Abstract
Introduction
Hyperoxaluria is a risk factor for kidney stone formation and chronic kidney disease progression. The microbiome is an important protective factor against oxalate accumulation through the activity of its oxalate-degrading enzymes (ODEs). In this cross-sectional study, we leverage multiomics to characterize the microbial community of participants with primary and enteric hyperoxaluria, as well as idiopathic calcium oxalate kidney stone (CKS) formers, focusing on the relationship between oxalate degrading functions of the microbiome.
Methods
Patients diagnosed with type 1 primary hyperoxaluria (PH), enteric hyperoxaluria (EH), and CKS were screened for inclusion in the study. Participants completed a food frequency questionnaire recording their dietary oxalate content while fecal oxalate levels were ascertained. DNA and RNA were extracted from stool samples and sequenced. Metagenomic (MTG) and metatranscriptomic (MTT) data were processed through our bioinformatics pipelines, and microbiome diversity, differential abundance, and networks were subject to statistical analysis in relationship with oxalate levels.
Results
A total of 38 subjects were recruited, including 13 healthy participants, 12 patients with recurrent CKS, 8 with PH, and 5 with EH. Urinary and fecal oxalate were significantly higher in the PH and the EH population compared to healthy controls. At the community level, alpha-diversity and beta-diversity indices were similar across all populations. The respective contributions of single bacterial species to the total oxalate degradative potential were similar in healthy and PH subjects. MTT-based network analysis identified the most interactive bacterial network in patients with PH. Patients with EH had a decreased abundance of multiple major oxalate degraders.
Conclusion
The composition and inferred activity of oxalate-degrading microbiota were differentially associated with host clinical conditions. Identifying these changes improves our understanding of the relationships between dietary constituents, microbiota, and oxalate homeostasis, and suggests new therapeutic approaches protecting against hyperoxaluria.
Keywords: hyperoxaluria, kidney stones, microbiome, multiomics, oxalate
Graphical abstract
Hyperoxaluria is a known risk factor for nephrolithiasis,1 which affects ∼9% of US adults and is increasing in prevalence.2 Most urinary tract stones are composed of calcium oxalate (CaOx).3 Oxalate in the urine originates from 2 sources; up to half comes from the diet whereas the remainder is an endogenous end product of hepatic metabolism.4 High urinary oxalate levels are also a risk factor for chronic kidney disease progression.5 Hepatic overproduction or intestinal overabsorption of oxalate increases urinary excretion, with hyperoxaluria defined as excretion >40 mg/24h.6 The PHs are a group of very rare inherited conditions, with estimated prevalence of 1 to 2 per million in the US, in which defective hepatic metabolism of oxalate precursors leads to oxalate overproduction.7 Type 1 PH (PH1) due to pathologic changes in the AGXT gene has on average the highest urinary oxalate excretion and more severe long term outcomes.8 These genetic diseases are distinguished from EH, which can result from any cause of intestinal malabsorption of fat including Roux-en-Y gastric bypass (RYGB)9 or inflammatory bowel disease.10 In EH, fat malabsorption leads to higher bioavailability of unbound dietary oxalate that is mostly absorbed passively in the gastrointestinal tract.11
The trillions of microorganisms that constitute the gut microbiome have emerged as an important determinant of health and disease, with bacterial metabolic activity often complementing missing host functions.12 Humans lack the enzymes for degrading oxalate, whereas oxalate-degrading bacteria (ODB), termed the oxalobiome, colonize the human intestinal track.13 Whereas earlier microbiome studies relied on 16S rRNA sequencing for determining taxonomic abundance, there is increasing focus on bacterial functional potential as inferred from MTG data. Furthermore, to date few studies have evaluated bacterial function in vivo using MTT sequencing.14 ODB abundances have been described in the microbiome of healthy participants and patients with inflammatory bowel disease15 and in CKS formers.16 However, the oxalobiome of patients with PH, which as a group is characterized by hepatic overproduction of oxalate, has not been defined beyond the prevalence of Oxalobacter formigenes colonization.17 Furthermore, there has not been sufficient study of the oxalobiome of patients with EH. Previous studies, based on 16S rRNA and MTG sequencing of the microbiome, do not accurately represent in vivo oxalate degradation activity.16 To address this gap, we previously described transcription of ODEs using MTT sequencing to understand oxalate degradation in vivo.15
We hypothesize that the diversity and functions of the microbiome are impacted by diseases that vary in their mechanism of oxalate excess. Thus, in this cross-sectional study, we characterized the microbiome composition of different populations at risk for hyperoxaluria and kidney stones. We then focused on the oxalobiome and assessed the link between diet, oxalobiome structure and function, and fecal oxalate.
Methods
Study Population Recruitment
Between August 2016 and September 2018, we recruited participants with PH1 from the Rare Kidney Stone Consortium Oxalosis and Hyperoxlauria PH registry at the Mayo Clinic, Rochester Minnesota. Healthy, EH, and CaOx stone-forming participants were recruited from the kidney stone clinics at Mayo Clinic and New York University Langone Health. Eligibility criteria included the following: age between 18 and 80 years and relatively preserved kidney function (estimated glomerular filtration rate >50 ml/min per 1.73 m2). PH1 was confirmed via pathologic AGXT mutations18 and patients with EH had undergone RYGB surgery at least 6 months previously. Idiopathic CKS formers were screened for a history of a CKS either passed spontaneously or removed by urological intervention within 5 years of recruitment, with stone composition determined by infrared spectroscopy or x-ray crystallography. The control group consisted of healthy participants with no history of kidney or bowel disease. Patients who received antibiotics within 6 months of recruitment or had a history of kidney or liver transplant were excluded from the study.
We reviewed the clinical records and collected data on patient age, sex, race, ethnicity, weight, gastrointestinal symptoms, height, body mass index, and estimated glomerular filtration rate within 2 years of the stool collection. Results of 24-hour urine study collection in the patients before enrollment allowed the confirmation of hyperoxaluria in patients with EH and those with PH (urinary oxalate >45 mg/24h). To quantify the average dietary intake of calcium, oxalate, and vitamin C, participants completed a food frequency questionnaire using the Viocare software within 1 month after the stool collection.19
This study was conducted in compliance with the Declaration of Helsinki. Written informed consent was obtained from the patients for enrollment and approval was granted by the institutional review board of both institutions (Protocol Number: NCT02794649).
Data and Sample Collection
Urine chemistry analytes was obtained from the 24-hour urine measurements in the clinical records before recruitment. Urine chemistry analytes included oxalate, calcium, citrate, sodium, potassium, and pH, together with volume and supersaturation calculations by EQUIL220 (Supplementary Table S1). For healthy subjects, urine oxalate was imputed based on normal distributions from a large US population.21 For the 2 patients with CKS with missing urine data, the primary outcomes were imputed from the means of urine oxalate from the other CKS participants in our study of the same sex, as previously described.22 Subjects with missing dietary oxalate data were excluded from the affected analyses.
All participants were provided with stool collection kits with written instructions for fecal sample collection in Norgen stool stabilizer that were shipped to New York University. Norgen has been shown to stabilize RNA for up to 1 week and DNA for up to 1 month at room temperature.23 These aliquots were stored at −70 °C until processing. Fecal oxalate levels were measured using the Abcam kit (Abcam, Cambridge UK/Catalog # ab196990)15 and normalized by weight. For the measurement of total fecal oxalate content (as micromoles/mg of feces), fecal samples were vigorously mixed with ultrapure 2 M HCl (ratio of 1 mg to 9 μl) for 20 minutes at room temperature to ensure complete dissolution of all oxalate crystals.24
DNA was extracted from fecal samples using the DNeasy PowerSoil Pro Kit (Qiagen, Germantown, MD/Catalog # 47016) per protocol. RNA extraction was performed using the QIAGEN RNeasy PowerMicrobiome Kit per protocol (Qiagen/Catalog # 26000-50). Library preparation was performed using Kapa Hyper library prep. Whole MTG and MTT sequencing was performed using the Illumina HiSeq4000 sequencing platform at the New York University Genome technology center. All 38 samples except 1 were sequenced in the same run to reduce batch effects.
