Abstract
The alphaherpesvirus tegument protein VP22 has been characterized with multiple traits including microtubule reorganization, nuclear localization, and nonclassical intercellular trafficking. However, all these data were derived from studies using herpes simplex virus type 1 (HSV-1) and may not apply to VP22 homologs of other alphaherpesviruses. We compared subcellular attributes of HSV-1 VP22 (HVP22) with bovine herpesvirus 1 (BHV-1) VP22 (BVP22) using green fluorescent protein (GFP)-fused VP22 expression vectors. Fluorescence microscopy of cell lines transfected with these constructs revealed differences as well as similarities between the two VP22 homologs. Compared to that of HVP22, the BVP22 microtubule interaction was much less pronounced. The VP22 nuclear interaction varied, with a marbled or halo appearance for BVP22 and a speckled or nucleolus-bound appearance for HVP22. Both VP22 homologs associated with chromatin at various stages of mitosis and could traffic from expressing cells to the nuclei of nonexpressing cells. However, distinct qualitative differences in microtubule, nuclear, and chromatin association as well as trafficking were observed. The differences in VP22 homolog characteristics revealed in this study will help define VP22 function within HSV-1 and BHV-1 infection.
As with those of other alphaherpesviruses, the bovine herpesvirus 1 (BHV-1) virion contains a complex structure called the tegument located between the nucleocapsid and the virus envelope (21). Limited data elucidating the functions of the various BHV-1 tegument proteins exist. In fact, practically all information about this crucial alphaherpesvirus virion structure has been obtained from studies with herpes simplex virus type 1 (HSV-1). Tegument proteins are first to encounter the intracellular environment and provide essential functions to subjugate the host cell (20). The tegument can assemble into a stable structure without capsid interaction, and its assembly or dissociation depends on the phosphorylation state of its structural proteins (15, 21). However, the site and mechanisms of tegument assembly and the functions of its protein components are largely unknown.
Homologs of the major HSV-1 tegument proteins VP13/14, VP16, and VP22 are found in BHV-1. Available information suggests similar as well as distinct roles for each BHV-1 and HSV-1 tegument homolog. The BHV-1 homolog to HSV-1 VP13/14 (HVP13/14) is BHV-1 VP8 (BVP8), the most abundant BHV-1 protein (14). Like HVP13/14, BVP8 contains O-linked carbohydrates acquired during transport of tegumented nucleocapsids through the Golgi (27). Compared to HVP13/14, BVP8 has less affinity for the nucleocapsid (19). Whether BVP8 can modulate alpha gene expression as does HVP13/14 is not known (30). HVP16 and BVP16 are both transcription activators that can recruit host homeodomain proteins Oct-1 and HCF into a transcriptional regulatory complex. However, they differ in DNA recognition, binding to HSV-1- or BHV-1-specific response element sequences (13). BVP22, like HVP22, is a phosphoprotein that can associate with the nuclear matrix (17). Nevertheless, BVP22 predominantly localizes to the nucleus during BHV-1 infection (17), while HVP22 localizes primarily to the cytoplasm early during HSV-1 infection and accumulates in the nucleus late in HSV-1 infection (8, 23). Further, the BVP22 gene is not considered an essential gene for viral replication (16), while the HVP22 gene is considered essential (12). These distinctions between the VP22 homologs suggest that BVP22 and HVP22 have different functional properties.
Although virtually no data on the functional properties of BVP22 exist, recent reports have shown HVP22 to possess several varied and fascinating characteristics. HVP22 expressed in cells transiently transfected or HSV-1 infected associates with and reorganizes the host cell microtubule network (6). In addition, HVP22 can traffic intercellularly through unknown, nonclassical export and import mechanisms. After trafficking to the surrounding cells, HVP22 targets the nuclei of these cells (5). Though the function of HVP22 during HSV-1 infection remains unknown, the fact that HVP22 can exploit the host cytoskeleton as well as accumulate in the nucleus suggests that this unusual tegument protein may have an important role in herpesvirus infection, replication, and pathogenesis.
Because of the potential importance of VP22 in herpesvirus infection and the lack of knowledge concerning the properties of the BHV-1 homolog, BVP22, our objective was to identify and compare functional characteristics of BVP22 and HVP22. Mammalian expression vectors of BVP22 or HVP22 fused to a green fluorescent protein (GFP) variant were transiently transfected into cell lines, and cellular localization was analyzed using fluorescence microscopy. We report that, like HVP22, BVP22 has varied properties including microtubule association, nuclear/chromatin association, and intercellular trafficking. However, within each of these common properties are variations between the homologs that could help explain the role of VP22 in both BHV-1 and HSV-1 maturation.
MATERIALS AND METHODS
Plasmid engineering.
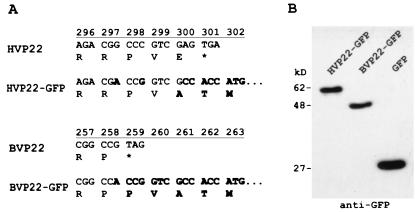
Supernatant from HSV-1 (strain 17)-infected Vero cells (ATCC CCL-81) or BHV-1 (Cooper's)-infected Madin-Darby bovine kidney (MDBK) cells (ATCC CCL-22) was amplified by PCR utilizing primers designed with restriction enzyme sites BamHI (5′) and AgeI (3′) for in-frame cloning of VP22 homologs into the GFP variant mammalian expression vector pEYFP-N1 (Clontech, Palo Alto, Calif.). Primers (Oligos Etc., Bethel, Maine) were as follows: HVP22, 5′ CGT GGA TCC ATG ACC TCT CGC CGC and 3′ TCG ACC GGT CGT CTG GGG CG; BVP22, 5′ GAC GGA TCC GCC ATG GCC CGG and 3′ TCG ACC GGT GGC CGG GCC CGC T. The PCR mixture consisted of Tth DNA polymerase, buffer, 3× PCR enhancer (Epicentre Technologies, Madison, Wis.), 1 mM MgCl2, a 0.2 mM concentration of each deoxynucleoside triphosphate, and a 1 μM concentration of each primer. DNA thermal cycler 480 (PE Applied Biosystems, Foster City, Calif.) parameters were 1 cycle of 95°C for 5 min and 30 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 2 min. The amplified product was gel purified (Qiagen, Valencia, Calif.), digested with AgeI/BamHI restriction enzyme, purified again, and ligated into AgeI/BamHI sites of pEYFP-N1. Positive clones were selected by restriction fragment length polymorphism analysis and transient transfection analysis using fluorescence microscopy and Western blotting. Sequencing confirmed VP22 from HSV-1 and BHV-1, and clones were designated pEYFP-HVP22 and pEYFP-BVP22. Figure 1A indicates the sequence of the fusion site between the VP22 homologs and GFP. Figure 1B demonstrates the transcription and translation of the constructs in transfected cells using Western blot analysis. Construct pEBFP-BVP8 was similarly engineered. Primers (Oligos Etc.) for BVP8 were 5′ CTA GGA TCC CTT AGA CGC CAT GGA CGC CGC and 3′ CCT ACC GGT CCG CCC AGG CGC GGG CC. The amplified product from the BHV-1 template was cloned into the AgeI/BamHI sites of pEBFP-N1 (Clontech). The VP22 gene GenBank accession number is U21137 (17); the HVP22 gene GenBank accession number is D10879 (18).
