Abstract
We constructed human immunodeficiency virus (HIV) mutants by replacing the matrix domain with sequences encoding the viral protease or p6* and protease. The chimeras retaining matrix myristylation and processing signals underwent efficient autoprocessing with severely defective particle budding. The budding defects of the chimeras were rescued by suppressing the chimera protease activity either through addition of an HIV protease inhibitor or through inactivating the chimera protease via a substitution mutation of the catalytic aspartic acid residue. This resulted in the release of chimeric virus-like particles with the density of a wild-type retrovirus particle. In addition, the assembly-competent but processing-defective chimeras produced proteolytically processed particles with significant reverse transcriptase activity when a downstream native pol gene was present. These results suggest that HIV has the potential to adapt heterologous sequences in place of the matrix sequence without major effects on virus-like particle budding. In addition, the positions of the protease and substrate accessibility may contribute significantly toward avoiding a premature Gag or Gag-Pol process, which leads to severe defects in both particle budding and incorporation.
The structural proteins of all retroviruses, including human immunodeficiency virus (HIV), are encoded by the gag genes (2, 3, 5, 23). During or shortly after virus budding, the HIV Gag precursor Pr55 is cleaved by the pol-encoded protease (PR) into four major products: the matrix (p17; MA), capsid (p24; CA), nucleocapsid (p7; NC), and C-terminal p6 protein (4, 7, 9, 11). The pol product is translated as a Pr160gag-pol fusion protein by a ribosomal frameshifting mechanism that occurs at a frequency of about 5% during translation of Gag (6). The relatively low level of Gag-Pol is thought to avoid premature Gag processing so that Gag assembly can proceed. Mechanisms of PR activation are unclear; it is proposed that PR dimerization, a prerequisite for PR activation, is promoted by the Gag domains (10, 20, 26). PR, once activated, autocleaves from Gag-Pol and subsequently processes Gag and Pol into mature products. Within Pr160gag-pol, C-terminal p6 is truncated and replaced by a domain referred to as p6* (13). p6*, adjacent to PR, separates NC from Pol. A number of studies suggest that p6* may be functionally involved in the regulation of PR activity (14, 17, 27).
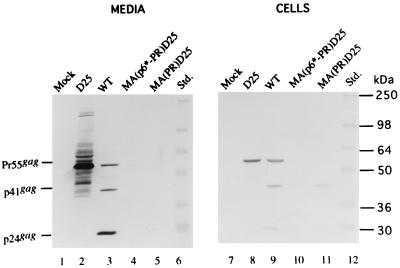
It has been demonstrated that chimeras derived from a replacement of the HIV or Rous sarcoma virus C-terminal gag sequences by foreign protein sequences can still direct virus-like particle assembly and release (21, 22, 25). In this study, we substituted the HIV-1 PR coding sequence for MA and analyzed the assembly and processing of the resultant chimeric proteins. The p6*-PR and PR sequence fragments were amplified by PCR using primers containing a ClaI and SalI restriction site in the 5′ and 3′ primers, respectively. The PCR-generated fragments then were treated with ClaI and SalI and used to replace the fragment from ClaI (HIV nucleotide [nt] 831) to SalI (nt 1147) of an HIV gag mutant that contained a SalI linker at nt 1147 (18). To assess the proteolytic activities of the inserted PR domains, the chimeric constructs were subcloned into an HIV PR-defective mutant, D25, of which the PR catalytic residue Asp was replaced with Asn. The resultant construct was designated MA(p6*-PR)D25 or MA(PR)D25 (Fig. 1A). The backbone of all mutant constructs was HIV gpt, which carries simian virus 40 ori and gpt genes in the env region (12). Wild-type (WT) and mutant HIVgpt plasmids were transfected into 293T cells. Expression and release of HIV Gag proteins were probed by immunoblotting using an anti-p24gag monoclonal antibody (1, 19). As shown in Fig. 2, the WT Pr55, the p41, and the mature p24gag proteins were detected in the medium and in cell samples (lanes 3 and 9). A major band representing Pr55gag was seen in the medium and cell samples of D25 (lanes 2 and 8). In contrast, chimeric proteins derived from processed MA(p6*-PR)D25 or MA(PR)D25 were detected only in the cell samples (lane 10 or 11, respectively). A faint band corresponding to Pr55gag observed in the MA(p6*-PR)D25 medium sample (Fig. 2, lane 4) may have resulted from a spillover from the adjacent WT sample because it was not seen in any repeat experiments.