Bioinformatics Pipeline
Analysis of the microbiome community was performed by the New York University Microbiomic Informatics Laboratory of the Center for Health Informatics and Bioinformatics. The raw data obtained from shotgun MTG and MTT sequencing underwent upstream processing for quality control, including adapter trimming performed via trim-galore; quality scores <30 were trimmed. Human-associated gene removal was performed by KneadData v.0.7.4 with default settings. Microbial taxonomic profiles were generated with Kraken2.25 The extensive microbiome and metagenome data compiled as part of the Human Microbiome Project26 served as reference data for our analysis.
Using the MTG and MTT data, we performed targeted identification of microbial enzymes involved in oxalate degradation (formyl-CoA:oxalate CoA transferase [FRC] and oxalyl-CoA decarboxylase [OXC]) using our targeted pipeline, as described previously.15 Two protein homolog families of FRC and OXC were obtained from Uniprot InterPro via protein family number IPR017659 and IPR017660. The MTG and MTT analysis focused on exploiting differential genomic content and transcripts via a gene-targeted approach.
The MTG and MTT reads were aligned against these 2 protein homologs via diamond BLASTX, with the best hit (--max-target-seqs 1). Using reads mapped via BLASTX, we built contigs, against KEGG sequences of the targeted oxalate genes, performed on a per-sample basis over the proposed set of total samples.
Statistical Analysis
All statistical analysis was performed using R software (Version 4.3). We conducted descriptive analyses to compare clinical variables across disease groups, using analysis of variance for continuous variables, and Fisher exact test for categorical variables. Pairwise comparisons between any 2 disease condition populations using t-test for continuous variables and Fisher’s exact test for categorical variables. Pearson correlation was employed to assess the associations between dietary variables and fecal oxalate. Alpha (Observed, Chao1, Shannon, and Simpson indices) and beta diversities (Bray–Curtis dissimilarity and Jensen–Shannon divergence) were calculated at the species rank to estimate microbial community-level diversity in both MTG and MTT data. Linear regression was used to check whether alpha diversities vary across different populations, while adjusting for age and sex. Wilcoxon signed-rank tests were used for pairwise group comparisons of alpha diversity. Spearman correlation tests assessed associations between alpha diversity and both dietary factors (calcium and/or oxalate) and continuous clinical outcomes (urinary or fecal oxalate). Permutational multivariate analysis of variance27 was used to assess group beta diversity differences, adjusted for age and sex.
We then assessed the population-level contribution of different taxa to total ODEs, as described.15 The abundance of each ODE protein homolog was calculated as reads/kilobase/million. By deconvoluting ODE genes into taxa at the species level, we then evaluated the contribution of each species to both ODE genetic material (MTG; DNA) and their transcription (MTT; RNA) relative to the full oxalobiome. Species with prevalence less than 10% were excluded from the following analysis, as previously described.28,29
For both MTG and MTT data, we calculated gamma diversity, reflecting the total number of unique taxa across all samples within each population. We used ANCOM-BC28 to assess the differential abundance of a particular taxa harboring an ODE, between one of the populations of interest and healthy controls, while also adjusting for age and sex. This evaluation in the transcriptomic dataset informs about the differential transcriptional activity within the oxalate degrading community.
In microbial ecology, bacterial networks allow the representation of the interactions within the community based on rates of cooccurrence or coexclusion of the species involved in the network.30 Network analyses were conducted to assess interactions among ODB in the 4 populations based on the taxon-taxon correlations obtained using SparCC31 in both the MTG and MTT data; every correlation between 2 ODB constitutes an edge between 2 nodes.31 A prespecified correlation cut-off of 0.2 was used for network construction and its graphical representation. We used permutation tests to compare PH, EH, and CKS networks of ODB with healthy controls, based on the following 2 invariance measures: the global strength and the structure invariance.32 The former measure tests whether the overall level of connectivity is the same across populations and the latter measure tests whether any edges are different. We also considered 2 scenarios for the weight of edges used in both measures. Weights were either defined at 1 or considered in their observed correlation coefficients if the absolute correlation between 2 taxa was ≥0.2; in both cases, weight was attributed a value of 0 if absolute correlation was <0.2.
Furthermore, we explored whether the species contributing to the network as a community were associated with dietary components and fecal oxalate. Specifically, for a given constructed network, we treated the member species as a subcommunity and calculated its community diversities using Shannon index, total abundance, and the number of present taxa, respectively.33 Spearman correlation was then employed to investigate the relationship between these community diversities and the dietary components and fecal oxalate. The Benjamini-Hochberg procedure was applied to correct for multiple comparison testing.34 The q value, a false discovery rate-adjusted P value, is calculated using the Benjamini-Hochberg procedure for multiple testing correction. Due to the small sample size, P/q values <0.10 were considered as statistical significance.
Results
Population Characteristics
In total, 38 participants were included in the study as follows: 8 with PH1; 5 with EH that developed post-RYGB; 12 who were idiopathic CKS formers, and 13 who were healthy participants (Supplementary Figure S1). Their baseline characteristics are summarized in Table 1 and a head-to-head comparison can be found in Supplementary Tables S2 to S4, with q-values summarized in Supplementary Table S5.
Table 1.
Characteristics of the 38 subjects in the study population
| Characteristics | Population |
Healthy control |
Enteric hyperoxaluria |
Primary hyperoxaluria |
CKS former |
P-value∗ |
|---|---|---|---|---|---|---|
|
n |
13 |
5 |
8 |
12 |
||
| Age | 39.1 (11.8) | 55.6 (14.5) | 33.0 (13.6) | 47.5 (13.8) | 0.017 | |
| Sex | Female | 7 (53.8%) | 4 (80%) | 6 (75%) | 4 (33.3%) | 0.222 |
| Male | 6 (46.2%) | 1 (20%) | 2 (25%) | 8 (66.7%) | ||
| Ethnicity | Hispanic or Latino | 2 (15.4%) | 0 (0%) | 0 (0%) | 1 (8.3%) | 0.852 |
| not Hispanic or Latino | 11 (84.6%) | 5 (100%) | 8 (100%) | 11 (91.7%) | ||
| Race | American Indian | 0 (0%) | 0 (0%) | 1 (12.5%) | 0 (0%) | 0.834 |
| White | 11 (84.6%) | 5 (100%) | 6 (75.0%) | 10 (83.3%) | ||
| Other | 2 (15.4%) | 0 (0%) | 1 (12.5%) | 2 (16.7%) | ||
| Body Measures | Height (cm) | 166 (10.5) | 167 (7.82) | 170 (4.60) | 174 (8.62) | 0.165 |
| Weight (kg) | 70.4 (13.7) | 99.4 (19.6) | 72.3 (8.40) | 83.5 (21.2) | 0.016 | |
| BMI | 25.2 (3.43) | 35.7 (5.88) | 24.9 (1.76) | 27.2 (5.43) | <0.001 | |
| Nutritional Data | Dietary calcium (mg) | 1160 (629) | 1250 (426) | 1320 (464) | 1300 (742) | 0.938 |
| Dietary oxalate (mg) | 236 (316) | 331 (275) | 333 (323) | 212 (167) | 0.741 | |
| Ox/Ca ratio | 0.280 (0.233) | 0.275 (0.232) | 0.250 (0.196) | 0.184 (0.144) | 0.832 | |
| Dietary vitamin C | 80.4 (37.6) | 133 (152) | 152 (61.5) | 156 (129) | 0.249 | |
| Outcome | Fecal oxalate (uM/mg) | 0.240 (0.0944) | 0.932 (0.117) | 0.998 (0.140) | 0.742 (0.192) | <0.001 |
| Urinary oxalate (mg/24h) | 31.9 (7.73) | 62.5 (12.7) | 99.9 (57.0) | 33.8 (12.2) | <0.001 |
BMI, body mass index.
