Abstract
A number of recent studies have demonstrated the significance of detergent-insoluble, glycolipid-enriched membrane domains or lipid rafts, especially in regard to activation and signaling in T lymphocytes. These domains can be viewed as floating rafts composed of sphingolipids and cholesterol which sequester glycosylphosphatidylinositol (GPI)-linked proteins, such as Thy-1 and CD59. CD45, a 200-kDa transmembrane phosphatase protein, is excluded from these domains. We have found that human immunodeficiency virus type 1 (HIV-1) particles produced by infected T-cell lines acquire the GPI-linked proteins Thy-1 and CD59, as well as the ganglioside GM1, which is known to partition preferentially into lipid rafts. In contrast, despite its high expression on the cell surface, CD45 was poorly incorporated into virus particles. Confocal fluorescence microscopy revealed that HIV-1 proteins colocalized with Thy-1, CD59, GM1, and a lipid raft-specific fluorescent lipid, DiIC16(3), in uropods of infected Jurkat cells. CD45 did not colocalize with HIV-1 proteins and was excluded from uropods. Dot immunoassay of Triton X-100-extracted membrane fractions revealed that HIV-1 p17 matrix protein and gp41 were present in the detergent-resistant fractions and that [3H]myristic acid-labeled HIV Gag showed a nine-to-one enrichment in lipid rafts. We propose a model for the budding of HIV virions through lipid rafts whereby host cell cholesterol, sphingolipids, and GPI-linked proteins within these domains are incorporated into the viral envelope, perhaps as a result of preferential sorting of HIV Gag to lipid rafts.
Glycolipid-enriched membrane (GEM) domains are organized areas on the cell surface enriched in cholesterol, sphingolipids, and glycosylphosphatidylinositol (GPI)-linked proteins. A recent review has described them as “rafts” that serve as moving platforms on the cell surface (44). These domains exist in a more ordered state, conferring resistance to Triton X-100 detergent treatment at 4°C (42). Many proteins have been shown to be associated with lipid rafts, including GPI-linked proteins, Src family kinases, protein kinase C, actin and actin-binding proteins, heterotrimeric and small G proteins, and caveolin (2, 5, 17, 34, 39, 45, 47). Saturated acyl chains of the GPI anchor have been shown to be a determinant for the association of GPI-linked proteins with lipid rafts (35, 41). Lipid rafts have been shown to exclude certain transmembrane molecules, specifically the membrane phosphatase CD45 (2, 36). Exclusion of CD45 results in the accumulation of phosphorylated signaling molecules in lipid rafts (36), and T-cell activation has recently been shown to require clustering of signaling molecules in these membrane domains (reviewed in reference 21).
Previous studies in our laboratory indicated that human immunodeficiency virus type 1 (HIV-1) excludes CD45 from its membrane, despite its abundance on the cell surface. This was in contrast to other membrane proteins, some expressed at lower levels than CD45, that were efficiently incorporated by the virus (29). CD45 is a large, heavily glycosylated, multiply spliced transmembrane protein that has two cytoplasmic tyrosine phosphatase domains. Extracellularly, it may extend as much as 40 nm from the cell surface, while intracellularly it has a large cytoplasmic tail of 707 amino acids. CD45 is one of the most highly expressed leukocyte surface proteins. Indeed, as much as 10 to 25% of the lymphocyte cell surface may be covered with CD45 (4). If HIV-1 incorporated host proteins randomly, a significant number of CD45 molecules should be present on the virus given its abundance on the cell surface. Our previous data demonstrated that this is not the case, and suggest that CD45 may be specifically excluded from the viral envelope. We therefore examined the possibility that CD45 is excluded from HIV-1 particles as a result of virus budding from lipid rafts which also exclude this protein.
We report here that HIV-1 incorporates the lipid raft-specific ganglioside, GM1, as well as GPI-linked proteins Thy-1 and CD59, and confirm our earlier report that CD45 is excluded from the virus. Confocal fluorescence microscopy showed that viral proteins colocalize with Thy-1, CD59, GM1, and 1 , 1′ - dihexadecyl - 3 , 3 , 3′ , 3′ - tetramethylindocarbocyanine [DiIC16(3)], a fluorescent dye that partitions to ordered domains, in uropods on infected cells. In contrast, CD45 is excluded from these GPI-linked protein-rich membrane projections. Upon membrane fractionation, HIV matrix (MA) and gp41, the transmembrane subunit of envelope (Env), are present in detergent-resistant, GPI-linked protein-rich fractions, confirming their association with lipid rafts. Specifically, myristylated Gag localizes predominantly to the detergent resistant lipid rafts. We propose that HIV-1 budding occurs through lipid rafts, thereby accounting for the cholesterol-rich, sphingolipid-rich virus membrane, which bears GPI-linked proteins such as Thy-1 and CD59 but lacks CD45.
MATERIALS AND METHODS
Cells and antibodies.
Jurkat cells were obtained from the American Type Culture Collection (Rockville, Md.) and maintained in complete medium, cRPMI, consisting of RPMI 1640 (Gibco BRL/Life Technologies, Gaithersburg, Md.) containing 10% fetal calf serum (HyClone, Logan, Utah) and 10 mM HEPES. Monoclonal antibodies (MAbs) to Thy-1 (5E10) and CD59 (p282/H19) were obtained from Pharmingen (San Diego, Calif.). Mouse MAb against HIV p17 was obtained from Advanced Biotechnologies, Inc. (Columbia, Md.). Goat anti-cholera toxin B (CTB) MAb was purchased from Calbiochem (La Jolla, Calif.). Rabbit anti-GM1 polyclonal antibody was purchased from Metraya, Inc. (Pleasant Gap, Pa.). T32 MAb specific for gp41 was kindly provided by Robert Siliciano, Johns Hopkins University School of Medicine. Biotinylated human anti-HIV polyclonal antibodies were produced from pooled human HIV+ sera. Soluble recombinant CD4-immunoglobulin Fc chimera (CD4Ig) was a generous gift from Tim Gregory (Genentech, South San Francisco, Calif.). Control mouse myeloma immunoglobulin G1 (IgG1) and rabbit anti-mouse IgG (Fc specific) were purchased from Jackson Immunoresearch (West Grove, Pa.). Fluorescein isothiocyanate (FITC)-conjugated sheep anti-human IgG was purchased from Cappel Research Products (Durham, N.C.). MAbs to major histocompatibility complex I (MHCI) antigen (MHM.5), HIV-1 Gag (Gag.M1), and CD45 (H5A5) were produced in our laboratory and were purified from ascites fluids (9, 15).
Virus production.
HIV-1RF (obtained from R. Gallo through the NIH AIDS Research and Reference Reagent Program) was used to chronically infect Jurkat cells. Viruses used for the capture assay were produced by washing 1 × 106 to 2 × 106 chronically infected cells with phosphate-buffered saline (PBS), resuspending cells in complete medium, and culturing for 1 to 3 days before collecting culture supernatants. Virus production was measured by p24 enzyme-linked immunosorbent assay (ELISA) after detergent lysis of supernatant.
Flow cytometry.
Flow cytometry was performed as previously described (29). Briefly, 2 × 105 cells in 100 μl of PBS containing 5% normal goat serum (NGS) were added to 100 μl of MAb (1 to 5 μg) and incubated for 30 min on ice. Cells were washed with PBS, resuspended in 100 μl of PBS plus 5% NGS containing 2 μg of FITC-goat anti-mouse IgG (FITC-GAM), and incubated 1 h on ice. Cells were then washed with PBS and fixed with 2% paraformaldehyde, followed by analysis on an EPICS Profile II (Coulter, Hialeah, Fla.) flow cytometer.
