Abstract
The replicase gene (gene 1) of the coronavirus mouse hepatitis virus (MHV) encodes two co-amino-terminal polyproteins presumed to incorporate all the virus-encoded proteins necessary for viral RNA synthesis. The polyproteins are cotranslationally processed by viral proteinases into at least 15 mature proteins, including four predicted cleavage products of less than 25 kDa that together would comprise the final 59 kDa of protein translated from open reading frame 1a. Monospecific antibodies directed against the four distinct domains detected proteins of 10, 12, and 15 kDa (p1a-10, p1a-12, and p1a-15) in MHV-A59-infected DBT cells, in addition to a previously identified 22-kDa protein (p1a-22). When infected cells were probed by immunofluorescence laser confocal microscopy, p1a-10, -22, -12, and -15 were detected in discrete foci that were prominent in the perinuclear region but were widely distributed throughout the cytoplasm as well. Dual-labeling experiments demonstrated colocalization of the majority of p1a-22 in replication complexes with the helicase, nucleocapsid, and 3C-like proteinase, as well as with p1a-10, -12, and -15. p1a-22 was also detected in separate foci adjacent to the replication complexes. The majority of complexes containing the gene 1 proteins were distinct from sites of accumulation of the M assembly protein. However, in perinuclear regions the gene 1 proteins and nucleocapsid were intercalated with sites of M protein localization. These results demonstrate that the complexes known to be involved in RNA synthesis contain multiple gene 1 proteins and are closely associated with structural proteins at presumed sites of virion assembly.
The coronaviruses are positive-strand RNA viruses that perform their entire replication program in the cytoplasm of infected cells. The replication strategies used by the coronaviruses are of particular interest since they utilize the most complex patterns of replicase protein expression and viral RNA transcription and processing of any positive-strand RNA viruses. Coronaviruses express the largest known replicase polyproteins, which in turn are proteolytically processed to yield a large number of mature proteins. The patterns of coronavirus polyprotein expression and processing have become more well defined in the past several years, but many of the predicted mature replicase gene products remain to be characterized in infected cells. More important, with the exception of well-defined motifs (helicase and RNA-dependent RNA polymerase) and two experimentally confirmed proteinases, none of the remaining identified or predicted replicase gene products have known functions. Thus, determination of the expression, processing, intracellular localization, and interactions of the replicase proteins is an essential step in understanding the unique features of coronavirus replication.
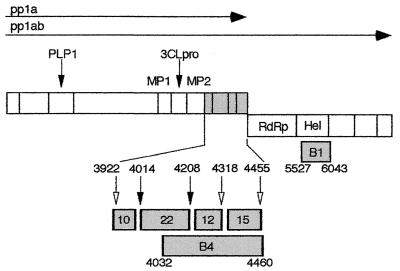
The coronavirus mouse hepatitis virus (MHV) contains a 32-kb single-stranded, positive-sense genomic RNA. The replicase gene, gene 1, of MHV strain A59 (MHV-A59) is 22 kb in length and contains two overlapping open reading frames (ORF1a and ORF1b) connected by a ribosomal frameshift (7, 8, 25). Translation of gene 1 results in two co-amino-terminal polyproteins with predicted masses of 495 and 803 kDa, corresponding to the ORF1a polyprotein (pp1a) or the ORF1a-1b fusion polyprotein (pp1ab) (Fig. 1). Two MHV ORF1a-encoded proteinases, the papain-like proteinase and 3C-like proteinase (3CLpro), have been experimentally confirmed (1, 2, 29, 34) and together are predicted to cleave the gene 1 polyprotein into at least 15 mature products (1, 2, 14, 25, 29, 30). Eleven of the proposed mature gene 1 proteins are known or predicted to be cleaved by 3CLpro. In addition to cleaving itself and helicase, MHV-A59 3CLpro has been experimentally shown to cleave a 22-kDa protein (p1a-22) from the carboxy-terminal region of pp1a (13, 27). Analyses of 3CLpro cleavage products in vitro along with putative 3CLpro cleavage sites suggested that p1a-22 was one component of a cassette consisting of four small proteins of 10, 22, 12, and 15 kDa (p1a-10, -22, -12, and -15, respectively) (27) (Fig. 1). Although the predicted cleavage sites for each of these proteins are conserved among murine (MHV), human (229E), avian (infectious bronchitis virus), and porcine (transmissible gastroenteritis virus) strains, none has significant sequence similarity to known proteins or expressed sequence tags outside the family Coronaviridae.
FIG. 1.
MHV-A59 gene 1 organization, putative cleavage sites, and cloned protein domains. The organization and arrangement of protein domains comprising polyprotein 1a (pp1a, ORF1a) and polyprotein 1ab (pp1ab, ORF1a/b) are shown. The locations of the papain-like proteinase 1 (PLP1) and 3CLpro are indicated by arrows above the schematic, and the predicted or confirmed cleavage products are demarcated by vertical bars. The location of the membrane protein domains (MP1 and MP2) in pp1a and the RNA-dependent RNA polymerase (RdRp) and helicase (Hel) in pp1ab are also shown. The cassette of four proteins in the carboxy-terminal portion of pp1a is indicated in gray. In the lower schematic, the p1a-10, -22, -12, and -15 protein domains are shown enlarged with filled arrows denoting MHV-A59 3CLpro cleavage sites that have been confirmed in vitro and open arrows indicating predicted 3CLpro cleavage sites. The cloned fragments used for production of antisera against the mature proteins (αp1a-10, -22, -12, and -15, designated by mass in kilodaltons) or against a fusion protein spanning the p1a-22 through p1a-15 domains (the B4 antibody) are also shown in gray. The fusion protein used for generating the B1 (α-helicase) antibody is similarly indicated. Relevant MHV-A59 amino acid numbers at the termini of each protein product are noted.
Recent confocal microscopy studies of MHV-infected cells have demonstrated that the gene 1-encoded helicase localizes to sites of viral RNA synthesis in large cytoplasmic structures (13). Immunoelectron microscopy studies confirmed the colocalization of helicase and viral RNA and showed accumulation of the pp1a cleavage product, p1a-22, at sites of viral RNA synthesis, suggesting functional roles for these 3CLpro cleavage products in coronavirus replication (46). Two ORF1b-encoded proteins of the related MHV-JHM strain, the putative RNA-dependent RNA polymerase and a 35-kDa protein, have also been shown to colocalize with newly synthesized viral RNA (38). In addition, an antiserum directed against the protein domain amino-terminal to the ORF1a-encoded papain-like proteinase detected protein that colocalized with viral RNA (38), and the RNA- and gene 1-containing foci were similar in appearance and localization to those previously observed for gene 1 proteins examined by single-labeling nonconfocal indirect immunofluorescence microscopy (4, 5, 17, 37, 46, 47). Thus, all studies to date indicate that the focal accumulations of viral proteins and viral RNA observed by confocal and electron microscopy are membrane-associated viral replication complexes. The term “complex” will therefore be used throughout this report to refer to viral proteins that colocalize in discrete foci in cyto.