FIG. 1.
Construction and functional expression of VP22-GFP fusions. HVP22 and BVP22 gene homologs were amplified by PCR and subcloned into a GFP variant-encoding mammalian expression vector. (A) Nucleic acid and amino acid changes to the wild-type VP22 homolog (boldface) through the start codon for the fused GFP. (B) Western immunoblot of expressed protein from these constructs utilizing anti-GFP.
Western blot analysis.
Transfected cell lysate and a broad-range, prestained protein marker (New England Biolabs, Beverly, Mass.) were separated by 10% polyacrylamide gel electrophoresis using a minigel apparatus (Hoefer, San Francisco, Calif.) following the manufacturer's protocol. Protein from the gel was then transferred to a nitrocellulose membrane using an electroblotting system (Bio-Rad Laboratories, Hercules, Calif.). A chemiluminescent Western blotting kit (Pierce Chemical Company, Rockford, Ill.) was used along with antibody to GFP (Clontech) to detect expression of VP22-GFP fusion proteins (Fig. 1B).
Cell culture and transient transfections.
All cells were cultured in a 37°C, humidified, 5% CO2 incubator with RPMI 1640 containing 2 mM l-glutamine, 1.5 g of sodium bicarbonate/liter, and 10% fetal bovine serum. Cell lines CCF-STTG1 (CRL-1718), D17 (CRL-6248), HeLa (CCL-2), MDBK (CCL-22), NMU (CRL-1743), and Vero (CCL-81) were obtained from the American Type Culture Collection. Primary F17 fibroblasts were isolated from the skin of a Holstein cow from the University of Wisconsin-Madison dairy herd.
Transient transfections were performed using the cationic lipid method. Cells were plated onto six-well plates containing sterile glass coverslips, grown until about 70% confluent, and then washed with 2 ml of OPTI-MEM reduced-serum medium (Life Technologies, Gaithersburg, Md.)/well. After discarding the wash, 0.8 ml of OPTI-MEM/well was added. For each transfection (well), 1 μg of plasmid DNA in 100 μl of OPTI-MEM was mixed with 6 μl of Lipofectamine (Life Technologies) in 100 μl of OPTI-MEM in a 12- by 75-mm culture tube. This mixture (0.2 ml) was left to incubate at room temperature for at least 15 min before being added to the cells. After a 3-h incubation at 37°C in a humidified 5% CO2 incubator, 1 ml of growth medium/well was added. In certain experiments, Colcemid (Life Technologies) was added to transfected cells (10 pg/ml) for 4 h prior to analysis.
Fluorescence microscopy.
An Axiovert S100 (Carl Zeiss, Inc., Thornwood, N.Y.) microscope was used for epifluorescence analysis of cells transfected with GFP-expressing constructs or stained by indirect immunofluorescence or with fluorescent probes. Images were recorded digitally and processed using Adobe Photoshop, version 5.0, software. For direct analysis, cells were washed two times with phosphate-buffered saline (PBS) and the coverslip was mounted on a glass microscope slide for immediate examination by fluorescence microscopy. For fixed cells, 2 ml of freshly made 4% paraformaldehyde-PBS was added to the PBS-washed cells and the cells were incubated at room temperature for 30 min. Cells were then washed twice in PBS.
An analysis of the translocation of VP22 from plasma membrane lysed transfected cells was performed using a cytoskeleton stabilization buffer (PHEM). This buffer consisted of 60 mM PIPES (piperazine-N,N′-bis(2-ethanesulfonic acid), 25 mM HEPES, 10 mM EGTA, 2 mM MgCl2, and 0.1% digitonin, pH 7.9. Transfected cells were washed two times with PBS and examined by fluorescence microscopy. Then, PBS was removed, and PHEM buffer was added during microscopy.
Fluorescence staining of cells.
For indirect immunofluorescence or fluorescent-probe staining, the following procedure was used. Fixed cells were extracted with 0.1% Triton X-100–PBS for 3 to 5 min at room temperature and washed twice in PBS. Then, cells were incubated in 1% bovine serum albumin (BSA)-PBS for 20 to 30 min. Primary antibody (α-tubulin [Molecular Probes Inc., Eugene, Oreg.] or phosphohistone H3 [Upstate Biotechnology, Lake Placid, N.Y.]) at 1:200 dilution in 1% BSA-PBS or fluorescent probe BODIPY FL taxol, BODIPY TR-X phallacidin, or MitoTracker Red CMXRos (Molecular Probes) at 80 nM, 15 U/ml, or 100 nM, respectively, in 1% BSA-PBS was added to the coverslip for 1 h at room temperature. After two washes with PBS, the fluorescent probe-labeled cells were examined by epifluorescence microscopy. A fluorescently labeled secondary antibody (Alexa 546 goat anti-mouse immunoglobulin G [IgG] or BODIPY FL goat anti-rabbit IgG [Molecular Probes]) at 1:500 dilution in 1% BSA-PBS was added to primary antibody (α-tubulin or phosphohistone H3)-labeled cells, and the cells were incubated for 1 h at room temperature and protected from light. After two washes with PBS, these cells were examined by fluorescence microscopy.
Propidium iodide (PI) fluorescence staining of DNA was performed by adding 1 ml of PI staining solution (50 μg of PI/ml, and 100 U of RNase A/ml in PBS)/well to PBS-washed cells for 30 min at room temperature. Cells were then washed with PBS three to five times and examined by fluorescence microscopy.
RESULTS
There is structural variation between VP22 of BHV-1 and HSV-1.
Although they are homologs, BVP22 and HVP22 are encoded by genes with considerably different open reading frame sizes (777 and 906 bp, respectively). Thus, we compared sequences by using computer algorithms to determine similarities and differences in alignment, possible functional motifs, and subcellular targeting signals. Interestingly, the two proteins have only 28.7% homology based on amino acid alignment results (Fig. 2). Further, while neither has an identifiable N-terminal signal sequence, BVP22 has internal consensus sequences that imply subcellular targeting different from that of HVP22. For example, using PROSITE II (PSORT World Wide Web server; revision date, 1 December 1998; PSORT II program; http://psort.nibb.ac.jp:8800/; last date accessed, 30 December 1999). HVP22 has two classic nuclear localization signals, pat4 at position 295 and pat7 at position 82; however, BVP22 does not contain any classic nuclear localization signal. Nevertheless, the program predicts, based on the high percentage of basic residues, that BVP22 will target the nucleus. These structural differences likely translate into localization and functional differences.
FIG. 2.
Alignment of BVP22 and HVP22. BVP22 (259 amino acids) and HVP22 (301 amino acids) amino acid sequences were optimally aligned using a computer program (ALIGN) available from GeneStream. The results revealed only 28.7% homology. :, identity; · , similar acidity; global alignment score, 273.
There are general similarities between VP22 of BHV-1 and HSV-1.