Data for body measurements, nutrition, and outcomes are presented as Mean (± SD).
P-value from the ANOVA test for the continuous variables, and chi-squared test for the categorical variables.
This study’s participants were mostly (84%) White, and 55% were females. Patients with PH (aged 33.0 ± 13.6 years) were significantly younger than in the other groups of stone formers CKS (aged 47.5 ± 13.8 years) and those with EH (aged 55.6 ± 14.6 years). All groups had similar body mass index except the patients with EH who had higher mean body mass index (35.7 ± 5.88 kg/m2), reflecting their history of obesity and indication for RYGB.
The main dietary exposures of interest, calcium and oxalate intake, were similar across the 4 populations (Table 1). Dietary vitamin C intake among patients with PH was higher than in healthy controls (152 ± 61.5 vs. 80.4 ± 37.6 mg, P = 0.02) (Supplementary Table S2).
Urine oxalate from chart review was similar between the CKS stone formers and the healthy controls in the US population (33.8 ± 12.2 mg/24h vs. 31.9 ± 7.73 mg/24h; P = 0.66).21 Compared to the controls, patients with EH (62.5 ± 12.7 mg/24h; P = 0.003) and PH1 (99.9 ± 57 mg/24h; P = 0.012) had significantly higher levels of urine oxalate, thus confirming the hyperoxaluria phenotype. Other urine parameters were similar in all 3 stone-forming groups, except for 24-hour urine volume, which was highest in PH1 participants (Supplementary Table S1).
Measured fecal oxalate levels were significantly higher in patients with PH1 (0.998 ± 0.140 μM/mg), those with EH (0.932 ± 0.117 μM/mg) or CKS formers (0.742 ± 0.192 μM/mg) compared with the healthy controls (0.240 ± 0.094 μM/mg; P < 0.001 in all 3 cases).
To address whether dietary intake patterns were associated with measured fecal oxalate, we examined dietary calcium, oxalate, and vitamin C intake within each disease subgroup (Supplementary Figure S2). There were no significant associations except that vitamin C was positively correlated with fecal oxalate in CKS formers.
Bacterial Community Level Analysis
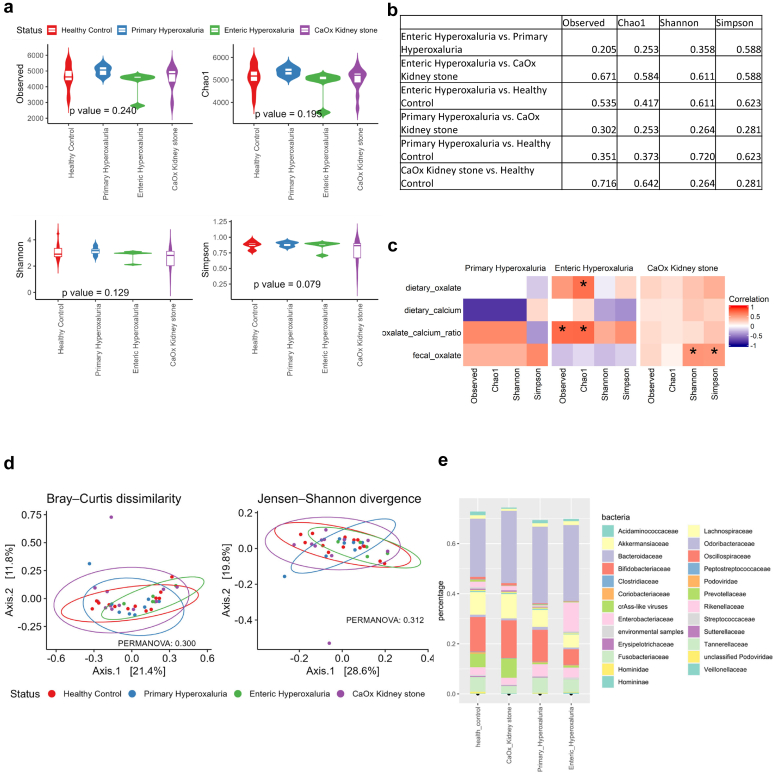
Alpha diversity measures the taxonomic richness and the evenness of the microbiome. There were no differences in alpha diversity between the groups of stone-formers and the healthy controls (Figure 1a and b). We then explored the correlation between alpha-diversity and dietary covariates and fecal oxalate (Figure 1c). None of the comparisons were significant, but the oxalate to calcium ratio was positively correlated with alpha diversity in PH and EH, and calcium intake was negatively associated with alpha diversity in PH and EH. Analysis of beta-diversity, examining differences in community population structure between the clinical groups, showed no significant differences (Figure 1d). Therefore, at the global level of microbiota analysis there were no consistent differences between the clinical groups.
Figure 1.
Metagenomic findings, based on Kraken output. (a) Alpha diversity comparison between groups, with p-value based on analysis of variance adjusted for age and sex. (b) Pairwise group comparison for alpha diversity: q-value based on Benjamini-Hochberg correction adjusted for age and sex. (c) Association analysis of alpha diversity with clinical variables using a heat map of metagenomic alpha diversity correlation with outcomes; ∗P-value < 0.1. (d) Beta-diversity based on the Bray-Curtis dissimilarity and the Jensen-Shannon divergence, with P-value based on permutational multivariate analysis of variance, adjusted for age and sex. (e) Taxonomic abundance in the different groups of study subjects. CaOx, calcium oxalate.
At the taxonomic family level, the Bacteroidaceae represented the largest single group in all populations. The Prevotellaceae, which were highly abundant in the healthy and CKS population, were not as abundant in the PH and EH populations. The Enterobacteriaceae accounted for a bigger part of the microbiome population in patients with EH than in the other groups (Figure 1e).
ODEs
Two oxalate degradation pathways have been described in bacteria. The 2-step pathway involving sequential reactions catalyzed by FRC and OXC is most frequently seen in human-associated microbiota, whereas the pathway involving the oxalate oxidase/decarboxylase enzyme is rarely detected in the human gut microbiota.15
High throughput sequencing, by evaluating genes related to oxalate degradation in all 38 subjects, led to 2 complementary outputs. MTG sequencing provides a normalized abundance of bacterial genes, allowing quantitation of the genetic potential of the microbiome for a specific function. The MTT output assesses the aggregate and specific transcriptional activity level of the microbiota. We focused on FRC in all further analyses for 2 reasons. First, FRC has been better characterized with a higher number of reference proteins in databases than OXC.35 Second, the profiles based on FRC and OXC mirrored each other in our datasets (Figure 2a and Supplementary Figure S3A).
