Abstract
The natural history of type D simian retrovirus (SRV) infection is poorly characterized in terms of viral load, antibody status, and sequence variation. To investigate this, blood samples were taken from a small cohort of mostly asymptomatic cynomolgus macaques (Macaca fascicularis), naturally infected with SRV type 2 (SRV-2), some of which were followed over an 8-month period with blood taken every 2 months. Provirus and RNA virus loads were obtained, the samples were screened for presence of antibodies to SRV-2 and neutralizing antibody titers to SRV-2 were assayed. env sequences were aligned to determine intra- and intermonkey variation over time. Virus loads varied greatly among cohort individuals but, conversely, remained steady for each macaque over the 8-month period, regardless of their initial levels. No significant sequence variation was found within an individual over time. No clear picture emerged from these results, which indicate that the variables of SRV-2 infection are complex, differ from those for lentivirus infection, and are not distinctly related to disease outcome.
The simian retroviruses (SRVs) are type D retroviruses only distantly related to the lentiviruses. They infect various Asian macaque species and can cause a fatal immune deficiency (7, 11, 12, 13, 22, 30), similar to that induced by simian immunodeficiency virus (SIV) in macaques. Of the five simian retrovirus neutralization serotypes identified (SRV-1 to SRV-5), three (SRV-1 to SRV-3) have been molecularly cloned and genomically sequenced (27, 29, 34). Disease caused by the more commonly found SRV-2 infection in macaques is characterized by diarrhea, fever, chronic weight loss, anemia, and sometimes retroperitoneal fibromatosis, a tumor of connective tissue origin (21). As in SIV infection, secondary opportunistic infections often develop in diseased monkeys (13, 25).
Type D retroviruses emerged as serious pathogens associated with immune deficiency between 1983 and 1985 to devastating effect in primate centers across the United States, including those in New England, California, Oregon, and Washington (7, 21, 30). The prevalence of type D retrovirus infection in these breeding colonies reached epidemic proportions; in the California Primate Center, for example, almost all adult macaques were infected with either SRV-1 or SRV-2 and the mortality rate among juveniles less than 2 years of age approached 50% (17). This was particularly disturbing since these monkeys represented a large proportion of primates used for biomedical research. Thus, considering the severity and frequency of disease caused by SRV-2 infection in macaque breeding populations, it is surprising that so few data exist on the probable correlates of disease, such as proviral copy numbers, RNA plasma levels, and antibody status. These variables are critical in determining the course of other retroviral disease therapy in humans, such as human immunodeficiency virus (HIV)-infected individuals (5, 6, 26). We have therefore hypothesized in this investigation that the course of SRV-2 induced disease will be determined by the same factors.
Data from SRV-1 experimentally infected macaques suggest that pathogenesis-associated parameters follow three profiles in which monkeys (i) died shortly after presenting with symptoms of disease, were viremic, but lacked detectable serum antibodies; (ii) remained alive after developing a mild form of disease, with low-grade viremia, and transient initial antibody response; and (iii) were asymptomatic, with high levels of serum antibodies and transient viremia (15, 23). While these studies correlate SRV-1 disease progression with the above-mentioned parameters, no quantitative data exist on virus loads. Likewise, comparative data on SRV-2 viral load over time in animals or even a range of viral loads between animals have not been reported. Additionally, the relationships between antibody status, plasma and cellular viral load, and sequence variation in SRV-2-infected macaque individuals remain unclear.
To investigate the natural history of SRV-2 infection, virus load, antibody status, and sequence variation were measured in a cohort of naturally infected but clinically stable asymptomatic cynomolgus macaques (Macaca fascicularis). Blood samples were taken from eight macaques on four occasions over the course of 8 months. To establish a range of viral loads for asymptomatic macaques, we took additional blood samples from other naturally infected monkey individuals with no signs of disease. Blood samples taken just before or at the time of death from diseased macaques served as controls. This analysis on a cohort of largely asymptomatic monkeys, a population in which it is difficult to identify correlates of disease, allowed us to confirm and extend data reported for SRV-1 infection and to compare SRV-2 virus load with that of other retroviruses.
MATERIALS AND METHODS
Study samples.
Seventeen cynomolgus macaques were identified by virus isolation as type D positive at the National Institute of Biological Standards and Control during the course of their ongoing type D retrovirus screening program to eliminate infected animals from AIDS research. PCR specifically determined the isolates to be SRV-2. Three-milliliter samples of whole blood were taken from all animals as part of their veterinary care. Of the 17 monkeys, 14 were asymptomatic for clinical signs of disease and 3, from which a blood sample was collected near or at the time of death, died of SRV-2-related illness. Clinical signs of disease included weight loss (L121 and L34), massive enlargement of the mesenteric lymph nodes (L121), neoplasia (L34), and enlargement of lymph nodes, spleen, and liver (L34 and 845). Eight of the 16 animals were followed over an 8-month period, and whole blood samples were collected from this cohort at 2- or 3-month intervals on four occasions. Samples from two uninfected macaques, as determined by virus isolation and PCR, served as negative controls.