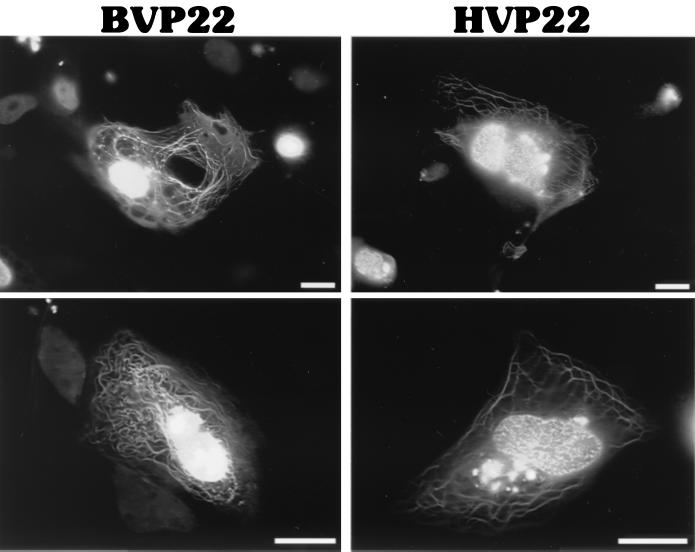
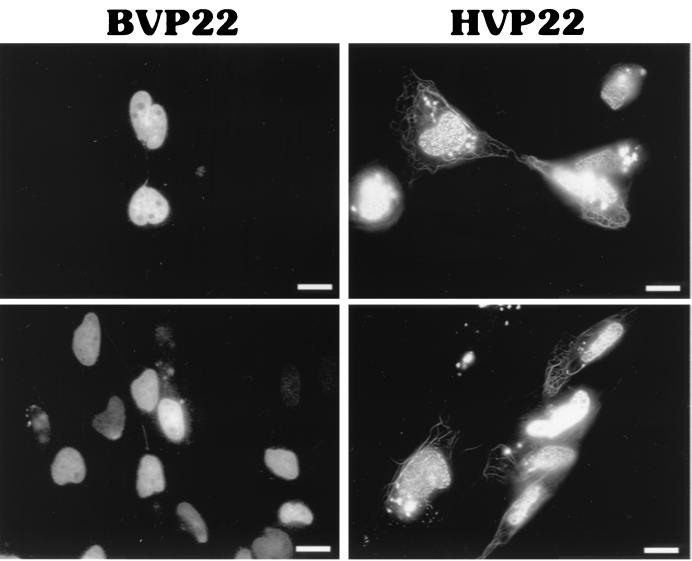
To directly compare subcellular targeting of BVP22 and HVP22, constructs were engineered to express a GFP variant fused to the carboxyl terminus of each VP22 homolog. Then cells were transiently transfected, and the results were assayed using fluorescence photomicroscopy. As shown in Fig. 1A, few alterations from the native HVP22 or BVP22 sequences were made at the GFP fusion site. The microscopy observations made by others (3) were confirmed by noting a mixed pattern of HVP22 subcellular localization, including filamentous cytoplasmic and nuclear compartmentalization. BVP22 displayed a varied pattern of filamentous cytoplasmic and nuclear localization as well (Fig. 3). Cells transfected with both VP22 homologs consisted of a heterogeneous population, with some cells having filamentous and nuclear staining while other cells had only nuclear staining. Frequently, staining of two or more nuclei attached by a thick filament was observed. This phenomenon was seen in both HVP22- and BVP22-transfected cells but appeared more pronounced with BVP22. Similar results, as shown in Fig. 3, were seen for all HVP22- or BVP22-transfected cell lines tested, including CCF-STTG1 (human astrocytoma), D17 (dog osteosarcoma), F17 (primary cow fibroblasts), HeLa (human epithelioid), MDBK (bovine kidney), NMU (rat mammary carcinoma), NXS2 (mouse neuroblastoma), and Vero (monkey kidney).
FIG. 3.
Intracellular localization of VP22 homologs. D17 cells were transiently transfected with BVP22 or HVP22 and analyzed by fluorescence microscopy. Scale bar, 2 μm.
The filamentous cytoplasmic intracellular staining pattern is rarer in BVP22-transfected cells than in HVP22-transfected cells.
Whereas both homologs had mixed nuclear and cytoplasmic targeting within transfected-cell populations, a striking difference between the extent of nuclear or filamentous cytoplasmic staining by HVP22 and that by BVP22 was noted. HVP22-transfected cells were invariably identified by intense labeling of thick filamentous bundles and less-notable florescence of nuclei. In contrast, BVP22-transfected cells were conspicuous by pronounced labeling of nuclei, and although cells with filamentous labeling were found, they were less numerous than such HVP22-transfected cells. In fact, within a population of BVP22-transfected cells, only 2% (15 ± 3 of 1,000) displayed a filamentous cytoplasmic pattern, whereas 20% (183 ± 3 of 1,000) of HVP22-transfected cells displayed this pattern. These data were collected from three transfection experiments using D17 cells, with the peak ratio of cells with filamentous labeling/total transfected cells observed 36 h after transfection. Cotransfections of the GFP-labeled BVP22 or HVP22 with blue fluorescent protein (BFP) demonstrated that BVP22- or HVP22-expressing cells could have either the filamentous and nuclear or nuclear-only phenotype (data not shown). This was observed for all the different cell types listed in the paragraph above.
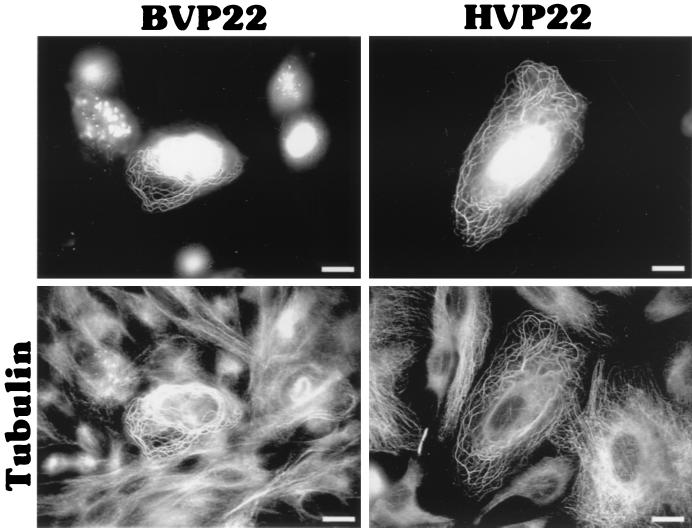
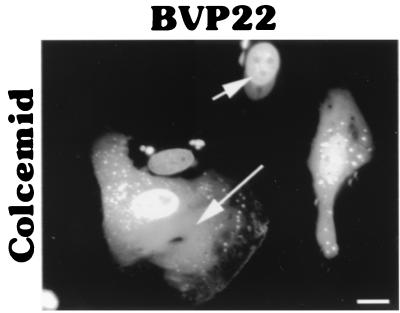
Others (6) have demonstrated that HVP22 exhibits the properties of a classical microtubule-associated protein, reorganizing and stabilizing the host cell microtubule network. We established by indirect immunofluorescence and fluorescent probe staining that BVP22 also associates with the transfected-cell microtubules. Figure 4 shows α-tubulin costained with BVP22 filaments. Further, the microtubule organization in BVP22-transfected filamentous cells was different from that in nontransfected cells or cells displaying nuclear BVP22 staining only. The microtubule organizing center was eliminated in filamentous BVP22-transfected cells, and thick bundles of microtubules were not seen in nontransfected cells or in cells with BVP22 staining only in the nuclei. A fluorescent phallotoxin probe specific for filamentous actin did not costain with BVP22 filaments (data not shown), confirming results obtained with HVP22 (6). Addition of Colcemid, a disrupter of microtubules, abolished the BVP22 filamentous pattern, resulting in fluorescent cytoplasmic particulate, but did not affect nuclear staining (Fig. 5). Since the PROSITE II computer algorithm predicted that HVP22 would target mitochondria, BVP22- and HVP22-transfected cells were costained with a fluorescent mitochondrial probe. No correlation between either VP22 homolog and mitochondria was evident (data not shown).
