Abstract
The majority of JAK2V617F-negative myeloproliferative neoplasms (MPNs) have disease-initiating frameshift mutations in calreticulin (CALR), resulting in a common carboxyl-terminal mutant fragment (CALRMUT), representing an attractive source of neoantigens for cancer vaccines. However, studies have shown that CALRMUT-specific T cells are rare in patients with CALRMUT MPN for unknown reasons. We examined class I major histocompatibility complex (MHC-I) allele frequencies in patients with CALRMUT MPN from two independent cohorts. We observed that MHC-I alleles that present CALRMUT neoepitopes with high affinity are underrepresented in patients with CALRMUT MPN. We speculated that this was due to an increased chance of immune-mediated tumor rejection by individuals expressing one of these MHC-I alleles such that the disease never clinically manifested. As a consequence of this MHC-I allele restriction, we reasoned that patients with CALRMUT MPN would not efficiently respond to a CALRMUT fragment cancer vaccine but would when immunized with a modified CALRMUT heteroclitic peptide vaccine approach. We found that heteroclitic CALRMUT peptides specifically designed for the MHC-I alleles of patients with CALRMUT MPN efficiently elicited a CALRMUT cross-reactive CD8+ T cell response in human peripheral blood samples but not to the matched weakly immunogenic CALRMUT native peptides. We corroborated this effect in vivo in mice and observed that C57BL/6J mice can mount a CD8+ T cell response to the CALRMUT fragment upon immunization with a CALRMUT heteroclitic, but not native, peptide. Together, our data emphasize the therapeutic potential of heteroclitic peptide–based cancer vaccines in patients with CALRMUT MPN.
INTRODUCTION
Philadelphia chromosome-negative myeloproliferative neoplasms (MPNs) are myeloid blood cancers arising from hematopoietic stem cells (1, 2) and are characterized by hyperactivated Janus kinase–signal transducer and activator of transcription (JAK-STAT) signaling (3). The majority of JAK2V617F-negative MPN tumors have an insertion or deletion (INDEL) mutation in the C-terminal region of calreticulin (CALR), creating a +1–base pair (bp) frameshift (4, 5). Whereas multiple unique INDELs are found, nearly all generate a 44–amino acid common peptide. A few rare cases generate a shorter 36–amino acid fragment (4, 5). Mutant CALR (CALRMUT) develops a pathogenic binding interaction with the extracellular portion of the thrombopoietin receptor (myeloproliferative leukemia protein; MPL), inducing ligand-independent constitutive JAK-STAT signaling pathway activation and oncogenesis (6–8). Consequently, the oncogenic CALRMUT fragment is an attractive source of frameshift-derived neoantigens for cancer vaccines in patients with CALRMUT MPN (9). However, in the few studies examining CALRMUT fragment immunogenicity, T cells from patients with CALRMUT MPN had less immunoreactivity to CALRMUT-derived peptides compared with healthy individuals (10–14), even though many immunogenic peptides are predicted (14, 15). On the other hand, T cells from healthy donors display a stronger and more frequent response to CALRMUT peptides compared with T cells from patients with CALRMUT MPN (13). In addition, several of the healthy donor T cell responses were elicited by memory T cells (13). This indicates that CALRMUT peptides are immunogenic in normal donors and suggest that CALRMUT-specific immune responses may be a mechanism of immunosurveillance eliminating the early tumor before its clinical manifestation. However, it is not clear at which extent tumors could escape this control in patients with clinical disease and why T cells from these patients did not respond to the CALRMUT fragment.
For protein antigens to be recognized by CD8+ T cells, they must first be processed into smaller peptides, translocate to the endoplasmic reticulum (ER), and, if conditions are met, bind to the class I major histocompatibility complex (MHC-I) to form a peptide:MHC-I complex (pMHC-I) capable of reaching the cell surface and being recognized by the T cell receptor. However, not all peptides fit the stoichiometric requirements for MHC-I binding. Successful pMHC-I binding requires peptides to be the correct length and to have the appropriate anchor residues at specific locations to stabilize binding to pockets of the MHC-I molecules (16). MHC genes have evolved to be extremely polymorphic across individuals. Human cells encode two copies of three different MHC-I genes, human leukocyte antigen A (HLA-A), HLA-B, and HLA-C, with each having hundreds or thousands of polymorphisms (17). Different polymorphic residues alter the MHC-I pocket affinity for peptide anchor residues such that peptides that bind to MHC-I molecules encoded by some alleles may not bind to others (18, 19). As a result, the sum of presented peptides can vary greatly across individuals. We therefore hypothesized that the suboptimal immune reactivity to the CALRMUT fragment in patients with CALRMUT MPN compared to healthy individuals could be due to differential expression of MHC-I alleles able to efficiently present CALRMUT-derived neopeptides. More specifically, individuals harboring MHC-I presenting CALRMUT-derived peptides would be able to eliminate early CALRMUT-positive MPNs, in contrast to those harboring other MHC-I alleles, who would be more likely to develop the disease.
Peptide binding affinities to MHC-I can range from strong to weak with many intermediate affinities possible. This affinity affects the number of surface-bound pMHC-I available for recognition by CD8+ T cells (20) and, ultimately, their activation. For productive stimulation, naïve T cells require interaction with a greater number of pMHC-I complexes than antigen-experienced memory T cells (21–26), and some antigens may be unable to activate naïve T cells. However, peptides can be synthetically optimized by altering one or two residues to increase MHC binding affinity. These resulting strong MHC binding peptides, called “heteroclitic” peptides (or anchor optimized or anchor improved), robustly activate naïve T cells and are similar enough to the original peptides that the activated T cells cross-react with the original antigen (27–30), assuming that the original antigen has an adequate intermediate binding affinity.
In this study, we find that six MHC-I alleles predicted to efficiently bind to multiple CALRMUT-derived peptides are less frequently observed in patients with CALRMUT MPN in two independent cohorts. This points to a higher risk of patients developing MPN with CALRMUT when lacking these MHC-I alleles. It also suggests that individuals with these MHC-I alleles can potentially control primordial CALRMUT-expressing tumors as part of the immunoediting process. In addition, this suggests that patients with CALRMUT MPN may generate suboptimal responses to cancer vaccines composed of the CALRMUT fragment. This supports the investigation of heteroclitic CALRMUT peptide–based vaccines, eventually in combination with immune checkpoint blockade, as a new therapeutic approach for patients with CALRMUT MPN.
RESULTS
Skewing of MHC-I allele frequencies in patients with CALRMUT MPN is associated with peptide binding affinity
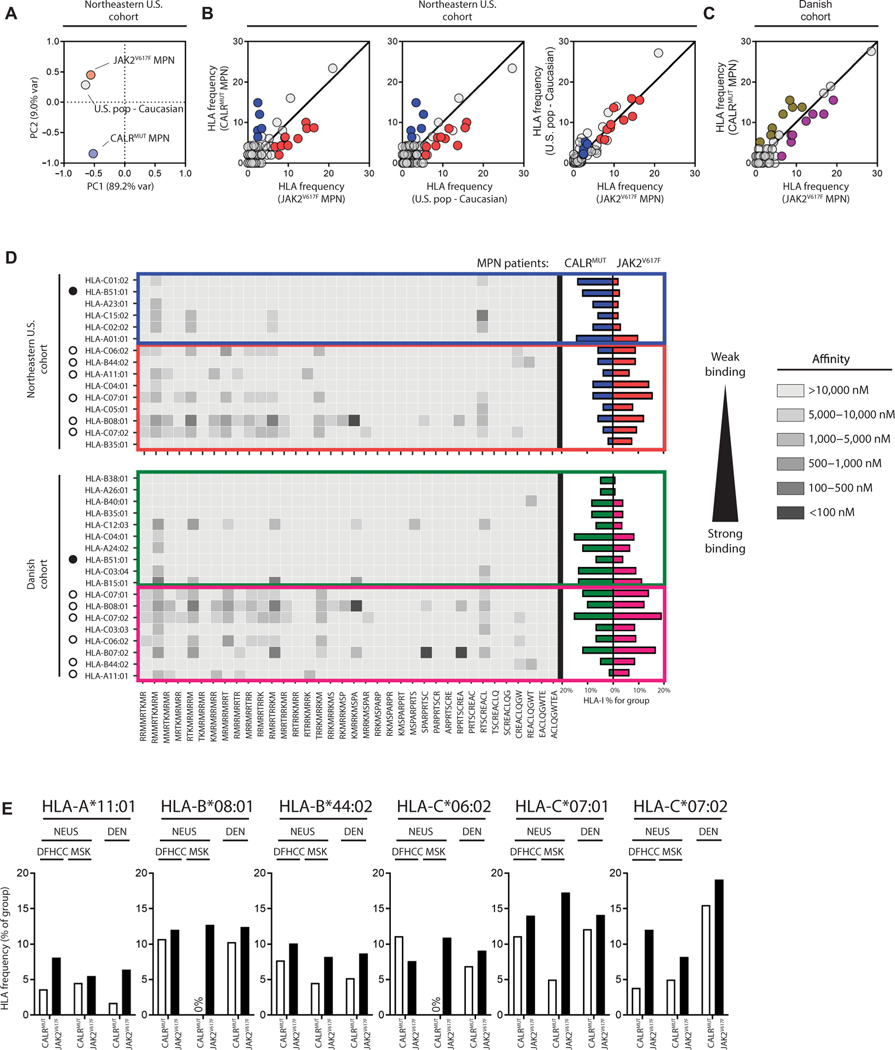
To test for MHC-I allele skewing in patients with CALRMUT MPN, MHC-I allele haplotypes from two medical centers in the northeastern United States (NEUS) and from eight medical centers in Denmark (Table 1 and tables S1 and S2) were investigated. MHC-I allele frequencies were initially calculated by group, such as HLA-A*02, for under- or overrepresentation Boschloo’s test (fig. S1A). Representative MHC-I alleles from significant (P < 0.05) or trending (P < 0.25) MHC-I groups were then assessed bioinformatically for predicted binding to peptides derived from the CALRMUT sequence (fig. S1B) using the NetMHCpan (v3) algorithm. We observed that MHC-I alleles that were less likely to be found in patients with CALRMUT MPN had good predicted binding to CALRMUT peptides, whereas overrepresented alleles had poor predicted binding.
Table 1. Distribution of patients with MPN according to mutation.
Total patient numbers (and relative frequency) associated with each cohort and center, and type of MPN, for which MHC-I haplotypes were collected.
| Cohort/institution | CALRMUT (N = 54) | JAK2V617F (N = 254) | Total (N = 308) |
|---|---|---|---|
| Northeastern U.S. cohort | 25 (19.2) | 105 (80.8) | 130 |
| Memorial Sloan Kettering Cancer Center | 11 (16.7) | 55 (83.3) | 66 |
| Dana-Farber Harvard Cancer Center | 14 (21.9) | 50 (78.1) | 64 |
| Danish cohort | 29 (16.3) | 149 (83.7) | 178 |
To refine our analysis, we examined MHC-I allele frequencies at the protein level, such as HLA-A*02:01 (tables S3 and S4). Furthermore, because MHC allele frequencies vary greatly by geographic location (31), we analyzed each cohort separately as well as together. Last, because the NEUS cohort is 88% Caucasian (table S5), we also compared MHC-I frequencies to those found in the U.S. Caucasian population from the National Marrow Donor Program (17). Unfortunately, MHC-I allele frequencies from the general Danish population were not available for comparison (31). To test for differences in MHC-I allele frequencies, we performed a principal components analysis (PCA) comparing frequencies from both NEUS MPN groups and the general U.S. Caucasian population. We observed that the JAK2V617F MPN group clusters in proximity to the U.S. Caucasian group, whereas the CALRMUT MPN group is isolated (Fig. 1A), again suggesting distinct MHC-I allele representation specifically in patients with CALRMUT MPN. Unfortunately, because of the large number of possible alleles and the modest number of total patients from each cohort, statistical power was further reduced (tables S4 and S5). We therefore, opted to analyze alleles over a minimal frequency of 5% (in either MPN groups) with a fold change above or below a set threshold of ±0.2-fold frequency change in the NEUS cohort and ±0.125-fold frequency change in the Danish cohort (Fig. 1, B and C).
Fig. 1. MHC-I alleles with predicted binding to CALRMUT-derived peptides are less frequent in CALRMUT MPNs.
(A) Principal components analysis of MHC-I allele frequencies from patients with CALRMUT MPN, patients with JAK2V617F MPN, and U.S. Caucasian population. (B) Comparison of MHC-I allele frequencies from patients with CALRMUT MPN, patients with JAK2V617F MPN, and U.S. Caucasian population compared to each other in the NEUS cohort and (C) the Danish cohort. Frequencies are expressed as percentages. MHC-I alleles that are overrepresented in patients with CALRMUT are in blue (NEUS cohort) and green (Danish cohort), whereas MHC-I alleles that are underrepresented in patients with CALRMUT are in red (NEUS cohort) and pink (Danish cohort). For the NEUS cohort, only MHC-I alleles differentially expressed between patients with CALRMUT MPN compared to both patients with JAK2V617F MPN and U.S. Caucasian population were considered for further analysis. (D) Heatmap of predicted binding of each CALRMUT-derived peptide to each MHC-I allele from (B) (left). MHC-I alleles that are underrepresented or overrepresented in patients with CALRMUT MPN in both cohorts are noted with white or black circles, respectively. Actual MHC-I allele frequencies in CALRMUT and JAK2V617F MPN patient populations are also noted (right). (E) Cohort breakdown of CALRMUT MPN MHC-I allele frequencies of individual institution consisting of the NEUS cohort for the six less frequent MHC-I alleles.