Virus phenotyping.
Virus phenotyping was carried out as previously described with some minor differences (29). Briefly, Costar ELISA plates (Costar, Cambridge, Mass.) were coated for 4 h at 37°C with 1.5 μg of rabbit anti-mouse IgG (Fc fragment specific) per well in 50 mM Tris (pH 9.5). The wells were then blocked with 3% bovine serum albumin (BSA) in PBS for 2 h at 37°C before adding 1 to 2 μg of the MAbs. The plates were then incubated overnight at room temperature before washing them six times with PBS–0.05% Tween 20. Viral supernatants were collected and clarified through 0.45-μm (pore-size) filters. The viral supernatants at 466 ng/ml of p24 were then added to the antibody-coated wells and incubated at 37°C for 1.5 h before washing them six times with RPMI. The bound viruses were then lysed with 1% Triton X-100 in cRPMI for 1 h at 37°C. Detergent-solubilized viral proteins were then transferred to a second plate to measure released p24 in a standard p24 ELISA.
Cell capture assay.
Costar ELISA plates were coated overnight at room temperature with 1.0 μg of GAM IgG (Fc specific) per well in 50 mM Tris (pH 9.5). Wells were blocked with 3% BSA in PBS for 1 h at 37°C before adding 1 to 2 μg of the MAbs. Plates were then incubated for 2 h at 37°C before washing them three times with RPMI. Wells were blocked again with 5% NGS in PBS for 1 h at 37°C before washing them three times with RPMI. Jurkat cells (107) were labeled with horseradish peroxidase (HRP; Sigma) at 1 mg/ml in cRPMI for 30 min at 37°C, washed once with cRPMI, and then resuspended in cRPMI to make 2.5 × 106 cells/ml. Cells (100 μl) were added to the wells and allowed to settle for 2 h at 37°C. Wells were washed three times with Hanks balanced salt solution (Gibco BRL) and then treated with lysis-substrate buffer (1% Triton X-100, 0.015% H2O2, 0.24 mg of tetramethyl benzidine per ml, 0.2 M sodium acetate-citric acid; pH 4.0) for 20 min before the addition of 0.5 M H2SO4 to stop the reaction. Absorbances at a 450-nm wavelength were determined on a plate reader, and cell number values were extrapolated from a linear curve.
β-Cyclodextrin treatment and virus precipitation.
Infected Jurkat cells (3 × 106) were treated with 20 mM hydroxypropyl-β-cyclodextrin (βCD; Cyclodextrin Technologies Development, Inc., Gainesville, Fla.) in 3 ml of cRPMI or with cRPMI alone for 1 h at 37°C. Cells were washed with PBS and then allowed to produce virus in 3 ml of cRPMI at 37°C for 2 h. Viral supernatants were clarified through a 0.45-μm filter, and 100 μl was added to 100 μl of MAb (10 μg/ml) in 5% NGS-PBS, and the mixture was incubated for 1 h on ice. Pansorbin (SaC) (50 μg; Calbiochem, San Diego, Calif.) was then added to the solution and incubated for 20 min on ice. Complexes were washed sequentially with 10× and 1× PBS. Precipitated virus was then lysed with 400 μl of 1% Triton X-100 in cRPMI. Lysates were diluted, and p24 was quantitated by standard p24 ELISA.
Cholera toxin capture of HIV-1.
HIV-1RF viral supernatant from an infected Jurkat cell line was collected and clarified through a 0.45-μm filter. Virus supernatant (100 μl) was added to 100 μl of CTB (Calbiochem, La Jolla, Calif.) dilutions (0 to 20 μg/ml) in cRPMI. The mixtures were incubated for 1 h at 37°C before adding 50 μl of goat anti-CTB at 10 μg/ml in 5% NGS-PBS. The mixture was then incubated for 1 h on ice before adding 50 μl of SaC, mixed well, and then incubated on ice for another 1 h with intermittent mixing. The SaC was then washed twice with PBS. The SaC-precipitated virus was then lysed with 400 μl of 1% Triton X-100 in cRPMI at room temperature for 30 min. Released p24 was measured with a standard p24 ELISA after pelleting the SaC.
Immunomicroscopy.
Cell surface staining of chronically infected cells and uninfected cells was performed under saturating conditions. Jurkat cells (3 × 105) were washed in cold PBS and preincubated on ice for 15 min in 5% NGS-PBS. Uninfected cells were incubated with 1 to 5 μg of MAb in 5% NGS-PBS for 30 min on ice, washed with PBS, and then incubated with 2 μg of Texas red-conjugated GAM IgG. Infected cells were incubated with biotinylated human anti-HIV polyclonal antibody (10 μg/ml in 5% NGS-PBS) for 30 min on ice and washed with PBS before incubating them with 2 μg of Texas red-streptavidin conjugate. Both cell types were then incubated with the second primary MAb at 1 to 5 μg in 5% NGS-PBS 30 min on ice, washed with PBS, and then incubated with 2 μg of FITC-GAM in 5% NGS-PBS. The cells were then fixed with 2% paraformaldehyde in PBS and cytospun onto poly-l-lysine-coated slides by using Cyto Funnels (Shandon, Pittsburgh, Pa.). The pellets were overlaid with 50 μl of 25% glycerol in PBS, and a coverslip was positioned over the droplet. The edges of the slides were sealed with nail polish before storing them at 4°C. This staining procedure was also performed with cells prefixed with 2% paraformaldehyde in PBS prior to MAb staining. Viewing of slides was performed with an Olympus IX50 confocal microscope under oil immersion at a ×100 magnification. Micrographs were analyzed on a Silicon Graphics Workstation with Intervision software. Final images were enhanced on the Silicon Graphics Workstation by two-dimensional deconvolution, and brightness and contrast were adjusted for viewing.
Cell lysis and equilibrium centrifugation.
Protein extraction and equilibrium centrifugation were performed as previously described with slight modifications (18). Briefly, 2 × 107 cells were washed twice in PBS and once in TKM buffer (50 mM Tris-HCl, pH 7.4; 25 mM KCl; 5 mM MgCl2; 1 mM EDTA). Cells were extracted on ice for 30 min in 500 μl of lysis buffer (TKM, 1% Triton X-100, 2 μg of aprotinin per ml). Lysates were centrifuged at 8,000 × g for 10 min at 4°C, and the supernatants were stored at −20°C. For equilibrium centrifugation, extracts were adjusted to 40% sucrose in TKM and loaded into SW41 tubes. The extracts were then overlaid with 6 ml of 38% sucrose-TKM, followed by 4.5 ml of 5% sucrose-TKM. Tubes were centrifuged at 100,000 × g for 18 h at 4°C. Eleven 1-ml fractions were collected from the bottom of the tube and stored at −20°C.
Dot immunoassay.
Dot immunoassays were performed as described previously with minor modifications (18). Briefly, 100-μl portions of each fraction diluted 1 to 10 in PBS (2 × 105 cell equivalents) were added to wells of a Bio-Dot apparatus (Bio-Rad, Hercules, Calif.), gently suctioned onto nitrocellulose membranes, and allowed to air dry. These membranes were cut into strips and stored at −20°C in plastic bags. Before blotting, strips were blocked with 5% nonfat milk powder in TBST (10 mM Tris-HCl, pH 7.5; 100 mM NaCl; 0.1% Tween 20) for 1 h at room temperature. Strips were then incubated with primary antibodies in TBST–0.5% milk powder for 1 h and washed 10 min three times with TBST, followed by incubation with HRP-conjugated GAM for 45 min. The strips were then washed five times and developed with an enhanced chemiluminescence (ECL) assay (Amersham Life Science, Arlington Heights, Ill.) before exposure to Hyper-Film ECL.