Cells and plasma separation.
Peripheral blood mononuclear cells (PBMCs) from each of the 17 monkeys were fractionated from 3 to 5 ml of EDTA-collected blood by Ficoll-Hypaque (Sigma) gradient centrifugation, washed in phosphate-buffered saline (PBS), and aliquoted at 106 cells per ml. The PBMCs were either cocultured with Raji cells (a Burkitt lymphoma B-cell line) (8), for virus isolation, after which DNA was extracted, or stored frozen in liquid nitrogen in 10% dimethyl sulfoxide (Sigma)–50% fetal calf serum (FCS) (Gibco)–40% RPMI 1640 (Gibco). Additionally, aliquots of 1 ml of plasma were frozen in liquid nitrogen for RNA extractions and neutralizing antibody assays.
Virus isolation.
Raji cells (8) were maintained in RPMI 1640 medium supplemented with 10% heat-inactivated FCS, 2 mM l-glutamine, and 1% penicillin and streptomycin. One million PBMCs were cocultured with 2 × 105 Raji cells in 24-well plates. Cells were passaged every 4 days and observed daily for 3 weeks for syncytium induction.
IFM.
SRV-2-infected human lung carcinoma cells (A549) (18) were maintained in Dulbecco's modified Eagle medium supplemented with FCS and antibiotics as described above. For immunofluorescence microscopy (IFM) assays, coverslips were seeded in six-well plates with 5 × 105 SRV-2-infected A549 cells and incubated for 16 to 20 h at 37°C. Cells were fixed on coverslips in 50:50 methanol and acetone for 10 min on ice, washed in PBS, and air dried at room temperature. Cells were incubated with antiserum (diluted 1:50 in PBS) for 1 h at room temperature, and unbound antibody was removed by three washes in PBS–3% FCS. Fluorescein isothiocyanate-conjugated anti-macaque immunoglobulin G (Sigma) diluted 1:40 in PBS–3% FCS was added for 20 min at room temperature. Coverslips were washed once in PBS–3% FCS and three times in PBS, counterstained in PBS–0.5% Evans blue (Sigma), and mounted onto slides in PBS–50% glycerol. Fluorescent cells were observed microscopically for cytoplasmic and envelope staining. Human and monkey sera negative for type D retrovirus by culture assays were included as negative controls.
Virus neutralization assay.
Seven days postinfection, neutralizing antibody titers to SRV-2 were assayed by syncytium induction in Raji cells. Fifty-microliter plasma samples, serially diluted twofold with RPMI 1640 containing 10% FCS, were incubated at 4°C for 30 min with 50 μl of cell-free virus (100 50% tissue culture infectious doses) in duplicate wells of a 96-well plate. Following incubation, 104 Raji cells (50 μl) were added to each well, plates were incubated at 37°C for 24 h, and 150 μl of medium was added. Four wells of cells received an equivalent amount of virus without the addition of the plasma sample (virus control).
DNA and RNA isolation.
PBMCs were counted and resuspended in TST buffer (10 mM Tris-HCl [pH 7.5], 0.32 M sucrose, 1% Triton X-100). Nuclei and cell debris were pelleted by centrifugation at 12,000 × g for 30 s and resuspended in an appropriate volume of TENT buffer (10 mM Tris-HCl [pH 8.3], 1 mM EDTA, 0.5% Nonidet P-40, 0.5% Tween 20) and proteinase K (200 g/ml). The lysate was incubated overnight at 56°C, heat inactivated at 85°C for 10 min, and stored at −20°C. RNA was isolated from plasma by using a commercially available kit (Qiagen), resuspended in nuclease-free water, and immediately converted to cDNA by a reverse transcriptase.
cDNA synthesis.
To synthesize cDNA, 5 μl of plasma-isolated RNA was added to a 15-μl cocktail containing, at initial concentrations, 10 U of avian myeloblastosis virus reverse transcriptase (Promega), 2.5 mM deoxynucleoside triphosphates (Advanced Biotechnologies), 5 μM antisense primer (Oswel DNA), and 20 U of RNAsin RNase inhibitor (Promega) and incubated in a thermocycler (Perkin-Elmer/Cetus) at 42°C for 30 min, followed by a 95°C inactivation step for 3 min. cDNA was amplified as described below.
In vitro enzymatic amplification.
Proviral DNA and plasma RNA were quantified by limiting dilution and nested PCR carried out as described for HIV (28). First, DNA from a known quantity of uncultured PBMCs and cDNA from plasma were serially diluted down a 1:4 gradient with deionized water and then amplified by nested PCR to determine that the last dilution at which a positive signal could be detected (endpoint dilution). For all in vitro amplification reactions, diluted and undiluted samples of DNA were added to a cocktail of buffers containing initial concentrations of 25 mM MgCl2, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 5 μmol each of dATP, dCTP, dGTP, and dTTP, 20 pmol each of the sense and antisense primers, and 2.5 U of Taq DNA polymerase (Gibco) in a total reaction volume of 50 μl. The reaction mixture was the same for first- and second-round amplifications.