FIG. 1.
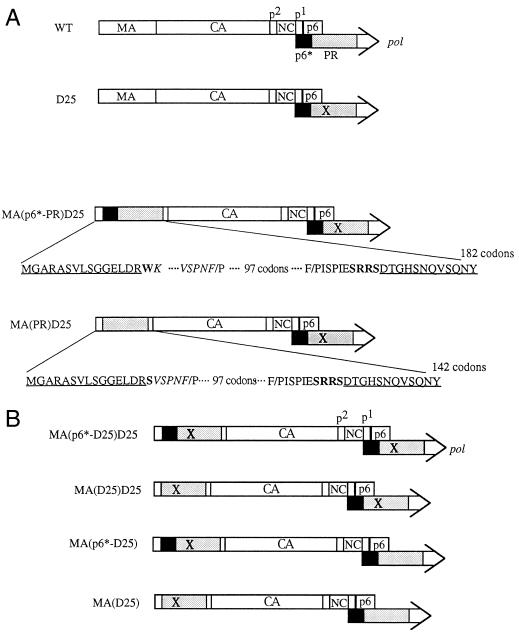
Schematic presentation of the WT and mutant HIVgpt constructs. Mature WT processed Gag proteins and the p6∗ (black) and PR (stippled) domains of pol are indicated. The “X” indicates a PR-defective point mutation (D25→Asn). (A) The D25 mutant, which contains a substitution of an Asn residue for the PR catalytic Asp residue, is defective in Gag processing. The MA(p6∗-PR)D25 mutant contains a deletion of 105 codons and a replacement of the p6∗-PR coding sequence in the MA protein. The myristylation signal residues and a few residues in the C terminus of MA remain intact (underlined). Changed or added codons (boldfaced) and residues in the N and C termini of the PR domain are indicated. Upstream of PR, there are 45 codons (italics) of the p6∗ domain starting from the N-terminal 12th codon, K. The MA(PR)D25 mutant is identical to the MA(p6∗-PR)D25 mutant except that it contains only five C-terminal codons of p6∗. Instead of having 132 codons as in the WT MA protein, the MA(p6∗-PR) and the MA(PR) constructs contain a total of 182 and 142 codons in their MA regions, respectively. (B) Mutant constructs were derived from the constructs shown in panel A. MA(p6∗-D25)D25 and MA(D25)D25 were identical to MA(p6∗-PR)D25 and MA(PR)D25, respectively, except that the former two contain the PR-defective mutations (D25) in their chimera PR fragments. Recombination of the WT with MA(p6∗-D25)D25 and MA(D25)D25 yielded MA(p6∗-D25) and MA(D25), respectively.
FIG. 2.
Expression and processing of the chimeric proteins. 293T cells were transfected with the designated constructs. At 48 h posttransfection, cells and supernatants were collected for protein analysis. Supernatant samples (lanes 1 to 5) corresponding to 50% of the total samples and cell samples (lanes 7 to 11) corresponding to 5% of the total samples were fractionated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and electroblotted onto a nitrocellulose filter. HIV p24gag and p24gag-associated chimeric proteins were detected with mouse anti-p24gag monoclonal antibody at a 1:5,000 dilution, followed by a secondary alkaline phosphatase-conjugated sheep anti-mouse antibody at a 1:5,000 dilution, and alkaline phosphatase activity was determined. Positions of standard (Std.) molecular size markers (lanes 6 and 12) are indicated on the right, and those of HIV Gag proteins Pr55, p41, and p24 are shown on the left.