A number of important questions concerning the formation and function of the gene 1 protein-containing complexes remain to be addressed. Since no direct dual-labeling immunofluorescence confocal studies of gene 1 proteins (e.g., hel and p1a-22) have yet been attempted, it is not known if all gene 1 proteins are tightly associated in a single type of membranous complex. In addition, the relationship of replication complexes to sites of virus assembly has not been investigated. It has been shown that viral replication complexes may form on endosomal membranes (46), whereas virion assembly has been shown to occur at sites of accumulation of the viral membrane protein (M) in the late endoplasmic reticulum, intermediate compartment (IC), and early Golgi (10, 13, 16, 20–22, 31, 36, 42–45). It remains unknown how viral RNAs synthesized in the periphery of the cell might gain access to sites of virion assembly.
This report describes the in cyto expression and processing of the four gene 1 proteins cleaved from the carboxy-terminal portion of pp1a, their localization in infected cells, and their relationship to each other and to the gene 1 proteins 3CLpro and hel. We have confirmed the expression of three small proteins of 10, 12, and 15 kDa flanking the previously identified p1a-22 in the MHV-A59 gene 1 polyprotein and have demonstrated that these proteins are proteolytically processed with kinetics similar to those of other 3CLpro cleavage products. Using confocal microscopy, we have demonstrated colocalization of p1a-22 with hel, 3CLpro, and the p1a-10, -12, and -15 cleavage products. In addition, p1a-22 was detected in cytoplasmic foci that were independent from the regions of p1a-22 colocalization with other gene 1 proteins and N. Finally, the gene 1 protein- and N-containing complexes were widely distributed throughout the infected cell but were almost entirely distinct from sites of M accumulation in the IC and Golgi, except late in infection when increasing interdigitation of gene 1 proteins, N, and M was observed. Together, these results demonstrate that coronavirus replication complexes contain multiple gene 1 proteins, that replication complexes interface with M at presumed sites of virion assembly, and that a subpopulation of p1a-22 may function independently of replication and assembly complexes.
MATERIALS AND METHODS
Generation of antisera.
To generate polyclonal antisera against the predicted mature p1a-10, -22, -12, and -15 domains, the protein-encoding regions were independently subcloned and expressed in Escherichia coli for subsequent immunization of New Zealand White rabbits (Fig. 1). All immunizations were performed by Cocalico, Inc. Reverse transcription-PCRs were performed using MHV-A59 genome RNA as template. All nucleotide and amino acid numbers correspond to the MHV-A59 sequence as modified by Bonilla et al. (7). The p1a-10 PCR product spanned nucleotides (nt) 11975 to 12253 (amino acids [aa] S3922 to Q4014), and primer-generated restriction sites were used to introduce a 5′ EcoRI site as well as a 3′ HindIII site followed by a translational stop codon. The p1a-10 fragment was then subcloned into the pMAL-C2 plasmid at the EcoRI and HindIII sites downstream of the maltose binding protein coding sequence (New England Biolabs). The resulting plasmid was expressed in E. coli according to the manufacturer's instructions, and the mature p1a-10 antigen was purified by amylose resin chromatography and factor Xa cleavage of the fusion protein. Prior to immunization of rabbits, the antigen was further purified by electroelution from a sodium dodecyl sulfate (SDS)–12% polyacrylamide gel in buffer containing 25 mM Tris base, 192 mM glycine, and 0.1% SDS. The p1a-22, -12, and -15 coding sequences were each subcloned into pET-23 vectors to produce six histidine-tagged proteins for purification by nickel resin chromatography and SDS-polyacrylamide gel electrophoresis electroelution. The p1a-22 PCR product spanned nt 12253 to 12833 (aa A4015 to Q4208) and was ligated into the pET-23d plasmid using primer-generated 5′ NcoI and 3′ XhoI sites. The p1a-12 amplification product from nt 12831 to 13160 (aa N4209 to Q4318) and the p1a-15 product from nt 13161 to 13571 (aa A4319 to Q4455) were both kinase treated prior to double-blunt ligation into HincII-digested pET-23b plasmids. The Sp9 (anti-3CLpro) and B1 (antihelicase) antisera have been previously described (13, 28). The αM and αN monoclonal antibodies (J.1.3 and J.3.3, respectively) were generously provided by John Fleming at the University of Wisconsin at Madison.
Radiolabeling of ORF1a proteins and immunoprecipitation.
MHV-A59 infections of DBT cells, pulse-label and pulse-chase experiments, and in vitro transcription-translation reactions were performed as previously described (14), with the following modifications. At the times indicated in the individual experiments, cells were moved to ice, washed with 150 mM Tris-HCl (pH 7.4), and swollen in 10 mM Tris-HCl (pH 7.4) for 30 s on ice. After removal of the 10 mM Tris, the cells were lysed in a solution containing 1% NP-40, 1% sodium deoxycholate, 150 mM sodium chloride, and 10 mM Tris (pH 7.4) (750 μl per 7.5 × 106 cells). Cellular debris was pelleted by centrifugation at 1,000 × g for 5 min at 4°C, and the supernatant was transferred to a fresh tube. SDS was added to a final concentration of 0.1% prior to freezing the samples at −20°C. One hundred microliters of cell lysate (106 cells) was subsequently used per 1 ml of immunoprecipitation reaction buffer. Lysate was combined with protein A-Sepharose beads and a 1:100 dilution of antibody in lysis buffer containing 0.1% SDS and 80 μg of phosphonomethylsulfonyl fluorone (Sigma). After incubation at 4°C for 2 h, beads were pelleted and washed with low-salt lysis buffer (150 mM NaCl) followed by high-salt lysis buffer (1 M NaCl) and a final low-salt wash. After rinsing, 2× Laemmli buffer (23) was added to the pelleted beads and boiled for 5 minutes prior to electrophoresis of the supernatant on SDS–10 to 20% gradient polyacrylamide gels.
Immunofluorescence assays.
DBT cells on glass coverslips were mock infected or infected with MHV-A59 at a multiplicity of infection of 10 PFU/cell in Dulbecco modified Eagle medium (pH 6.8) containing 6% fetal calf serum. At the various times indicated after infection, the cells were fixed with −20°C 100% methanol and were stored at −20°C under methanol until use. For immunofluorescence assays, the cells were rehydrated in phosphate-buffered saline (PBS) with 5% bovine serum albumin prior to blocking in the diluent solution containing 1% bovine serum albumin, 2% goat serum, and 0.05% NP-40 in PBS. Gene 1 antisera were precleared in PBS at room temperature for 1 h on fixed DBT cells prior to incubation with infected cell monolayers. In single-labeling experiments, cells were incubated with a 1:100 dilution of the precleared primary rabbit anti-gene 1 antiserum, washed, and incubated with a 1:1,000 dilution of Cy2-conjugated goat anti-rabbit secondary antiserum prior to mounting in Aquapolymount (Polysciences, Inc.). Immunofluorescence was detected at 488 nm using a Zeiss LSM 410 confocal microscope in the Vanderbilt University Molecular Imaging Core. Red and green colors were artificially assigned to all gray scale images using the hue-saturation option in Adobe Photoshop 5.0.
For dual labeling of gene 1 proteins, cells were fixed and incubated with a 1:100 dilution of precleared gene 1 antibody (αp1a-10, αp1a-15, Sp9, or B1) followed by Cy3-conjugated anti-rabbit secondary antisera. p1a-22 was then detected using αp1a-22 directly conjugated to Cy2 according to the manufacturer's instructions (Amersham). Dual-labeled cells were imaged at 586 and 488 nm for red and green, respectively. For Fig. 5, the brightness and contrast of each image were identically modified in Adobe Photoshop 5.0 so as not to alter the relative fluorescence of p1a-22 at each time point.