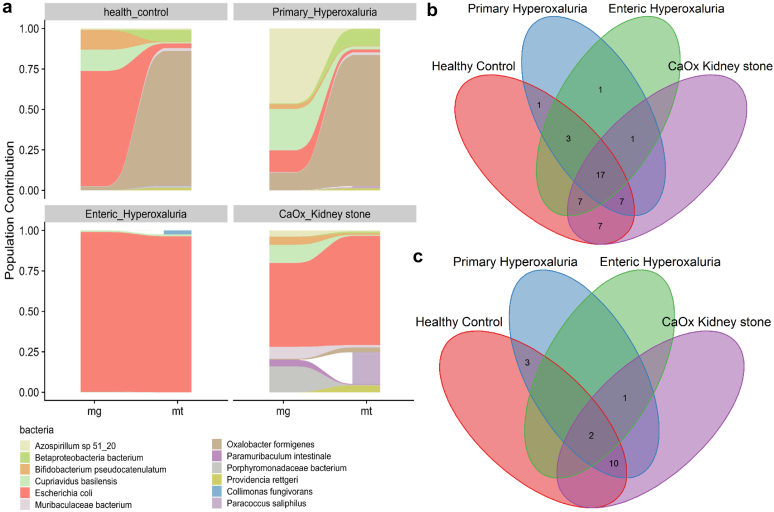
Figure 2.
Population level contribution of the formyl-CoA:oxalate CoA transferase oxalate-degrading enzyme. (a) Population level contribution based on formyl-CoA:oxalate CoA transferase metagenomic and metatranscriptomic level of activity. (b) Gamma diversity of the oxalobiome based on formyl-CoA:oxalate CoA transferase in the metagenomic analysis. (c) Gamma diversity of the oxalobiome based on formyl-CoA:oxalate CoA transferase in the metatranscriptomic analysis. CaOx, calcium oxalate.
In total, we identified 183 taxa encoding FRC in the MTG and 106 taxa transcribing FRC in the MTT data. These results confirm our previous findings that not all taxa with ODG transcribe those genes in vivo.14 Computing the contribution of each bacterial taxa to overall oxalate degradation from the MTG to the MTT levels in every population allowed identification of the most active contributors to oxalate degradation in each of the groups in vivo (Figure 2a).
In healthy participants, the contribution of O formigenes to total ODEs increased from less than 10% at the gene level to approximately >70% at the transcripts level. A similar pattern was observed in PH1, with the O formigenes contribution accounting for most of the ODEs at the transcriptional level. Bacteria that predominated at the gene level, like Escherichia coli in healthy participants or Azospirillum sp 51_20 in patients with PH1, had a more modest contribution when the transcriptome was assessed. Importantly, the E coli contribution to the ODEs was predominant in CKS formers and patients with EH at both the gene and transcript level, suggesting a disruption of the normal oxalate degrading community in these diseases, with little or no contribution of O formigenes.
In summary, patients with PH1 had a similar pattern of oxalate-degrading activity across their microbiome when compared with healthy participants, with predominant O formigenes contribution at the transcriptional level, whereas oxalate degradation was via other bacteria including E Coli in EH and CKS.
At the MTG level, the healthy controls had the highest number of taxa with oxalate-degrading potential, (42) followed by CKS, PH, and EH (Figure 2b). A total of 17 taxa were shared by the 4 groups (Supplementary Table S6). At the MTT level, PH1 had the highest number of taxa transcribing FRC; followed by healthy controls; then CKS; and EH, which only has 3 taxa transcribing FRC (Figure 2c). These pattern differences point toward microbiome associations with illness status that may have substantial clinical significance.
Differential Abundance Analysis
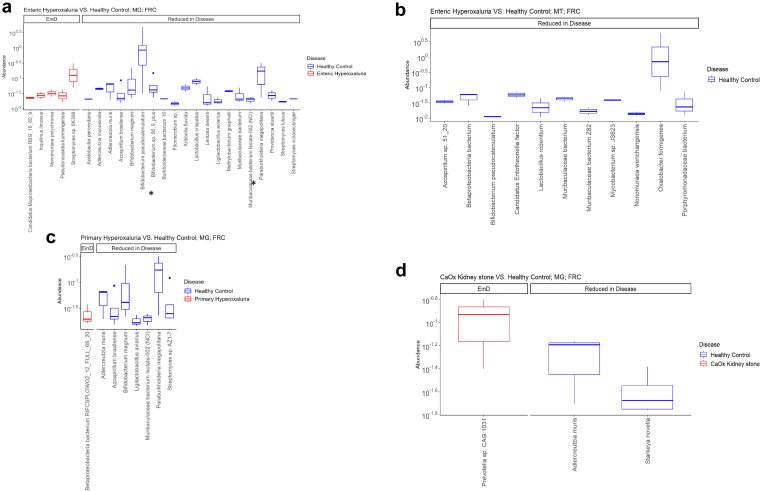
At the species level, in the MTG patients with data EH had a decreased abundance of several oxalate-degraders such as Bifidobacteria and Muribaculaceae (Figure 3a). In the MTT dataset, the differential abundance analysis reflects the differential activity of a member of the gut microbiome community. Notably, O formigenes was noted to be depleted in the EH population, as was Bifidobacterium pseudocatenulatum (Figure 3b). At the MTG level, Adlercreutzia muris had decreased abundance in the 3 disease populations (Figure 3).
Figure 3.
Differential abundance analysis using ANCOM-BC in primary hyperoxaluria, enteric hyperoxaluria, and CaOx kidney stone patients compared with healthy controls, based on formyl-CoA:oxalate CoA transferase, adjusted for age and sex. (a) enteric hyperoxaluria versus healthy controls at metagenomic level; species found to be concordantly differentially abundant in the metagenomic and metatranscriptomic datasets are highlighted with an ∗. (b) Enteric hyperoxaluria versus healthy controls at metatranscriptomic level. (c) Primary hyperoxaluria versus healthy controls at metagenomic level. (d) CKS versus healthy controls at metagenomic level. CaOX, calcium oxalate; EinD, Enriched in disease.
Network Analysis
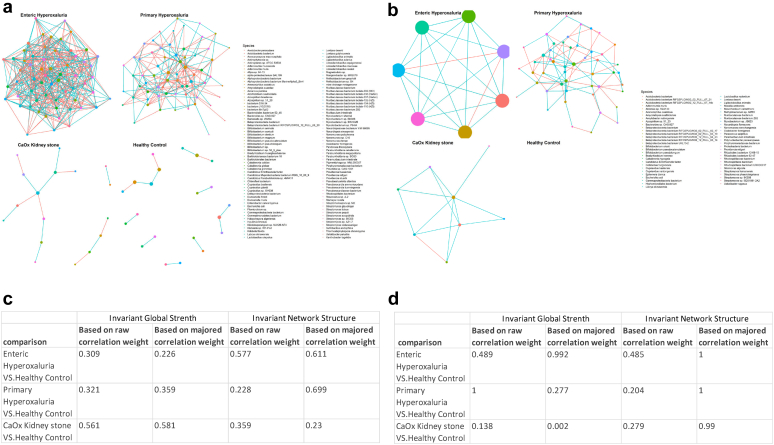
In the MTG analysis, patients with EH had the most active bacterial interaction network whereas patients with PH1 demonstrated a less interactive network at the gene level (Figure 4a and c). In contrast, the transcriptional data revealed an opposite pattern with a dense network in patients with PH1 as opposed to a scarce network interconnection in the EH group (Figure 4b and d). ODB interaction networks in CKS formers and healthy controls were poorly developed based on both the MTG and MTT datasets. Moreover, CKS formers manifest a significantly different network than healthy controls, based on the invariant global strength of the FRC transcriptomic network (P = 0.002). Although OXC-based networks were less developed for all groups, they showed similar patterns as the FRC networks (Supplementary Figure S4A–D).