We observed that HLA-B*51:01 is the only allele overrepresented in patients with CALRMUT MPN from both cohorts, whereas six MHC-I proteins are similarly underrepresented: HLA-A*11:01, HLA-B*08:01, HLA-B*44:02, HLA-C*07:01, HLA-C*07:02, and HLA-C*06:02. This trend is also reflected in the fraction of patients who are positive for these alleles (tables S6 and S7). Five of the six MHC-I alleles that met these requirements for underrepresentation had a moderate predicted affinity (<10,000 nM) to about a quarter of all 9-mer (Fig. 1D) or, to a lesser extent, 10-mer (fig. S2) peptides, of which many had <500 nM predicted affinity. On the other hand, all MHC-I alleles that were overrepresented in patients with CALRMUT MPN had poor binding to almost all CALRMUT-derived peptides with the exception of HLA-B*15:01 and HLA-C*12:03, which only had a high frequency in the Danish cohort (Fig. 1D). The HLA-B*07:02 allele, which has good predicted binding to CALRMUT peptides, was underrepresented in both cohorts but only met the designated threshold for underrepresentation in the Danish cohort. When the NEUS cohort was separated into the two original cohorts [Memorial Sloan Kettering (MSK) and Dana-Farber Harvard Cancer Center], the same MHC-I skewing was confirmed except for HLA-C*06:02, which was slightly elevated in patients from MSK (Fig. 1E). Considering that five of the six underrepresented MHC-I alleles happened to have a good peptide binding suggests a trend worthy of further examination.
MHC-I skewing in CALRMUT MPN is specific for CALRMUT-derived peptides
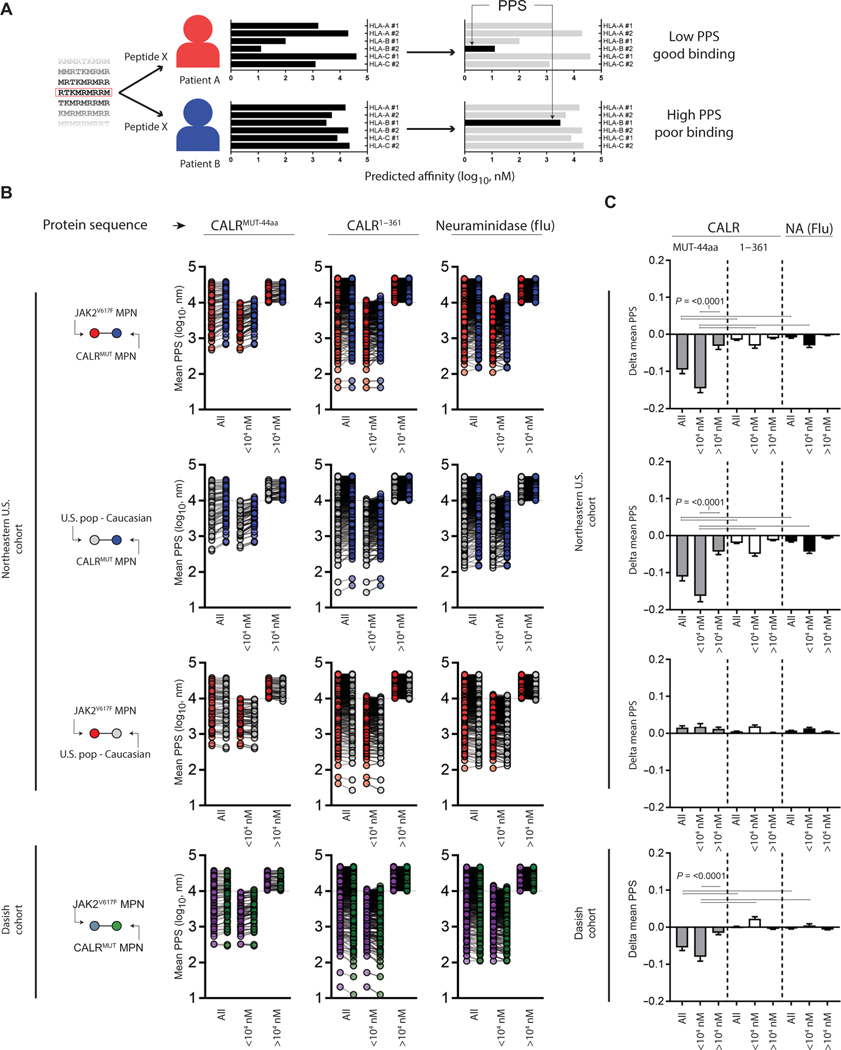
Prediction algorithms for pMHC-I binding based on neural networks like NetMHC are generally accepted to be useful yet imperfect tools, and their biases are typically hard to capture. To control for the prediction algorithm, we hypothesized that MHC-I allele frequency bias should not be observed for proteins or protein fragments that are not under selective immune pressure. To test this, we scored the predicted binding affinity of each CALRMUT-derived peptide or other irrelevant proteins in each individual patient with MPN to generate what we have termed the patient:peptide score (PPS). Briefly, the PPS of a peptide in a patient is equal to the binding score (nM) of that peptide against the MHC-I allele with the strongest predicted binding affinity of the six possible MHC-I alleles for that peptide (Fig. 2A). Furthermore, because statistically significant differences for any single MHC-I allele frequency was difficult to achieve, taking each patient’s six MHC-I proteins into consideration would offer a more robust statistical analysis of MHC-I protein skewing. As expected based on our initial findings, the average PPS of CALRMUT-derived peptides was elevated in patients with CALRMUT MPN from both cohorts compared to control groups, where a higher score is associated with worse predicted binding (Fig. 2, B and C). However, the average PPS of peptides derived from the wild-type (WT) portion of CALR was barely changed in the NEUS cohort and completely unchanged in the Danish cohort for the patients with CALRMUT MPN compared to control groups (Fig. 2, B and C). Similarly, the capacity to bind peptides derived from the irrelevant foreign protein neuraminidase (NA) from the influenza virus did not substantially change when comparing patients with CALRMUT or JAK2V617F MPN and the general U.S. Caucasian populations (Fig. 2, B and C). Furthermore, when CALRMUT-derived peptides were subdivided based on predicted binding (<10,000 nM) and nonbinding (>10,000 nM) scores, the greatest shift in average PPS occurred in predicted binding peptides (<10,000 nM), suggesting selection pressure against the capacity to efficiently bind those peptides in patients with CALRMUT MPN (Fig. 2, B and C). A generous cutoff of 10,000 nM, as opposed to a much stricter 500 nM threshold, was used here to account for any bias in the prediction algorithm. Together, our findings suggest that patients with CALRMUT MPN have a skewed MHC-I allele repertoire that is less immunologically responsive to the CALRMUT protein fragment. The implication of this observation also suggests that these patients would be less likely to respond to cancer vaccines consisting of the CALRMUT fragment.
Fig. 2. MHC-I bias is selective against the CALRMUT fragment.
(A) Schema depicting calculation of patient:peptide score (PPS). Briefly, single peptides derived from longer sequences were applied to NetMHCpan v3 to predict class-I HLA binding against any given patient’s six possible class-I HLAs. In each patient-peptide combination, the strongest affinity was registered as the PPS. (B) Mean PPS of individual peptides derived from indicated protein sequence (9- to 10-mers) from patients with CALRMUT and JAK2V617F MPN and pseudopopulation created from expected frequencies of U.S. Caucasian population (NEUS only). Protein sequences are the 44–amino acid CALRMUT sequence (CALRMUT−44aa), the wild-type CALR sequence upstream of CALRMUT−44aa (CALR1–361), and the irrelevant foreign antigen neuraminidase (NA) from influenza. Also shown are peptides broadly subdivided from predicted possible binding (<104 nM) and predicted nonbinding (>104 nM) peptides. (C) Difference in mean PPS of each group from (B). The Student’s t test was performed to calculate significance. The horizontal bars represent the combination where the statistical test resulted in the noted P value.
Underrepresented MHC-I alleles potentiate response to CALRMUT peptides
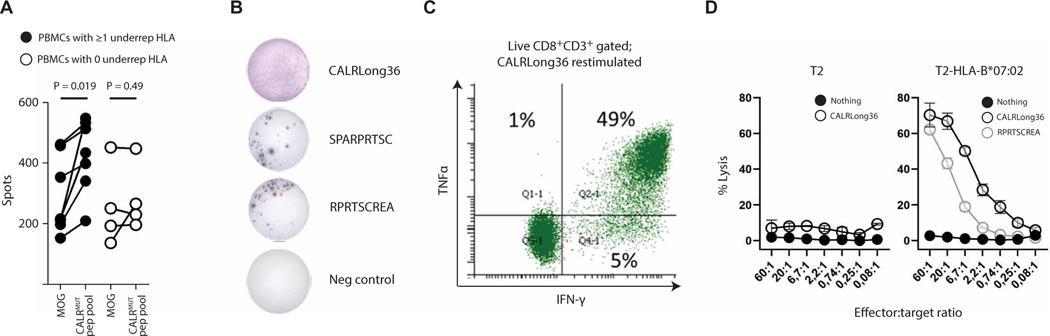
To test whether underrepresented MHC-I alleles were more likely to potentiate an immune response against the CALRMUT fragment, peripheral blood mononuclear cells (PBMCs) from seven healthy donors positive for at least one of the underrepresented MHC-I alleles, and four healthy donors that were negative for these MHC-I alleles (table S8) were stimulated in vitro with peptides covering the entire CALRMUT fragment and examined for reactivity by interferon-γ (IFN-γ) enzyme-linked immunospot (ELISpot) after a final round of peptide restimulation. In the donors positive for the underrepresented MHC-I alleles, seven of seven (100%) responded, whereas a response was observed in only one of four (25%) of donors negative for the underrepresented MHC-I alleles (Fig. 3A). Separately, to confirm MHC-I specificity, we generated CD8+ T cell cultures specific for the CALRLong36 peptide from a patient with HLA-B*07:02–positive CALRMUT post–essential thrombocythemia primary myelofibrosis who was enrolled in the previously reported CALRMUT vaccine trial (13). Reactivity was validated by IFN-γ ELISpot (Fig. 3B) and flow cytometry (Fig. 3C). After this, CALRLong36-primed– specific CD8+ T cells were analyzed in a standard Cr51 cytotoxicity assay with peptide-pulsed normal T2 cells or HLA-B*07:02– transduced T2 cells as target cells. In this context, only the peptide-pulsed T2-HLA-B*07:02 but not the peptide-pulsed T2 cell or the unpulsed T2-HLA-B*07:02 was killed (Fig. 3D), demonstrating that CD8+ T cells were restricted to HLA-B*07:02, one of the underrepresented alleles from the Danish cohort. Together, we show that patients with CALRMUT-positive MPNs were less likely to have an MHC-I allele predicted to bind with high affinity to peptides derived from the CALRMUT fragment. This may be due to the fact that individuals with MHC-I alleles that can efficiently present CALRMUT-derived peptides are less likely to develop CALRMUT MPN.
Fig. 3. Underrepresented MHC-I alleles potentiate response to CALRMUT peptides.
(A) IFN-γ ELISpot of PBMCs from healthy donors that had either at least one (black circles) or zero (white circles) underrepresented MHC-I allele expanded with a peptide pool (15-mers) derived from the entire CALRMUT fragment and restimulated after 10 days with either the irrelevant peptide (MOG) or the same CALRMUT fragment peptide pool. (B) Reactivity of CALRLong36-specific CD8+ T cells derived from an HLA-B*07:02+ patient with post–essential thrombocythemia myelofibrosis. Cells restimulated with different CALRMUT peptides in an IFN-γ ELISpot, (C) intracellular IFN-γ and TNFα cytokine staining on the CALRLong36-specific CD8+ T cells stimulated with CALRLong36 (D) target cell lysis of wild-type or HLA-B*07:02–transduced T2 cells pulsed with different CALRMUT peptides.
Full-length CALRMUT antigen is processed and presented by human primary myeloid cells
A necessary requirement for immune-mediated targeting of CALRMUT-positive MPN tumor cells by CALRMUT-specific CD8+ T cells is the endogenous processing and presentation of CALRMUT peptide by MHC-I. Whereas many in the field presumed this to be happening, it has not yet been proven directly. To test whether endogenous processing and presentation of CALRMUT peptide by MHC-I is occurring in human primary myeloid cells, we used two different strategies. In the first set of experiments, the aforementioned patient-derived CALRLong36 specific CD8+ T cells were enriched and cloned using a limiting dilution. The CD8+ T cell clones’ ability to recognize CALRMUT target cells was assessed by IFN-γ ELISpot after restimulation with autologous CALRMUT-positive CD14+ monocytes that were subjected to CALR-specific siRNA or control knockdown (fig. S3A). Reactivity to the CALR siRNA–transfected target cells was diminished in all clones. In a separate experiment, primary human myeloid cells from healthy donors were infected by adenovirus (AdV) expressing the full-length 44–amino acid antigen, and these cells were used to activate autologous PBMCs for 11 days in vitro, whereupon CALRMUT-specific reactivity was assessed by secondary peptide restimulation followed by flow cytometry. In 7 out of 20 donors, there was a clear priming of CALRMUT-specific CD8+ T cells (fig. S3, B and C). Furthermore, almost all of the donors that had reactivity had at least one of the previously discussed underrepresented MHC-I allele (fig. S3D). We therefore conclude that the CALRMUT antigen can be processed and presented endogenously by human primary myeloid cells, further supporting the model that CALRMUT MPN can be subject to immune-mediate eradication, either endogenously or vaccine mediated, presuming a favorable MHC-I allele haplotype.
