in the NS1 gene. dNS1.1, dNS1.2, and dNS1.3 RNAs contain deletions of 295, 334, and 341 NS1 codons, respectively, out of a total 352 codons (Fig. 1A). (A) Selected fields of repBHK cells transfected with deleted RNAs and stained with anti-E antibodies at 2, 4, and 6 days (2d, 4d, and 6d) after transfection. (B) Northern blot analysis with a radioactive prM-E cDNA probe of ∼5 μg of total RNA isolated from repBHK cells transfected with deleted RNAs. The arrow in panel B indicates the position in the gel of RNA of about 11 kb, determined relative to migration in the same gel of an ethidium bromide stained 1 Kb Plus DNA Ladder (GibcoBRL); the control lane contains ∼10 ng of in vitro-transcribed full-length KUN RNA.
TABLE 1.
Complementation of KUN RNAs with in-frame deletions and frameshift mutation in the nonstructural coding region
| Construct | Deleted amino acidsa | Complementation by IFb | Titer in CFc (IU/ml) |
|---|---|---|---|
| dNS1.1 | 795–1088 | ++++ (4) | ∼6 × 106 (4) |
| dNS1.2 | 795–1127 | +++ (6) | ∼3 × 104 (6) |
| dNS1.3 | 795–1134 | ++ (6) | ∼3 × 102 (6) |
| dNS2A | 1178–1371 | − | ND |
| dNS2A'B3' | 1320–1570 | − | ND |
| dNS3.1 | 1884–2072 | + (4) | <10 |
| dNS4A | 2126–2241 | − | ND |
| dNS4B | 2395–2503 | − | ND |
| ns5Age*d | 2602–3433e | − | ND |
| FLdSAMf | 2603–2613 | ++++ (7) | ∼2 × 104 (7) |
| ns5dEBd | 2675–3433 | − | ND |
| ns5dBsBd | 2755–3433 | ++ (4) | ∼102 (4) |
| ns5dNBd | 2926–3433 | ++++ (2) | ∼(3–5) × 105 (4) |
| ns5dABd | 3122–3433 | ++++ (2) | ∼5 × 106 (4) |
| FLdGDDf | 3194–3197 | ++++ (5) | ∼5 × 105 (5) |
| ns5dSBd | 3389–3433 | ++++ (2) | ∼(3–5) × 105 (4) |
| dNS1.1/5AB | 795–1088, 3122–3433 | ++++ (4) | ∼106 (4) |
| dNS1.1/5NB | 795–1088, 2926–3433 | +++ (4) | ∼8 × 104 (4) |
| dNS1.1/3.2/5AB | 795–1088, 1684–1762, 3122–3433 | + (4) | <10 |
| dNS1.1/3.3/5AB | 795–1088, 1684–2072, 3122–3433 | + (4) | <10 |
Positions of deleted amino acids in KUN polyprotein (11).
Relative efficiencies of complementation were determined at the earliest time when the maximum number of transfected repBHK cells were positive in IF with anti-E antibodies (indicated in parentheses in days; see text): ++++, 100% positive cells; +++, ∼50 to 70% positive cells; ++, ∼10 to 15% positive cells; +, 1 to 2%; −, no positive cells.
Titer of complemented virus in 4-, 6-, or 7-day (as indicated in parentheses) CF from repBHK cells transfected with the corresponding RNA and determined by infection of repBHK cells with serial dilutions followed by counting of anti-E IF positive foci at 2 days after infection. ND, not determined.
Data from reference 23.
Amino acids unable to be translated in the correct reading frame due to a frameshift mutation.
FIG. 4.
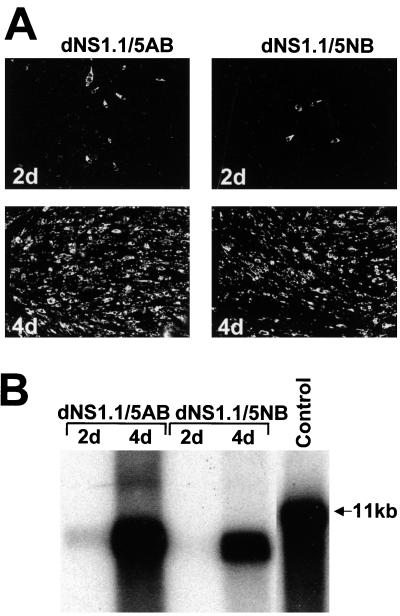
Complementation of KUN RNAs with double deletions in the NS1 and NS5 genes. Both dNS1.1/5AB and dNS1.1/5NB RNAs have a deletion of 295 codons in the NS1 gene and a C-terminal deletion of either 313 or 506 codons in the NS5 gene (KUN polyprotein amino acids 3122 to 3433 and 2926 to 3433, respectively [Fig. 1B]). (A) Selected fields of repBHK cells transfected with deleted RNAs and stained with anti-E antibodies at 2 and 4 days (2d and 4d) after transfection. (B) Northern blot analysis with a radioactive prM-E cDNA probe of the total RNA isolated from repBHK cells transfected with deleted RNAs. The arrowhead in panel B indicates the position in the gel of RNA of about 11 kb, determined as in Fig. 1B; the control lane contains ∼10 ng of in vitro-transcribed full-length KUN RNA.
FIG. 6.