Dialkylindocarbocyanine labeling.
Three million Jurkat HIV-1RF-infected cells were washed with PBS and resuspended in 1 ml of cRPMI. DiIC16(3) or DiIC12(3) (1,1′-didodecyl-3,3,3′,3′-tetramethylindocarbocyanine) (both from Molecular Probes, Eugene, Oreg.) in 0.1 mg of ethanol per ml was added to make a final concentration of 1 to 10 μM. Cells were incubated on ice for 15 min to allow the incorporation of dyes. The cells were washed with PBS and fixed with 2% paraformaldehyde in PBS before further MAb labeling for confocal microscopy as described above.
[3H]myristic acid labeling and immunoprecipitation.
HIV-1-infected Jurkat cells (2 × 107) were labeled in 2 ml of cRPMI containing 1 mCi of [9,10(n)-3H]myristic acid (40 to 60 Ci/mmol; Amersham Pharmacia Biotech, Piscataway, N.J.) for 4 h at room temperature. Labeled cells were lysed and subjected to sucrose gradient equilibrium centrifugation as described above. GEM domain (lipid raft) fractions 3, 4, and 5 were pooled as were soluble fractions 8, 9, and 10. Samples were precleared by incubation with 20 μl of normal human serum for 1 h at 4°C before adding 100 μl of SaC and incubating them an additional 30 min. The preimmune complexes were removed, and samples were then incubated with excess IgG1 myeloma or Gag.M1 MAb for 1.5 h at 4°C, followed by the addition of 2 μg of RAM (Fc specific). After 1 h 50 μl of SaC was added, followed by incubation for 30 min. Immune complexes were washed twice with PBS and resuspended in 200 μl of PBS. Samples were then boiled, and the supernatant was blotted onto a nitrocellulose membrane with a Bio-Dot apparatus. The membrane was treated with En3Hance Spray (DuPont, Wilmington, Del.) and then exposed to Hyperfilm-MP (Amersham) for 5 days. Dots were quantitated by densitometry analysis by MacBAS software version 2.5, and the percent distribution in GEM domains was determined by using the following formula: (GagGEM − IgGGEM)/[(GagGEM − IgGGEM) + (GagSol − IgGSol)].
RESULTS
Microfluorimetry of infected Jurkat cells shows high expression of CD45 and low expression of Thy-1 and CD59.
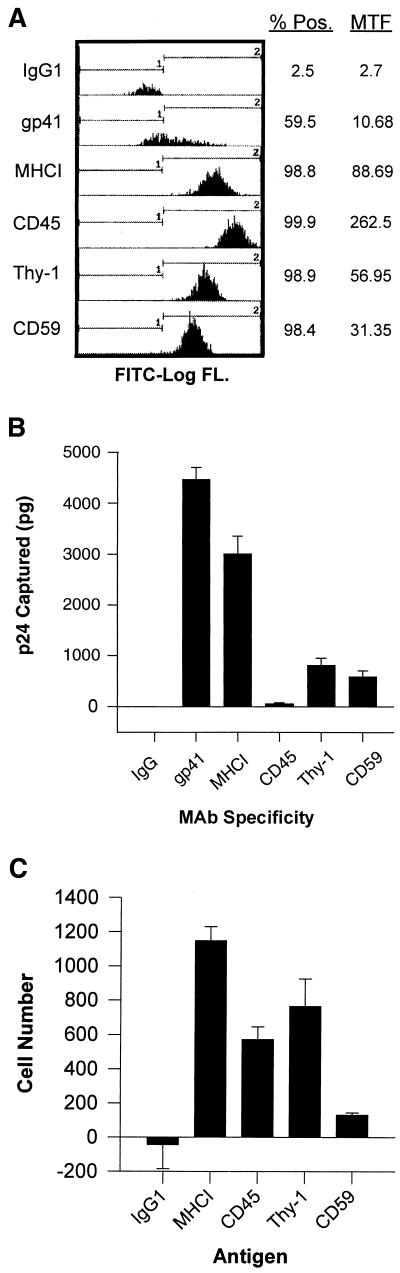
Flow cytometry under saturating conditions was used to determine the expression of CD45, Thy-1, and CD59 on the surface of infected Jurkat cells. The results are shown in Fig. 1A, which displays the percent positive cells and the mean total fluorescence (brightness of staining) obtained with the antibodies indicated. As observed in previous studies, CD45 is highly expressed on Jurkat cells. An antibody against MHCI was used as a positive control, while mouse myeloma immunoglobulin (IgG1) was used as a negative control. As seen in the figure, the expression of Thy-1 and CD59 is significantly lower than that of CD45 and MHCI. These data correlate with previous surface expression analyses (29) and was corroborated by conventional immunofluorescence staining (data not shown).
FIG. 1.
Flow cytometry and virus capture. (A) Jurkat cells were fixed with 2% paraformaldehyde and stained with MAbs specific for gp41, MHCI, CD45, Thy-1, and CD59. Antibodies were then detected with FITC-conjugated GAM IgG. FITC log fluorescence intensity is plotted on the x axis, and the number of counts is plotted on the y axis. % Pos, percentage of cells with >7.15 fluorescence intensity; MTF, mean total fluorescence of all cells in the sample. (B) Virus capture immunoassays to determine relative viral phenotypes were performed with MAbs specific for gp41, MHCI, CD45, Thy-1, and CD59 as described in Materials and Methods. (C) Jurkat cells labeled with soluble HRP were captured with the indicated MAbs in an assay similar to the virus capture as described in the Materials and Methods. Error bars represent the standard deviation in panels B and C.
HIV incorporates the GPI-linked proteins, Thy-1 and CD59, and ganglioside GM1.
We used our previously reported virus phenotyping assay to determine the relative host protein phenotype of HIV-1 particles (29). In this assay HIV-1 particles are captured by MAbs through host proteins present on the surface of the particles; Fig. 1B shows the relative p24 captured with the MAbs indicated. This experiment was performed three times with similar results. MAbs to gp41 and MHCI capture virus efficiently, as expected from previous results (3, 29). Thy-1 and CD59 support efficient viral capture despite low expression on the host cell surface. As seen in a previous study (29) very little HIV-1 was captured through CD45 despite very high expression on the cell surface. The failure of this anti-CD45 MAb (H5A5) to capture HIV-1 is not due to low MAb affinity or failure to bind to the capture plate (29). We found that H5A5 MAb was capable of capturing HRP-labeled Jurkat cells in a similar assay as efficiently as MAbs against other membrane proteins (Fig. 1C). Thus, the failure of anti-CD45 MAb to capture virus is not due to a failure of the MAb to work in capture assays. These data indicate a significant preference for HIV-1 incorporation of GPI-linked proteins compared to CD45. The high expression of CD45 on the cell surface and its low incorporation into virus particles is consistent with exclusion of this molecule from budding particles.