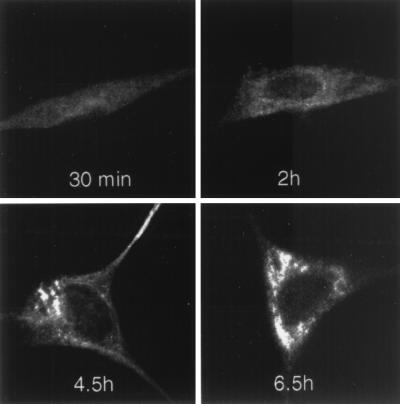
FIG. 5.
Time course of p1a-22 localization during MHV-A59 infection. Infected DBT cells were fixed in methanol at the times p.i. as indicated prior to preparation for immunofluorescence using αp1a-22. Confocal images were acquired and modified as described for Fig. 4. All images were identically processed to allow direct comparison of the extent of p1a-22 expression.
Dual labeling of p1a-22 and helicase with M was accomplished using monoclonal αM (J.1.3). Infected or mock-infected DBT cells were incubated with a 1:1,000 dilution of αM followed by Cy3- or Cy2-conjugated anti-mouse secondary antisera. Cells were then washed and incubated with a 1:100 dilution of αp1a-22 or B1 antiserum prior to successive washing and incubation with Cy2- or Cy3-conjugated anti-rabbit antibodies. For dual labeling of nucleocapsid and M, cells were incubated with a 1:1,000 dilution of monoclonal αN (J.3.3) prior to incubation with αM directly conjugated to Cy2 (Amersham).
Image reconstruction.
Three-dimensional reconstructions of cells were performed following z sectioning on the Zeiss 410 LSM confocal microscope. Cells were fixed on glass coverslips, prepared for immunofluorescence, and subjected to a series of 4-s laser scans at 488 or 586 nm. For each coverslip, the scans were performed at 0.233-μm intervals in the z dimension beginning at the bottom of the cell (coverslip) and progressing up through the nucleus and over the top of the cell. Typically, between 40 and 60 images were obtained using a 63× objective with 1.7× zoom and a pinhole of 45 Airy units. The image series for each cell were then transferred into NIH Image 1.61 (W. Rasband, NIH Image, 1.55 ed., 1994, zippy.nimh.nih.gov.), stacked, and digitally merged to examine protein localization throughout the intact infected cell. The settings for brightness and contrast in each section for a given cell were constant.
RESULTS
Detection of gene 1 proteins in MHV-infected DBT cells.
We have previously identified an MHV-A59-encoded 22-kDa protein (p1a-22) that is cleaved from the MHV-A59 gene 1 polyprotein at the LQ_S4014 and LQ_N4208 sites (27). The B4 antibody used to detect p1a-22 in the previous study was induced using a fusion protein spanning aa 4032 to 4460 and incorporating the coding sequence downstream of p1a-22 to the end of ORF1a. Although p1a-22 was the predominant and confirmed precipitation product of the B4 antiserum, in extended pulse-chase two proteins between 12 and 15 kDa were also detected (27). The proteins were thought to result from cleavage of the gene 1 polyprotein at LQ_A4318 and FQ_S4455. We therefore proposed that the carboxy-terminal region of the ORF1a polyprotein was cleaved by 3CLpro into mature products of 10, 22, 12, and 15 kDa (Fig. 1), designated p1a-10, p1a-22, p1a-12, and p1a-15, respectively. To define the entire complement of the pp1a carboxy-terminal region cleavage products, a series of monospecific polyclonal antisera were generated. The αp1a-10, αp1a-12, and αp1a-15 antisera were induced in rabbits using E. coli-expressed fusion proteins incorporating aa 3922 to 4013 (αp1a-10), aa 4208 to 4317 (αp1a-12), or aa 4318 to 4454 (αp1a-15) of the ORF1a polyprotein (7) (Fig. 1). These protein domains were chosen based on the known 3CLpro cleavage specificity as well as the identification of mature cleavage products corresponding to these cleavage sites in other coronavirus strains (26, 37, 47). In addition, a monospecific antiserum was generated against the previously identified p1a-22 (aa 4014 to 4207) for direct comparison of the mature proteins.
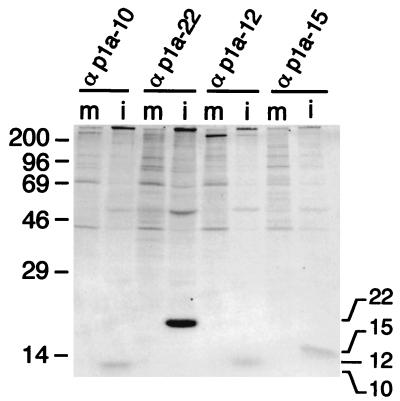
When whole-cell lysates of MHV-infected DBT cells were immunoprecipitated using the region-specific antisera, three previously unidentified MHV-A59 proteins were identified (Fig. 2). The proteins were not detected in uninfected whole-cell lysates and had apparent masses of 10, 12, and 15 kDa, respectively, corresponding precisely with the predicted protein masses. The immunoprecipitated proteins also comigrated on SDS-polyacrylamide gels with the E. coli-expressed proteins used to generate the antibodies, and the detection of each of the proteins in infected cells was eliminated by the addition of E64d, a known inhibitor of 3CLpro (data not shown).
FIG. 2.
Identification of proteins processed from the carboxy-terminal portion of the ORF1a polyprotein in MHV-A59-infected cells. MHV-A59-infected (i) or mock-infected (m) DBT cell lysates were immunoprecipitated with the antisera directed against the individual protein domains as indicated above each lane and analyzed by SDS–10 to 20% gradient polyacrylamide gel electrophoresis and fluorography. Molecular mass markers (kilodaltons) are shown to the left of the gel.
Kinetics of p1a-10, p1a-12, and p1a-15 expression in cyto.
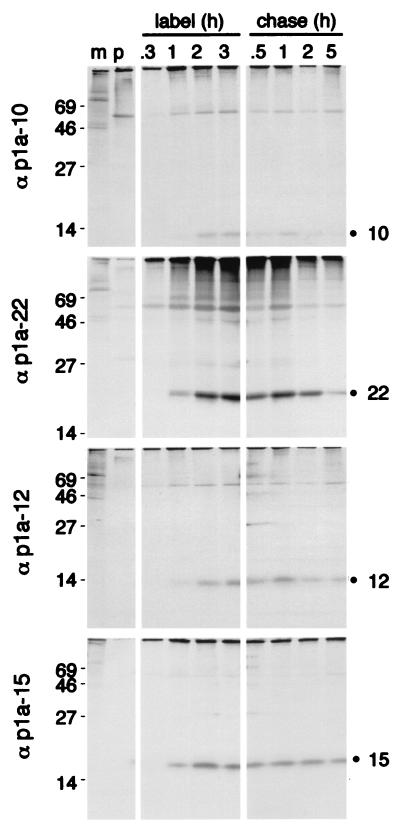
To determine if the proteins exhibited different patterns of cleavage or degradation, infected cells were subjected to pulse-label and pulse-chase analyses (Fig. 3). During pulse-label experiments, DBT cells were infected with MHV-A59 for 5.5 h and were labeled with [35S]Met for up to 180 min. Similar to the previously identified p1a-22, the 10-, 12-, and 15-kDa proteins were each first detectable by 60 min of labeling at approximately 6.5 h postinfection (p.i.) and continued to accumulate throughout the 180-min labeling period (8.5 h p.i.). The protein bands for p1a-22 and p1a-15 were more dense than those of p1a-10 and p1a-12, suggesting nonequivalent amounts of protein. This pattern was reproducible and was not an artifact of protein loading. However, when the proteins were analyzed by densitometry and normalized for the deduced methionine and cysteine content, the proteins were shown to be present in similar amounts.