A region within the gp70 SRV-2 envelope gene was targeted with a nested set of conserved oligonucleotide primer pairs as previously described (10). Template strands were subjected to a total of 30 amplification cycles, yielding a 440-bp product between base positions 6247 and 6687. After the second round of amplification, appropriately sized products (440 bp) were identified by electrophoresis on a 1% agarose (Sigma) gel, followed by ethidium bromide staining under UV light. Negative and positive controls were included in all experiments. Extraction and storage of DNA, nested PCR, and gel electrophoresis were performed in separate laboratories to avoid PCR carryover and sample contamination (16).
Nucleic acid quantification.
The virus load was estimated from the last dilution at which a positive signal was detected. The endpoint dilution concentration was treated as a single sample replicated 92 times in a 96-well plate along with four negative controls. The 92 replicates were amplified by PCR, and resulting products were examined for the target fragment as described above. The mixture of positive and negative results in a 92-sample replicate was used to infer a maximum likelihood estimator of the mean number of viral particles per reaction that were present preamplification. (The mean number of virus particles per reaction is equal to −ln[F], where F is the fraction of negative reactions in the 92-sample replicate, as previously described [28].) The average, corrected for sample and dilution volumes, was then used to estimate the number of DNA or RNA molecules in the original sample solution. Advantages of using this method of virus load estimation include its statistical accuracy as well as its likely distribution of a single molecule per well (28). Since a single SRV-2 virus molecule is usually present in positive wells, the need for cloning is eliminated. This method allows for the analysis of sequence variability between individuals and within populations of SRV-2 without problems of sequence population mixture.
DNA sequencing.
Multiple samples from each time point for each individual were selected for sequencing to infer intramonkey variation. To increase the likelihood of single-molecule distribution, SRV-2 cDNA and provirus were isolated by limiting dilution, amplified by nested PCR, and directly sequenced. Amplified product DNA was eluted through a spin column (Qiagen) to remove salts and unincorporated oligonucleotides and deoxynucleoside triphosphates. For the sequencing reaction, around 50 ng of purified amplified product was added to 3.2 pmol of either the forward or the reverse primer. The mixture was denatured at 100°C for 2 min and then kept on ice until the addition of 8 μl of Terminator Ready Reaction mix, supplied with the dRhodamine Terminator Cycle Sequencing Ready Reaction kit (ABI). The 20-μl reaction volume was overlaid with a top layer of white light oil to prevent evaporation, pulse spun, and subjected to an automated thermocycler program on a DNA Thermal Cycler 480 (Perkin-Elmer/Cetus) of 25 cycles of denaturation at 96°C for 10 s, primer annealing at 50°C for 5 s, and extension at 60°C for 4 min. Unincorporated dye terminators and excess primers were removed from the extension products by standard ethanol precipitation. Purified products were allowed to dry at 40°C for 3 to 5 min. Sequencing products were reconstituted with template suppression reagent (ABI), resolved by electrophoresis, and analyzed by an Applied BioSystems model 310 genetic analyzer and automated sequencing software.
Quantitative sequence data analysis.
Sequences were aligned by eye and by using CLUSTAL (14) against the homologous region of an SRV-2 sequence previously generated (accession no. M16605) (34). The alignments were entered into the PAUP (Phylogenetic Analysis Using Parsimony) 3.1.1 computer program (31) to produce a data matrix consisting of 45 samples and 311 characters. Gaps were considered as a fifth character state and deleted from all analyses. After removal of positions in the alignment in which gaps had been inserted, DNA distance matrices were constructed for all 45 samples, using a correction for multiple substitutions and relatively rapid rate of divergence (32). Genetic diversity was compared by averaging distance estimates from multiple isolates for the same individual.
Nucleotide sequence accession numbers.
The viral sequences obtained in this study have been submitted to GenBank under nucleotide accession no. AF191840 to AF191905.
RESULTS
RNA and DNA virus load.
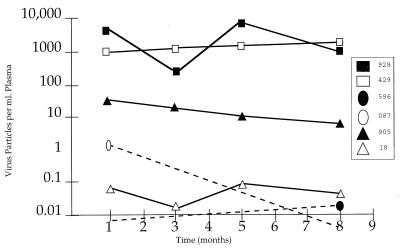
The SRV-2 virus load in a single individual or its range across infected individuals was investigated. Quantitatively, cellular viremia is expressed as provirus copies per 2 × 106 cells, while plasma viremia is expressed as RNA copies per milliliter of plasma. Viral loads were estimated by endpoint dilution analysis and by PCR amplification. The last dilution at which a positive signal could be detected was treated as a 92-sample set; after amplification by nested PCR, the positive signals were counted to estimate virus load. Viral load calculations were based on the null class of a Poisson distribution, described previously (28). To assess the reproducibility of our assay, virus loads were replicated on many of these samples and found to be similar. Table 1 compares viral loads from plasma RNA and those from cellular DNA for all monkeys. For the cohort of monkeys followed over time (Fig. 1), only loads from the final time point are compared (Table 1). The number of RNA copies in 1 ml of plasma varied greatly among asymptomatic individuals. In five monkeys (087, 631, 176, LR68, and LR78), plasma viral RNA could not be detected in the 92-sample replicates amplified by nested PCR. Similarly, low but detectable estimates of RNA copies were calculated for monkeys 18, 596, and 203 (<1 RNA copy/ml) and 905 (6.2 RNA copies/ml). In contrast, higher loads were estimated for monkeys 928 (1,271.6 RNA copies/ml), 429 (1,448.6 RNA copies/ml), LR50 (2,497.3 RNA copies/ml), LR70 (6,905.5 RNA copies/ml), and LR83 (10,383.9 RNA copies/ml). Plasma viremia ranged from undetectable to over 10,000 RNA copies per ml over all time points (Table 1; Fig. 1).
