Abstract
Background
In Colombia and worldwide, breast cancer (BC) is the most frequently diagnosed neoplasia and the leading cause of death from cancer among women. Studies predominantly involve hereditary and familial cases, demonstrating a gap in the literature regarding the identification of germline mutations in unselected patients from Latin-America. Identification of pathogenic/likely pathogenic (P/LP) variants is important for shaping national genetic analysis policies, genetic counseling, and early detection strategies. The present study included 400 women with unselected breast cancer (BC), in whom we analyzed ten genes, using Whole Exome Sequencing (WES), know to confer risk for BC, with the aim of determining the genomic profile of previously unreported P/LP variants in the affected population. Additionally, Multiplex Ligation-dependent Probe Amplification (MLPA) was performed to identify Large Genomic Rearrangements (LGRs) in the BRCA1/2 genes. To ascertain the functional impact of a recurrent intronic variant (ATM c.5496 + 2_5496 + 5delTAAG), a minigene assay was conducted.
Results
We ascertained the frequency of P/LP germline variants in BRCA2 (2.5%), ATM (1.25%), BRCA1 (0.75%), PALB2 (0.50%), CHEK2 (0.50%), BARD1 (0.25%), and RAD51D (0.25%) genes in the population of study. P/LP variants account for 6% of the total population analyzed. No LGRs were detected in our study. We identified 1.75% of recurrent variants in BRCA2 and ATM genes. One of them corresponds to the ATM c.5496 + 2_5496 + 5delTAAG. Functional validation of this variant demonstrated a splicing alteration probably modifying the Pincer domain and subsequent protein structure.
Conclusion
This study described for the first time the genomic profile of ten risk genes in Colombian women with unselected BC. Our findings underscore the significance of population-based research, advocating the consideration of molecular testing in all women with cancer.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40246-024-00623-7.
Keywords: Unselected breast cancer, Whole exome sequencing, Pathogenic germline variants, Minigene assay
Background
Breast cancer (BC) is the most frequently diagnosed malignant neoplasm and the leading cause of death from cancer in women [1].
BC incidence in Latin-American (LATAM) countries is generally lower when compared to high-income countries (HIC). However, unlike in HIC, BC-related mortality has not shown a declining trend in LATAM and has, in fact, increased in some countries over the past decade (https://gco.iarc.fr/). In Colombia, the observed 5-year survival rate for BC was 72% according to the CONCORD3 trial [2], highlighting a significant disparity between LATAM and HIC. This disparity can be attributed to various factors, including disease characteristics, healthcare system issues, and the availability of early diagnosis programs, among others.
An essential factor in risk assessment and early diagnosis is the recognition of hereditary BC risk, which may account for as much as 10% of all BC cases [3].
Germline cancer risk study can have various approaches, among them, the study of selected populations based on pedigrees with hereditary and familial cancer segregation analysis to identify genes linked with specific risks, or the study of unselected cases. In the latter approach, unselected cases, which involve individuals without consideration of family history or age at diagnosis, enable the calculation of estimates related to germline mutation prevalence, assessment of cancer risk genes, and identification of at-risk relatives, free from ascertainment bias [4].
Worldwide Breast Cancer Association Consortium (BCAC) reported BRCA1, BRCA2, ATM, PALB2, CHEK2, BARD1, RAD51C, RAD51D, CDH1, and TP53, as the main genes for the prediction of hereditary BC risk [5]. Furthermore, BCAC and Cancer Risks Estimates Related to Susceptibility Consortium (CARRIERS), suggested BRCA1, BRCA2, ATM, PALB2, and CHEK2 genes as highly penetrant; both consortiums pinpointed that 10% of BC patients have cancer susceptibility germline mutation [3].
The mutational spectrum of some of these genes has been assessed in Latin American populations in studies of unselected BC, indicating carrier frequencies ranging from 10.7% in Argentina to 25.2% in Brazil [6, 7]. These differences may reflect the high ethnic variability attributed to Latin American populations.
Colombia has a mixed population composed of Amerindian (descendants of indigenous people), European immigrants (mostly Spanish), and Africans, with recent waves of settlements that have included individuals from the Middle East, Romanies, Germans (around World War I and II), and Asian populations [8], although these represent a small minority.
To date, no study in Colombia has comprehensively examined all the genes considered highly significant by the BCAC and CARRIERS consortia. Additionally, there is a lack of data on the mutational spectrum of these genes within the unselected Colombian population. Our research aims to describe for the first time in our country the genomic profile of ten genes risk for breast cancer, in 400 unselected Colombian women with BC, using whole exome sequencing (WES). This population is particularly noteworthy since most studies in our and other Latin American countries have primarily focused on hereditary cases. Our findings uncovered both new and recurrent pathogenic variants. Furthermore, through functional validation, we propose molecular mechanisms that are linked to the etiology of the disease.
Methods
Patients
From March 2019 to May 2022 women with BC were included in the trial, in cancer centers located throughout Colombia. The study included women with a diagnosis of invasive BC (within one year from diagnosis) supported with biopsy and immunohistochemical test. Women or their relatives with known BRCA1 or BRCA2 mutations were excluded. Patients older than 18 years were invited to participate in this study and those who accepted signed an informed consent.
This study was performed in compliance with the Helsinki Declaration and all experimental procedures were approved by Fundación Cardioinfantil–Instituto de Cardiología and Universidad del Rosario Ethics Committee (approval numbers: 402018 7–11-2018, DVO005 1805-CV1469 3–12-2021, Pfizer: WI241988 – Investigator initiate research, independent review board: 28–08-2018, GF1147 2018).
Clinical data collection
The clinical and sociodemographic variables collected have been described in supplementary methods.
Genomic DNA extraction
The quality and quantity of the DNA were evaluated through the measurement of absorbance with a Nanodrop (OD260/280 and OD260/230).
MLPA (multiplex ligation-dependent probe amplification—MLPA)
MLPA was performed using the commercial kit SALSA MLPA Probemix P002-D1 for BRCA1 and P090-C1 for BRCA2 (MRC-Holland, Amsterdam). Experimental details have been included as supplementary methods.
Next generation sequencing (NGS–WES)
Genomic DNA was extracted from peripheral blood samples according to the protocol of the Quick-DNA Miniprep plus kit (Zymo Research, Orange, California, USA). Experimental details of library preparation, WES, bioinformatic analysis and germline variant classification have been included as supplementary methods.
Segregation analysis
All families with an index case carrier of a pathogenic or likely pathogenic germline variant classified according to the ACMG/AMP, ClinGen, or ENIGMA criteria and confirmed by Sanger sequencing were invited to participate in a segregation analysis and all relatives of the index case (with or without cancer at any age), who were willing to participate in the study, were tested. A total of 36 relatives were included in the family segregation analysis.
Functional validation of the recurrent intronic variant in ATM gene (minigene assay)
Experimental details of minigene assay have been included as supplementary methods.
Statistical analysis
Qualitative variables are summarized as frequencies and percentages while quantitative variables as medians and interquartile ranges were reported. To assess possible associations with mutation status Kruskal–Wallis test for quantitative variables and the Chi-square independence test for qualitative were used. All statistical analyses were done in software R version 4.3.0 [9].
Results
Population of study
We enrolled 400 patients in the study, the median age of diagnosis was 53 years, 55.5% of them were post-menopausal and 60.3% were overweight or obese. The main histologic diagnosis was ductal carcinoma (85.5%), the prevalence of triple-negative BC (TNBC) was 11.5%, the prevalence of metastatic disease was 4%, and 61.1% of the patients met NCCN criteria for hereditary BC testing. Table 1 summarizes the main data obtained from the 400 women with unselected BC.
Table 1.