Characterization of secreted complemented viruses by RT-PCR analysis. (A) Schematic representation of the KUN full-length (wild-type [wt]) and replicon genomes and predicted sizes of the RT-PCR products. The numbers represent nucleotide positions in the KUN RNA sequence (11, 21) plus additional nucleotides (numbers shown in parentheses) incorporated into the RT-PCR primers a and b (see below). These numbers were used to calculate the size of RT-PCR fragments shown above the lines. Primers a and b used in RT-PCRs for the E-NS1 region were 5′-CCCCCGCGGCACCCTCTTACACTCTTAAGCT-3′ (nucleotides 1475 to 1505 of the KUN sequence) and 5′-gctggatcctaGGCATTCACCTGTGA-3′ (minus sense, complementary to nucleotides 3511 to 3525 of the KUN sequence), respectively, with nucleotides in lowercase representing 11 nucleotides not present in the KUN sequence. Primer c (which included an additional nine nucleotides) and primer d for RT-PCR of the region containing NS5 deletions were described previously (23). Note that primers a and d cannot bind to the helper replicon RNA because they represent the sequences in the structural region and in the 3′UTR, respectively, of the KUN genome which are deleted in the replicon construct C20DXrepNeo used for generation of repBHK cells (21, 22). (B) RT-PCR analysis of recovered defective viral RNAs. KUN virus particles secreted from cells after complementation of the defective RNAs were treated with RNase A and DNase and immunoprecipitated with anti-E antibodies; the virion RNA was extracted and used in RT-PCR analysis using the SuperScript One-Step RT-PCR system (GibcoBRL) and primer pairs a-b and c-d as described above. Lanes shown as wt in panels B and C represent RT-PCRs with ∼10 ng of KUN virion RNA purified as described previously (20); M lanes show 1 Kb Plus DNA Ladder (GibcoBRL).
Complementation of the defective KUN RNAs with single deletions in other nonstructural genes.
In continuation of our search for trans-acting elements in the nonstructural coding region, we prepared KUN full-length RNAs containing large in-frame deletions in each of the other NS genes (Fig. 1B) and attempted their trans complementation in repBHK cells. None of the dNS2A, dNS4A, and dNS4B RNAs was complemented despite our numerous attempts (results not shown). We also could not complement dNS2A'B3' RNA in trans; this construct has an in-frame deletion extending from the last 56 codons of NS2A through the entire NS2B and to the first 65 codons of NS3 which included the conserved histidine (codon 1557) of the catalytic triad in the serine protease (16) (see Fig. 1B for construct; complementation results not shown).
In contrast, RNA dNS3.1, with a single in-frame deletion of 190 codons in the C-terminal region of the NS3 gene which removed domain VI (NS3 amino acids 378 to 567) of the helicase motif (17), was complemented in repBHK cells, albeit with low efficiency. We detected ∼5% E-positive cells by IF (Fig. 3A, 2d) and a weak band in Northern blots (Fig. 3B, lane 2d) at 2 days after transfection. We showed previously that KUN RNA is degraded quickly after transfection and can be detected by Northern blotting or by expression of encoded proteins using IF only if it is amplified (21). Therefore, detection of E-positive cells and prM-E specific RNA at 2 days after transfection of dNS3.1 RNA clearly demonstrates that the latter RNA is amplified in repBHK cells. However, unlike our previous results on complementation of NS1 and NS5 deleted RNAs (22–24), no increase in the proportion of E-positive cells and no multicellular E-positive foci were observed at day 4 after transfection (Fig. 3A, 4d). Similarly, no increase in the amount of complemented RNA from day 2 to day 4 after transfection was detected by Northern blot analysis (Fig. 3B, compare lanes 2d and 4d). Moreover, we observed a decrease in the number of E-positive cells and in the amount of detected complemented RNA from days 4 to 6 after transfection (4d in Fig. 3A and B, respectively). Intrigued by this unusual decrease in the amount of complemented RNA later in transfection not previously observed in any of our complementation experiments with deletions in the NS1 and NS5 genes, we decided to examine the total cell RNA samples for the presence of helper replicon RNA by rehybridizing the same blot with the probe to EMCV IRES sequence (see Materials and Methods). The results of this rehybridization confirmed the presence of similar amounts of helper replicon RNA and thus the total number of the replicon-expressing cells in the RNA samples at all three time points (Fig. 3C). No E-positive cells were detected after transfection of dNS3.1 RNA into normal BHK cells (results not shown). Hence, these transfection results suggested that although replication of dNS3.1 RNA was complemented in repBHK cells, this complementation apparently did not result in the secretion and spread of complemented virus. Indeed, when undiluted CFs from transfected cells collected at 2, 4, and 6 days after transfection were used to infect fresh repBHK cells, no E-positive cells were detected even by day 3 after infection (results not shown), thus confirming the absence of complemented virus in the CF of dNS3.1-transfected repBHK cells. Furthermore, no NS3-positive normal BHK cells were detected after infection with these CFs (data not shown), indicating the absence of secreted virus particles containing packaged helper replicon RNA. We concluded that although the deletion in the C-terminal region of NS3 gene coding for NS3 amino acids 378 to 567 can be complemented for RNA replication, it abrogates the assembly and/or release of complemented defective virus as well as of virus particles containing encapsidated helper replicon RNA.
Complementation of the defective KUN RNAs with double deletions in the NS1 and NS5 genes.
Having established that replication of KUN RNAs with large deletions in either the NS1 gene (see above) or the NS5 gene (23) could be complemented in repBHK cells, we wished to determine whether we could complement RNA with deletions in both genes. Two double-deletion constructs, dNS1.1/5AB and dNS1.1/5NB, containing the same dNS1.1 deletion in the NS1 gene and two different deletions ns5dAB and ns5dNB in the NS5 gene (23), respectively, were prepared (Fig. 1B). We recently showed that RNAs containing either of these deletions in the NS5 gene (312 C-terminal codons deleted in ns5dAB or 508 C-terminal codons in ns5dNB) were complemented in repBHK cells (23). Thus, the new constructs dNS1.1/5NB and dNS1.1/5AB contained total deletions of 803 and 607 codons, representing 30 and 23%, respectively, of the total nonstructural coding region. Transfection of dNS1.1/5NB and dNS1.1/5AB RNAs in repBHK cells resulted in their replication and apparent secretion of complemented defective viruses, as detected by the dramatic increase in the numbers of E-positive cells in IF analyses and in the amount of complemented RNAs in Northern blot analyses from days 2 to 4 after transfection (Fig. 4). The titers of secreted complemented viruses with double deletions in 4-day CFs were ∼8 × 104 IU/ml for dNS1.1/5NB and ∼106 IU/ml for dNS1.1/5AB (Table 1), in accord with the difference in the complementation efficiencies observed by IF and Northern blot analysis (Fig. 4). The lower complementation efficiency of dNS1.1/5NB RNA than of dNS1.1/5AB RNA is also in accord with our previous complementation results with RNAs containing corresponding single deletions in the NS5 gene (23) in which the titers of secreted viruses in 4-day CFs were ∼3 × 105 to 5 × 105 IU/ml for ns5dNB and ∼5 × 106 IU/ml for ns5dAB. In summary, we have now demonstrated that trans complementation of both RNAs with double deletions in the NS1 and NS5 genes can be achieved with reasonably high efficiency.