FIG. 3.
Pulse-label and pulse-chase translation of p1a-10, -22, -12, and -15. For pulse-label translation (label), DBT cells were mock infected (m) or infected with MHV-A59 for 5.5 h and were incubated with [35S]Cys-Met for 0.3, 1, 2, or 3 h prior to immunoprecipitation with the antibodies as indicated to the right of each gel. p denotes immunoprecipitation with preimmune serum. For pulse-chase translation (chase), DBT cells were radiolabeled beginning at 5.5 h p.i. with [35S]Cys-Met for 90 min prior to addition of excess cold methionine and cycloheximide for the times indicated above each well. Following harvesting and immunoprecipitation of the cells, label and chase samples for each antibody were run on the same SDS–10 to 20% gradient polyacrylamide gel, followed by fluorography. Molecular mass markers (kilodaltons) are shown to the left of the gels, and the locations of the p1a-10, -22, -12, and -15 proteins are indicated to the right of the gels.
To further examine cleavage and stability of the mature proteins, pulse-chase experiments were performed (Fig. 3). A 90-min pulse period following 5.5 h of infection was chosen since this corresponded to a time of active [35S]Met-Cys incorporation in the pulse-label experiment. The mature p1a-10, -22, -12, and -15 proteins were easily detectable after 30 min of chase (7.5 h p.i.) and continued to accumulate up to 60 min of chase, indicating that the proteins were cleaved from a pool of precursors for at least 60 min after translation. The proteins remained detectable through 5 h of chase (12 h p.i.). We have previously demonstrated that addition of cycloheximide and excess cold methionine (chase) results in immediate cessation of incorporation of new [35S]Met-Cys into nascent protein (13). Hence, accumulation of p1a-10, -22, -12, and -15 for an hour after addition of the chase medium was consistent with continued proteolytic processing and/or the maintenance of stable protein populations. After overnight incubation in the chase medium, the cell monolayers were destroyed and the gene 1 proteins were no longer detectable. These results were similar to our previous studies of p1a-22 using the B4 antibody (27). Each of the antibodies precipitated products that were present at the top of the resolving gel, but it was not possible to determine their mass or resolve them on the 10 to 20% gradient gels designed for maximum separation of low-molecular-mass proteins. Due to the almost complete amino acid conservation of the carboxy-terminal portion of pp1a between MHV-A59 and MHV-JHM, it is likely that the 150-kDa intermediate precursor identified in MHV-JHM-infected cells may also be present in A59 and that the small cleavage products of 10, 22, and 12 kDa will be conserved between the strains, as has already been shown for p1a-15 (37).
Intracellular localization of p1a-10, -22, -12, and -15.
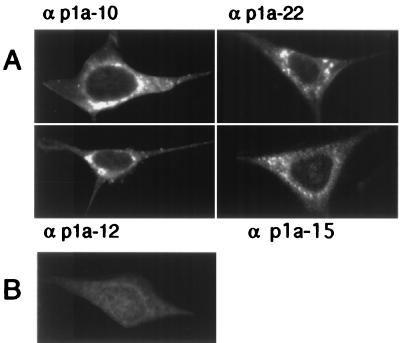
To define the intracellular localization of p1a-10, -22, -12, and -15, DBT cells were infected with MHV-A59 and prepared for immunofluorescence studies as described in Materials and Methods. In infected cells that were not fused in syncytia, each of the four proteins was detected in cytoplasmic foci that were largely perinuclear, often forming perinuclear rings or unilateral crescents adjacent to the nucleus (Fig. 4). These protein populations were specific to infected cells and were not present in mock-infected cells probed with immune antisera or in infected cells probed with preimmune antisera. The observed pattern of localization for each of the proteins at 6 h p.i. varied according to the chosen focal plane, with some planes revealing a primarily perinuclear distribution of the gene 1 proteins and other planes revealing a punctate pattern throughout the cytoplasm. To determine if gene 1 protein localization changed during the course of infection, cells were harvested at 30 min or 2, 4.5, or 6.5 h and analyzed by confocal microscopy. Gene 1 proteins were not detectable prior to 4.5 h p.i. (Fig. 5). Once detectable, the pattern of localization was predominantly punctate and discrete. Late in infection (6 to 7 h p.i.), the protein populations became abundant and merged into what appeared to be, at the resolution of light microscopy, large confluent complexes. Accumulation of protein-containing complexes within the extended cell processes was apparent at later times of infection, especially with the αp1a-22 antibody (Fig. 5).
FIG. 4.
Localization of p1a-10, -22, -12, and -15 in MHV-infected cells. MHV-A59-infected or mock-infected DBT cells were fixed and prepared for immunofluorescence microscopy as described in Materials and Methods, using the antibodies as noted by each frame. Cells were imaged on a Zeiss LSM 410 confocal microscope using a 488-nm laser with acquisition in the LSM software. The images are single confocal slices using a 63× objective. Image processing (brightness and contrast) was performed in Photoshop 5.0. A representative mock-infected cell probed with αp1a-22 is shown in panel B. αp1a-10, -12, and -15 resulted in a similar lack of background staining in the mock-infected cells.
Dual labeling of gene 1 proteins in infected cells.
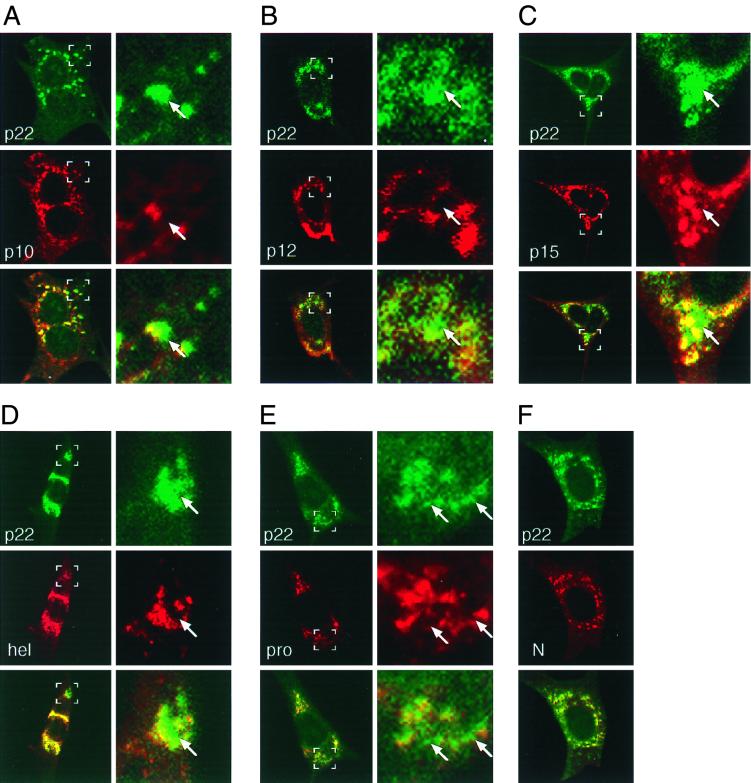
The initial single-labeling experiments demonstrated striking similarities in the localization patterns of the four ORF1a carboxy-terminal proteins and similarities to the previously described complexes containing hel, N, and new viral RNA (13, 38, 46). We next sought to determine if the four proteins colocalized with each other and the previously characterized gene 1 proteins. Since all available gene 1 protein antisera were derived from rabbits, dual-labeling studies of gene 1 proteins required direct conjugation of fluorescent label to one or more primary antibodies. αp1a-22 was selected for direct conjugation because of its ease of detection in both biochemical and imaging studies and because it has been shown by immunoelectron microscopy to localize to sites of RNA synthesis (46). Cy2-labeled αp1a-22 was first used in dual-labeling studies with the well-characterized hel and N proteins. p1a-22 colocalized with both hel and N to a large extent (Fig. 6), confirming association of p1a-22 with sites of viral RNA synthesis. p1a-22 also colocalized with the majority of the 3CLpro and the newly identified 10-, 12-, and 15-kDa cleavage products. Our results therefore directly demonstrate significant overlap of six independent gene 1 proteins.