FIG. 4.
VP22 homolog filamentous pattern costains with microtubules. BVP22- or HVP22-transfected D17 cells were costained with α-tubulin antibody and analyzed by fluorescence microscopy. The same fields are shown in upper and lower panels using different filter sets. Scale bar, 2 μm.
FIG. 5.
Colcemid treatment disrupted the filamentous cytoplasmic pattern of BVP22. Addition of Colcemid to BVP22-transfected D17 cells resulted in a diffuse cytoplasmic pattern (long arrow). However, the nuclear association pattern (short arrow) remained unchanged. Scale bar, 2 μm.
There are prominent nuclear staining pattern differences between VP22 of BHV-1 and HSV-1.
Besides the extent of microtubule staining, another marked difference between HVP22- and BVP22-transfected cell populations was the nature of nuclear staining. As is evident in Fig. 6, often nuclei of HVP22-transfected cells had a speckled appearance. Speckled nuclei were rarely observed with BVP22-transfected cells; nuclei of BVP22-transfected cell populations generally had a marbled appearance (in Fig. 4, the gain was increased on the image to enhance VP22-stained microtubules, overwhelming the marbled [BVP22] and speckled [HVP22] nuclear patterns). BVP22 association with the nucleus has been established (17). However, whether this association is with the nuclear membrane, nuclear lamina, or chromatin is unknown. Thus, the nuclear membranes of VP22-transfected cells were disrupted to determine whether BVP22 or HVP22 was bound to the membrane—thereby dispersing when the membrane was lysed—or was bound to the protein matrix and chromatin of the nucleus.
FIG. 6.
Distinctions in the nuclear localization patterns of BVP22 and HVP22. Fluorescence microscopy of transiently transfected D17 cells revealed a marbled pattern of nuclear staining by BVP22 and a speckled nuclear staining by HVP22. Frequently, BVP22-stained nuclei would be attached by filaments. Scale bar, 2 μm.
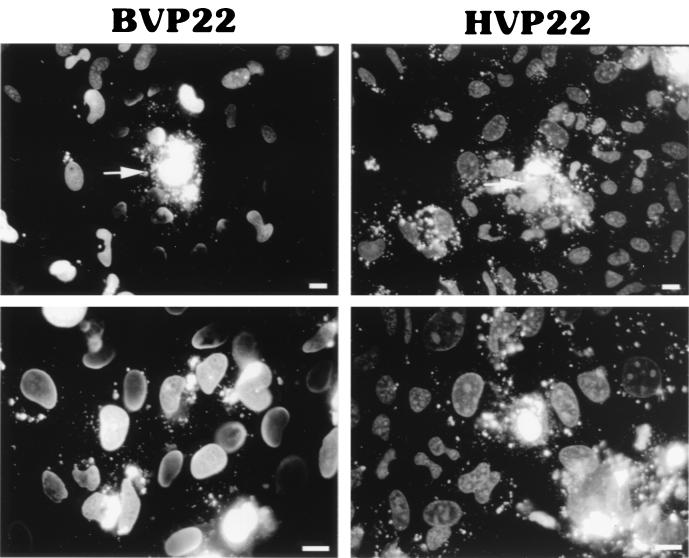
Plasma and nuclear membrane lysis of transfected cells, utilizing a cytoskeleton stabilization buffer, accentuated the nuclear association differences between HVP22 and BVP22. Figure 7 shows that both VP22 homologs bind to a nonmembrane fraction of the nucleus. However, comparing the transfected cells with intact nuclear membranes of Fig. 6 with those with disrupted nuclear membranes (Fig. 7) indicates altered nuclear localization patterns, especially for HVP22. The speckled pattern of HVP22 disappeared, revealing a nucleolus binding pattern. Nucleolus binding by BVP22 was not as notable, nor was there much change in the generally marbled pattern between lysed-membrane and intact-membrane transfected cell nuclei. Although both HVP22 and BVP22 have bright nuclear rim binding, BVP22 has a marked halo appearance. Strikingly, membrane lysis immediately following buffer addition resulted in HVP22 or BVP22 labeling of every cell nucleus in the monolayer. The transfected cell (Fig. 7, top) released VP22-GFP, and a gradient of stained nuclei resulted. This was especially remarkable for BVP22, where a “crescent moon” effect on nuclei facing the BVP22-transfected cell was evident.
FIG. 7.
Nuclear membrane lysis of VP22 homolog-transfected D17 cells accentuates differences in nuclear protein fraction labeling. BVP22-transfected cultures have a halo appearance, whereas HVP22-transfected cultures have accentuated nucleolus labeling. Lysis of VP22-expressing cells (arrows; upper panels) resulted in nuclear labeling of the entire monolayer for both homologs. Scale bar, 2 μm.
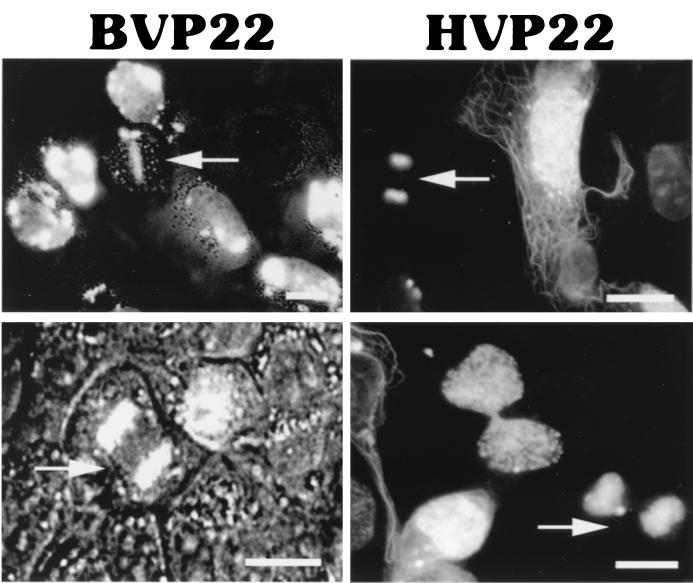
The lack of a halo appearance, the prominence of nucleoli, and the weaker nuclear staining of HVP22 than of BVP22 suggest that HVP22 may have a greater affinity for intranuclear or chromatin binding, whereas BVP22 has a greater affinity for nuclear lamina. HVP22 is capable of binding chromatin during mitosis (5). As seen in Fig. 8, chromatin readily costained for BVP22 during mitosis, as well as for HVP22. Thus, BVP22 binds chromatin and is carried to daughter cells. However, chromatin binding does not exclude an affinity for nuclear lamina by either HVP22 or BVP22.
FIG. 8.
