Abstract
An immunodominant antigen, p35, is expressed on the envelope of intracellular mature virions (IMV) of vaccinia virus. p35 is encoded by the viral late gene H3L, but its role in the virus life cycle is not known. This report demonstrates that soluble H3L protein binds to heparan sulfate on the cell surface and competes with the binding of vaccinia virus, indicating a role for H3L protein in IMV adsorption to mammalian cells. A mutant virus defective in expression of H3L (H3L−) was constructed; the mutant virus has a small plaque phenotype and 10-fold lower IMV and extracellular enveloped virion titers than the wild-type virus. Virion morphogenesis is severely blocked and intermediate viral structures such as viral factories and crescents accumulate in cells infected with the H3L− mutant virus. IMV from the H3L− mutant virus are somewhat altered and less infectious than wild-type virions. However, cells infected by the mutant virus form multinucleated syncytia after low pH treatment, suggesting that H3L protein is not required for cell fusion. Mice inoculated intranasally with wild-type virus show high mortality and severe weight loss, whereas mice infected with H3L− mutant virus survive and recover faster, indicating that inactivation of the H3L gene attenuates virus virulence in vivo. In summary, these data indicate that H3L protein mediates vaccinia virus adsorption to cell surface heparan sulfate and is important for vaccinia virus infection in vitro and in vivo. In addition, H3L protein plays a role in virion assembly.
Vaccinia virus is the prototypic member of the poxvirus family of large DNA viruses. Vaccinia virus replicates in the cytoplasm of infected cells and produces multiple forms of infectious particles: intracellular mature virions (IMV), intracellular enveloped virions (IEV), and extracellular enveloped virions (EEV) (1, 26, 58). Each form of vaccinia virion contains different membrane structures and is capable of using different routes for infection (1, 40, 52).
IMV is the most abundant virion form, and many of its surface envelope proteins have been identified (30, 57). Some of these envelope proteins play a role in virus entry into the cell. For example, A27L protein binds to cell surface heparan sulfate (HS) and is required for fusion of virus-infected cells (9, 17, 22, 43, 44). In addition, A27L protein is associated with A17L protein and is important for Golgi membrane wrapping of IMV during formation of IEV and EEV (46, 48). Inactivation of A27L protein expression resulted in a small plaque phenotype and reduced EEV production (44). Another envelope protein, D8L, binds to cell surface chondroitin sulfate (CS); inactivation of D8L expression reduces IMV binding to cells in vitro and attenuates virus virulence in mice (23, 34–36). Another envelope protein, L1R, is important for virus penetration, since a monoclonal antibody (MAb) recognizing L1R protein blocked virus entry at a postbinding step (27, 28, 41, 63).
The product of the H3L gene, p35, is another envelope protein that is an immunodominant antigen found on vaccinia IMV (66). Strong immune responses to p35 protein have been detected in mice, sheep, rabbits, and humans (6, 21, 62). Amino acid sequencing of p35 purified from the LIVP strain of vaccinia virus revealed that it was encoded by the H3L gene (50, 66). The H3L gene is present in the genome of different strains of vaccinia virus as well as in sheep poxvirus, variola, and orf viruses (6, 16, 20, 50, 53). Despite its wide presence in poxvirus family viruses, the function of H3L protein in the virus life cycle is not known.
Glycosaminoglycans (GAGs) are ubiquitously expressed in many cell types. Many viruses, such as herpesvirus, dengue virus, and foot-and-mouth disease virus, bind to GAGs during virus infections (7, 29, 54). Although vaccinia virus binds to cell surface GAGs through A27L and D8L proteins, other viral envelope proteins may contribute to virion binding and penetration into various cell types (9, 22, 23). This study investigates the role(s) of H3L protein in virus attachment to mammalian cells and in the life cycle of vaccinia virus. A H3L− mutant virus was constructed and characterized. The properties of the H3L− mutant virus indicate that H3L protein is important for virus infectivity in vitro and in vivo as well as for virus morphogenesis.
MATERIALS AND METHODS
Reagents and viruses.
Soluble heparin (HP), CS, and dermantan sulfate (DS) were purchased from Sigma Inc. Spurr resin was purchased from Electron Microscopy Sciences. Wild-type (WT) vaccinia virus (WR strain) was grown in BSC40 cells. VMJ360, a recombinant vaccinia virus that expresses lacZ from a synthetic early promoter, was obtained from B. Moss (12). IMV were purified by centrifugation of cell lysates through a 25 to 40% sucrose gradient, and the pellets were stored for use as virus stocks (31). EEV were collected from fresh media of the infected cell cultures and used directly without freezing.
Protein expression and purification.
Full-length recombinant H3L protein was used as an antigen to generate rabbit antisera. The full-length H3L gene was amplified by PCR with two primers, as follows: a 5′ primer (5′-ATAGGATCCATGGCGGCGGC-3′) and a 3′ primer (5′-TCAAAGCTTAGATAAATGCGGTAACG-3′). The PCR product was cloned into the pVE02 expression vector fused with a six-histidine tag at the N terminus. Full-length H3L protein was expressed in Escherichia coli BL21(DE3)/pLysS and was purified in the presence of Triton X-100 as described previously (20).
The recombinant ectodomain region of H3L protein was expressed for biological assays. The H3L gene region encoding the soluble ectodomain was amplified with the following two PCR primers: a 5′ primer (5′-ATAGGATCCACATTTCCTAATGTTCAT-3′) and a 3′ primer (5′-GTGAAGCTTTCCTGGATAACGTTTAGC-3′). The DNA fragment was digested with BamHI and HindIII and cloned into pET21a (Novagen). The resulting plasmid expressed an ectodomain of H3L protein from amino acids (aa) 21 to 270 fused with a T7 tag peptide at the N terminus and six-histidine sequences at the C terminus, as previously described (9, 22). This plasmid was transformed into E. coli BL21(DE3), and the transformant was used for expression of soluble H3L ectodomain protein. Cultures were induced with 0.2 mM isopropyl-β-d-thiogalactoside (IPTG) for 30 min at 37°C; cells were harvested, resuspended, and lysed by sonication; the lysate was centrifuged; and the supernatant was loaded onto a nickel column and purified as described previously (22).
Rabbit antisera and neutralization assays.
Two New Zealand White rabbits were inoculated intramuscularly with 250 μg of the purified recombinant full-length H3L protein in Freund's incomplete adjuvant, boosted 3 and 5 weeks after the first inoculation, and bled 1 week after the second boost. The immune response of the animals was checked by Western immunoblot analysis.
For neutralization assays, preimmune or postimmune sera (B&C) at various dilutions were mixed with 200 PFU of WT vaccinia virus at 4°C for 30 min. The mixtures were added to BSC40 cells in 60-mm-diameter dishes in duplicate and incubated at 37°C for 1 h. Cells were washed with phosphate-buffered saline (PBS) and overlaid with 1% agar. Plaque numbers were determined after 3 days. Control plaque numbers were between 100 and 150 plaques per 60-mm-diameter dish. The percent plaque formation was determined by the following calculation: [(number of plaques in the presence of antiserum)/(number of plaques in the absence of antiserum)] × 100.
Biotinylation of soluble H3L protein.
Biotinylation of soluble ectodomain of H3L protein was performed with an ECL biotinylation system from Amersham Life Science, Inc. In brief, 1 mg of purified recombinant H3L ectodomain protein was mixed with 40 μl of biotinylation reagent N-hydroxysuccinamide ester in 40 mM bicarbonate buffer (pH 8.6) at room temperature for 1 h according to the manufacturer's instructions. The mixture was loaded on a 9-ml Sephadex G25 column previously equilibrated with PBS. Biotinylated H3L protein was collected in 500-μl aliquots from fractions 5 to 9, and the extent of biotinylation was confirmed by Western blot analysis with alkaline phosphatase-conjugated streptavidin, as described previously (9).
Cell binding assays.
BSC40 cells (106) were washed with cold PBS and incubated with biotinylated H3L protein (10 μg/100 μl) in staining medium (PBS–4% fetal bovine serum–10 mM HEPES [pH 7.2]). In some experiments, GAGs (50 μg/ml) were also added as competitors. The mixture was incubated at 4°C for 60 min with gentle rotation. Cells were washed with cold staining medium, and phycoerythrin-conjugated streptavidin (1:100) was added for another 60 min at 4°C. Cells were washed three times with cold PBS and analyzed with a fluorescence-activated cell sorter (FACS) (excitation, 488 nm; emission, 578 nm) as described previously (22).
Virion binding assays.
To test if the soluble ectodomain of H3L protein blocks vaccinia virus infection at the binding step, 4 × 105 BSC40 cells were incubated with various amounts of soluble H3L protein (0, 1, 10, or 50 μg in 300 μl) at 4°C for 30 min. The cultures were subsequently infected with WT vaccinia virus at a multiplicity of infection (MOI) of 5 PFU per cell at 4°C for 30 min. After washing, cells were immediately harvested for plaque assays on BSC40 cells. The number of plaques was 100 to 150 per 60-mm-diameter dish. The percent inhibition was determined by the following calculation: [1 − (number of plaques in the presence of H3L protein)/(number of plaques in the absence of H3L protein)] × 100.
Viral early gene expression was measured in BSC40 cells infected with vMJ360 that expresses lacZ from a synthetic early promoter at a MOI of 5 PFU per cell (12). The infected cells were harvested at 2 h postinfection (p.i.) for β-galactosidase (β-gal) assays as described previously (22).
Construction of H3L− mutant vaccinia virus.