Characterization of complemented secreted viruses with deletions in the NS1 and NS5 genes.
Initial characterization and determination of the titers of viruses recovered in CF of repBHK cells transfected with defective RNAs containing either single deletions in the NS1 gene or combined double deletions in the NS1 and NS5 genes were performed by IF analysis. Diluted or undiluted CFs collected at 4 or 6 days after transfection of corresponding RNAs into repBHK cells were used to infect repBHK cells, and replication of complemented viruses was detected by IF analysis with anti-E antibodies at day 2 after infection (Fig. 5). The titer of complemented viruses was determined by counting E-positive foci of infected repBHK cells as described in Materials and Methods. The relative number of E-positive cells after infection with corresponding dilutions of CFs (Fig. 5, repBHK panels) as well as titers of complemented viruses (Table 1) correlated well with the efficiencies of complementation observed in transfection experiments (Fig. 4). The results of IF analysis of infected repBHK cells with anti-E antibodies clearly demonstrated the production of defective secreted viruses in complementation experiments in which all of the defective RNAs contained deletions either in the NS1 gene or in both the NS1 and NS5 genes.
Similar to our previous results on complementation of NS5-deleted RNAs in repBHK cells (22, 23), infection of normal BHK cells with some secreted complemented viruses resulted in detection of rare E-positive cells (Fig. 5, dNS1.1/5AB BHK panel) which were also labeled with anti-NS3 antibodies in the dual-IF analysis (results not shown). We previously demonstrated that such detection did not involve recombinant self-replicating virus but was a result of coinfection of individual normal BHK cells with two types of particles, one containing packaged replicon RNA from repBHK cells (NS3 positive only) and the other containing packaged complemented full-length RNA (NS3 and E positive) (22, 23). This coinfection event would allow complementation of replication of the defective RNA by replicon RNA and thus would lead to the detection of E-positive cells. Importantly, no significant increase in the amount of E-positive cells was observed in the present study after longer (4 days) incubation of infected BHK cells (results not shown), thus once again confirming the absence of recombinant self-replicating viruses in the recovered CFs.
To confirm the defective genotype of complemented secreted viruses, we performed a reverse transcription (RT)-PCR analysis of RNAs isolated from the RNase- and DNase-treated defective virus particles immunoprecipitated from corresponding CFs using anti-E antibodies (see Materials and Methods). We used two sets of primers for detection of deletions in the NS1 and NS5 regions (Fig. 6A). To facilitate RT-PCR amplification of only deleted RNAs and to eliminate RT-PCR amplification of helper replicon RNA, one primer in each primer set (primers a and d) was designed so that it would not bind to the replicon RNA (see the legend to Fig. 6A). The results of RT-PCR amplification of dNS1.1, dNS1.2, and dNS1.3 viral RNAs in the secreted viral particles demonstrated the presence of corresponding deletions in the NS1 gene, as judged by the size of the amplification products (Fig. 6B). RT-PCR analysis of double-deletion dNS1.1/5AB and dNS1.1/5NB viral RNAs was performed with two sets of primers (Fig. 6A) and resulted in detection of corresponding deletions in both NS1 and NS5 genes (Fig. 6C). Thus, the results of IF and RT-PCR analysis demonstrated and confirmed the presence of secreted complemented viruses in the CFs collected after transfection of repBHK cells with defective RNAs containing corresponding single or double deletions in the NS1 and NS5 genes.
Complementation of the defective KUN RNAs with triple deletions in the NS1, NS3, and NS5 genes.
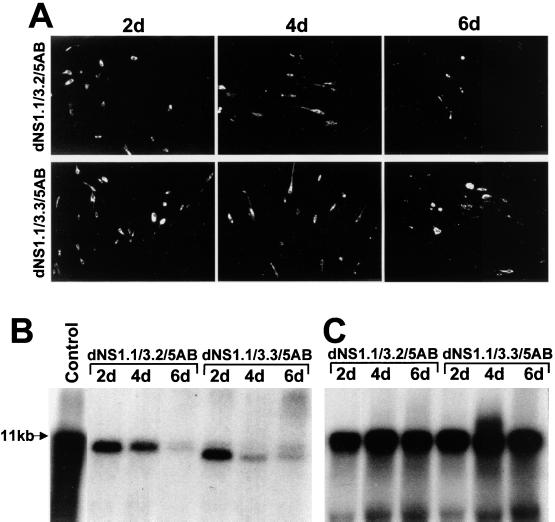
Encouraged by the positive complementations of RNA with a single deletion in the C-terminal part of the helicase region of NS3 gene and complementation of RNAs with double deletions in the NS1 and NS5 genes, we prepared triple-deletion constructs containing large deletions in the NS1 and NS5 genes as well as a smaller deletion in the N-terminal part of the helicase region of NS3 gene (Fig. 1B, dNS1.1/3.2/5AB) or a large deletion of the entire helicase region of NS3 gene (Fig. 1B, dNS1.1/3.3/5AB). Replication of both RNAs in repBHK cells was detected by IF and by Northern blot analyses at day 2 after transfection (Fig. 7A and B). Similar to the previous results with dNS3.1 RNA (Fig. 3), IF analysis of repBHK cells transfected with dNS1.1/3.2/5AB or dNS1.1/3.3/5AB RNAs showed no apparent increase in the proportion of E-positive cells from days 2 to 4 after transfection and a decrease in the proportion of positive cells from days 4 to 6 after transfection (Fig. 7A). The amounts of accumulated complemented RNA detected by Northern blotting decreased either slightly (Fig. 7B, dNS1.1/3.2/5AB) or significantly (Fig. 7B, dNS1.1/3.3/5AB) at day 4 after transfection relative to day 2. The difference in the amounts of accumulated complemented dNS1.1/3.2/5AB and dNS1.1/3.3/5AB RNAs may be due to the different initial rates of replication/complementation caused by the size and the position of the deletion in the NS3 gene. The amounts of complemented RNAs subsequently decreased and were barely detected at day 6 after transfection (Fig. 7B, 6d lanes). Importantly, as observed previously with complementation of dNS3.1 RNA, the amounts of helper replicon RNA and therefore total amounts of the replicon-expressing cells were similar at all three time points of analysis, as confirmed by Northern blotting with the probe to the EMCV IRES sequence present in the replicon RNA (Fig. 7C). Noticeably, replication of all RNAs with deletions in the NS3 gene in repBHK cells was relatively efficient early in transfection (first 2 to 4 days) but decreased dramatically later in transfection (4 to 6 days) (Fig. 3B and 7B). This decrease in the amount of detected complemented RNA later in transfection coincided with an increase in the number of dead E-positive cells (Fig. 3A and 7A, 6d).