Both VP22 homologs bound chromatin during mitosis. Arrows, BVP22 or HVP22 bound to chromatin in metaphase (upper left panel) or telophase (all other panels) in transfected D17 cells. BVP22-transfected cells were viewed under both bright-field and fluorescence microscopy. HVP22-transfected cells were viewed under fluorescence microscopy only. Scale bar, 2 μm.
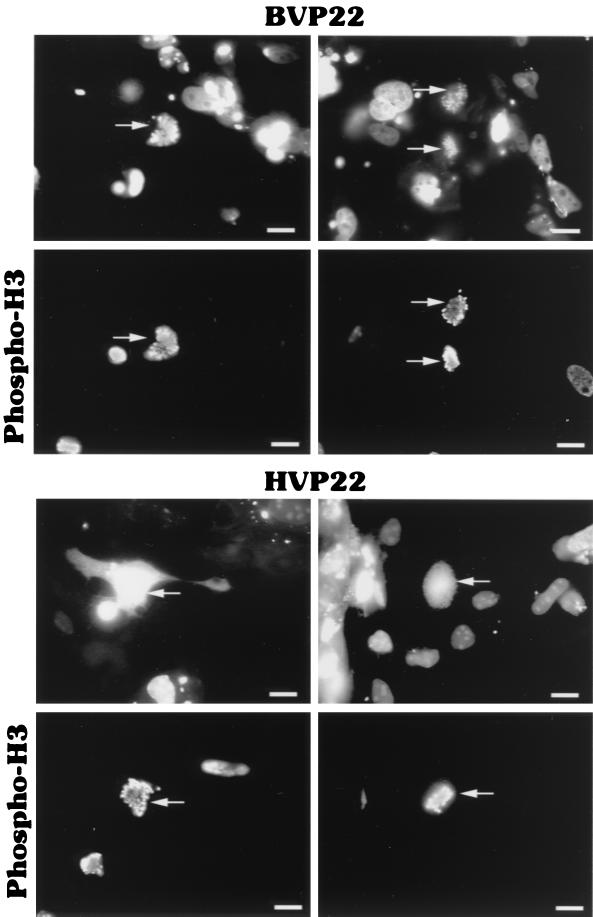
To determine more precisely the nuclear localization of BVP22, Colcemid-treated BVP22- and HVP22-transfected cells were costained with phosphohistone H3 antibody. Phosphorylation of histone H3 correlates strongly with mitosis in all mammals and is required for proper chromosome coiling and segregation (28). Interestingly, VP22 homolog costaining with the mitosis marker phosphohistone H3 revealed another difference between BVP22 and HVP22. As shown in Fig. 9, BVP22 labeling correlated with that of phosphohistone H3. However, HVP22 appeared to be dispersed throughout the cell (although HVP22 binding to condensed chromosomes cannot be discounted). This staining pattern difference between the VP22 homologs may only be at an early stage of mitosis since HVP22, like BVP22, is completely bound to chromatin during the telophase as shown in Fig. 8. Further, Fig. 9 is representative of cells treated with Colcemid and thus arrested in metaphase.
FIG. 9.
Difference in the colocalizations of VP22 homologs and the mitosis marker phosphohistone H3 (phospho-H3). BVP22- or HVP22-transfected D17 cells were treated with Colcemid (4 h) to arrest cells in metaphase. Subsequently, transfects were stained with phosphohistone H3 antibody and colocalization was analyzed by fluorescence microscopy. The same fields are shown in upper and lower panels using filter sets for green (upper panels) and red (lower panels). Arrows, cells costaining for the VP22 homolog and phosphohistone H3. Scale bar, 2 μm.
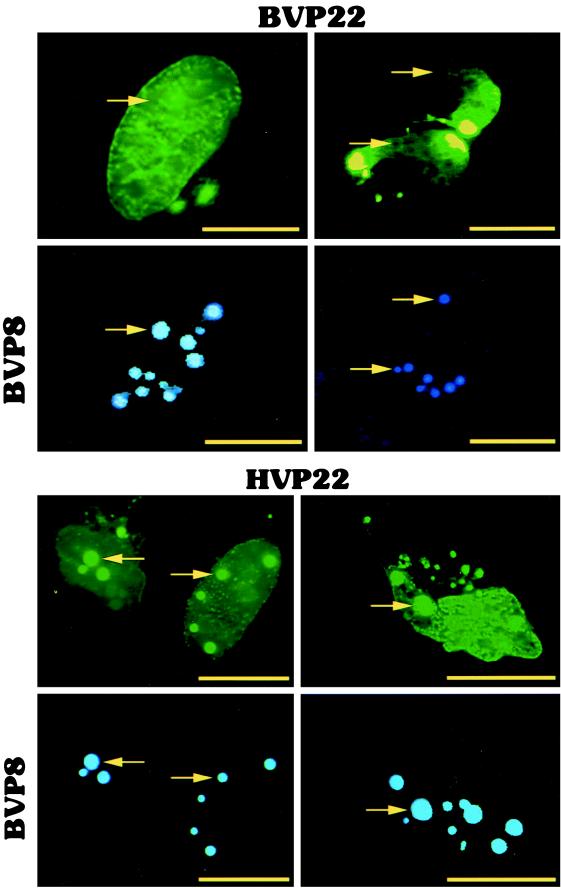
Another striking difference between BVP22 and HVP22 was observed during cotransfection studies with the VP22 homologs and the BHV-1 tegument protein VP8. Cotransfection of BVP22 and BVP8 did not result in any noticeable alteration in the intracellular localization of either protein (Fig. 10). However, cotransfection of HVP22 and BVP8 resulted in BVP8 partially sequestering HVP22. VP22 homolog cotransfection with BVP8 resulted in a display of bright spheres only in HVP22- and BVP8-cotransfected cells, and these spheres correlated exactly with BVP8 localization.
FIG. 10.
Difference in the colocalizations of VP22 homologs and BVP8. D17 cells were cotransfected with BVP22-GFP (BVP22) or HVP22-GFP (HVP22) and BVP8-BFP (BVP8) and analyzed using fluorescence microscopy. The same fields are shown in upper and lower panels using filter sets for green (upper panels) and blue (lower panels). Arrows, location of BVP8 spheres. BVP8 and BVP22 do not costain, whereas BVP8 costains with HVP22, indicating that BVP8 can partially sequester HVP22. Scale bar, 2 μm.
There are intercellular nuclear trafficking differences between VP22 of BHV-1 and HSV-1.
We observed that as time progressed, transfected monolayers contained increasing numbers of BVP22-GFP-labeled nuclei, whereas the numbers of GFP-transfected cells remained the same. Thus, we hypothesized that BVP22, like HVP22, trafficked intercellularly. HVP22 has been shown to spread from the synthesizing cell to surrounding cell nuclei (3, 4, 22, 29), but whether VP22 homologs from other alphaherpesviruses, including BVP22, have the same remarkable transport property is unknown.
