Abstract
The glycoproteins I and E of pseudorabies virus are important mediators of cell-to-cell spread and virulence in all animal models tested. Although these two proteins form a complex with one another, ascribing any function to the individual proteins has been difficult. We have shown previously, using nonsense mutations, that the N-terminal ectodomain of the gE protein is sufficient for gE-mediated transsynaptic spread whereas the cytoplasmic domain of the protein is required for full expression of virulence. These same studies demonstrated that the cytoplasmic domain of gE is also required for endocytosis of the protein. In this report, we describe the construction of viruses with nonsense mutations in gI that allowed us to determine the contributions of the gI cytoplasmic domain to protein expression as well as virus neuroinvasion and virulence after infection of the rat eye. We also constructed double mutants with nonsense mutations in both gE and gI so that the contributions of both the gE and gI cytoplasmic domains could be determined. We observed that the gI cytoplasmic domain is required for efficient posttranslational modification of the gI protein. The gE cytoplasmic domain has no effect on gE posttranslational glycosylation. In addition, we found that infection of all gE-gI-dependent anterograde circuits projecting from the rat retina requires both ectodomains and at least one of the cytoplasmic domains of the proteins. The gI cytoplasmic domain promotes transsynaptic spread of virus better than the gE cytoplasmic domain. Interestingly, both gE and gI cytoplasmic tails are required for virulence; lack of either one or both results in an attenuated infection. These data suggest that gE and gI play differential roles in mediating directional neuroinvasion of the rat; however, the gE and gI cytoplasmic domains most likely function together to promote virulence.
Pseudorabies virus (PRV) is a neurotropic alphaherpesvirus of swine that also infects and causes lethal disease in most mammals (except higher primates) and some birds (3, 32). In adult pigs, PRV infects the peripheral nervous system after primary infection of the mucosal epithelium lining the nasal cavity (40). Infection of sensory and autonomic ganglia through the nerve termini leads to establishment of life-long latency of the virus in the natural host (34). After infection of other susceptible animals, or young piglets, PRV usually does not establish latency in peripheral nerves but rather invades the central nervous system (CNS), causing lethal encephalitis.
At least three nonessential membrane proteins encoded by wild-type PRV are important for directional spread of virus from neuron to neuron after PRV infection of the rat CNS. The type I transmembrane glycoproteins E (gE) and I (gI) and the type II membrane protein Us9 are required for spread from presynaptic retinal ganglion cells to postsynaptic neurons in the superior colliculus (SC) and geniculate complex after infection of the rat eye (4, 5, 39). In pigs, gE and gI are required for PRV to spread from presynaptic olfactory neurons to postsynaptic neurons in the olfactory bulb after infection of the nasal olfactory mucosa (19, 24, 26, 30, 31). In mice, gE is required for transmission of virus to postsynaptic second order neurons after intranasal inoculation (1). In the rat CNS, gE, gI, and Us9 are required for efficient transmission of virus to postsynaptic neurons in the striatum after direct prefontal cortex injection. In general, all viruses deleted for these genes maintain the ability to spread from postsynaptic to presynaptic neurons in most models (4, 5, 41).
The precise functions of gE, gI, and Us9 in promoting directional spread of virus in the CNS have not yet been defined. gE and gI form a hetero-oligomer that facilitates the maturation and intracellular transport of both proteins to the plasma membrane of cells (39, 43). There is no evidence that Us9 forms a complex with gE or gI, suggesting that it may act at another step to promote transneuronal spread of PRV (our unpublished observations). Infection of certain nonneuronal (e.g., Madin-Darby bovine kidney [MDBK]) cells in culture by gE and gI null mutants, but not Us9 null mutants, leads to formation of small plaques, suggesting that gE and gI are required for efficient cell-to-cell spread in these cells (reviewed in references 4 and 17). However, spread as measured by plaque size in tissue culture does not always correlate with the ability of a virus to invade the CNS (4, 36, 38). Coinfection studies show that gE, gI, and Us9 null mutants are not defective in entering the primary retinal ganglion cells after intravitreal injection (4, 14). These observations suggest that the mutants are defective either in transport of the virus from the cell body to the axon terminal or in transfer of virus to the second-order neuron (discussed in reference 4). Analysis of viral mutants expressing truncated gE proteins has shown that the N-terminal extracellular domain of gE is sufficient for gI-gE complex formation as well as sufficient to mediate wild-type anterograde invasion after rat retina infection (36, 38). The C-terminal cytoplasmic domain of gE is dispensable for invasion of the CNS through anterograde and retrograde transport of virus. One hypothesis to explain gI-gE-mediated spread is that the ectodomain of the complex binds to a cellular ligand to allow passage of virus from an infected cell to an uninfected cell (10).
