Abstract
Gene-editing technologies promise to create a new class of therapeutics that can achieve permanent correction with a single intervention. Besides eliminating mutant alleles in familial disease, gene-editing can also be used to favorably manipulate upstream pathophysiologic events and alter disease-course in wider patient populations, but few such feasible therapeutic avenues have been reported. Here we use CRISPR-Cas9 to edit the last exon of amyloid precursor protein (App), relevant for Alzheimer’s disease (AD). Our strategy effectively eliminates an endocytic (YENPTY) motif at APP C-terminus, while preserving the N-terminus and compensatory APP-homologues. This manipulation favorably alters events along the amyloid-pathway – inhibiting toxic APP-β-cleavage fragments (including Aβ) and upregulating neuroprotective APP-α-cleavage products. AAV-driven editing ameliorates neuropathologic, electrophysiologic, and behavioral deficits in an AD knockin mouse model. Effects persist for many months, and no abnormalities are seen in WT mice even after germline App-editing; underlining overall efficacy and safety. Pathologic alterations in the glial-transcriptome of App-KI mice, as seen by single nuclei RNA-sequencing (sNuc-Seq), are also normalized by App C-terminus editing. Our strategy takes advantage of innate transcriptional rules that render terminal exons insensitive to nonsense-decay, and the upstream manipulation is expected to be effective for all forms of AD. These studies offer a path for a one-time disease-modifying treatment for AD.
Keywords: Amyloid precursor protein, amyloid beta, CRISPR, gene-editing, Alzheimer’s disease
INTRODUCTION
The ability to treat diseases at a genetic level with a single intervention and bring about a permanent cure is poised to transform the practice of medicine (1). Recent trials in hematologic and systemic disorders have reported unprecedented clinical outcomes (2, 3), leading to the first approvals of CRISPR therapies (4); raising hopes that similar strategies can be employed for neurodegenerative diseases like AD where traditional therapeutics have been largely disappointing (5). Although recent trials using monoclonal antibodies to Aβ assemblies show efficacy (6), clinical benefits are modest and the drugs need to be repeatedly administered throughout life, increasing risks of intracranial hemorrhage (6, 7). Moreover, many studies have shown that a number of APP-β-cleavage products – other than Aβ – can also cause pathologic deficits such as endo-lysosomal dysfunction (8–10), and removing extracellular Aβ would not be effective against these toxic intermediates. With the promise of targeting etiology and achieving “permanent correction” (11), gene-based therapies offer an alternative.
Recent studies have begun to explore gene-editing in AD. CRISPR-based strategies have been used to selectively inactivate disease-associated mutant alleles in AD, while keeping WT alleles intact (12–14). Such allele-specific inactivation reverses biochemical abnormalities in cells (13), and one study has shown that AAV-mediated delivery of CRISPR-components by local injections can ameliorate some deficits in a transgenic (“5x familial AD”) mouse model (14). Besides caveats of CRISPR-editing in over-expression-based models where unknown transgene-copies are expressed, at best, these strategies would only be effective in a small number of familial AD patients with known gene mutations [< 1% (15)]. In fact, few mutation-independent CRISPR-based therapeutic approaches have ever been reported (16). Local brain injections of nanoparticles carrying CRISPR-Cas9 have also been used to inactivate β-site APP-cleaving enzyme 1 (BACE1) in mice (17), but there is broad consensus that BACE1 is not a good clinical target (18). Moreover, local brain injection is unlikely to be effective in in AD, given the widespread pathology. An exploration of CRISPR-based germline App-edits in mice revealed a 3’UTR-deletion that decreased App expression (19), but safety and therapeutic implications have not been evaluated.
Our translational efforts have centered around APP, that has an established role in AD pathogenesis (20, 21); and remarkably, a single amino-acid APP variant (A673T ‘Icelandic mutation’) is protective for sporadic AD (22). One therapeutic approach is to attenuate the entire APP gene, but APP (or APP-like) genes are highly conserved across species, implying an important physiologic role (23). Moreover, protective APP-cleavage products – that are physiologically expressed – would also be eliminated in this scenario. While deposition of APP-β/γ-cleavage products is a hallmark of AD, under normal conditions, APP is largely cleaved by an alternative α-cleavage pathway, generating distinct APP-fragments that are neuroprotective and neuroregenerative (24). APP cleavage by α-secretases preclude β-cleavage (20, 24), and in principle, this shift in the balance of cleavage (β/γ → α) can be therapeutically manipulated for both sporadic and familial AD. Interestingly, the protective Icelandic APP mutation is also thought to make APP a less favorable substrate for β-cleavage and Aβ production (22, 25). Our current studies follow up on previous work from us and others, showing that the YENPTY-motif at the APP C-terminus is critical in mediating the trafficking of APP into endosomes enriched in BACE1 and triggering APP/BACE1 interaction, which is the rate-limiting step initiating the β/γ-cleavage pathway (26–30). More recently, we showed that elimination of the APP C-terminus containing the YENPTY motif attenuated β-cleavage and augmented α-cleavage (29). Though our proof-of-principle experiments showed that this approach can work in cells, effects on AD pathology and behavior are unknown. Moreover, safety of this approach has not been examined in vivo, and in general, potential therapeutic feasibility of this strategy is unclear.
RESULTS
Our approach is based on CRISPR-Cas9 editing by non-homologous end joining (NHEJ), which is the most studied of all editing approaches, and also the basis of almost all clinically-relevant applications to date (2, 3, 31, 70). A conceptual schematic demonstrating altered trafficking of APP after C-terminus deletion is shown in Figure 1A. Note that loss of the YENPTY-motif (post-editing) blocks internalization of surface APP into endosomes containing BACE1, attenuating APP-β-cleavage. Consequently, APP-α-cleavage is augmented – presumably due to increased dwelling of edited APP on membranes where α-cleavage is predominant (32) – leading to a shift of APP cleavage pattern upon gene-editing. In conventional applications, NHEJ leads to premature stop-codons and nonsense decay of the transcript, resulting in a loss of the protein (33). However, our strategy takes advantage of innate transcriptional rules that prevent nonsense decay when premature termination-codons are installed in the last exon and ~ 50–55 nucleotides upstream of the last exon-exon junction – reviewed in (34) (also see discussion). Note that the YENPTY motif and flanking regions within the APP C-terminus are encoded by the last exon (exon 18) of APP (Fig. 1B).
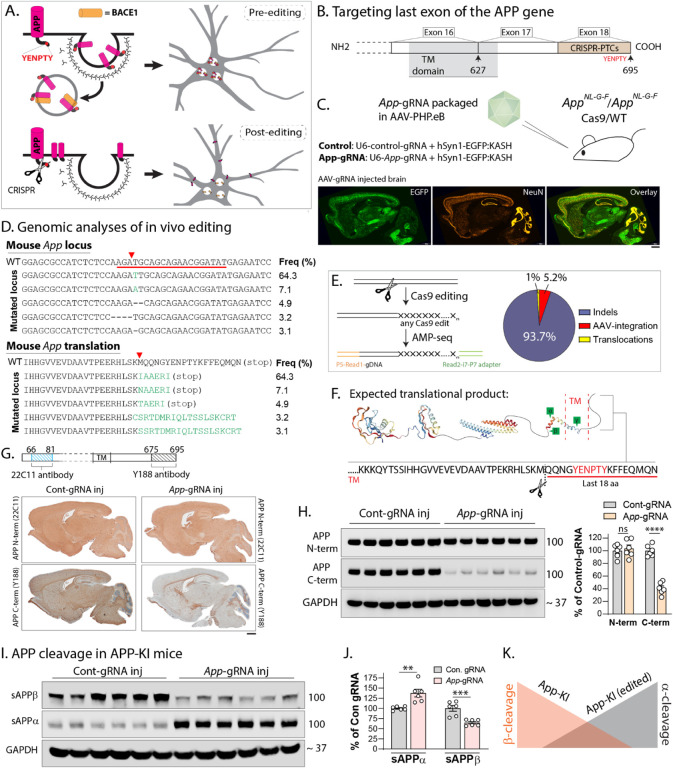
Figure 1: AAV-CRISPR-editing of App C-terminus favorably alters APP β/α cleavage pattern in vivo.
A) Schematic showing internalization of APP into endosomes containing BACE1 (top). CRISPR-mediated effective deletion of the APP-YENPTY motif (scissors, below – see results for details) prevents the endocytosis of APP and its interaction with BACE1, which is the rate-limiting step triggering APP-β-cleavage pathway (schematic attribution: Alec Nabb).
B) Note that the YENPTY motif lies within the region encoded by the last exon (exon 18) of APP, and CRISPR-induced premature termination codons (PTCs) installed within the last exon are expected to generate a truncated protein (see results and discussion for details, TM = transmembrane domain).
C) A construct including a gRNA targeting the last App exon – along with an EGFP marker – was packaged into the AAV-PHP.eB capsid and intravenously injected into AppNL-G-F/Cas9-KI mouse brains. Representative images of AAV-injected mouse brains (below) showing broad transduction (also see Supp. Fig. 1A). Scale bar = 1 mm.
D) Major mutated App loci resulting from above in vivo CRISPR-editing, and their relative frequencies (top), with expected translational products (bottom). Note that most of the editing outcomes predict short indels (also see Supp. Figs. 1B–D). E) Analysis of editing outcomes using quantitative AMP-seq (left) to identify insertion/deletion (indel), translocation, or AAV-integration at Cas9-cleavage sites. Quantification of editing outcomes – expressed as percentages of total edits – in AAV/App-gRNA injected mice. Note that indels are the most common outcome of editing.
F) Schematic showing segment of APP (last 18 amino-acids, containing he YENPTY motif) that is expected to be eliminated after CRISPR-editing.
G) Schematic on top shows recognition sites for APP N-and C-terminus antibodies. Note that upon editing, The C-terminus antibody (Y188) is not able to recognize the translational product, which is used as a surrogate marker for editing in our experiments. Immunostains of mouse brains (bottom panels) show selective attenuation of Y188-antibody signal in App-gRNA injected mice (also see Supp. Figs. 2A–B). Scale bar = 1 mm.
H) Western blots from mouse brains also show selective attenuation of the Y188-antibody signal, quantified on right (N=6 mice per condition, ns=non-significant, ****p<0.0001 – see full blots in Supp. Fig. 2C).
I) Western blots from soluble AppNL-G-F/Cas9-KI mouse brain fractions to evaluate β/α cleavage products (see full blots in Supp. Fig. 2D). Note reversal of the β/α cleavage pattern in App-gRNA injected mice; quantified in J) (N=6 mice per condition, ** p<0.01, ***p<0.001). K) Conceptual schematic showing changes in β/α cleavage after App-editing. All quantitative data presented as mean +/− SEM.