FIG. 6.
Dual-labeling immunofluorescence confocal microscopy of MHV-infected cells. At 6.5 h p.i. MHV-infected DBT cells were fixed and prepared for immunofluorescence using αp1a-22 as a primary antibody and other gene 1 protein antibodies or the αN monoclonal antibody as the second primary antibody. p1a-22 (p22) is shown in green in each panel, and the other proteins are shown in red as indicated. The merged images are shown with areas of colocalization in yellow. The right column of each panel is a higher magnification (×5.3) of the area demarcated by the white boxes in the left image. Arrows indicate areas of intercalation of p1a-22 with the other gene 1 proteins. (A) p1a-10 (p10); (B) p1a-12 (p12); (C) p1a-15 (p15); (D) helicase (hel); (E) 3CLpro (pro); (F) nucleocapsid (N).
When cells were imaged at higher magnification, areas were observed where the signals for p1a-22 and the other gene 1 proteins were discrete but tightly interdigitated. This pattern of areas of intercalated as well as coincident signal was detected in all dual-labeling studies using αp1a-22 (Fig. 6), both in individual cells (Fig. 6, p1a-12 and 3CLpro) and in syncytia (Fig. 6, p1a-10 and -22 and hel). Because the nominal resolution of confocal microscopy in our study was 0.2 μm, an observation of noncolocalization required separation of fluorescent foci of at least 0.2 μm (Fig. 6). The majority of the p1a-22-containing complexes were contiguous with areas of p1a-22 colocalization with other proteins, but infrequently the foci of noncolocalized p1a-22 were distanced from these regions. In all cases, multiple sections in the z dimension were obtained, and uniformly the areas of colocalization and also areas of noncolocalization of p1a-22 were confirmed in individual slices and in the context of three-dimensional reconstructions.
Dual labeling of gene 1 proteins with M.
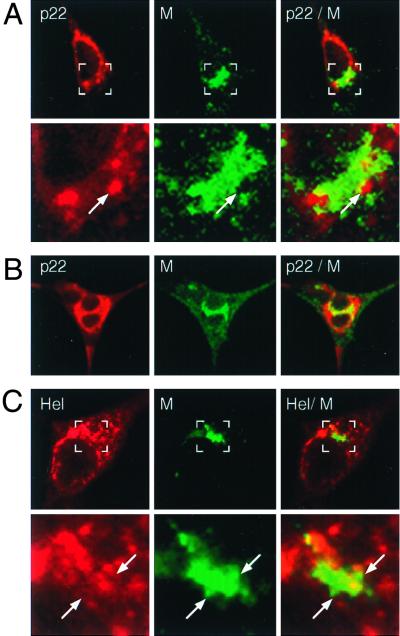
Having defined the localization of the gene 1 proteins, we next sought to determine if any of the observed gene 1 protein-containing complexes involved the presumed sites of virion assembly. The MHV membrane protein (M) is known to be required for the assembly of progeny virions and has been well documented as a marker of virion assembly (21). To determine the relationship of replication and assembly complexes during a peak phase of virion formation, we used dual-labeling studies to define the localization of p1a-22 and helicase with respect to M at 6.5 h p.i. (Fig. 7). The complexes containing either p1a-22 or hel were almost entirely distinct from those containing M, both in single discrete cells and in virus-induced syncytia. However, a small amount of labeling for p1a-22 or hel colocalized with M in perinuclear structures. Colocalization of p1a-22 and helicase with M was most easily seen in syncytia, where examination of infected cells at high magnification revealed intercalation of p1a-22 and hel-containing complexes in the perinuclear region with complexes containing M, presumably in the IC and Golgi (Fig. 7B). In contrast, the gene 1 protein-containing foci in the cell periphery were uniformly devoid of M. These relationships between p1a-22, hel, and M were prominent and characteristic of the majority of infected cells at late times of infection. Combined with studies showing colocalization of hel and p1a-22 with de novo-synthesized viral RNA (13, 46), the data indicated that viral RNAs were synthesized in replication complexes in the cell periphery as well as in perinuclear regions but that detectable colocalization of replication and assembly complexes was concentrated at perinuclear sites of abundant M accumulation.
FIG. 7.
Dual-labeling imaging of M and p1a-22 or helicase. DBT cells were infected for 6 h prior to fixation and preparation for dual-labeling immunofluorescence using αM (green in all images) and either αp1a-22 (p22) (A and B) or B1 αhel (Hel) (C). (A and C) The bottom panels show higher magnifications of the region marked by the white box in the corresponding upper panel. Arrows indicate regions of intercalation of p1a-22 or hel with M. (B) Localization of p1a-22 and M in a virus-induced syncytium.
Dual labeling of M with nucleocapsid.
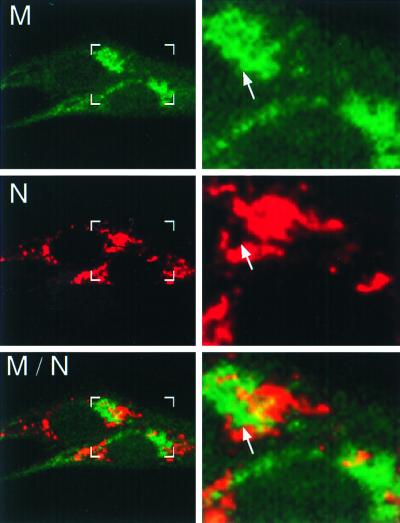
We next defined the relationship of nucleocapsid (N) and M in MHV-infected cells. We have previously demonstrated that N is tightly associated with hel in replication complexes in DBT cells but surprisingly does not appear to be distributed diffusely throughout the cell or to localize in complexes distinct from hel (13). We have shown a similar association of N with p1a-22 in this report (Fig. 6F). However, since previous studies have suggested a role for N in viral RNA synthesis (3, 12, 33) and since N is also a structural component of the virion (6, 9, 18, 19, 35, 39–41), we expected that N would be abundant both at sites of replication and at sites of assembly. To define the relationship between N-containing replication complexes and sites of virion assembly, we conjugated αM to Cy2 to allow dual labeling of M and N in MHV-infected DBT cells (Fig. 8). N was detected in discrete, punctate cytoplasmic complexes throughout the cell, with a pattern characteristic of that seen with the gene 1 proteins, whereas M was detected in a perinuclear Golgi-like accumulation similar to that seen during dual-labeling studies with gene 1 proteins (Fig. 7) (11). The patterns of N and M localization closely resembled that between the gene 1 proteins and M, with N and M being primarily distinct except for an intercalated pattern of N and M signals in perinuclear regions (Fig. 8). Areas of N-M colocalization were also observed at the margins of the N- and M-containing complexes, presumably representing sites of assembly of the two structural proteins into progeny virions. In all infected cells, N was detected in discrete complexes, and there were no cells where N demonstrated diffuse cytoplasmic distribution or extensive colocalization with M.