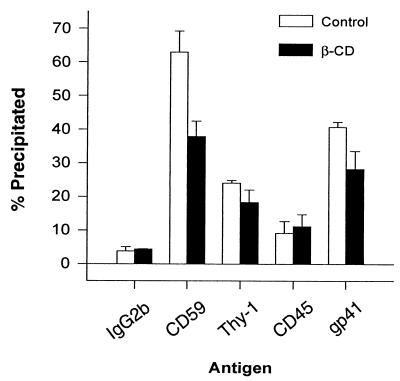
To corroborate the MAb plate capture assay results, we also performed HIV-1 immunoprecipitations with MAbs as described in Materials and Methods. This assay allows for the potential interaction of all the virions in solution with the MAbs in contrast to the plate virus capture assay in which only a small fraction of the particles make contact with the MAbs. As seen in Fig. 2, the anti-gp41 MAb, T32, which was used as a positive control for intact virions, can precipitate up to 60% of the p24 in the supernatant, depending on the virus preparation. In this assay, the anti-CD59 MAb precipitates as much p24 as anti-gp41 MAb T32. Even in this assay anti-CD45 MAb captures very little virus. We also examined the effects of βCD, a cellular cholesterol efflux inducing molecule, on the incorporation of host molecules into virions. By removing cholesterol, βCD is believed to partially perturb organized lipid rafts resulting in dispersal of their components (19). As seen in Fig. 2, the capture of HIV-1 by MAbs against CD59 and gp41 decreased substantially (P < 0.05) after treating cells with βCD, as measured by the percentage of total p24. The decrease in Thy-1 was not statistically significant (P = 0.08). CD45 capture remained mostly unaffected. The effects on virus precipitation through gp41 imply that intact lipid rafts are required for efficient gp41 incorporation into virions since the overall cellular release of p24 actually increased after βCD treatment (data not shown).
FIG. 2.
βCD effects on GPI-linked protein incorporation. Jurkat cells were treated with or without 20 mM βCD in cRPMI for 1 h at 37°C. Virions produced for 2 h posttreatment were precipitated with MAbs to CD59, Thy-1, CD45, and gp41. IgG2b was used as a nonspecific control. The captured p24 was measured and expressed as a percentage of the total p24 in the supernatant. Error bars represent the standard deviation.
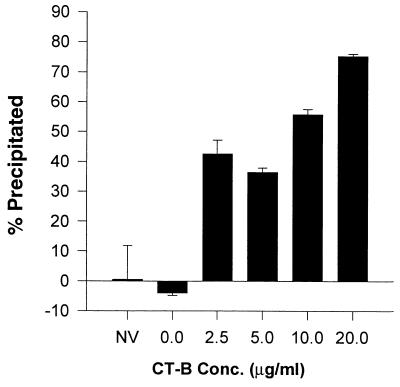
We then assayed the relative incorporation of GM1, a ganglioside marker specific for lipid rafts (18, 30). Using a soluble CTB binding assay described in Materials and Methods, we determined that as much as 75% of HIV-1 could be precipitated by using goat anti-CTB and SaC after treating the virus with GM1-specific CTB (Fig. 3). The CTB binding to virus was specific and dose dependent, and no virus was precipitated in the absence of CTB as measured by p24 ELISA. These data show that the majority of HIV-1 particles have incorporated the lipid raft-specific marker GM1.
FIG. 3.
CTB captures GM1 on HIV-1. CTB in cRPMI was added to viral supernatants to the indicated concentrations. Goat anti-CTB and SaC were added to precipitate viral CTB complexes. The captured virus was lysed and p24 was measured and expressed as a percentage of the total p24 from the original supernatant. NV, no virus control. Error bars represent the standard deviation.
Thy-1, CD59, and GM1 colocalize with HIV-1 proteins on infected cell uropods which exclude CD45.
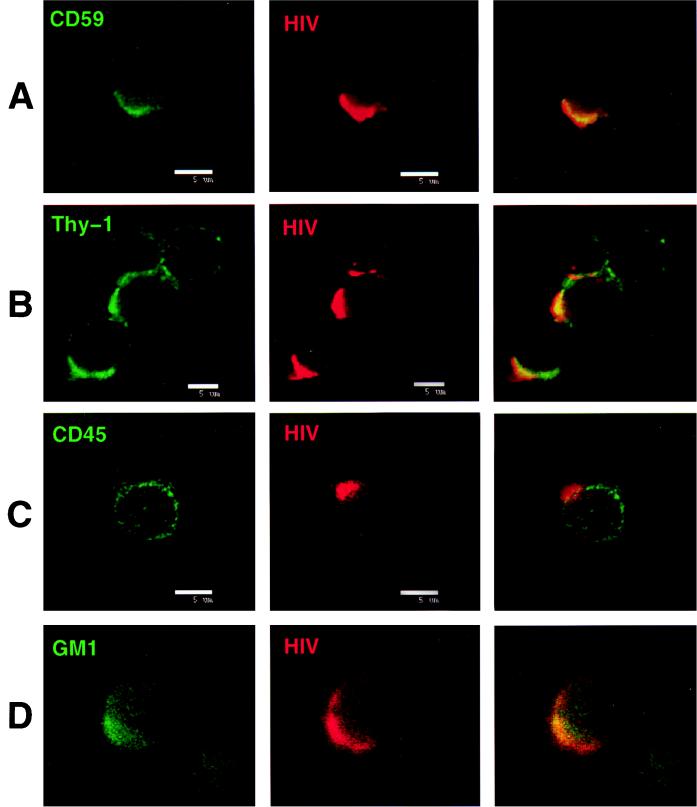
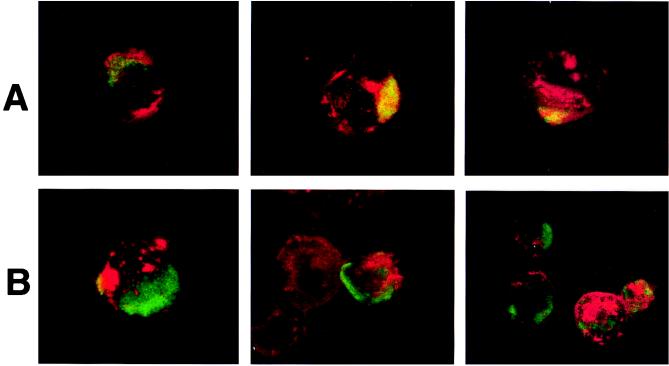
To determine the distribution of HIV-1 proteins relative to GPI-linked proteins that serve as lipid raft markers, infected cells were subjected to immunofluorescence staining followed by confocal microscopy. Expression of HIV-1 proteins was localized to uropods projecting from one end of the cell. This capping pattern was seen on most cells in the infected cell culture. Uropods protruding from HIV-1-infected cells have been previously described for adherent T cells (11, 32). Thy-1 and CD59 both colocalized with cell surface HIV-1 proteins, as shown by the superimposed green (Thy-1 or CD59) and red (HIV-1 proteins) fluorescence, as indicated by the yellow color in the third panels of Fig. 4A and B. Cells prefixed with 2% paraformaldehyde before staining showed a similar appearance, indicating that the colocalization was not due to antibody cross-linking of viral and GPI-linked proteins. Since the cells were not permeabilized before staining, the HIV proteins seen in these studies are most likely gp41 and gp120. This was confirmed in studies with anti-gp41 MAb T32 in the colocalization studies (data not shown). Uninfected cells showed no capping of Thy-1 or CD59 (data not shown). CD45 did not localize to areas of HIV-1 protein expression and was excluded from uropods (Fig. 4C). The distribution of CD45 was unaffected by HIV-1 infection, and the molecule remained evenly dispersed in patches all over the cell surface. These data are entirely consistent with the results obtained in the virus phenotyping studies.