AAV-driven App C-terminus editing favorably alters the balance of APP cleavage
To edit App in vivo, we packaged a App-gRNA (“676-gRNA”, numbering based on predicted amino-acid cut-site) – or control-gRNA not targeting to any known sequence (Supp. Table 1) – and a fluorescent marker into AAV-PHP.eB, which is an engineered capsid that allows widespread transduction into mouse brains after intravenous injections (35). This AAV-cargo was delivered into homozygous App-KI mice crossed with Cas9-KI mice, leading to broad transduction of mouse brains (Fig. 1C; also see Supp. Fig. 1A and Supp. Table 2). The App-KI mice in our experiments have a humanized Aβ domain, carry three familial AD mutations (Swedish, Arctic, and Iberian – AppNL-G-F), and accrue Aβ plaques and other deficits over time (36). Unlike most other AD mouse models, the AppNL-G-F mice do not overexpress APP, and the gene is driven and regulated by native transcriptional and translational elements. Notably, APP overexpression is known to induce pathologic changes in various model-systems (37, 38), and generates abnormal APP fragments that can confound interpretation (39) – concerns that are not applicable in the App-KI mice.
First we asked if our AAV-CRISPR injections edited the mouse App gene as expected. Genomic analysis of the AAV injected App-KI mice showed that edits occurred in the expected App loci and led to premature stop codons within the last exon, with relatively short insertions and deletions (indels, Fig. 1D). Quantification of editing outcomes in these samples using amplicon-sequencing [AMP-seq (40)] showed that indels were by far the most common outcome at the cut-site, followed by AAV-integration and translocations, without detectable off-target effects (Fig. 1E, also see Supp. Fig. 1B–D and Supp. Table 1). Note that the last 18 amino-acids – including the pentapeptide YENPTY motif – are expected to be deleted by our editing strategy (Fig. 1F).
To verify that our App editing generated a translational product that was truncated at the C-terminus, we used an antibody (clone Y188) that only recognizes the last 20 amino-acids of APP, combining this with antibodies against APP N-terminus (Fig. 1G, top). While full-length APP is recognized by the Y188 antibody, if the APP C-terminus is missing, this antibody will not recognize the truncated protein [see further characterization of these antibodies in our previous study(29)]. As shown in the mouse brain section in Figure 1G – top panels, the N-terminus APP antibody showed widespread staining of APP in both control- and App-676-gRNA injected brains. However, Y188 staining was selectively attenuated in App-676-gRNA injected mice, compared to mice injected with the control-gRNA (Fig. 1G – bottom panels). Two-color immunofluorescence showed that most of the AAV-transduced neurons (~ 90%) also had attenuated Y188-antibody staining, suggesting efficient editing (Supp. Fig. 2A–B). Western blots using N- and C-terminus antibodies also confirmed the selective C-terminal truncation of APP. While signals from the Y188 antibody were significantly attenuated in App edited samples, band-intensities in western blots using an APP N-terminus antibody were unchanged (Fig. 1H, see full blots in Supp. Fig. 2C).
Next, we asked if editing the App C-terminus led to a shift in the balance of APP β/α cleavage in vivo. Cleavage of APP by β/α secretases lead to secreted APP-cleavage products (sAPPβ/α) that can be biochemically detected in soluble brain fractions. As expected, brains from App-KI mice with control-gRNA injections had a substantial increase in sAPPβ, compared to sAPPα (Fig. 1I – left lanes). However, brains of App-KI mice injected with the AAV-CRISPR payload had a marked reduction in β-cleavage products, with an increase in α-cleavage fragments (Fig. 1I – right lanes, quantified in Fig. 1J; see full blots in Supp. Fig. 2D). Overall, the data indicate that somatic editing of the App C-terminus using AAV vectors led to a shift in the APP-β/α cleavage pattern (Fig. 1K – also see our previous study (29) for further characterization of β/α cleavage post-editing in mouse and human cells). Note that these changes in β/α cleavage are not due to alterations in levels of the secretases ADAM10 (A Disintegrin And Metalloprotease domain-containing protein 10) and BACE1, which are not affected by our editing strategy (Supp. Fig. 2E–F).
AAV-driven App C-terminus editing rescues multiple deficits in App-KI mice
Next we asked if altering the balance of β/α APP-cleavage using our approach also attenuated pathologic phenotypes in the App-KI mice. Schematic in Figure 2A shows the experimental plan of AAV injections and evaluation timepoints. Note that the mice were injected just before the emergence of Aβ pathology in this model (~ 2 months(36)). As shown in exemplary images (Fig. 2B), Aβ plaques were attenuated after App-676-gRNA injection. Evaluation of Aβ pathology over the course of our experiments showed that while plaque deposition continued in animals injected with the control-gRNA, the Aβ-attenuating effect of the App-676-gRNA was additive over time, likely due to ongoing dampening of APP β-cleavage for several months after a single injection (Fig. 2C). Substantial reductions in insoluble Aβ were also seen in the brains of one-year old animals (Fig. 2D). Endo-lysosomal pathology is a prominent feature of AD (10), and such abnormalities in the App-KI mice were also diminished by App-676-gRNA injections (Fig. 2E – quantified in Fig. 2F). Recent studies have revealed an important role of neuroinflammation in the progression of AD pathology. Neuroinflammatory markers are increased in AD, and human genetics have uncovered several AD-risk genes that have functions in innate immunity (41). Innate immune responses in the brain are primarily mediated by glia, and an increase in neuroinflammation has also been documented in AppNL-G-F mice (39). Astrocytosis and microglial activation is also attenuated in the App-KI mice after a single App-676-gRNA injection, and these attenuating effects were also additive over time (Figs. 2G–H and Supp. Fig. 2G).
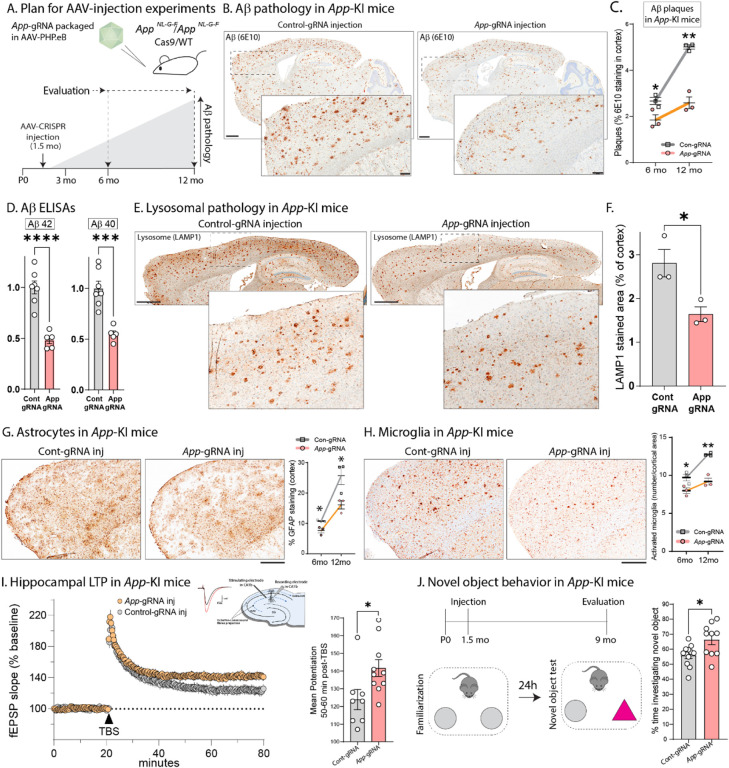
Figure 2: AAV-driven CRISPR editing of the App C-terminus ameliorates multiple deficits in App-KI mice.
A) Schematic showing timepoints of AAV-App-gRNA injections and evaluation of neuropathology in App-KI/Cas9-KI mice.
B) Representative sections showing Aβ pathology in one-year old mice, following protocol in A). Note decreased Aβ plaques in App-gRNA injected animals. Scale bars: main = 1 mm, zoomed inset = 200 μm.
C) Quantification of Aβ progression. While Aβ deposits increase over time in control-gRNA injected mouse brains as expected, there is a marked attenuation in App-gRNA injected animals (N=3 mice per condition; *p<0.05, ** p<0.01).
D) Aβ ELISAs from GuHCl soluble brain fractions in one-year old mice. Note marked Aβ attenuation in App-gRNA injected mice (N=5–7 animals for each condition, ***p<0.001, ****p<0.0001).
E) Representative sections showing lysosomal pathology in one-year old App-KI mice. Note attenuated lysosomal staining in App-gRNA injected animals, quantified in F) (N=3 animals for each condition, *p<0.05). Scale bars = 1 mm.
G, H) Representative sections showing staining of astrocytes G) and microglia H) in App-KI mice injected with control-gRNA or App-gRNA, with quantification on right. Note sustained attenuation of glial pathology after a single App-gRNA injection (N=3 mice per condition; *p<0.05, ** p<0.01; also see Supp. Fig. 2G). Scale bars = 500 μm.
I) LTP recordings from acute hippocampal slices of App-KI mice (10-month-old) injected with control or App-gRNA at 1.5 months; data quantified on right. Note augmentation in LTP after App-gRNA injections. (N=8–10 slices per condition, *p<0.05 – also see Supp. Fig. 3).
J) Novel object test behavior in 9-month-old old mice (schematic on top). Note that the App-gRNA injected mice spend more time exploring the novel object, compared to controls. (N=10–11 animals per condition, *p<0.05). All quantitative data presented as mean +/− SEM.
Previous studies have shown that long-term potentiation (LTP) is impaired in acute hippocampal slices from older AppNL-G-F mice (42). LTP reflects the persistent strengthening of synapses in response to recent activity and is thought to be the cellular mechanism underlying learning and memory. Thus, LTP impairments in the AppNL-G-F mice are especially relevant in the context of a memory disorder such as AD. We compared LTP in acute hippocampal slices from 10-month-old AppNL-G-F/Cas9 mice that were injected with AAV-App-676-gRNA (or control-gRNA) at 1.5 months. As shown in Figure 2I, there was an augmentation of LTP in the App-676-gRNA injected group compared to the control-gRNA injected group (editing in slices was verified post-hoc, see Supp. Fig. 3). Impairments in the novel-object recognition test have also been reported in older AppNL-G-F mice (43). Novel-object recognition is a measure of innate exploratory behavior in the absence of external cues, and based on the logic that cognitively intact mice show a natural preference for new objects in their environment. Previous studies have shown that older AppNL-G-F mice have deficits in this test, showing a decreased preference for novel objects (43). Accordingly, we performed this test in 9-month-old AppNL-G-F/Cas9 mice that were injected with AAVs carrying App-676-gRNA (or control-gRNA) at 1.5 months. As shown in Figure 2J, the App-676-gRNA injected mice spent significantly more time exploring the novel object compared to controls, suggesting rescue of cognitive function upon App editing. Comparable results were obtained with another gRNA that edited App slightly upstream of the 676-position, but still within the last exon (App-659-gRNA, see Supp. Fig. 4).