FIG. 8.
M and N are distinct but closely associated in perinuclear regions. At 6 h p.i., infected BHK-R cells were fixed and labeled with αN (J.3.3) followed by Cy3-conjugated anti-mouse secondary antibody and Cy2-conjugated αM (J.1.3). In the left panel are shown two infected cells with boxes corresponding to the increased-magnification panels to the right. Arrows indicate areas of intercalation between M and nucleocapsid (N). Areas of colocalization are shown in yellow.
DISCUSSION
The carboxy-terminal portion of the ORF1a polyprotein (pp1a) is cleaved into four mature proteins.
In this study, we have shown that the carboxy-terminal 59 kDa of the MHV-A59 ORF1a polyprotein (pp1a) is processed to yield four proteins of 10, 22, 12, and 15 kDa. In contrast to the proteins processed from the first 300 kDa of pp1a-pp1ab, the p1a-10, -22, -12, and -15 proteins together comprise a cassette of low-molecular-mass protein products that are conserved among coronavirus strains in their location and relative sizes. We have previously shown that p1a-22 is cleaved at a conserved 3CLpro cleavage site at its amino terminus and at its carboxy terminus at a novel noncanonical Q_N 3CLpro cleavage site that is completely conserved among the coronaviruses (27). The masses of the newly detected proteins (10, 12, and 15 kDa) correspond precisely with the predicted masses of products obtained if cleavage occurred at consensus 3CLpro cleavage sites. In addition, the gene 1-encoded proteins corresponding to MHV-A59 p1a-10, -12, and -15 have recently been detected in cells infected with human coronavirus 229E, and homologs of p1a-10 and p1a-15 have also been identified in infectious bronchitis virus- and MHV-JHM-infected cells, respectively (26, 37, 47). Thus, it appears that these four proteins comprise the mature cleavage products of the final 59 kDa of the MHV ORF1a polyprotein and likely of all coronaviruses and that the cleavages of the mature proteins from the polyprotein are mediated by 3CLpro.
The four cleavage products accumulated in infected cells at similar rates and were detectable for prolonged periods during exhaustive chases. These results were similar to those for other gene 1 proteins, including 3CLpro, p1a-22, and hel (13, 27, 28), and were interesting in light of in vitro analyses of 3CLpro cleavage of the human coronavirus 229E polyprotein demonstrating significant differences in cleavage efficiency between the analogous 5-, 23-, 12-, and 16-kDa proteins of 229E (47). There is enough conservation among coronaviruses at the cleavage sites flanking and downstream of 3CLpro to presume that differences in 3CLpro cleavage efficiency detected in human coronavirus 229E (27) would also be present in MHV. Possible explanations to account for the equivalent amounts of the proteins detected in MHV-A59-infected cells include strain-dependent differences in 3CLpro cleavage of the small proteins or, more likely, differences in the context of the proteins within the polyprotein in vitro versus in cyto. It is also possible that differential cleavage specificity may play a critical role in the initial stages of replication complex formation or function but that at later times of infection there is an accumulation of the proteins in membranous complexes that provide some protection from degradation. This might in turn imply less need for regulation of gene 1 protein amounts at later times of infection or alternatively that some of the proteins detected are no longer serving roles in virus replication. The development and use of mutants in the individual proteins or control in the amounts of expression will be necessary to answer these questions.
Localization of gene 1 proteins in cytoplasmic complexes.
Previous immunofluorescence microscopic studies of gene 1 proteins have relied on similarities in patterns of localization of individual proteins and their colocalization with a common marker, such as viral RNA, to draw conclusions about their interactions. Thus, there have been no direct comparisons using immunofluorescence of more than one gene 1 protein in individual infected cells. By conjugating the αp1a-22 antisera to a fluorescent dye, we were able to define the in cyto relationships of pairs of gene 1 proteins in multiple planes of individual infected cells. Our dual-labeling experiments demonstrated that the majority of p1a-22 was colocalized with N, hel, 3CLpro, and p1a-10, -12, and -15, confirming the incorporation of multiple structural and nonstructural proteins into complexes at sites of viral RNA synthesis. Detection of additional p1a-22 at sites independent of other gene 1 proteins also indicates that gene 1 proteins may be targeted to at least two populations of membranous complexes. The areas of independent p1a-22 localization were distinct in all dual-labeling combinations, and it will be interesting to determine if p1a-22 is unique in its separate localization. Our study did not define the localization of all known mature gene 1 proteins or investigate all possible dual-labeling combinations. It is therefore possible that the separate p1a-22-containing regions also contain other gene 1 proteins. Confirmation of the precise relationship of all the gene 1 proteins will require biochemical studies as well as multiple-labeling studies using several dye-conjugated anti-gene 1 antibodies. However, the current data indicate that coronavirus RNA synthesis is mediated by multiprotein replication complexes containing gene 1 proteins and N.
Gene 1 proteins and N do not colocalize with M but are closely approximated at presumed sites of virion assembly.
Coronavirus assembly has been shown to occur primarily in the membranes of the rough endoplasmic reticulum and IC, followed by transport to or through the Golgi prior to exocytic release of progeny virions (10, 15, 21, 22, 36, 42–45). Our dual labeling using directly dye-conjugated αM in combination with either αN or a number of anti-gene 1 antibodies clearly demonstrated that a majority of signal for the gene 1 proteins or N had no overlap with M. Yet, there were areas of intercalation and interface of gene 1 proteins and N with M such that there was a margin of coincident signal at the periphery of sites of M accumulation. These data are consistent with early electron microscopy studies demonstrating dense aggregates of N in foci immediately adjacent to immature virions budding from IC-Golgi membranes (32). Our results suggest that the aggregates of N visualized by electron microscopy may have been components of functional replication complexes in close association with M. We propose that accumulation of N and gene 1 proteins in perinuclear membranes is likely important for virion assembly, potentially providing a mechanism for the targeting of RNA-nucleocapsids to sites of assembly.
Models of replication complex accumulation in perinuclear membranes.