Because gE and gI can be found in a complex, and deletion of either gene alone shows a similar phenotype in the infected animal, it has often been asserted that gE and gI act together as a single functional unit to mediate cell-to-cell and directional transneuronal spread of infection. Several lines of evidence, however, suggest that gE and gI proteins may function on their own. gI null viruses are more virulent than gE null mutants in pigs and day-old chicks (28). Furthermore, a gI-negative virus spreads more extensively than a gE null mutant in the CNS of pigs after intranasal infection (25). In addition, PRV recombinants expressing either the bovine herpesvirus 1 (BHV-1) gE or gI proteins in exchange for the PRV gE and gI homologs have unexpected phenotypes (22, 23). A PRV recombinant expressing BHV-1 gI alone spreads anterograde to visual centers in some animals after retina infection. This is not seen with a PRV recombinant expressing BHV-1 gE. In this paradigm, the expression of BHV-1 gI without its obligate partner, BHV-1 gE, still allows for spread to areas requiring expression of both PRV gE and gI. Furthermore, a gE-negative virus produces significantly smaller plaques than a gI null mutant (17). Finally, several groups have shown that herpes simplex virus type 1 (HSV-1), varicella-zoster virus, and PRV gE-gI complexes have Fc receptor activity (15, 20, 21, 27). HSV-1 gE protein binds immunoglobulin G (IgG) aggregates but not monomers, while HSV-1 gI is not able to bind to any form of IgG. When expressed together, the HSV-1 gE-gI complex binds both aggregates and monomers. Thus, gI acts to modify gE in its affinity for binding to IgG (11, 12, 20, 21). Taken together, these observations suggest that gE and gI mediate similar functions but can do so independently in certain circumstances. From the Fc receptor experiments, it appears that gE provides the primary activity that is modified when complexed with gI.
In this report, we describe the construction of viruses with nonsense mutations in gI. We also constructed double mutants in which the contributions of both the gE and gI cytoplasmic domains could be determined. We found that the gI mutants lacking the cytoplasmic domain exhibit a novel neuroinvasion defect after infection of the rat visual system markedly different from that observed for gE mutants lacking the cytoplasmic domain. In addition, the gE and gI double mutants lacking both cytoplasmic domains demonstrate a more severe spread defect than either single gI or gE mutant alone. These data are consistent with the hypothesis that the ectodomains of the proteins do not always act in concert with one another in mediating transneuronal spread in the rat CNS. Two other unexpected results were obtained. First, all animals infected with any of the gE or gI cytoplasmic domain mutants lived to extended times postinfection and exhibited only mild symptoms of disease. These observations indicate that both cytoplasmic domains are required for full gE- and gI-mediated virulence. Second, the gI cytoplasmic domain is required for efficient posttranslational modification of the gI protein.
MATERIALS AND METHODS
Virus strains and cells.
Table 1 comprises a list of viruses used in this study. Wild-type PRV strain Becker (PRV Be) and the isogenic strains PRV 26, PRV 107, PRV 91, PRV 98, and PRV 99 have been previously described (36, 38, 39). Cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), while viral infections were performed in DMEM supplemented with 2% FBS. All PRV strains were propagated in pig kidney epithelial (PK15) cells. Plaque size phenotypes were analyzed on MDBK cells grown in 1% Methocel in DMEM supplemented with 2% FBS.
TABLE 1.
Viruses used in this study
| Virus | Descriptiona |
|---|---|
| PRV Be | Wild type |
| PRV 91 | gE deletion |
| PRV 98 | gI deletion |
| PRV 99 | gE and gI deletions |
| PRV 107 | Am457-gE (anchored gE) |
| PRV 26 | Am428-gE (secreted gE) |
| PRV 108 | Am310-gI (anchored gI) |
| PRV 109 | Am281-gI (secreted gI) |
| PRV 110 | Am457-gE; Am310-gI (double anchored) |
| PRV 111 | Am428-gE; Am281-gI (double secreted) |
| PRV 108R | Rescue PRV 108 (wild type) |
| PRV 109R | Rescue PRV 109 (wild type) |
| PRV 110R | Rescue PRV 110 (wild type) |
Am refers to the replacement of the indicated arginine residue with an amber stop codon.
Antisera.