Germline editing of the App last-exon in App-KI and WT mice
To evaluate the safety of our gene-editing approach, we generated mice where the last-exon of App was edited in the germline of WT mice. We used the App-659-gRNA in these experiments, reasoning that since these germline-edited mice lacked the C-terminus of APP from birth – with the 659-gRNA editing a larger segment of the APP C-terminus – examining older mice from this group would be a stringent test of safety. Accordingly, we injected App-659-gRNA/Cas9 ribonucleoprotein complexes into zygotes from WT mice (and also App-KI mice, see next), and selected two germline-edited strains with indels within the last exon, expected to effectively delete ~ 35 amino acids from the APP C-terminus throughout the brain (Supp. Fig. 5A–B - named “Δ5” and “insert-T” based on generated indels. Western blots of brain homogenates were consistent with a selective C-terminal truncation of APP in genomically edited mice (Supp. Fig. 5C). Importantly, our App-editing strategy had no effect on the App homologues APLP1/2 due gRNA target-sequence mismatch (Supp. Fig. 5D). Note that APLP1/2 are known to compensate for loss of App function in mice, and also contain YENPTY domains [reviewed in (44)]. Interestingly, WT App editing also led to a decrease in endogenous APP β-cleavage and an increase in α-cleavage (Supp. Fig.5E), suggesting that our CRISPR-driven manipulation of β/α cleavage can favorably alter cleavage pattern of WT APP in vivo – relevant in the context of sporadic AD without APP mutations. For further details on genotyping strategies, see Supp. Fig. 5F–H, Supp. Table 2, and Methods. Examination of brains from WT germline-edited mice did not show any gross deficits in neurons, synapses, astrocytes and microglia, when compared to their WT counterparts (Supp. Fig. 6A–C), and both groups performed similarly in memory tests (Supp. Fig. 6D).
Next we tested our germline-editing approach in App-KI mice, generating App-KI-Δ5/InsT mice that lacked the APP C-terminus from birth (Fig. 3A). As expected, there was a marked suppression of APP-β-cleavage and augmentation of APP-α-cleavage in germline-edited App-KI mice (Fig. 3B). In all experiments so far, we used homozygous (AppNL-G-F/AppNL-G-F) mice, mainly because the relatively faster development of pathology in these animals is more amenable to experimentation. To test our editing strategy in a more plausible clinical scenario, we performed germline App editing in heterozygous (AppNL-G-F/WT) mice and evaluated Aβ pathology in older (10.5-month) animals. Strikingly, Aβ plaques were almost absent in the genomically deleted AppNL-G-F/WT mouse strains (Fig. 3C and Supp. Fig. 6E). Since almost all familial AD patients are heterozygous, and the magnitude of neuropathology is generally comparable in familial and sporadic AD brains, these data suggest that APP C-terminus editing at the earliest possible stages in AD may potentially have profound therapeutic effects.
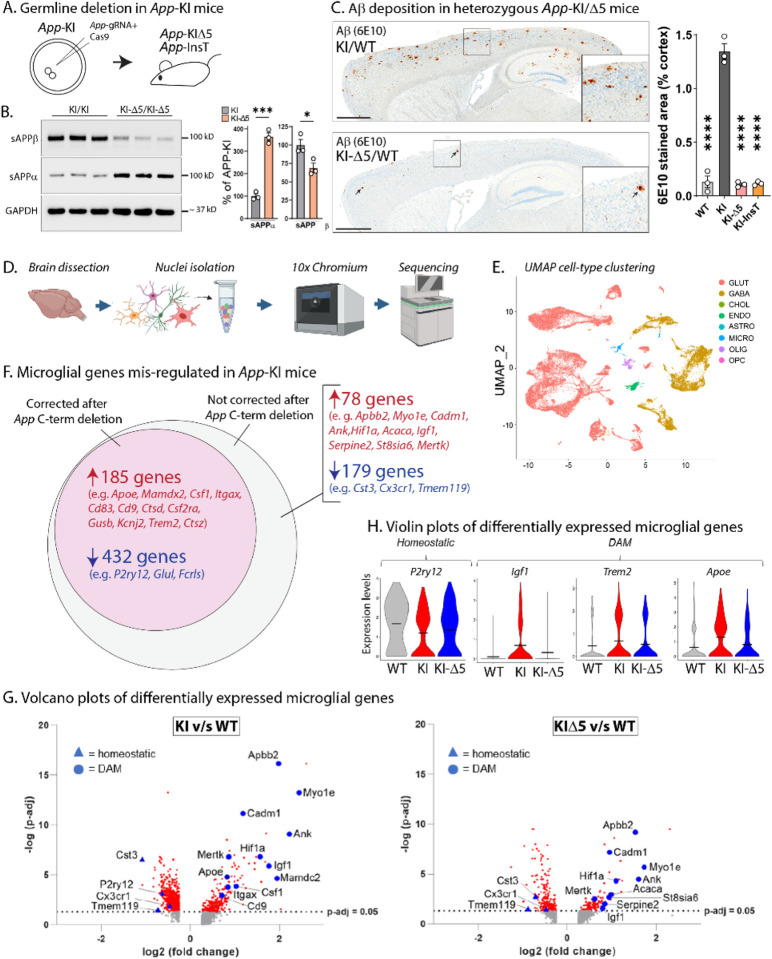
Figure 3: Germline App C-terminus editing in App-KI and WT mice.
A) Strategy for generating App-KIΔ5/InsT mice by germline CRISPR-deletion. Zygotes from App-KI mice were injected with App-gRNA/Cas9 ribonucleoprotein complexes targeting the App C-terminus, and resulting founders were screened and outbred to produce stable heterozygous and homozygous lines (also see Supp. Figs. 5–6).
B) Reversal of APP β/α cleavage pattern in germline-edited App-KI mice, quantified on right.
C) Representative sections of Aβ staining in germline-edited heterozygous App-KI mice. Note that Aβ plaques are almost absent in the edited mice (bottom panel, small arrowheads point to rare plaques, also see Supp. Fig. 6E), quantified on right. Scale bar = 1mm, data shown as mean +/− SEM. N = 3/condition. * p < 0.05, ***p < 0.001, **** p < 0.0001 and ns = non-significant.
D) Workflow for sNuc-Seq from App-KI and App-KIΔ5 mouse brains (N = 2 brains per genotype). Transcriptomic profiles of isolated nuclei were generated with 10x chromium sNucSeq, followed by deep sequencing of cDNA libraries.
E) UMAP (Uniform Manifold Approximation and Projection) visualization (right) shows efficient sorting into different cell-types (also see Supp. Fig. 6A).
F) Venn-diagrams showing mis-regulated microglial genes in App-KI mice (p-adj > 0.05) that were either corrected after gene-editing (pink), or not (grey). Genes in red were upregulated, and genes in blue were downregulated; some examples of DAM/homeostatic genes are listed (see Supp. Tables 3–5 for full lists). Note that mis-expression of ~ 71% of microglial genes was corrected after App C-terminus editing.
G) Volcano plots of differentially expressed microglial genes in App-KI and App-KIΔ5 mouse brains (compared to WT brains) showing the – log10 (adjusted p-value) and the log2 fold change (FC) in expression for all genes with log2FC > 0.25. Note that several homeostatic (blue triangles) or DAM (blue circles) microglial genes are up-regulated or down-regulated in App-KI brains, compared to WT brains (left graph). Similar analyses on App-KIΔ5 mouse brains shows that mis-regulation of many of these microglial genes is substantially attenuated in the App-KIΔ5 setting, suggesting restoration of homeostasis upon gene-editing.
H) Violin plots showing the distribution of expression levels for key DAM and homeostatic genes. Wider sections of the violin plot represent higher probability of expression at a given level, thinner sections indicate a lower probability, and short horizontal lines indicate mean values. Note mis-regulation in App-KI, and shift towards WT in App-KIΔ5 mouse brains.
App C-terminus editing rescues microglial transcriptomic changes in App-KI mice
Neuroinflammation and microglial activation have emerged as important players in the pathologic progression of AD (41). Besides human genetics, the relevance of microglia in AD pathology has been further bolstered by studies using single-cell/nuclei sequencing that can overcome limitations posed by cell-type heterogeneity and diversity of disease phenotypes. Transcriptomic studies of microglia in AD mouse models show gradual transitions from a homeostatic to a disease-linked state, revealing a set of disease associated microglial (DAM) genes that are altered with disease progression (45), which are also seen in the AppNL-G-F knockin model (46). Accordingly, we asked if our App editing strategy also ameliorated some of these disease-linked alterations in microglia. To ensure a homogenous background of edited cells, we used the homozygous App-KI germline-deletion (App-KI-Δ5) mice for these experiments. As expected, glial activation was significantly attenuated in 10-month-old App-KI mice with germline-deletion of the C-terminus (Supp. Fig. 7A–B), along with Aβ plaques and associated pathologies. The experimental flow of the sNuc-Seq experiments is shown in Figure 3D. Briefly, ~ 19,000–23,000 high-quality nuclei from 10-month-old mouse brains were analyzed to compare cellular-molecular maps between App-KI and App-KI-Δ5 animals. The nuclei were partitioned into multiple cell-types following standard protocols (47) (Fig. 3E, also see Supp. Fig. 7C), and sNuc-Seq data from App-KI-Δ5/App-KI mouse brains were compared to brains from WT animals.
For data-analyses, our two-step goal was to first identify glial genes that were differentially altered in the App-KI brains when compared to WT brains, and then ask if these alterations were abrogated in the App-KI-Δ5 animals. Additionally, we also compared our sNuc-Seq data to published RNA-seq databases from AD mouse models(46). As expected, several microglial genes were altered in App-KI mice, but interestingly, ~ 71% of these were corrected in the App-KI-Δ5 animals (Fig. 3F, see Supp. Tables 3–5 for full lists of genes). Figure 3G shows volcano plots of differentially expressed microglial genes in KI v/s WT and KI-Δ5 v/s WT animals. Note that several homeostatic and DAM genes are mis-regulated in the App-KI mice (Fig. 3G, left panel), but many of these are normalized in the App-KI-Δ5 animals (Fig. 3G, right panel). Violin plots in Figure 3H show the individual expression-patterns of a few of the key homeostatic and DAM genes thought to be associated with AD.