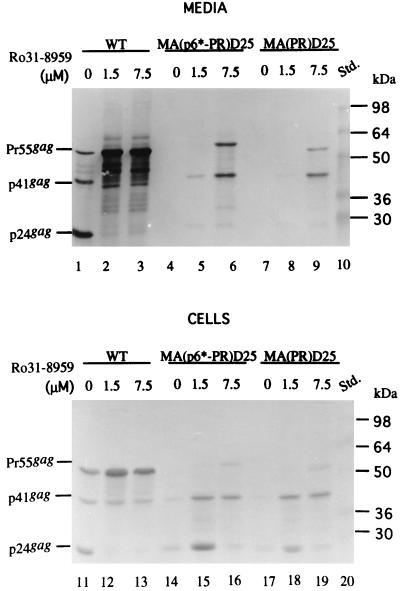
To test whether the inability of the chimeras to release from cells is due to PR-mediated premature autoprocessing (8, 24), we added an HIV-1 PR inhibitor, Ro31-8959 (15), to the WT and chimera transfectants. Figure 3 shows that proteolytic Pr55gag processing was significantly suppressed in the presence of the PR inhibitor (lanes 2 to 3 and 12 to 13) compared with that of untreated samples (lanes 1 and 11). The levels of released chimeric proteins correlated with the degree of the PR activity suppression (Fig. 3, lane 6 versus lane 5, and lane 9 versus lane 8). The expected chimera intermediates p6*, PR, and CA are absent in Fig. 2 and 3; instead, a band migrating with WT p41gag is readily observed. This might result from altered PR preferential cleavage sites. Alternatively, the p41gag chimera was derived from the incompletely cleaved product CA-NC-p6 (11). Further experiments are required to test this proposition.
FIG. 3.
Release of the chimeras into the medium in the presence of an HIV PR inhibitor. 293T cells grown on 10-cm-diameter dish plates were transfected with the WT, MA(p6∗-PR)D25, and MA(PR)D25 HIVgpt constructs. At 18 h posttransfection, cells were split equally onto three 10-cm-diameter dishes and treated, respectively, with 0 μM (lanes 1, 4, 7, 11, 14, and 17), 1.5 μM (lanes 2, 5, 8, 12, 15, and 18), and 7.5 μM (lanes 3, 6, 9, 13, 16, and 19) concentrations of the HIV PR inhibitor Ro31-8959. Four hours later, the culture supernatants were removed and replaced with medium plus the designated concentration of the PR inhibitor. At 48 h after addition of the PR inhibitor, culture supernatants and cells were collected for protein analysis. Samples were fractionated by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis and subjected to immunoblot analysis with anti-p24gag antibody. Std., standards (lanes 10 and 20). Positions of the molecular size markers are indicated on the right, and those of the HIV Gag proteins Pr55, p41, and p24 are shown on the left.