FIG. 7.
Complementation of KUN RNAs with triple deletions in the NS1, NS3, and NS5 genes. Both dNS1.1/3.2/5AB and dNS1.1/3.3/5AB RNAs contain deletions of most of the NS1 gene (295 codons) and most of the RNA polymerase region in the NS5 gene (C terminal 313 codons), as well as a small deletion in the N-terminal region of the helicase domain (79 codons, polyprotein amino acids 1684 to 1762) and deletion of the entire helicase domain (389 codons, polyprotein amino acids 1684 to 2072) in the NS3 gene, respectively (Fig. 1B and 8). (A) Selected fields of repBHK cells transfected with the deleted RNAs (as shown) and stained with anti-E antibodies at 2, 4, and 6 days (2d, 4d, and 6d) after transfection. (B and C) Northern blot analysis with a radioactive prM-E probe (B) and EMCV IRES (C) probe of the same blot containing samples of total RNA (∼10 μg) isolated from repBHK cells transfected with deleted RNAs. Northern blot hybridization was performed first with the prME probe, and the blot was then rehybridized with the EMCV IRES probe as described in Materials and Methods. The arrow in panel B indicates the position in the gel of RNA of about 11 kb, determined as in Fig. 1B; the control lane contains ∼10 ng of in vitro-transcribed full-length KUN RNA. The part of the gel with the control lane was exposed to X-ray film for 3 h, while the rest of the gel was exposed for 2.5 days. The gel in panel C was exposed for 16 h.
CFs of transfected cells were then examined for the presence or absence of (i) complemented viruses by infection of fresh repBHK cells and IF analysis with anti-E antibodies and (ii) packaged replicon virus particles by infection of normal BHK cells and IF analysis with anti-NS3 antibodies. No foci of E-positive repBHK cells or NS3-positive BHK cells were detected at 3 days after infection with any of the CFs (results not shown). In view of these negative results on detection of secreted viruses in the CFs and an increase in the proportion of dead E-positive cells, the observed decrease in the amount of complemented RNA later in transfection can probably be explained by the absence of further amplification of complemented RNA due to the absence of viral spread, as well as by washing away dead cells containing replicating complemented full-length RNAs.
DISCUSSION
In this study we demonstrated successful trans complementation of KUN genomic RNAs with large in-frame deletions in the NS1 and NS3 genes by providing corresponding helper proteins from KUN replicon RNA persistently replicating in repBHK cells. Previously we showed trans complementation of KUN genomic RNAs with C-terminal deletions of more than half of the NS5 gene (23). By combining these individual deletions in the same RNA molecule, we were able to demonstrate trans complementation of RNAs containing double deletions in the NS1 and NS5 genes or triple deletions in the NS1, NS3, and NS5 genes. This is the first demonstration of trans complementation of replication of flavivirus RNAs containing deletions of as much as 84 to 97% of the NS1 gene, or of any deletion in the NS3 gene, or of deletions in two or three NS genes in the same RNA molecule.
In this and our previous studies we have attempted complementation of deletions introduced into over 80% of the nonstructural region of the infectious KUN genome, using assays for expression of viral antigens, amplification of genomic RNAs containing deletions, and recovery of transmissible complemented viruses. The only other comparable work with flaviviruses has been with YF virus. An in-frame deletion of 260 codons in the NS1 gene (amino acids 12 to 271, inclusive) was efficiently complemented by YF helper NS1 expressed from a Sindbis virus replicon vector, but a deletion of 340 codons (amino acids 12 to 351, inclusive) was not complemented (28). In this study we demonstrated that KUN RNAs dNS1.1, dNS1.2, and dNS1.3 with in-frame deletions of 295, 334, and 341 codons, respectively, in the 352 codons of the NS1 gene, all retaining only the first 3 codons and the last 54, 15, and 8 codons, respectively (Fig. 1A), were complemented with diminishing efficiency by the helper NS1 expressed from KUN replicon RNA (Fig. 2 and Table 1). The dramatic decrease in the efficiency of complementation between dNS1.1 and dNS1.2 RNAs as well as the very low efficiency of complementation of dNS1.3 RNA demonstrated that the C-terminal region of the NS1 gene commencing from amino acid codon 298 may need to be translated in cis for efficient replication of the defective (NS1-deleted) RNA in the helper repBHK cells. Noticeably, the flavivirus NS1 region between codons 298 (end of deletion in dNS1.1) and 337 (end of deletion in dNS1.2) contains two highly conserved amino acid sequences CRxCx(M/L)PP(L/V) (codons 308 to 317) and CWY(G/A)MEIRP (codons 329 to 337) (Fig. 1A) (6, 11). We suggest that these conserved lumenal peptides (still present in dNS1.1) may be involved in the proposed interactions of NS1 with other components of the viral replicase such as NS4A during assembly and/or targeting of the RC to cellular membranes (23, 24, 28, 29, 45). Importantly, the conserved octapeptide upstream of the KUN NS1-NS2A junction was retained (13, 18, 35), ensuring correct N-terminal cleavage of NS2A (see Results).