Amelioration Alzheimer’s pathology by a single AAV-vector carrying all CRISPR-components
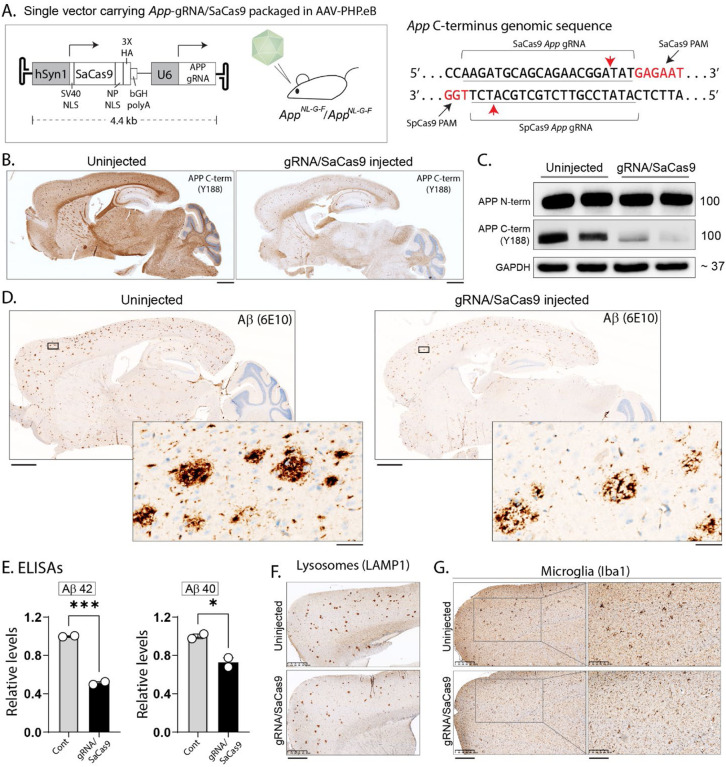
Finally, we designed a single AAV-vector carrying all CRISPR-components to edit the C-terminus of the mouse App gene. The overall design of the vector is shown in Figure 4A. Note that packaging these components with the smaller human synapsin-1 (hSyn1) promoter, and the compact SaCas9 editor (31) allowed us to fit all required elements into one vector. A HA-tag was added to identify transduced neurons, and we chose a PAM-site/genomic-target that was close to the App-676-gRNA used in our previous AAV-injection experiments. The single-vector AAV-CRISPR payload was intravenously injected into homozygous App-KI mice at ~1.5 months, and brains were evaluated at 5–6 months for transduction efficiency and editing of the App C-terminus (using Y188 immunoreactivity as a surrogate marker, see Fig. 1G). To simulate a real-world clinical scenario in these pilot experiments, we compared the AAV-App-gRNA/SaCas9 cohort with age-matched un-injected mice. As shown in Fig. 4B–C (also see Supp. Fig. 7D), there was widespread brain-transduction, and APP-Y188 (C-terminus) immunoreactivity was attenuated in App-gRNA/SaCas9 injected brains, with relative preservation of the N-terminus. Representative Aβ stains (6E10 antibody) are shown in Figure 4D. Note reduced Aβ deposition, and less compacted plaques in mice injected with AAV-gRNA/SaCas9. ELISAs from the brains of these mice confirm the reduction of insoluble Aβ (Fig. 4E). Lysosomal and microglial pathology was also attenuated in the App-KI mice after AAV-gRNA/SaCas9 injections (Fig. 4F–G), though there was an increase in reactive astrocytes (Supp. Fig. 7E), likely due to AAV-driven inflammatory reaction. All statistical data from main figures is shown in Supplementary Table 6.
Figure 4: A single AAV carrying all CRISPR components attenuates neuropathology in App-KI mice.
A) Schematic showing the construct that was packaged between the AAV-ITRs, consisting of the smaller SaCas9 driven by the human synapsin-1 promoter, App-gRNA driven by the U6 promoter, and a HA-tag to identify transduced neurons. App-KI mice (homozygous) were intravenously injected with the single AAV carrying all CRISPR-components at ~ 1.5 months, and brains were evaluated at 5–6 months. Note that the position of the SaCas9 PAM/cut-site (red arrowheads) in the App C-terminus are close to the SpCas9 cut-site (below).
B-C) Representative sections and western blots from un-injected and AAV-injected brains [from A)] stained with APP antibodies indicate widespread editing (also see Fig. 1F–H and Supp. Fig. 7D). Scale bars = 1 mm.
D) Representative Aβ stained sections of control and App-gRNA/SaCas9 injected mouse brains. Note attenuated Aβ staining in the gRNA-injected brain. Scale bars: main = 1 mm, zoomed inset = 50 μm.
E) Aβ ELISAs from brains (GuHCl soluble fractions) show marked reduction in gRNA/Cas9 injected brains, compared to controls (N=2 per condition, data presented as mean +/− SEM, * p < 0.05, ***p < 0.001).
F-G) Representative sections showing attenuation of lysosomal and microglial pathology App-gRNA/SaCas9 injected mouse brains.
DISCUSSION
Here we demonstrate a CRISPR-Cas9 based therapeutic strategy to effectively delete a pentapeptide endocytic motif at the C-terminus of APP and favorably alter the balance of APP cleavage – attenuating toxic APP-β-cleavage, while upregulating neuroprotective APP-α-cleavage. This manipulation rescues neuropathologic, electrophysiologic, behavioral, and transcriptomic changes in an AD knockin mouse model, and a single injection of AAVs carrying CRISPR-components rescued deficits for many months. Early germline editing of the App C-terminus in a more AD-relevant heterozygous App-KI background essentially eliminated Aβ pathology, suggesting that when combined with early intervention, this approach may have substantial therapeutic effects. Similar germline editing in WT mice had no detectable anatomic or behavioral consequences in vivo, attesting to the safety of this approach. Collectively, our studies offer a path for developing a one-time disease-modifying therapeutic for AD.
Incidental features in the APP gene that favor our editing strategy
Indels generated by CRISPR-Cas mediated NHEJ usually lead to premature stop codons in the DNA (33). Typically, eukaryotic cells see these synthetic termination signals – proximal to the native stop codon – as abnormal, and consequently, innate repair mechanisms are triggered to eliminate the mRNA via nonsense-mediated decay, leading to a loss of the protein. However, recognition of a stop codon as ‘abnormal’ depends on the position of the termination codon relative to the last exon-exon junction, and extensive previous work has shown that premature stop codons within the last exon do not lead to decay of the transcript [reviewed in (34)]. Briefly, transcriptional rules dictate that if a premature termination signal lies within the last exon – or within ~50 nucleotides upstream of the last exon-exon junction – innate mechanisms do not recognize this transcript as abnormal, and instead, translation is terminated at the premature stop signal, which is expected to generate a truncated protein. Fortunately, about half of the APP C-terminus, including the YENPTY-domain and flanking regions, is encoded by the last exon (exon 18), allowing us to manipulate APP in a way that preserves the transmembrane-domain and N-terminus, permitting physiologic α-cleavage and generating soluble APPα fragments. The precision of CRISPR-editing also allows normal production of APP-like proteins (APLP1 and APLP2, see Supp. Fig. 5D and (29)). APLP1/2 are encoded by separate genes, and also have YENPTY-domains (44), and it is expected that protein-protein interactions mediated by this domain will continue to occur post-editing. Extensive in-vivo studies in mice have also established that Aplp1/2 can compensate for App function (44), and the lack of any detectable abnormality in WT App C-terminus edited mice (Supp. Figs. 5–6) may also be attributable to this fortuitous scenario.
A one-time gene editing therapeutic for all forms of Alzheimer’s disease
In principle, our upstream manipulation of APP-cleavage is expected to be therapeutic for sporadic AD, as well as familial AD with APP and presenilin (PSEN) mutations. The etiology of sporadic AD is unclear, and likely complex. While studies suggest that there is deficient Aβ clearance in AD patients (48), human genetics also implicate upstream genes in the amyloid pathway like APP, PSEN1/2 and ADAM10 (15, 49), and BACE1 activity is also reportedly higher in sporadic AD brains (50) – incriminating Aβ overproduction. Regardless, our upstream manipulation to tilt APP cleavage towards a more physiologic state is expected to be effective for sporadic AD, though, in the absence of a reliable model for sporadic AD, this cannot be directly tested. Notably, a shift in APP processing towards the non-amyloidogenic pathway is also thought to be the basis for neuroprotection in the APP A673T Icelandic mutation that is seen in sporadic AD population (22, 25), further suggesting that a similar approach could work in sporadic AD. Studies have also shown that secreted α-cleavage products sAPPα – but not sAPPβ – can rescue hippocampal LTP deficits in APP knockout mice (51), implying that a sustained increase in sAPPα may be effective in AD.
Our previous studies have demonstrated the efficacy of our approach in isogenic human iPSC-lines carrying the APP-London mutation (29). Taken together with the data shown here in the App-KI mice (efficacy in a mouse model with humanized Aβ-domain containing three familial AD APP-mutations), the collective evidence strongly suggests that our approach would be effective for familial AD with APP mutations. Regarding familial AD with PSEN mutations, it is thought that the increased Aβ aggregation is due to the relative abundance of longer Aβ peptides that are more aggregation-prone (52). More recent studies suggest that PSEN mutations may lead to a loss of function in processing APP-β-CTF (C-terminal fragments) (53). Regardless, since our approach attenuates APP-β-CTF fragments that are required for subsequent cleavage by the presenilin/γ-secretase complex, it is expected that our strategy would be therapeutic in this scenario. However, there are ~ 300 presenilin mutations in familial AD, and further focused work is needed to test therapeutic efficacy and safety of APP C-terminus editing in this setting. Widespread delivery into larger primate brains remains a challenge for all gene-therapy approaches targeting the brain. Nevertheless, several ongoing clinical trials are relying on intracisternal injection of AAVs into the cerebrospinal fluid for broad transduction (NCT04747431, NCT04408625, NCT03634007, NCT04127578), and emerging technologies are expected to offer better solutions in the near future (1).
In summary, our studies demonstrate the potential of a one-time CRISPR-Cas9 based gene-editing treatment for AD. Though further work is needed for human translation, our vision is to develop a universal ‘one-and-done’ gene therapy for AD. Potentially, our gene-editing therapeutic could also be used as a combination-therapy in elderly patients; delivered after initial doses of amyloid-attenuating agents (such as lecanemab) and abrogating the need for frequent intravenous injections throughout life that substantially increases risks of intracerebral hemorrhage.