TABLE 1.
Plasma RNA and proviral loads in SRV-2-infected monkeys
| Animal | Clinical signs | RNA (particles/ml of plasma) | DNA (molecules/ 2 × 106 cells) |
|---|---|---|---|
| 905 | None | 6.2 | 115,073 |
| 928 | None | 1,271.6 | 2.8 × 108 |
| 18 | None | 0.06 | UDa |
| 087 | None | UD | 57.4 |
| 429 | None | 1,448.6 | 4.3 × 106 |
| 631 | None | UD | 554 |
| 176 | None | UD | UD |
| 596 | None | 0.02 | 55.5 |
| 203 | None | 0.02 | 2,876.8 |
| LR50 | None | 2,497.3 | 320,000 |
| LR68 | None | UD | 65.6 |
| LR70 | None | 6,905.5 | 1.2 × 106 |
| LR78 | None | UD | 8.4 |
| LR83 | None | 10,383.9 | 5.6 × 106 |
| 845 | Disease | 11.9 | 27.7 |
| L34b | |||
| T1 | Disease | NA | 6.9 |
| T2 | Disease | NA | 2.5 × 106 |
| L121b | |||
| T1 | Disease | NA | 10,317 |
| T2 | Disease | NA | 304.4 |
UD, undetectable.
Results at two time points (T1 and T2) are shown. Plasma was not available (NA) for them.
FIG. 1.
RNA load measurements from asymptomatic monkeys as measured from four time points over an 8-month period. Viral loads of <0.01 particles/ml of plasma, indicated by dashed lines, were undetectable.
In monkeys where plasma RNA virus loads were over 1,000 particles/ml of plasma (e.g., animals 928, 429, LR50, LR60, and LR83), provirus loads were also extremely high (Table 1). There was little agreement between the amount of virus found in plasma compared to proviral DNA (Table 1). Provirus loads tended to be much higher than plasma virus loads, and disparities between the two measurements varied from no essential difference (e.g., animals 18, 176, and 845) to up to 4-logs-higher proviral load (e.g., animals 928, 905, LR70, and LR83) (Table 1). Not only did proviral loads vary immensely among monkeys with disease, but there were disparities between those measured at two time points in each of two individuals (samples taken 1 month [L34] and 5 months [L121] apart) (Table 1). Very high provirus loads found in both asymptomatic and monkeys with disease (>100 copies per cell) may be artificially elevated by the detection of provirus DNA along with circular, unintegrated viral DNA.
While plasma virus load varied greatly among individuals, it largely remained steady in each macaque over the 8-month period, with one exception (Fig. 1). The viral load in animal 928 had a 2-log variation over the 8-month period, in contrast with those in the rest of the cohort, whose loads remained within the same log value, regardless of the virus load measurement at time point 1 (Fig. 1).
Antibody detection.
The antibody response to plasma virus load was evaluated in all animals by IFM (data not shown) and compared with RNA viral load for each time point (Table 2). Most samples were clearly positive, but a few remained indeterminate (monkeys 928 at time point 1, 18 at time point 1, 631 at time points 1 and 2, and 176 at time point 4) even after repeated testing (Table 2). In general, seropositive monkeys also had low (or undetectable) plasma viral load levels over all time points (905, 087, 176, and 596 [Table 2]). Conversely, in monkeys where the viral loads were high, antibodies were undetectable (e.g., animal 928 at time points 3 and 4 and animal 429 at time points 2 and 4) (Table 2).
TABLE 2.
IFM results, RNA viral loads, and neutralizing antibody titers for SRV-2 naturally infected monkeysa
| Animal | Time point 2
|
Time point 3
|
Time point 4
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| IFM | RNA (particles/ml of plasma) | Neutralizing antibody titer | IFM | RNA | Neutralizing antibody | IFM | RNA (particles/ml of plasma) | Neutralizing antibody titer | |
| 905 | + | 16 | 1:32 | + | 11 | 1:64 | + | 6 | 1:16 |
| 928 | +/− | 195 | 1:64 | − | 8,951 | 1:4 | − | 1,271.6 | 1:4 |
| 18 | +/− | 0.01 | 1:32 | + | 0.08 | 1:32 | + | 0.06 | 1:16 |
| 087 | + | 0.38 | 1:16 | + | UD | 1:16 | + | UD | 1:8 |
| 429 | − | 1,150 | 1:4 | NA | 1,232 | NA | − | 1,448.6 | — |
| 631 | +/− | UD | 1:32 | +/− | UD | 1:8 | − | UD | 1:16 |
| 176 | + | UD | 1:64 | − | UD | 1:8 | +/− | UD | 1:8 |
| 596 | + | UD | 1:16 | + | UD | 1:64 | + | 0.02 | 1:16 |
| 845 | NA | NA | NA | NA | NA | NA | + | 11.9 | — |
| 203 | NA | NA | NA | NA | NA | NA | + | 0.02 | 1:16 |
NA, sample not available; UD, undetectable; —, none detected.