Numerous reports are available on the effects of mutations on the cleavage efficiency of the flavivirus NS2B and NS3 proteins supplied separately or as a protease complex in cis and in trans when expressed in vitro or in cells infected with recombinant vaccinia viruses or transfected with plasmids (see for example references 4 and 5; for a review, see reference 37). However, there are no reports on whether mutations or deletions in the serine protease domain of NS3 can be complemented in full-length genomic RNA. Flavivirus helicase activity has been shown recently only for dengue type 2 virus NS3 in an in vitro assay (27). We deleted the polyprotein codons 1884 to 2072 in KUN NS3, which removed helicase motifs V and VI (17) producing the lethally mutated construct dNS3.1. The mutant RNA was complemented in trans but after 4 days had almost disappeared from the Northern blot. Only a low proportion of transfected helper repBHK cell were E positive by IF; there was no spread of complemented virus in helper cells, and no complemented virus was recovered in the CF (Fig. 3 and Table 1). It is reasonable to assume that absence of a subsequent increase in accumulation of complemented RNA (from 4 to 6 days) was caused by the lack of production of secretable virus particles and subsequent virus spread. Such results are in contrast to those observed previously in all our complementation experiments with RNAs containing deletions in the NS1 and NS5 genes, where secretion of complemented virus into the CF was invariably detected even with very inefficiently complemented RNAs (Fig. 4 and 5; references 22 to 24). Later experiments with complementation of RNAs containing triple deletions in the NS1, NS3, and NS5 genes showed that deletion of as many as 389 codons in the NS3 gene (63%) representing the entire helicase region could be complemented in trans (see results for dNS1.1/3.2/5AB and dNS1.1/3.3/5AB), but again no secreted virus was detected.
One of the possible reasons for the lack of virus secretion noted above could be the inability of these RNAs with deletions in the helicase domain of NS3 to be packaged due to the absence of a putative packaging signal normally located (say) in the deleted NS3 region. However, this seems to be unlikely because no secreted particles containing packaged replicon RNA were also detected even though E protein and presumably the other structural proteins were produced from the replicating complemented RNAs in equimolar amounts. Moreover, secreted viruses were not detected in complementation experiments with RNAs containing deletions in different parts of the NS3 gene situated at a distance of ∼360 nucleotides from each other (Fig. 1B, dNS3.1 and dNS1.1/3.2/5AB constructs). It seems unlikely that the flavivirus packaging signal would be so large compared to the average size (ranging from 58 to 160 nucleotides) of the reported packaging signals for other positive-strand RNA viruses (14, 15, 47). Alternatively, lack of virion assembly/secretion could be the result of the conformational changes in the NS2B-NS3 protease complex triggered by the helicase-deleted NS3. For example, the deletion may inhibit efficiency of cleavage at the dibasic site preceding the C terminus of core protein (39). This cleavage event is critical for release of core protein and subsequent assembly and release of viral particles (1, 40, 48). Inefficient cleavage of core protein would also result in accumulation of unprocessed C-prM intermediate and of free E protein. This accumulation is likely to produce severe cytopathic effects leading to the cessation of RNA replication in dying cells, which would explain the decrease in the amount of complemented RNA detected in the remaining attached cells late in transfection. Although a helper NS2B–full-length NS3 protease complex is obviously produced in trans from the replicon RNA in repBHK cells, it is possible that translation in cis of an NS2B–full-length NS3 complex is required for proper cleavage of core protein translated upstream from the same mRNA and for subsequent release of secreted mature virions. The NS3 deletions in the helicase domain also removed the conserved dibasic cleavage site QRR↓GR (in the helicase motif VI) which generates the truncated NS3′ product of ∼460 residues shown to be present in tick-borne encephalitis virus- and dengue virus-infected cells (2, 12, 36, 42). Since the function of this truncated NS3′ protein has not been established, NS3′ may play some role in virus assembly and/or secretion when produced from the same RNA molecule. Clearly, more experimental work is needed to determine how the absence of translation of the helicase region of NS3 in cis may influence assembly and/or secretion of complemented viruses as well as of virus-like particles containing encapsidated helper replicon RNA.
Interestingly, when the large deletions in the NS1 and NS5 genes were made in the same RNA molecule (dNS1.1/5AB and dNS1.1/5NB [Fig. 1B and Table 1]), they were still tolerated and efficient trans complementation occurred. For the functions of NS5 to be rendered noncomplementable, it was necessary to delete all sequences downstream from the MT motif (ns5dEB construct) or introduce a frameshift mutation very close to the N terminus (construct ns5Age*), respectively, as described previously (23). Deletion of the MT motif alone (in FLdSAM) still permitted readily detectable trans-complementation (22). In regard to trans complementation of double- or triple-deletion mutants, it is of interest that the results reflect the combined effects previously observed with single-deletion mutants. For example, NS1 and NS5 with large deletions were still complemented efficiently when combined, and inclusion of NS3 with a large deletion in the helicase domain was also complemented but no complemented virus was secreted. Thus, the largest total of in-frame deletions in the nonstructural region still allowing relatively efficient replication of corresponding RNA in presence of helper replicon RNA was 995 codons, or nearly 3 kb (dNS1.1/3.3/5AB RNA).