Methods
List of antibodies used in this study
| Antibody | Usage | Source | Catalog # |
|---|---|---|---|
| APP C-terminus (Y188) | WB/IHC/IF* | Abcam | ab32136 |
| APP N-terminus (22C11) | IHC | Thermo Fisher | 14–9749-82 |
| HA | IHC | Cell signaling | #3724 |
| APP N-terminus | WB | LS Bio | LS-B2970 |
| ADAM10 | WB | Abcam | ab124695 |
| GAPDH | WB | cell signaling | 2118S |
| a-tubulin | WB | cell signaling | 3873S |
| sAPPα (2B3) | WB (AppNL-G-F background) | IBL America | 11088 |
| sAPPβ-Swe | WB (AppNL-G-F backgorund) | IBL America | 10321 |
| sAPPα (M3.2) | WB (WT background) | Biolegend | 805701 |
| sAPPβ | WB (WT background) | Biolegend | 813401 |
| APLP1 | WB | Abcam | ab291070 |
| APLP2 | WB | GeneTex | GTX33014 |
| BACE1 | WB | Abcam | ab183612 |
| LAMP1 | IHC | cell signaling | 46483 |
| GFAP | IHC | Dako | Z0334 |
| Iba1 | IHC | Wako | 019–19741 |
| NeuN | IHC/IF | Abcam | ab177487 |
| Synaptophysin | IHC | Invitrogen | MAI-213 |
| GFP | IF | Abcam | ab183734 |
| 6E10 | IHC | Biolegend | 803001 |
| PSD 95 | WB | Abcam | ab18258 |
| anti-mouse secondary | WB | Thermo Fisher | ab205719 |
| anti-rabbit secondary | WB | Thermo Fisher | ab205718 |
| anti-mouse secondary | IHC/IF | Roche | Omni (05266548001); Ultra (05269717001) |
| anti-rabbitt secondary | IHC/IF | Roche | Omni (05269652001) |
WB = western blot, IHC = immunohistochemistry, IF = immunofluorescence
Mouse breeding and CRISPR deletion of App C-terminus in vivo
General procedures
All animal procedures were done according to guidelines set by the University of California, San Diego. Mice were euthanized by CO2 asphyxiation and the brains were rapidly dissected out. Tissue was snap-frozen on dry ice (for ELISAs/biochemistry) or drop-fixed in 4% paraformaldehyde (for histology).
Generation of AppNL-G-F/Cas9 knockin mice
AppNL-G-F (App-KI) mice harboring a humanized Aβ domain and three familial AD mutations – Swedish, Arctic and Iberian(36) – were obtained from RIKEN Brain Science Institute (Japan). KI mice were then crossed with CAG-driven SpCas9 knockin mice from Jackson labs (stock # 028239(54)) to produce App-KI/SpCas9 KI mice. Mice used for AAV-driven SpCas9/App-gRNA injection experiments were homozygous for the App knock-in and hemizygous for SpCas9 knockin. Mice used for SaCas9/App-gRNA (single-vector) experiments were only homozygous for the App knock-in allele. For AAV-injection experiments, littermates (both males and females) were randomly selected for transduction with AAVs carrying scrambled (non-targeting) control-gRNA or APP-gRNA. For genotyping of the App-KI/SpCas9 KI mice, genomic DNA was isolated from ear clips using QuickExtract buffer (Lucigen) following manufacturer’s instructions. Knock-in alleles were then PCR amplified from genomic DNA templates using RedExtract PCR mix (Sigma), and amplicons were visualized on agarose gels with SybrSafe (Thermo Fisher).
Generation of App genomic-deletion mice
These mice were generated at the University of Wisconsin-Madison Genome Editing and Animal Models Core. One-cell C57BL/6J or App-KI fertilized embryos were microinjected with 50 ng/μl App C-terminus targeting guide-RNA and 40 ng/μl Cas9 protein (PNA Bio, Newbury Park, California). Injected embryos were transplanted into pseudo-pregnant B6D2 F1 females. Pups were sequenced at weaning to screen for gene-edits. Two APP C-terminus indels, a T base pair insertion (insT) and a 5bp deletion (Δ5), were selected during screening of founders and selected for colony propagation. Founders were crossed with C57BL/6J mates and F1 mice were sequenced. F1 WT-Δ5/InsT or App-KI-Δ5/InsT mice were then crossed with C57BL/6J mice for an additional 2 generations. Finally, F3 generation mice were crossed to produce homozygous WT-Δ5/InsT or App-KI-Δ5/InsT mice. For genotyping of Δ5 mice, genomic DNA was isolated from ear clips using QuickExtract buffer (Lucigen) following manufacturer’s instructions. Genomic region containing the CRISPR-edit was PCR-amplified using OneTaq polymerases in Standard Buffer (New England Biolabs). After PCR, 1μl of AciI restriction enzyme was added to 50μl of PCR reaction sample and samples were incubated at 37°C for 1 hr. DNA bands were then visualized agarose gels with SybrSafe (Thermo Fisher).
AAV vectors – cloning, production, and intravenous injections
Cloning, production and purification of AAV vectors
For experiments in App-KI/Cas9 mice, scrambled control or App-targeting gRNAs were cloned into pAAV9-U6sgRNA(SapI)_hSyn-GFP-KASH-bGH vectors at Sap1 site as previously described(29). Experiments in APP-KI mice used pX601-AAV-CMV::NLS-SaCas9-NLS-3xHA-bGHpA;U6::BsaI-sgRNA (Addgene #61591), in which the CMV-promoter was replaced with the human Synapsin1 promoter using Gibson cloning. This AAV-SaCas9 backbone system was used to clone sgRNAs targeting the App C-terminus. Briefly, the vector was digested using the BsaI restriction enzyme, and a pair of annealed oligos was cloned scarlessly into the vector upstream of the sgRNA scaffold. Oligos were designed based on the target site sequence (21–22bp), with NGRRT or NGRRN PAM sequences on the 3’ end. Potential off-target effects were assessed using the online tool CRISPROR (http://crispor.tefor.net/), and a guide with minimal off-target effects was selected for experiments. All CRISPR sgRNA sequences were analyzed with the National Center for Biotechnology Information’s (NCBI) Basic Local Alignment Search Tool (BLAST) to ensure specificity. All AAVs were packaged at the UCSD viral vector core. Helper virus-free AAV.PHPeB vectors were produced by transient transfection of HEK293T cells maintained in Dulbecco’s modified Eagle’s medium (DMEM) with 10% fetal calf serum. HEK293T cells in 150 mm dishes were co-transfected (using Polyethylenimine) with the vector plasmid pRep2/CapPHPeB (obtained from Addgene(35), and pAd-Helper plasmid(55). Cell lysates prepared 72 hrs after transfection were treated with benzonase, and viruses were pelleted through 30% sucrose-cushion ultracentrifugation. Pellets were resuspended, and viruses were further purified through anion-exchange column chromatography (Q-Sepharose, GE Health Science)(56), followed by concentration through 30% sucrose-cushion ultracentrifugation. The final pellets were resuspended in a solution containing 10mM Tris-HCl, pH7.9, 1mM MgCl2, and 3% sucrose. Virus titers were measured by real-time Q-PCR to determine genome copy number of the vector preparations (gc/ml) as a measure of AAV particles with full genome content.
AAV injections in vivo
Mice (age: ~ 5–7-weeks) were anesthetized by isoflurane, and AAVs were injected retro-orbitally using an 8mm, 31G insulin syringes (BD Medical Systems) as described previously. (57) A dose of 3–10^11 VG/mouse was used for injections in App-KI/Cas9 mice. APP KI mice treated with SaCas9 vectors received a dose of 3×10^12 VG/mouse. Mice were maintained 4–10.5mo post-injection for behavioral testing and tissue collection.
Histology and related procedures
General procedures
Mouse hemibrains were quickly dissected, and immediately drop fixed in 4% paraformaldehyde. Tissue was then embedded into paraffin blocks, and 5μm thick sections were collected on Super Frost glass slides (Fisher Scientific). For immunohistochemistry and immunofluorescence, tissue sections were stained with antibodies to appropriate epitopes on a Ventana Discovery Ultra platform (Ventana Medical Systems, Tucson, AZ, USA). Antigen retrieval was performed using CC1 (Tris-EDTA based; pH 8.6) for 40 minutes at 95 °C. Primary antibodies were incubated on the sections for 32 minutes at 37 °C, and were detected using the OmniMap systems (Ventana). Antibody-presence was visualized used DAB as a chromagen followed by hematoxylin as a counterstain. Dual Y188/HA stain: In some cases, dual chromagenic for Y188 (abcam ab32136) was performed using anti-Y88 antibody and the OmniMap detection system horseradish-peroxidase (HRP)-coupled goat anti-rabbit (Ventana) followed by chromogenic labeling with Ventana Purple kit according to the manufacturer’s instructions. For dual immunohistochemistry, the antibodies were fully denatured, inactivated, and removed from the tissue by treatment in CC2 citrate-based, pH 6.5 (Ventana) for 20 min at 100 °C. Subsequently, the second antibody (anti-HA, Cell Signaling #3724) was applied and detected by the OmniMap system (Ventana). Chromogenic labeling for HA-expressing cells used the Ventana Green kit followed by hematoxylin counterstain. Two color immunofluorescence: Staining with appropriate antibodies was performed using the OmniMap detection system HRP-coupled secondary antibodies (Ventana) followed by immunofluorescence-labeling according to the manufacturer’s with tyramide signal amplification (TSA)-Alexa 488 or TSA-Alexa 594 tyramide-conjugated fluorophores (Invitrogen). To perform dual staining, the antibodies were fully denatured, inactivated, and removed from the tissue by treatment in CC2 citrate-based, pH 6.5 (Ventana) for 20 min at 100 °C. Subsequently, the second antibody was applied and detected by the OmniMap system (Ventana) using TSA-Alexa 488 or TSA-Alexa 594 tyramide-conjugated fluorophores. After immunofluorescence staining, sections were rinsed and coverslipped with Vectashield containing DAPI (Vectorlabs). Slides were imaged at the UCSD microscopy core using a VS200 (Olympus) or Nanozoomer (Hamamatsu) slide scanner.
Quantification of Aβ (6E10), LAMP1 and GFAP staining
After histological sections were scanned at 20x zoom using NanoZoomer slide scanner, the subsequent .ndpi files were opened in QuPath and converted to .tiff files using a pixel-to-micron ratio of 1:0.4527 to maintain optimal resolution. (58) These high-resolution files were then processed in Adobe Photoshop. After deleting the background, the polygonal lasso tool was used to select the anatomical area of interest (i.e. cortex), and images were further processed in MATLAB (version R201A), where the stain of interest was isolated using the “color thresholding” feature. After a threshold was generated for each stain, the same threshold was used to process all images within that group. Data generated from thresholding was processed through custom codes (available upon request) to identify % area stained. At least 6–8 tissue sections from each animal were analyzed per stain.