Baseline characteristics of the patients
| Variable | n = 400 | % | |
|---|---|---|---|
| Age at diagnosis (median–range) | 53 | 43–64 | |
| Tumor size (median–range) | 20 | 12–30 | |
| Positive nodes (median–range) | 0 | 0–1 | |
| Ki67 (median–range) | 25 | 13–45 | |
| Histologic diagnosis | Ductal | 342 | 85,50 |
| Lobular | 21 | 5,25 | |
| Other | 35 | 8,75 | |
| ND | 2 | 0,50 | |
| ER status | Negative | 87 | 21,75 |
| Positive | 308 | 77 | |
| ND | 5 | 1,25 | |
| PR status | Negative | 117 | 29,25 |
| Positive | 278 | 69,5 | |
| ND | 5 | 1,25 | |
| HER-2 status | Negative | 298 | 74,5 |
| Positive | 95 | 23,75 | |
| ND | 7 | 1,75 | |
| TNBC status | No | 349 | 87,25 |
| Yes | 46 | 11,50 | |
| ND | 5 | 1,25 | |
| Nodal stage | 0 | 204 | 51 |
| 1 | 134 | 33,50 | |
| 2 | 39 | 9,75 | |
| 3 | 14 | 3,50 | |
| ND | 9 | 2,25 | |
| Tumoral stage | I | 91 | 22,75 |
| II | 183 | 45,75 | |
| III | 104 | 26 | |
| IV | 16 | 4 | |
| ND | 6 | 1,50 | |
| Age of menarche (median–range) | 13 | 12–14 | |
| Born children (median–range) | 2 | 1,50–3 | |
| Age first born child (median–range) | 23 | 19–28 | |
| Lactation | No | 76 | 19 |
| Yes | 322 | 80,50 | |
| ND | 2 | 0,50 | |
| Menopause | No | 173 | 43,25 |
| Yes | 222 | 55,50 | |
| ND | 5 | 1,25 | |
| Age of menopause (median–range) | 50 | 46–52 | |
| Weight (median–range) | 65 | 58–73,30 | |
| Height (median–range) | 1,58 | 1,55–1,63 | |
| BMI (median–range) | 25,96 | 23,57–29,14 | |
| Overweight–obesity | No | 153 | 39 |
| Yes | 241 | 60,25 | |
| ND | 3 | 0,75 | |
| Hormonal contraception exposure | No | 201 | 50,25 |
| Yes | 196 | 49 | |
| ND | 3 | 0,75 | |
| HRT | No | 372 | 93 |
| Yes | 24 | 6 | |
| ND | 4 | 1 | |
| Current or past smoking | No | 296 | 74 |
| Yes | 102 | 25,50 | |
| ND | 2 | 0,50 | |
| Alcohol consumption | No | 347 | 86,75 |
| Yes | 50 | 12,50 | |
| ND | 3 | 0,75 | |
| Radiation exposure | No | 346 | 86,50 |
| Yes | 25 | 6,25 | |
| ND | 29 | 7,25 | |
| Personal history of cancer | No | 377 | 94,25 |
| Yes | 21 | 5,25 | |
| ND | 2 | 0,50 | |
| Family history of cancer | No | 120 | 30 |
| Yes | 275 | 68,75 | |
| ND | 5 | 1,25 | |
| fulfill of NCCN criteria v1.2023 | No | 155 | 38,75 |
| Yes | 243 | 60,75 | |
| ND | 2 | 0,50 | |
| Inheritance | Sporadic | 158 | 39,50 |
| Familial | 187 | 46,75 | |
| Hereditary | 50 | 12,50 | |
| ND | 5 | 1,25 | |
| P/LP variant | No | 376 | 94 |
| Yes | 24 | 6 | |
| Mutation status | No mut | 376 | 94 |
| BRCA | 13 | 3,25 | |
| noBRCA | 11 | 2,75 |
Tumor size was measured in millimeters (mm); weight is given in kilograms (kg); height is given in meters (m); BMI: body mass index (kg/m2); other (mixed, medullary, mucinous, metaplastic, tubular, micropapillary, papillary, adenocarcinoma, apocrine, and cribriform); HRT: hormonal replacement therapy; NCCN criteria v1.2023 [10]; hereditary breast cancer: autosomal dominant inheritance pattern involving at least three generations, consider cancers associated with Lynch syndrome [11]; familial breast cancer: breast cancer with a family history of one or more first- or second-degree relatives with breast cancer that does not fit the hereditary breast cancer definition [11]; P/LP variant: presence of pathogenic or likely pathogenic germline variant; no_mut: absence of P/LP variant; BRCA: P/LP variant identified in genes BRCA1 or BRCA2; noBRCA: P/LP variant identified in genes ATM, BARD1, CHEK2, PALB2 or RAD51D
Germline mutations identified in women with unselected BC
All 400 women with unselected BC were assessed with ten known cancer genes as follows: BRCA1, BRCA2, ATM, PALB2, CHEK2, BARD1, RAD51C, RAD51D, CDH1, and TP53 which were sequenced by WES. 24 (6%) patients had pathogenic or like pathogenic variants (P/LP variants) identified. 18 germline pathogenic variants were identified in 19 individuals (11 in BRCA1/2 genes and seven in ATM, BARD1, CHEK2, PALB2, and RAD51D genes). From these variants, 12 were frameshift (67%), four nonsense (22%), and two missense (11%). PALB2 gene showed two molecular changes that were not reported in ClinVar nor dbSNP database, designated as novel. Three likely pathogenic variants were identified in five women in ATM, CHEK2, and PALB2 genes. Likely pathogenic variants were represented by one missense (33%) and two intronic (67%). All the P/LP variants were in a heterozygous state. The variants are summarized in Tables 2, 3, and supplementary methods Fig. 1.
Table 2.
Frequency of P/LP variants in the population of study and their distribution by genes
| Genes | Population of study (n = 400). Frequency of patients with mutation (%) | P/LP variants (n = 21). Frequency of P/LP variants (%) |
|---|---|---|
| BRCA2 | 10 (2.50%) | 8 (38.10%) |
| ATM | 5 (1.25%) | 3 (14.28%) |
| BRCA1 | 3 (0.75%) | 3 (14.28%) |
| PALB2 | 2 (0.50%) | 3 (14.28%) |
| CHEK2 | 2 (0.50%) | 2 (9.52%) |
| BARD1 | 1 (0.25%) | 1 (4.76%) |
| RAD51D | 1 (0.25%) | 1 (4.76%) |
Table 3.
Molecular and clinicopathological description of germline mutations
| Gene | Variant (HGVS nomenclature) | AF exomes (gnomAD v2.1.1) | AF latino (gnomAD v2.1.1) | AF population of study | ACMG/AMP classification | ENIGMA classification | SNP ID | Molecular subtype | Personal history of cancer | Family history of cancer | Number of patients |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BRCA1 | NM_007294.3:c.5123C > A (p.Ala1708Glu) | 0.00001990 | 0.00005784 | 0.00125 | NA | Pathogenic (BRCAexchange) | rs28897696 | TNBC | No | No | 1 |
| BRCA1 | NM_007294.3:c.5324 T > G (p.Met1775Arg) | 0.00001193 | 0 | 0.00125 | NA | Pathogenic (BRCAexchange) | rs41293463 | ER-,PR + ,HER2 + | No | No | 1 |
| BRCA1 | NM_007294.3:c.1674delA (p.Gly559Valfs*13) | NR | NR | 0.00125 | NA | Pathogenic (BRCAexchange) | rs80357600 | TNBC | No | Yes | 1 |
| BRCA2 | NM_000059.3:c.2380dupA (p.Met794Asnfs*8) | NR | NR | 0.00125 | NA | Pathogenic (BRCAexchange) | rs730881602 | ER + ,PR-,HER2- | No | No | 1 |
| BRCA2 | NM_000059.3:c.3860delA (p.Asn1287Ilefs*6) | 0.0000153412 | 0 | 0.00125 | NA | Pathogenic (BRCAexchange) | rs80359406 | ER-,PR-,HER2 + | No | Yes | 1 |
| BRCA2 | NM_000059.3:c.4889C > G (p.Ser1630Ter) | 0.00000803232 | 0 | 0.00125 | NA | Pathogenic (BRCAexchange) | rs80358711 | ER-,PR + ,HER2- | No | Yes | 1 |
| BRCA2 | NM_000059.3:c.5773C > T (p.Gln1925Ter) | 0.00000398902 | 0 | 0.00125 | NA | Pathogenic (BRCAexchange) | rs80358806 | ER + ,PR + ,HER2- | No | Yes | 1 |
| BRCA2 | NM_000059.3:c.5851_5854delAGTT (p.Ser1951Trpfs*11) | NR | NR | 0.00125 | NA | Pathogenic (BRCAexchange) | rs80359543 | ER + ,PR + ,HER2 + | Yes | No | 1 |
| BRCA2 | NM_000059.3:c.2808_2811delACAA (p.Ala938Profs*21) | 0.00000797283 | 0 | 0.0025 | NA | Pathogenic (BRCAexchange) | rs80359351 | ER + ,PR + ,HER2-; ER + ,PR + ,HER2- | No; No | Yes; Yes | 2 |
| BRCA2 | NM_000059.4:c.1763_1766delATAA (p.Asn588Serfs*25) | NR | NR | 0.0025 | NA | Pathogenic (BRCAexchange) | rs80359303 | ER + ,PR + ,HER2-; TNBC | No; Yes | Yes; Yes | 2 |
| BRCA2 | NM_000059.4:c.9097dupA (p.Thr3033Asnfs*11) | NR | 0 | 0.00125 | NA | Pathogenic (BRCAexchange) | rs397507419 | ER + ,PR + ,HER2- | No | No | 1 |
| ATM | NM_000051.3:c.4507C > T (p.Gln1503Ter) | 0.00000795817 | 0.00005783 | 0.00125 |
Pathogenic (PVS1 + PS4 moderate + PM2 supporting) |
NA | rs2227945 | ER + ,PR + ,HER2- | No | No | 1 |
| ATM | NM_000051.3:c.3510dupA (p.Gln1171Thrfs*8) | 0.00000397874 | 0.00002891 | 0.00125 |
Pathogenic (PVS1 + PS4 supporting + PM2) |
NA | rs876658899 | ER + ,PR + ,HER2- | No | Yes | 1 |
| ATM | NM_000051.3:c.5496 + 2_5496 + 5delTAAG | NR | NR | 0.00375 |
Likely pathogenic (PVS1 moderate + PS4 moderate + PM2) |
NA | rs1565479572 | ER + ,PR + ,HER2-; ER + ,PR + ,HER2-; ER + ,PR-,HER2- | No; Yes; No | No; Yes; Yes | 3 |
| BARD1 | NM_000465.3:c.176_177delAG (p.Glu59Alafs*8) | NR | NR | 0.00125 |
Pathogenic (PVS1 + PM1 + PM2) |
NA | rs1057517589 | TNBC | No | No | 1 |
| CHEK2 | NM_007194.3:c.1100delC (p.Thr367Metfs*15) | 0.00204432 | 0.00005649 | 0.00125 |
Pathogenic (PVS1 + PS3 + PS4 moderate + PM1) |
NA | rs555607708 | ER + ,PR + ,HER2 + | No | Yes | 1 |
| CHEK2 |
NM_007194.3:c.349A > G (p.Arg117Gly) |
0.00011941 | 0.0001129 | 0.00125 |
Likely pathogenic (PS3 + PS4 moderate + PM1 + PP3) |
NA | rs28909982 | ER + ,PR + ,HER2- | No | Yes | 1 |
| PALB2 | NM_024675.3:c.984delT (p.Leu329Terfs) | NR | NR | 0.00125 |
Pathogenic (PVS1 + PM2) |
NA | Novel | ER + ,PR + ,HER2 + | No | No | 1* |
| PALB2 | NM_024675.3:c.986delT (p.Leu329Glnfs*17) | NR | NR | 0.00125 |
Pathogenic (PVS1 + PM2) |
NA | Novel | ER + ,PR + ,HER2 + | No | No | 1* |
| PALB2 | NM_024675.3:c.3350 + 4A > G | 0.000003977 | 0 | 0.00125 |
Likely pathogenic (PS3 very strong + PM2 + BP4) |
NA | rs180177136 | TNBC | No | Yes | 1 |
| RAD51D | NM_002878.3:c.556C > T (p.Arg186Ter) | 0.00004011 | 0 | 0.00125 |
Pathogenic (PVS1 + PS4 supporting + PM1) |
NA | rs387906843 | ER + ,PR + HER2- | No | No | 1 |
NR Not reported, NA Not applied, *variants identified in the same patient.