Patients with CALRMUT MPN exhibit skewed MHC-II allele frequencies
We also examined MHC-II allele frequencies for skewing using haplotype data from the NEUS cohort. Unfortunately, MHC-II haplotypes were unavailable for the Danish cohort. Similar to MHC-I alleles, MHC-II alleles also exhibited distinct frequencies by PCA (fig. S4A). No single allele had a statistically significant decrease in frequency in patients with CALRMUT MPN (table S9), and the same fold change threshold approach as for HLA-I alleles was therefore applied. We observed that HLA-DRB1*03:01, HLA-DRB1*04:01, HLA-DRB1*07:01, HLA-DRB1*13:01, HLA-DQB1*02:01, and HLA-DQB1*06:03 were less frequent in patients with CALRMUT MPN compared to patients with JAK2V617F MPN and U.S. individuals of European descent, whereas HLA-DRB1*11:01, HLA-DRB1*11:04, and HLA-DQB1*03:01 were more frequent (fig. S4B). Both of the HLA-DRB1 MHC-II alleles found at higher frequency in patients with CALRMUT MPN that could be assessed for predicted binding were predicted to bind strongly to more than half of 15-mer peptides, whereas only one of the four HLA-DRB1 MHC-II alleles found at lower frequency appears to do so (fig. S4C). When we examined the MHC-II alleles of the healthy donors from which PBMCs were used to generate memory CD4+ T cell lines against long CALRMUT peptides in a previous study (13), both donors were positive for HLA-DRB1*13:01, and one of these was also positive for HLA-DRB1*04:01 (table S10).
Nonresponding human PBMCs can cross-react with CALR9p2 if first primed with heteroclitic peptides
We hypothesized that one approach to elicit an immune response against CALRMUT MHC-I skewed patients is to use a vaccine consisting of MHC-I binding-optimized heteroclitic peptides to increase the initial activation of CALRMUT-specific CD8+ T cells. To test this hypothesis, we examined the peptides with the lowest PPS in the patients with CALRMUT MPN as possible candidates. We observed that the top peptide in both cohorts was the 9mer CALRMUT peptide starting at position 2 (CALR9p2) RMMRTKMRM (fig. S5A), which is predominantly a function of its predicted binding to frequently observed MHC-I alleles such as HLA-A*02:01 and HLA-A*03:01 (fig. S5, B and C), which are not underrepresented in patients with CALRMUT MPN (table S3). Therefore, based on the mean PPS of CALR9p2 in both cohorts, we reasoned that this peptide was less likely to directly activate naïve CD8+ T cells, but could be recognized and targeted by CD8+ T cells preprimed with peptides of similar specificities and improved/optimized MHC-I binding, such as heteroclitic peptides. We therefore investigated whether heteroclitic peptides could be used to induce cross-reactivity against CALRMUT-derived peptides in human samples.
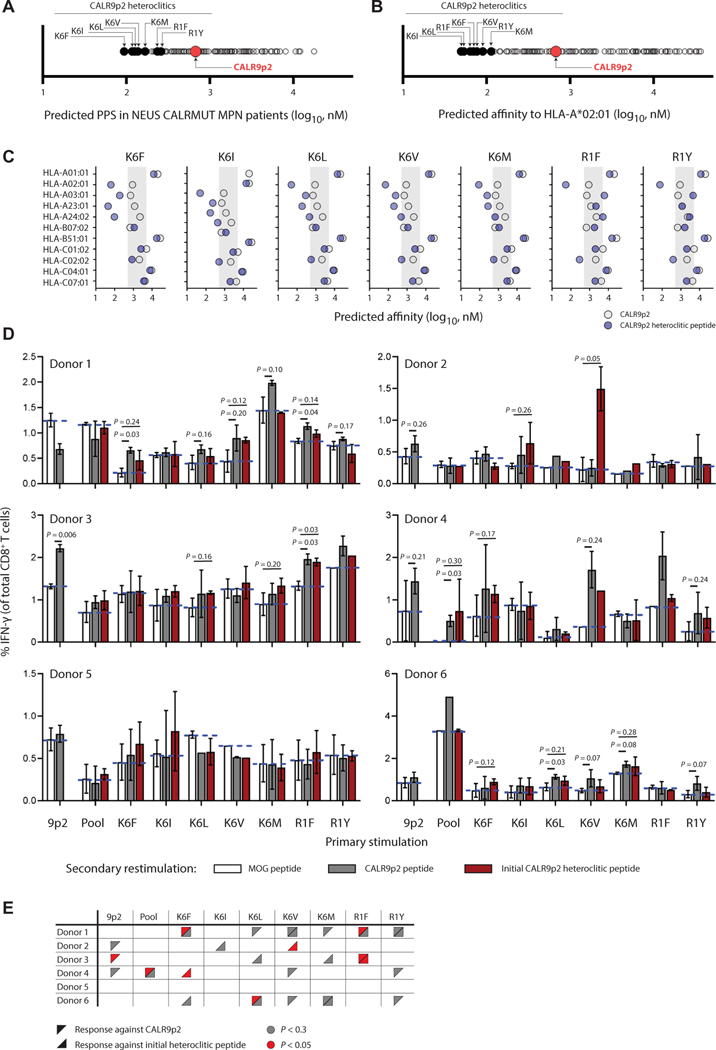
To identify the best candidate CALR9p2 heteroclitic peptide, we examined the mean predicted PPS score of the NEUS CALRMUT MPN cohort to every possible CALR9p2 peptide variant containing a single amino acid substitution (Fig. 4A). Seven of the top heteroclitic CALR9p2 were selected for testing based on their predicted binding to HLA-A*02:01 (Fig. 4B). Five peptides were selected with a substitution at position 6 (K6), and two were selected with a substitution at position 1 (R1). Accordingly, alterations of the residues at positions 1 and 6 were predicted to affect HLA-A*02:01 binding through its minor anchor sites instead of the main anchor sites at positions 2 and 9, where the existing native residues have predicted weakly favorable binding conditions (fig. S6) (32, 33). A closer examination of binding predictions of each heteroclitic candidate to the top 10 most frequent MHC-I alleles in patients with CALRMUT MPN revealed that the K6 heteroclitic peptides could target multiple MHC-I alleles with predicted binding of the CALR9p2 ranging from 500 to 5000 nM, whereas the R1 heteroclitic peptides mostly affected HLA-A*02:01 binding affinity (Fig. 4C). We thus tested the seven candidate heteroclitic CALR9p2 peptides for their ability to bind to and stabilize HLA-A*02:01 on TAP (transporter associated with antigen processing)2-deficient HLA-A*02:01–positive T2 cells and confirmed that they have greater binding capacity than native CALR9p2 peptide, which had weak binding signal compared with dimethyl sulfoxide (DMSO) control (fig. S7). However, all of the heteroclitic CALR9p2 peptides had weaker binding potential than the MART1-A2 peptide positive control.
Fig. 4. Human CD8+ T cells activated with heteroclitic CALR9p2 peptides cross-react with CALR9p2 peptide.
(A) Predicted mean PPS in patients with CALRMUT MPN of all single amino acid substitution of CALR9p2. (B) Predicted affinity of all single amino acid substitution of CALR9p2 to HLA-A*02:01. Seven heteroclitic peptides were chosen for further testing and are identified here only by their amino acid substitution in the CALR9p2 peptide. (C) Binding affinity of CALR9p2 and indicated heteroclitic peptide to the most common MHC-I in the CALRMUT MPN patient cohorts. Shadowed area indicates predicted binding affinity range of 5000 to 500 nM. (D) Percent IFN-γ+ CD8+ T cells after primary in vitro stimulation of PBMCs with CALR9p2 or CALR9p2 heteroclitic peptides (noted on x axis) followed by secondary restimulation with either control (MOG, median represented by dotted blue line) peptide, CALR9p2, or initial CALR9p2 heteroclitic peptide (noted by color of bar). (E) Summary of responding donor PBMCs to CALR9p2 or CALR9p2 heteroclitic peptides.
To test the ability of these peptides to elicit a T cell response, we stimulated PBMCs from six healthy HLA-A*02:01 individuals with known MHC-I haplotypes (table S11) and PPS (table S12) in the presence of native CALR9p2, using each heteroclitic peptide individually, as well as using a pool of all heteroclitic peptides. Cells were then exposed to a secondary restimulation with either control peptides, the same peptides used in the initial stimulation, or the native CALR9p2 peptide and tested for IFN-γ production (Fig. 4, D and E). As controls, some cells were left unstimulated (DMSO) or were stimulated with a positive control peptide pool of T cell epitopes from cytomegalovirus (CMV), Epstein-Barr virus (EBV), influenza, and Clostridium tetani (CEFT), or with phorbol myristate acetate (PMA) and ionomycin (fig. S8). We found that four of six donors responded to at least one heteroclitic peptide (Fig. 4, D and E). Three of the six healthy donors had significant (P < 0.05) or trending (P < 0.30) CD8+ T cell responses to CALR9p2 alone (donors 2, 3, and 4), and two of them also showed significant CD8+ T cell cross-reactivity against CALR9p2 if primed with a heteroclitic peptide (donors 3 and 4; P < 0.05). In two of the samples that did not respond to CALR9p2 alone (donors 1 and 6), there was significantly detectable CD8+ T cell cross-reactivity with CALR9p2 when the PBMCs were primed with heteroclitic peptides (P < 0.05). To test whether the heteroclitic peptides were promoting cross-reactivity through HLA-A*02:01, we again activated healthy donor HLA-A*02:01– positive PBMCs using pooled heteroclitic peptides, but the final restimulation was provided using the peptide-pulsed K562-HLA-A*02:01 cell line. In the four additional donors tested, two had a cross-reactive response against the CALR9p2 peptides, demonstrating that HLA-A*02:01 was at least one of the HLA alleles through which the heteroclitic peptides were providing a cross-reactive response (fig. S9). Together, our results suggest that cross-reactive immunity to CALRMUT can be achieved in human cells, especially if multiple different heteroclitic peptides are used.
Modeling CALRMUT MPN patient MHC-I allele skewing in mice
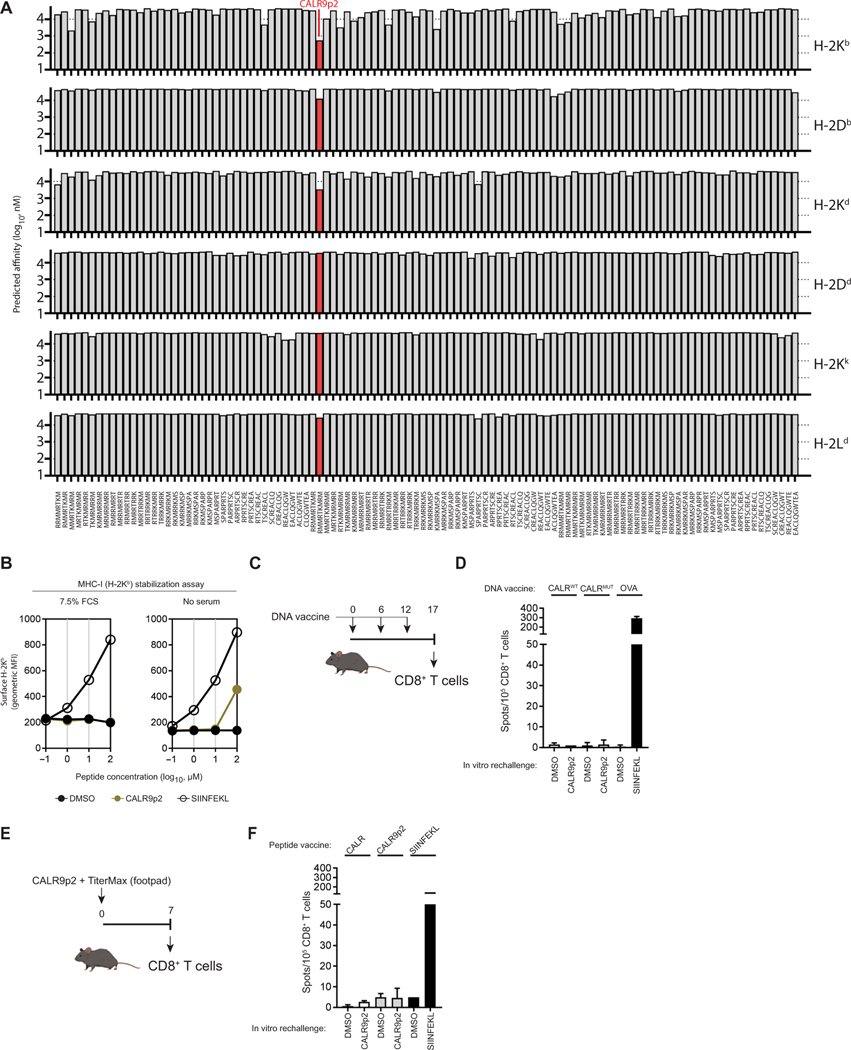
To determine whether a heteroclitic peptide cancer vaccine is a viable strategy against the CALRMUT fragment, we tested this approach in a preclinical mouse model mimicking CALRMUT MPN MHC-I allele skewing. We analyzed the predicted binding of all CALRMUT-derived peptides against all murine MHC-I alleles for which predictions were possible. We found no strong binding peptide (<500 nM) to any murine MHC-I alleles (Fig. 5A) but did observe that H-2Kb had weak predicted binding affinity to CALR9p2. When tested for its ability to stabilize MHC-I in the H-2Kb–expressing TAP-deficient RMA/S cell line, CALR9p2 did not elicit detectable H-2Kb stabilization compared to the control strong binding chicken ovalbumin (OVA)–derived peptide SIINFEKL (Fig. 5B). However, when serum was omitted from the assay, H-2Kb was stabilized only with the highest concentration of peptide (100 μg/ml), suggesting poor but still detectable binding (Fig. 5B). To investigate whether the CALRMUT protein fragment is immunogenic in vivo, we immunized H2-Kb–expressing C57BL/6J (B6) mice with a DNA vaccine (Fig. 5C) encoding the full-length 52-bp deletion variant of the CALRMUT sequence (4–6) and analyzed CALR9p2 peptide– specific CD8+ T cells from draining lymph nodes. Compared to the immunogenic CD8+ T cell response against the SIINFEKL peptide in OVA-immunized mice, mice immunized with the fulllength CALRMUT sequence did not elicit any CD8+ T cell response against CALR9p2 (Fig. 5D). Likewise, mice immunized in the footpad (Fig. 5E) with the CALR9p2 peptide emulsified in the TiterMax adjuvant did not elicit any CALR9p2-s pecific CD8+ T cell response compared to SIINFEKL-immunized mice either (Fig. 5F). Because we observed poor but detectable binding of CALR9p2 to H-2Kb but no vaccine-induced CALR9p2-specific CD8+ T cell responses in B6 mice, this mouse model constitutes a relevant preclinical setting resembling patients with CALRMUT MPN with an MHC-I skewed haplotype in which to test the in vivo efficacy of CALRMUT heteroclitic peptide-based vaccines.