Quantification of activated microglia using the HALO module
Images of Iba-1 stained tissue were analyzed using the Microglial Activation module (Version 3.5.3577.214) on the HALO Image Analysis Platform (Indica Labs, Inc.). The Halo module parameters used for detecting activated microglia were: Min Microglia Cell Body Diameter = 0.46, Microglia Contrast Threshold = 0.258, Minimum Microglia Process Radius = 7.8, Microglia Max Fragmentation Length = 12.58, Activation Process Thickness: 3.31.
Quantification of editing efficiency (Y188/GFP dual immunofluorescence)
To determine the number of GFP-positive cells positive for APP C-terminus staining in cortices, tissue sections from one-year-old control and APP-gRNA AAV-injected mice were dual-IF stained with APP C-terminus (Y188) and GFP antibodies. For each tissue section, 7 cortical areas were selected and the number of GFP-positive cells positive for APP C-terminus staining were counted. Cells were counted in 4 different tissue sections/mouse.
Quantification of transduction efficiency (GFP/NeuN dual immunofluorescence)
To determine the number of NeuN positive cells transduced by AAVs, tissue sections from APP-gRNA AAV-treated mice were dual-IF stained with NeuN and GFP antibodies. GFP positivity was quantified in two dual-labeled sections from each mouse (N = 3 mice). A total of 8 cortical regions were analyzed per section. To quantify NeuN/GFP overlap, images were processed in MetaMorph (Molecular Devices). Images were thresholded to remove background pixels and regions of positive signal were designated. The ratio of NeuN positive regions overlapping with GFP regions was used to determine transduction efficiency.
Biochemical assays and evaluation
Western blots of brain tissue – procedures and analyses
Protein isolation:
Frozen tissue was homogenized in 0.15 % Triton X-100 with protease inhibitors, pH 7.4 (Pierce) in PBS. The resulting suspension was centrifuged at 21,000 RCF (relative centrifugal force) at 4 °C for 30 minutes, and the resultant supernatant was collected for western blotting.
Preparation of WB samples:
Protein concentrations of lysates were determined by DC protein assay (BioRad). After protein assays, samples for western-blots were made using 4X LDS buffer (Invitrogen) and reducing agent (Invitrogen) Samples were added to NuPage 4%−12% acrylamide gradient gels (Thermo Fisher) and gel electrophoresis performed using NuPage MES SDS running buffer with antioxidant solution.
Blotting:
Proteins were transferred to PVDF membranes (0.2um pore) using 20% methanol in Nupage transfer buffer (Thermo Fisher) with antioxidant (Thermo Fisher). Membranes blocked with 5% fat-free milk in PBS and Primary antibodies were diluted in 5% fat-free milk and added to membranes overnight at 4 °C. Membranes were then washed 3X 5minutes with 0.1% Tween 20 in PBS, and incubated in HRP-conjugated secondary antibodies diluted into 5% fat-free milk for 1 hour at room temperature. Membranes were washed again 3x for 5 minutes, incubated in chemiluminescent development solution (Thermo Fisher) and imaged with Chemidoc imager (BioRad).\Western blots were processed and quantified using Image Lab software (version 6.1, BioRad). For quantification, blot images (.scn files) were opened in Image Lab and sample bands delineated using image tools. Band volumes were then quantified by the software.
ELISAs of brain tissue – procedures and analyses
For ELISA, brains were quickly dissected and snap frozen on dry ice. Frozen brain tissue was then homogenized in 50 mM TBS with protease inhibitors (pH 7.4) and centrifuged at 200,000x g for 22 min at 4 °C. The resulting supernatant was removed, and the remaining pellet was washed once with 50 mM TBS, centrifuged (200,000x g, 22 min at 4 °C) and then solubilized in 6 M GuHCl (Sigma) with protease inhibitors. Samples were sonicated for 30 sec, vortexed at 1800 RPM for 5 minutes, and incubated at 25 °C for 60 min. Samples were centrifuged once more at 200,000x g for 22 min at 25 °C. Finally, the supernatant was collected for ELISA of GuHCl-soluble fractions. Human Aβ40 and Aβ42 were detected using ELISA kits, according to the manufacturer’s instructions (WAKO 296–64401 for Aβ42, WAKO 298–64601 for Aβ40). Briefly, diluted GuHCl-soluble lysates were added to ELISA plates overnight at 4 °C. Plates were then washed 5x, human Aβ40/42 detection antibodies were added into wells, and plates were incubated for 3 h at 4 °C. After further washing (5x), the stabilized chromogen was added, and incubated for another 30 min at room-temperature in the dark. Reaction was stopped, and absorbance at 450 nm was read using a luminescence microplate reader.
Procedures for hippocampal LTP recordings
Preparation of hippocampal slices and electrophysiology recordings
Male and female scrambled-gRNA (control) and APP-gRNA injected mice were anesthetized with isoflurane, decapitated, and the brains were processed for acute hippocampal recordings as described previously. (59) Briefly, brains were removed and submerged in ice-cold, oxygenated dissection medium containing (in mM): 124 mM NaCl, 3 mM KCl, 1.25 mM KH2PO4, 5 mM MgSO4, 0 mM CaCl2, 26 mM NaHCO3, and 10 mM glucose. Coronal hippocampal slices were prepared using a Leica vibrating tissue slicer, before being transferred to an interface recording containing preheated artificial cerebrospinal fluid (aCSF) composed of 124 mM NaCl, 3 mM KCl, 1.25 mM KH2PO4, 1.5 mM MgSO4, 2.5 mM CaCl2, 26 mM NaHCO3, and 10 mM glucose and maintained at 31 ± 10 °C. Slices were continuously perfused with this solution at a rate of 1.75–2 ml/min while the surfaces were exposed to warm, humidified 95 % O2/5% CO2. Recordings began after at least 2 h of incubation. Field excitatory postsynaptic potentials (fEPSPs) were recorded from CA1b stratum radiatum apical dendrites using a single glass pipette filled with 2 M NaCl (2–3 MΩ) in response to stimulation (twisted nichrome wire, 65 μm diameter) of Schaffer collateral-commissural projections in CA1c stratum radiatum. Pulses were administered at 0.05 Hz using a current that elicited a 50% maximal spike-free response. After establishing a 20-minute stable baseline, LTP was induced by delivering a single episode of 5 ‘theta’ bursts. Each burst consisted of four pulses at 100 Hz, and were separated by 200 ms (i.e., theta burst stimulation or TBS). The stimulation-intensity did not increase during TBS. To ensure that female mice were in the diestrus phase of the estrus cycle at the time of testing, uteri were removed, examined for vascularization and the presence of fluid (estrus phase), patted dry and weighed. (60) Mice with uteri in estrus weighing double (~200–250 mg) that of uteri obtained from mice in diestrus (70–120 mg) were omitted from the study. The fEPSP slope was measured at 10–90% fall of the slope and data in figures on LTP were normalized to the last 20 min of baseline. Electrophysiological measures were analyzed using a one-way ANOVA, and the level of significance was set at p ≤ 0.05.
Confirmation of gene-editing in LTP slices
Brain slices were immediately frozen after LTP experiments. Genomic DNA was extracted from frozen slices using QuickExtract buffer (Lucigen). The genomic region containing CRISPR-edits was PCR amplified from genomic DNA template using RedExtract PCR mix (Sigma). Amplicons were purified from PCR buffer using Monarch PCR cleanup kit (New England Biolabs) and submitted for Sanger sequencing (EtonBio).
Genomic analyses and evaluation
Next-generation AMP-sequencing
Library preparation and PCRs:
The AMP-seq (anchored multiplex PCR sequencing) library preparation protocol was followed as described. (61) Briefly, genomic DNA (gDNA) from scrambled-control/App-gRNA AAV-injected cortices were used for AMP-seq analysis. For each sample, 30 ng of gDNA was enzymatically fragmented using the KAPA HyperPlus Kit (Roche). Samples were purified at 0.8X using AMPure XP beads (Beckman Coulter), and end-repaired and A-tailed (KAPA HyperPlus Kit). Adapter primers containing amplicons for sequencing, as well as an 8-mer barcode and a random 8-mer for de-depulication during analysis, were synthesized by Integrated DNA Technologies (IDT; F: 5’-CAAGCAGAAGACGGCATACGAGATTATCCTCTNNWNNWNNGTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT*T-3’; R: 5’-/5Phos/AGATCGGAAGAG*G*T-3’), annealed, and ligated to fragmented DNA. The first round of PCR was performed using a primer complementary to the adapter (5’-CAAGCAGAAGACGGCATACGAGATTATCCTCT-3’) and a primer complementary to the genome (5’-TGCCTACGAGTACTGTGCTCC-3’). Following AMPure XP bead cleanup, a second round of PCR was performed using the same adapter primer and a nested primer complementary to an upstream region of the genome (5’- AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTAACCCGGATCTCTGTACCTG-3’) and also containing a common sequencing adapter to create an amplicon ready for next-generation sequencing. Library concentrations were quantified using the KAPA library quantification kit (Roche), bioanalyzed for quality control (Agilent 2100 Bioanalyzer), and sequenced on an Illumina MiniSeq using the MiniSeq Mid Output Kit with standard sequencing primers (Illumina #FC-420–1002; 150 cycles; paired-end reads). Data analysis: Reads were demultiplexed using Je suite(62) and then filtered such that at least 90% of bases had a Phred quality score ≥ 20 (99.0% accuracy). Reads were then analyzed and binned into editing event categories based on deviation from gRNA reference sequence ± 20 bp from expected Cas9-cleavage site. Reads that were 100% complementary to the reference genome were filtered out as unedited reads. Reads that contained fusion of reference sequence and adapter sequence within ± 20 bp from expected Cas9 cleavage site were also filtered out. We then removed any existing adapter using CUTADAPT and used STAR 2.7.0(63) (default parameters) to retain only on-target reads that aligned to a 1000 bp window surrounding the gRNA locus. Reads were then aligned to a custom AAV reference genome to identify host genomic reads that were fused to AAV genome within ± 20 bp from expected Cas9 cleavage site. Remaining reads were then re-aligned to the gRNA locus of interest to isolate reads containing insertions or deletions (indels). Substitutions were ignored as presumed sequencing errors, as previously reported. (64, 65) Remaining reads were then aligned to an mm10 reference genome (STAR; default parameters) to identify reads containing extra-chromosomal sequence ± 20 bp from the expected Cas9-cleavage site (translocations). All bins were manually spot-checked for accuracy and visualized through CRISPResso2(66) to confirm accuracy. CRISPResso2 parameters were as follows: CRISPResso -r1 <fastq_file.fastq> -a <amplicon_sequence> -g <gRNA_sequence> -q 20 -amas 50 -ignore_substitutions --quantification_window_size 20 --quantification_window_center −3 --exclude_bp_from_left 20 --exclude_bp_from_right 20 --plot_window_size 40. Data was then quantified as percentage of reads with each editing event category to the total number of reads with any category of editing event.