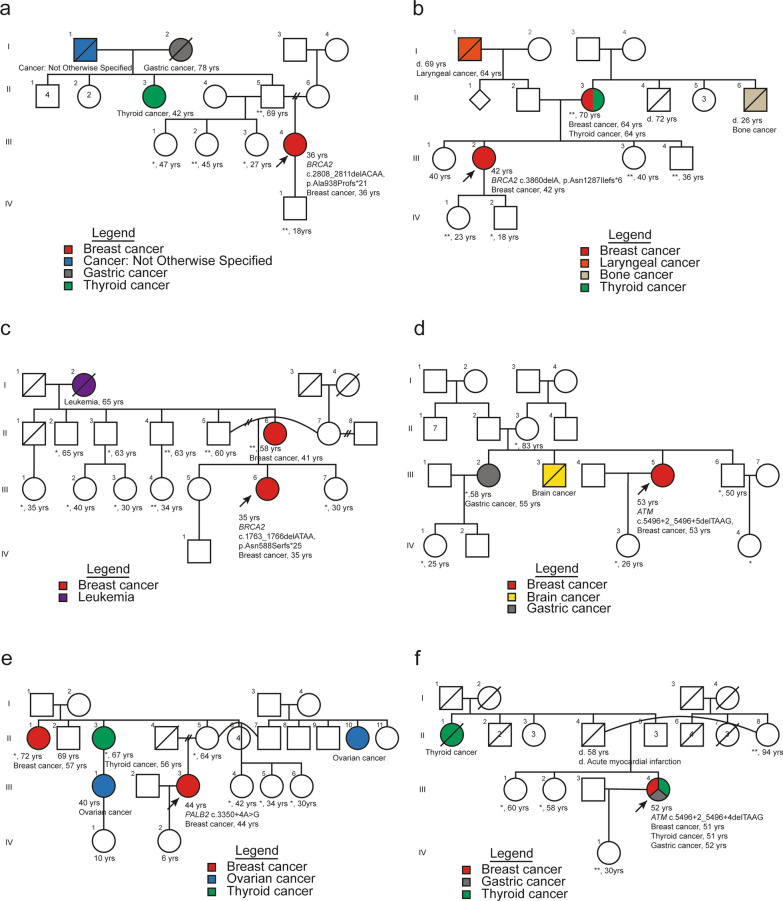
Fig. 1.
Pedigrees of index cases and their relatives assessed in the segregation analysis. a Germline mutation located in BRCA2 gene: c.2808_2811delACAA, p.Ala938Profs*21; b Germline mutation located in BRCA2 gene: c.3860delA, p.Asn1287Ilefs*6; c Germline mutation located in BRCA2 gene: c.1763_1766delATAA, p.Asn588Serfs*25; d and f Germline mutation located in ATM gene: c.5496 + 2_5496 + 5delTAAG; e Germline mutation located in PALB2 gene: c.3350 + 4A > G; **individuals tested harboring germline mutation; * individuals tested not harboring germline mutation
Estrogen and/or progesterone positive hormonal receptors (HR) were identified in 75% (18/24) of unselected women carrying P/LP variants. From them, germinal mutations distribution was: BRCA2, 44.4% (8/18); ATM, 27.8% (5/18); CHEK2, 11.1% (2/18); BRCA1, 5.5% (1/18); PALB2 5.5% (1/18) and RAD51D, 5.5% (1/18).
P/LP variants were found in 13/24 (54.2%) women without HER-2 amplification (independently of HR status), and their gene distribution was: BRCA2, 46.2% (6/13); ATM, 38.5% (5/13); CHEK2, 7.7% (1/13) and RAD51D, 7.7% (1/13). In contrast, oncoprotein HER-2 amplification (independently of HR status) was detected in 20,8% of the tumors. 40% (2/5) of the P/LP variants were identified in BRCA2 gene and 60% (3/5) in BRCA1, CHEK2 and PALB2 genes.
TNBC tumors (according to the absence of HR expression and HER-2 amplification) were identified in 5/24 affected women (20.8%), from them, 40% (2/5) of the variants were detected in BRCA1 gene, and 60% (3/5) in BRCA2, BARD1, and PALB2 genes.
Ductal BC was present in 95.8% of the patients carrying a P/LP variant; metaplastic BC was observed in 1/24 (4.2%) women with a heterozygous PALB2 gene mutation.
Interestingly, two patients (2/24) suffered more than one primary cancer. One woman was diagnosed with lymphoma with a prior ductal BC, and she had a heterozygous BRCA2 c.1763_1766delATAA, p.Asn588Serfs*25; the other one had three cancers, thyroid, gastric, and ductal BC harboring an ATM c.5496 + 2_5496 + 5delTAAG, a likely pathogenic mutation.
Family history of cancer was documented in 14/24 women (58.3%), in their first, second, and third consanguinity-degree relatives. BRCA2 accounts for 50% (7/14), with P/LP variants including a variety of tumors in relatives such as breast, thyroid, bone, gastric, prostate, leukemia, and esophagus, followed by ATM, in 3/14 (21.4%), which referred on thyroid, gastric, and brain cancers. Two women (14.3%) have P/LP variants in the CHEK2 gene, and their relatives have breast and pancreas cancer. One patient (1/14) who had a pathogenic variant in the BRCA1 gene, had several relatives with BC. Lastly, an affected woman with the PALB2 gene intronic variant, described relatives with ovary, breast, and thyroid cancers.
Three recurrent germline mutations were detected: two in BRCA2 (c.2808_2811delACAA, p.Ala938Profs*21 and c.1763_1766delATAA, p.Asn588Serfs*25); the other one in ATM (c.5496 + 2_5496 + 5delTAAG). From them, BRCA2 c.1763_1766delATAA, p.Asn588Serfs*25, and ATM c.5496 + 2_5496 + 5delTAAG were not previously reported in the gnomAD v2.1.1 database (https://gnomad.broadinstitute.org/).
Regarding MLPA analysis for BRCA1/2 genes, there were no large genomic rearrangements (LGRs) in the sample of 400 unselected women with BC.