The full-length H3L gene was amplified by PCR using a 5′ primer (5′-AATAAGCTTATGGCGGCGGTGAAAACT-3′) and a 3′ primer (5′-CTCGGATCCTTAGATAAATGCGGTAAC-3′) as described previously (16). The PCR product was digested with BamHI and HindIII and cloned into pBluescript KS (Stratagene), resulting in pBS-H3L. A lacZ expression cassette was obtained from pSC11-5 by restriction digestion with SalI and PstI (5). The lacZ cassette was blunt-ended and cloned into an EcoRV site within the H3L region of pBS-H3L so that the lacZ cassette insertionally inactivated the H3L gene.
The resulting plasmid, pBS-H3L/lacZ, was transfected into CV-1 cells which were infected with WT vaccinia virus at an MOI of 0.1 PFU per cell. The lysates were harvested after 3 days, and virus was titered on agar containing 5-bromo-4-chloro-3-indolyl-β-d-glucoronic acid (X-Gal) (150 μg/ml). Pure recombinant H3L− virus expressing β-gal was isolated after three rounds of plaque purification. To confirm that the mutant virus contained only a single insertion, two independent H3L− virus isolates were characterized. Viral DNA was purified from these isolates, and restriction digestions were performed. The results indicated that the lacZ gene was inserted into the H3L locus in both isolates and that both isolates had identical restriction patterns. Subsequent experiments were carried out with one isolate.
H3L protein expression was examined in IMV (2 × 106 PFU) and cell lysates infected with WT and H3L− mutant viruses. Protein samples were separated by sodium dodecyl sulfate (SDS)–12% polyacrylamide gel electrophoresis (PAGE) and transferred for Western blot analysis with antiserum B (1:1,000) against H3L protein.
Cell fusion assay induced by low pH treatment.
BSC40 cells were infected with WT or H3L− mutant virus at an MOI of 5 PFU per cell and incubated at 37°C for 24 h. Cells were washed three times with PBS (pH 7.2), treated with PBS (pH 4.8) for 3 min at room temperature, and washed again, and then the PBS was replaced with normal medium. These cells were incubated for another 3 h and photographed with a Nikon inverted microscope.
Pulse-chase labeling for viral protein synthesis and proteolytic processing of P4a and P4b core proteins.
BSC40 cells were infected with WT or H3L− mutant virus at an MOI of 10 PFU per cell, and the cultures were incubated at 37°C. At 8 h p.i. viral protein synthesis was monitored with 35S-methionine (50 μCi/ml) for 30 min and chased with normal growth medium for various times (0, 15, and 30 min and 1, 2, 4, 12, and 24 h). At each time one set of samples was harvested, lysed in SDS-containing buffer, separated by SDS–10% PAGE, and analyzed by autoradiograms as described previously (60).
Electron microscopy of virus morphogenesis and virion particle determination.
BSC40 cells were seeded on round coverslips and infected with WT or H3L− mutant virus at an MOI of 20 PFU per cell. These cells were directly fixed on coverslips at 24 h p.i. in 2.5% glutaraldehyde in 0.1 M sodium phosphate saline buffer (pH 7.2) at room temperature for 1 h and rinsed in three 15-min changes of the same buffer. Cells were then treated with 1% OsO4 in 0.1 M sodium phosphate (pH 7.2) at room temperature for 60 min and subsequently washed three times in the same buffer. Cells were dehydrated using an ethanol series, and Spurr's resin was used for infiltration and embedding, as described previously (55). After embedding, cells were separated from coverslips and used for thin sectioning with an Ultracut ultramicrotome. Thin sections of 90 nm were stained with uranyl acetate and lead citrate and analyzed with a Zeiss 902 transmission electron microscope (42).
For negative staining of IMV, 1 μl of serially diluted purified vaccinia IMV was spotted onto 300-mesh parlodion-coated grids and stained with 2% phosphotunstic acid for 15 s. The total number of particles was determined with a Zeiss 902 transmission electron microscope, as previously described (19).
Assays for virulence.
Male BALB/c mice 5 to 6 weeks old were individually weighed on a daily basis for a week prior to infection. They were anesthetized and infected intranasally with 5 μl of diluted IMV for infection with 105, 106, and 107 PFU per mouse. Each day, mice were individually weighed and monitored for signs of illness. The average weight change of the five mice in each group was calculated.
RESULTS
Antisera against H3L protein neutralize vaccinia virus IMV infection, and soluble H3L protein blocks vaccinia virus IMV adsorption to cells.
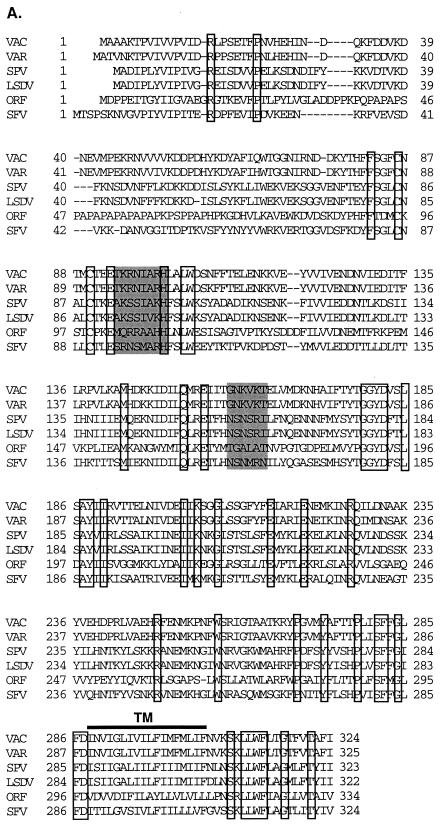
The H3L gene of vaccinia virus encodes a protein of 324 aa with a potential transmembrane region close to the C terminus (21). The H3L gene is conserved among poxviruses (Fig. 1A). An alignment of known vaccinia H3L gene and its homologues in other poxviruses is shown in Fig. 1A, which reveals highly conserved regions. The middle region, from aa 80 to 286, is more conserved than the N-terminal region, from aa 1 to 80. Two cysteine residues at positions 86 and 90 are identical in all poxviruses. Furthermore, two short regions, aa 94 to 101 and aa 159 to 164, have sequences similar to the consensus GAG-binding motifs (X-B-B-B-X-X-B-X and X-B-B-X-B-X, where B is a basic residue and X is a hydropathic residue) (4). Since H3L protein is expressed on the IMV envelope, the presence of potential GAG-binding sites suggests a role for H3L protein in virus adsorption to cells during infection (20). Experiments described below explore this possibility.
FIG. 1.
(A) Alignment of amino acid sequences deduced from vaccinia virus H3L gene and its homologues in poxvirus family. VAC, vaccinia virus H3L gene (SP/P07240); VAR, variola virus (IND3, isolated in India in 1967) immunodominant envelope protein P35 gene (accession no. sp/P33059) (53); SPV, sheep poxvirus P32 antigen gene (accession no. gb/AF124517) (20); LSDV, lumpy skin disease virus P32 antigen gene (accession no. gb/AF124516) (20); ORF, orf virus (strain NZ2) immunodominant envelope antigen F1L gene (accession no. gi/AF097215) (21); and SFV, shope fibroma virus S071L gene (D. H. Evans, personal communication). The boxed areas indicate identical amino acids, and the shaded areas are potential GAG-binding sites, as described previously (4). TM, transmembrane region. (B) Neutralization of vaccinia virus infection by antisera against H3L protein. BSC40 cells were infected with vaccinia virus in the absence or the presence of various dilutions of antisera, as indicated, and an agar overlay was added for plaque determination. The number of plaques obtained in the absence of sera, approximately 150 PFU per 60-mm-diameter dish, was used as the 100% value. (C) Purified soluble H3L protein (sH3L) stained by Coomassie blue after SDS–12% PAGE. Protein marker masses, in kilodaltons, are shown at the left side of the gel. (D) sH3L protein blocked viral early gene expression. BSC40 cells were infected with vMJ360 expressing lacZ at an MOI of 10 PFU per cell in the presence of various amounts of sH3L (0, 1, 10, or 50 μg) and harvested at 2 h p.i. for β-gal assays as described previously (12). Cells infected with vaccinia virus without sH3L protein were used as a control. (E) sH3L protein blocked vaccinia virus adsorption to cells. BSC40 cells were infected with vaccinia virus at an MOI of 10 PFU per cell at 4°C for 30 min. After washing, these cells were immediately harvested and cell-associated virions were determined by plaque assay on BSC40 cells (22). Controls were the same as described for panel D.
Antisera against a full-length recombinant H3L protein was prepared in rabbits and used to investigate the role of H3L protein during vaccinia virus infection (20). Antisera B and C were diluted serially and tested for neutralization during infections of BSC40 cells (Fig. 1B). At a 1:100 dilution sera B and C reduced plaque formation levels to 65 and 42%, respectively, of the level of the control without antiserum. The percent plaque formation was further reduced to 40 and 10% at 1:50 dilutions of antisera B and C, respectively. Preimmune serum had no effect. These results indicate that H3L protein is important for vaccinia virus infection in cell culture.