The monoclonal antibody (MAb) specific for gE complexed to gI (MAb 1/14), the polyclonal rabbit PRV-specific antiserum (Rb133), the polyclonal rabbit antiserum to gI (Rb1544), and the polyclonal goat antiserum to gC (282) have all been previously described (16, 33, 39). T. Ben-Porat kindly provided the MAb pool to gE (M133, M156, and M138). Rabbit polyclonal antiserum to gE was a generous gift from K. Bienkowska-Szewczyk (University of Gdansk).
Rabbit polyclonal antiserum to the cytoplasmic domain of gE was made against a fusion protein in which this gE protein domain was fused to the glutathione S-transferase (GST) protein (pRS45, described below). Escherichia coli DH5α cells transformed with pRS45 were induced to express the GST-gE fusion protein by the addition of isopropyl-β-d-thiogalactopyranoside (IPTG). Following an inclusion body preparation (35), the fusion protein was found in the soluble supernatant. The fusion protein was affinity purified using GST-agarose (Sigma) following the GST-Gene Fusion System protocol (Pharmacia) and dialyzed against phosphate-buffered saline (PBS); 250 μg of protein was injected into New Zealand White rabbits. Complete Freund's adjuvant was used in the first injection, while incomplete Freund's adjuvant was used in all subsequent injections. Injections were performed every 2 weeks for 6 weeks. Serum was collected at 2-week intervals beginning 1 month after the initial injection and was tested by Western blot analysis for reactivity against gE. This antiserum was also found to be reactive in immunoprecipitations and immunofluorescence studies.
For some immunoprecipitations, antibody Rb1544 was cross-linked to protein A-agarose beads (50% slurry; Sigma) in the following manner. An equal volume of Rb1544 antibody was mixed for 1 h at room temperature with gentle rocking with protein A-agarose, creating a 25% final slurry of beads. The beads were washed twice with 10 volumes of 0.2 M sodium borate (pH 9.0) and centrifuged at 3,000 × g for 5 min before removal of the supernatant. The beads were resuspended in 10 volumes of 0.2 M sodium borate, and dimethyl pimelimidate (Sigma) was added to a final concentration of 20 mM. After rocking for 30 min at room temperature, the reaction was stopped by one wash as before with 10 volumes of 0.2 M ethanolamine (pH 8.0) and then incubation for 2 h at room temperature in 10 volumes of 0.2 M ethanolamine with gentle mixing. After the wash, the beads were resuspended in PBS containing 0.01% thimerosal (Sigma) to make a final suspension of 50% beads.
Alexa-568-conjugated goat anti-mouse IgG was purchased from Molecular Probes. Horseradish peroxidase-conjugated donkey anti-mouse, anti-rabbit, and anti-goat IgGs were purchased from Kirkegaard & Perry Laboratories, Inc.
Plasmids and viruses.
All amplified sequences were determined using Sequenase version 2.0 (United States Biochemical) to confirm the presence of only desired mutations.
Construction of a plasmid encoding the GST-gE cytoplasmic domain fusion.
The sequences encoding the cytoplasmic domain of gE were PCR amplified (5′-GGGGAATTCCGCCGCCGGGCGGCCTCGCGG-3′; 5′-GGGAGATCTTAAGCGGGGCGGGCATTCAACAGG-3′) and cloned into pGEX-4T-1 (Pharmacia), using EcoRI and XhoI restriction sites. This plasmid was named pRS45.
Mutation of arginine 281 and arginine 310 of gI to amber stop codons.
The StuI-XcmI fragment of BamHI-7 of PRV viral DNA was cloned into a pAlter-1 plasmid to which an oligonucleotide linker was added (Promega), creating pPH3. Site-directed mutagenesis was performed on pPH3 (Altered Sites kit; Promega), using an oligonucleotide that results in the substitution of amino acid 281 or 310 of the protein with an amber stop codon (5′-ACGCCACGGCGGGCGCCTAGGGCCCCGGGAAGATAGC-3′; 5′-GGGGTCGCCTGCGCGGCCCGCTAGTGCGCGCGCGGAATCGCATC-3′). The plasmids were designated pRS37 (am281-gI) and pRS38 (am310-gI). The addition of the stop codons creates novel BfaI restriction sites. The 730-bp StuI-XcmI restriction fragment from either pRS37 or pRS38 containing the site-directed mutation was introduced into the transfer vector pPH2 (36), resulting in pRS39 (am281-gI) or pRS40 (am310-gI). Transfer of the appropriate fragments was confirmed by restriction analysis of the resulting plasmids with BfaI.