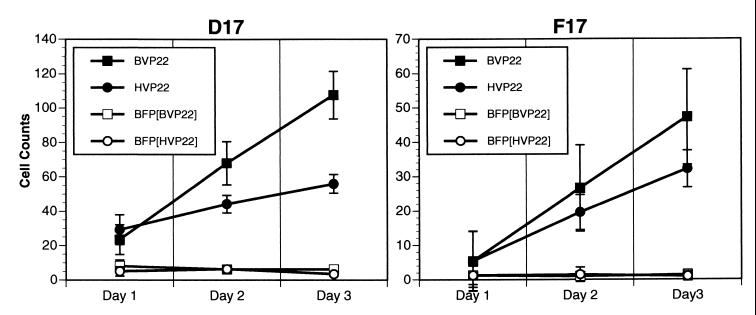
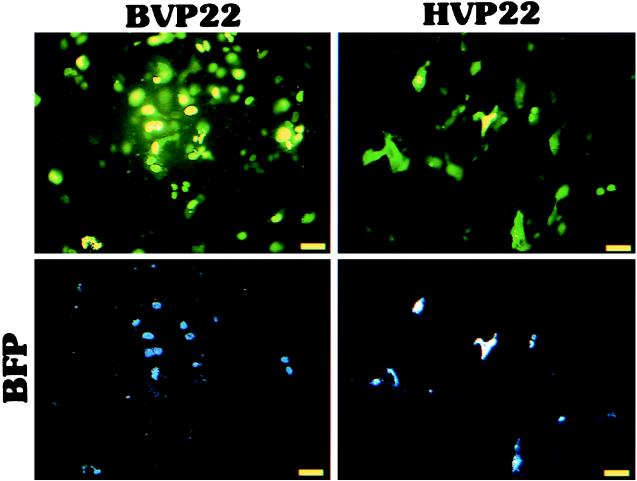
To test our hypothesis supporting BVP22 trafficking, cell monolayers were cotransfected with BVP22 or HVP22 fused to GFP and a vector expressing BFP and then numbers of green and blue cells were scored daily for 3 days. BVP22-GFP was compared to HVP22-GFP for intercellular trafficking and to BFP (a nontrafficking protein) as an internal control for cell division and trafficking. As graphed in Fig. 11, the numbers of green BVP22-labeled cells increased dramatically over 3 days compared with the numbers of cotransfected BFP-labeled cells, confirming the transport capability of BVP22. Further, BVP22 trafficking appeared more proficient than that of HVP22. Figure 12 demonstrates this trafficking with a low-magnification epifluorescence image of nonfixed D17 cells cotransfected with the VP22 homolog and BFP for 3 days.
FIG. 11.
BVP22 traffics to nontransfected cells in a monolayer. D17 or F17 cell monolayers were cotransfected with BVP22-GFP (BVP22) or HVP22-GFP (HVP22) and BFP (BFP[BVP22]; BFP[HVP22]). Nine random fields per coverslip of triplicate sets of nonfixed transfected cells were counted daily for numbers of blue fluorescing cells (BFP) and green fluorescing cells (BVP22 or HVP22). Data are expressed as means ± standard deviations and are representative of at least three experiments.
FIG. 12.
Demonstration of VP22 homolog trafficking in live cells. D17 cell monolayers were cotransfected with BVP22-GFP (BVP22) or HVP22-GFP (HVP22) and BFP. After 3 days, cells were washed in PBS and analyzed by epifluorescence microscopy. Identical images are shown in the upper (green) and lower (blue) panels for BVP22-BFP- and HVP22-BFP-cotransfected cells using corresponding filter sets. Scale bar, 5 μm.
DISCUSSION
Our research demonstrates that BHV-1 tegument protein VP22, like its HSV-1 homolog, exhibits unusual functional characteristics including microtubule association, nuclear localization, and nonclassical intercellular trafficking (5); however, significant trait differences between the homologs exist. The microtubule association pattern within transfected populations of cells is 10 times rarer in BVP22-transfected cells than in HVP22-transfected cells. Nuclear association shows a marbled pattern for BVP22 and a speckled pattern for HVP22. When nuclear membrane is lysed, BVP22 stains in a crescent moon pattern whereas HVP22 stains prominently with nucleoli. BVP8 does not alter the intracellular localization of BVP22 but does alter HVP22 intracell localization by partially sequestering HVP22 into perinuclear spheres. Observations made with costained phosphohistone H3 may indicate that BVP22 specifically binds condensed chromatin at an earlier stage in mitosis than does HVP22. Finally, BVP22 traffics intercellularly to neighboring cell nuclei more efficiently than HVP22. These trait differences of the VP22 homologs suggest functional differences between BVP22 and HVP22 within BHV-1 or HSV-1 infection.
Interestingly, the BVP22 gene is not considered an essential BHV-1 survival gene (17), but the HVP22 gene is essential for HSV-1 replication (12). A BVP22-deleted BHV-1 mutant was capable of replication in cell culture although at a significantly reduced yield (17). Nevertheless, infection with this BVP22-deleted BHV-1 mutant was unable to induce clinical disease, nor was there any viral shedding (16). Hence, BVP22 is an important virulence factor. The difference in VP22 status as being either essential or nonessential for HSV-1 or BHV-1 maturation implies that these VP22 homologs have different functional properties despite the general similarities reported here. In fact, the observed variations within the overall similarities between BVP22 and HVP22 could help further define their respective roles in herpesviral pathogenesis.
Within each of the categories of VP22 properties, i.e., microtubule association, nuclear localization, and nonclassical intercellular trafficking, we show that BVP22 has particular traits distinct from those of HVP22. The unique traits of the VP22 homologs can be attributed to their relatively low amino acid sequence homology of 28.7%. In addition, using algorithms such as PROSITE II and MotifFinder (GenomeNet World Wide Web server; revision date, 1 April 1999; MOTIF program; Institute for Chemical Research, Kyoto University; Human Genome Center, Institute of Medical Science, University of Tokyo; http://www.genome.ad.jp/; last date accessed, 30 December 1999) made it evident that there were differences between BVP22 and HVP22 motifs. Studies with HVP22 have demonstrated phosphorylation by casein kinase II (CKII) and an unidentified cellular kinase (9, 10). This phosphorylation has been implicated in the dissociation of HVP22 from the tegument (21). Though similar studies were not performed with BVP22, MotifFinder identified the CKII sequence as well as other cellular kinase consensus sequences. Whether these sites have a functional effect is not known. The posttranslationally modified state of the VP22 homologs can undoubtedly affect the function and intracellular localization of VP22 (21, 23). The “tight” association of BVP22 and phosphorylated histone H3, contrasted with a seemingly “loose” association of HVP22 and phosphohistone H3, could be due to a difference in the phosphoregulation or to some other modification of the two VP22 homologs. Further, BVP8, the homolog of HVP13/14, is a protein kinase (24) that does not alter the intracellular localization of BVP22 when coexpressed in a cell. However, BVP8 is able to partially sequester HVP22. Finally, mutation of tyrosine kinase phosphorylation sites in BVP22 alters the pattern of microtubule association in BVP22-transfected cells (X. Ren, unpublished data). Additional mutation studies will help define the varied characteristics of both VP22 homologs. The PROSITE II algorithm indicates that HVP22 has two classic nuclear localization signals, whereas BVP22 has none. In fact, one HVP22 nuclear localization signal, pat4 beginning at amino acid 295, was within the carboxyl-terminal 34 residues of an HVP22 mutant for which only a filamentous cytoplasm pattern was found; the signal was not found in the nuclei of surrounding cells (5). Aligning this HVP22 carboxyl-terminal 34-amino-acid region with BVP22 resulted in 8.9% identity (GeneStream II website; copyright 1999; ALIGN program; Institut de Génétique Humaine, Montpellier, France; http://xylian.igh.cnrs.fr/; last date accessed, 30 December 1999). However, a stretch of six amino acids, including the pat4 nuclear localization signal of HVP22, aligned partially (four of six amino acids) with BVP22 at position 234 (data not shown). Both the pat4 nuclear localization signal of HVP22 and the related site of BVP22 will be targets for future mutation studies to help identify the nuclear localization signals in both VP22 homologs.