Fig. 5. CALRMUT sequence is not immunogenic in C57BL/6 J mice.
(A) The predicted binding affinity of CALRMUT-derived peptides (8- to 10-mers shown) against all available murine MHC-I alleles. The strongest binding peptide CALR9p2 is identified in red. (B) H-2Kb stabilization assay using TAP-deficient RMA/S cells was performed for CALR9p2 in the presence and absence of serum. The chicken ovalbumin (OVA)–derived peptide SIINFEKL was used as a positive control. (C) Timeline of DNA immunization schedule and CD8+ T cell collection for the experiment in (D). (D) IFN-γ ELISpot depicting secondary reactivity of CD8+ T cells isolated from draining lymph nodes of mice DNA immunized with full-length CALRWT, CALRMUT, and OVA. (E) Timeline of peptide immunization and CD8+ T cell collection for the experiment in (F). (F) IFN-γ ELISpot depicting secondary reactivity of CD8+ T cells isolated from draining lymph nodes of mice peptide immunized with adjuvant and DMSO, CALR9p2, or SIINFEKL. Data shown represent results from one repeat of experiments performed at least three times.
Full-length CALRMUT variant does not have dominant-negative activity
One possible issue with a CALRMUT vaccine is the uncertainty as to whether the CALRMUT fragment itself can inhibit antigen presentation. Wild-type CALR is required for antigen presentation in healthy cells (34, 35), and the full-length CALRMUT is reported to be non-functional with respect to peptide loading (35). However, nearly all CALRMUT-positive MPN tumors are heterozygous (4, 5) and therefore have one WT copy of CALR, yet it is unknown whether CALRMUT acts as a dominant-negative with respect to its role in antigen presentation. To exclude this possibility, we cotransfected the murine B16F10 cells with the DNA sequences encoding OVA and either CALRWT-mCherry, CALRMUT-mCherry, or the mCherry constructs, and measured surface expression of H-2Kb–presented SIINFEKL peptide. We observed that cells transfected with the CALRMUT variant had an equal percentage of H-2Kb-SIINFEKL peptide–expressing cells (fig. S10, A and B), suggesting that the CALRMUT variant did not inhibit antigen presentation. We observed a marginal decrease in total surface MHC-I expression (fig. S10C), suggesting some effect of the CALRMUT variant, although it is unclear whether such a small reduction in MHC-I would have any functional effect. As a result, we conclude that CALRMUT does not prevent tumor cells from presenting antigens and suggest that CALRMUT tumors could respond to cancer vaccines if the correct antigen is selected and is expressed by the tumor.
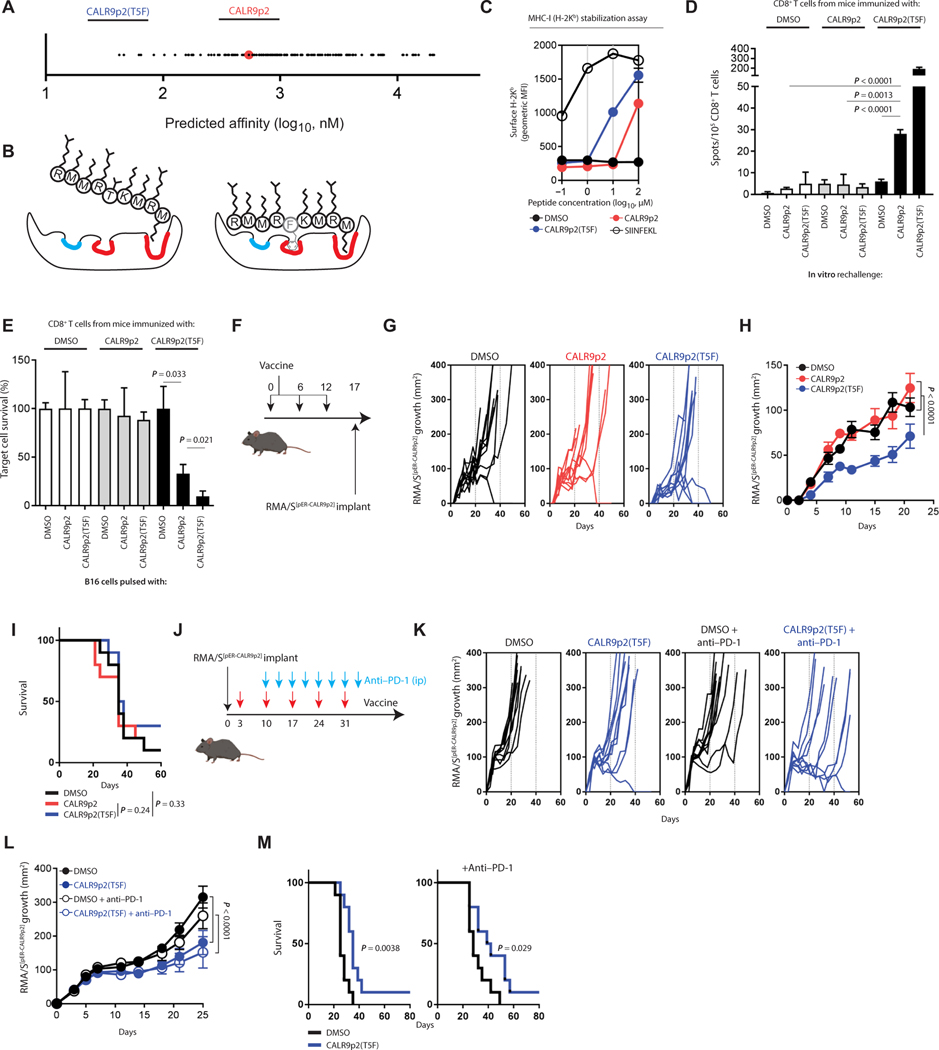
C57BL/6J-optimized heteroclitic CALR9p2 peptide elicits cross-reactive immunity in mice
To identify the best candidate CALR9p2 heteroclitic peptide in B6 mice, we examined the predicted binding affinity of every possible CALR9p2 peptide variant containing a single amino acid substitution to H-2Kb. We observed that the variant with the strongest predicted affinity had a threonine (T)–to–phenylalanine (F) substitution at position 5 (T5F) of the CALR9p2 peptide (Fig. 6A). This is consistent with previous studies showing that this site is an important anchor residue for H-2Kb (Fig. 6B and fig. S11) (19, 36, 37). Specifically, for 9-mers, in addition to the major anchor sites at positions 2 and 9 of H-2Kb, which bind to the nonoptimal but weakly favorable M2 and M9 residues of CALR9p2, the T residue is predicted to have a net negative binding effect at the other important position 5 anchor site, where the F residue instead favors stronger binding. When we investigated the ability of CALR9p2(T5F) to stabilize H-2Kb in RMA/S cells, this heteroclitic peptide demonstrated about a 10-fold greater H-2Kb stabilization compared to CALR9p2 (Fig. 6C). We then tested whether a CALR9p2(T5F) peptide vaccine could elicit a cross-reactive CALR9p2 immune response. Mice immunized with a single dose of CALR9p2(T5F) peptide vaccine elicited a CD8+ T cell response capable of cross-reacting to CALR9p2 in vitro (Fig. 6D) and killing tumor cells pulsed with the CALR9p2 peptide (Fig. 6E). Likewise, mice immunized with a DNA vaccine against the full-length CALRMUT sequence encoding the CALR9p2(T5F) variant (fig. S12A) also developed a cross-reactive response against the native CALR9p2 peptide (fig. S12B). The ability of the full-length antigen to induce a specific response demonstrates again that the full-length CALRMUT sequence can be endogenously processed and presented. As expected, immunizing mice with the heteroclitic CALR9p2(T5F) variant, by peptide or DNA vaccine, elicited an even stronger IFN-γ and cytotoxic T cell response against the same CALR9p2(T5F) heteroclitic peptide, which served as positive control in these experiments (Fig. 6, D and E, and fig. S12B). When we monitored CALR9p2(T5F)-specific CD8+ T cell responses in mice by tetramer staining upon vaccination with either the CALRMUT DNA encoding the CALR9p2(T5F) heteroclitic peptide (fig. S12, C and D) or directly with the CALR9p2(T5F) heteroclitic peptide (fig. S13, A and B), we found that these cells were expanded only by the heteroclitic sequence, and not by WT CALR. Furthermore, CD8+ T cells from heteroclitic-immunized mice displayed higher surface expression of the activation markers CD44, TIM3 (T cell immunoglobulin domain and mucin domain 3), and PD-1 (programmed cell death protein 1) compared to tetramer-negative CD8+ T cells from the same animals (fig. S12, E and F). Only the CALR9p2(T5F) tetramer-positive CD8+ T cell fraction responded to the CALR9p2 native peptide upon in vitro secondary restimulation, as measured by up-regulating IFN-γ and tumor necrosis factor–α (TNFα) production (fig. S13, C and D), indicating that the same T cell clones could recognize both CALR9p2(T5F) and CALR9p2. In addition, a second restimulation with either CALR9p2(T5F) or CALR9p2 peptides of in vivo CALR9p2(T5F)-primed CD8+ T cells induced similar up-regulation of PD-1 and TIM3 in CALR9p2(T5F) tetramer-positive CD8+ T cells (fig. S13, E and F).
Fig. 6. Heteroclitic CALR9p2 peptide vaccine elicits cross-reactive CD8+ T cell response against CALR9p2 and controls tumor growth in mice.
(A) Predicted binding affinity to H-2Kb of all single amino acid substitution variants of CALR9p2. Top predicted peptide CALR9p2(T5F) is shown. (B) Cartoon of the expected effect of T5F substitution in CALR9p2 peptide conformational binding into H-2Kb. Known dominant anchor sites and minor anchor sites are depicted in red and blue, respectively. (C) MHC-I stabilization assay using TAP-deficient RMA/S cells was performed for CALR9p2 and CALR9p2(T5F) in the absence of serum. SIINFEKL was used as a positive control. Representative results from one repeat of an experiment performed at least three times. (D) IFN-γ ELISpot depicting secondary reactivity of CD8+ T cells isolated from draining lymph nodes of mice peptide immunized with adjuvant and DMSO, CALR9p2, or CALR9p2(T5F). Representative results from one repeat of an experiment performed at least three times. The Student’s t test was performed to calculate significance. (E) Killing assay of peptide-pulsed B16 cells by CD8+ T cells isolated from peptide-immunized mice. Representative results from one repeat of an experiment performed at least three times. The Student’s t test was performed to calculate statistical significance. (F) Timeline of peptide immunization prior to tumor implantation for prophylactic vaccine. (G) RMA/SpER-CALR9p2 tumor growth over time after prophylactic peptide immunization for individual tumors or (H) average growth until the second mouse reached the end point of a tumor size of 400 mm2. (I) Survival of mice after the prophylactic vaccine. (J) Timeline of peptide immunization and tumor implantation for therapeutic vaccine and in combination with anti–PD-1 therapy. ip, intraperitoneally. (K) Tumor growth over time after therapeutic vaccine for individual tumors or (L) averaged tumor growth until the second mouse reached the end point. (M) Survival of mice after the therapeutic vaccine, with or without anti–PD-1 therapy. Data from tumor growth experiments represent results from one repeat of experiments performed twice. Significance for tumor growth experiments was calculated by performing a Student’s t test on the area under the curve of each tumor. Significance for survival was calculated by performing a log-rank test.