If H3L protein plays a role in vaccinia virus entry into the cell, then it is predicted that H3L protein might compete for cell surface binding sites and inhibit infection when present as a soluble factor. The full-length recombinant H3L protein is only soluble in the presence of 0.5% Triton X-100 and thus was not suitable for competition studies or other biological assays (20). To test the ability of H3L to compete with virus particles, a soluble recombinant form of H3L protein including the extracellular domain from aa 21 to 270 was prepared. The extracellular domain of H3L protein was fused with a T7 tag at the N terminus and a six-histidine tag at the C terminus for purification by nickel column chromatography, enabling purification to near homogeneity with only minor degradation (Fig. 1C). BSC40 cells were infected with virus in the presence or absence of soluble H3L protein, and expression of a viral early marker gene, lacZ, was determined at 2 h p.i. (Fig. 1D). lacZ expression was reduced in a dose-dependent manner when H3L protein was added. Furthermore, in the presence of soluble H3L protein, virion binding was significantly reduced (Fig. 1E). The inhibitory effect of H3L protein was not due to the T7 or the His tag sequences, since other proteins with identical tags had no effect on vaccinia virus binding (9, 22). These results indicate that H3L protein plays a role in IMV adsorption to cells.
Soluble H3L protein bound to cell surface HS.
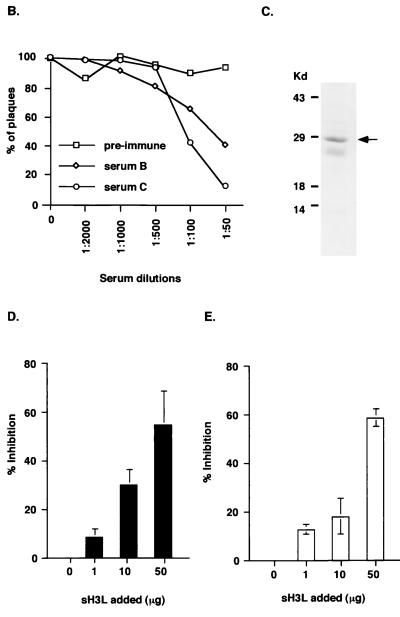
The above results suggest that H3L protein may bind to the cell surface. This idea was tested by incubating soluble biotinylated H3L protein with BSC40 cells and analyzing them with a FACS (Fig. 2). H3L protein bound to BSC40 cells, and a significant shift in cell surface staining was detected (red lines in Fig. 2A to C). Soluble GAGs were also added as competitors for H3L protein binding. HP at concentrations of 10 and 100 μg/ml reduced H3L protein binding to cells and shifted the fluorescence intensity more than 10-fold (Fig. 2A). Addition of HP at a concentration of 1,000 μg/ml did not reduce the fluorescence staining further, suggesting that 100 μg of GAGs per ml was adequate to saturate the available HP binding sites (data not shown). Addition of CS at concentrations of 10 and 100 μg/ml had no effect on H3L protein binding (Fig. 2B), whereas 10 μg of DS per ml (Fig. 2C) did not inhibit binding but 100 μg of DS per ml caused partial inhibition.
FIG. 2.
sH3L protein bound to cell surface HS. BSC40 (A to C), L (D), gro2C (E), or sog9 (F) cells were incubated with PBS (black line) or biotinylated H3L (red line) and analyzed with a FACS as described previously (22). Alternatively, as shown in panels A to C, biotinylated H3L protein was mixed with 10 μg/ml (results shown by green line) or with 100 μg/ml (results shown by blue line) of soluble GAGs and analyzed with a FACS. The soluble GAGs used in competitions were HS (A), CS (B), or DS (C).
Competition assays with soluble GAGs have been widely used in cell binding assays, but they are indirect measurements of cell surface interactions. To provide direct evidence of H3L protein interaction with cell surface HS, H3L protein binding to the cell surface was tested using three cell lines expressing different GAGs: mouse L cells and two mutant lines derived from L cells, gro2C and sog9 (2, 3, 18). Mouse L cells express both HS and CS on the cell surface, gro2C expresses only CS, and sog9 expresses neither HS nor CS (18). These mutant cells have been used for studies of herpes simplex virus type 1 gB and gC proteins and their interaction with cell surface GAGs (2, 3, 14). We also used them to demonstrate vaccinia virus D8L protein binding to cell surface CS (23).
When biotinylated H3L protein was incubated with L cells, a strong cell surface staining was detected (Fig. 2D). In contrast, H3L protein bound weakly to gro2C cells, indicating that HS is required on the cell surface for strong binding (Fig. 2E). H3L protein bound equally well to gro2C and sog9 cells, indicating that CS is not important for H3L protein interaction with the cell surface (Fig. 2E and F). The possibility that H3L protein binds to other non-GAG recognition elements on gro2C and sog9 cells cannot be completely excluded, because a low level of H3L protein staining (3 to 4% of the signal on L cells) was detected with these two cell lines. Nevertheless, these results suggest that HS is important for the H3L protein interaction with the cell surface. Earlier studies showed that vaccinia virus A27L and D8L proteins bind to cell surface HS and CS, respectively (8, 19). Thus, vaccinia virus IMV includes three envelope proteins that recognize cell surface GAGs: A27L and H3L proteins bind to HS, and D8L protein binds to CS.
Inactivation of H3L gene expression affects vaccinia virus growth in cell culture.
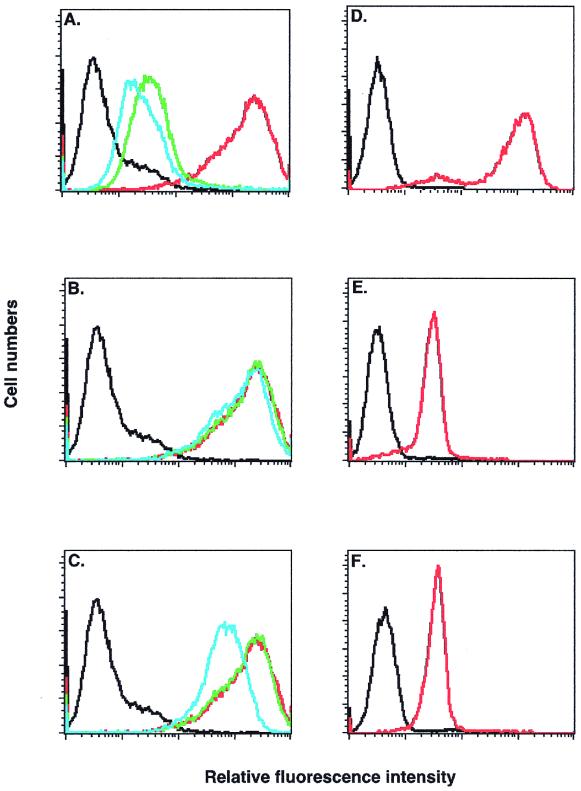
To investigate the biological importance of H3L protein, an H3L− mutant virus was constructed by homologous recombination, as described in Materials and Methods. The mutant had a lacZ cassette inserted into the H3L gene so that H3L protein synthesis was abolished. The H3L− mutant virus was viable, and a pure stock producing blue plaques was isolated after three rounds of plaque purification on BSC40 cells. Thus, the H3L gene is not essential for vaccinia virus plaque formation on BSC40 cells. However, the plaques formed by the H3L− mutant virus were smaller than plaques formed by the WT virus (Fig. 3A). Two independent mutant virus clones were isolated, and their behavior was comparable in cell culture. One isolate of the H3L− mutant virus was used for all subsequent experiments.
FIG. 3.
(A) H3L− mutant virus formed smaller plaques on BSC40 cells. BSC40 cells were infected with WT (left) or H3L− mutant virus (right) for plaque assay with neutral red as described above, and photographed at 3 days p.i. (B) Expression of H3L gene was inactivated in H3L− mutant virus. Purified virions from WT (lane 2) and H3L− mutant (lane 4) virus as well as infected cell lysates from WT (lane 3) and H3L− mutant (lane 5) virus were loaded onto a SDS–12% PAGE gel and transferred for Western blot analysis with antiserum B (1:1,000) against H3L protein. Mock lysates (lane 1) were used as a negative control. (C) H3L− mutant virus was resistant to antiserum C. BSC40 cells were infected with WT or H3L− mutant virus in the presence of various dilutions of an antiserum against H3L protein (serum C) or a MAb against L1R protein (2D5), as indicated at the bottom of the figures, followed by plaque determination (39). The number of plaques obtained in the absence of sera, around 150 PFU per 60-mm-diameter dish, was used as the 100% value.
H3L gene expression was characterized in H3L− mutant and WT virus using Western blot analyses of purified IMV and virus-infected cell lysates (Fig. 3B). A viral protein of 35 kDa was detected in purified virions and cell lysates from a WT virus infection (Fig. 3B, lanes 2 and 3), but no protein was detected in samples from an H3L− mutant virus infection (Fig. 3B, lanes 4 and 5), indicating that expression of H3L protein is completely interrupted in the H3L− mutant virus. Consistent with this fact, the H3L− mutant virus is resistant to neutralization by antibodies against H3L protein (Fig. 3C, top panel). The acquisition of resistance to these antibodies was specifically due to the absence of H3L protein on the virion, since antibody against other envelope proteins such as L1R neutralized H3L− mutant virus and WT virus to a similar extent (Fig. 3C, lower panel).
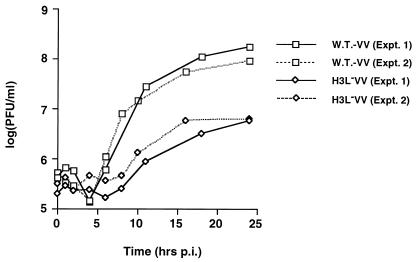
Growth of the H3L− mutant virus in cell culture was examined by one-step growth curve analysis (Fig. 4). BSC40 cells were infected with WT or H3L− mutant viruses at an MOI of 5 PFU per cell and harvested at various times to monitor IMV production. The titer of WT virus increased more than two log units, reaching 1 × 108 PFU/ml; in contrast, the titers for H3L− mutant virus only increased to 6 × 106 PFU/ml. H3L protein deficiency apparently reduced the yield of vaccinia virus IMV more than 10-fold. In addition, the titer of EEV produced during H3L− mutant virus infection was also reduced (Table 1).