Neutralizing antibody responses.
Neutralizing antibody titers against SRV-2 were assessed at all time points in eight macaques for which samples were available and then compared to plasma virus load (Table 2). Neutralizing activity fluctuated in each individual over the study period. In general, neutralizing antibody inversely mirrored plasma viremia levels for each monkey. Neutralizing activity was consistently higher in monkeys whose plasma RNA viral loads were undetectable (631, 176, 596, and 203) or <100 copies/ml of plasma (905, 18, and 087) (Table 2). This trend was clearly shown in animal 928, which had low neutralizing antibody levels and high virus loads at time points 3 and 4 but high neutralizing activity and low virus load at time point 2 (Table 2). Monkey 845 was the only one to have both a low virus load and undetectable neutralizing antibodies. This monkey, however, was showing signs of disease whereas the others tested for neutralizing activity were asymptomatic. Antibodies to SRVs were found in serum samples taken prior to viral load testing from diseased monkey L121 but not from monkey L34. These monkeys were not tested for neutralizing antibodies.
Sequence variation.
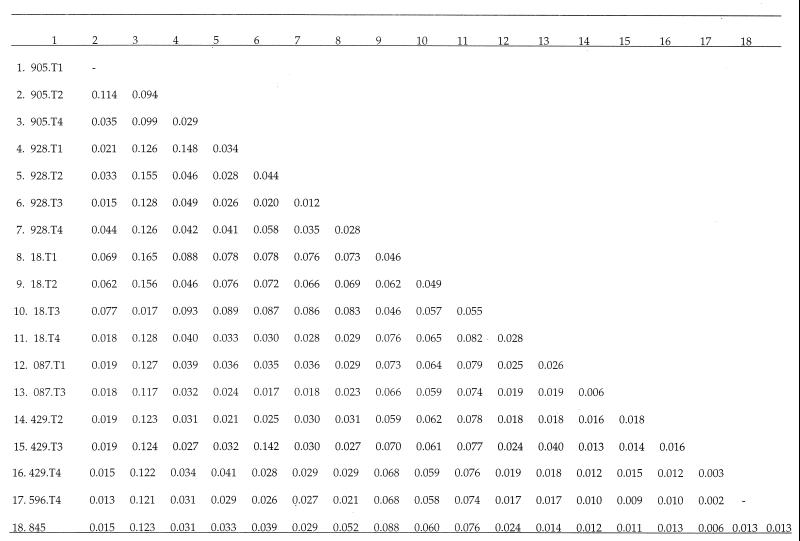
We sought to determine whether more genetic variation in env exists between individuals who have high viral loads than in those whose loads are almost undetectable. Three to five multiple isolates from each animal were sequenced directly from cDNA present in vivo which is not affected by the in vitro selection of potential virus variants. Distance estimates were averaged in order to infer genetic variation between monkeys and between time points for the same individual (Fig. 2). An appreciable amount of variation was found, and average distances among SRV-2 isolates within individuals (range, 0.003 to 0.094) were similar to those found among individuals (range, 0.002 to 0.165) (Fig. 2). To assess load-specific variation, nucleotide divergence between high- and low-virus-load populations was assessed. Nucleotide divergence is the component of diversity not explained by polymorphism within virus populations (24). To estimate nucleotide divergence, nucleotide distances were averaged among all time points in each monkey. Very little average nucleotide divergence ± standard error was found to exist between monkeys 928 (high load) and 087 (low load) (0.006 ± 0.008), 928 and 18 (low load) (0.016 ± 0.008), 928 and 596 (low load) (0.014 ± 0.009), and 928 and diseased monkey 845 (0.012 ± 0.008). Similar low divergence estimates were found between monkeys 429 (high load) and 18 (0.016 ± 0.012) and monkeys 429 and 596 (0.001 ± 0.003). Together, these results indicate that no significant difference in variation exists between populations of high and low virus load.
FIG. 2.
Nucleotide distance estimates averaged for each individual over four time points.
DISCUSSION
Unlike other retrovirus infections, almost nothing is known about the natural history of type D infection. This study focused on a cohort of largely asymptomatic monkeys, naturally infected with SRV-2, in order to understand the roles of variables such as viral load, antibody status, and sequence variation in the natural history of infection.