FIG. 4.
Colocalization of Thy-1, CD59, and GM1 into areas of HIV protein expression and the exclusion of CD45 from the site of HIV surface protein localization. Infected Jurkat cells were labeled with a primary antibody to a host antigen, followed by an appropriate FITC-conjugated secondary (GAM or goat anti-rabbit) IgG, and then labeled with biotinylated anti-HIV human polyclonal antibody and detected with streptavidin-Texas red. Superimposed FITC and Texas red images are viewed in column 3 of all panels. Colocalization is indicated by the yellow color. Each panel was labeled with the following antibodies: A, anti-CD59 MAb p282(H19); B, anti-Thy-1 MAb 5E10; C, anti-CD45 MAb H5A5; and D, rabbit anti-GM1 polyclonal antibody. The figure was prepared on a Silicon Graphics Workstation with Showcase software.
We wanted to determine if GM1 colocalized on the cell surface with HIV-1 proteins in support of our finding that GM1 was present on virions. We found that GM1 staining was relatively faint with rabbit anti-GM1 antibody, but confocal microscopy did show colocalization of this molecule with HIV-1 labeled cells (Fig. 4D).
Lipid raft-partitioning lipid analog, DiIC16(3), colocalizes with HIV-1 proteins on uropods of infected cells.
In order to evaluate the localization of lipids in lipid rafts, we used two forms of dialkylindocarbocyanine, a fluorescent lipid analog. DiIC16(3) partitions preferentially to lipid-ordered domains due to its two 16-carbon saturated fatty acid chains, while DiIC12(3) with its two 12-carbon saturated fatty acid chains partitions to fluid domains (25, 46). Infected Jurkat cells were labeled with the dyes for 15 min on ice, washed with PBS, and fixed with 2% paraformaldehyde in PBS. Cells were then stained with soluble CD4Ig or human anti-HIV polyclonal antibody and FITC-labeled sheep anti-human IgG to detect surface gp120/41. Confocal microscopy showed that cells labeled with DiIC16(3), indicated in red, extensively colocalized with HIV-1 proteins (Fig. 5A; the overlap of FITC and dialkylindocarbocyanine is indicated by a yellow staining). In contrast, DiIC12(3) did not preferentially label uropods and was specifically excluded from areas with HIV-1 staining (Fig. 5B). As expected, CD45 staining was also excluded from uropods that stained positive for DiIC16(3) and HIV-1 (data not shown).
FIG. 5.
DiIC16(3) colocalizes with HIV, while DiIC12(3) does not. Infected Jurkat cells were incubated with DiIC16(3) (A) or DiIC12(3) (B) for 15 min on ice before being fixed in 2% paraformaldehyde-PBS. Cells were then labeled with CD4Ig or anti-HIV IgG and FITC-labeled sheep anti-human IgG. Dialkylindocarbocyanine lipids are shown in red, FITC staining is shown in green, and the overlap is indicated in yellow. All of the micrographs in this figure were labeled with CD4Ig, except the last micrograph in panel A, which was labeled with anti-HIV IgG.
HIV proteins are detected in isolated lipid raft fractions.
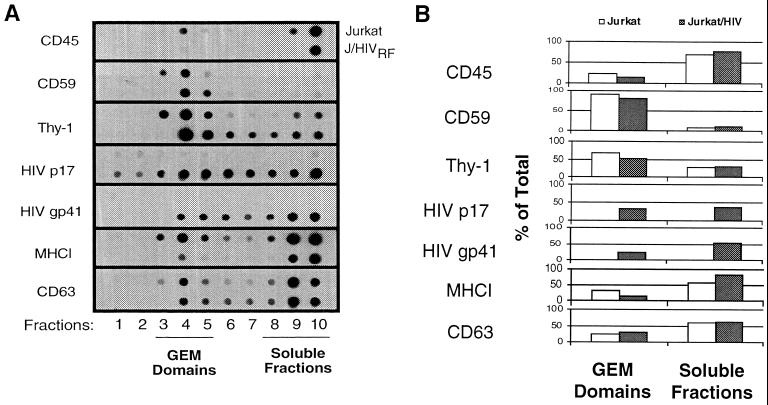
Lipid rafts were purified by cell lysis and equilibrium centrifugation as previously published (18) in order to confirm the presence of HIV-1 proteins in these membrane structures. The fractions were then assayed for the presence of viral and host proteins by immunoblot analysis. The separation of detergent-resistant lipid rafts was confirmed by the abundance of Thy-1 and CD59 in fractions 3 through 5, while CD45 was present only in the bottom fractions 9 and 10, as previously described (18, 48) (Fig. 6A). Immunoblot detection of membrane fractions revealed that the HIV MA protein, p17, and gp41 are both present in the detergent-insoluble lipid rafts of infected cells (Fig. 6A). The distribution between GEM domains and soluble fractions was quantitated by MacBAS software version 2.5 (Fig. 6B). Since lysates were prepared from whole cells, the anti-MA MAb could bind not only the membrane associated MA protein but also non-membrane-associated forms of the group-specific antigen precursor protein (Gag) and MA. This may account for the abundance of p17 detected in the soluble fractions of the blot. We have identified a possible mechanism for the targeting of the HIV Env protein, gp41, to lipid rafts involving palmitylation of two cysteines in its cytoplasmic tail (50). Dual acylation of host proteins involving palmitate and myristate have been shown to target proteins to lipid rafts (34). As expected, a substantial portion of the transmembrane subunit of Env, gp41, is also present in the GEM fractions (Fig. 6A). Our preliminary data indicate that palmitylated gp41 partitions specifically to lipid rafts (data not shown). Since palmitylation is a reversible posttranslational modification, it is not likely that all of the gp41 present in the cell would be palmitylated at any given time (26). This could account for the large proportion present in soluble domains. We have also found that other transmembrane proteins, such as MHCI and CD63, previously shown to be incorporated into HIV-1 (29), are detected in lipid rafts as well, although the majority of both molecules are in the solubilized fractions (Fig. 6).
FIG. 6.
HIV matrix, p17, and gp41 are colocalized with GEM domains upon equilibrium centrifugation of Triton X-100-treated Jurkat cells. Jurkat cells were treated with 1% Triton X-100 at 4°C as described in Materials and Methods. The lysates were equilibrium centrifuged on a discontinuous sucrose gradient, and 1-ml fractions were collected from the bottom and labeled 1 through 10. (A) Fraction 10 is the bottom fraction, while fraction 1 corresponds to the top of the tube. GEM domains are indicated as fractions 3 through 5 as previously identified (36). For immunoblots, each MAb was incubated with lysates from uninfected Jurkat cells (top row) and infected Jurkat cells (bottom row). The figure was prepared from scanned blots in Adobe Photoshop on a Power Macintosh 7600. (B) Blots were quantitated as described in Materials and Methods, and the amounts in the GEM domains or soluble fractions were determined as a percentage of the total of all the dots.
Myristylated HIV-1 Gag partitions to GEM domains.