We then tested the ability of these cross-reacting immune responses induced by CALRMUT heteroclitic vaccines to induce antitumor effects in vivo. To generate a suitable tumor model for this, we used the newly developed PresentER antigen minigene system (38). Here, the nucleotide sequence of the CALR9p2 peptide is cloned downstream of an ER signal sequence (SS) and virally transduced into TAP-deficient RMA/S lymphoma cell line (RMA/SpER-CALR9p2). Once expressed, the peptide-SS is shuttled into the ER and the peptide is cleaved from the SS, releasing CALR9p2 into the ER, where it can be loaded into MHC-I, assuming binding is possible. When mice were given three doses of the heteroclitic peptide vaccine prior to tumor implantation, RMA/SpER-CALR9p2 tumors grew significantly slower than those injected into mice given adjuvant alone in the same vehicle (DMSO) or preimmunized with the native CALR9p2 peptide, which both grew at the same rate (P < 0.0001; Fig. 6, F and G), although differences in survival were not significant (Fig. 5H). However, when RMA/SpER-CALR9p2 tumors were allowed to grow before mice received multiple therapeutic doses of the heteroclitic peptide vaccine (Fig. 6I), tumors grew significantly slower (P < 0.0001; Fig. 6, J and K) and mice had improved survival (Fig. 6L) compared to the adjuvant alone condition. The effect of the vaccine was even more prominent when the immunization was administered with PD-1 immune checkpoint blockading antibody (Fig. 6, I to K). The therapeutic vaccine had greater efficacy than the prophylactic vaccine. As this was unexpected, we hypothesized that CALR9p2-specific cross-reactive CD8+ T cells were diminishing in efficacy over time and that the available CALR9p2 antigen present in the tumor cells was not generating a strong memory response. To investigate this further, we immunized mice with three doses of the heteroclitic CALR9p2(T5F) peptide vaccine and compared cross-reactive potential in conditions where mice instead received CALR9p2 peptide boosts after an initial CALR9p2(T5F) priming dose. Consistent with the prophylactic vaccine results, mice that received an initial CALR9p2(T5F) followed by two CALR9p2 boosts had no detectable cross-reactive CD8+ T cell responses to CALR9p2 in vitro and a very small response to the CALR9p2(T5F) peptide (fig. S14). On the other hand, mice that received two doses of CALR9p2(T5F) and a final CALR9p2 boost had no detectable CALR9p2 cross-reactive CD8+ T cells but an intermediate response to CALR9p2(T5F). Therefore, it appears that cross-reactive immunity to the CALR9p2 peptide in the context of B6 mice is a function of time-from-last CALR9p2(T5F) dose. Together, this provides a proof of principle that a heteroclitic peptide cancer vaccine strategy in a murine model is a viable option for tumor cells expressing weakly binding tumor epitopes, such as those predicted to be presented in CALRMUT MPN tumors.
DISCUSSION
It has long been postulated that the immune system shapes tumors by eliminating clones expressing immunogenic antigens in a process called immunoediting (39–41). We have also recently shown that there is a trade-off between tumorigenicity of a mutation and its immunogenicity (42). Clones that evade elimination would either enter a state of equilibrium with the immune system or manage to evade it completely, such that it manifests clinically. However, by its very nature, the elimination of primordial tumor clones has been difficult to visualize experimentally and in patients. Patients with MPNs with the CALRMUT frameshift fragment offer a simple yet elegant example of the elimination phase in vivo. In this tumor type, the CALRMUT frameshift is a driver truncal mutation and may constitute a neoantigen (15). Although this indicates that an effective immune response against CALRMUT can be potentially generated, not everyone can develop immunity against CALRMUT MPN. To unravel the molecular mechanisms explaining the lack of immunogenicity of CALRMUT in patients developing MPN, we investigated potential differences in MHC-I and MHC-II allele frequencies in this patient population.
In this study, we observe that patients with CALRMUT MPN from two independent cohorts are less likely to carry the MHC-I alleles HLA-A*11:01, HLA-B*08:01, HLA-B*44:02, HLA-C*06:02, HLA-C*07:01, and HLA-C*07:02. Of these six MHC-I alleles, all except HLA-B*44:02 are predicted to bind to multiple CALRMUT-derived peptides. This contrasts with MHC-I alleles that are more frequent in patients with CALRMUT MPN, which are predicted to not bind efficiently to CALRMUT-derived peptides. We therefore posit that individuals with a negatively skewed MHC-I allele, which is predicted to bind to CALRMUT, are more likely to undergo immune-mediated tumor rejection such that the tumors never manifest clinically.
In a study examining CALRMUT immunoreactivity in healthy donors, HLA-C*07:01 was expressed in four of five healthy donors that responded to multiple CALRMUT-derived peptides (14). In another study, the MHC-I allele HLA-B*07:02, which is underrepresented in patients with CALRMUT MPN in the Danish but not the NEUS cohort, was found to elicit an immune response to multiple CALRMUT-derived peptides (12). In another example, in a rare case where CD8+ T cells from a CALRMUT MPN patient did respond to a long CALRMUT peptide in vitro, this patient was identified to be positive for the HLA-C*06 allele (10, 11). Therefore, many of the MHC-I alleles that we find to be negatively skewed in patients with CALRMUT MPN have been reported to be immunogenic against some CALRMUT-derived peptides, which supports our main hypothesis.
Our findings suggest that oncogenic CALR mutations are more frequent than it is currently observed, because we hypothesize that individuals with advantageous MHC-I haplotypes can eliminate the disease before it manifests and develop specific immunological memory. This would imply that many healthy individuals can mount an immune response against the CALRMUT fragment. Evidence for such MHC-I allele–specific immunity against recurring mutations in blood cancers has been observed elsewhere. Specifically, it was observed that MHC-I alleles that have shown to bind to the BCR-ABL fusion protein are less frequent in patients with BCR-ABL+ chronic myeloid leukemia (43), and likewise, MHC-I alleles predicted to bind to nucleophosmin 1 frameshift mutations are less frequently observed in patients with acute myeloid leukemia (44).
As we still observe patients with CALRMUT MPN with the negatively skewed predicted immunogenic MHC-I alleles, it is clear that MHC-I haplotype alone is insufficient for the elimination of the disease. It is currently unclear why negatively skewed predicted immunogenic MHC-I–expressing patients with CALRMUT MPN still develop the disease, but many explanations are plausible: (i) Many tumors evade immune-mediated rejection by inhibiting the antigen processing machinery itself. This includes the epigenetic down-regulation of MHC-I, or by acquiring mutations in TAP or β2M, which are frequently observed in tumors under immune pressure (45, 46). (ii) It is possible that the tumor establishes an immunosuppressive microenvironment such that priming of the immune system or the elimination of the tumor is inhibited. A recent study found that T cells from patients with CALRMUT MPN had elevated surface expression of PD-1 and cytotoxic T lymphocyte-associated antigen-CTLA-4 compared to healthy donors, as well as increased numbers of myeloid-derived suppressor cells and regulatory T cells in the blood (14), all of which are associated with immunosuppression (47). It is possible that negatively skewed MHC-I–expressing patients with CALRMUT MPN may have defects in the antigen presentation machinery or a higher than normal immunosuppressive phenotype preventing tumor control. The same study showed that CALRMUT MPN PBMCs, which were unresponsive to multiple CALRMUT-derived peptides in vitro, could elicit a response when a PD-L1 antibody was added to the coculture (14). Therefore, negatively skewed MHC-I–expressing patients with CALRMUT MPN might be promising candidates for immune checkpoint blockade therapy as well as a heteroclitic peptide cancer vaccine, because it could favor immune-mediated recognition of CALRMUT peptides correctly presented in proper MHC-I.
Our study had three main limitations. First, the limited number of patients with CALRMUT MPN resulted in trending but nonsignificant statistical power when calculating whether any individuals MHC-I alleles were underrepresented. We tried to address this limitation by examining two different independent patient cohorts for similar patterns of MHC-I allele frequencies, as well as showing that patients with CALRMUT MPN have significantly lower global MHC-I binding affinities to all CALRMUT-derived peptides when all six MHC-I alleles are considered. Second, we were unable provide direct evidence that the full-length CALRMUT antigen could be endogenously processed and presented in CALRMUT MPN tumor cells in vivo. However, we showed that antigen-presenting cells (APCs) expressing the full-length CALRMUT antigen can activate CD8+ T cells in an experimental setting, strongly suggesting that endogenous processing and presenting in CALRMUT MPN tumor cells in vivo in patients are possible. Last, whereas we have shown that a heteroclitic peptide vaccine is effective in our mouse model of a tumor presenting a CALRMUT antigen, it is important to note that heteroclitic peptide-based cancer vaccines have had limited success in past human clinical trials (48). However, all of these trials were targeting nonmutated tumor-associated antigens or cancer germline antigens where the cross-reactive pool of CD8+ T cells may have been subject to some immunological tolerance, which is not the case in the context of the CALRMUT fragment.
Should a vaccine prove effective in patients with CALRMUT MPN, it would provide a valuable nonredundant benefit in these patients, because there are currently no rationally designed treatments specifically targeting the CALR mutation. Although the JAK1/JAK2 inhibitor ruxolitinib is approved by the U.S. Food and Drug Administration for the treatment of patients with MPN, this approval was granted primarily based on symptomatic benefits (49, 50). Although patients with CALRMUT MPN demonstrate clinical responses to ruxolitinib, there is no reduction in CALRMUT allele burden after JAK2 inhibition, and as a result, ruxolitinib does not have substantial disease-modifying activity in MPN and is not curative (51). A longer-term goal of peptide vaccination should be to treat patients with CALRMUT MPN early in the course of their disease before genetic and clonal evolution has occurred. By directing autologous immune responses specifically against CALRMUT, peptide vaccination offers the potential to preferentially target the disease-initiating MPN stem cell in patients, which is a deficiency of current MPN drugs, including JAK inhibitors.
In the first human clinical phase 1 vaccination trial with a CALRMUT peptide fragment with the adjuvant Montanide, it was shown that although the majority of patients displayed a marked CALRMUT-specific CD4+ T cell response, only 2 of 10 patients mounted a CD8+ T cell response to the vaccine and none had a clinical response (52). Although limited patient responses suggest that CALRMUT presentation by peptide vaccination is suboptimally immunogenic, the possibility exists that native CALRMUT immunization combined with strong immunostimulatory adjuvancy could result in successful priming in vivo. In addition, our findings indicate that these patients could further benefit from a vaccine approach incorporating optimized heteroclitic CALRMUT peptides. Last, as we show that this strategy can be enhanced by PD-1 blockade in our mouse model, we propose that an off-the-shelf vaccine composed of one or more CALRMUT heteroclitic peptides in combination with anti–PD-1 or other immune checkpoint inhibitor therapies can be a viable treatment for patients with CALRMUT MPN.
METHODS
Study design
The primary objective of this study was to investigate associations between the frequency of MHC-I alleles in patients with CALRMUT MPN compared to control groups (patients with JAKV617F MPN and, if possible, the general population) and the likelihood that these specific MHC-I alleles were contributing to anti-CALRMUT MPN immunity. The only criteria for admission into this study were for a patient to be diagnosed with MPN and having a known MHC-I haplotype. All patients analyzed in this study were diagnosed with MPN and tested positive for the CALRMUT or JAKV617F mutations. Approval was obtained for the use of patient-derived specimens and access to clinical data extracted from patient charts by the Institutional Review Boards at MSK Cancer Center, the Dana-Farber Cancer Institute, and the Massachusetts General Hospital, as well as by the Danish Regional Science Ethics Committee. Our analysis supported the hypothesis that MHC-I alleles that were predicted to induce anti-CALRMUT MPN immunity were underrepresented in patients with CALRMUT MPN. Therefore, the secondary objective of this study was to develop a strategy to counteract the skewing of MHC-I haplotypes in patients with CALRMUT MPN using heteroclitic peptides. Eight- to 10-week-old female C57BL/6J mice (strain #000664) were purchased from the Jackson Laboratory. Mouse experiments were performed in accordance with institutional guidelines under a protocol approved by the MSK Cancer Center Institutional Animal Care and Use Committee. All mice were maintained in a pathogen-free facility according to the National Institutes of Health (NIH) Animal Care guidelines. For prophylactic cancer vaccines, animals were randomized for the tumor inoculation. For therapeutic cancer vaccines, animals were randomized after tumor inoculation. No mice were excluded from the experiment. All in vivo experiments consisted of at least 10 mice per group.
MHC allele frequencies
For the NEUS cohort, MHC genotypes were manually extracted from patient charts. For the Danish cohort, HLA class I genotypes were determined by next-generation sequencing performed on biobanked samples collected for a previous study (53) using recently described methods (54). In cases where National Marrow Donor Program (NMDP) allele codes were used instead of the World Health Organization nomenclature, the conversion was done according to https://bioinformatics.bethematchclinical.org/hla-resources/allele-codes/allele-code-lists/. If multiple alleles were plausible for a given NMDP allele code, we selected the most likely allele based on ethnicity [typically around 99% confidence, based on known frequencies in the general population (17)]. MHC allele frequency for each HLA gene (A, B, C, DQ, and DR) was broadly calculated as the number of each specific allele divided by the number of the total allele in that cohort (2n per individual). In rare samples, certain patients had incomplete haplotype information where one or more alleles were unknown or incomplete. If the allele was missing, it was censored from the number of total alleles. If locus and group were known but the exact protein was unknown, this allele was censored from the frequency calculation only for alleles from the same group. For an MHC allele to be considered positively skewed, it was required to have an allele frequency of >0.05 in CALRMUT MPN cases and have >0.2-fold frequency increase compared to both the JAK2V617F and U.S. Caucasian population group allele frequency in the NEUS cohort, or a >0.125-fold frequency increase compared to the JAK2V617F group allele frequency in the Danish cohort. Likewise, for an MHC allele to be negatively skewed, it was required to have a frequency of at least 0.05 in both the JAK2V617F and U.S. Caucasian population groups for the NEUS cohort, or just the JAK2V617F group for the Danish cohort, and have a >0.2-fold decrease in the CALRMUT MPN compared to both the JAK2V617F and U.S. Caucasian population group allele frequency in the NEUS cohort, or a >0.125-fold frequency decrease compared to the JAK2V617F group allele frequency in the Danish cohort. PCA was calculated in R and plotted in GraphPad Prism v7. All data processing and analysis were performed using the R version 3.3.2 Sincere Pumpkin Patch and GraphPad Prism v7.