Construction of double-mutant plasmids.
The BstEII-SphI fragment of pRS25, which contains an amber stop codon in place of amino acid 457 of gE (36), was transferred to pRS40, containing am310-gI, resulting in pRS43 (am310-gI; am457-gE). To confirm the transfer of the appropriate fragment, the plasmid was restricted with BfaI. This plasmid contains stop codons after the sequences encoding the transmembrane domains of both the gI and gE proteins. To construct a plasmid containing stop codons just before the sequences encoding the transmembrane domains of both gI and gE, the BstEII-SphI fragment of pRT20 (am428-gE) (38) was transferred into pRS39 (which contains am281-gI), creating pRS44 (am281-gI; am428-gE). Transfer of this fragment was confirmed by restriction analysis of the plasmid with AluI and BfaI.
Construction of a plasmid encoding gI signal sequence-GFP-gI transmembrane domain and cytoplasmic domain hybrid.
To construct a fusion of the sequences encoding these protein segments, the sequences encoding the gI transmembrane domain and cytoplasmic domain were amplified by PCR (5′-GAAGATCTCGGGGCCCCGGGAAGATAGCCATGGTG-3′; 5′-CGGAATTCTGGCGAAGCTCGGCCAACGTCATC-3′) and cloned into pEGFP-C1 (Clontech), using the BglII and EcoRI restriction sites resulting in pRS35. The sequences encoding the gI signal sequence were then PCR amplified (5′-CGGCTAGCCCGGTCCGTAGCCTCCGCAGTACC-3′; (5′-ATACCGGTCTGAAGAGGACGCCCCCGACGCGCGG-3′) and cloned into pRS35, using the NheI and AgeI restriction sites, which places these sequences just 5′ of the enhanced green fluorescent protein (GFP) cassette. This plasmid was designated pRS36. The RsrII-XcmI restriction fragment from pRS36 was then cloned into pPH2 to create pRS48 (gI-GFP hybrid transfer vector).
Construction of a plasmid encoding gE signal sequence-GFP-gE transmembrane domain and cytoplasmic domain hybrid.
The gE-GFP hybrid was constructed in the same manner as the gI-GFP hybrid. The sequences encoding the gE transmembrane domain and cytoplasmic domain were amplified by PCR (5′-GAAGATCTCTGTTTGTGCTGGCGCTGGGCTCCT-3′; 5′-CGGAATTCGCCGGTTCTCCCGGTATTTAAGCG-3′) and cloned into pEGFP-C1, using the BglII and EcoRI sites, creating pRS2. The signal sequence and upstream intergenic sequences of gE were PCR-amplified (5′-TTAGCTAGCCAACCCCGTCGCCGGGGCGCC-3′; 5′-ATACCGGTGGGGTCGTCTCGGCGGAGAGGCTCG-3′) and cloned into pRS2, using the NheI and AgeI restriction sites, resulting in pRS22. The XcmI-SphI restriction fragment of pRS22 was then transferred to pPH2, resulting in pRS27 (gE-GFP hybrid transfer vector).
Construction of mutant viruses.
PRV 99 DNA (which is deleted for the sequences encoding both gE and gI) was cotransfected by the calcium phosphate precipitation method into PK15 cells with either pRS39 (am281-gI) or pRS40 (am310-gI). After a complete cytopathic effect was observed, the infected cells were harvested, frozen, thawed, and replated onto PK15 cells to allow plaque formation. Plaques formed by recombinant virus were screened for gE expression by a black plaque assay (described below) using a pool of monoclonal antisera against gE. Recombinants that expressed gE protein were picked and purified by four rounds of plaque purification. The resulting viruses were named PRV 108 (am310-gI) and PRV 109 (am281-gI).
PRV 91 DNA (which lacks the sequences for gE) was cotransfected into PK15 cells with pRS27 (gE-GFP hybrid). Recombinant virus was purified as described above by screening for GFP expression by standard epifluorescence. Recombinants that expressed GFP were picked and purified by four rounds of plaque purification, resulting in PRV 103.
PRV 103 DNA (gE-GFP hybrid) was cotransfected into PK15 cells with either pRS43 (am310-gI; am457-gE) or pRS44 (am281-gI; am428-gE), and recombinant virus was plaque purified by screening for lack of GFP expression by epifluorescence. Recombinants that did not express GFP resulted in PRV 110 (am310-gI; am457-gE) and PRV 111 (am281-gI; am428-gE).
Construction of revertant viruses.