In view of the success in trans-complementation experiments involving large deletions in NS1, NS3, and NS5, we were surprised that deletions in the four small NS proteins were invariably noncomplementable. These are all hydrophobic proteins, and the deletions represented 84% of NS2A, 83% of NS4A, and 42% of NS4B. The deletion in construct NS2A'B3' included C-terminal 56 codons of NS2A, 100% of NS2B, and the first 65 codons of NS3. NS2B and NS3 have been shown to retain protease function in vitro when expressed in trans (for a review, see reference 39); hence it was unexpected that our construct dNS2A'B3' could not be complemented in trans by replicon RNA in vivo. The inability to complement deletions in NS2A, NS4A, and NS4B, like NS2B, may be associated with their hydrophobic interaction with the membranes induced during virus replication (see the introduction). It may be that their necessary hydrophobic interactions in the intracellular environment of infected cells can occur only when they are translated in cis with NS1, NS3, and/or NS5. Possibly the helper small hydrophobic proteins cannot individually exchange or be inserted in the membranes involved in translation/cleavage, or in replication of deleted RNA, even though the lumenal (NS1) and cytosolic (NS3 and NS5) proteins can apparently exchange with their cognate mutant products in the same environment. The deletion in mutant NS2A'B3' obviously eliminates cleavage of NS3 and NS5 during their translation in cis; however, it might still permit commencement of assembly of the RC on the just translated RNA as per our model (23). The involvement of polyprotein intermediates in KUN RNA replication seem to be unlikely in view of our previous results (i) on inhibition of protein synthesis in KUN-infected cells by treatment with cycloheximide for several hours (46), demonstrating that once established, viral RNA synthesis does not require de novo protein synthesis, and (ii) on translational mapping in KUN-infected cells synchronized in initiation, showing that all KUN nonstructural proteins were translated in about 17 min and all were apparently correctly cleaved within a chase period of 30 min (38).
The map in Fig. 8 indicates those regions in the nonstructural proteins which can be complemented in trans, while the open boxes cover the regions which are not complemented and hence must be cis-acting elements. There are still areas in the nonstructural region (shown by question marks) representing only 18% which we have not examined for complementation. While small functional domains such as the deleted MT and GDD motifs in NS5, and cysteine mutants in the C-terminal region of NS1, can be complemented efficiently in trans (22, 24), it remains to be discovered whether small specific deletions in the protease domain of NS3 or in the four small hydrophobic nonstructural proteins can also be complemented in trans. In preliminary experiments, we were unable to complement small deletions in NS4A or point mutations in NS4B (Khromykh, unpublished data); hence we suspect that use of selected temperature-sensitive mutants of NS4A and/or NS4B, as well as of NS2A and NS2B, may be required to demonstrate complementation in trans, if it can actually occur. Because of the lack of obvious conserved amino acid sequences among them, assessing how function is effected for NS2A, NS2B, NS4A, and NS4B may require careful consideration of their membrane-associated topology in relation to their protein-protein interactions, as discussed for the association of the membrane-bound NS2B-NS3 dengue type 2 virus protease complex (3). It is significant in this regard that even small (three to four amino acids) deletions in the nonconserved hydrophobic regions of the YF NS2B protein completely abolished infectivity of the corresponding full-length RNA transcripts, even though each of three such constructs retained NS2B-NS3 protease activity in an in vitro cell-free assay (5).
FIG. 8.
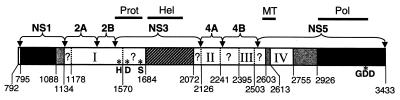
Map of cis- and trans-acting elements in the nonstructural region of KUN virus RNA. Numbers represent amino acid positions in the KUN polyprotein (11) and show boundaries of introduced deletions. Prot, Hel, MT, and Pol indicate corresponding functional domains (as in Fig. 1B). H, D, and S with asterisks show locations of the amino acids of the catalytic triad of the serine protease, and GDD with an asterisk shows the location of the characteristic RNA polymerase motif. Black boxes show trans-acting sequences efficiently complemented singly in NS1 (84% of NS1; yield, 6 × 106 IU/ml at day 4), in NS5 (C-terminal 56% of NS5 including entire polymerase region; yield, 3 × 105 to 5 × 105 IU/ml at day 4), or in RNA deleted in both NS1 and NS5 (yield, 8 × 104 IU/ml at day 4). Gray boxes show trans-acting sequences complemented inefficiently in NS1 (97% of NS1; yield, 3 × 102 IU/ml at day 6), in MT of NS5 (11 amino acids; yield, 2 × 104 IU/ml at day 7), and in NS5 (C-terminal 75% of NS5; yield, 102 IU/ml at day 6). Striped box shows the trans-acting sequence in NS3 (63% of NS3 including entire helicase region) complemented very inefficiently, and no secreted complemented virus was recovered by day 6. Numbered open boxes represent cis-acting sequences apparently not complemented in trans for any deletions, e.g., within box I (NS2A, NS2B, or N terminus of NS3), box II (NS4A), box III (C-terminal half of NS4B), or box IV (region between MT and Pol motifs at the N terminus of NS5). Question marks denote regions that have not been analyzed in complementation assays and represent only 18% of the entire nonstructural coding sequence. Boundaries of the boxes were determined from the complementation data summarized in Table 1.
In conclusion, we believe that the map of cis- and trans-acting elements shown in Fig. 8, together with all of our complementation data (this report; references 22 to 24) and our model of formation of the RC discussed above, will greatly assist further studies on flavivirus complementation and analyses of the RC.
ACKNOWLEDGMENTS
We are grateful to R. Hall for supplying KUN anti-E monoclonal antibodies.
This work was supported by the grant N981442 from the National Health and Medical Research Council of Australia.
Footnotes
Publication no. 102 from the Sir Albert Sakzewski Virus Research Centre.