FIG. 4.
One-step growth curve analysis of WT vaccinia virus (vv) and H3L− mutant virus. BSC40 cells were infected with WT and H3L− mutant viruses at an MOI of 5 PFU per cell and harvested at various times for plaque assays as described previously (22). Expt., experiment.
TABLE 1.
IMV and EEV titers of WT and H3L− mutant viruses after single-round infections in BSC40 cells
| Vaccinia virus | Experiment | IMV titer (PFU/ml)
|
EEV titer (PFU/ml) at 24 h | ||
|---|---|---|---|---|---|
| 0 h | 24 h | Fold increase after 24 h | |||
| WT | 1 | 7.9 × 105 | 1.8 × 108 | 228 | 6.7 × 103 |
| H3L− | 2.9 × 105 | 4.4 × 106 | 15 | 1 × 103 | |
| WT | 2 | 6.5 × 105 | 1.7 × 108 | 262 | 5 × 103 |
| H3L− | 1.4 × 105 | 3.3 × 106 | 23 | 2.6 × 102 | |
Inactivation of H3L gene expression affects virion morphogenesis.
There are two possible explanations for reduced virus titers during infection by the H3L− mutant virus. The H3L− mutant virus may be defective at some stage of the virus life cycle such as during viral gene expression or regulation or virion assembly. Alternatively, the H3L− mutant virus may grow well but produce virion particles of lower infectivity than WT virus. Viral gene expression was examined in cells infected by the H3L− mutant virus. BSC40 cells were infected with WT or H3L− mutant virus at an MOI of 5 PFU per cell, and viral protein synthesis was monitored at various times p.i. with 35S-methionine. Shutoff of host protein synthesis was evident at 8 h p.i., and the overall pattern of viral protein synthesis was comparable up to 24 h p.i. for H3L− mutant and WT virus (data not shown). This result indicates that the H3L gene is not important for viral gene expression or regulation.
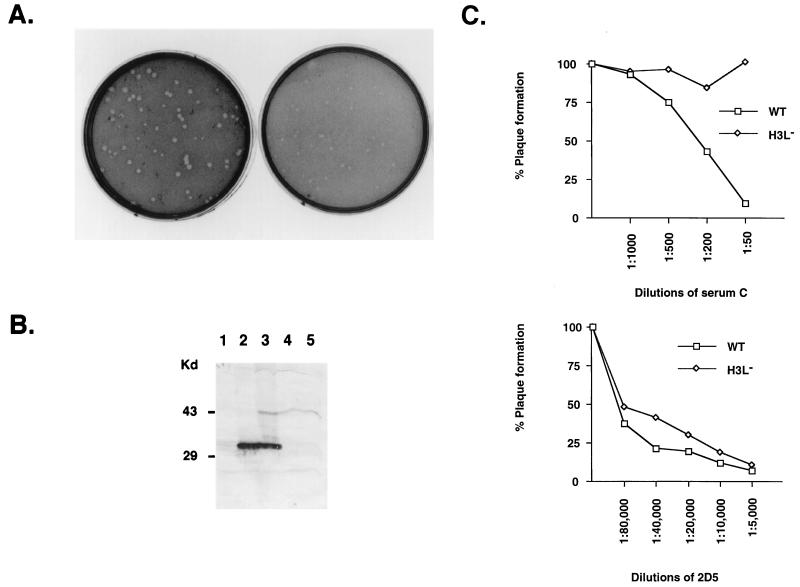
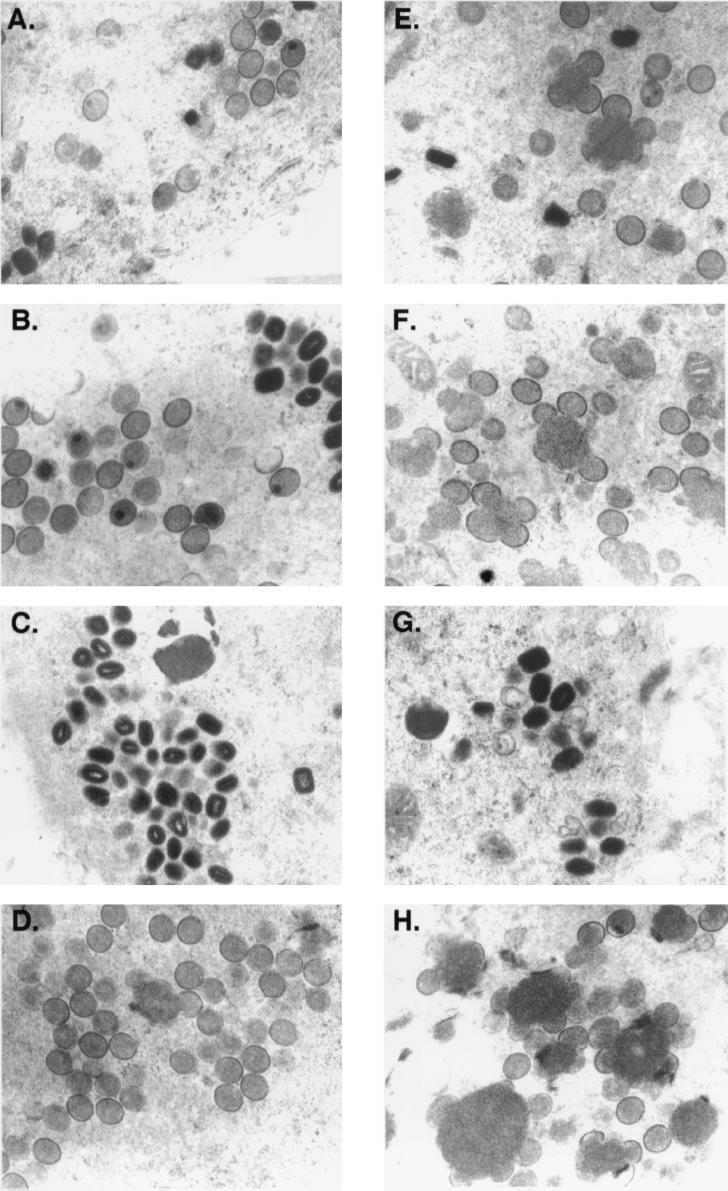
To investigate the role of H3L protein in virion morphogenesis, the intracellular virus structures in the infected cells were analyzed by electron microscopy (Fig. 5). At 12 h p.i., cells infected by WT virus or H3L− mutant virus contained mostly intermediate viral structures such as crescents associated with viral factories and immature virions (IV) (Fig. 5A and E) in the cytoplasm, with only a slight difference in the assembly process, i.e., cells infected by the mutant virus seemed to accumulate more nucleoprotein mass. However, at 24 h p.i., progression of virion assembly for WT and H3L− mutant virus was dramatically different. In cells infected by WT virus, IMV and some IV were present (Fig. 5B); in cells infected by mutant virus, there were more viral crescents associated with nucleoprotein mass and unclosed viral membrane structures were predominant (Fig. 5F). This difference became more pronounced at 48 h p.i., when most cells infected by WT virus contained more IMV (Fig. 5C), but the number of IMV in cells infected by mutant virus was greatly reduced (Fig. 5G). Few cells infected by WT virus still contained IV at 48 h p.i. (Fig. 5D), but most cells infected with H3L− mutant virus contained many viral crescents and IV (Fig. 5H). These data indicate that deficiency in H3L protein interrupts the IMV assembly process, most likely affecting IV conversion to IMV.
FIG. 5.
Electron micrographs of vaccinia virion morphogenesis. BSC40 cells were infected with WT (A to D) and H3L− mutant viruses (E to H), fixed at 12 h (A and E), 24 h (B and F), or 48 h (C, D, G, and H) p.i. and analyzed with a Zeiss 902 transmission electron microscope. Magnification, ×13,000.
Proteolytic cleavage of two viral core precursor proteins, P4a and P4b, into their mature forms, 4a and 4b, has been shown to coincide with the late morphological changes that occur during IV conversion to IMV (60). Processing of P4a and P4b proteins has been thought to be essential for the formation of IMV, since chemicals that inhibit such cleavages, directly or indirectly, blocked virion production (8, 33, 61). Inactivation of F18R, I7L, and L1R gene expression resulted in no core protein processing, and morphogenesis was blocked at IV conversion to IMV (15, 41, 65).