Binding affinity predictions and PPS
pMHC-I binding predictions and binding motifs Seq2Logo graphs were collected using NetMHCpan v3 (32) for human MHC-I alleles and NetMHC v4 (36) for murine MHC-I alleles. pMHC-II binding predictions were collected using NetMHCIIpan v3.2 (55). To calculate the PPS, peptide affinities for all six possible MHC-I alleles were identified, and only the lowest affinity value was retained. The protein fragment RRMMRTKMRMRRMRRTRRKMRRKMSPARPRTSCREACLQGWTEA was used for the CALRMUT. For the CALRWT sequence, amino acids 1 to 361 of the UniProt sequence P27797 were used. For the influenza NA sequence, the full UniProt sequence D7ED91 was used. All subsequent data processing and analyses were performed using the R version 3.3.2 Sincere Pumpkin Patch and GraphPad Prism v7.
MHC-I stabilization assay
To test peptide binding to murine H-2Kb or human HLA-A*02:01, TAP-deficient RMA/S and T2 cells were used, respectively, as previously reported (56, 57). Briefly, RMA/S cells (58) were serum starved in serum-free RPMI 1640 media overnight at 31°C, and peptides were added at indicated final concentrations, followed by 30 min at 31°C and another 3 hours at 37°C before measuring H-2Kb by flow cytometry (BD Biosciences, clone AF6–88.5). T2 cells (59) were serum starved in serum-free RPMI 1640 media overnight, and cells were added at indicated final concentrations for 16 hours before measuring HLA-A*02:01 surface expression by flow cytometry (BD Biosciences, clone BB7.2). The SIINFEKL peptide was acquired as a custom order from GenScript. The MART1-A2 peptide (ELAGIGILTV) was purchased from JPT Peptide Technologies.
Murine immunizations and subsequent antigen-specific analysis
All peptides were purchased as custom peptide synthesis orders from GenScript at a purity of >98% and resuspended at 10 mg/ml in DMSO (Sigma-Aldrich). For peptide vaccines, peptides were diluted in phosphate-buffered saline (PBS) and emulsified with TiterMax (TiterMax USA Inc.) at a 1:1 ratio immediately prior to immunization, such that each dose was composed of 10 μg of peptide in a total volume of 25 μl. In control (DMSO) immunization, an equivalent volume of DMSO is substituted for the diluted peptide in the TiterMax emulsion. For subsequent in vitro assays, draining inguinal and popliteal lymph nodes were collected at indicated time points, and CD8+ T cells were isolated using positive magnetic sorting using mouse CD8a (Ly-2) MicroBeads (Miltenyi Biotec). DNA vaccines were performed using a gene gun as previously described (27, 60) according to indicated time points. Briefly, mice received four injections (400 lb/inch2) of DNA-coated gold particles into the abdominal region of the skin for a total 4 μg of DNA per dose. DNA plasmids encoding the WT or 52-bp deletion CALRMUT sequences fused with the flag sequence are as previously described (61) and were cloned into the pING plasmid (62). For pING-OVA, the full-length chicken OVA sequence was also cloned into the pING plasmid (62). For pING-CALRMUT-CALR9p2(T5F), the sequence was directly modified from pING-CALRMUT by site-directed mutagenesis using Q5 Site-Directed Mutagenesis Kit using the following primers: GGCGAATGATGCGGTTCAAAATGCGCA and TTGTTCTCTGCTCTTCGTCT. CD8+ T cells were collected as before, but only from draining inguinal lymph nodes. To test for antigen specificity, the mouse IFN-γ ELISpot set (BD Biosciences) was used according to the manufacturer’s instructions. Briefly, CD8+ T cells were frozen immediately after purification in fetal bovine serum (FBS) containing 10% DMSO, thawed 1 day prior to restimulation, and allowed to recover overnight in interleukin-1 (IL-2; 20 U/ml) (PeproTech) RPMI 1640 medium containing 10% FBS, Na-pyruvate, l-glutamine, and penicillin/streptomycin (pen/strep). As a source of APCs, splenocytes from naïve mice were depleted of T cells using magnetic microbeads for CD8a (Ly2) and CD4 (L3T4) (Miltenyi Biotec), pulsed for 1 hour with peptide (100 μg/ml) in serum-free media at 37°C followed by a wash. For each well, 105 CD8+ T cells were cocultured with 1 × 105 to 3 × 105 peptide-pulsed APCs and incubated for about 18 hours. Spots were counted using the ImmunoSpot analyzer (Cellular Technology Limited). For tetramer-specific immune characterization, cells were stained with the CALR9p2(T5F) tetramers (NIH Tetramer Core Facility), CD8a (BioLegend, clone 53–6.7), TIM3 (BioLegend, clone RMT3–2.3), CD3e (eBioscience, clone 17A2), CD4 (BD Biosciences, clone RM4–5), CD279/PD-1 (eBioscience, clone J43; Invitrogen, clone RMP1–30), CD44 (Invitrogen, clone IM7), IFN-γ (eBioscience, clone XMG1.2), and TNFα (BD Biosciences, clone MP6-XT22), and the fixable viability dye (Thermo Fisher Scientific), assessed using the BD LSR II Flow cytometer and analyzed by FlowJo v10.
SIINFEKL:H-2Kb expression
B16F10 cells were cotransfected with equal parts pING-OVA and pCMV-Sport6-CALR constructs fused to mCherry, which are previously described (61), using the Megatran 1.0 transfection reagent (Origene). Each construct was mixed with the transfection reagent separately such that all cells received the same amount of pING-OVA construct. B16F10 cells were originally obtained from I. Fidler (M. D. Anderson Cancer Center) and cultured in RPMI 1640 medium supplemented with 7.5% inactivated FBS, 1× nonessential amino acids, and 2 mM l-glutamine. One day after transfection, cells were stained by flow cytometry with H-2Kb (BD Biosciences, clone AF6–88.5) and H-2Kb-SIINFEKL (BioLegend; Clone 25-D1.16).
B16 killing assay
A total of 104 B16F10 cells were seeded in 24-well plates 1 day before coculture. On the day of the killing assay, B16F10 cells were washed and peptide pulsed with peptide (100 μg/ml) for 2 hours at 37°C before a final wash. CD8+ T cells from peptide-immunized mice were thawed and allowed to recover as before, and 106 CD8+ T cells were added to wells containing the peptide-pulsed B16F10 cells in the presence of recombinant murine interleukin(IL)-2 (rmIL-2) (20 U/ml) (PeproTech) and cocultured for 72 hours at 37°C. After the coculture, supernatants and floating cells were carefully removed, and the remaining B16F10 cells were washed and collected after trypsinization. Countbright counting beads (Invitrogen) were added to the samples, and the cell population was stained with LIVE/DEAD Fixable Green Dead cell stain kit (Invitrogen) and CD45-APC-Cy7 (BD Biosciences, clone 30-F11) before being analyzed by Guava easyCyte 8HT (Millipore). Live CD45-negative cells were normalized to bead counts and normalized by sample and target cells.
Tumor growth experiments
RMA/S cells were maintained in RPMI 1640 medium supplemented with 7.5% inactivated FBS, 1× nonessential amino acids, and 2 mM l-glutamine. The DNA sequence encoding the CALR9p2 peptide (bold) was cloned into the PresentER-IRES-GFP (38) construct using the following oligo 5′-GGCCGTATTGGCCCCGCCACCTGTGAGCGGGAGGATGATGAGGACAAAGATGA.
GGATGTAAGGCCAAACAGGCC-3′ after Sfi I digestion and T4 ligation (New England Biolabs). The resulting construct was used to generate retrovirus by cotransfection with pCL-Ampho into ecotropic Phoenix cells (American Type Culture Collection). PresentER-IRES-GFP and pCL-Ampho were generously provided by D. Scheinberg’s group. Viral supernatants were collected at 48 and 72 hours, and pooled and Retro-X Concentrator (Takara Bio)– concentrated retrovirus was used to transduce RMA/S cells by spinoculation using polybrene (Sigma-Aldrich). GFP-positive cells were FACS (fluorescence-activated cell sorting)–sorted (BD FACSAria III) and cultured in puromycin (4 μg/ml) (Gibco) media. A total of 5 × 106 cells were injected subcutaneously in the flank of mice. For anti–PD-1 treatment, 250 μg of RMP1–14 was injected intraperitoneally in PBS at indicated time points, starting 10 days after tumor inoculation.
Human PBMC in vitro restimulation
For the experiment described in Figs. 3A and 4 (D and E), freshly isolated or thawed cryopreserved healthy donor PBMCs were restimulated with cytokines and peptides as previously described (14). Briefly, on day 0, PBMCs were resuspended in X-VIVO15 media (Lonza) and seeded at 105 per well of a 96 U-bottom plate with granulocyte-macrophage colony-stimulating factor (GM-CSF) (1000 IU/ml) (Sanofi), IL-4 (500 IU/ml) (R&D Systems), and Flt3L (50 ng/ml) (R&D Systems). On day 1, media were refreshed with lipopolysaccharide (0.1 μg/ml) (InvivoGen), 10 μM R848 (InvivoGen), IL-1β (5 μg/ml) (R&D Systems), and indicated peptides (1 μg/ml), and incubated for 24 hours. The CMV, EBV, flu, tetanus (CEFT), and the myelin oligodendrocyte glycoprotein (MOG) peptide pools (JPT Technologies) were used as positive and negative controls, respectively. On days 2 and 5, half the media was refreshed with RPMI 1640 (Gibco) containing 10% human serum (Gemini Bio-Products), gentamycin (10 μg/ml) (Gibco), Hepes (Gibco), GlutaMAX (Gibco), and human(h) IL2 (hIL-2) and hIL-7 to a final concentration of 10 IU/ml and 10 ng/ml, respectively (R&D Systems). On day 8, the media was refreshed without cytokines. On day 10, PBMCs were restimulated with corresponding peptides in the presence of anti-hCD28 and anti-hCD49d (1 μg/ml) (BD Biosciences). As controls, some cells were stimulated with PMA (50 ng/ml; Sigma-Aldrich) and ionomycin (1 μg/ml; Sigma-Aldrich). All antibodies were purchased from BioLegend. LIVE/DEAD Fixable Blue Dead Cell Stain Kit by Thermo Fisher Scientific was used for live and dead cell discrimination. Data were acquired using the BD LSRFortessa, and the data were analyzed on FlowJo V10 (TreeStar). For the ELISpot, methods are as previously described (14) using the follower 15-mer peptides that cover the entire CALRMUT fragment: RTRRMMRTKMRMRRM, MMRTKMRMRRMRRTR, TKMRMRRMRRTRRK, RMRRMRRTRRKMRRK, MRRTRRKMRRKMSPA, RRKMRRKMSPARPRT, RRKMSPARPRTSCRE, SPARPRTSCREACLQ, PRTSCREACLQGWTE, TSCREACLQGWTEA.
For the experiments described in Fig. 3 (B to D) and fig. S3A, generation of specific T cell cultures and subsequent ELISpot and flow cytometry is as described previously (13), whereas Chromium-51 release cytotoxic assay, autologous CD14+ purification, culture, and siRNA-mediated CALR silencing are as described previously (11).
For experiments described in fig. S3 (B and C), frozen dendritic cells purified by magnetic bead positive selection specific to CD1c surface antigen were thawed and cultured in 5% human serum Iscove’s modified Dulbecco’s medium (IMDM) (Gibco) with pen/strep, l-glutamine, and Hepes (henceforth I5 media) with IL-4 (80 ng/ml) (PeproTech) and GM-CSF (Gibco) for 1 day, and subsequently infected by empty, CMV, EBV and influenza (CEF) encoding or the 44–amino acid long CALRMUT antigen-encoding AdV5 in human lactoferrin (1 mg/ml) (Sigma-Aldrich) I5 media at a multiplicity of infection of 5000 for 24 hours, followed by a PBS wash. Autologous pan T cells were thawed and cultured with the transduced dendritic cells at 10:1 T:DC ratio in I5 media with IL-15 (10 ng/ml) (PeproTech) and IL-21 (30 ng/ml) (R&D Systems) for 2 days. Media was refreshed every 2 to 3 days with I5 media containing IL-15 (10 ng/ml) and IL-2 (10 IU/ml). For peptide restimulation, T cells were cultured overnight in fresh IL-15/IL-2 I5 media containing a CALRMUT peptide pool (95% purity Vivitide) at 2 μg/ml and eBioscience transport inhibitor cocktail (Thermo Fisher Scientific). As controls, human PepTivator AdV5 Hexon Peptide Pool (Miltenyi) or CEF-MHC Class I Control Peptide Pool “Plus” (Cellular Technologies) was used. Cells were stained with LIVE/DEAD Fixable Aqua Dead Cell Stain (V500) (Thermo Fisher Scientific), CD3 PerCP Cy5.5 (BioLegend), CD4 BV421 (BioLegend), CD8 APC-Cy7 (BioLegend), TNFα fluorescein isothiocyanate (FITC) (R&D Systems), and IFN-γ BV785 (BioLegend). The eBioscience Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific) was used to fix and permeabilize the cells. Samples were assessed by BD FACSCelesta flow cytometer and analyzed by FlowJo v10.