Figure 4.
Oxalate-degrading bacteria network based on formyl-CoA:oxalate CoA transferase levels. (a) Oxalate-degrading bacterial interaction network based on the metagenomic abundances of formyl-CoA:oxalate CoA transferase. (b) Oxalate-degrading bacterial interaction network based on the metatranscriptomic abundances of formyl-CoA:oxalate CoA transferase. (c) Network comparison in the metagenomic-based formyl-CoA:oxalate CoA transferase networks. (d) Network comparison in the metatranscriptomic-based formyl-CoA:oxalate CoA transferase networks. A correlation cut-off of 0.2 is used for graphical representation. Each red edge is a negative correlation, and a blue edge is a positive correlation. Network comparisons are summarized in a table of P-values, using 2 metrics with 2 types of comparison, one involving the raw correlation value and the other a majored value. Significance is shown in bold. CaOx, calcium oxalate.
The node degree (ND) of a species in network analyses refers to its number of connections to other species and estimated betweenness (EB) to the number of times a node lies on the shortest path between other nodes. Both ND and EB infer the importance of species’ involvement in a specific network. Species with the highest ND and EB are central to a network because they act as bridges between multiple nodes (Supplementary Tables S7–S10). In the PH1 group, O formigenes involvement in the oxalate-degraders network seemed peripheral based on the MTG analysis of FRC, whereas analysis of the MTT dataset instead establishes O formigenes as the most crucial species in the network, having both the highest ND and EB of the network. B pseudocatenulatum, Muribaculaceae, and E coli were also among the major species. In patients with EH, networks were only identifiable on the gene level (MTG), with the major species being Ligilactobacillus animalis, Pseudonocardiaceae bacterium, Caballeronia grimmiae and Actinophytocola sp., whereas species such as B dentium, O formigenes, and Porphyromonadaceae bacterium had intermediate importance in the network. ND and EB were not computed for the CKS and healthy participants because the networks were underdeveloped.
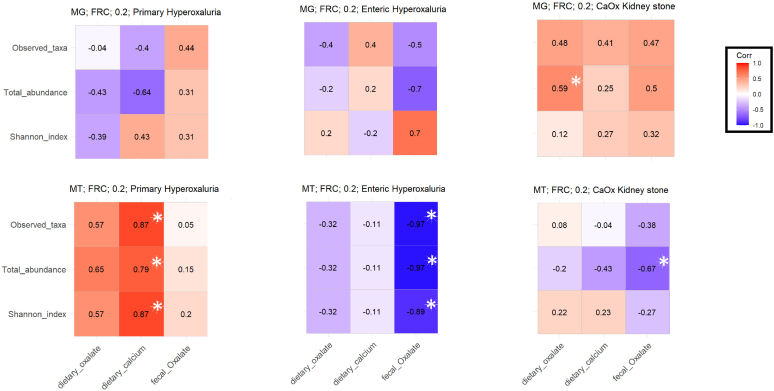
Next, we examined correlations between the taxa identified as members of a network and either dietary factors or fecal oxalate in EH, PH1 and CKS participants (Figure 5). In the EH and PH1 cohorts, 3 microbial metrics, namely total abundance, Shannon index, and observed taxa from the transcriptomic MTT data, had consistent correlations, whereas there was none at the MTG level. In patients with PH1, the oxalate-degrading network was positively correlated with dietary calcium and oxalate exposure. In patients with EH, the network of oxalate degraders was negatively correlated with fecal oxalate levels. For the CKS formers, the MTG and MTT datasets yielded opposite trends in every analysis.
Figure 5.
Network correlation with dietary factors and outcomes of interests in patients with primary hyperoxaluria patients with enteric hyperoxaluria, and CKS formers. Correlation of FRC activity at the metagenomic and metatranscriptomic levels to dietary factors and urinary and fecal oxalate. Spearman correlation is performed. ∗P 0.05. CKS, calcium oxalate kidney stone; FRC, formyl-CoA:oxalate CoA transferase; MG, metagenomics; MT, metatranscriptomics.
Discussion
Our study provides novel insights into the characteristics of the gut microbiome and ODB in 3 distinct cohorts of kidney stone formers. Although the microbiome of stone formers has been explored in previous studies,16 the gut microbiome and specifically the function of ODB have not been examined in patients with PH1 or those with EH. The differences we identified between the MTG and MTT outputs highlight the importance of focusing on functional analysis rather than just taxonomic composition. Our results show that although overall community compositions were comparable in the healthy and other KS-forming conditions, analysis of bacterial networks demonstrated the highest bacterial interaction density in patients with PH on an MTT level. By comparing the MTG and MTT analyses, we further support the notion that functional level analysis provides a more comprehensive understanding of microbiome activity and its implications for health outcomes. Our approach also typifies a step toward personalized medicine whereby in the case of kidney stone formers, based on their microbiome, we argue that not all populations would benefit equally from a probiotic. High fecal oxalate levels were found in the 3 stone forming groups (EH, PH1, and CKS) compared with healthy controls despite no significant difference in dietary calcium or oxalate intake. In PH1, this finding is consistent with observations in PH1 murine models that suggested a gut oxalate clearance route in chronic kidney disease.36,37 We also identified enrichment of the normal oxalate-degrading community and function in the PH1 cohort and speculate that this derives from increased substrate availability due to oxalate secretion into the gut lumen. Enrichment of the oxalobiome in patients with PH1 could explain the lack of apparent effect on urinary oxalate excretion in human trials that supplemented O formigenes.38,39 The gut microbiome response to a high oxalate milieu was different in CKS formers and patients with EH, suggesting a dysbiotic oxalobiome.
The interactions between diet, the microbiome, and host are major determinants of health and disease.40 Human oxalate homeostasis is maintained by the interplay between dietary oxalate gut absorption and secretion, its endogenous production, and renal excretion.41 Oxalate is ubiquitous in the human diet,42 and urinary excretion levels are related to dietary amounts.43,44 PH1 is an exception because dietary effects on urinary oxalate are overwhelmed by hepatic overproduction. The dietary oxalate intakes we measured were similar in the healthy controls and patients with kidney stone, confirming previous reports demonstrating that stone-formers often have difficulty complying with a low oxalate diet.45,46
EH and PH1 are rare conditions characterized by a disruption of oxalate homeostasis.11,47 In PH1, the most severe form of PH, an increase in endogenous oxalate production leads to severe hyperoxaluria frequently requiring kidney and/or liver transplant. In EH, increased bioavailability of dietary oxalate secondary to fat malabsorption causes hyperoxaluria, kidney stones, and in severe cases, kidney failure. An important modulator of intestinal oxalate absorption is bacterial oxalate degradation.41 It has been inferred from studies in CKS formers that high oxalate loads could affect the gut microbiome. Inhibition of multiple species including O formigenes was attributed to increased fecal oxalate concentrations, a claim that should be cautiously interpreted in the absence of fecal oxalate measurements.48 A similar potentially toxic effect of high intestinal oxalate concentrations on the gut microbiome has been inferred from animal studies where bacterial diversity steeply decreased once dietary oxalate supplementation reached 12%.49 The translation of this finding to humans is questionable because of the use of supraphysiological dietary oxalate. Interestingly, the same study described increased microbiota diversity and oxalate-degrading activity with lesser (0.5%–3%) oxalate supplementation.49
Despite previous attempts to reduce gastrointestinal oxalate absorption via oral administration of oxalate degraders to both patients with PH1 and murine PH1 models,38,39,50,51 to our knowledge, no study has attempted to investigate the effect of high oxalate on the oxalobiome in PH. We now have established that the gut microbiome of patients with PH1 is exposed to high oxalate concentrations, with a fecal oxalate level 4 times higher than in healthy controls; this increases the activity of the bacterial community to counter the increased load and increase oxalate degradation. Network analysis of the metatranscriptome showed a densely interconnected network in PH1, with O formigenes identified as the core species of the oxalate-degrading network. In this case, the observation that patients with PH1 and healthy controls share many species in the gamma diversity analysis suggests a simple response to increased substrate in the PH1 group.