Correlation between mutation status and baseline characteristics of women with unselected BC
For women harboring germline mutations, statistical association tests were performed, comparing baseline characteristics of the population of study among 3 groups based on mutation status, such as absence of germline mutations (no mut), presence of germline mutations in BRCA1/2 genes (BRCA), and germline mutations in noBRCA genes (ATM, BARD1, CHEK2, PALB2, and RAD51D) (Table 4). Women with germline mutations in BRCA1/2 genes had an earlier age at diagnosis in comparison with no mut group (median age 36 vs 54, p = 0.0003), and 15.38% of women in the group BRCA had menopause in contrast with the no mut group were 57.68% of the patients had menopause (p = 0.009). The variable nodal stage (specifically nodal stage 2) had a higher frequency in the BRCA group (30.77% vs 9.26%, p = 0.0425) showing an association with the spread of cancer to a higher number of lymph nodes in this particular mutation status. No association with statistical significance was established for the noBRCA group.
Table 4.
BRCA and noBRCA mutations statistical association test with clinical and pathological data
| No mut | BRCA | noBRCA | BRCA vs No mut | noBRCA vs No mut | p-value | ||
|---|---|---|---|---|---|---|---|
| Age at diagnosis | 54 (43–65) | 36 (30–44) | 51 (45–56) | − 15.54 (− 20.92;− 10.16) | − 3.54 (− 9.1; 2.02) | 0.0003 | |
| Tumor size | 20 (12–30) | 20 (15–25) | 30 (16–39.5) | 1.44 (− 14.61;17.49) | 5.81 (− 7.41;19.02) | 0.5662 | |
| Positive nodes | 0 (0–1) | 0 (0–1) | 1 (0.25–4.25) | − 0.66 (− 1.07;-0.25) | 2.71 (− 0.66;6.07) | 0.0469 | |
| Ki67 | 25 (12–43.8) | 35 (20–65) | 25 (15–45) | 8.59 (− 5.57;22.76) | 2.77 (− 11.87;17.23) | 0.3644 | |
| Histologic diagnosis | Ductal | 319 (85, 29%) | 13 (100%) | 10 (90.91%) | – | – | 0.5784 |
| Lobular | 21 (5.61%) | 0 | 0 | 0 (0.03;9.58) | 0 (0.04;12.50) | ||
| Other | 34 (9.09%) | 0 | 1 (9.09%) | 0 (0.02;5.90) | 0.83 (0.23;7.59) | ||
| ER status | Pos | 291 (78.44%) | 8 (61.54%) | 9 (81.82%) | 0.36 (0.14;1.28) | 0.82 (0.25;4.32) | 0.3353 |
| PR status | Pos | 261 (70.35%) | 9 (69.23%) | 8 (72.73%) | 0.76 (0.28;2.80) | 0.84 (0.29;3.63) | 0.9814 |
| HER-2 status | Pos | 91 (24.66%) | 2 (15.38%) | 2 (18.18%) | 0.50 (0.16;2.65) | 0.60 (0.19;3.29) | 0.8696 |
| TNBC status | Yes | 41 (11.05%) | 3 (23.08%) | 2 (18.18%) | 2.14 (0.76;9.28) | 1.57 (0.50;8.76) | 0.1744 |
| Nodal stage | 0 | 195 (53.13%) | 6 (46.15%) | 3 (27.27%) | – | – | 0.0425 |
| 1 | 126 (34.33%) | 3 (23.08%) | 5 (45.45%) | 0.66 (0.22;3.11) | 1.92 (0.62;9.45) | ||
| 2 | 34 (9.26%) | 4 (30.77%) | 1 (9.09%) | 3.18 (1.12;13.77) | 1.39 (0.35;17.01) | ||
| 3 | 12 (3.27%) | 0 | 2 (18.18%) | 0 (0.06;22.59) | 7.5 (2.00;62.44) | ||
| Tumoral stage | I | 88 (23.78%) | 3 (23.08%) | 0 | - | - | 0.3523 |
| II | 171 (46.22%) | 5 (38.46%) | 7 (63.64%) | 0.64 (0.21;3.17) | 3.58 (0.44;137.09) | ||
| III | 96 (25.95%) | 5 (38.46%) | 3 (27.27%) | 1.13 (0.37;5.68) | 2.72 (0.33;126.04) | ||
| IV | 15 (4.05%) | 0 | 1 (9.09%) | 0 (0.04;16.58) | 5.5 (0.67;439.91) | ||
| Age of menarche | 13 (12–14) | 14 (13–15) | 13 (11–13.5) | 0.95 (− 0.50;2.40) | − 0.75 (− 2.28;0.80) | 0.2672 | |
| Parity | 2 (2–3) | 2 (2–3) | 2 (1–2.75) | − 0.18 (− 0.76;0.39) | − 0.64 (− 1.31;0.04) | 0.4164 | |
| Born children | 2 (2–3) | 2 (1.5–2) | 1.5 (1–2.75) | − 0.56 (− 1.01;-0.11) | − 0.57 (− 1.27;0.13) | 0.2465 | |
| Age first born child | 23 (19–28) | 20 (18.5–23.5) | 26 (21.5–33.5) | − 2.95 (− 5.63;-0.27) | 2.59 (− 1.44;6.63) | 0.1001 | |
| Lactation | Yes | 301 (80.48%) | 11 (84.62%) | 10 (90.91%) | 0.89 (0.30;4.51) | 1.21 (0.30;9.62) | 0.6466 |
| Menopause | Yes | 214 (57.68%) | 2 (15.38%) | 6 (54.55%) | 0.12 (0.04;0.64) | 0.73 (0.27;2.75) | 0.009 |
| Age of menopause | 50 (46–52) | 52.5 (51.75–53.25) | 50.5 (46–54.25) | 3.27 (0.26;6.28) | 1.27 (− 3.21;5.75) | 0.4365 | |
| Weight | 65 (58–73) | 68 (57–75) | 73.3 (65.8–79.5) | 2.95 (− 5;10.9) | 6.07 (-0.05;12.2) | 0.1377 | |
| Height | 158 (155–163) | 164 (157–165) | 158 (151.5–160) | 3.25 (− 0.35;6.84) | − 2.25 (− 5.76;1.26) | 0.1143 | |
| BMI | 25.84 (23.44–29.02) | 25.33 (24.14–29) | 29.4 (25.96–33.95) | − 0.07 (− 2.33;2.20) | 3.14 (0.71;5.57) | 0.0666 | |
| Overweight | Yes | 226 (60.59%) | 7 (53.85%) | 8 (72.73%) | 0.65 (0.26;2.19) | 1.29 (0.45;5.60) | 0.6297 |
| Hormonal contraception | Yes | 186 (49.87%) | 5 (38.46%) | 5 (45.45%) | 0.64 (0.18;1.98) | 0.84 (0.23;2.92) | 0.8011 |
| HRT | Yes | 24 (6.45%) | 0 | 0 | 0 (0.03;9.13) | 0 (0.03;10.81) | 1 |
| Smoking | Yes | 95 (25.40%) | 3 (23.08%) | 4 (36.36%) | 0.79 (0.28;3.34) | 1.45 (0.53;5.78) | 0.6582 |
| Alcohol consumption | Yes | 47 (12.60%) | 2 (15.38%) | 1 (9.09%) | 1.13 (0.37;6.07) | 0.62 (0.17;5.58) | 0.8771 |
| Radiation exposure | Yes | 23 (6.61%) | 1 (8.33%) | 1 (9,09%) | 1.13 (0.31;10.42) | 1.23 (0.34;11.52) | 0.4583 |
| Personal history cancer | Yes | 19 (5.08%) | 1 (7.69%) | 1 (9.09%) | 1.36 (0.38;12.64) | 1.61 (0.44;15.30) | 0.3868 |
| Family history cancer | Yes | 260 (70.08%) | 9 (69.23%) | 6 (54.55%) | 0.77 (0.29;2.83) | 0.42 (0.16;1.61) | 0.5433 |
| NCCN criteria v1.2023 | Yes | 224 (59.89%) | 11 (84.62%) | 8 (72.73%) | 2.44 (0.77;12.30) | 1.33 (0.46;5.75) | 0.1439 |
| Inheritance | Sporad | 148 (39.89%) | 5 (38.46%) | 5 (45.45%) | - | - | 0.8491 |
| Fam | 175 (47.17%) | 6 (46.15%) | 6 (54.55%) | 0.84 (0.32;3.18) | 0.84 (0.32;3.18) | ||
| Her | 48 (12.94%) | 2 (15.38%) | 0 | 1.01 (0.30;6.43) | 0 (0.02;5.13) |
Qualitative variables are summarized as frequencies and percentages, and quantitative variables are reported as medians and interquartile ranges. Effect measure and IC95% were determined with means difference and OR (odds ratio) for quantitative and qualitative variables, respectively. P-values calculated using the Kruskal-Wallis test for quantitative variables and the Chi-square test for qualitative variables. Associations with statistical significance are shown in bold (p-value < 0.05). sporad: sporadic fam: familial her: hereditary
Segregation analysis
Analysis was performed in six families which were ascertained by an index case: three families for three different pathogenic variants in the BRCA2 gene, two with a likely pathogenic variant in the ATM gene, and one family with a likely pathogenic variant in the PALB2 gene (Fig. 1). In total, 13 relatives tested positive for the mutations assessed (11 in BRCA2 and two in ATM). Particularly, two BRCA2 families with a pathogenic variant were tested, one of the relatives had been diagnosed with BC at 41 years (c.1763_1766delATAA, p.Asn588Serfs*25; age diagnosis index case: 35 years), and another relative was diagnosed with breast and thyroid cancer at 64 years (c.3860delA, p.Asn1287Ilefs*6; age diagnosis index case: 42 years). All of the relatives who tested positive received genetic counseling.