Recovery from artificially induced SRV-1 infection is related to the disappearance of virus from plasma and PBMCs (23). Because the animals in this study remained mainly asymptomatic, failing to progress to disease in the study period, whether or not virus is immunologically cleared from plasma and PBMCs cannot be inferred from our data. However, our results indicate that low plasma viral load is associated with a higher neutralizing antibody response (monkeys 631, 176, 596, and 203 [Table 2]), perhaps indicating some protective effect as reported by others for SRV-1 (23). Additionally, animals with high viral loads had little to no antibody response (Table 2), a pattern which was also found in monkey 928 at time points 2, 3, and 4, where viral loads and antibody response varied (Table 2). These data are consistent with others (15) in which SRV-1-challenged monkeys survived with persistent viremia (measured by culture) but had declining antibody levels as viremia became more persistent. In contrast, studies on HIV-infected healthy individuals have suggested that differences in viral load levels are not associated with the presence of neutralizing antibodies (3). Only monkey 845 was found to have both nonneutralizing antibodies and a low viral load (Table 2). This monkey, whose blood was taken before death, was also the only animal to show continuous signs of disease. Thus, when type D testing programs in macaque breeding colonies are being considered, it should be noted that antibody testing alone is an insufficient determinant of infection (19).
It is possible that the profiles seen for SRV-2 viral loads and neutralizing antibody responses may be indicative of those seen in HIV infection over time, involving initial viremia and rapid antibody production followed by some viral clearance (1, 9). However, whereas in HIV there is no humoral protective immunity, in SRV-2 infection there does seem to be some protection, as evidenced by neutralizing antibody production and correspondingly low plasma viral loads. In general, the strength of the antibody response was inversely related to the viral load estimates, although these results were not statistically significant by Spearman's rank coefficient (P < 0.01) in our limited sample.
It should be noted that others have found it difficult to coculture SRV-1 in PBMCs from animals having high levels of antibodies (15). In contrast, and in common with HIV (35), SRV-2 was easily cocultured from PBMCs regardless of serum antibody levels (data not shown).
While there was a wide range in plasma viral burden between monkeys, the load within monkeys (estimated only for the last time point) over the 8-month study period remained relatively constant. In two of these animals (928 and 429), provirus load estimates indicated that there were multiple copies per cell, a situation which also might extend to animals with low proviral load. These estimates may be due to the fact that a large number of proviruses is in linear, unintegrated form, as seen in HIV (4). Thus, most of the proviruses detected are probably either defective or uninfectious virus particles.
The gp70 region in the env gene is the most likely region to find genetic variability both among the three sequenced serotypes of SRV type D viruses (34) and specifically between different isolates of SRV-2 (20). Thus, we chose a region of gp70 to estimate the quantity and level of sequence variation among isolates. There was some overall sequence variation. Interestingly, the range of SRV-2 variation found within (0.30 to 9.4%) and among (0.20 to 16.5%) monkeys is similar to the variation found in a cohort of hemophiliacs infected from a single source (mean intrapatient [5.5%] and interpatient [8.3%] variation) (2). On the other hand, the monkeys are imported from various populations, kept in different colonies, and infected from multiple sources. Thus, unexpectedly, SRV-2 sequences vary to the same extent as HIV-1 under certain circumstances.
Importantly, there was just as much variation distributed among isolates in an individual as there was averaged among individuals (Fig. 2), indicating that there is no structure to the variation. Divergence estimates, which showed little difference in sequences obtained from high-viral-load individuals compared to low-viral-load individuals, confirmed this assumption.
In conclusion, our results from mainly clinically stable, asymptomatic animals, a population in which it is difficult to identify correlates of disease, suggest that no clear pattern emerged among factors that contribute to the natural history of SRV-2 infection on a molecular level. RNA and proviral loads varied greatly among both diseased and asymptomatic individuals. However, RNA virus loads remained steady for each individual throughout the study period, regardless of initial levels. In the asymptomatic individuals, viral loads tended to have an inverse relationship to neutralizing antibody levels so that higher neutralizing activity was found in monkeys with low viral loads. However, the diseased monkey had antibodies, but none that were detectably neutralizing, coupled with low viral loads. Finally, genetic variation was not structured between low-viral-load and high-viral-load isolates.
ACKNOWLEDGMENTS
We thank the NIBSC, United Kingdom, for providing blood samples. In particular, we are grateful to Kirsty Silvera and Rebecca Sangster at the NIBSC. We thank Richard Pitman for statistical help and Ximena Patino for generating some of the sequence data. A549 cells containing an SRV-2 plasmid were a generous gift from Margaret Thouless (University of Washington, Seattle).
We are grateful to the Wellcome Trust and the Jefferiss Research Trust for their support.