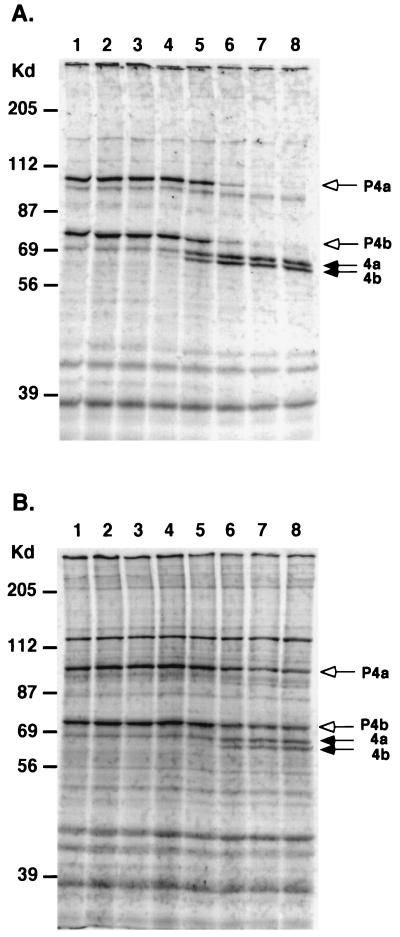
To determine whether proteolytic processing of these core proteins occurs in cells infected by H3L− mutant virus, BSC40 cells were infected by each virus, pulsed at 8 h p.i., and chased for periods of up to 24 h p.i. (Fig. 6). In cells infected by WT virus, the processing of P4a and P4b was initiated after 60 min of chasing time. Most of the precursor processing was accomplished after a 4-h chasing period (Fig. 6A). In cells infected by H3L− mutant virus, little processing was observed within the first 2-h chasing period. Cleavages of P4a and P4b proteins were observed after 4 h of chasing time, although the processing was only partial even after 24 h of chasing (Fig. 5B). Thus, in the absence of H3L protein the vaccinia virus life cycle proceeds beyond the core protein processing step and is arrested at a step downstream of where it is a arrested in F18R−, I7L−, and L1R− mutants.
FIG. 6.
Processing of P4a and P4b precursor core proteins in H3L− mutant virus. BSC40 cells were infected with WT (A) or H3L− mutant virus (B) at an MOI of 10 PFU per cell, and the cultures were pulsed with 35S-methionine (50 μCi/ml) for 30 min for viral late protein synthesis (lane 1) and chased with normal growth medium for various times, as follows: 15 min (lane 2), 30 min (lane 3), 1 h (lane 4), 2 h (lane 5), 4 h (lane 6), 12 h (lane 7), and 24 h (lane 8). At each time one set of samples was harvested, lysed in SDS-containing buffer, separated on a SDS–10% PAGE gel, and analyzed by autoradiography. The white arrows mark the positions of two precursor core proteins, P4a and P4b, and the black arrows indicate the processed forms of core proteins 4a and 4b as described previously (60).
Interruption of IMV morphogenesis significantly reduced the number of IMV during infection with the H3L− mutant. Blockage of the morphogenesis pathway might also alter accumulation of other virions forms such as EEV and IEV. EEV are derived from Golgi wrapping of IMV particles. A decrease in EEV production was observed for the H3L− mutant, as shown in Table 1. While the amount of IEV in cells is difficult to quantitate, its importance for cell-to-cell transmission in plaque formation has been demonstrated (51). The fact that the H3L− mutant virus formed small plaques implies that the number of IEV with actin tails is also affected (10, 11).
In summary, deficiency in H3L protein expression significantly blocks IV maturation to IMV particles in cells; consequently, the production of IEV and EEV were also affected in the H3L− mutant.
Previously, two IMV envelope proteins, A27L and D8L, were shown to bind to GAGs during vaccinia virus entry into cells. However, neither of these two proteins is required for IMV assembly and only A27L is essential for formation of EEV (46). Thus, H3L protein is unique in that it plays dual roles in the vaccinia virus life cycle; H3L protein is a GAG-binding protein during virus entry, and it mediates the virion assembly process.
H3L− mutant IMV exhibit structural alterations and have low infectivity compared to WT virus.
As indicated above, it is possible that H3L− mutant particles are less infectious than WT virus particles. Since H3L protein also mediates virion binding to cell surface HS, the absence of H3L protein on the IMV envelope may result in defective virion adsorption. To test if H3L− mutant virus is as infectious as WT virus, the particle-to-PFU ratio (i.e., the specific infectivity) of both IMV virions was determined. Purified WT and H3L− mutant virions were counted under the electron microscope, and biological titer assays were performed as described previously. The particle-to-PFU ratio was 4.5 ± 2.7 for WT virions and was 28 ± 6.9 for the H3L− mutant (values are means ± standard deviations). This result demonstrates that the H3L− mutant virion has sixfold lower infectivity than WT.
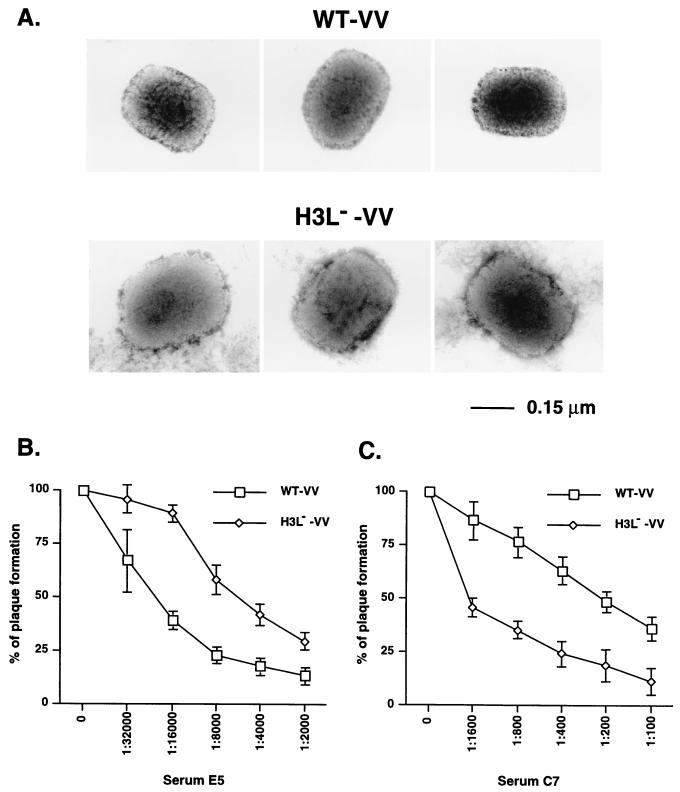
One possible mechanism for reduced infectivity might be a reduced ability of H3L− mutant virus to bind to cells. To test this idea, WT and H3L− mutant virions were incubated with BSC40 cells at 4°C for various times, cells were harvested, and the number of cell-associated viruses were determined by plaque assays. Binding of WT virus to cells increased with incubation time and was saturated after 4 to 5 h. However, binding of H3L− mutant IMV to cells was lower than that of WT IMV even after a 5-h incubation (data not shown). These data are consistent with the soluble protein competition experiments and indicate that H3L protein acts to help IMV adsorption to HS on the cell surface. In addition, other variables may also contribute to the low infectivity of H3L− mutant virus. For example, H3L protein may play a structural role to help maintain of the shape of IMV and its absence could have a pleiotropic effect on the virion structure. Indeed, by electron microscopy, the purified H3L− mutant virus appeared slightly bigger than WT virus, with approximately a 10% increase in length in both dimensions (300 ± 13 nm by 235 ± 11 nm versus 271 ± 11 nm by 211 ± 7 nm) (Fig. 7A). It thus appears that H3L protein is important for virion rigidity and that H3L− mutant virions either are loose in native structure or are more vulnerable to environmental alterations such as osmotic change and become swollen during virion preparation or the staining process.
FIG. 7.
Structures of H3L− mutant IMV and the neutralization sensitivity to antibodies against A27L and D8L proteins. (A) Electron microscope images of purified WT and H3L− mutant virions after negative staining as described in Materials and Methods. (B and C) BSC40 cells were infected with WT or H3L− mutant vaccinia virus (vv) in the presence of various dilutions of antisera against A27L (B) or D8L (C) proteins. After infection, cells were washed and overlaid with 1% agar, and plaque numbers were determined. The plaque numbers obtained in the absence of antiserum were used as the 100% values.
Although the above-described structural alteration of H3L− mutant virions did not change the ability of antibody 2D5 to neutralize L1R protein on these mutant virions (Fig. 3C), it did not exclude the possibility that other envelope proteins are affected. It is conceivable that if the above-mentioned virion structural alterations indirectly influence the conformation of other GAG-binding proteins, such as A27L and D8L proteins, the virion infectivity could be affected as well. Western blot analysis confirmed that both A27L and D8L proteins are still present on purified H3L− mutant virions (data not shown). To analyze the native structures of A27L and D8L proteins on IMV we compared H3L− mutant virus and WT virus for their sensitivity to neutralization by anti-A27L and anti-D8L antisera, respectively. As shown in Fig. 7B, anti-A27L serum E5, at a 1:24,000 dilution, neutralized 50% of WT virus plaque formation, whereas a lower dilution of 1:6,000 was required to inhibit H3L− mutant virus to the same extent, indicating that H3L− mutant became more resistant to the anti-A27L serum. In contrast, anti-D8L serum exerted an opposite effect on these two viruses, since a dilution of 1:200 led to 50% inhibition of the plaques produced by WT virus, whereas a higher dilution of 1:1,600 was needed for H3L− mutant virus (Fig. 7C).
Taken together, the above data revealed a possible role of H3L protein for virion architecture. Consequently, the lower infectivity of H3L− mutant virus may not be due to the lack of H3L protein per se. Instead, the side effects on other viral GAG-binding proteins due to H3L deletion need to be considered as well.
H3L protein is not required for fusion of virus-infected cells under acidic treatment.