On- and off-target analysis of gene-editing
gDNA from was PCR amplified using primers listed in Table 1, PCR purified (Qiagen), and Sanger sequenced using Genewiz (Azenta Life Sciences). Data from Cas9- and control-gRNA treated .ab1 files were uploaded to the Synthego ICE CRISPR analysis tool(67) using default settings to estimate editing frequencies at on- and off-target sites for SpCas9 APP gRNA. Results were plotted as predicted insertion and deletion sizes, as a percentage to total sequences.
Behavioral assays and analyses
Novel Object Recognition test
Novel object recognition testing was performed as previously described. (68) Briefly, on day one, mice were placed into an enclosure with two identical objects. Thereafter, the mice were allowed to explore freely until they reached 20 seconds of total exploration-time of the objects, or 10 minutes total-time in the enclosure, whichever came first. 24 hours later, mice were placed in the same enclosure with one of the objects replaced with a novel object. These mice were free to explore the objects until 20-seconds of total exploration-time, or 10 minutes total-time in the enclosure was reached. The percentage of total exploration-time spent investigating the novel object was calculated.
Morris water maze test
A circular pool was filled with ~23 °C water and made opaque with non-toxic, water-based paint, and the pool was filled until water-level reached 2 cm above the target platform. Visual cues were placed at each quadrant. For each trial of the training phase, mice were placed into the pool and allowed to swim freely for up to 60 sec. Mice that did not find the platform before 60 sec were manually placed onto the platform. For all trials, mice remained on the platform for 20 sec before being removed from the pool. Each mouse completed four trials per day for 8 consecutive days. For probe trial, the target platform was removed on the 9th day of testing. Subsequently, mice were placed into the pool and allowed to swim freely for 60 seconds and the time spent in each quadrant was recorded. Trials were recorded with a webcam (Logitech) and videos analyzed with ANYMaze software.
Single-nucleus RNA-seq – procedures and analyses
Nuclear isolation:
Brains from 9-month-old WT, App-KI and App-KIΔ5 mice were quickly dissected and cut into hemispheres with a single sagittal midline cut, and the hemispheres were flash-frozen and stored at −80 °C. Nuclei were isolated as described in(69) with some modifications. Briefly, frozen hemispheres were transferred directly to 5 ml of chilled lysis buffer (10 mM Tris-HCl, 10 mM NaCl, 3 mM MgCl2, and 0.1% Nonidet™ P40 Substitute in Nuclease-Free Water) in a 7 ml glass Dounce homogenizer. Tissues were incubated in the lysis buffer on ice for 5 minutes followed by 3–5 passes with the A pestle, a second 10-minute incubation on ice, and 3–5 passes with the B pestle. Homogenates were then filtered through a 30mm MACs SmartStrainer and centrifuged at 500 rcf for 5 minutes at 4 °C to isolate the nuclear fraction. Nuclear fractions were washed twice with 10 ml of resuspension buffer (1X PBS with 1.0% BSA and 0.2 U/ml Protector RNAse inhibitor (Sigma-Aldrich)). Myelin debris was removed using magnetic separation with Miltenyi Myelin Removal Beads, LS columns and centrifugation. Nuclear pellets were then resuspended in resuspension buffer at a concentration of 1000 nuclei/ml.
Single cell RNA sequencing:
Single-nucleus RNA libraries were generated by the UW-Madison Biotechnology Center Gene Expression Center using the Chromium Next GEM Single Cell 3′ Library (10X Chromium). These libraries were subjected to paired end sequencing on the NovaSeq 6000 system (Illumina). Demultiplexed FASTQ files were aligned to the GRCm39 Mus musculus reference genome using Cell Ranger 6.1.2 with the include-introns flag selected. Following alignment, default quality control settings in Cell Ranger were used to filter reads and sort them into cell-associated matrixes. A second round of quality control excluding nuclei with >5% mitochondrial genes, > 50,000 unique molecular identifiers, < 100 or > 7500 genes, as well as all other downstream analyses were performed in Seurat version 4.1.1 (R Studio version 4.2.0).
Cell-type identification by dimensionality reduction:
Data sets from each sample were integrated following the workflow described in the Seurat guided analysis. Filtered matrices were log-normalized and highly variable features were identified for each sample using the FindVariableFeatures function with default parameters. The FindIntegrationAnchors function was then used to identify variable features conserved across all datasets. These anchors were used to integrate the datasets using the IntegrateData function. The integrated matrix was then scaled and linear dimensional reduction was performed using the RunPCA function. An elbow plot was used to visualize the percentage of variance explained by each principal component and opted to use the first 20 principal components for graph-based clustering. Next, UMAP and K-nearest neighbor clustering was performed using the function RunUMAP with the parameter dims = 20 and the functions FindNeighbors and FindClusters with the parameter resolution = 1. Cell-type identities were assigned to each cluster based on expression of known celltype markers.
Microglia differential-expression analysis:
Transcriptome profiles of microglia from App-KI and App-KIΔ5 mice were compared to WT microglial transcriptomes by the Wilcoxon rank-sum test using the FindMarkers function with default parameters. P-values were adjusted using the Benjamini-Hochberg procedure. The level of statistical significance was set at an adjusted p- value < 0.05 and |log2FC| > 0.25.
Supplementary Material
Acknowledgements:
This work was supported by grants to SR from the NINDS (R01AG048218, R21AG052404, UF1NS134063), the Epstein Foundation, and the Farmer Family Foundation. Brent Aulston was supported by a postdoctoral fellowship from the Alzheimer’s Association. The work was also supported by the following grants: R35GM119644 (KS), NHGRI T32HG002760 (KG), NINDS R01NS109304 (MJZ), NIA AG076835 (MAW), GM134865 and NS124165 (AA), and a grant to the UCSD microscopy core (NINDS P30NS047101). We thank Kathy Krentz and Dustin Rubinstein at the University of Wisconsin Genome Editing and Animal Model core for use of their facilities and services; and Sushmita Roy at the Wisconsin Institute for Discovery for the use of electronic resources. We also thank Atsushi Miyanohara from the UCSD Vector Development Core, Rohan Sharma (UCSD) for help with image analysis, and Brian Head (VA San Diego Health care system/UCSD) for help obtaining mice.
Footnotes
Competing interest declaration:
S.R. is the scientific co-founder and owns equity in CRISPRAlz.
REFERENCES
- 1.Sun J., Roy S., Gene-based therapies for neurodegenerative diseases. Nat Neurosci 24, 297–311 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frangoul H. et al. , CRISPR-Cas9 Gene Editing for Sickle Cell Disease and beta-Thalassemia. N Engl J Med 384, 252–260 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Gillmore J. D. et al. , CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N Engl J Med 385, 493–502 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Wong C., UK first to approve CRISPR treatment for diseases: what you need to know. Nature 623, 676–677 (2023). [DOI] [PubMed] [Google Scholar]
- 5.Cummings J. et al. , Alzheimer’s disease drug development pipeline: 2023. Alzheimers Dement (N Y) 9, e12385 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Dyck C. H. et al. , Lecanemab in Early Alzheimer’s Disease. N Engl J Med 388, 9–21 (2023). [DOI] [PubMed] [Google Scholar]
- 7.Honig L. S. et al. , ARIA in patients treated with lecanemab (BAN2401) in a phase 2 study in early Alzheimer’s disease. Alzheimers Dement (N Y) 9, e12377 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwart D. et al. , A Large Panel of Isogenic APP and PSEN1 Mutant Human iPSC Neurons Reveals Shared Endosomal Abnormalities Mediated by APP beta-CTFs, Not Abeta. Neuron 104, 1022 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Nixon R. A., Amyloid precursor protein and endosomal-lysosomal dysfunction in Alzheimer’s disease: inseparable partners in a multifactorial disease. FASEB J 31, 2729–2743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Small S. A., Simoes-Spassov S., Mayeux R., Petsko G. A., Endosomal Traffic Jams Represent a Pathogenic Hub and Therapeutic Target in Alzheimer’s Disease. Trends Neurosci 40, 592–602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keeler C. E., Gene therapy. J Hered 38, 294–298 (1947). [PubMed] [Google Scholar]
- 12.Gyorgy B. et al. , CRISPR/Cas9 Mediated Disruption of the Swedish APP Allele as a Therapeutic Approach for Early-Onset Alzheimer’s Disease. Mol Ther Nucleic Acids 11, 429–440 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Konstantinidis E. et al. , CRISPR-Cas9 treatment partially restores amyloid-beta 42/40 in human fibroblasts with the Alzheimer’s disease PSEN 1 M146L mutation. Mol Ther Nucleic Acids 28, 450–461 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duan Y. et al. , Brain-wide Cas9-mediated cleavage of a gene causing familial Alzheimer’s disease alleviates amyloid-related pathologies in mice. Nat Biomed Eng 6, 168–180 (2022). [DOI] [PubMed] [Google Scholar]
- 15.Holstege H. et al. , Exome sequencing identifies rare damaging variants in ATP8B4 and ABCA1 as risk factors for Alzheimer’s disease. Nat Genet 54, 1786–1794 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kemaladewi D. U. et al. , A mutation-independent approach for muscular dystrophy via upregulation of a modifier gene. Nature 572, 125–130 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Park H. et al. , In vivo neuronal gene editing via CRISPR-Cas9 amphiphilic nanocomplexes alleviates deficits in mouse models of Alzheimer’s disease. Nat Neurosci 22, 524–528 (2019). [DOI] [PubMed] [Google Scholar]
- 18.Zhang Y., Chen H., Li R., Sterling K., Song W., Amyloid beta-based therapy for Alzheimer’s disease: challenges, successes and future. Signal Transduct Target Ther 8, 248 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nagata K. et al. , Generation of App knock-in mice reveals deletion mutations protective against Alzheimer’s disease-like pathology. Nat Commun 9, 1800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hampel H. et al. , The Amyloid-beta Pathway in Alzheimer’s Disease. Mol Psychiatry 26, 5481–5503 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fortea J. et al. , Alzheimer’s disease associated with Down syndrome: a genetic form of dementia. Lancet Neurol 20, 930–942 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonsson T. et al. , A mutation in APP protects against Alzheimer’s disease and age-related cognitive decline. Nature 488, 96–99 (2012). [DOI] [PubMed] [Google Scholar]
- 23.Steubler V. et al. , Loss of all three APP family members during development impairs synaptic function and plasticity, disrupts learning, and causes an autism-like phenotype. EMBO J 40, e107471 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller U. C., Deller T., Korte M., Not just amyloid: physiological functions of the amyloid precursor protein family. Nat Rev Neurosci 18, 281–298 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Maloney J. A. et al. , Molecular mechanisms of Alzheimer disease protection by the A673T allele of amyloid precursor protein. J Biol Chem 289, 30990–31000 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perez R. G. et al. , Mutagenesis identifies new signals for beta-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Abeta42. J Biol Chem 274, 18851–18856 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Koo E. H., Squazzo S. L., Evidence that production and release of amyloid beta-protein involves the endocytic pathway. J Biol Chem 269, 17386–17389 (1994). [PubMed] [Google Scholar]