Fluorescence microscopy of the VP22 homologs fused to GFP transfected into cell lines revealed fascinating contrasts between BVP22 and HVP22. Although both homologs could associate with host microtubules, reorganizing them into thick bundles, HVP22-transfected cells had a much greater propensity for this filamentous cytoplasmic pattern. Notably, few filamentous BVP22-transfected cells were evident, whereas filamentous HVP22-transfected cells were easily located. Others have asserted that filamentous cytoplasmically stained cells are VP22-transfected cells while nucleus-stained cells are VP22 trafficked cells (6). Our observations concur that trafficked cells display only VP22 nuclear staining. However, our cotransfection studies clearly showed that BVP22-expressing cells can also have a nuclear-only staining pattern. We speculate that some BVP22-transfected cells display filamentous cytoplasmic and nuclear patterns, while most BVP22-transfected cells display a nuclear pattern due to posttranslational modification differences. The various modified states of BVP22 are possibly regulated by the host cell growth phase. The number of BVP22-transfected cells with filamentous staining could be increased by cotransfecting BVP22 with the BHV-1 transcription enhancer, BICP0 (unpublished observation). BICP0 may up-regulate many cellular genes along with BVP22 (26) and may indirectly alter BVP22. Microtubule reorganization is important for viral exocytosis (2) and for transport of HSV-1 capsids to the nucleus (25). Whether these functions are mediated by HVP22 is not known, and whether there are similar roles for VP22 in BHV-1 infection is also undetermined.
The extent of the cytoplasmic filamentous pattern in BVP22 (2%) compared to that in HVP22 (20%) does not affect nuclear trafficking to surrounding cells. BVP22 and HVP22 can traffic from expressing, transfected cells to nontransfected cells, where they localize to the nucleus. Our data independently confirm HVP22-GFP trafficking, which had been questioned by others (11). Besides being the first to report BVP22 trafficking, we now confirm VP22-GFP transport in living cells (3, 29). Previous studies had described HVP22-GFP transport utilizing methanol-fixed cells (7). In fact, others could not detect HVP22-GFP trafficking in living cells, only in fixed cells, and hypothesized that this difference resulted from a concentration effect or removal of interfering components (1, 7). However, our detection system may be more sensitive through use of a yellow (EYFP) variant of GFP and different cell lines that may induce higher expression from the cytomegalovirus promoter. Methanol treatment of cells results in a gradient in VP22-GFP staining in nontransfected-cell nuclei around the transfected cell. Interestingly, this gradient effect is seen only in cells transfected with VP22-GFP and not in cells transfected with GFP. Methanol fixation of GFP-transfected cells does not change the GFP staining pattern. This observation has been confirmed by others (29). The spread of VP22 to surrounding cell nuclei following methanol fixation of a transfected-cell monolayer was similar to results observed with the cell membrane disruption buffer (Fig. 7). However, cell membrane disruption buffer treatment of GFP-transfected cells resulted in complete loss of GFP cell staining (data not shown). We did not observe this methanol-induced VP22 gradient effect using paraformaldehyde fixation. Others have also noted that, under methanol fixation and permeabilization, VP22 seeps out of infected cells and is retained in the nuclei of adjacent cells and that this seepage is not observed with paraformaldehyde fixation (23). Hence, VP22 spread using methanol-fixed cells does not represent VP22 trafficking. In fact, we found that methanol fixation of VP22-GFP-transfected cells results in nuclear staining of every cell in the monolayer. To eliminate fixation artifacts, our VP22 trafficking data were obtained from counts of living cells. We did not see VP22 trafficking to every cell in the monolayer in living cells as is seen in methanol-fixed cells. Thus, the ratio of VP22-GFP to GFP for methanol-fixed transfected cells would be much greater than the same ratio for living transfected cells depending on transfection efficiency and the time after transfection that trafficking was assayed. In vivo trafficking of BVP22 and HVP22 was seen in all cell lines we tested (CCF-STTG1, D17, F17, HeLa, MDBK, NMU, NXS2, and Vero), suggesting that transport of both BVP22 and HVP22 is not restricted to certain tissues. Others have also demonstrated HVP22-GFP trafficking in cell lines of various tissues and species (29). HVP22 has been considered a potent biotherapeutic delivery agent for cancer suicide gene therapy (4, 22). Our studies comparing VP22s of BHV-1 and HSV-1 indicate that distinctions within BVP22 could make it a more effective therapeutic transporter than HVP22. Future BVP22 and HVP22 comparison studies should identify the nonclassical import and export motif(s) involved in VP22 trafficking.
Although both VP22 homologs bound chromatin at various stages of mitosis, qualitative differences in the nuclear association of BVP22 and that of HVP22 were observed. The speckled appearance of HVP22-labeled nuclei, resembling nuclear pores, disappeared upon nuclear membrane lysis, illuminating a nucleolus binding pattern. In contrast, the marbled pattern of BVP22 remained after nuclear membrane lysis; however, a conspicuous halo around BVP22-labeled nuclei appeared. The significance of these nuclear labeling distinctions may reflect observed VP22 nuclear localization differences between HSV-1 and BHV-1 infection. Vero cells infected with VP22-GFP-expressing HSV-1 exhibited cytoplasmic, perinuclear HVP22 staining in a Golgi apparatus-like pattern (8). In contrast, MDBK cells infected with BHV-1 exhibited primarily nuclear localization of VP22 (17).
Our findings demonstrating trait differences between VP22 homologs of BHV-1 and HSV-1 provide valuable information to help determine the specific role each homolog has in BHV-1 or HSV-1 maturation.
ACKNOWLEDGMENTS
This work was supported by a grant from the Robert Draper Technology Innovation Fund of the University of Wisconsin-Madison to J.S.H. and by USDA grant 99-35204-7933.