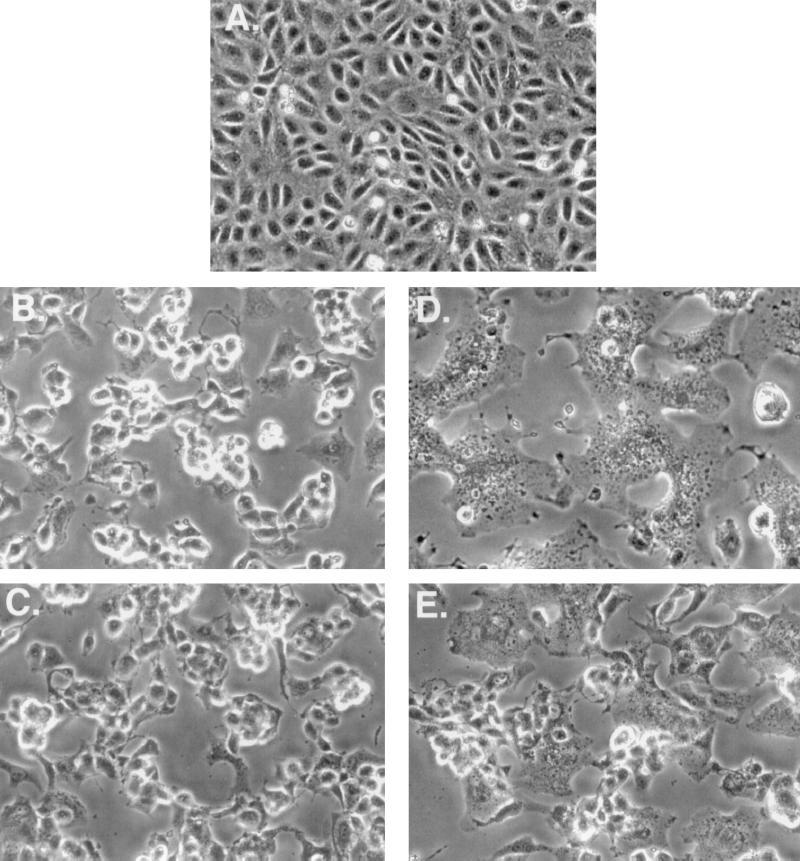
Cells infected by vaccinia virus undergo cell fusion when they are briefly incubated in acidic buffer with a pH level below 6 (13, 17). Previously, it was shown that IMV envelope protein A27L is required for fusion of infected cells and the N-terminal HS-binding region is necessary for the fusion activity (17, 22, 44). Although parameters mediating virus and cell fusion are not necessarily identical with those for fusion of infected cells, the cell fusion assay has been widely used to investigate virus and cell fusion. Since H3L protein also binds to HS, its potential role in cell fusion was examined (Fig. 8). BSC40 cells infected with WT virus and maintained at a neutral pH level did not develop cell fusion at 24 h p.i. (Fig. 8B). However, when these cells were briefly treated with acidic buffer they developed cell fusion (Fig. 8D). Cell fusion progressed to form giant cells containing multiple nuclei which ultimately floated up and died. Similarly, cells infected by H3L− mutant virus rounded up (Fig. 8C) and fused together when exposed to acidic buffer (Fig. 8E). Although for these cells the kinetics of fusion was slow and the extent of cell fusion was less complete compared to those of cells infected by WT virus, we conclude that H3L protein is not required for cell fusion.
FIG. 8.
H3L protein is not required for fusion of virus-infected cells. BSC40 cells were either mock infected (A) or infected with WT (B and D) or H3L− mutant virus (C and E) at an MOI of 5 PFU per cell. At 24 h p.i., cells were briefly treated with PBS at a pH of 7.2 (B and C) or with PBS at a pH of 4.8 (D and E), incubated for another 3 h, and photographed with a Nikon inverted microscope.
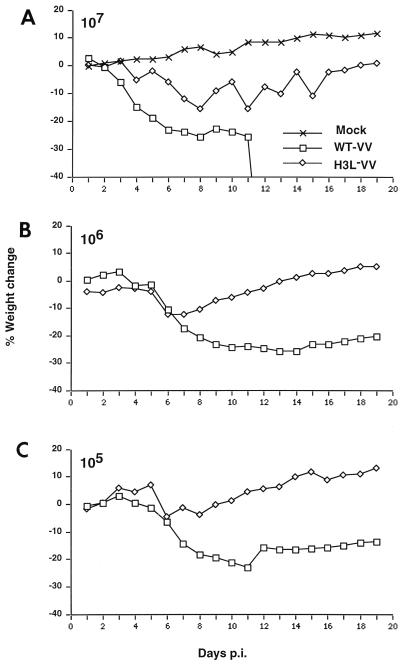
H3L− mutant virus has attenuated virulence in vivo.
Since the H3L− mutant virus is defective in cell culture growth in vitro, the biological importance of H3L protein was determined during vaccinia virus infection of mice. BALB/c mice were infected with WT and H3L− mutant viruses intranasally, and body weight was measured on a daily basis (56, 59). As shown in Fig. 9, at the highest dosage of 107 PFU per animal all the mice infected with WT virus lost a significant amount of weight and died after 10 days (Fig. 9A). In contrast, mice infected with H3L− mutant virus lost weight initially, but then they regained weight gradually and recovered after 2 weeks. At the lower dosage of 106 or 105 PFU per animal, the body weight loss of mice infected with WT virus was more severe than that of mice infected by H3L− mutant virus (Fig. 9B and C). These data provide direct evidence that H3L protein is important for vaccinia virus virulence in vivo.
FIG. 9.
Virulence of H3L− mutant virus is attenuated in vivo. BALB/c mice were either mock infected or intranasally infected with WT or H3L− mutant virus doses of 107 PFU (A), 106 PFU (B), and 105 PFU (C) at day 0 as indicated in each graph. Body weight was measured on a daily basis. There were five mice per group. All (100%) of the mice infected with 107 PFU of the WT virus 50% of mice infected with 106 PFU of the WT virus died between 10 to 14 days p.i.
DISCUSSION
Vaccinia virus has a wide host range and binds to cell surface GAGs during virus entry (9). Previously, two envelope proteins on IMV, A27L and D8L, were shown to mediate virus-GAG interactions with different specificity (9, 22, 23). A27L protein binds to cell surface HS, whereas D8L protein recognizes cell surface CS (9, 23). Mechanistically, D8L protein plays a role in IMV adsorption to cells, whereas A27L protein is more involved in virus penetration (23, 45, 47).
Since GAGs are widely expressed on cells, these studies indicate that GAG recognition helps recruit vaccinia virus to interact with a wide variety of cell types. However, mutant viruses defective for A27L and/or D8L gene expression have been constructed and were viable in cell culture (23, 38, 46, 49). One possible explanation is that other viral proteins substitute for the GAG-binding function in these mutant viruses. Here, we report the identification of a third IMV envelope protein, H3L, that binds to cell surface HS during virus entry. We show that soluble H3L protein binds to cell surface HS and blocks IMV adsorption to cells. In addition, inactivation of the H3L gene results in a sixfold reduction of virion infectivity in vitro and significant attenuation of virulence in vivo. Although H3L, like A27L protein, binds to HS, our data suggest that it mediates the initial adsorption of IMV to cells rather than the penetration stage (9, 22, 46).
From a biochemical point of view, both A27L and H3L proteins may bind to HS through similar motifs. Short hexapeptide sequences rich in basic amino acids have been identified as consensus GAG-binding sites; A27L protein has one such sequence and H3L protein has at least two (4). The consensus GAG-binding site on A27L protein has been shown to be critical for A27L protein binding to cell surface HS and cell fusion (22). Therefore, the two putative GAG-binding sites on H3L protein may be responsible for HS binding. Future studies with various mutant H3L protein constructs are needed to explore this issue.
In addition to its role as a GAG-binding protein during virus entry, H3L protein also participates in virion morphogenesis, since deficiency of H3L protein severely interrupts assembly of IMV in cells. The assembly defect is not due to a polar effect on neighboring gene expression, since H4L− mutant was reported with a different phenotype (32, 64). Furthermore, a revertant virus expressing H3L protein was constructed from the H3L mutant virus. In cells infected by the revertant virus, both H3L protein expression and virion morphogenesis was restored, supporting the idea that H3L protein has a specific role in virion assembly (data not shown). Our data with core protein processing further indicate that H3L protein is not required for P4a and P4b processing but is required for IV conversion to IMV during virion morphogenesis.
The electron microscopic images of H3L− mutant-infected cells revealed unusually large amounts of dense nucleoprotein mass with unclosed crescents. After literature searches we found that mutations of two other vaccinia virus genes such as A8L and D6R resulted in a similar phenotype (24, 25). Vaccinia virus A8L and D6R proteins are subunits of viral early transcription factors of 82 and 70 kDa and are assembled into mature virions. Cells infected by A8L− or D6R− mutant virus accumulated immature particles and granular masses associated with viral crescents, reminiscent of what was observed for H3L− mutant virus. Why would the mutations of early transcription factors result in morphogenesis blockage similar to that observed with H3L mutation? One possibility is that encapsidation of A8L and D6R proteins somehow requires H3L protein. Alternatively, these early transcription factors may regulate certain late genes such as H3L that are required for morphogenesis. If the first hypothesis is correct, we would expect that the specific activity of permeabilized H3L− mutant virions in transcription in vitro would be lower than WT virions. If the second hypothesis is right, we would expect that no H3L expression occurs in cells infected by A8L− or D6R− mutant virus. These interesting possibilities will be sorted out in the future.
The blockage of IMV formation due to H3L mutation also affects subsequent formation of IEV and EEV indirectly. Such pleiotropic effects on all three forms of virions may explain the phenotype of the H3L− mutant virus. For example, the titer of EEV in cells infected by the H3L− mutant was reduced to 10% of the WT level. In addition, the plaque size was previously shown to be correlated with the ability for actin tail formation on the tips of IEV (10, 11). The small plaque phenotype of H3L− mutant virus may reflect the fact that fewer IEV are formed in cells (37, 51).