- 28.Das U. et al. , Visualizing APP and BACE-1 approximation in neurons yields insight into the amyloidogenic pathway. Nat Neurosci 19, 55–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sun J. et al. , CRISPR/Cas9 editing of APP C-terminus attenuates beta-cleavage and promotes alpha-cleavage. Nat Commun 10, 53 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ring S. et al. , The secreted beta-amyloid precursor protein ectodomain APPs alpha is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J Neurosci 27, 7817–7826 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maeder M. L. et al. , Development of a gene-editing approach to restore vision loss in Leber congenital amaurosis type 10. Nat Med 25, 229–233 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Rajendran L., Annaert W., Membrane trafficking pathways in Alzheimer’s disease. Traffic 13, 759–770 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Sander J. D., Joung J. K., CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol 32, 347–355 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kurosaki T., Maquat L. E., Nonsense-mediated mRNA decay in humans at a glance. J Cell Sci 129, 461–467 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chan K. Y. et al. , Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito T. et al. , Single App knock-in mouse models of Alzheimer’s disease. Nat Neurosci 17, 661–663 (2014). [DOI] [PubMed] [Google Scholar]
- 37.Kreis A. et al. , Overexpression of wild-type human amyloid precursor protein alters GABAergic transmission. Sci Rep 11, 17600 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Torroja L., Chu H., Kotovsky I., White K., Neuronal overexpression of APPL, the Drosophila homologue of the amyloid precursor protein (APP), disrupts axonal transport. Curr.Biol. 9, 489–492 (1999). [DOI] [PubMed] [Google Scholar]
- 39.Sasaguri H. et al. , Recent Advances in the Modeling of Alzheimer’s Disease. Front Neurosci 16, 807473 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Breton C., Clark P. M., Wang L., Greig J. A., Wilson J. M., ITR-Seq, a next-generation sequencing assay, identifies genome-wide DNA editing sites in vivo following adeno-associated viral vector-mediated genome editing. BMC Genomics 21, 239 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Griciuc A., Tanzi R. E., The role of innate immune genes in Alzheimer’s disease. Curr Opin Neurol 34, 228–236 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Latif-Hernandez A. et al. , The two faces of synaptic failure in App(NL-G-F) knock-in mice. Alzheimers Res Ther 12, 100 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mehla J. et al. , Age-dependent behavioral and biochemical characterization of single APP knock-in mouse (APP(NL-G-F/NL-G-F)) model of Alzheimer’s disease. Neurobiol Aging 75, 25–37 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Muller U. C., Zheng H., Physiological functions of APP family proteins. Cold Spring Harb Perspect Med 2, a006288 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keren-Shaul H. et al. , A Unique Microglia Type Associated with Restricting Development of Alzheimer’s Disease. Cell 169, 1276–1290 e1217 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Chen W. T. et al. , Spatial Transcriptomics and In Situ Sequencing to Study Alzheimer’s Disease. Cell 182, 976–991 e919 (2020). [DOI] [PubMed] [Google Scholar]
- 47.Lacar B. et al. , Nuclear RNA-seq of single neurons reveals molecular signatures of activation. Nat Commun 7, 11022 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mawuenyega K. G. et al. , Decreased clearance of CNS beta-amyloid in Alzheimer’s disease. Science 330, 1774 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cruchaga C. et al. , Rare variants in APP, PSEN1 and PSEN2 increase risk for AD in late-onset Alzheimer’s disease families. PLoS One 7, e31039 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rossner S., Sastre M., Bourne K., Lichtenthaler S. F., Transcriptional and translational regulation of BACE1 expression--implications for Alzheimer’s disease. Prog Neurobiol 79, 95–111 (2006). [DOI] [PubMed] [Google Scholar]
- 51.Hick M. et al. , Acute function of secreted amyloid precursor protein fragment APPsalpha in synaptic plasticity. Acta Neuropathol 129, 21–37 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Chavez-Gutierrez L. et al. , The mechanism of gamma-Secretase dysfunction in familial Alzheimer disease. EMBO J 31, 2261–2274 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Devkota S. et al. , Familial Alzheimer mutations stabilize synaptotoxic gamma-secretase-substrate complexes. Cell Rep 43, 113761 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiou S. H. et al. , Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev 29, 1576–1585 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zolotukhin S. et al. , Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods 28, 158–167 (2002). [DOI] [PubMed] [Google Scholar]
- 56.Gao G. et al. , Purification of recombinant adeno-associated virus vectors by column chromatography and its performance in vivo. Hum Gene Ther 11, 2079–2091 (2000). [DOI] [PubMed] [Google Scholar]
- 57.Yardeni T., Eckhaus M., Morris H. D., Huizing M., Hoogstraten-Miller S., Retro-orbital injections in mice. Lab Anim (NY) 40, 155–160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bankhead P. et al. , QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong T. N. et al. , Temporal endurance of exercise-induced benefits on hippocampus-dependent memory and synaptic plasticity in female mice. Neurobiol Learn Mem 194, 107658 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hall J. A., Cantley T. C., Galvin J. M., Day B. N., Anthony R. V., Influence of ovarian steroids on relaxin-induced uterine growth in ovariectomized gilts. Endocrinology 130, 3159–3166 (1992). [DOI] [PubMed] [Google Scholar]
- 61.Zheng Z. et al. , Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 20, 1479–1484 (2014). [DOI] [PubMed] [Google Scholar]
- 62.Girardot C., Scholtalbers J., Sauer S., Su S. Y., Furlong E. E., Je, a versatile suite to handle multiplexed NGS libraries with unique molecular identifiers. BMC Bioinformatics 17, 419 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A. et al. , STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hanlon K. S. et al. , High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat Commun 10, 4439 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lackner M., Helmbrecht N., Paabo S., Riesenberg S., Detection of unintended on-target effects in CRISPR genome editing by DNA donors carrying diagnostic substitutions. Nucleic Acids Res 51, e26 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Clement K. et al. , CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 37, 224–226 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Conant D. et al. , Inference of CRISPR Edits from Sanger Trace Data. CRISPR J 5, 123–130 (2022). [DOI] [PubMed] [Google Scholar]
- 68.Leger M. et al. , Object recognition test in mice. Nat Protoc 8, 2531–2537 (2013). [DOI] [PubMed] [Google Scholar]
- 69.Krishnaswami S. R. et al. , Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc 11, 499–524 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70., Pierce E. A. et al. , Gene Editing for CEP290-Associated Retinal Degeneration. New England Journal of Medicine May 6 doi: 10.1056/NEJMoa2309915. Online ahead of print (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
REFERENCES for Methods
- 1.Saito T. et al. Single App knock-in mouse models of Alzheimer’s disease. Nat Neurosci 17, 661–663, doi: 10.1038/nn.3697 (2014). [DOI] [PubMed] [Google Scholar]
- 2.Chiou S. H. et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev 29, 1576–1585, doi: 10.1101/gad.264861.115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun J. et al. CRISPR/Cas9 editing of APP C-terminus attenuates beta-cleavage and promotes alpha-cleavage. Nat Commun 10, 53, doi: 10.1038/s41467-018-07971-8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan K. Y. et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci 20, 1172–1179, doi: 10.1038/nn.4593 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zolotukhin S. et al. Production and purification of serotype 1, 2, and 5 recombinant adeno-associated viral vectors. Methods 28, 158–167, doi: 10.1016/s1046-2023(02)00220-7 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Gao G. et al. Purification of recombinant adeno-associated virus vectors by column chromatography and its performance in vivo. Hum Gene Ther 11, 2079–2091, doi: 10.1089/104303400750001390 (2000). [DOI] [PubMed] [Google Scholar]
- 7.Yardeni T., Eckhaus M., Morris H. D., Huizing M. & Hoogstraten-Miller S. Retro-orbital injections in mice. Lab Anim (NY) 40, 155–160, doi: 10.1038/laban0511-155 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bankhead P. et al. QuPath: Open source software for digital pathology image analysis. Sci Rep 7, 16878, doi: 10.1038/s41598-017-17204-5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dong T. N. et al. Temporal endurance of exercise-induced benefits on hippocampus-dependent memory and synaptic plasticity in female mice. Neurobiol Learn Mem 194, 107658, doi: 10.1016/j.nlm.2022.107658 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hall J. A., Cantley T. C., Galvin J. M., Day B. N. & Anthony R. V. Influence of ovarian steroids on relaxin-induced uterine growth in ovariectomized gilts. Endocrinology 130, 3159–3166, doi: 10.1210/endo.130.6.1597136 (1992). [DOI] [PubMed] [Google Scholar]
- 11.Zheng Z. et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med 20, 1479–1484, doi: 10.1038/nm.3729 (2014). [DOI] [PubMed] [Google Scholar]
- 12.Girardot C., Scholtalbers J., Sauer S., Su S. Y. & Furlong E. E. Je, a versatile suite to handle multiplexed NGS libraries with unique molecular identifiers. BMC Bioinformatics 17, 419, doi: 10.1186/s12859-016-1284-2 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dobin A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hanlon K. S. et al. High levels of AAV vector integration into CRISPR-induced DNA breaks. Nat Commun 10, 4439, doi: 10.1038/s41467-019-12449-2 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lackner M., Helmbrecht N., Paabo S. & Riesenberg S. Detection of unintended on-target effects in CRISPR genome editing by DNA donors carrying diagnostic substitutions. Nucleic Acids Res 51, e26, doi: 10.1093/nar/gkac1254 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clement K. et al. CRISPResso2 provides accurate and rapid genome editing sequence analysis. Nat Biotechnol 37, 224–226, doi: 10.1038/s41587-019-0032-3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Conant D. et al. Inference of CRISPR Edits from Sanger Trace Data. CRISPR J 5, 123–130, doi: 10.1089/crispr.2021.0113 (2022). [DOI] [PubMed] [Google Scholar]
- 18.Leger M. et al. Object recognition test in mice. Nat Protoc 8, 2531–2537, doi: 10.1038/nprot.2013.155 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Krishnaswami S. R. et al. Using single nuclei for RNA-seq to capture the transcriptome of postmortem neurons. Nat Protoc 11, 499–524, doi: 10.1038/nprot.2016.015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.