PRV 108 and PRV 109 were reverted in a two-step process. First, either PRV 108 or PRV 109 DNA was cotransfected with a fragment encoding the gI-GFP hybrid from pRS48 (gI-GFP hybrid). Recombinants were isolated by screening for plaques that expressed GFP and were plaque purified four times. The resulting viruses were designated PRV 108GR and PRV 109GR. PRV 108GR and PRV 109GR were reverted to wild type by cotransfecting DNA from these viruses with a wild-type restriction fragment (SalI-AgeI from BamHI-7). Wild-type recombinants were screened for lack of GFP expression, subjected to four rounds of plaque purification, and named PRV 108R and PRV 109R.
PRV 110 was reverted by cotransfecting PRV 110 DNA with the wild-type StuI-BspEII restriction fragment from BamHI-1. Wild-type recombinants were screened by plaque size phenotypes (large plaques) on MDBK cells. One isolate was plaque purified four times and was named PRV 110R.
Verifying genotypes of recombinant viruses.
The presence or absence of the desired mutations in recombinant viral DNA was confirmed by Southern blot analysis as follows. The amber mutation in PRV 108 creates a novel BfaI restriction site in the BamHI-7 fragment of PRV that alters a 6,612-bp fragment to a 2,987- and a 3,625-bp fragment following DNA digestion with BamHI and BfaI. The amber mutation in PRV 109 creates a novel BfaI restriction site in the BamHI-7 fragment of PRV that alters a 6,612-bp fragment to a 3,074- and a 3,538-bp fragment following DNA digestion with BamHI and BfaI. The amber mutations in PRV 110 both create novel BfaI sites in the BamHI-7 fragment of PRV that alter a 6,612-bp fragment to a 3,631-, a 1,648-, and a 1,333-bp fragment following DNA digestion with BamHI and BfaI. The amber mutations in PRV 111 create a novel BfaI site and introduce another AluI site in the BamHI-7 fragment of PRV. Digestion with BamHI and BfaI changes a 6,612-bp fragment to a 3,074- and a 3,538-bp fragment, while digestion with BamHI and AluI changes a 1,458-bp restriction fragment to 1,300 bp. The replacement of wild-type gE with the gE-GFP-hybrid fusion construct in PRV 103 was confirmed using a GFP-specific probe and by digesting the DNA with BamHI and SphI. The gI-GFP-hybrid fusion in PRV 108GR and PRV 109GR introduces a novel BglII restriction site in the BamHI-7 fragment of PRV that alters a 6,612-bp fragment to a 3,546- and a 3,066-bp fragment following DNA digestion with BamHI and BglII. The rescued viruses, PRV 108R, PRV 109R, and PRV 110R, displayed wild-type restriction patterns as predicted.
Indirect immunofluorescence and endocytosis assays.
Indirect immunofluorescence assays were performed on fixed, permeabilized PK15 cells that had been infected for 4 h using a MAb that specifically recognizes gE complexed to gI (MAb 1/14), as previously described (37). All endocytosis assays were performed at 4 h postinfection with MAb 1/14 as previously described (37). Single optical sections were taken through the centers of the cells, using a Nikon MRC600 confocal microscope mounted on an Optiphot II, which utilizes an argon-krypton laser.
Black plaque and plaque size analysis.
For black plaque analysis, PK15 cells were infected and overlaid with 1% Methocel for 48 h. Plaques were reacted with a 1:1:1 mixture of a gE MAb pool diluted 1:10 as previously described (38). Plaque sizes were measured on MDBK cells at 72 h postinfection as previously described (36). The diameter of 20 plaques was measured per virus, and the results were averaged.
Immunoprecipitation analysis.
For steady-state experiments, PK15 cells were infected at a multiplicity of infection (MOI) of 10 for 5 h prior to labeling with [35S]methionine-[35S]cysteine (Dupont-NEN). Lysates were collected 16 h postinfection in TNX buffer (10 mM Tris [pH 7.4], 150 mM NaCl, 1% Triton X-100), and both denatured and native immunoprecipitations were performed as previously described (39). For immunoprecipitations with protein A-coupled antibody Rb1544 20 μl of beads (50% slurry) was added to denatured extracts and allowed to incubate overnight at 4°C. All washes were performed as previously described (39). For pulse-chase analysis, PK15 cells were infected at an MOI of 10. At 5.5 h postinfection, the cells were incubated in cysteine- and methionine-free medium for 30 min and pulsed for 7 min with 125 μCi of label in 1 ml, and then radioactive medium was removed and replaced with nonradioactive medium. Samples were taken at the times indicated. Endoglycosidase H (endo H) digestions were performed in the following manner. The Staphylococcus aureus cells containing the immunoprecipitated proteins were resuspended in buffer H (50 mM Tris [pH 6.5], 1% sodium dodecyl sulfate [SDS], 1% β-mercaptoethanol) plus phenylmethylsulfonyl fluoride and boiled for 3 min to remove the bound proteins. The supernatant was removed, and sodium citrate (pH 5.5) was added to a final concentration of 100 mM; 1 mU of endo H (Boehringer Mannheim) was added, and the samples were incubated overnight at 37°C. The samples were acetone precipitated prior to analyzing on an SDS-polyacrylamide gel.