Reduced oxalate-degrading activity has been found in fecal cultures of patients who underwent jejunoileal bypass compared to healthy individuals52; however, assessment of oxalobiome function has not been previously performed in patients after RYGB surgery. Exploring the association of the known dysbiosis in RYGB patients with higher fecal oxalate concentration was one of our primary aims. This is particularly relevant in the context of the ongoing worldwide increase in obesity prevalence,53 especially because obesity is associated with both CKS and hyperoxaluria.54 In previous studies, hyperoxaluria before RYGB surgery did not correlate with fecal fat excretion; however, urinary oxalate excretion and fecal fat output were strongly correlated 1 year after surgery.9 This observation confirms that malabsorption after bariatric surgery is a main driver of hyperoxaluria in this situation.55 Our EH patient population demonstrated differential species abundance compared with healthy participants, most notably depletion of Bifidobacteria and O formigenes. Given that both are ODB, supplementation with these taxa might be beneficial in patients with EH. Depletion of ODB (O formigenes and Bifidobacteria sp.) could be explained by the steatorrhea associated with EH leading to the thriving of bacteria that degrade fat for their energy metabolism; this could be potentially an environment overwhelming ODB. Another possibility is that during the RYGB procedure, patients receive antibiotics, which can suppress those bacteria and our data and others have shown that O formigenes is sensitive to several commonly used antibiotics.56 Bile acid malabsorption resulting from the RYGB could also affect the viability of the gut microbiota, particularly O formigenes.57
Although several studies evaluated ODB abundance in stone formers at the gene level,16,58 we clearly see that an MTT assessment is needed to infer the functional aspect along the microbiome community. This is particularly relevant because the Oxalobacter genus has not been previously associated with the occurrence of nephrolithiasis in this population.59 We found that the fecal microbiome population in CKS formers differed significantly from healthy individuals in relation to abundance patterns for ODE, thus representing a dysbiosis. This finding implies that CKS formers rely more heavily on oxalate reduction by E coli, a pattern similar to patients with EH. E coli performs oxalate degradation for acid tolerance rather than as an energy substrate.60 ODE network analyses in CKS formers revealed a lower interaction density than in either patients with EH or those with PH, probably reflecting the enriched substrate sources in both hyperoxaluria illnesses. MTG and MTT datasets demonstrated opposite associations between members of the ODB networks in relation to the fecal oxalate concentrations which highlights the importance of assessing transcriptional activity.
Finally, we note that the population contribution of species to oxalate degradation at the gene and transcript level in the healthy controls is in agreement with our previous analysis of a larger cohort of healthy adults, highlighting the stability of the composition and its functional characteristics across healthy individuals.15 The network of oxalate-degraders in healthy participants was rudimentary compared with patients with EH or those with PH, suggesting that with lower fecal oxalate and a nondysbiotic microbiome, there is little selection for ODB community interaction. An underdeveloped network could also be explained by the contribution of individual species to total oxalate degrading activity, which suggested that >70% of oxalate degradation is performed by O formigenes. Although the concepts of niche partitioning and functional redundancy are highly relevant in shaping the microbiome in chronic kidney disease,61 the development of the healthy oxalobiome appears to follow these concepts with multiple species encoding genes for oxalate degradation and only a few species performing the function under healthy conditions.
We acknowledge that our study has limitations. The sample size is small, which is a common limitation of studies in rare diseases. However, because our patient populations have clear phenotypes, we believe our findings are significant. Given that RYGB results in multiple gut and metabolic changes, our findings may not be generalizable to all forms of EH. The imputation of some variables and timing variation in their collection are study limitations. Although diet is a known modulator of the gut microbiome, and we considered many dietary factors and urinary lithogenic features involved in CKS, diet was evaluated via a questionnaire. Even though our cross-sectional analysis identified salient features, causality cannot be inferred. Any significant relationships identified in statistical modeling must be confirmed experimentally. Interpretation of the identified networks, essential to an understanding of bacterial ecology, should also be cautious because these exploratory analyses were performed on relatively small sample sizes.
In conclusion, we now report on the PH1 gut microbiome and assess oxalate-degrading community function in different hyperoxaluric and stone-forming conditions. Identifying the species associated with the outcomes of interests creates a framework for further investigations in the role of the microbiome in affecting kidney diseases. MTT results, more closely reflecting the oxalobiome activity and identifying the most active species or clusters, should guide future attempts to manipulate the microbiome. A more extensive characterization of the microbiome in hyperoxaluric conditions could guide development of future targeted therapies.
Disclosure
All the authors declared no competing interests.
Acknowledgments
Funding for this study was from the Rare Kidney Stone Consortium U54DK083908; R01: 1R01LM014085, R01DK129675; and U01: AI22285.
Data Availability Statement
Data will be deposited into NCBI upon manuscript acceptance.
Author Contributions
DSG and LN conceived the study. JCL, DM, BS, DSG, and LN screened and recruited patients for the study. MH performed the assays. CW, ZC, HL, KVR, and NZ analyzed the data. NZ and LN wrote the first draft of the manuscript. NZ, CW, DM, JCL, HL, KVR, DSG, MB, and LN edited the manuscript. All authors approved the final version of the manuscript.
Footnotes
Figure S1. Study enrolment by clinical group.
Figure S2. Correlation heatmap between dietary components and fecal oxalate.
Figure S3. Population level contribution of the oxalyl-CoA decarboxylase oxalate-degrading enzyme.
Figure S4. Oxalate-degrading bacterial network based on oxalyl-CoA decarboxylase levels.
Table S1. 24-hour urinalysis.
Table S2. Group comparisons of clinical outcomes: primary hyperoxaluria versus healthy control.
Table S3. Group comparisons of clinical outcomes: enteric hyperoxaluria versus healthy control.
Table S4. Group comparisons of clinical outcomes: calcium oxalate kidney stone versus healthy control.
Table S5. q-value across 6 pairwise comparisons based on Benjamini-Hochberg correction.
Table S6. Gamma diversity: Venn diagram taxa.
Table S7. Node betweenness and degree for every species involved in formyl-CoA:oxalate CoA transferase metagenome dataset of patients with enteric hyperoxaluria.
Table S8. Node betweenness and degree for every species involved in formyl-CoA:oxalate CoA transferase metagenome dataset of patients with primary hyperoxaluria.
Table S9. Node betweenness and degree for every species involved in formyl-CoA:oxalate CoA transferase metatranscriptome dataset of patients with enteric hyperoxaluria.
Table S10. Node betweenness and degree for every species involved in in formyl-CoA:oxalate CoA transferase metatranscriptome dataset of patients with primary hyperoxaluria.
Supplementary Materials
Figure S1. Study enrolment by clinical group.
Figure S2. Correlation heatmap between dietary components and fecal oxalate.
Figure S3. Population level contribution of the oxalyl-CoA decarboxylase oxalate-degrading enzyme.
Figure S4. Oxalate-degrading bacterial network based on oxalyl-CoA decarboxylase levels.
Table S1. 24-hour urinalysis.
Table S2. Group comparisons of clinical outcomes: primary hyperoxaluria versus healthy control.