For experiments involving K562 cell restimulation from fig. S9, PBMCs were restimulated in complete IMDM media + 10% human serum + 4 mM l-glutamine + 55 μM β-mercaptoethanol + rhIL-2 (100 U/ml) (Chiron), rhIL-7 (25 ng/ml), and rhIL-15 (25 ng/ml) (PeproTech) with DMSO or 10 μg/mL per peptide for 14 days. Cytokines were refreshed twice a week, and peptides were refreshed at day 7. For the secondary restimulation, HLA-null K562 cells were transduced to express HLA-A*02:01 using the retroviral construct SFG-HLA-A*02:01 (provided by Dr. R. O’Reilly, MSKCC), were peptide pulsed with indicated peptides (100 μg/ml in serum-free RPMI 1640 for 2 hours at 37C), and used as target cells in the presence of anti-hCD28 and anti-hCD49d (1 μg/ml). For intracellular staining, monensin and brefeldin A (BD Biosciences) were added 1 hour after restimulation and culture were left to incubate for another 12 hours. Cells were then stained for CD3 (clone: OKT3, FITC), CD4 (clone: RPA-T8, APC), and CD8a (clone: RPA-T4, BV785), permeabilized, and fixed with BD Cytofix/Cytoperm reagents according to the manufacturer’s protocol and subsequently stained for IFN-γ [clone: B27, phycoerythrin (PE)] and TNFα (clone: Mab11, PE/Cy7). Samples were assessed by BD LSR II Flow cytometer and analyzed by FlowJo v10.
Statistical analysis
Details of the study outline, sample size, and statistical analysis are shown in the main text, figures, tables, and figure legends. To calculate significance in distribution of MHC frequencies, Barnard’s unconditional test and the chi-square test were used as indicated in R using the barnard.test() function (two-tail) from the Barnard package and as well as the base chisq.test() function. The R version 3.3.2 Sincere Pumpkin Patch was used. For unpaired Student’s t tests, area under the curve calculations, log-rank survival test, and GraphPad Prism v7 were used.
Supplementary Material
Acknowledgments:
We are thankful to R. Gejman and D. Scheinberg for providing the PresentER-IRES-GFP construct along with their technical suggestions during the cloning process, to R. O’Reilly for providing the SFG-HLA-A*02:01 construct, to the NIH Tetramer Core Facility for generating the tetramers, and to M. Rashkovan for critically reading the manuscript. AM is a Scholar of the Leukemia and Lymphoma Society.
Funding:
A portion of this research was funded in part through the Starr Cancer Consortium Award I9-A9-059 (to M.G., R.Z., S.P., T.M., J.D.W., A.M., and T.C.), the NIH/NCI Cancer Center Support Grant P30 CA008748/NCI R01 CA056821, the Swim Across America, Ludwig Institute for Cancer Research, Ludwig Center for Cancer Immunotherapy at Memorial Sloan Kettering, Cancer Research Institute, and Parker Institute for Cancer Immunotherapy (to M.G., R.Z., S.P., L.M.B.M., S.V., S.S., M.M.G., D.V., A.G., Z.M., T.C., R.R., O.A-W., J.D.W., and T.M.), through the NHLBI/NIH (R01HL131835 to A.M.). A.E.M. receives funding from the U.S. Department of Defense (Horizon Award W81XWH-20-1-0904) through the Parker Institute for Cancer Immunotherapy (to C.C.B. and N.B.) and through the Danish Cancer Society (R149-A10159 to M.H.A.).
Footnotes
Data and materials availability: All data associated with this study are present in the paper or the Supplementary Materials.
Competing interests: M.G., J.D.W., and T.M. are inventors on patent/patent application (WO 2021/188619 A1 and PCT/US2021/022679) held/submitted by Memorial Sloan Kettering Cancer Center that covers heteroclitic peptide cancer vaccines targeting CALRMUT. R.Z. is an inventor on patent applications related to work on GITR, PD-1, and CTLA-4 antibodies, which is unrelated to this study, and is a consultant for Leap Therapeutics. S.P. is currently employed at Regeneron. S.S. has filed a provisional patent for work on phosphatidylserine-targeting antibodies, which is unrelated to this study. A.G. has consulted for Adivo Associates and Group H. G.S.H. has received funding from Merck, Bayer, Incyte, and Constellation; is member of the advisory boards for Jazz, Agios, Merck, Incyte, Celgene, and BMS; has received grant support from ASH-AMFDP, K-12 Paul Calabresi Award, and Sanchez Ferguson Award for work unrelated to this study; and has consulted for Abbvie, Novartis, Pfizer, Pharmaxis, Incyte, Constellation, and Blueprint Medicine. D.L.H. has received grant support from Alexion Pharma Nordic AB, Novartis Healthcare, and the A. P. Møller and Chastine Mc-Kinney Møller Foundation for work unrelated to this study. R.C.L. has received grant support from MedImmune and Jazz Pharmaceuticals and has consulted for Takeda Pharmaceuticals, Bluebird Bio, Qiagen, Nuprobe, and Thermo Fisher Scientific. H.H. has received grant support from Novartis A/S. M.H.A. is a shareholder and board member of IO Biotech ApS. T.C. is a cofounder of and holds equity in Gritstone Oncology, as well as holds equity in An2H; has received grant support from Bristol-Myers Squibb, AstraZeneca, Illumina, Pfizer, An2H, and Eisai for work unrelated to this study; has served as an advisor for Bristol-Myers, MedImmune, Squibb, Illumina, Eisai, AstraZeneca, and An2H; and holds ownership of intellectual property on using tumor mutation burden to predict immunotherapy response, which is unrelated to this study. O.A.-W. has served as a consultant for H3B Biomedicine, Foundation Medicine Inc., Merck, and Janssen; is on the Scientific Advisory Board of Envisagenics Inc.; and has received prior research funding from H3B Biomedicine for work unrelated to this study. P.W. and M.A.S. hold shares and stock options in Johnson & Johnson. R.R. has consulted for Morphosys/Constellation, Incyte, CTI Celgene/BMS, Novartis, Promedior, Blueprint Stemline, Galaecto, Pharmessentia Abbvie, Sierra Oncology, and Disc Medicines. N.B. serves as an advisor and a board member for Neon Therapeutics, Tempest, Checkpoint Sciences, Curevac, Primevax, Novartis, Array BioPharma, Roche, Avidea, Boehringer Ingelheim, and Parker Institute for Cancer Immunotherapy, and has received grant support from Parker Institute for Cancer Immunotherapy, Novocure, Celldex, Ludwig Institute, Genentech, Oncovir, Melanoma Research Alliance, Leukemia & Lymphoma Society, NYSTEM, and Regeneron. J.D.W. has consulted for Adaptive Biotech, Advaxis, Amgen, Apricity, Array BioPharma, Ascentage Pharma, Astellas, Bayer, Beigene, Bristol Myers Squibb, Celgene, Chugai, Dragonfly, Elucida, Eli Lilly, F Star, Genentech, Imvaq, Janssen, Kleo Pharma, Kyowa Hakko Kirin, Linneaus, MedImmune, Merck, Neon Therapuetics, Northern Biologics, Ono, Polaris Pharma, Polynoma, Psioxus, Puretech, Recepta, Takara Bio, Trieza, Sellas Life Sciences, Serametrix, Surface Oncology, Syndax, and Syntalogic; has received grant support from Bristol Myers Squibb, Medimmune, Merck Pharmaceuticals, and Genentech; has equity in Potenza Therapeutics, Tizona Pharmaceuticals, Adaptive Biotechnologies, Elucida, Imvaq, Beigene and Trieza, and Linneaus; has received honoraria from Esanex; and holds ownership, has licensed, receives royalties, or is pending approval on intellectual property related to alpha virus–based vaccine, oncolytic viral therapy, neo-antigen modeling, anti-CD40 agonist mAb fused to monophosphoryl lipid A (MPL), CAR+ T cells targeting differentiation antigens as means to treat cancer, identifying and treating subjects at risk for checkpoint blockade therapy associated colitis, immunosuppressive follicular helper–like T cells modulated by immune checkpoint blockade, phosphatidylserine targeting agents, and uses thereof for adoptive T cell therapies, xenogeneic DNA vaccines, myeloid-derived suppressor cell assay, and GITR, PD-1, and CTLA-4 antibodies, all of which are unrelated to this study. A.M. has consulted for Janssen, PharmaEssentia, Constellation, Relay Therapeutic, Aclaris Therapeutics, and Cellarity. A.M. has received research support from Janssen, Actuate Therapeutics, and Relay Therapeutics. T.M. has consulted for Immunos Therapeutics and Pfizer; is cofounder and has equity in IMVAQ therapeutics; has received grant support from Bristol-Myers Squibb, Surface Oncology, Kyn Therapeutics, Infinity Pharmaceuticals Inc., Peregrine Pharmaceuticals Inc., Adaptive Biotechnologies, Leap Therapeutics Inc., and Aprea; and has intellectual property ownership on applications related to oncolytic viral therapy, alpha virus–based vaccine, neo-antigen modeling, CD40, GITR, OX40, PD-1, and CTLA-4. The other authors declare that they have no competing interest.
REFERENCES AND NOTES
- 1.Levine RL, Gilliland DG, Myeloproliferative disorders. Blood 112, 2190–2198 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Campbell PJ, Green AR, The myeloproliferative disorders. N. Engl. J. Med. 355, 2452–2466 (2006). [DOI] [PubMed] [Google Scholar]
- 3.Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel JP, Brunel JP, Mermel CH, Bass AJ, Pretz J, Ahn J, Hricik T, Kilpivaara O, Wadleigh M, Busque L, Gilliland DG, Golub TR, Ebert BL, Levine RL, Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood 123, e123–e133 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, Avezov E, Li J, Kollmann K, Kent DG, Aziz A, Godfrey AL, Hinton J, Martincorena I, Van Loo P, Jones AV, Guglielmelli P, Tarpey P, Harding HP, Fitzpatrick JD, Goudie CT, Ortmann CA, Loughran SJ, Raine K, Jones DR, Butler AP, Teague JW, O’Meara S, McLaren S, Bianchi M, Silber Y, Dimitropoulou D, Bloxham D, Mudie L, Maddison M, Robinson B, Keohane C, Maclean C, Hill K, Orchard K, Tauro S, Du MQ, Greaves M, Bowen BJP Huntly CN Harrison NC Cross P, Ron D, Vannucchi AM, Papaemmanuil, Campbell PJ, Green AR, Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N. Engl. J. Med. 369, 2391–2405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, Them NC, Berg T, Gisslinger B, Pietra D, Chen D, Vladimer GI, Bagienski K, Milanesi C, Casetti IC, Sant’Antonio E, Ferretti V, Elena C, Schischlik F, Cleary C, Six M, Schalling M, Schonegger A, Bock C, Malcovati L, Pascutto C, Superti-Furga G, Cazzola M, Kralovics R, Somatic mutations of calreticulin in myeloproliferative neoplasms. N. Engl. J. Med. 369, 2379–2390 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Elf S, Abdelfattah NS, Chen E, Perales-Paton J, Rosen EA, Ko A, Peisker F, Florescu N, Giannini S, Wolach O, Morgan EA, Tothova Z, Losman JA, Schneider RK, Al-Shahrour F, Mullally A, Mutant calreticulin requires both its mutant C-terminus and the thrombopoietin receptor for oncogenic transformation. Cancer Discov. 6, 368–381 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chachoua I, Pecquet C, El-Khoury M, Nivarthi H, Albu RI, Marty C, Gryshkova V, Defour JP, Vertenoeil G, Ngo A, Koay A, Raslova H, Courtoy PJ, Choong ML, Plo I, Vainchenker W, Kralovics R, Constantinescu SN, Thrombopoietin receptor activation by myeloproliferative neoplasm associated calreticulin mutants. Blood 127, 1325–1335 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Araki M, Yang Y, Masubuchi N, Hironaka Y, Takei H, Morishita S, Mizukami Y, Kan S, Shirane S, Edahiro Y, Sunami Y, Ohsaka A, Komatsu N, Activation of the thrombopoietin receptor by mutant calreticulin in CALR-mutant myeloproliferative neoplasms. Blood 127, 1307–1316 (2016). [DOI] [PubMed] [Google Scholar]
- 9.How J, Hobbs GS, Mullally A, Mutant calreticulin in myeloproliferative neoplasms. Blood 134, 2242–2248 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holmstrom MO, Riley CH, Svane IM, Hasselbalch HC, Andersen MH, The CALR exon 9 mutations are shared neoantigens in patients with CALR mutant chronic myeloproliferative neoplasms. Leukemia 30, 2413–2416 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Holmstrom MO, Martinenaite E, Ahmad SM, Met O, Friese C, Kjaer L, Riley CH, Thor Straten P, Svane IM, Hasselbalch HC, Andersen MH, The calreticulin (CALR) exon 9 mutations are promising targets for cancer immune therapy. Leukemia 32, 429–437 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Tubb VM, Schrikkema DS, Croft NP, Purcell AW, Linnemann C, Freriks MR, Chen F, Long HM, Lee SP, Bendle GM, Isolation of T cell receptors targeting recurrent neoantigens in hematological malignancies. J. Immunother. Cancer 6, 70 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holmstrom MO, Ahmad SM, Klausen U, Bendtsen SK, Martinenaite E, Riley CH, Svane IM, Kjaer L, Skov V, Ellervik C, Pallisgaard N, Hasselbalch HC, Andersen MH, High frequencies of circulating memory T cells specific for calreticulin exon 9 mutations in healthy individuals. Blood Cancer J. 9, 8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cimen Bozkus C, Roudko V, Finnigan JP, Mascarenhas J, Hoffman R, Iancu-Rubin C, Bhardwaj N, Immune checkpoint blockade enhances shared neoantigen-induced T-cell immunity directed against mutated calreticulin in myeloproliferative neoplasms. Cancer Discov. 9, 1192–1207 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schischlik F, Jager R, Rosebrock F, Hug E, Schuster MK, Holly R, Fuchs E, Milosevic Feenstra JD, Bogner E, Gisslinger B, Schalling M, Rumi E, Pietra D, Fischer GF, Fae I, Vulliard L, Menche J, Haferlach T, Meggendorfer M, Stengel A, Bock C, Cazzola M, Gisslinger H, Kralovics R, Mutational landscape of the transcriptome offers putative targets for immunotherapy of myeloproliferative neoplasms. Blood 134, 199–210 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Madden DR, The three-dimensional structure of peptide-MHC complexes. Annu. Rev. Immunol. 13, 587–622 (1995). [DOI] [PubMed] [Google Scholar]