Minigene assay
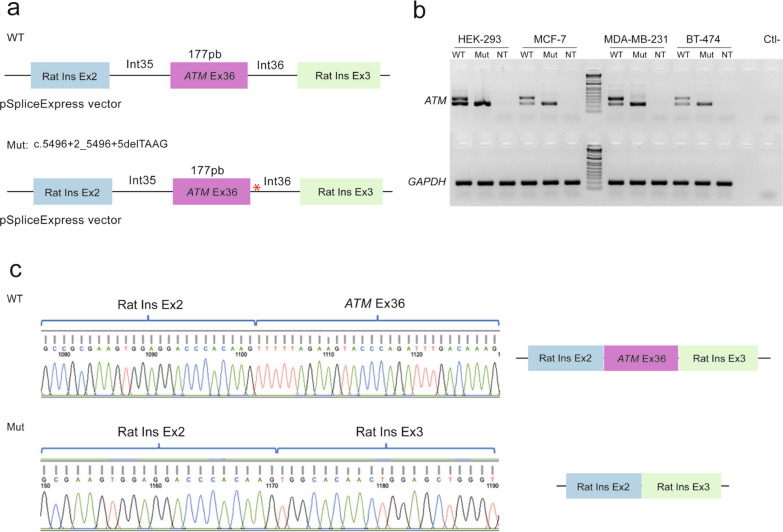
Three affected and unrelated women showed heterozygous ATM c.5496 + 2_5496 + 5delTAAG variant. A minigene assay was performed to identify the alternative splicing effect in mRNA. This assay evidenced an exon 36 skipping which was confirmed by Sanger sequencing (Fig. 2).
Fig. 2.
Exon skipping of exon 36 of the ATM gene due to germline mutation c.5496 + 2_5496 + 5delTAAG. a Diagram of the minigene pSpliceExpress vectors, WT which is constituted by exon 36 of ATM and exons 2 and 3 from Rat insulin (Rat Ins Ex2 and Rat Ins Ex3), and Mut which represents the presence of the germline mutation of interest. b RT-PCR, performed after transfection of the WT and Mut plasmids, in HEK-293, MCF-7, MDA-MB-231, and BT-474 cell lines, showed exon skipping in all cell lines, negative control was not transfected cells (NT). c Sanger sequencing was performed to confirm the effect in splicing observed in RT-PCR
Discussion
Identifying germline mutations in high and moderate-risk BC genes is of paramount importance for establishing genetic screening programs that facilitate early diagnosis and development of national public health policies. Implementation of genomic analysis through NGS and incorporation of noBRCA genes has proven to be an adequate strategy to increase sensibility regarding recurrent mutation analysis restricted only to BRCA1/2 genes [12, 13].
Globally, germline mutation cancer prevalence, can be estimated from hereditary, familial, or unselected BC cases. European, North American, and Asian populations have been the primary focus to obtain this data.
To our knowledge, this is the first report on the prevalence of mutations in the top 10 clinically impactful genes, identified by WES in 400 women with unselected BC from various regions of Colombia.
We evaluated NCCN criteria [10] in the women studied. Significantly, 20.8% of them with a P/LP variant did not fulfill those criteria. This finding demonstrates that molecular testing should be considered in all women with BC regardless of the age of diagnosis, molecular subtype, and personal or family history of cancer.
Our findings determined that 6% of the Colombian women with unselected BC had germline mutations in seven of the genes studied, being BRCA2 the gene with the highest frequency of variants and women affected, followed by ATM, BRCA1, PALB2, CHEK2, BARD1 and RAD51D genes. No P/LP variants were detected in CDH1, RAD51C, and TP53 genes. BC prevalence of germline mutations and their frequency in cancer risk genes, varies thoroughly depending on the selection criteria of the population studied.
Interestingly, to date at least, 41 articles have been described that analyze genes related to BC in the Latin American population. This includes a diversity of patients from Argentina, Brazil, Chile, Guatemala, Colombia, Peru, Puerto Rico, and Mexico, covering 40% of the countries considered in the region through genetic analysis (Supplementary Table 1, and the references therein). These studies have examined approximately 51,000 Latin American patients, which have provided insights into the frequencies of molecular variants of interest in the analyzed genes (BRCA1/2 and noBRCA) (Supplementary Table 1 and the references therein). Concerning the mutational spectrum exhibited by the BRCA1/2 genes, a range from 10.1% to 37.2% has been noted across the populations. This variation is estimated to be strongly linked to the migration history of the Latin American populations, including the overlap of some mutations determined by shared events and exchanges that characterize the migration history of each geographical region [14]. Additionally, within the same population, such as Brazil, there is high variability in the mutation frequencies of the BRCA genes (10.1% vs 22.4%), supporting the observation that the genetic background of Latin American populations results from events leading to unique population structures within and between countries [14–16]. Specifically, the highest frequencies for the BRCA genes reported in the Latin American population are described in patients with breast and ovarian cancer from Afro-Colombian families, in whom 33.3% of pathogenic variants were identified [17], demonstrating the impact of patient selection criteria on the variability of reported data.
Unlike our study, most studies reported in Latin America have involved patients with hereditary BC, in whom the representation of pathogenic variants in the BRCA genes is substantially higher than in cases of unselected BC. For this latter group, frequencies between 1.2 and 14.5% have been reported (with eight studies in Latin America) [6, 18–24], which is consistent with the findings identified in the present study (Table 2 and supplementary Table 1). The analysis of unselected populations has been recommended to avoid the overestimation of the true prevalence of germline cancer-related P/LP variants in the general population [25].
In Colombia, previous reports described mutations in BRCA1/2 genes focused on hereditary/familial cases [17, 21, 26, 27]. Even though, few studies analyzed mutation prevalence in BRCA1/2 genes from unselected BC patients, finding that their frequency ranges from 0.4 to 3.3% [20, 21], which is concordant with our results, since the frequency of women with mutations in BRCA1/2 is 3.25%.
Beyond BRCA genes in Latin America NGS multigene analysis has been conducted in 78% of studies, including the current study, which has enabled the identification of P/LP variants in moderate and low cancer-risk genes, potentially actionable [28]. Our study demonstrated that while 52.3% of the P/LP variants were associated with BRCA1/2 genes, nearly 50% of the women had mutations in noBRCA genes. These findings are similar to those reported in unselected BC populations from countries such as Argentina and Guatemala, where the contribution of noBRCA genes was described as 4.7% and 3.2%, respectively [6, 19]. Similar to studies concerning hereditary BC cases, the frequency of P/LP mutations in noBRCA genes constitutes a significant proportion (Supplementary Table 1). Paixão et al. (2022) found P/LP variants from 9.6% (BRCA1/2) to 25.2% (noBRCA) analyzing 321 Brazilian patients with a panel of 94 genes [7]. Additionally, Cock-Rada and colleagues assessed 25 cancer susceptibility genes in 85 women from Medellin, who met the criteria for HBOC molecular testing; this study identified mutations in six genes: BRCA2, BRCA1, PALB2, ATM, MSH2, and PMS2 [29]. All these findings describe germline mutation profiles which, like our results, demonstrate the contribution to the genetic variability in BC of genes such as ATM, PALB2, and CHEK2, and should be taken into consideration. This finding is consistent with reports from other Latin American populations, where mutations in PALB2 or RAD51C explain a significant proportion of cases. The present results, along with others previously published, demonstrate that the analysis of genes other than BRCA1/2 increases the detection rate of P/LP variants, which maximizes the identification of germline variants in patients with hereditary and unselected BC.