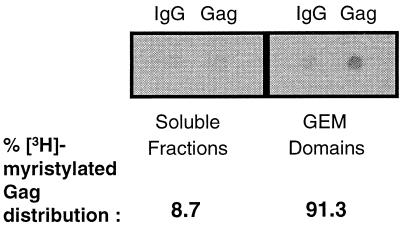
The myristylation of Gag is necessary for membrane association, proteolytic processing, and virus budding (12, 22). So it can be inferred that with a mixed population of myristylated and nonmyristylated Gag within a cell, only the myristylated forms will be responsible for membrane association and potentially determining the site of virus budding. To ensure that we analyzed cellular Gag and not virus-associated Gag, we used a MAb specific for p24 and p55. To determine which areas of the membrane myristylated Gag would bind, we labeled cells with [3H]myristic acid and isolated lipid rafts as described in Materials and Methods. Lipid raft fractions and soluble fractions were pooled separately and immunoprecipitated with an anti-Gag MAb. Blotting of the precipitated proteins showed that myristylated Gag protein is present predominantly in the lipid raft fractions (Fig. 7). The blots were quantitated by densitometry, and the IgG backgrounds were subtracted from each to determine the distribution of the myristic acid label expressed as a percentage of the total. The results showed that more than 90% of the cellular myristylated Gag is in lipid rafts. This result is consistent with the observation that myristylation targets a number of host proteins, such as Src, Lck, Lyn, and HCK protein tyrosine kinases, to lipid rafts as well (2, 34, 35).
FIG. 7.
[3H]myristic acid-labeled Gag partitions to GEM domains. Jurkat cells labeled with [3H]myristic acid were treated with 1% Triton X-100 at 4°C and then equilibrium centrifuged. GEM domain fractions 3 to 5 were pooled and precipitated for Gag. IgG was used as a negative control. Soluble fractions 8 to 10 were pooled and precipitated for Gag as well. Precipitated proteins were blotted onto nitrocellulose and exposed to film. Blots were quantitated as described in Materials and Methods and expressed as a percentage of the total.
DISCUSSION
In this study we examined the hypothesis that exclusion of CD45 from the viral membrane of HIV-1 is the result of budding of the virus from lipid rafts on the cell surface. We found that lipid raft-associated molecules, including the GPI-anchored proteins Thy-1 and CD59 and the ganglioside GM1, colocalize with HIV-1 proteins on the cell surface as determined by confocal fluorescence microscopy. Virus phenotyping with MAbs also indicated that these molecules were indeed incorporated into HIV-1 particles. In contrast, CD45 was excluded from HIV-1 protein-rich uropods and was also found to be excluded from the viral membrane. Similarly, DiIC16(3), a lipid analog that sorts preferentially to lipid rafts, colocalized with HIV-1 proteins, while DiIC12(3), a lipid analog that prefers fluid membrane domains, was excluded from these areas. Dot blot immunoassays of membrane fractions confirmed the presence of HIV-1 gp41 and MA proteins in lipid rafts. Labeling of cells with [3H]myristic acid and immunoprecipitation showed the partitioning of myristylated Gag to lipid rafts.
It has been previously reported that HIV-1 acquires CD55 (DAF) and CD59 that inhibit steps in the complement pathway (23, 38). Both CD55 and CD59 are GPI-linked proteins that have been found to be enriched in GEM domains (14). CD55 and CD59 together provide an advantage for the virus by shielding it from lysis and from neutralization by complement (37). Our data are in complete agreement with previous studies and confirm the incorporation of GPI-anchored proteins, which preferentially sort to lipid rafts, by HIV-1.
Previous studies have shown that cholesterol and sphingomyelin are enriched in the HIV-1 membrane relative to their levels in the host cell membrane (1). Those authors suggested the possibility that viruses are budding from microdomains that have increased cholesterol. Our data provide strong evidence that lipid rafts are the cell membrane microdomains from which HIV-1 buds. The high concentration of both cholesterol and sphingolipids in lipid rafts would explain the high levels of these lipids in the membrane of HIV-1 and provides support for this model of HIV-1 budding. Interestingly, inhibition of cholesterol synthesis has been shown to decrease the production of virus from infected cells (24). Since it is unlikely that viral proteins are able to aggregate individual cholesterol and sphingolipid molecules, the Gag (MA) protein may preferentially interact with existing lipid rafts where aggregation of Gag (MA) molecules may initiate virus budding. In this manner, sphingolipid and cholesterol-rich lipid rafts would be efficiently taken up by new viruses during budding. This model apparently holds true for other viruses as well. Thus, Scheiffele et al. have demonstrated the selective budding of an influenza family virus, fowl plague virus, from ordered lipid domains (40). The requirement for cholesterol and sphingolipids in target membranes for Semliki Forest virus fusion has been established and may be relevant to HIV-1 fusion as well (28, 33). We have not yet investigated the interactions of lipid rafts with accessory HIV-1 molecules such as Vif and Nef that may also have important roles in virus budding. The interactions of myristylated HIV and simian immunodeficiency virus Nef with Lck, known to be in lipid rafts, and its incorporation into virions have already been established (6, 10, 49).
The incorporation of Thy-1, CD59, and other GPI-linked proteins into the viral envelope may have a number of consequences for virus infection and pathogenicity. Thy-1, CD59, and CD55 have cell-signaling capabilities, and the transfer of these highly concentrated proteins into the host cell by HIV-1 particles may trigger an activation signal leading to interleukin-2 production and T-cell proliferation (7, 20). GPI-linked proteins have been shown to be physically associated with the α-subunit of G proteins that are important in signal transduction (45), while other signaling molecules, such as Src family kinases, have been found to be associated with lipid rafts (35). Delivery of these signal transduction molecules to the host cells by the virus may have important effects on virus infectivity, depending on the cell type and its state of activation. Among other effects, GPI-linked molecules acting through G proteins can activate integrins such as LFA-1, which we and others have shown can contribute greatly to HIV-1 infectivity and syncytium formation (13, 16).
A recent model proposed by Shaw and Dustin (43) suggests that CD45 is driven out of cap sites that serve as zones for cellular adhesion and activation between a T cell and an antigen-presenting cell. In their model, short, low-affinity molecules, such as the T-cell receptor, are clustered into the cap site, enhancing the two-dimensional affinity of these molecules for their ligands. This same mechanism results in exclusion of CD45 and capping of GPI-linked proteins and lipid rafts into the areas of cell-to-cell contact. Viral protein targeting through association with lipid rafts into cap sites may facilitate virus particle formation at that site on the surface by directing myristylated matrix proteins and accessory molecules.
The exclusion of CD45 from virions may be an important aspect of HIV assembly. Since the cytoplasmic tail of CD45 is so large (more than 700 amino acids), incorporation of CD45 might hinder critical interactions between gp41 and matrix proteins or other molecules. Furthermore, the long, highly negatively charged extracellular domain of CD45, determined to be as long as 41 nm (4), might sterically hinder viral binding to target cells if it were to be incorporated, considering that a virus particle is only about 100 nm in diameter. The results presented here do not definitively show that the exclusion of CD45 from HIV-1 is due solely to its exclusion from lipid rafts. Critical factors such as Gag membrane binding and assembly, as well as the structure of the CD45 cytoplasmic tail, need to be further examined to fully determine the mechanism for the apparent exclusion of CD45.
Based on the data presented here and that of other studies mentioned above, we propose that HIV-1 buds through lipid rafts. During the course of infection, the cell becomes activated and polarization occurs, capping normally dispersed lipid rafts along with GPI-linked proteins and associated intracellular signaling molecules. Membrane areas containing CD45 are cleared out of the cap site. The newly translated viral Gag precursor protein associated with lipid rafts are then directed to the capped pole, where assembly and budding occurs. Palmitylated gp41 (gp160) is also directed into lipid rafts, and the interaction of its cytoplasmic tail with MA in lipid rafts prevents its internalization, allowing for the incorporation of gp160 into virions only at the site of budding (8, 51). Individual targeting of Gag and Env to the same site at the membrane could be very important in delivering these proteins to the site of budding since Gag and Env are processed and transported in different pathways within the cell (31). The host membrane becomes the new viral coat, resulting in the incorporation of cholesterol, sphingolipids, Thy-1, and CD59 and in the exclusion of CD45. This model does not rule out the incorporation of molecules from neighboring membrane areas, which could account for the incorporation of some host proteins not normally associated with lipid rafts.