Although mature H3L− mutant IMV were greatly reduced in number, they were not completely eliminated in the infected cells. The reason for incomplete interruption of virion morphogenesis is not understood, but it is not likely that the interruption is due to low-level expression of H3L protein expression in cells infected by the mutant virus. Western blot analyses and neutralization assays indicate that no detectable level of H3L protein is expressed by the mutant virus.
Finally, H3L protein may also be important for virion structure maintenance, based on electron microscopy and neutralization analyses. When purified H3L− IMV were examined by electron microscopy, they still appeared brick-shaped but were slightly bigger than WT virions. At first we assumed that the lack of H3L protein did not cause gross structure abnormality in IMV, since both WT and the mutant IMV are equally sensitive to 2D5, a neutralizing MAb against L1R protein (39). However, the mutant virus exhibits an altered sensitivity to neutralization effects of antibodies against A27L and D8L proteins, indicating possible structural alterations of these two GAG-binding proteins on H3L− mutant virions. Thus, the interpretation of lower infectivity of H3L− mutant virions needs to be done very carefully, for it may not be a direct consequence of the lack of H3L protein per se.
The attenuation of H3L− mutant virulence in vivo could be due to several factors. The mutant IMV were less efficient in initiating infections in new hosts since binding to GAGs may not be optimal. In addition, the infected cells produced fewer progeny, including IMV and EEV. Consequently, the severity of secondary infections transmitted to distant organs through the bloodstream in infected animals may be reduced.
In summary, this study indicates that vaccinia virus uses multiple envelope proteins to bind to cell surface GAGs. This may be an adaptation in cell culture due to extended passages in vitro. It appears that vaccinia virus possesses a battery of GAG-binding proteins to expand its ability to infect different cells and to maximize its binding capacity by recognizing different GAG-containing structures on the cell surface. For example, if a cell expresses both HS and CS on the surface, vaccinia virus could simultaneously recognize both structures and effectively engage virus adsorption onto the cells. On the other hand, some cells may only express a particular type of GAG. In that case, the presence of arrays of GAG-binding proteins on the virion may ensure that vaccinia virus binds to diverse cell populations. The interactions of envelope protein with GAGs only initiate the first step in virus entry. Further investigation will be required for understanding additional steps of vaccinia virus entry into cells.
ACKNOWLEDGMENTS
We thank Sue-Ping Lee for excellent techniques in electron microscopy. We also thank F. Tufaro for L, gro2C, and sog9 cells and R. Condit for suggestions regarding the manuscript.
This work was supported by grants from Academia Sinica and the National Science Council (NSC89-2311-B-001-080) of the Republic of China.
REFERENCES
- 1.Appleyard G, Hapel A J, Boulter E A. An antigenic difference between intracellular and extracellular rabbitpox virus. J Gen Virol. 1971;13:9–17. doi: 10.1099/0022-1317-13-1-9. [DOI] [PubMed] [Google Scholar]
- 2.Banfield B W, Leduc Y, Esford L, Schubert K, Tufaro F. Sequential isolation of proteoglycan synthesis mutants by using herpes simplex virus as a selective agent: evidence for a proteoglycan-independent virus entry pathway. J Virol. 1995;69:3290–3298. doi: 10.1128/jvi.69.6.3290-3298.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Banfield B W, Leduc Y, Esford L, Visalli R J, Brandt C R, Tufaro F. Evidence for an interaction of herpes simplex virus with chondroitin sulfate proteoglycans during infection. Virology. 1995;208:531–539. doi: 10.1006/viro.1995.1184. [DOI] [PubMed] [Google Scholar]
- 4.Cardin A D, Weintraub H J R. Molecular modeling of protein-glycosaminoglycan interactions. Arteriosclerosis. 1989;9:21–32. doi: 10.1161/01.atv.9.1.21. [DOI] [PubMed] [Google Scholar]
- 5.Chakrabarti S, Brechling K, Moss B. Vaccinia virus expression vector: coexpression of β-galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol. 1985;5:3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chelyapov N V, Chernos V I, Andzhaparidze O G. Analysis of antibody production to vaccinia virus in man and rabbits in response to inoculation of a recombinant vaccinia-hepatitis B vaccine. Vopr Virusol. 1988;33:175–179. [PubMed] [Google Scholar]
- 7.Chen Y, Maguire T, Hileman R E, Fromm J R, Esko J D, Linhardt R J, Marks R M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–868. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 8.Child S J, Franke C A, Hruby D E. Inhibition of vaccinia virus replication by nicotinamide: evidence for ADP-ribosylation of viral proteins. Virus Res. 1988;9:119–132. doi: 10.1016/0168-1702(88)90027-5. [DOI] [PubMed] [Google Scholar]
- 9.Chung C S, Hsiao J C, Chang Y S, Chang W. A27L protein mediates vaccinia virus interaction with cell surface heparan sulfate. J Virol. 1998;72:1577–1585. doi: 10.1128/jvi.72.2.1577-1585.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 11.Cudmore S, Reckmann I, Griffiths G, Way M. Vaccinia virus: a model sustem for actin-membrane interactions. J Cell Sci. 1996;109:1739–1747. doi: 10.1242/jcs.109.7.1739. [DOI] [PubMed] [Google Scholar]
- 12.Davison A J, Moss B. Structure of vaccinia virus early promoters. J Mol Biol. 1989;210:749–769. doi: 10.1016/0022-2836(89)90107-1. [DOI] [PubMed] [Google Scholar]
- 13.Doms R W, Blumenthal R, Moss B. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J Virol. 1990;64:4884–4892. doi: 10.1128/jvi.64.10.4884-4892.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyer A P, Banfield B W, Martindale D, Spanner D-M, Tufaro F. Dextran sulfate can act as an artificial receptor to mediate a type-specific herpes simplex virus infection via glycoprotein B. J Virol. 1997;71:191–198. doi: 10.1128/jvi.71.1.191-198.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ericsson M, Cudmore S, Shuman S, Condit R C, Griffiths G, Krijnse Locker J. Characterization of ts16, a temperature-sensitive mutant of vaccinia virus. J Virol. 1995;69:7072–7086. doi: 10.1128/jvi.69.11.7072-7086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goebel S J, Johnson G P, Perkus M E, Davis S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 17.Gong S C, Lai C F, Esteban M. Vaccinia virus induces cell fusion at acid pH and this activity is mediated by the N-terminus of the 14-kDa virus envelope protein. Virology. 1990;178:81–91. doi: 10.1016/0042-6822(90)90381-z. [DOI] [PubMed] [Google Scholar]
- 18.Gruenheid S, Gatzke L, Meadows H, Tufaro F. Herpes simplex virus infection and propagation in a mouse L cell mutant lacking heparan sulfate proteoglycans. J Virol. 1993;67:93–100. doi: 10.1128/jvi.67.1.93-100.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayat M A. Principles and techniques of electron microscopy. Hong Kong, China: MacMillan Press, Ltd.; 1989. [Google Scholar]
- 20.Heine H G, Stevens M P, Foord A J, Boyle D B. A capripoxvirus detection PCR and antibody ELISA based on the major antigen P32, the homolog of the vaccinia virus H3L gene. J Immunol Methods. 1999;227:187–196. doi: 10.1016/s0022-1759(99)00072-1. [DOI] [PubMed] [Google Scholar]
- 21.Housawi F M T, Roberts G M, Gilray J A, Pow I, Reid H W, Nettleton P F, Sumption K J, Hibma M H, Mercer A A. The reactivity of monoclonal antibodies against orf virus with other parapoxviruses and the identification of a 39 kDa immunodominant protein. Arch Virol. 1998;143:2289–2303. doi: 10.1007/s007050050461. [DOI] [PubMed] [Google Scholar]
- 22.Hsiao J-C, Chung C-S, Chang W. Cell surface proteoglycans are necessary for A27L protein-mediated cell fusion: identification of the N-terminal region of A27L protein as the glycosaminoglycan-binding domain. J Virol. 1998;72:8374–8379. doi: 10.1128/jvi.72.10.8374-8379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiao J-C, Chung C-S, Chang W. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J Virol. 1999;73:8750–8761. doi: 10.1128/jvi.73.10.8750-8761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X, Carroll L J, Wolffe E J, Moss B. De novo synthesis of the early transcription factor 70-kilodalton subunit is required for morphogenesis of vaccinia virions. J Virol. 1996;70:7669–7677. doi: 10.1128/jvi.70.11.7669-7677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu X, Wolffe E J, Weisberg A S, Carroll L J, Moss B. Repression of the A8L gene, encoding the early transcription factor 82-kilodalton subunit, inhibits morphogenesis of vaccinia virions. J Virol. 1998;72:104–112. doi: 10.1128/jvi.72.1.104-112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichihashi Y, Dales S. Biogenesis of poxviruses: interrelationship between hemagglutinin production and polykaryocytosis. Virology. 1971;46:533–543. doi: 10.1016/0042-6822(71)90057-2. [DOI] [PubMed] [Google Scholar]
- 27.Ichihashi Y, Oie M. Neutralizing epitope on penetration protein of vaccinia virus. Virology. 1996;220:491–494. doi: 10.1006/viro.1996.0337. [DOI] [PubMed] [Google Scholar]
- 28.Ichihashi Y, Takahashi T, Oie M. Identification of a vaccinia virus penetration protein. Virology. 1994;202:834–843. doi: 10.1006/viro.1994.1405. [DOI] [PubMed] [Google Scholar]