In our study, we identified recurrent mutations in 1.75% of the population analyzed, indicating that most P/LP variants are private. This finding is consistent with previous reports in other LATAM populations, where the recurrence of mutations is low [13]. Three recurrent variants were identified: two in the BRCA2 gene (c.2808_2811delACAA, p.Ala938Profs*21 and c.1763_1766delATAA, p.Asn588Serfs*25), and one in the ATM gene (c.5496 + 2_5496 + 5delTAAG). Two women carrying recurrent mutations in BRCA2 (c.1763_1766delATAA, p.Asn588Serfs*25) and ATM (c.5496 + 2_5496 + 5delTAAG) genes had the diagnosis of other types of cancer that is, lymphoma and, thyroid and gastric, respectively. Co-occurrence between BC and other types of cancer has been pinpointed in the literature [30, 31]. Specifically, P/LP variants in the ATM gene are associated with gastric and thyroid cancers, and risk estimates have also been described; for gastric cancer, several studies associated ATM mutations with OR (odds ratio) ranging from 2.97 to 4.74 [32–34]. Recently, the association between H. pylori infection and germline P variants in genes as BRCA1, BRCA2, ATM, and PALB2, has been described; people with H. pylori infection and germline mutations in those genes have a higher gastric cancer cumulative risk at 85 years of 45.5% (95% CI, 20.7 to 62.6); in contrast, the risk in people with H. pylori infection alone is 14.4% (95% CI, 12.2 to 16.6) [35]. Thyroid cancer (TC) has also been associated with the presence of germline mutations in BRCA2 and ATM genes [36, 37]. Interestingly, a published study showed an increased oncogenic SNPs burden in cases with co-occurrence of BC and TC. In patients with double cancers, germline variants were found in PALB2, BRCA1, BRCA2, ATM, and CHEK2 genes, which are known risk genes associated with BC [38].
Recurrent variants could also be considered founder mutations. The prevalence of founder mutations has been extensively documented for the BRCA1 and BRCA2 genes. These pathogenic variants represent the majority of observed mutations in specific populations and have been confirmed as true founders through analysis of common ancestral haplotypes [39]. In our population of study three Colombian founder mutations, previously described [21], were identified, one in BRCA1 c.5123C > A (A1708E), and two in BRCA2 c.1763_1766delATAA (1991del4) and c.2808_2811delACAA (3034del4).
Identification of recurrent pathogenic variants in the ATM gene is of importance, as previous studies have demonstrated that women carrying mutations in this gene have a significantly increased risk of developing BC with a risk similar to that conferred by germline mutations in the BRCA2 gene [40]. Interestingly, the allelic frequency of the ATM variant c.5496 + 2_5496 + 5delTAAG was 0.375%, although it has not been previously reported in the population database gnomAD, the variant has been identified in cases related to ataxia-telangiectasia syndrome, familial breast cancer, and hereditary cancer predisposition syndrome. These findings are not supported by population-based studies but have been submitted by molecular diagnostic centers such as Color Diagnostics (2019), Fulgent Genetics (2021), Baylor Genetics (2022), Invitae (2022), Ambry Genetics (2023), and Myriad Genetics (2024) (https://www.ncbi.nlm.nih.gov/clinvar/variation/VCV000565770.15 (accessed May 7, 2024)). In all instances, the variant has been determined to be germline. However, due to the unknown origin, the number of affected individuals, or the lack of familial segregation analysis, we cannot make comparisons with the data from the current study. It is noteworthy to date, this variant has been attributed to a significant impact on RNA splicing, although this has not been experimentally proven, hence its classification according to ACMG criteria is likely pathogenic. Functional validation of this recurrent variant demonstrated an exon skipping, leading to a predicted deletion of 59 amino acids located in the Pincer domain of the ATM protein [41]. The splicing process is an event that most eukaryotes genes go through and is regulated by RNA-Binding Proteins (RBPs), cis-regulatory elements, and trans-acting factors [42]. Alternative splicing is dysregulated in cancerous cells in comparison with healthy cells, and carcinogenesis has been associated with alterations in direct and indirect regulators, leading to altered splicing profiles [43]. In the present study, the minigene assay resulted in an exon skipping, caused by a cis-regulatory element (c.5496 + 2_5496 + 5delTAAG) on the ATM gene. This molecular finding added to the absence of this mutation in the gnomAD database, supports the pathogenic effect of the mutation in the function of the ATM protein and the possible role in BC development. Dysregulation of alternative splicing in cancer has made it a therapeutic target and several therapeutic strategies are currently under study; that is, targeting RNA splicing factors, splicing factors regulated by blocking kinases, and antiRNA molecules [44].
Although 58.3% of women with a mutation had several relatives with various types of cancer, segregation analysis was performed in some families with index cases having P/LP variants in BRCA2, ATM, and PALB2 genes. Interestingly, segregation of P/LP variant and phenotype was observed in two families tested for BRCA2 mutations (Fig. 1). Index cases of these families had an earlier age of onset compared with their relatives who suffered BC as well, suggesting anticipation phenomena. This finding may be associated with the greater penetrance of the BRCA1/2 genes, compared to other genes with moderate penetrance such as ATM, but some authors have proposed the interference of non-genetic factors as an explanation for this anticipation [45, 46].
Germline pathogenic small indels and LGRs contribute to the development of breast and ovarian cancers [47]. Ratios of BRCA1/2 LGRs germline mutations are population dependent [22, 48–56]. To our knowledge, in Colombia, BRCA1/2 LGRs have been tested in two studies. Vargas and colleagues tested 60 Afro-Colombian families with HBOC, they did not find LGRs in that population [17]. Torres and colleagues tested 221 breast/ovarian cancer families, finding a LGR in the BRCA2 (ex1-14del) gene in two unrelated patients (0,9%) [21]. Considering the three Colombian cohorts of patients assessed for BRCA1/2 LGRs (Vargas et al., Torres et al., and ours), the prevalence of this type of rearrangement in BRCA1/2 genes would be 0,3% (2/681). Pondering the frequencies described previously, LGRs prevalence in BRCA1/2 genes is low in Colombian BC patients, regardless of hereditary or family history.
This study has some limitations. The germline variants analyzed are rare and although they are located in high and moderate-risks genes, common SNPs also contribute to the development of BC. LGRs were only studied in BRCA1/2, although this type of rearrangement has been found in genes including CHEK2 and ATM, in BC patients [57].
In conclusion, molecular analysis via WES enabled the establishment of the genomic profile of P/LP variants in ten clinically significant genes related to BC risk in the analyzed population. Additionally, this investigation was conducted in a population of women with unselected BC, which has been less addressed in the global literature compared to the vast amount of research conducted on individuals with hereditary cancer. Based on the information described and our study results, the germline mutation profile exhibits variation in genes and frequencies, contingent upon the region and characteristics of the population assessed. This underscores the importance of conducting population-based studies and determining the prevalence of clinically impactful genes. Such efforts can aid in the identification of mutations and facilitate the implementation of national genetic analysis policies, genetic counseling, and early detection strategies. Our study also highlights the utility of WES as an appropriate method for identifying germline variants located in coding and exon–intron boundary regions of genes that are clinically relevant in BC. WES analysis has the potential to detect rare, novel, and infrequently studied P/LP variants, including intronic mutations.
Supplementary Information
Acknowledgements
We thank the patients and their families for their participation in the study. We would like to thank the medical students of Universidad del Rosario, in particular Mariana Angulo-Aguado, Valentina Balaguera and Kevin Llinás-Caballero for their support in sample recollection.
Abbreviations
- ACMG/AMP
American College of Medical Genetics and Genomics/American Molecular Pathology
- BC
Breast cancer
- BMI
Body mass index (kg/m2)
- ER
Estrogen receptor
- HBOC
Hereditary breast and ovary cancer
- HER-2
Human epidermal growth factor receptor 2
- HR
Hormonal receptors
- HRT
Hormonal replacement therapy
- LATAM
Latin-American
- LGRs
Large genomic rearrangements
- MLPA
Multiplex ligation dependant probe amplification
- NCCN
National cancer comprehensive network
- NGS
Next generation sequencing
- P/LP
Pathogenic/likely pathogenic
- PR
Progesterone receptor
- SNP
Single nucleotide polymorphism
- TNBC
Triple negative breast cancer
- WES
Whole exome sequencing
Author contributions
MB, IM, ML, HI, DT, NS, AIO, DL, JG, GR, PALR, RM, WR, JP, MCQ and WM contributed to samples and clinico-pathologic data collection and curation. DCSD, AM, DJFM, NCB carried out the molecular biology experiments. DCSD, MB and CMR performed segregation analysis and genetic counseling. DCSD, AM and RC made bioinformatics analysis and variant classification. NMG performed statistical analysis. CMR and WM contributed to the study conception and design. The first draft of the manuscript was written by DCSD, DJFM, WM and CMR. All authors commented on previous version the manuscript and all of them read and approved the final version.
Funding
The study was financially supported by Pfizer, Universidad del Rosario (ABN-062), Fundación Cardioinfantil–Instituto de Cardiología, and Hospital Universitario Mayor-Méderi (QANBG073).
Availability of data and materials
Further data and the datasets supporting this study are available from the corresponding author upon justified demand.
Declarations
Ethics approval consent to participate
This study was performed in compliance with the Helsinki Declaration and all experimental procedures were approved by Fundación Cardioinfantil–Instituto de Cardiología and Universidad del Rosario Ethics Committee (approval numbers: 402018 7-11-2018, DVO005 1805-CV1469 3-12-2021, Pfizer: WI241988—Investigator initiate research, independent review board: 28-08-2018, GF1147 2018).