REFERENCES
- 1.Amberg S M, Rice C M. Mutagenesis of the NS2B-NS3-mediated cleavage site in the flavivirus capsid protein demonstrates a requirement for coordinated processing. J Virol. 1999;73:8083–8094. doi: 10.1128/jvi.73.10.8083-8094.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arias C F, Preugschat F, Strauss J H. Dengue 2 virus NS2B and NS3 form a stable complex that can cleave NS3 within the helicase domain. Virology. 1993;193:888–899. doi: 10.1006/viro.1993.1198. [DOI] [PubMed] [Google Scholar]
- 3.Brinkworth R I, Fairlie D P, Leung D, Young P R. Homology model of the dengue 2 virus NS3 protease: putative interactions with both substrate and NS2B cofactor. J Gen Virol. 1999;80:1167–1177. doi: 10.1099/0022-1317-80-5-1167. [DOI] [PubMed] [Google Scholar]
- 4.Chambers T J, Grakoui A, Rice C M. Processing of the yellow fever virus nonstructural polyprotein: a catalytically active NS3 proteinase domain and NS2B are required for cleavages at dibasic sites. J Virol. 1991;65:6042–6050. doi: 10.1128/jvi.65.11.6042-6050.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers T J, Nestorowicz A, Amberg S M, Rice C M. Mutagenesis of the yellow fever virus NS2B protein: effects on polyprotein processing, NS2B-NS3 complex formation, and viral replication. J Virol. 1993;67:6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang G-J. Molecular biology of dengue viruses. In: Gubler D J, Kuno G, editors. Dengue and dengue hemorrhagic fever. Wallingford, United Kingdom: CAB International; 1997. pp. 175–198. [Google Scholar]
- 7.Chen C-J, Kuo M-D, Chien L-J, Hsu S-L, Wang Y-M, Lin J-H. RNA-protein interactions: involvement of NS3, NS5, and 3′ noncoding regions of Japanese encephalitis virus genomic RNA. J Virol. 1997;71:3466–3473. doi: 10.1128/jvi.71.5.3466-3473.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu P W, Westaway E G. Replication strategy of Kunjin virus: evidence for recycling role of replicative form RNA as template in semiconservative and asymmetric replication. Virology. 1985;140:68–79. doi: 10.1016/0042-6822(85)90446-5. [DOI] [PubMed] [Google Scholar]
- 9.Chu P W, Westaway E G. Characterization of Kunjin virus RNA-dependent RNA polymerase: reinitiation of synthesis in vitro. Virology. 1987;157:330–337. doi: 10.1016/0042-6822(87)90275-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chu P W, Westaway E G. Molecular and ultrastructural analysis of heavy membrane fractions associated with the replication of Kunjin virus RNA. Arch Virol. 1992;125:177–191. doi: 10.1007/BF01309636. [DOI] [PubMed] [Google Scholar]
- 11.Coia G, Parker M D, Speight G, Byrne M E, Westaway E G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specified proteins. J Gen Virol. 1988;69:1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- 12.Falgout B, Pethel M, Zhang Y-M, Lai C-J. Both nonstructural proteins NS2B and NS3 are required for the proteolytic processing of dengue virus nonstructural proteins. J Virol. 1991;65:2467–2475. doi: 10.1128/jvi.65.5.2467-2475.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Falgout B, Markoff L. Evidence that flavivirus NS1-NS2A cleavage is mediated by a membrane-bound host protease in the endoplasmic reticulum. J Virol. 1995;69:7232–7243. doi: 10.1128/jvi.69.11.7232-7243.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fosmire J A, Hwang K, Makino S. Identification and characterization of a coronavirus packaging signal. J Virol. 1992;66:3522–3530. doi: 10.1128/jvi.66.6.3522-3530.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frolova E, Frolov I, Schlesinger S. Packaging signals in alphaviruses. J Virol. 1997;71:248–258. doi: 10.1128/jvi.71.1.248-258.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorbalenya A E, Donchenko A P, Koonin E V, Blinov V. N-terminal domains of putative helicases of flavi- and pestiviruses may be serine proteases. Nucleic Acids Res. 1989;17:3889–3897. doi: 10.1093/nar/17.10.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V. Two related superfamilies of putative helicases involved in replication, recombination, repair and expression of DNA and RNA genomes. Nucleic Acids Res. 1989;17:4713–4730. doi: 10.1093/nar/17.12.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hori H, Lai C-J. Cleavage of dengue virus NS1-NS2A requires an octapeptide sequence at the C terminus of NS1. J Virol. 1990;64:4573–4577. doi: 10.1128/jvi.64.9.4573-4577.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kapoor M, Zhang L, Ramachandra M, Kusukawa J, Ebner K E, Padmanabhan R. Association between NS3 and NS5 proteins of dengue virus type 2 in the putative RNA replicase is linked to differential phosphorylation of NS5. J Biol Chem. 1995;270:19100–19106. doi: 10.1074/jbc.270.32.19100. [DOI] [PubMed] [Google Scholar]
- 20.Khromykh A A, Westaway E G. Completion of Kunjin virus RNA sequence and recovery of an infectious RNA transcribed from stably cloned full-length cDNA. J Virol. 1994;68:4580–4588. doi: 10.1128/jvi.68.7.4580-4588.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khromykh A A, Westaway E G. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J Virol. 1997;71:1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khromykh A A, Kenney M T, Westaway E G. trans-complementation of flavivirus RNA polymerase gene NS5 by using Kunjin virus replicon-expressing BHK cells. J Virol. 1998;72:7270–7279. doi: 10.1128/jvi.72.9.7270-7279.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khromykh A A, Sedlak P L, Westaway E G. trans-complementation analysis of flavivirus Kunjin NS5 gene reveals an essential role for translation of its N-terminal half in RNA replication. J Virol. 1999;73:9247–9255. doi: 10.1128/jvi.73.11.9247-9255.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khromykh A A, Sedlak P L, Guyatt K J, Hall R A, Westaway E G. Efficient trans-complementation of the flavivirus Kunjin NS5 protein but not of the NS1 protein requires its coexpression with other components of the viral replicase. J Virol. 1999;73:10272–10280. doi: 10.1128/jvi.73.12.10272-10280.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koonin E V. The phylogeny of RNA-dependent RNA polymerases of positive-strand RNA viruses. J Gen Virol. 1991;72:2197–2206. doi: 10.1099/0022-1317-72-9-2197. [DOI] [PubMed] [Google Scholar]