- 17.Gragert L, Madbouly A, Freeman J, Maiers M, Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum. Immunol. 74, 1313–1320 (2013). [DOI] [PubMed] [Google Scholar]
- 18.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee HG, Allele-specific motifs revealed by sequencing of self-peptides eluted from MHC molecules. Nature 351, 290–296 (1991). [DOI] [PubMed] [Google Scholar]
- 19.Rammensee HG, Friede T, Stevanoviic S, MHC ligands and peptide motifs: First listing. Immunogenetics 41, 178–228 (1995). [DOI] [PubMed] [Google Scholar]
- 20.Boulanger DSM, Eccleston RC, Phillips A, Coveney PV, Elliott T, Dalchau N, A mechanistic model for predicting cell surface presentation of competing peptides by MHC class I molecules. Front. Immunol. 9, 1538 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ericsson PO, Orchansky PL, Carlow DA, Teh HS, Differential activation of phospholipase C-gamma 1 and mitogen-activated protein kinase in naive and antigen0primed CD4 T cells by the peptide/MHC ligand. J. Immunol. 156, 2045–2053 (1996). [PubMed] [Google Scholar]
- 22.Kimachi K, Croft M, Grey HM, The minimal number of antigen-major histocompatibility complex class II complexes required for activation of naive and primed T cells. Eur. J. Immunol. 27, 3310–3317 (1997). [DOI] [PubMed] [Google Scholar]
- 23.London CA, Lodge MP, Abbas AK, Functional responses and costimulator dependence of memory CD4+ T cells. J. Immunol. 164, 265–272 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Pihlgren M, Dubois PM, Tomkowiak M, Sjogren T, Marvel J, Resting memory CD8+ T cells are hyperreactive to antigenic challenge in vitro. J. Exp. Med. 184, 2141–2152 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rogers PR, Dubey C, Swain SL, Qualitative changes accompany memory T cell generation: Faster, more effective responses at lower doses of antigen. J. Immunol. 164, 2338–2346 (2000). [DOI] [PubMed] [Google Scholar]
- 26.Slifka MK, Whitton JL, Functional avidity maturation of CD8+ T cells without selection of higher affinity TCR. Nat. Immunol. 2, 711–717 (2001). [DOI] [PubMed] [Google Scholar]
- 27.Dyall R, Bowne WB, Weber LW, LeMaoult J, Szabo P, Moroi Y, Piskun G, Lewis JJ, Houghton AN, Nikolic-Zugic J, Heteroclitic immunization induces tumor immunity. J. Exp. Med. 188, 1553–1561 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rosenberg SA, Yang JC, Schwartzentruber DJ, Hwu P, Marincola FM, Topalian SL, Restifo NP, Dudley ME, Schwarz SL, Spiess PJ, Wunderlich JR, Parkhurst MR, Kawakami Y, Seipp CA, Einhorn JH, White DE, Immunologic and therapeutic evaluation of a synthetic peptide vaccine for the treatment of patients with metastatic melanoma. Nat. Med. 4, 321–327 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.England RD, Kullberg MC, Cornette JL, Berzofsky JA, Molecular analysis of a heteroclitic T cell response to the immunodominant epitope of sperm whale myoglobin. Implications for peptide partial agonists. J. Immunol. 155, 4295–4306 (1995). [PubMed] [Google Scholar]
- 30.Boehncke WH, Takeshita T, Pendleton CD, Houghten RA, Sadegh-Nasseri S, Racioppi L, Berzofsky JA, Germain RN, The importance of dominant negative effects of amino acid side chain substitution in peptide-MHC molecule interactions and T cell recognition. J. Immunol. 150, 331–341 (1993). [PubMed] [Google Scholar]
- 31.Gonzalez-Galarza FF, Takeshita LY, Santos EJ, Kempson F, Maia MH, da Silva AL, Teles e Silva AL, Ghattaoraya GS, Alfirevic A, Jones AR, Middleton D, Allele frequency net 2015 update: New features for HLA epitopes, KIR and disease and HLA adverse drug reaction associations. Nucleic Acids Res. 43, D784–D788 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nielsen M, Andreatta M, NetMHCpan-3.0; improved prediction of binding to MHC class I molecules integrating information from multiple receptor and peptide length datasets. Genome Med. 8, 33 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hoof I, Peters B, Sidney J, Pedersen LE, Sette A, Lund O, Buus S, Nielsen M, NetMHCpan, a method for MHC class I binding prediction beyond humans. Immunogenetics 61, 1–13 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gao B, Adhikari R, Howarth M, Nakamura K, Gold MC, Hill AB, Knee R, Michalak M, Elliott T, Assembly and antigen-presenting function of MHC class I molecules in cells lacking the ER chaperone calreticulin. Immunity 16, 99–109 (2002). [DOI] [PubMed] [Google Scholar]
- 35.Arshad N, Cresswell P, Tumor-associated calreticulin variants functionally compromise the peptide loading complex and impair its recruitment of MHC-I. J. Biol. Chem. 293, 9555–9569 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andreatta M, Nielsen M, Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics 32, 511–517 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nielsen M, Lundegaard C, Worning P, Lauemoller SL, Lamberth K, Buus S, Brunak S, Lund O, Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 12, 1007–1017 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gejman RS, Chang AY, Jones HF, DiKun K, Hakimi AA, Schietinger A, Scheinberg DA, Rejection of immunogenic tumor clones is limited by clonal fraction. eLife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn GP, Old LJ, Schreiber RD, The three Es of cancer immunoediting. Annu. Rev. Immunol. 22, 329–360 (2004). [DOI] [PubMed] [Google Scholar]
- 40.Teng MW, Galon J, Fridman WH, Smyth MJ, From mice to humans: Developments in cancer immunoediting. J. Clin. Invest. 125, 3338–3346 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanahan D, Weinberg RA, Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 42.Hoyos D, Zappasodi R, Schulze I, Sethna Z, de Andrade KC, Bajorin DF, Bandlamundi C, Callahan MK, Funt SA, Hadrup SR, Holm JS, Rosenberg JE, Shah SP, Vazquez-Garcia I, Weigelt B, Wu M, Zamarin D, Campitelli LF, Osborne EJ, Klinger M, Robins HS, Khincha PP, Savage SA, Balachandran VP, Wolchok JD, Hellmann MD, Merghoub T, Levine AJ, Luksza M, Greenbaum BD, Fundamental immune-oncogenicity trade-offs define driver mutation fitness. Nature, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Posthuma EF, Falkenburg JH, Apperley JF, Gratwohl A, Roosnek E, Hertenstein B, Schipper RF, Schreuder GM, D’Amaro J, Oudshoorn M, van Biezen JH, Hermans J, Willemze R, Niederwieser D, HLA-B8 and HLA-A3 coexpressed with HLA-B8 are associated with a reduced risk of the development of chronic myeloid leukemia. The Chronic Leukemia Working Party of the EBMT. Blood 93, 3863–3865 (1999). [PubMed] [Google Scholar]
- 44.Kuzelova K, Brodska B, Fuchs O, Dobrovolna M, Soukup P, Cetkovsky P, Altered HLA class I profile associated with type A/D nucleophosmin mutation points to possible anti-nucleophosmin immune response in acute myeloid leukemia. PLOS ONE 10, e0127637 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A, Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168, 707–723 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodig SJ, Gusenleitner D, Jackson DG, Gjini E, Giobbie-Hurder A, Jin C, Chang H, Lovitch SB, Horak C, Weber JS, Weirather JL, Wolchok JD, Postow MA, Pavlick AC, Chesney J, Hodi FS, MHC proteins confer differential sensitivity to CTLA-4 and PD-1 blockade in untreated metastatic melanoma. Sci. Transl. Med. 10, (2018). [DOI] [PubMed] [Google Scholar]
- 47.O’Donnell JS, Teng MWL, Smyth MJ, Cancer immunoediting and resistance to T cell-based immunotherapy. Nat. Rev. Clin. Oncol. 16, 151–167 (2019). [DOI] [PubMed] [Google Scholar]
- 48.Klebanoff CA, Acquavella N, Yu Z, Restifo NP, Therapeutic cancer vaccines: Are we there yet? Immunol. Rev. 239, 27–44 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrison C, Kiladjian JJ, Al-Ali HK, Gisslinger H, Waltzman R, Stalbovskaya V, McQuitty M, Hunter DS, Levy R, Knoops L, Cervantes F, Vannucchi AM, Barbui T, Barosi G, JAK inhibition with ruxolitinib versus best available therapy for myelofibrosis. Engl. J. Med. 366, 787–798 (2012). [DOI] [PubMed] [Google Scholar]
- 50.Verstovsek S, Mesa RA, Gotlib J, Levy RS, Gupta V, DiPersio JF, Catalano JV, Deininger M, Miller C, Silver RT, Talpaz M, Winton EF, Harvey JH Jr., Arcasoy MO, Hexner E, Lyons RM, Paquette R, Raza A, Vaddi K, Erickson-Viitanen S, Koumenis IL, Sun W, Sandor V, Kantarjian HM, A double-blind, placebo-controlled trial of ruxolitinib for myelofibrosis. N. Engl. J. Med. 366, 799–807 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guglielmelli P, Rotunno G, Bogani C, Mannarelli C, Giunti L, Provenzano A, Giglio S, Squires M, Stalbovskaya V, Gopalakrishna P, Vannucchi AM; COMFORT-II Investigators, Ruxolitinib is an effective treatment for CALR-positive patients with myelofibrosis. Br. J. Haematol. 173, 938–940 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Handlos Grauslund J, Holmstrom MO, Jorgensen NG, Klausen U, Weis-Banke SE, El Fassi D, Schollkopf C, Clausen MB, Gjerdrum LMR, Breinholt MF, Kjeldsen JW, Hansen M, Koschmieder S, Chatain N, Novotny GW, Petersen J, Kjaer L, Skov V, Met O, Svane IM, Hasselbalch HC, Andersen MH, Therapeutic cancer vaccination with a peptide derived from the calreticulin exon 9 mutations induces strong cellular immune responses in patients with CALR-mutant chronic myeloproliferative neoplasms. Front. Oncol. 11, 637420 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Knudsen TA, Hansen DL, Ocias LF, Bjerrum OW, Brabrand M, El Fassi D, Frederiksen M, Kjær L, Kristensen TK, Kruse TA, Mourits-Andersen T, Møller P, Overgaard UM, Severinsen MT, Skov V, Stentoft J, Starklint J, de Stricker K, Thomassen M, Larsen TS, Hasselbalch HC, Long-term efficacy and safety of recombinant interferon alpha-2 vs. hydroxyurea in polycythemia vera: Preliminary results from the three-year analysis of the daliah trial - A randomized controlled phase III clinical trial. Blood 132, 580 (2018). [Google Scholar]
- 54.Jan M, Leventhal MJ, Morgan EA, Wengrod JC, Nag A, Drinan SD, Wollison BM, Ducar MD, Thorner AR, Leppanen S, Baronas J, Stevens J, Lane WJ, Kekre N, Ho VT, Koreth J, Cutler CS, Nikiforow S, Alyea III EP, Antin JH, Soiffer RJ, Ritz J, Lindsley RC, Ebert BL, Recurrent genetic HLA loss in AML relapsed after matched unrelated allogeneic hematopoietic cell transplantation. Blood Adv. 3, 2199–2204 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jensen KK, Andreatta M, Marcatili P, Buus S, Greenbaum JA, Yan Z, Sette A, Peters B, Nielsen M, Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 154, 394–406 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Houghton CS, Engelhorn ME, Liu C, Song D, Gregor P, Livingston PO, Orlandi F, Wolchok JD, McCracken J, Houghton AN, Guevara-Patino JA, Immunological validation of the EpitOptimizer program for streamlined design of heteroclitic epitopes. Vaccine 25, 5330–5342 (2007). [DOI] [PubMed] [Google Scholar]
- 57.Guevara-Patino JA, Engelhorn ME, Turk MJ, Liu C, Duan F, Rizzuto G, Cohen AD, Merghoub T, Wolchok JD, Houghton AN, Optimization of a self antigen for presentation of multiple epitopes in cancer immunity. J. Clin. Invest. 116, 1382–1390 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hosken NA, Bevan MJ, An endogenous antigenic peptide bypasses the class I antigen presentation defect in RMA-S. J. Exp. Med. 175, 719–729 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salter RD, Howell DN, Cresswell P, Genes regulating HLA class I antigen expression in T-B lymphoblast hybrids. Immunogenetics 21, 235–246 (1985). [DOI] [PubMed] [Google Scholar]
- 60.Weber LW, Bowne WB, Wolchok JD, Srinivasan R, Qin J, Moroi Y, Clynes R, Song P, Lewis JJ, Houghton AN, Tumor immunity and autoimmunity induced by immunization with homologous DNA. J. Clin. Invest. 102, 1258–1264 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Elf S, Abdelfattah NS, Baral AJ, Beeson D, Rivera JF, Ko A, Florescu N, Birrane G, Chen E, Mullally A, Defining the requirements for the pathogenic interaction between mutant calreticulin and MPL in MPN. Blood 131, 782–786 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wolchok JD, Gregor PD, Nordquist LT, Slovin SF, Scher HI, DNA vaccines: An active immunization strategy for prostate cancer. Semin. Oncol. 30, 659–666 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.