Informed consent
Informed consent was obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflicts of interest.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Arnold M, Morgan E, Rumgay H, Mafra A, Singh D, Laversanne M, et al. Current and future burden of breast cancer: global statistics for 2020 and 2040. Breast. 2022;66:15–23. doi: 10.1016/j.breast.2022.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. The Lancet marzo de. 2018;391(10125):1023–1075. doi: 10.1016/S0140-6736(17)33326-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassan MM, Cyr AE, Hagemann IS. Estimating the breast cancer risk conferred by germline mutations. Clin Chem. 2022;68(3):382–384. doi: 10.1093/clinchem/hvab252. [DOI] [PubMed] [Google Scholar]
- 4.Easton DF, Pharoah PDP, Antoniou AC, Tischkowitz M, Tavtigian SV, Nathanson KL, et al. Gene-panel sequencing and the prediction of breast-cancer risk. N Engl J Med. 2015;372(23):2243–2257. doi: 10.1056/NEJMsr1501341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorling L, Carvalho S, Allen J, González-Neira A, Luccarini C, Wahlström C, et al. Breast cancer risk genes—association analysis in more than 113,000 women. N Engl J Med. 2021;384(5):428–439. doi: 10.1056/NEJMoa1913948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cerretini R, Mercado G, Morganstein J, Schiaffi J, Reynoso M, Montoya D, et al. Germline pathogenic variants in BRCA1, BRCA2, PALB2 and RAD51C in breast cancer women from Argentina. Breast Cancer Res Treat. 2019;178(3):629–636. doi: 10.1007/s10549-019-05411-9. [DOI] [PubMed] [Google Scholar]
- 7.Paixão D, Torrezan GT, Santiago KM, Formiga MN, Ahuno ST, Dias-Neto E, Tojal Silva I, Foulkes WD, Polak P, Carraro DM. Characterization of genetic predisposition to molecular subtypes of breast cancer in Brazilian patients. Front Oncol. 2022;31(12):976959. doi: 10.3389/fonc.2022.976959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Castro M, Restrepo CM. Genetics and genomic medicine in Colombia. Mol Genet Genomic Med. 2015;3(2):84–91. doi: 10.1002/mgg3.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R Core Team. No Title. R foundation for statistical computing, Vienna, Austria; 2023.
- 10.Daly MB, Pal T, Maxwell KN, Churpek J, Kohlmann W, AlHilli Z, Arun B, Buys SS, Cheng H, Domchek SM, Friedman S. Genetic/familial high-risk assessment: breast, ovarian, and pancreatic, version 2.2024 featured updates to the NCCN guidelines. JNCCN J Nat Compr Cancer Netw. 2023;21(10):1001–1010. doi: 10.6004/jnccn.2023.0051. [DOI] [PubMed] [Google Scholar]
- 11.Lynch HT, Marcus JN, Lynch J, Snyder CL, Rubinstein WS. Breast cancer genetics. In: Kirby I, Bland EMC, editors. The breast. Elsevier; 2009. pp. 371–415. [Google Scholar]
- 12.Tung N, Battelli C, Allen B, Kaldate R, Bhatnagar S, Bowles K, et al. Frequency of mutations in individuals with breast cancer referred for BRCA 1 and BRCA 2 testing using next-generation sequencing with a 25-gene panel. Cancer. 2015;121(1):25–33. doi: 10.1002/cncr.29010. [DOI] [PubMed] [Google Scholar]
- 13.Torres-Mejía G, Royer R, Llacuachaqui M, Akbari MR, Giuliano AR, Martínez-Matsushita L, et al. Recurrent BRCA1 and BRCA2 mutations in Mexican women with breast cancer. Cancer Epidemiol Biomark Prev. 2015;24(3):498–505. doi: 10.1158/1055-9965.EPI-13-0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dutil J, Golubeva VA, Pacheco-Torres AL, Diaz-Zabala HJ, Matta JL, Monteiro AN. The spectrum of BRCA1 and BRCA2 alleles in Latin America and the Caribbean: a clinical perspective. Breast Cancer Res Treat. 2015;154(3):441–453. doi: 10.1007/s10549-015-3629-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guindalini RSC, Viana DV, Kitajima JPFW, Rocha VM, López RVM, Zheng Y, et al. Detection of germline variants in Brazilian breast cancer patients using multigene panel testing. Sci Rep. 2022;12(1):4190. doi: 10.1038/s41598-022-07383-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silva FC, Lisboa BC, Figueiredo MC, Torrezan GT, Santos ÉM, Krepischi AC, et al. Hereditary breast and ovarian cancer: assessment of point mutations and copy number variations in Brazilian patients. BMC Med Genet. 2014;15(1):55. doi: 10.1186/1471-2350-15-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vargas E, de Deugd R, Villegas VE, Gil F, Mora L, Viaña LF, et al. Prevalence of BRCA1 and BRCA2 germline mutations in patients of African descent with early-onset and familial Colombian breast cancer. Oncologist. 2022;27(2):e151–e157. doi: 10.1093/oncolo/oyab026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abugattas J, Llacuachaqui M, Allende YS, Velásquez AA, Velarde R, Cotrina J, et al. Prevalence of BRCA1 and BRCA2 mutations in unselected breast cancer patients from Peru. Clin Genet. 2015;88(4):371–375. doi: 10.1111/cge.12505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren M, Orozco A, Shao K, Albanez A, Ortiz J, Cao B, et al. Germline variants in hereditary breast cancer genes are associated with early age at diagnosis and family history in Guatemalan breast cancer. Breast Cancer Res Treat. 2021;189(2):533–539. doi: 10.1007/s10549-021-06305-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hernández JEL, Llacuachaqui M, Palacio GV, Figueroa JD, Madrid J, Lema M, et al. Prevalence of BRCA1 and BRCA2 mutations in unselected breast cancer patients from Medellín, Colombia. Hered Cancer Clin Pract. 2014;12(1):11. doi: 10.1186/1897-4287-12-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Torres D, Bermejo JL, Rashid MU, Briceño I, Gil F, Beltran A, et al. Prevalence and penetrance of BRCA1 and BRCA2 germline mutations in Colombian breast cancer patients. Sci Rep. 2017;7(1):4713. doi: 10.1038/s41598-017-05056-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Villarreal-Garza C, Alvarez-Gómez RM, Pérez-Plasencia C, Herrera LA, Herzog J, Castillo D, et al. Significant clinical impact of recurrent BRCA1 and BRCA2 mutations in Mexico. Cancer. 2015;121(3):372–378. doi: 10.1002/cncr.29058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.González-Rivera M, Lobo M, López-Tarruella S, Jerez Y, Del Monte-Millán M, Massarrah T, et al. Frequency of germline DNA genetic findings in an unselected prospective cohort of triple-negative breast cancer patients participating in a platinum-based neoadjuvant chemotherapy trial. Breast Cancer Res Treat. 2016;156(3):507–515. doi: 10.1007/s10549-016-3792-1. [DOI] [PubMed] [Google Scholar]
- 24.Godinez Paredes JM, Rodriguez I, Ren M, Orozco A, Ortiz J, Albanez A, et al. Germline pathogenic variants associated with triple-negative breast cancer in US Hispanic and Guatemalan women using hospital and community-based recruitment strategies. Breast Cancer Res Treat. 2024 doi: 10.1007/s10549-024-07300-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poliani L, Greco L, Barile M, Buono AD, Bianchi P, Basso G, et al. Canonical and uncanonical pathogenic germline variants in colorectal cancer patients by next-generation sequencing in a European referral center. ESMO Open. 2022;7(6):100607–100607. doi: 10.1016/j.esmoop.2022.100607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cifuentes-C L, Rivera-Herrera AL, Barreto G. BRCA1 and BRCA2 mutations in breast and ovarian cancer families from south west Colombia. Colombia Méd. 2019;50(3):163–175. doi: 10.25100/cm.v50i3.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Torres D, Rashid MU, Gil F, Umana A, Ramelli G, Robledo JF, et al. High proportion of BRCA1/2 founder mutations in Hispanic breast/ovarian cancer families from Colombia. Breast Cancer Res Treat. 2007;103(2):225–232. doi: 10.1007/s10549-006-9370-1. [DOI] [PubMed] [Google Scholar]