We have previously demonstrated that HIV-1 can acquire functional adhesion molecules from host cells (29). We and others have shown that these host-acquired proteins can significantly affect the biology of HIV-1 (11, 16). Therefore, understanding the process of budding and the mechanism by which this virus acquires host proteins is essential to a complete understanding of viral pathogenicity. The budding of HIV-1 from lipid rafts may allow prediction of virus phenotypes and may also allow novel strategies for drug development. Indeed, we have demonstrated that acquisition of host GPI-linked proteins from lipid rafts makes HIV-1 extremely sensitive to a bacterial toxin, the first demonstration that a pathogenic human virus can be targeted in this manner (27).
ACKNOWLEDGMENTS
We thank Michael Delannoy and Sujatha Iyengar for assistance with confocal microscopy. We thank Richard Hampton and Scott Plafker for technical assistance. We also thank Wade Gibson for use of his equipment. We are greatly appreciative of the present and past members of the Leukocyte Immunochemistry Laboratory for their discussions and comments.
This work was supported by a grant from the National Institute of Health (RO1 AI31806). D.H.N. was supported by a training grant from the National Institute of General Medical Sciences (ST32 GM07626).
REFERENCES
- 1.Aloia R C, Tian H, Jensen F C. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci USA. 1993;90:5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arni S, Ilangumaran S, van Echten-Deckert G, Sandhoff K, Poincelet M, Briol A, Rungger-Brandle E, Hoessli D C. Differential regulation of Src-family protein tyrosine kinases in GPI domains of T lymphocyte plasma membranes. Biochem Biophys Res Commun. 1996;225:801–807. doi: 10.1006/bbrc.1996.1254. [DOI] [PubMed] [Google Scholar]
- 3.Arthur L O, Bess J W J, Sowder R C, Benveniste R E, Mann D L, Cherman J-C, Henderson L E. Cellular proteins bound to immunodeficiency viruses: implication for pathogenesis and vaccines. Science. 1992;258:1935–1938. doi: 10.1126/science.1470916. [DOI] [PubMed] [Google Scholar]
- 4.Barclay A N, Beyers A D, Birkeland M L, Brown M H, Davis S J, Somoza C, Williams A F. The leukocyte antigen facts book. New York, N.Y: Academic Press; 1993. [Google Scholar]
- 5.Cinek T, Horejsi V. The nature of large noncovalent complexes containing glycosyl-phosphatidylinositol-anchored membrane glycoproteins and protein tyrosine kinases. J Immunol. 1992;149:2262–2270. [PubMed] [Google Scholar]
- 6.Collette Y, Dutartre H, Benziane A, Ramos-Morales F, Benarous R, Harris M, Olive D. Physical and functional interaction of Nef with Lck. J Biol Chem. 1996;271:6333–6341. doi: 10.1074/jbc.271.11.6333. [DOI] [PubMed] [Google Scholar]
- 7.Davis L S, Patel S S, Atkinson J P, Lipsky P E. Decay-accelerating factor functions as a signal transducing molecule for human T cells. J Immunol. 1988;141:2246–2252. [PubMed] [Google Scholar]
- 8.Egan M A, Carruth L M, Rowell J F, Yu X, Siliciano R F. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of Pr55gag precursor protein. J Virol. 1996;70:6547–6556. doi: 10.1128/jvi.70.10.6547-6556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis S A, Taylor C, Hildreth J E K, August J T. An HLA class I specific monoclonal antibody that fails to bind to all HLA-A antigens. Hum Immunol. 1985;13:13–19. doi: 10.1016/0198-8859(85)90023-0. [DOI] [PubMed] [Google Scholar]
- 10.Flaherty M T, Barber S A, Clements J E. Neurovirulent simian immunodeficiency virus incorporates a nef-associated kinase activity into virions. AIDS Res Hum Retrovir. 1998;14:163–170. doi: 10.1089/aid.1998.14.163. [DOI] [PubMed] [Google Scholar]
- 11.Fortin J F, Cantin R, Lamontagne G, Tremblay M. Host-derived ICAM-1 glycoproteins incorporated on human immunodeficiency virus type 1 are biologically active and enhance viral infectivity. J Virol. 1997;71:3588–3596. doi: 10.1128/jvi.71.5.3588-3596.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Freed E O. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 13.Gomez M B, Hildreth J E K. Antibody to adhesion molecule LFA-1 enhances plasma neutralization of human immunodeficiency virus type 1. J Virol. 1995;69:4628–4632. doi: 10.1128/jvi.69.8.4628-4632.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hannan L A, Edidin M. Traffic, polarity, and detergent solubility of a glycosylphosphatidylinositol-anchored protein after LDL-deprivation of MDCK cells. J Cell Biol. 1996;133:1265–1276. doi: 10.1083/jcb.133.6.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hildreth J E K, August J T. The human lymphocyte function-associated (HLFA) antigen and a related macrophage differentiation antigen (Hmac-1): functional effects of subunit-specific monoclonal antibodies. J Immunol. 1985;134:3272–3280. [PubMed] [Google Scholar]
- 16.Hildreth J E K, Orentas R J. Involvement of a leukocyte adhesion receptor (LFA-1) in HIV-induced syncytium formation. Science. 1989;244:1075–1078. doi: 10.1126/science.2543075. [DOI] [PubMed] [Google Scholar]
- 17.Hoessli D C, Rungger-Brandle E. Isolation of plasma membrane domains from murine T lymphocytes. Proc Natl Acad Sci USA. 1983;80:439–443. doi: 10.1073/pnas.80.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ilangumaran S, Arni S, Chicheportiche Y, Briol A, Hoessli D C. Evaluation by dot-immunoassay of the differential distribution of cell surface and intracellular proteins in glycosylphosphatidylinositol-rich plasma membrane domains. Anal Biochem. 1996;235:49–56. doi: 10.1006/abio.1996.0090. [DOI] [PubMed] [Google Scholar]
- 19.Ilangumaran S, Hoessli D C. Effects of cholesterol depletion by cyclodextrin on the sphingolipid microdomains of the plasma membrane. Biochem J. 1998;335:433–440. doi: 10.1042/bj3350433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korty P E, Brando C, Shevach E M. CD59 functions as a signal-transducing molecule for human T cell activation. J Immunol. 1991;146:4092–4098. [PubMed] [Google Scholar]
- 21.Lanzavecchia A, Iezzi G, Viola A. From TCR engagement to T cell activation: a kinetic view of T cell behavior. Cell. 1999;96:1–4. doi: 10.1016/s0092-8674(00)80952-6. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y M, Tian C J, Yu X F. A bipartite membrane-binding signal in the human immunodeficiency virus type 1 matrix protein is required for the proteolytic processing of gag precursors in a cell type-dependent manner. J Virol. 1998;72:9061–9068. doi: 10.1128/jvi.72.11.9061-9068.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marschang P, Sodroski J, Wurzner R, Dierich M P. Decay-accelerating factor (CD55) protects human immunodeficiency virus type 1 from inactivation by human complement. Eur J Immunol. 1995;25:285–290. doi: 10.1002/eji.1830250147. [DOI] [PubMed] [Google Scholar]