- 29.Jackson T, Ellard F M, Ghazaleh R A, Brookes S M, Blakemore W E, Corteyn A H, Stuart D I, Newman J W I, King A M Q. Efficient infection of cells in culture by type O foot-and-mouth disease virus requires binding to cell surface heparan sulfate. J Virol. 1996;70:5282–5287. doi: 10.1128/jvi.70.8.5282-5287.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen O N, Houthaeve T, Shevchenko A, Cudmore S, Ashford T, Mann M, Griffiths G, Krijnse Locker J. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J Virol. 1996;70:7485–7497. doi: 10.1128/jvi.70.11.7485-7497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Joklik W K. The purification of four strains of poxvirus. Virology. 1962;18:9–18. doi: 10.1016/0042-6822(62)90172-1. [DOI] [PubMed] [Google Scholar]
- 32.Kane E M, Shuman S. Temperature-sensitive mutations in the vaccinia virus H4 gene encoding a component of the virion RNA polymerase. J Virol. 1992;66:5752–5762. doi: 10.1128/jvi.66.10.5752-5762.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz E, Margalith E, Winer B, Lazar A. Characterization and mixed infections of three strains of vaccinia virus: wild type IBT-resistant and IBT-dependent mutants. J Gen Virol. 1973;21:469–475. doi: 10.1099/0022-1317-21-3-469. [DOI] [PubMed] [Google Scholar]
- 34.Lai C F, Gong S C, Esteban M. The 32-kilodalton envelope protein of vaccinia virus synthesized in Escherichia coli binds with specificity to cell surfaces. J Virol. 1991;65:499–504. doi: 10.1128/jvi.65.1.499-504.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai C F, Gong S C, Esteban M. Structural and functional properties of the 14-kDa envelope protein of vaccinia virus synthesized in Escherichia coli. J Biol Chem. 1990;265:22174–22180. [PubMed] [Google Scholar]
- 36.Maa J S, Rodriguez J F, Esteban M. Structural and functional characterization of a cell surface binding protein of vaccinia virus. J Biol Chem. 1990;265:1569–1577. [PubMed] [Google Scholar]
- 37.McIntosh A A, Smith G L. Vaccinia virus glycoprotein A34R is required for infectivity of extracellular enveloped virus. J Virol. 1996;70:272–281. doi: 10.1128/jvi.70.1.272-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Niles E G, Seto J. Vaccinia virus gene D8 encodes a virion transmembrane protein. J Virol. 1988;62:3772–3778. doi: 10.1128/jvi.62.10.3772-3778.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oie M, Ichihashi Y. Neutralizing epitope on penetration protein of vaccinia virus. Virology. 1996;220:491–494. doi: 10.1006/viro.1996.0337. [DOI] [PubMed] [Google Scholar]
- 40.Payne L. Polypeptide composition of extracellular enveloped vaccinia virus. J Virol. 1978;27:28–37. doi: 10.1128/jvi.27.1.28-37.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravanello M P, Hruby D E. Conditional lethal expression of the vaccinia virus L1R myristylated protein reveals a role in virion assembly. J Virol. 1994;68:6401–6410. doi: 10.1128/jvi.68.10.6401-6410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Reynolds E. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J Cell Biol. 1963;55:541–552. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rodriguez D, Rodriguez J R, Esteban M. The vaccinia virus 14-kilodalton fusion protein forms a stable complex with the processed protein encoded by the vaccinia virus A17L gene. J Virol. 1993;67:3435–3440. doi: 10.1128/jvi.67.6.3435-3440.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez J F, Esteban M. Mapping and nucleotide sequence of the vaccinia virus gene that encodes a 14-kilodalton fusion protein. J Virol. 1987;61:3550–3554. doi: 10.1128/jvi.61.11.3550-3554.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez J F, Janeczko R A, Esteban M. Isolation and characterization of neutralizing monoclonal antibodies to vaccinia virus. J Virol. 1985;56:482–488. doi: 10.1128/jvi.56.2.482-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez J F, Smith G L. IPTG-dependent vaccinia virus: identification of a virus protein enabling virion envelopment by Golgi membrane and egress. Nucleic Acids Res. 1990;18:5347–5351. doi: 10.1093/nar/18.18.5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez J F, Paez E, Esteban M. A 14,000-Mr envelope protein of vaccinia virus is involved in cell fusion and forms covalently linked trimers. J Virol. 1987;61:395–404. doi: 10.1128/jvi.61.2.395-404.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rodriguez J R, Risco C, Carrascosa J L, Esteban M, Rodríguez D. Characterization of early stages in vaccinia virus membrane biogenesis: implication of the 21-kilodalton and a newly identified 15-kilodalton envelope protein. J Virol. 1997;71:1821–1833. doi: 10.1128/jvi.71.3.1821-1833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez J R, Rodriguez D, Esteban M. Insertional inactivation of the vaccinia virus 32-kilodalton gene is associated with attenuation in mice and reduction of viral gene expression in polarized epithelial cells. J Virol. 1992;66:183–189. doi: 10.1128/jvi.66.1.183-189.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rosel J L, Earl P L, Weir J P, Moss B. Conserved TAAATG sequence at the transcriptional and translational initiation sites of vaccinia virus late genes deduced by structural and functional analysis of the HindIII H genome fragment. J Virol. 1986;60:436–449. doi: 10.1128/jvi.60.2.436-449.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sanderson C M, Frischknecht F, Way M, Hollinshead M, Smith G L. Roles of vaccinia virus EEV-specific proteins in intracellular actin tail formation and low pH-induced cell-cell fusion. J Gen Virol. 1998;79:1415–1425. doi: 10.1099/0022-1317-79-6-1415. [DOI] [PubMed] [Google Scholar]
- 52.Schmelz M, Sodeik B, Ericsson M, Wolffe E J, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–147. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shchelkunov S N, Blinov V M, Totmenin A V, Marennikova S S, Kolykhalov A A, Frolov I V, Chizhikov V E, Gytorov V V, Gashukov P V, Belanov E F, Belavin P A, Resenchuk S M, Andzhaparidze O G, Sandakhchiev L S. Nucleotide sequence analysis of variola virus HindIII M, L, I genome fragments. Virus Res. 1993;27:25–35. doi: 10.1016/0168-1702(93)90110-9. [DOI] [PubMed] [Google Scholar]
- 54.Spear P G, Shieh M T, Herold B C, WuDunn D, Koshy T I. Heparan sulfate glycosaminoglycans as primary cell surface receptors for herpes simplex virus. Adv Exp Med Biol. 1992;313:341–353. doi: 10.1007/978-1-4899-2444-5_33. [DOI] [PubMed] [Google Scholar]
- 55.Spurr A. A low-viscosity epoxy resin embedding medium for electron microscopy. J Ultrastruct Res. 1969;26:31–43. doi: 10.1016/s0022-5320(69)90033-1. [DOI] [PubMed] [Google Scholar]
- 56.Symons J A, Alcami A, Smith G L. Vaccinia virus encodes a soluble type 1 interferon receptor of novel structure and broad species specificity. Cell. 1995;81:551–560. doi: 10.1016/0092-8674(95)90076-4. [DOI] [PubMed] [Google Scholar]
- 57.Takahashi T, Oie M, Ichihashi Y. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology. 1994;202:844–852. doi: 10.1006/viro.1994.1406. [DOI] [PubMed] [Google Scholar]
- 58.Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. Progeny vaccinia viruses and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol. 1993;60:163–178. [PubMed] [Google Scholar]
- 59.Turner G S. Respiratory infection of mice with vaccinia virus. J Gen Virol. 1967;1:399–402. doi: 10.1099/0022-1317-1-3-399. [DOI] [PubMed] [Google Scholar]
- 60.VanSlyke J K, Franke C A, Hruby D E. Proteolytic maturation of vaccinia virus core proteins: identification of a conserved motif at the N termini of the 4b and 25K virion proteins. J Gen Virol. 1991;72:411–416. doi: 10.1099/0022-1317-72-2-411. [DOI] [PubMed] [Google Scholar]
- 61.Villarreal E, Roseman N A, Hruby D E. Isolation of vaccinia virus mutants capable of replicating independently of the host cell nucleus. J Virol. 1984;51:359–366. doi: 10.1128/jvi.51.2.359-366.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilton S, Gordon J, Dales S. Identification of antigenic determinants by polyclonal and hybridoma antibodies during the course of infection by vaccinia virus. Virology. 1986;148:84–96. doi: 10.1016/0042-6822(86)90405-8. [DOI] [PubMed] [Google Scholar]
- 63.Wolffe E J, Vijaya S, Moss B. A myristylated membrane protein encoded by the vaccinia virus L1R open reading frame is the target of potent neutralizing monoclonal antibodies. Virology. 1995;211:53–63. doi: 10.1006/viro.1995.1378. [DOI] [PubMed] [Google Scholar]
- 64.Zhang Y, Ahn B-Y, Moss B. Targeting of a multicomponent transcription apparatus into assembling vaccinia virus particles requires RAP94, an RNA polymerase-associated protein. J Virol. 1994;68:1360–1370. doi: 10.1128/jvi.68.3.1360-1370.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Moss B. Vaccinia virus morphogenesis is interrupted when expression of the gene encoding an 11-kilodalton phosphorylated protein is prevented by the Escherichia coli lac repressor. J Virol. 1991;65:6101–6110. doi: 10.1128/jvi.65.11.6101-6110.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zinoviev V V, Tchikaev N A, Chertov O Y, Malygin E G. Identification of the gene encoding vaccinia virus immunodominant protein p35. Gene. 1994;147:209–214. doi: 10.1016/0378-1119(94)90067-1. [DOI] [PubMed] [Google Scholar]