REFERENCES
- 1.Allain J P, Laurian Y, Paul D A, Senn D. Serological markers in early stages of human immunodeficiency virus infection in haemophiliacs. Lancet. 1986;2:1233–1236. doi: 10.1016/s0140-6736(86)92673-5. [DOI] [PubMed] [Google Scholar]
- 2.Balfe P, Simmonds P, Ludlam C A, Bishop J O, Brown A J L. Concurrent evolution of human immunodeficiency virus type 1 in patients infected from the same source: rate of sequence change and low frequency of inactivating mutations. J Virol. 1990;64:6221–6233. doi: 10.1128/jvi.64.12.6221-6233.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker E, Mackewicz C E, Reyes-Teran G, Sato A, Stranford S A, Fujimura S H, Christopherson C, Chang S Y, Levy J A. Virological and immunological features of long-term human immunodeficiency virus-infected individuals who have remained asymptomatic compared with those who have progressed to acquired immunodeficiency. Blood. 1998;92:3105–3114. [PubMed] [Google Scholar]
- 4.Bukrinsky M I, Stanwick T L, Dempsey M P, Stevenson M. Quiescent T lymphocytes as an inducible virus reservoir in HIV-1 infection. Science. 1991;254:423–426. doi: 10.1126/science.1925601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun T-W, Carruth L, Finzi D, Shen X, DiGiuseppe J A, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn T C, Kuo Y-H, Brookmeyer R, Zeiger M A, Barditch-Crovo P, Siliciano R F. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 6.Coombs R W, Collier A C, Allain J-P, Nikora B, Leuther M, Gjerset G F, Corey L. Plasma viremia in human immunodeficiency virus infection. N Engl J Med. 1989;321:1626–1631. doi: 10.1056/NEJM198912143212402. [DOI] [PubMed] [Google Scholar]
- 7.Daniel M D, King N W, Letvin N L, Hunt R D, Sehgal P K, Desrosiers R C. A new type of D retrovirus isolated from macaques with an immunodeficiency syndrome. Science. 1984;223:602–605. doi: 10.1126/science.6695172. [DOI] [PubMed] [Google Scholar]
- 8.Epstein M A, Achong B G, Barr Y M, Zajac B, Henle G, Henle W. Morphological and virological investigations on cultured Burkitt tumor lymphoblasts (strain Raji) J Natl Cancer Inst. 1966;37:457–459. [PubMed] [Google Scholar]
- 9.Goudsmit J, de Wolf F, Paul D A, Epstein L G, Lange J M, Krone W J, Speelman H, Wolters E C, Van der Noord J, Oleske J M, van der Helm H J, Coutinho R A. Expression of human immunodeficiency virus antigen (HIV-Ag) in serum and cerebrospinal fluid during acute and chronic infection. Lancet. 1986;ii:177–180. doi: 10.1016/s0140-6736(86)92485-2. [DOI] [PubMed] [Google Scholar]
- 10.Grant R F, Malinak C J, Wu H, Sabo A, Tsai C-C. PCR amplification and DNA sequencing of SRV-2 from archived tumor tissues. Virus Res. 1995;36:187–200. doi: 10.1016/0168-1702(95)00003-9. [DOI] [PubMed] [Google Scholar]
- 11.Gravell M, London W T, Hamilton R S, Severe J L, Kapikian A Z, Murti G, Arthur L O, Gilden R V, Osborn K G, Marx P A, Hendrickson R V, Gardner M B. Transmission of simian AIDS with type D retrovirus isolate. Lancet. 1984;i:334–335. doi: 10.1016/s0140-6736(84)90376-3. [DOI] [PubMed] [Google Scholar]
- 12.Henrickson R V, Maul D H, Lerche N W, Osborn K G, Lowenstine L J, Prahalada S, Sever J L, Madden D L, Gardner M B. Clinical features of simian acquired immunodeficiency syndrome (SAIDS) in rhesus monkeys. Lab Anim Sci. 1984;34:140–145. [PubMed] [Google Scholar]
- 13.Henrickson R V, Osborn K G, Madden D L, Anderson J H, Maul D H, Sever J L, Ellingsworth L R, Lowenstine L J, Gardner M B. Epidemic of acquired immunodeficiency in rhesus monkeys. Lancet. 1983;i:388–390. doi: 10.1016/s0140-6736(83)91503-9. [DOI] [PubMed] [Google Scholar]
- 14.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 15.Kwang H-S, Pedersen N S, Lerche N W, Osborn K G, Marx P A, Gardner M B. Viremia, antigenemia, and serum antibodies in rhesus macaques infected with simian retrovirus type 1 and their relationship to disease course. Lab Investig. 1987;56:591–597. [PubMed] [Google Scholar]
- 16.Kwok S, Higuchi R. Avoiding false positives with PCR. Nature. 1989;339:237–238. doi: 10.1038/339237a0. [DOI] [PubMed] [Google Scholar]
- 17.Lerche N W, Hendrickson R V, Maul D H. Epidemiologic aspects of an outbreak of acquired immunodeficiency in rhesus monkeys (Macaca mulatta) Lab Anim Sci. 1984;34:146–150. [PubMed] [Google Scholar]