Table S3. Group comparisons of clinical outcomes: enteric hyperoxaluria versus healthy control.
Table S4. Group comparisons of clinical outcomes: calcium oxalate kidney stone versus healthy control.
Table S5. q-value across 6 pairwise comparisons based on Benjamini-Hochberg correction.
Table S6. Gamma diversity: Venn diagram taxa.
Table S7. Node betweenness and degree for every species involved in formyl-CoA:oxalate CoA transferase metagenome dataset of patients with enteric hyperoxaluria.
Table S8. Node betweenness and degree for every species involved in formyl-CoA:oxalate CoA transferase metagenome dataset of patients with primary hyperoxaluria.
Table S9. Node betweenness and degree for every species involved in formyl-CoA:oxalate CoA transferase metatranscriptome dataset of patients with enteric hyperoxaluria.
Table S10. Node betweenness and degree for every species involved in in formyl-CoA:oxalate CoA transferase metatranscriptome dataset of patients with primary hyperoxaluria.
References
- 1.Asplin J.R. Hyperoxaluric calcium nephrolithiasis. Endocrinol Metab Clin North Am. 2002;31:927–949. doi: 10.1016/s0889-8529(02)00030-0. [DOI] [PubMed] [Google Scholar]
- 2.Chewcharat A., Curhan G. Trends in the prevalence of kidney stones in the United States from 2007 to 2016. Urolithiasis. 2021;49:27–39. doi: 10.1007/s00240-020-01210-w. [DOI] [PubMed] [Google Scholar]
- 3.Khan S.R., Pearle M.S., Robertson W.G., et al. Kidney stones. Nat Rev Dis Primers. 2016;2 doi: 10.1038/nrdp.2016.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mitchell T., Kumar P., Reddy T., et al. Dietary oxalate and kidney stone formation. Am J Physiol Ren Physiol. 2019;316:F409–F413. doi: 10.1152/ajprenal.00373.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Waikar S.S., Srivastava A., Palsson R., et al. Association of urinary oxalate excretion with the risk of chronic kidney disease progression. JAMA Intern Med. 2019;179:542–551. doi: 10.1001/jamainternmed.2018.7980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hatch M. Gut microbiota and oxalate homeostasis. Ann Transl Med. 2017;5:36. doi: 10.21037/atm.2016.12.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochat P., Rumsby G. Primary hyperoxaluria. N Engl J Med. 2013;369:649–658. doi: 10.1056/NEJMra1301564. [DOI] [PubMed] [Google Scholar]
- 8.Zhao F., Bergstralh E.J., Mehta R.A., et al. Predictors of incident ESRD among patients with primary hyperoxaluria presenting prior to kidney failure. Clin J Am Soc Nephrol. 2016;11:119–126. doi: 10.2215/CJN.02810315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moreland A.M., Santa Ana C.A., Asplin J.R., et al. Steatorrhea and hyperoxaluria in severely obese patients before and after Roux-en-Y gastric bypass. Gastroenterology. 2017;152:1055–1067 e3. doi: 10.1053/j.gastro.2017.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Caudarella R., Rizzoli E., Pironi L., et al. Renal stone formation in patients with inflammatory bowel disease. Scan Microsc. 1993;7:371–379. [PubMed] [Google Scholar]
- 11.Witting C., Langman C.B., Assimos D., et al. Pathophysiology and treatment of enteric hyperoxaluria. Clin J Am Soc Nephrol. 2021;16:487–495. doi: 10.2215/CJN.08000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Backhed F., Ley R.E., Sonnenburg J.L., et al. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 13.Liu M., Nazzal L. Enteric hyperoxaluria: role of microbiota and antibiotics. Curr Opin Nephrol Hypertens. 2019;28:352–359. doi: 10.1097/MNH.0000000000000518. [DOI] [PubMed] [Google Scholar]
- 14.Ojala T., Kankuri E., Kankainen M. Understanding human health through metatranscriptomics. Trends Mol Med. 2023;29:376–389. doi: 10.1016/j.molmed.2023.02.002. [DOI] [PubMed] [Google Scholar]
- 15.Liu M., Devlin J.C., Hu J., et al. Microbial genetic and transcriptional contributions to oxalate degradation by the gut microbiota in health and disease. eLife. 2021;10 doi: 10.7554/eLife.63642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stanford J., Charlton K., Stefoska-Needham A., et al. The gut microbiota profile of adults with kidney disease and kidney stones: a systematic review of the literature. BMC Nephrol. 2020;21:215. doi: 10.1186/s12882-020-01805-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoppe B., Niaudet P., Salomon R., et al. A randomised Phase I/II trial to evaluate the efficacy and safety of orally administered Oxalobacter formigenes to treat primary hyperoxaluria. Pediatr Nephrol. 2017;32:781–790. doi: 10.1007/s00467-016-3553-8. [DOI] [PubMed] [Google Scholar]
- 18.Hoppe B. An update on primary hyperoxaluria. Nat Rev Nephrol. 2012;8:467–475. doi: 10.1038/nrneph.2012.113. [DOI] [PubMed] [Google Scholar]
- 19.Kristal A.R., Kolar A.S., Fisher J.L., et al. Evaluation of web-based, self-administered, graphical food frequency questionnaire. J Acad Nutr Diet. 2014;114:613–621. doi: 10.1016/j.jand.2013.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Werness P.G., Brown C.M., Smith L.H., Finlayson B. EQUIL2: a BASIC computer program for the calculation of urinary saturation. J Urol. 1985;134:1242–1244. doi: 10.1016/s0022-5347(17)47703-2. [DOI] [PubMed] [Google Scholar]
- 21.Curhan G.C., Willett W.C., Speizer F.E., Stampfer M.J. Twenty-four-hour urine chemistries and the risk of kidney stones among women and men. Kidney Int. 2001;59:2290–2298. doi: 10.1046/j.1523-1755.2001.00746.x. [DOI] [PubMed] [Google Scholar]
- 22.Gelman A., Hill J. Cambridge University Press; 2006. Data Analysis Using Regression and Multilevel/Hierarchical Models. [Google Scholar]
- 23.Kim W.S., Causarano J., Earle M., et al. Preservation of RNA in soil, plant and stool samples using Norgen’s RNA preserve. https://norgenbiotek.com/sites/default/files/resources/App%20Note%2092%20-%20Preservation%20of%20RNA%20using%20RNA%20Preserve%202019_2.pdf Published 2019. Accessed March 4, 2024.
- 24.Jiang J., Knight J., Easter L.H., et al. Impact of dietary calcium and oxalate, and Oxalobacter formigenes colonization on urinary oxalate excretion. J Urol. 2011;186:135–139. doi: 10.1016/j.juro.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20:257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson M.J. In: Wiley StatsRef: Statistics Reference Online. Balakrishnan N., Colton T., Everitt B., editors. John Wiley & Sons, Ltd; 2014. Permutational multivariate analysis of variance (PERMANOVA) pp. 1–15. [Google Scholar]
- 28.Lin H., Peddada S.D. Analysis of compositions of microbiomes with bias correction. Nat Commun. 2020;11:3514. doi: 10.1038/s41467-020-17041-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lloyd-Price J., Arze C., Ananthakrishnan A.N., et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature. 2019;569:655–662. doi: 10.1038/s41586-019-1237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matchado M.S., Lauber M., Reitmeier S., et al. Network analysis methods for studying microbial communities: a mini review. Comp Struct Biotechnol J. 2021;19:2687–2698. doi: 10.1016/j.csbj.2021.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedman J., Alm E.J. Inferring correlation networks from genomic survey data. PLOS Comput Biol. 2012;8 doi: 10.1371/journal.pcbi.1002687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van Borkulo C.D., van Bork R., Boschloo L., et al. Comparing network structures on three aspects: a permutation test. Psychol Methods. 2022;28:1273–1285. doi: 10.1037/met0000476. [DOI] [PubMed] [Google Scholar]
- 33.Wang C., Segal L.N., Hu J., et al. Microbial risk score for capturing microbial characteristics, integrating multi-omics data, and predicting disease risk. Microbiome. 2022;10:121. doi: 10.1186/s40168-022-01310-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Benjamini Y., Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- 35.IPR017659. (https://www.ebi.ac.uk/interpro/entry/InterPro/IPR017659/pathways/#table).