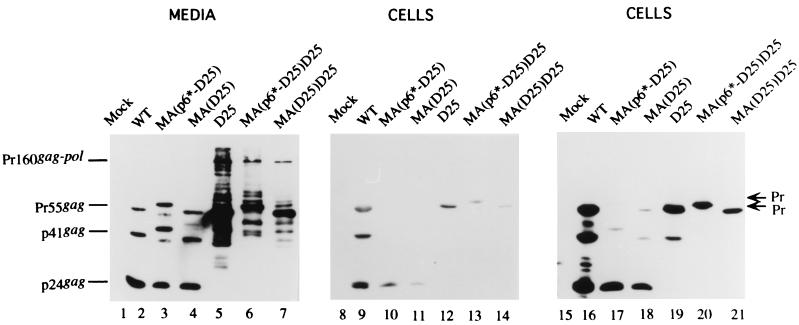
To further confirm that suppression of the PR activity promotes chimera release, the PR-inactivating mutation D25 was introduced into the chimeras MA(p6*-PR)D25 and MA(PR)D25, yielding constructs MA(p6*-D25)D25 and MA(D25)D25, respectively (Fig. 1B). To test whether a normal PR downstream of the chimeric mutations could functionally compensate for the proximal, catalytically inactivated chimera PR, chimeras MA(p6*-D25) and MA(D25) were constructed by placing the native HIV pol gene downstream of the chimeras (Fig. 1B). As shown in Fig. 4, chimeras MA(p6*-D25)D25 and MA(D25)D25 were assembled and released efficiently, at a level at least 1.7-fold higher than that of D25. Interestingly, chimeras MA(p6*-D25) and MA(D25) exhibited an efficient processing profile (Fig. 4, lanes 10, 11, 17, and 18) and had, respectively, three- and sevenfold (lanes 3 to 4 and 10 to 11) higher levels of release efficiency than the WT (lanes 2 and 9). Sucrose density gradient fractionation analysis indicated that all the mutants had a WT retrovirus particle density of 1.16 to 1.18 g/ml (data not shown). To further assess the particle incorporation of the chimera-Pol fusion proteins, the particle-associated RT activity of the assembly-competent chimeras was assayed using exogenous templates (19). Because particle processing can affect the RT assay (16), RT activities of the processing-defective chimeras MA(p6*-D25)D25 and MA(D25)D25 were compared in parallel with those of D25. As shown in Table 1, the chimeras MA(p6*-D25) and MA(D25) possessed significant RT activity at a level just over 50% of that of the WT. Surprisingly, the processing-defective chimeras, MA(p6*-D25)D25 and MA(D25)D25, exhibited relatively low RT activity; all levels were below 20% of the level shown by D25 in three independent experiments. Because the results shown in Fig. 4 indicate that the chimeric particles contained significant levels of chimera-Pol (lanes 6 and 7), the low RT activities of MA(p6*-D25)D25 and MA(D25)D25 were less likely due to insufficient Pol incorporation. Inaccessibility of substrates to the chimera-Pol construct and/or impaired enzymatic activity due to the chimeric mutations might account for the low RT activity.
FIG. 4.
Assembly and processing of chimeric particles. 293T cells were transfected with the designated plasmid. At 48 to 72 h posttransfection, supernatants and cells were prepared for Western immunoblotting. HIV-1 CA-associated proteins were detected by an enhanced-chemiluminescence detection system (Amersham). The primary antibody was an anti-p24gag monoclonal antibody used at a 1:5,000 dilution. The secondary antibody was a sheep anti-mouse horseradish peroxidase-conjugated antibody used at a 1:5,000 dilution. Positions of chimeric protein precursors are indicated on the right (arrows), and those of the HIV Gag proteins Pr55, p41, and p24 are shown on the left.
TABLE 1.
RT activities of HIV mutantsa
| Constsruct | Expt | cpm incorporated | Relative activity (%)b |
|---|---|---|---|
| WT | 1 | 29,227 | 100 |
| 2 | 146,976 | 100 | |
| 3 | 61,654 | 100 | |
| 4 | 97,524 | 100 | |
| MA(p6*-D25) | 1 | 7,715 | 67 |
| 2 | 66,830 | 72 | |
| 3 | 78,033 | 95 | |
| 4 | 94,678 | 86 | |
| MA(D25) | 2 | 46,275 | 93 |
| 3 | 77,703 | 95 | |
| 4 | 35,095 | 42 | |
| D25 | 1 | 29,589 | 100 |
| 2 | 131,614 | 100 | |
| 3 | 304,404 | 100 | |
| MA(p6*-D25)D25 | 1 | 2,409 | 19 |
| 2 | 8,522 | 7 | |
| 3 | 57,915 | 2 | |
| MA(D25)D25 | 1 | 2,689 | 9 |
| 2 | 5,892 | 5 | |
| 3 | 21,414 | 2 |
Supernatants were prepared and RT assays were performed as described in Materials and Methods. For each sample, virus-associated Gag or chimeric protein levels were quantitated by scanning mutant and WT Pr55, p41, and 24 or p24-associated band densities from immunoblots. Results of four separate transfection experiments are given.