- 18.Lieber M, Smith B, Szakal A, Nelson-Rees W, Todaro G. A continuous tumor-cell line from a human lung carcinoma with properties of type II alveolar epithelial cells. Int J Cancer. 1976;15:62–70. doi: 10.1002/ijc.2910170110. [DOI] [PubMed] [Google Scholar]
- 19.Liska V, Lerche N W, Ruprecht R M. Simultaneous detection of simian retrovirus type D serotypes 1, 2, and 3 by polymerase chain reaction. AIDS Res Hum Retroviruses. 1997;13:433–437. doi: 10.1089/aid.1997.13.433. [DOI] [PubMed] [Google Scholar]
- 20.Marracci G H, Kelley R D, Pilcher K Y, Crabtree L, Shiigi S M, Avery N, Leo G, Webb M C, Hallick L M, Axthelm M K, Machida C A. Simian AIDS type D serogroup 2 retrovirus: isolation of an infectious molecular clone and sequence analyses of its envelope glycoprotein gene and 3′ long terminal repeat. J Virol. 1995;69:2621–2628. doi: 10.1128/jvi.69.4.2621-2628.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Marx P A, Bryant M L, Osborn K G, Maul D H, Lerche N W, Moody P, Lowenstine L J, Kluge J D, Zaiss C P, Hendrickson R V, Shiigi S M, Wilson B J, Malley A, Olson L C, Arthur L O, Gilden R V, Barker C S, Hunter E, Munn R J, Heidecker-Fanning G, Gardner M B. Isolation of a new serotype of SAIDS type D retrovirus from Celebes black macaques (Macaca nigra) with immune deficiency and retroperitoneal fibromatosis. J Virol. 1985;56:571–578. doi: 10.1128/jvi.56.2.571-578.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marx P A, Maul D H, Osborn K G, Lerche N W, Moody P, Lowenstine L J, Hendrickson R V, Arthur L O, Gilden R V, Gravell M, London W T, Sever J L, Levy J A, Munn R J, Gardner M B. Simian AIDS: isolation of a type D retrovirus and disease transmission. Science. 1984;223:1083–1086. doi: 10.1126/science.6695196. [DOI] [PubMed] [Google Scholar]
- 23.Maul D H, Lerche N W, Osborn K G, Marx P A, Zaiss C, Spinner A, Kluge J D, MacKenzie M R, Lowenstine L J, Bryant M L, Blakeslee J R, Henrickson R V, Gardner M B. Pathogenesis of simian AIDS in rhesus macaques inoculated with SRV-1 strain of type D retrovirus. Am J Vet Res. 1986;47:863–868. [PubMed] [Google Scholar]
- 24.Nei M. Molecular evolutionary genetics. New York, N.Y: Columbia University Press; 1987. [Google Scholar]
- 25.Osborn K G, Prahalada S, Lowenstine L J, Gardner M B, Maul D H, Henrickson R V. The pathology of an epizootic of acquired immunodeficiency in rhesus macaques. Am J Pathol. 1984;114:94–103. [PMC free article] [PubMed] [Google Scholar]
- 26.Perelson A S, Essunger P, Cao Y, Vesanen M, Hurley A, Saksela K, Markowitz M, Ho D D. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature. 1997;387:188–191. doi: 10.1038/387188a0. [DOI] [PubMed] [Google Scholar]
- 27.Power M D, Marx P A, Bryant M L, Gardner M B, Barr P J, Luciw P A. Nucleotide sequence of SRV-1, a type D simian acquired immune deficiency syndrome retrovirus. Science. 1986;231:1567–1572. doi: 10.1126/science.3006247. [DOI] [PubMed] [Google Scholar]
- 28.Simmonds P, Balfe P, Peutherer J F, Ludlam C A, Bishop J O, Leigh Brown A. Human immunodeficiency virus-infected individuals contain provirus in small numbers of peripheral mononuclear cells at low copy numbers. J Virol. 1990;64:864–872. doi: 10.1128/jvi.64.2.864-872.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonigo, Barker C, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer Monkey virus: an immune-suppressive D-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- 30.Stromberg K, Benveniste R E, Arthur L O, Rabin H, Giddens W E, Ochs H D, Morton W R, Tsai C. Characterization of exogenous type D retrovirus from a fibroma of a macaque with simian AIDS and fibromatosis. Science. 1984;224:289–292. doi: 10.1126/science.6200929. [DOI] [PubMed] [Google Scholar]
- 31.Swofford D L. PAUP. Phylogenetic analysis using parsimony, version 3.1.1 Illinois Natural History Survey. 1993. Champaign, Ill. [Google Scholar]
- 32.Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- 33.Thayer R M, Power M D, Bryant M L, Gardner M B, Barr P J, Luciw P A. Sequence relationships of type D retroviruses which cause simian acquired immunodeficiency syndrome. Virology. 1987;157:317–329. doi: 10.1016/0042-6822(87)90274-1. [DOI] [PubMed] [Google Scholar]
- 34.Zagury D, Fouchard M, Vol J-C, Cattan A, Leibowitch J, Feldman M, Sarin P S, Gallo R C. Detection of infectious HTLV-III/LAV virus in cell-free plasma from AIDS patients. Lancet. 1985;2:505–506. doi: 10.1016/s0140-6736(85)90442-8. [DOI] [PubMed] [Google Scholar]