- 26.Koonin E V. Computer-assisted identification of a putative methyltransferase domain in NS5 protein of flaviviruses and lambda 2 protein of reovirus. J Gen Virol. 1993;74:733–740. doi: 10.1099/0022-1317-74-4-733. [DOI] [PubMed] [Google Scholar]
- 27.Li H, Clum S, You S, Ebner K E, Padmanabhan R. The serine protease and RNA-stimulated nucleoside triphosphatase and RNA helicase functional domains of dengue virus type 2 NS3 converge within a region of 20 amino acids. J Virol. 1999;73:3108–3116. doi: 10.1128/jvi.73.4.3108-3116.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindenbach B D, Rice C M. Trans-complementation of yellow fever virus NS1 reveals a role in early RNA replication. J Virol. 1997;71:9608–9617. doi: 10.1128/jvi.71.12.9608-9617.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindenbach B D, Rice C M. Genetic interaction of flavivirus nonstructural proteins NS1 and NS4A as a determinant of replicase function. J Virol. 1999;73:4611–4621. doi: 10.1128/jvi.73.6.4611-4621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mackenzie J M, Jones M K, Young P R. Immunolocalization of the dengue virus nonstructural glycoprotein NS1 suggests a role in viral RNA replication. Virology. 1996;220:232–240. doi: 10.1006/viro.1996.0307. [DOI] [PubMed] [Google Scholar]
- 31.Mackenzie J M, Khromykh A A, Jones M K, Westaway E G. Subcellular localization and some biochemical properties of the flavivirus Kunjin nonstructural proteins NS2A and NS4A. Virology. 1998;245:203–215. doi: 10.1006/viro.1998.9156. [DOI] [PubMed] [Google Scholar]
- 32.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Virus taxonomy: classification and nomenclature of viruses: Sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. [Google Scholar]
- 33.Muylaert I R, Galler R, Rice C M. Genetic analysis of the yellow fever NS1 protein: identification of a temperature-sensitive mutation which blocks RNA accumulation. J Virol. 1997;71:291–298. doi: 10.1128/jvi.71.1.291-298.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.O'Reilly E K, Kao C C. Analysis of RNA-dependent RNA polymerase structure and function as guided by known polymerase structures and computer predictions of secondary structure. Virology. 1998;252:287–303. doi: 10.1006/viro.1998.9463. [DOI] [PubMed] [Google Scholar]
- 35.Pethel M, Falgout B, Lai C-J. Mutational analysis of the octapeptide sequence motif at the NS1-NS2A cleavage junction of dengue type 4 virus. J Virol. 1992;66:7225–7231. doi: 10.1128/jvi.66.12.7225-7231.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pugachev K V, Yu N, Nomokonova, Morozova O V, Pletnev E G. A short form of the tick-borne encephalitis virus NS3 protein. FEBS Lett. 1992;297:67–69. doi: 10.1016/0014-5793(92)80329-f. [DOI] [PubMed] [Google Scholar]
- 37.Ryan M D, Monaghan S, Flint M. Virus-encoded proteinases of the Flaviviridae. J Gen Virol. 1998;79:947–959. doi: 10.1099/0022-1317-79-5-947. [DOI] [PubMed] [Google Scholar]
- 38.Schrader A P, Westaway E G. Translation mapping with the flavivirus Kunjin: gene order and anomalities in translation of NS5. Virus Res. 1988;9:323–334. doi: 10.1016/0168-1702(88)90091-3. [DOI] [PubMed] [Google Scholar]
- 39.Speight G, Westaway E G. Carboxy-terminal analysis of nine proteins specified by the flavivirus Kunjin: evidence that only the intracellular core protein is truncated. J Gen Virol. 1989;70:2209–2214. doi: 10.1099/0022-1317-70-8-2209. [DOI] [PubMed] [Google Scholar]
- 40.Stocks C E, Lobigs M. Signal peptidase cleavage at the flavivirus C-prM junction: dependence on the viral NS2B-3 protease for efficient processing requires determinants in C, signal peptide, and prM. J Virol. 1998;72:2141–2149. doi: 10.1128/jvi.72.3.2141-2149.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tan B-H, Fu J, Sugrue R J, Yap E-H, Chan Y-C, Tan Y H. Recombinant dengue type 1 virus NS5 protein expressed in Escherichia coli exhibits RNA-dependent RNA polymerase activity. Virology. 1996;216:317–325. doi: 10.1006/viro.1996.0067. [DOI] [PubMed] [Google Scholar]
- 42.Teo K F, Wright P J. Internal proteolysis of the NS3 protein specified by dengue virus 2. J Gen Virol. 1997;78:337–341. doi: 10.1099/0022-1317-78-2-337. [DOI] [PubMed] [Google Scholar]
- 43.Wengler G, Wengler G. The NS3 nonstructural protein of flaviviruses contains an RNA triphosphatase activity. Virology. 1993;197:265–273. doi: 10.1006/viro.1993.1587. [DOI] [PubMed] [Google Scholar]
- 44.Westaway E G, Khromykh A A, Kenney M T, Mackenzie J M, Jones M K. Proteins C and NS4B of the flavivirus Kunjin translocate independently into the nucleus. Virology. 1997;234:31–41. doi: 10.1006/viro.1997.8629. [DOI] [PubMed] [Google Scholar]
- 45.Westaway E G, Mackenzie J M, Kenney M T, Jones M K, Khromykh A A. Ultrastructure of Kunjin virus-infected cells: colocalization of NS1 and NS3 with double-stranded RNA, and of NS2B with NS3, in virus-induced membrane structures. J Virol. 1997;71:6650–6661. doi: 10.1128/jvi.71.9.6650-6661.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Westaway E G, Khromykh A A, Mackenzie J M. Nascent flavivirus RNA co-localized in situ with double-stranded RNA in stable replication complexes. Virology. 1999;258:108–117. doi: 10.1006/viro.1999.9683. [DOI] [PubMed] [Google Scholar]
- 47.White C L, Thomas M, Dimmock N J. Deletion analysis of a defective interfering Semliki Forest virus RNA genome defines a region in the nsP2 sequence that is required for efficient packaging of the genome into virus particles. J Virol. 1998;72:4320–4326. doi: 10.1128/jvi.72.5.4320-4326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamshchikov V F, Compans R W. Formation of the flavivirus envelope: role of the viral NS2B-NS3 protease. J Virol. 1995;69:1995–2003. doi: 10.1128/jvi.69.4.1995-2003.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]