RT activities of the processing-defective chimeric mutants MA(p6*-D25)D25 and MA(D25)D25 were compared with that of their parental PR-defective mutant D25. Relative activities were determined as percentages of WT or D25 activities (100%) by the equation 100 × [(mutant counts per minute − background)/mutant Gag protein × WT or D25 Gag protein densitometry units/(WT or D25 counts per minute − background)]. All RT activities with counts per minute were at least threefold over the background level (730 ± 239).
Acknowledgments
This work was supported by grant NSC88-2314-B010-075 from the National Science Council and, in part, by grant DOH88-DC-1020 from the Ministry of Health, Taipei, Taiwan.
The hybridoma clone 183 H12-5C was a gift provided by the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, from Bruce Chesebro. The HIV-1 protease inhibitor Ro31-8959 was kindly provided by Hoffmann-LaRoche (Switzerland).
REFERENCES
- 1.Chen Y-L, Ts'ai P-W, Yang C-C, Wang C-T. Generation of infectious virus particles by transient co-expression of human immunodeficiency virus type 1 gag mutants. J Gen Virol. 1997;78:2497–2501. doi: 10.1099/0022-1317-78-10-2497. [DOI] [PubMed] [Google Scholar]
- 2.Freed E O. HIV Gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 3.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–638. [PubMed] [Google Scholar]
- 4.Henderson L E, Bowers M A, Sowder II R C, Serabyn S A, Johnson D G, Bess J W, Jr, Arthur L O, Bryant D K, Fenselau C. Gag proteins of the highly replicative MN strain of human immunodeficiency virus type 1: posttranslational modifications, proteolytic processing, and complete amino acid sequences. J Virol. 1992;66:1856–1865. doi: 10.1128/jvi.66.4.1856-1865.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 6.Jacks T, Power M D, Masiarz F R, Luciw P A, Barr P J, Varmus H E. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature (London) 1988;331:280–283. doi: 10.1038/331280a0. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan A H, Manchester M, Swanstorm R. The activity of the protease of human immunodeficiency virus type 1 is initiated at the membrane of infected cells before the release of viral proteins and is required for release to occur with maximum efficiency. J Virol. 1994;68:6782–6786. doi: 10.1128/jvi.68.10.6782-6786.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krausslich H-G. Human immunodeficiency virus proteinase dimer as component of the viral polyprotein prevents particle assembly and viral infectivity. Proc Natl Acad Sci USA. 1991;88:3213–3217. doi: 10.1073/pnas.88.8.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Leis J, Baltimore D, Bishop J B, Coffin J, Fleissner E, Goff S P, Oroszlan S, Robinson H, Skalka A M, Temin H M, Vogt V. Standardized and simplified nomenclature for proteins common to all retroviruses. J Virol. 1988;62:1808–1809. doi: 10.1128/jvi.62.5.1808-1809.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Louis J M, Nashed N T, Parris K D, Kimmel A R, Jerina D M. Kinetics and mechanism of autoprocessing of human immunodeficiency virus type 1 protease from an analog of the Gag-Pol polyprotein. Proc Natl Acad Sci USA. 1994;91:7970–7974. doi: 10.1073/pnas.91.17.7970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mervis R J, Ahmad N, Lillehoj E P, Raum M G, Salazar F H R, Chan H W, Venkatesan S. The gag gene products of human immunodeficiency virus type 1: alignment within the gag open reading frame, identification of posttranslation modifications, and evidence for alternative gag precursors. J Virol. 1988;62:3993–4002. doi: 10.1128/jvi.62.11.3993-4002.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Page K A, Landau N R, Littman D R. Construction and use of a human immunodeficiency virus: vector for analysis of virus infectivity. J Virol. 