- 36.Costello J.F., Smith M., Stolarski C., Sadovnic M.J. Extrarenal clearance of oxalate increases with progression of renal failure in the rat. J Am Soc Nephrol. 1992;3:1098–1104. doi: 10.1681/ASN.V351098. [DOI] [PubMed] [Google Scholar]
- 37.Neumeier L.I., Thomson R.B., Reichel M., et al. Enteric oxalate secretion mediated by Slc26a6 defends against hyperoxalemia in murine models of chronic kidney disease. J Am Soc Nephrol. 2020;31:1987–1995. doi: 10.1681/ASN.2020010105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoppe B., Groothoff J.W., Hulton S.A., et al. Efficacy and safety of Oxalobacter formigenes to reduce urinary oxalate in primary hyperoxaluria. Nephrol Dial Transplant. 2011;26:3609–3615. doi: 10.1093/ndt/gfr107. [DOI] [PubMed] [Google Scholar]
- 39.Ariceta G., Collard L., Abroug S., et al. ePHex: a phase 3, double-blind, placebo-controlled, randomized study to evaluate long-term efficacy and safety of Oxalobacter formigenes in patients with primary hyperoxaluria. Pediatr Nephrol. 2023;38:403–415. doi: 10.1007/s00467-022-05591-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Leulier F., MacNeil L.T., Lee W.J., et al. Integrative physiology: at the crossroads of nutrition, microbiota, animal physiology, and human health. Cell Metab. 2017;25:522–534. doi: 10.1016/j.cmet.2017.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ermer T., Nazzal L., Tio M.C., et al. Oxalate homeostasis. Nat Rev Nephrol. 2023;19:123–138. doi: 10.1038/s41581-022-00643-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Holmes R.P., Kennedy M. Estimation of the oxalate content of foods and daily oxalate intake. Kidney Int. 2000;57:1662–1667. doi: 10.1046/j.1523-1755.2000.00010.x. [DOI] [PubMed] [Google Scholar]
- 43.Holmes R.P., Goodman H.O., Assimos D.G. Dietary oxalate and its intestinal absorption. Scan Microsc. 1995;9:1109–1118. [PubMed] [Google Scholar]
- 44.Holmes R.P., Goodman H.O., Assimos D.G. Contribution of dietary oxalate to urinary oxalate excretion. Kidney Int. 2001;59:270–276. doi: 10.1046/j.1523-1755.2001.00488.x. [DOI] [PubMed] [Google Scholar]
- 45.Siener R., Ebert D., Nicolay C., Hesse A. Dietary risk factors for hyperoxaluria in calcium oxalate stone formers. Kidney Int. 2003;63:1037–1043. doi: 10.1046/j.1523-1755.2003.00807.x. [DOI] [PubMed] [Google Scholar]
- 46.Taylor E.N., Curhan G.C. Oxalate intake and the risk for nephrolithiasis. J Am Soc Nephrol. 2007;18:2198–2204. doi: 10.1681/ASN.2007020219. [DOI] [PubMed] [Google Scholar]
- 47.Bhasin B., Urekli H.M., Atta M.G. Primary and secondary hyperoxaluria: understanding the enigma. World J Nephrol. 2015;4:235–244. doi: 10.5527/wjn.v4.i2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suryavanshi M.V., Bhute S.S., Gune R.P., Shouche Y.S. Functional eubacteria species along with trans-domain gut inhabitants favour dysgenic diversity in oxalate stone disease. Sci Rep. 2018;8 doi: 10.1038/s41598-018-33773-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miller A.W., Oakeson K.F., Dale C., Dearing M.D. Effect of dietary oxalate on the gut microbiota of the mammalian herbivore Neotoma albigula. Appl Environ Microbiol. 2016;82:2669–2675. doi: 10.1128/AEM.00216-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klimesova K., Whittamore J.M., Hatch M. Bifidobacterium animalis subsp. lactis decreases urinary oxalate excretion in a mouse model of primary hyperoxaluria. Urolithiasis. 2015;43:107–117. doi: 10.1007/s00240-014-0728-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoppe B., Pellikka P.A., Dehmel B., et al. Effects of Oxalobacter formigenes in subjects with primary hyperoxaluria type 1 and end-stage renal disease: a phase II study. Nephrol Dial Transplant. 2021;36:1464–1473. doi: 10.1093/ndt/gfaa135. [DOI] [PubMed] [Google Scholar]
- 52.Allison M.J., Cook H.M., Milne D.B., et al. Oxalate degradation by gastrointestinal bacteria from humans. J Nutr. 1986;116:455–460. doi: 10.1093/jn/116.3.455. [DOI] [PubMed] [Google Scholar]
- 53.NCD Risk Factor Collaboration (NCD-RisC) Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: a pooled analysis of 2416 population-based measurement studies in 128.9 million children, adolescents, and adults. Lancet. 2017;390:2627–2642. doi: 10.1016/S0140-6736(17)32129-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Taylor E.N., Stampfer M.J., Curhan G.C. Obesity, weight gain, and the risk of kidney stones. JAMA. 2005;293:455–462. doi: 10.1001/jama.293.4.455. [DOI] [PubMed] [Google Scholar]
- 55.Maalouf N.M., Tondapu P., Guth E.S., et al. Hypocitraturia and hyperoxaluria after Roux-en-Y gastric bypass surgery. J Urol. 2010;183:1026–1030. doi: 10.1016/j.juro.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nazzal L., Francois F., Henderson N., et al. Effect of antibiotic treatment on Oxalobacter formigenes colonization of the gut microbiome and urinary oxalate excretion. Sci Rep. 2021;11 doi: 10.1038/s41598-021-95992-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Duncan S.H., Richardson A.J., Kaul P., et al. Oxalobacter formigenes and its potential role in human health. Appl Environ Microbiol. 2002;68:3841–3847. doi: 10.1128/AEM.68.8.3841-3847.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Suryavanshi M.V., Bhute S.S., Jadhav S.D., et al. Hyperoxaluria leads to dysbiosis and drives selective enrichment of oxalate metabolizing bacterial species in recurrent kidney stone endures. Sci Rep. 2016;6 doi: 10.1038/srep34712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu M., Zhang Y., Wu J., et al. Causal relationship between kidney stones and gut microbiota contributes to the gut-kidney axis: a two-sample Mendelian randomization study. Front Microbiol. 2023;14 doi: 10.3389/fmicb.2023.1204311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fontenot E.M., Ezelle K.E., Gabreski L.N., et al. YfdW and YfdU are required for oxalate-induced acid tolerance in Escherichia coli K-12. J Bacteriol. 2013;195:1446–1455. doi: 10.1128/JB.01936-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shankaranarayanan D., Raj D.S. Gut microbiome and kidney disease: reconciling optimism and skepticism. Clin J Am Soc Nephrol. 2022;17:1694–1696. doi: 10.2215/CJN.04480422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be deposited into NCBI upon manuscript acceptance.