REFERENCES
- 1.Aints A, Dilber M S, Smith C I E. Intercellular spread of GFP-VP22. J Gene Med. 1999;1:275–279. doi: 10.1002/(SICI)1521-2254(199907/08)1:4<275::AID-JGM44>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 2.Avitabile E, Di Gaeta S, Torrisi M R, Ward P L, Roizman B, Campadelli-Fiume G. Redistribution of microtubules and Golgi apparatus in herpes simplex virus-infected cells and their role in viral exocytosis. J Virol. 1995;69:7472–7482. doi: 10.1128/jvi.69.12.7472-7482.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Derer W, Easwaran H P, Knopf C W, Leonhardt H, Cardoso M C. Direct protein transfer to terminally differentiated muscle cells. J Mol Med. 1999;77:609–613. doi: 10.1007/s001099900036. [DOI] [PubMed] [Google Scholar]
- 4.Dilber M S, Phelan A, Aints A, Mohamed A J, Elliott G, Edvard-Smith C I, O'Hare P. Intercellular delivery of thymidine kinase prodrug activating enzyme by the herpes simplex virus protein, VP22. Gene Ther. 1999;6:12–21. doi: 10.1038/sj.gt.3300838. [DOI] [PubMed] [Google Scholar]
- 5.Elliott G, O'Hare P. Intercellular trafficking and protein delivery by a herpesvirus structural protein. Cell. 1997;88:223–233. doi: 10.1016/s0092-8674(00)81843-7. [DOI] [PubMed] [Google Scholar]
- 6.Elliott G, O'Hare P. Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J Virol. 1998;72:6448–6455. doi: 10.1128/jvi.72.8.6448-6455.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott G, O'Hare P. Intercellular trafficking of VP22-GFP fusion proteins. Gene Ther. 1999;6:149–151. doi: 10.1038/sj.gt.3300850. [DOI] [PubMed] [Google Scholar]
- 8.Elliott G, O'Hare P. Live-cell analysis of a green fluorescent protein-tagged herpes simplex virus infection. J Virol. 1999;73:4110–4119. doi: 10.1128/jvi.73.5.4110-4119.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott G, O'Reilly D, O'Hare P. Phosphorylation of the herpes simplex virus type 1 tegument protein VP22. Virology. 1996;226:140–145. doi: 10.1006/viro.1996.0638. [DOI] [PubMed] [Google Scholar]
- 10.Elliott G, O'Reilly D, O'Hare P. Identification of phosphorylation sites within the herpes simplex virus tegument protein VP22. J Virol. 1999;73:6203–6206. doi: 10.1128/jvi.73.7.6203-6206.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fang B, Xu B, Koch P, Roth J A. Intercellular trafficking of VP22-GFP fusion proteins is not observed in cultured mammalian cells. Gene Ther. 1998;5:1420–1424. doi: 10.1038/sj.gt.3300741. [DOI] [PubMed] [Google Scholar]
- 12.Fawl R L, Roizman B. The molecular basis of herpes simplex virus pathogenicity. Semin Virol. 1994;5:267–271. [Google Scholar]
- 13.Huang C C, Herr W. Differential control of transcription by homologous homeodomain coregulators. Mol Cell Biol. 1996;16:2967–2976. doi: 10.1128/mcb.16.6.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laboissière S, Trudel M, Simard C. The bovine herpesvirus type 1 major tegument protein VP8 expressed in recombinant vaccinia virus does not induce significant immunity in mice. Virus Res. 1996;40:191–198. doi: 10.1016/0168-1702(95)01273-7. [DOI] [PubMed] [Google Scholar]
- 15.Leslie J, Rixon F J, McLauchlan J. Overexpression of the herpes simplex virus type 1 tegument protein VP22 increases its incorporation into virus particles. Virology. 1996;220:60–68. doi: 10.1006/viro.1996.0286. [DOI] [PubMed] [Google Scholar]
- 16.Liang X, Chow B, Babiuk L A. Study of immunogenicity and virulence of bovine herpesvirus 1 mutants deficient in the UL49 homolog, UL49.5 homolog and dUTPase genes in cattle. Vaccine. 1997;15:1057–1064. doi: 10.1016/s0264-410x(97)00008-x. [DOI] [PubMed] [Google Scholar]
- 17.Liang X, Chow B, Li Y, Raggo C, Yoo D, Attah-Poku S, Babiuk L A. Characterization of bovine herpesvirus 1 UL49 homolog gene and product: bovine herpesvirus 1 UL49 homolog is dispensable for virus growth. J Virol. 1995;69:3863–3867. doi: 10.1128/jvi.69.6.3863-3867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGeoch D J, Dalrymple M A, Davison A J, Dolan A, Frame M C, Mcnab D, Perry L J, Scott J E, Taylor P. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69:1531–1574. doi: 10.1099/0022-1317-69-7-1531. [DOI] [PubMed] [Google Scholar]
- 19.Meredith D M, Lindsay J A, Halliburton I W, Whittaker G R. Post-translational modification of the tegument proteins (VP13 and VP14) of herpes simplex virus type 1 by glycosylation and phosphorylation. J Gen Virol. 1991;72:2771–2775. doi: 10.1099/0022-1317-72-11-2771. [DOI] [PubMed] [Google Scholar]
- 20.Morrison E E, Stevenson A J, Meredith D M. Differences in the intracellular localization and fate of herpes simplex virus tegument proteins early in the infection of Vero cells. J Gen Virol. 1998;79:2517–2528. doi: 10.1099/0022-1317-79-10-2517. [DOI] [PubMed] [Google Scholar]
- 21.Morrison E E, Wang Y, Meredith D M. Phosphorylation of structural components promotes dissociation of the herpes simplex virus type 1 tegument. J Virol. 1998;72:7108–7114. doi: 10.1128/jvi.72.9.7108-7114.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Phelan A, Elliott G, O'Hare P. Intercellular delivery of functional p53 by the herpesvirus protein VP22. Nat Biotechnol. 1998;16:440–443. doi: 10.1038/nbt0598-440. [DOI] [PubMed] [Google Scholar]
- 23.Pomeranz L E, Blaho J A. Modified VP22 localizes to the cell nucleus during synchronized herpes simplex virus type 1 infection. J Virol. 1999;73:6769–6781. doi: 10.1128/jvi.73.8.6769-6781.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwyzer M, Ackermann M. Molecular virology of ruminant herpesviruses. Vet Microbiol. 1996;53:17–29. doi: 10.1016/s0378-1135(96)01231-x. [DOI] [PubMed] [Google Scholar]
- 25.Sodeik B, Ebersold M W, Helenius A. Microtubule-mediated transport of incoming herpes simplex virus 1 capsids to the nucleus. J Cell Biol. 1997;136:1007–1021. doi: 10.1083/jcb.136.5.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steinmann N A, Nuñez R, Köppel R, Ackermann M. Construction and characterization of a stably transformed HeLa cell line in which the expression of bovine herpesvirus 1 ICP0 (BICP0) is induced by tetracycline. Arch Virol. 1998;143:35–48. doi: 10.1007/s007050050266. [DOI] [PubMed] [Google Scholar]
- 27.Van Drunen Littel-Van Den Hurk S, Garzon S, Van Den Hurk J V, Babiuk L A, Tussen P. The role of the major tegument protein VP8 of bovine herpesvirus-1 in infection and immunity. Virology. 1995;206:413–425. doi: 10.1016/s0042-6822(95)80057-3. [DOI] [PubMed] [Google Scholar]
- 28.Wei Y, Yu L, Bowen J, Gorovsky M A, Allis C D. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- 29.Wybranietz W A, Prinz F, Spiegel M, Schenk A, Bitzer M, Gregor M, Lauer U M. Quantification of VP22-GFP spread by direct fluorescence in 15 commonly used cell lines. J Gene Med. 1999;1:265–274. doi: 10.1002/(SICI)1521-2254(199907/08)1:4<265::AID-JGM48>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 30.Zhang Y, Sirko D A, McKnight J L C. Role of herpes simplex virus type 1 UL46 and UL47 in αTIF-mediated transcriptional induction: characterization of three viral deletion mutants. J Virol. 1991;65:829–841. doi: 10.1128/jvi.65.2.829-841.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]