1990;64:5270–5276. doi: 10.1128/jvi.64.11.5270-5276.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Partin K, Krausslich H G, Ehrlich L, Wimmer E, Carter C. Mutational analysis of a native substrate of the human immunodeficiency virus type 1 proteinase. J Virol. 1990;64:3938–3947. doi: 10.1128/jvi.64.8.3938-3947.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Partin K, Zybarth G, Ehrlich L, DeCrombrugghe M, Wimmer E, Carter C. Deletion of sequences upstream of the proteinase improve the proteolytic processing of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1991;88:4776–4780. doi: 10.1073/pnas.88.11.4776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roberts N A, Martin J A, Kinchington D, Broadhurst A V, Craig J C, Duncan J B, Galpin S A, Handa B K, Kay J, Krohn A, Lambert R W, Merrett J H, Mills J S, Parkes K E B, Redshaw S, Ritchie A J, Tayor D L, Thomas G J, Machin P J. Rational design of peptide-based HIV proteinase inhibitors. Science. 1990;248:358–361. doi: 10.1126/science.2183354. [DOI] [PubMed] [Google Scholar]
- 16.Srinivasakumar N, Hammarskjold M-L, Rekosh D. Characterization of deletion mutations in the capsid region of human immunodeficiency virus type 1 that affect particle formation and Gag-Pol precursor incorporation. J Virol. 1995;69:6106–6114. doi: 10.1128/jvi.69.10.6106-6114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tessmer U, Krausslich H-G. Cleavage of human immunodeficiency virus type 1 proteinase from the N-terminally adjacent p6* protein is essential for efficient Gag polyprotein processing and viral infectivity. J Virol. 1998;72:3459–3463. doi: 10.1128/jvi.72.4.3459-3463.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang C-T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 Gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang C-T, Lai H-Y, Li J-J. Analysis of minimal human immunodeficiency virus type 1 gag coding sequences capable of virus-like particle assembly and release. J Virol. 1998;72:7950–7959. doi: 10.1128/jvi.72.10.7950-7959.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weber I T. Comparison of the crystal structures and inter subunit interactions of human immunodeficiency virus and Rous sarcoma virus protease. J Biol Chem. 1990;265:10492–10496. [PubMed] [Google Scholar]
- 21.Weldon R A, Jr, Erdie C R, Oliver M G, Wills J W. Incorporation of chimeric Gag protein into retroviral particles. J Virol. 1990;64:4169–4179. doi: 10.1128/jvi.64.9.4169-4179.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weldon R A, Wills J W. Characterization of a small (25-kilodalton) derivative of the Rous sarcoma virus Gag protein competent for particle release. J Virol. 1993;67:5550–5561. doi: 10.1128/jvi.67.9.5550-5561.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wills J W, Craven R C. Form, function, and use of retroviral gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 24.Xiang Y, Ridky T W, Krishna N K, Leis J. Altered Rous sarcoma virus Gag polyprotein processing and its effects on particle formation. J Virol. 1997;71:2083–2091. doi: 10.1128/jvi.71.3.2083-2091.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly functions of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zybarth G, Carter C. Domains upstream of the protease (PR) in human immunodeficiency virus type 1 Gag-Pol influence PR autoprocessing. J Virol. 1995;69:3878–3884. doi: 10.1128/jvi.69.6.3878-3884.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zybarth G, Krausslich H-G, Partin K, Carter C. Proteolytic activity of novel human immunodeficiency virus type 1 proteinase proteins from a precursor with a blocking mutation at the N terminus of the PR domain. J Virol. 1994;68:240–250. doi: 10.1128/jvi.68.1.240-250.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]