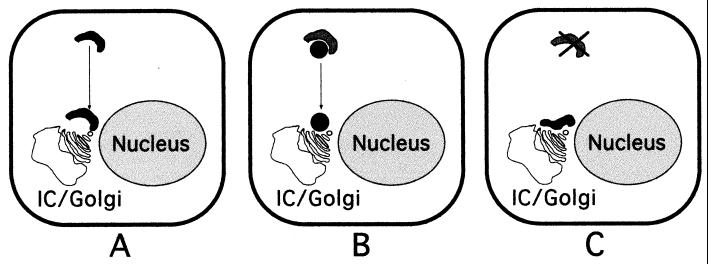
The results of our studies raise critical questions concerning the formation and function of replication complexes and their interaction with sites of virion assembly. How do nascent nucleocapsids containing genome RNA reach sites of virion assembly? Are only the replication complexes directly juxtaposed to sites of M accumulation functional in the production of packaged RNA-nucleocapsids? Our data suggest several possible models for interdigitation of replication complexes with sites of assembly (Fig. 9). Entire replication complexes from the cell periphery may be recruited to sites of assembly via cellular highways (Fig. 9A). Alternatively, genome RNAs may be shuttled to sites of assembly in transport complexes containing specific gene 1 proteins such as p1a-22 (Fig. 9B). Finally, it is possible that assembly may be initiated as the replication complexes are amplified in the cytoplasm, resulting in incorporation of nucleocapsids into virions only at the interface of complexes containing gene 1 proteins, N, and RNA with those containing other structural proteins (Fig. 9C). Answers to these questions will require biochemical, electron microscopic, and live-cell imaging approaches to determine the intracellular membranes involved in the establishment, maintenance, and potential transporting of gene 1 protein complexes during the course of infection. These studies will greatly enhance our understanding of how coronaviruses take advantage of the dynamic intracellular environment to mediate and integrate the processes of RNA synthesis and production of infectious progeny virions.
FIG. 9.
Possible models for interdigitation of replication complexes with sites of virion assembly. (A) Replication complexes throughout the cytoplasm (black) move along cytoskeletal pathways to sites juxtaposed to IC-Golgi membranes. (B) RNA-nucleocapsids move from replication complexes (dark grey) via separate transport complexes (black). (C) Only amplified or newly formed replication complexes (black) directly adjacent to the IC-Golgi membranes interact with sites of assembly. The model is not intended to describe the source of membranes for replication complexes.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI-26603 (M.R.D.) and AI01479 (M.R.D.) and the Turner Scholar's Program at Vanderbilt University.
We acknowledge critical reading of the manuscript by Amy Sims and Erik Prentice and the assistance of David Piston and Jonathan Sheehan in the Molecular Imaging Shared Resource of the Vanderbilt Cancer Center (IP30CA68485). We also thank John Fleming at the University of Wisconsin and Jennifer Lippincott-Schwartz at the National Institutes of Health for kindly providing the J.1.3 αM and J.3.3 αN antibodies and the hGalT-GFP construct, respectively. The BHK-R cells were graciously provided by Ralph Baric at the University of North Carolina, Chapel Hill.
REFERENCES
- 1.Baker S C, Shieh C-K, Soe L H, Chang M-F, Vannier D M, Lai M M C. Identification of a domain required for autoproteolytic cleavage of murine coronavirus gene A polyprotein. J Virol. 1989;63:3693–3699. doi: 10.1128/jvi.63.9.3693-3699.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker S C, Yokomori K, Dong S, Carlisle R, Gorbalenya A E, Koonin E V, Lai M M. Identification of the catalytic sites of a papain-like cysteine proteinase of murine coronavirus. J Virol. 1993;67:6056–6063. doi: 10.1128/jvi.67.10.6056-6063.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baric R S, Nelson G W, Fleming J O, Deans R J, Keck J G, Casteel N, Stohlman S A. Interactions between coronavirus nucleocapsid protein and viral RNAs: implications for viral transcription. J Virol. 1988;62:4280–4287. doi: 10.1128/jvi.62.11.4280-4287.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bi W, Bonilla P J, Holmes K V, Weiss S R, Leibowitz J L. Intracellular localization of polypeptides encoded in mouse hepatitis virus open reading frame 1A. Adv Exp Med Biol. 1995;380:251–258. doi: 10.1007/978-1-4615-1899-0_40. [DOI] [PubMed] [Google Scholar]
- 5.Bi W, Pinon J D, Hughes S, Bonilla P J, Holmes K V, Weiss S R, Leibowitz J L. Localization of mouse hepatitis virus open reading frame 1a derived proteins. J Neurovirol. 1998;4:594–605. doi: 10.3109/13550289809114226. [DOI] [PubMed] [Google Scholar]
- 6.Bingham R, Almeida J. Studies on the structure of a coronavirus-avian infectious bronchitis virus. J Gen Virol. 1977;36:495–502. doi: 10.1099/0022-1317-36-3-495. [DOI] [PubMed] [Google Scholar]
- 7.Bonilla P J, Gorbalenya A E, Weiss S R. Mouse hepatitis virus strain A59 RNA polymerase gene ORF 1a: heterogeneity among MHV strains. Virology. 1994;198:736–740. doi: 10.1006/viro.1994.1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breedenbeek P J, Pachuk C J, Noten A F H, Charite J, Luytjes W, Weiss S R, Spaan W J M. The primary structure and expression of the second open reading frame of the polymerase gene of the coronavirus MHV-A59; a highly conserved polymerase is expressed by an efficient ribosomal frameshifting mechanism. Nucleic Acids Res. 1990;18:1825–1832. doi: 10.1093/nar/18.7.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cavanagh D. Structural polypeptides of coronavirus IBV. J Gen Virol. 1981;53:93–103. doi: 10.1099/0022-1317-53-1-93. [DOI] [PubMed] [Google Scholar]
- 10.Chasey D, Alexander D. Morphogenesis of avian infectious bronchitis virus in primary chick kidney cells. Arch Virol. 1976;52:101–111. doi: 10.1007/BF01317869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cole N B, Smith C L, Sciaky N, Terasaki M, Edidin M, Lippincott-Schwartz J. Diffusional mobility of Golgi proteins in membranes of living cells. Science. 1996;273:797–801. doi: 10.1126/science.273.5276.797. [DOI] [PubMed] [Google Scholar]
- 12.Compton S R, Rogers D B, Holmes K V, Fertsch D, Remenick J, McGowan J J. In vitro replication of mouse hepatitis virus strain A59. J Virol. 1987;61:1814–1820. doi: 10.1128/jvi.61.6.1814-1820.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denison M R, Spaan J M, van der Meer Y, Gibson C A, Sims A C, Prentice E, Lu X T. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J Virol. 1999;73:6862–6871. doi: 10.1128/jvi.73.8.6862-6871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denison M R, Zoltick P W, Hughes S A, Giangreco B, Olson A L, Perlman S, Leibowitz J L, Weiss S R. Intracellular processing of the N-terminal ORF 1a proteins of the coronavirus MHV-A59 requires multiple proteolytic events. Virology. 1992;189:274–284. doi: 10.1016/0042-6822(92)90703-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dubois-Dalcq M, Doller E, Haspel M, Holmes K. Cell tropism and expression of mouse hepatitis viruses (MHV) in mouse spinal cord cultures. Virology. 1982;119:317–331. doi: 10.1016/0042-6822(82)90092-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hauri H P, Schweizer A. The endoplasmic reticulum-Golgi intermediate compartment. Curr Opin Cell Biol. 1992;4:600–608. doi: 10.1016/0955-0674(92)90078-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heusipp G, Harms U, Siddell S G, Ziebuhr J. Identification of an ATPase activity associated with a 71-kilodalton polypeptide encoded in gene 1 of the human coronavirus 229e. J Virol. 1997;71:5631–5634. doi: 10.1128/jvi.71.7.5631-5634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hogue B. Bovine coronavirus nucleocapsid protein processing and assembly. Adv Exp Med Biol. 1995;380:259–263. doi: 10.1007/978-1-4615-1899-0_41. [DOI] [PubMed] [Google Scholar]