- 24.Maziere J C, Landureau J C, Giral P, Auclair M, Fall L, Lachgar A, Achour A, Zagury D. Lovastatin inhibits HIV-1 expression in H9 human T lymphocytes cultured in cholesterol-poor medium. Biomed Pharmacother. 1994;48:63–7. doi: 10.1016/0753-3322(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee S, Soe T T, Maxfield F R. Endocytic sorting of lipid analogues differing solely in the chemistry of their hydrophobic tails. J Cell Biol. 1999;144:1271–1284. doi: 10.1083/jcb.144.6.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mumby S M. Reversible palmitoylation of signaling proteins. Curr Opin Cell Biol. 1997;9:148–154. doi: 10.1016/s0955-0674(97)80056-7. [DOI] [PubMed] [Google Scholar]
- 27.Nguyen D H, Liao Z, Buckley J T, Hildreth J E K. Neutralization of HIV-1 by a pore-forming bacterial toxin, aerolysin. Mol Microbiol. 1999;33:659–666. doi: 10.1046/j.1365-2958.1999.01518.x. [DOI] [PubMed] [Google Scholar]
- 28.Nieva J L, Bron R, Corver J, Wilschut J. Membrane fusion of semliki forest virus requires sphingolipids in the target membrane. EMBO J. 1994;13:2797–2804. doi: 10.1002/j.1460-2075.1994.tb06573.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Orentas R J, Hildreth J E K. Association of host cell surface adhesion receptors and other membrane proteins with HIV and SIV. AIDS Res Hum Retrovir. 1993;9:1157–1165. doi: 10.1089/aid.1993.9.1157. [DOI] [PubMed] [Google Scholar]
- 30.Orlandi P A, Fishman P H. Filipin-dependent inhibition of cholera toxin: evidence for toxin internalization and activation through caveolae-like domains. J Cell Biol. 1998;141:905–915. doi: 10.1083/jcb.141.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pal R, Mumbauer S, Hoke G M, Takatsuki A, Sarngadharan M G. Brefeldin A inhibits the processing and secretion of envelope glycoproteins of human immunodeficiency virus type 1. AIDS Res Hum Retrovir. 1991;7:707–712. doi: 10.1089/aid.1991.7.707. [DOI] [PubMed] [Google Scholar]
- 32.Pearce-Pratt R, Malamud D, Phillips D M. Role of the cytoskeleton in cell-to-cell transmission of human immunodeficiency virus. J Virol. 1994;68:2898–2905. doi: 10.1128/jvi.68.5.2898-2905.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phalen T, Kielian M. Cholesterol is required for infection by Semliki Forest virus. J Cell Biol. 1991;112:615–623. doi: 10.1083/jcb.112.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Robbins S M, Quintrell N A, Bishop J M. Myristoylation and differential palmitoylation of the HCK protein-tyrosine kinases govern their attachment to membranes and association with caveolae. Mol Cell Biol. 1995;15:3507–3515. doi: 10.1128/mcb.15.7.3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rodgers W, Crise B, Rose J K. Signals determining protein tyrosine kinase and glycosyl-phosphatidylinositol-anchored protein targeting to a glycolipid-enriched membrane fraction. Mol Cell Biol. 1994;14:5384–5391. doi: 10.1128/mcb.14.8.5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rodgers W, Rose J K. Exclusion of CD45 inhibits activity of p56lck associated with glycolipid-enriched membrane domains. J Cell Biol. 1996;135:1515–1523. doi: 10.1083/jcb.135.6.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saifuddin M, Crnich C, Long T, Saarloos M-N, Spear G T. Transfer of host T-cell membrane HLA-DR and CD25 to target cells by human retroviruses. J Acquir Immune Defic Syndr Hum Retrovirol. 1998;17:196–202. doi: 10.1097/00042560-199803010-00002. [DOI] [PubMed] [Google Scholar]
- 38.Saifuddin M, Hedayati T, Atkinson J P, Holguin M H, Parker C J, Spear G T. Human immunodeficiency virus type 1 incorporates both glycosylphosphatidylinositol-anchored CD55 and CD59 and integral membrane CD46 at levels that protect from complement-mediated destruction. J Gen Virol. 1997;78:1907–1911. doi: 10.1099/0022-1317-78-8-1907. [DOI] [PubMed] [Google Scholar]
- 39.Sargiacomo M, Sudol M, Tang Z, Lisanti M P. Signal transducing molecules and glycosyl-phosphatidylinositol-linked proteins form a caveolin-rich insoluble complex in MDCK cells. J Cell Biol. 1993;122:789–807. doi: 10.1083/jcb.122.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scheiffele P, Rietveld A, Wilk T, Simons K. Influenza viruses select ordered lipid domains during budding from the plasma membrane. J Biol Chem. 1999;274:2038–2044. doi: 10.1074/jbc.274.4.2038. [DOI] [PubMed] [Google Scholar]
- 41.Schroeder R, London E, Brown D. Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc Natl Acad Sci USA. 1994;91:12130–12134. doi: 10.1073/pnas.91.25.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schroeder R J, Ahmed S N, Zhu Y, London E, Brown D A. Cholesterol and sphingolipid enhance the Triton X-100 insolubility of glycosylphosphatidylinositol-anchored proteins by promoting the formation of detergent-insoluble ordered membrane domains. J Biol Chem. 1998;273:1150–1157. doi: 10.1074/jbc.273.2.1150. [DOI] [PubMed] [Google Scholar]
- 43.Shaw A S, Dustin M L. Making the T cell receptor go the distance: a topological view of T cell activation. Immunity. 1997;6:361–369. doi: 10.1016/s1074-7613(00)80279-4. [DOI] [PubMed] [Google Scholar]
- 44.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 45.Solomon K R, Rudd C E, Finberg R W. The association between glycosylphosphatidylinositol-anchored proteins and heterotrimeric G protein alpha subunits in lymphocytes. Proc Natl Acad Sci USA. 1996;93:6053–6058. doi: 10.1073/pnas.93.12.6053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spink C H, Yeager M D, Feigenson G W. Partitioning behavior of indocarbocyanine probes between coexisting gel and fluid phases in model membranes. Biochim Biophys Acta. 1990;1023:25–33. doi: 10.1016/0005-2736(90)90005-9. [DOI] [PubMed] [Google Scholar]
- 47.Stefanova I, Horejsi V, Ansotegui I J, Knapp W, Stockinger H. GPI-anchored cell-surface molecules complexed to protein tyrosine kinases. Science. 1991;254:1016–1019. doi: 10.1126/science.1719635. [DOI] [PubMed] [Google Scholar]
- 48.van den Berg C W, Cinek T, Hallett M B, Horejsi V, Morgan B P. Exogenous glycosyl phosphatidylinositol-anchored CD59 associates with kinases in membrane clusters on U937 cells and becomes Ca2+-signaling competent. J Cell Biol. 1995;131:669–677. doi: 10.1083/jcb.131.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Welker R, Kottler H, Kalbitzer H R, Krausslich H G. Human immunodeficiency virus type 1 Nef protein is incorporated into virus particles and specifically cleaved by the viral proteinase. Virology. 1996;219:228–236. doi: 10.1006/viro.1996.0240. [DOI] [PubMed] [Google Scholar]
- 50.Yang C, Spies C P, Compans R W. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc Natl Acad Sci USA. 1995;92:9871–9875. doi: 10.1073/pnas.92.21.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu X, Yuan X, Matsuda Z, Lee T H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]