- 28.O’Leary E, Iacoboni D, Holle J, Michalski ST, Esplin ED, Yang S, et al. Expanded gene panel use for women with breast cancer: identification and intervention beyond breast cancer risk. Ann Surg Oncol. 2017;24(10):3060–3066. doi: 10.1245/s10434-017-5963-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cock-Rada AM, Ossa CA, Garcia HI, Gomez LR. A multi-gene panel study in hereditary breast and ovarian cancer in Colombia. Fam Cancer. 2018;17(1):23–30. doi: 10.1007/s10689-017-0004-z. [DOI] [PubMed] [Google Scholar]
- 30.Zheng G, Yu H, Hemminki A, Försti A, Sundquist K, Hemminki K. Familial associations of female breast cancer with other cancers. Int J Cancer. 2017;141(11):2253–2259. doi: 10.1002/ijc.30927. [DOI] [PubMed] [Google Scholar]
- 31.Ramin C, Veiga LHS, Vo JB, Curtis RE, Bodelon C, Aiello Bowles EJ, et al. Risk of second primary cancer among women in the Kaiser permanente breast cancer survivors cohort. Breast Cancer Res. 2023;25(1):50. doi: 10.1186/s13058-023-01647-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall MJ, Bernhisel R, Hughes E, Larson K, Rosenthal ET, Singh NA, et al. Germline pathogenic variants in the ataxia telangiectasia mutated (ATM) gene are associated with high and moderate risks for multiple cancers. Cancer Prev Res. 2021;14(4):433–440. doi: 10.1158/1940-6207.CAPR-20-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Helgason H, Rafnar T, Olafsdottir HS, Jonasson JG, Sigurdsson A, Stacey SN, et al. Loss-of-function variants in ATM confer risk of gastric cancer. Nat Genet. 2015;47(8):906–910. doi: 10.1038/ng.3342. [DOI] [PubMed] [Google Scholar]
- 34.Yoshimura A, Imoto I, Iwata H. Functions of breast cancer predisposition genes: implications for clinical management. Int J Mol Sci. 2022;23(13):7481. doi: 10.3390/ijms23137481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Usui Y, Taniyama Y, Endo M, Koyanagi YN, Kasugai Y, Oze I, et al. Helicobacter pylori, homologous-recombination genes, and gastric cancer. N Engl J Med. 2023;388(13):1181–1190. doi: 10.1056/NEJMoa2211807. [DOI] [PubMed] [Google Scholar]
- 36.Miasaki FY, Saito KC, Yamamoto GL, Boguszewski CL, de Carvalho GA, Kimura ET, et al. Thyroid and breast cancer in 2 sisters with monoallelic mutations in the ataxia telangiectasia Mutated (ATM) gene. J Endocr Soc. 2022;6(4):26. doi: 10.1210/jendso/bvac026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu Y, Dong L, Li D, Chuai S, Wu Z, Zheng X, et al. Targeted DNA sequencing detects mutations related to susceptibility among familial non-medullary thyroid cancer. Sci Rep. 2015;5(1):16129. doi: 10.1038/srep16129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakos B, Kiss A, Árvai K, Szili B, Deák-Kocsis B, Tobiás B, et al. Co-occurrence of thyroid and breast cancer is associated with an increased oncogenic SNP burden. BMC Cancer. 2021;21(1):706. doi: 10.1186/s12885-021-08377-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rebbeck TR, Friebel TM, Friedman E, Hamann U, Huo D, Kwong A, et al. Mutational spectrum in a worldwide study of 29,700 families with BRCA1 or BRCA2 mutations. Hum Mutat. 2018;39(5):593–620. doi: 10.1002/humu.23406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goldgar DE, Healey S, Dowty JG, Da Silva L, Chen X, Spurdle AB, et al. Rare variants in the ATMgene and risk of breast cancer. Breast Cancer Res. 2011;13(4):R73. doi: 10.1186/bcr2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Warren C, Pavletich NP. Structure of the human ATM kinase and mechanism of Nbs1 binding. Elife. 2022;11:e74218. doi: 10.7554/eLife.74218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Ann Rev Biochem. 2003;72(1):291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 43.Sciarrillo R, Wojtuszkiewicz A, Assaraf YG, Jansen G, Kaspers GJL, Giovannetti E, et al. The role of alternative splicing in cancer: From oncogenesis to drug resistance. Drug Res Updates. 2020;53:100728. doi: 10.1016/j.drup.2020.100728. [DOI] [PubMed] [Google Scholar]
- 44.Peng Q, Zhou Y, Oyang L, Wu N, Tang Y, Su M, et al. Impacts and mechanisms of alternative mRNA splicing in cancer metabolism, immune response, and therapeutics. Mol Ther. 2022;30(3):1018–1035. doi: 10.1016/j.ymthe.2021.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kedmi A, Kadouri L, Sagy I, Hamburger T, Levin G, Zimhony-Nissim N, et al. Genetic anticipation of breast cancer among BRCA1/BRCA2 mutation carriers: a retrospective study. Int J Gynecol Obstet. 2022;159(2):537–543. doi: 10.1002/ijgo.14179. [DOI] [PubMed] [Google Scholar]
- 46.Noh JM, Choi DH, Baek H, Kim MJ, Park H, Huh SJ, et al. Genetic anticipation of familial breast cancer with or without BRCA mutation in the Korean population. Cancer Genetics abril de. 2014;207(4):160–163. doi: 10.1016/j.cancergen.2014.04.002. [DOI] [PubMed] [Google Scholar]
- 47.Bozsik A, Pócza T, Papp J, Vaszkó T, Butz H, Patócs A, et al. Complex characterization of germline large genomic rearrangements of the BRCA1 and BRCA2 genes in high-risk breast cancer patients—novel variants from a large national center. Int J Mol Sci. 2020;21(13):4650. doi: 10.3390/ijms21134650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petrij-Bosch A, Peelen T, van Vliet M, van Eijk R, Olmer R, Drüsedau M, et al. BRCA1 genomic deletions are major founder mutations in Dutch breast cancer patients. Nat Genet. 1997;17(3):341–345. doi: 10.1038/ng1197-341. [DOI] [PubMed] [Google Scholar]
- 49.Agata S, Viel A, Puppa LD, Cortesi L, Fersini G, Callegaro M, Palma MD, Dolcetti R, Federico M, Venuta S, Miolo G. Prevalence of BRCA1 genomic rearrangements in a large cohort of Italian breast and breast/ovarian cancer families without detectable BRCA1 and BRCA2 point mutations. Genes Chromosomes Cancer. 2006;45(9):791–797. doi: 10.1002/gcc.20342. [DOI] [PubMed] [Google Scholar]
- 50.Smith MJ, Urquhart JE, Harkness EF, Miles EK, Bowers NL, Byers HJ, et al. The contribution of whole gene deletions and large rearrangements to the mutation spectrum in inherited tumor predisposing syndromes. Hum Mutat. 2016;37(3):250–256. doi: 10.1002/humu.22938. [DOI] [PubMed] [Google Scholar]
- 51.Montagna M. Genomic rearrangements account for more than one-third of the BRCA1 mutations in northern Italian breast/ovarian cancer families. Hum Mol Genet. 2003;12(9):1055–1061. doi: 10.1093/hmg/ddg120. [DOI] [PubMed] [Google Scholar]
- 52.Hogervorst FBL, Nederlof PM, Gille JJP, McElgunn CJ, Grippeling M, Pruntel R, et al. Large genomic deletions and duplications in the BRCA1 gene identified by a novel quantitative method. Can Res. 2003;63(7):1449–1453. [PubMed] [Google Scholar]
- 53.Karami F, Mehdipour P. A comprehensive focus on global spectrum of BRCA1 and BRCA2 mutations in breast cancer. Biomed Res Int. 2013;2013:1–21. doi: 10.1155/2013/928562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sluiter MD, van Rensburg EJ. Large genomic rearrangements of the BRCA1 and BRCA2 genes: review of the literature and report of a novel BRCA1 mutation. Breast Cancer Res Treat. 2011;125(2):325–349. doi: 10.1007/s10549-010-0817-z. [DOI] [PubMed] [Google Scholar]
- 55.Concolino P, Rizza R, Mignone F, Costella A, Guarino D, Carboni I, et al. A comprehensive BRCA1/2 NGS pipeline for an immediate copy number variation (CNV) detection in breast and ovarian cancer molecular diagnosis. Clin Chim Acta. 2018;480:173–179. doi: 10.1016/j.cca.2018.02.012. [DOI] [PubMed] [Google Scholar]
- 56.Machado PM, Brandão RD, Cavaco BM, Eugénio J, Bento S, Nave M, et al. Screening for a BRCA2 rearrangement in high-risk breast/ovarian cancer families: evidence for a founder effect and analysis of the associated phenotypes. J Clin Oncol. 2007;25(15):2027–2034. doi: 10.1200/JCO.2006.06.9443. [DOI] [PubMed] [Google Scholar]
- 57.Dennis J, Tyrer JP, Walker LC, Michailidou K, Dorling L, Bolla MK, et al. Rare germline copy number variants (CNVs) and breast cancer risk. Commun Biol. 2022;5(1):65. doi: 10.1038/s42003-021-02990-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Further data and the datasets supporting this study are available from the corresponding author upon justified demand.