- 19.Hogue B, Brian D. Structural proteins of human respiratory coronavirus OC43. Virus Res. 1986;5:131–144. doi: 10.1016/0168-1702(86)90013-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klumperman J, Krijnse Locker J, Meijer A, Horzinek M C, Geuze H J, Rottier P J M. Coronavirus M protein accumulates in the Golgi complex beyond the site of virion budding. J Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krijnse-Locker J, Ericsson M, Rottier P J, Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krijnse-Locker J, Klumperman J, Oorschot V, Horzinek M, Geuze H, Rottier P. The cytoplasmic tail of mouse hepatitis virus M protein is essential but not sufficient for its retention in the Golgi complex. J Biol Chem. 1994;269:28263–28269. [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Lavi E, Wang Q, Weiss S R, Gonatas N K. Syncytia formation induced by coronavirus infection is associated with fragmentation and rearrangement of the Golgi apparatus. Virology. 1996;221:325–334. doi: 10.1006/viro.1996.0382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H-J, Shieh C-K, Gorbalenya A E, Koonin E V, LaMonica N, Tuler J, Bagdzhadhzyan A, Lai M M C. The complete sequence (22 kilobases) of murine coronavirus gene 1 encoding the putative proteases and RNA polymerase. Virology. 1991;180:567–582. doi: 10.1016/0042-6822(91)90071-I. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu D, Xu H, Brown T. Proteolytic processing of the coronavirus infectious bronchitis virus 1a polyprotein: identification of a 10-kilodalton polypeptide and determination of its cleavage sites. J Virol. 1997;71:1814–1820. doi: 10.1128/jvi.71.3.1814-1820.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu X T, Sims A C, Denison M R. Mouse hepatitis virus 3C-like protease cleaves a 22-kilodalton protein from the open reading frame 1a polyprotein in virus-infected cells and in vitro. J Virol. 1998;72:2265–2271. doi: 10.1128/jvi.72.3.2265-2271.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lu X T, Lu Y Q, Denison M R. Intracellular and in vitro translated 27-kDa proteins contain the 3C-like proteinase activity of the coronavirus MHV-A59. Virology. 1996;222:375–382. doi: 10.1006/viro.1996.0434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu Y, Lu X, Denison M R. Identification and characterization of a serine-like proteinase of the murine coronavirus MHV-A59. J Virol. 1995;69:3554–3559. doi: 10.1128/jvi.69.6.3554-3559.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lu Y Q, Denison M R. Determination of mouse hepatitis virus 3C-like proteinase activity. Virology. 1997;230:335–342. doi: 10.1006/viro.1997.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machamer C E, Mentone S A, Rose J K, Farquhar M G. The E1 glycoprotein of an avian coronavirus is targeted to the cis Golgi complex. Proc Natl Acad Sci USA. 1990;87:6944–6948. doi: 10.1073/pnas.87.18.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Massalski A, Coulter-Mackie M, Knobler R, Buchmeier M, Dales S. In vivo and in vitro models of demyelinating diseases. V. Comparison of the assembly of mouse hepatitis virus, strain JHM, in two murine cell lines. Intervirology. 1982;18:135–146. doi: 10.1159/000149316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mizutani T, Maeda A, Hayashi M, Isogai H, Namioka S. Both antisense and sense RNAs against the nucleocapsid protein gene inhibit the multiplication of mouse hepatitis virus. J Vet Med Sci. 1994;56:211–215. doi: 10.1292/jvms.56.211. [DOI] [PubMed] [Google Scholar]
- 34.Pinon J, Mayreddy R, Turner J, Khan F, Bonilla P, Weiss S. Efficient autoproteolytic processing of the MHV-A59 3C-like proteinase from the flanking hydrophobic domains requires membranes. Virology. 1997;230:309–322. doi: 10.1006/viro.1997.8503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Risco C, Anton I, Enjuanes L, Carrascosa J. The transmissible gastroenteritis coronavirus contains a spherical core shell consisting of M and N proteins. J Virol. 1996;70:4773–4777. doi: 10.1128/jvi.70.7.4773-4777.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Risco C, Muntion M, Enjuanes L, Carrascosa J. Two types of virus-related particles are found during transmissible gastroenteritis virus morphogenesis. J Virol. 1998;72:4022–4031. doi: 10.1128/jvi.72.5.4022-4031.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiller J J, Kanjanahaluethai A, Baker S C. Processing of the coronavirus MHV-JHM polymerase polyprotein: identification of precursors and proteolytic products spanning 400 kilodaltons of ORF1a. Virology. 1998;242:288–302. doi: 10.1006/viro.1997.9010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi S T, Schiller J J, Kanjanahaluethai A, Baker S, Oh J, Lai M M C. Colocalization and membrane association of murine hepatitis virus gene 1 products and de novo-synthesized viral RNA in infected cells. J Virol. 1999;73:5957–5969. doi: 10.1128/jvi.73.7.5957-5969.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siddell S. Coronavirus JHM: tryptic peptide fingerprinting of virion proteins and intracellular polypeptides. J Gen Virol. 1982;62:259–269. doi: 10.1099/0022-1317-62-2-259. [DOI] [PubMed] [Google Scholar]
- 40.Stohlman S, Lai M. Phosphoproteins of murine hepatitis viruses. J Virol. 1979;32:672–675. doi: 10.1128/jvi.32.2.672-675.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sturman L S, Holmes K V, Behnke J. Isolation of coronavirus envelope glycoproteins and interaction with the viral nucleocapsid. J Virol. 1980;33:449–462. doi: 10.1128/jvi.33.1.449-462.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tooze J, Tooze S, Fuller S. Sorting of progeny coronavirus from condensed secretory proteins at the exit from the trans-Golgi network of AtT20 cells. J Cell Biol. 1987;105:1215–1226. doi: 10.1083/jcb.105.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tooze J, Tooze S, Warren G. Replication of coronavirus MHV-A59 in sac-cells: determination of the first site of budding of progeny virions. Eur J Cell Biol. 1984;33:281–293. [PubMed] [Google Scholar]
- 44.Tooze J, Tooze S A. Infection of AtT20 murine pituitary tumour cells by mouse hepatitis virus strain A59: virus budding is restricted to the Golgi region. Eur J Cell Biol. 1985;37:203–212. [PubMed] [Google Scholar]
- 45.Tooze S, Tooze J, Warren G. Site of addition of N-acetyl-galactosamine to the E1 glycoprotein of mouse hepatitis virus-A59. J Cell Biol. 1988;106:1475–1487. doi: 10.1083/jcb.106.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.van der Meer Y, Snijder E J, Dobbe J C, Schleich S, Denison M R, Spaan W J M, Krijnse Locker J. The localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J Virol. 1999;73:7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ziebuhr J, Siddell S G. Processing of the human coronavirus 229E replicase polyproteins by the virus-encoded 3C-like proteinase: identification of proteolytic products and cleavage sites common to pp1a and pp1ab. J Virol. 1999;73:177–185. doi: 10.1128/jvi.73.1.177-185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]