Animal experiments, tissue processing, and immunohistochemistry.
Adult male Sprague-Dawley rats weighing 200 to 300 g at the time of the experiment were used in this study. Food and water were freely available during the course of the experiment, and the photoperiod was standardized to 14 h of light and 10 h of darkness. Experimental protocols were approved by the Princeton University Animal Welfare Committee and were consistent with the regulations stipulated by the American Association for Accreditation of Laboratory Animal Care and those in the Animal Welfare Act (Public Law 99-198). The animals were confined to a biosafety level 2 facility, and the experiments were conducted with specific safeguards as described previously.
For intraocular injections, 2.5 μl of virus suspension (approximately 1 × 108 to 2 × 108 PFU/ml) was injected into the vitreous humor of the left eye of an anesthetized animal. When symptoms of infection were overt, the animals were sacrificed and exsanguinated, and the brains were removed as described previously (13). Immunohistochemical analysis of coronal brain slices using rabbit polyclonal antiserum to whole PRV virus (Rb133) has been described previously (13). Tissues were taken for analysis just prior to the estimated time to death.
RESULTS
Construction of mutant viruses.
All viruses are described in Table 1. Anchored mutants express the extracellular and transmembrane domains of the indicated proteins, while secreted mutants express only the extracellular domains of the proteins. To facilitate comparison of secreted and anchored gI with secreted and anchored gE, and the double-anchored or -secreted mutants with the single-anchored or -secreted mutants, the data also include infections with PRV 107 (anc [anchored] gE) and PRV 26 (sec [secreted] gE), which have been described previously (36, 38). A diagram of the wild-type gI protein and constructed mutants is shown in Fig. 1. We will refer to the anchored and secreted gI (PRV 108 and 109) or gE (PRV 107 and 26) mutants as single-anchored or -secreted gI or gE truncation mutants and to the double-anchored gI and anchored gE (PRV 110) or double-secreted gI and -secreted gE (PRV 111) as the double-truncation mutants.
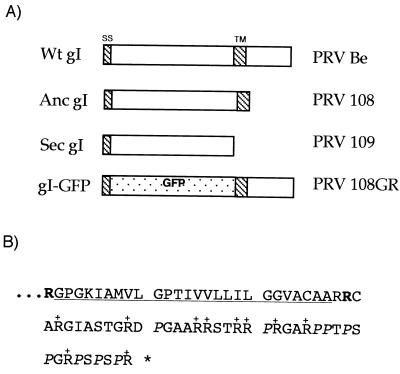
FIG. 1.
Wild-type (Wt) gI protein and products encoded by the mutant viruses. The gI protein is a type I transmembrane protein with an N-terminal signal sequence (SS), a 281-amino-acid ectodomain, a transmembrane domain (TM), and a short cytoplasmic domain (40 amino acids). (A) Depiction of various gI and gE proteins; (B) sequence of the gI cytoplasmic domain. Replacement of arginine residue 310 with an amber stop codon (shown in bold in panel B) should result in an anchored gI protein lacking the cytoplasmic domain [PRV 108 (anc gI)]. Likewise, a stop codon replacing the arginine at residue 281 (shown in bold in panel B) should result in a secreted gI protein lacking the transmembrane domain and the cytoplasmic domain [PRV 109 (sec gI)]. This same strategy has previously been used to construct anchored and secreted forms of the type I transmembrane protein, gE (36, 38). PRV 110 and PRV 111 are double mutants expressing anchored gI plus anchored gE and or secreted gI plus secreted gE, respectively. In panel B, the 10 arginine residues in the domain are indicated with +, the nine proline residues are in italics, and the transmembrane domain is underlined.
Secreted and anchored forms of gI and gE are expressed to high levels and retain the ability to form a complex.
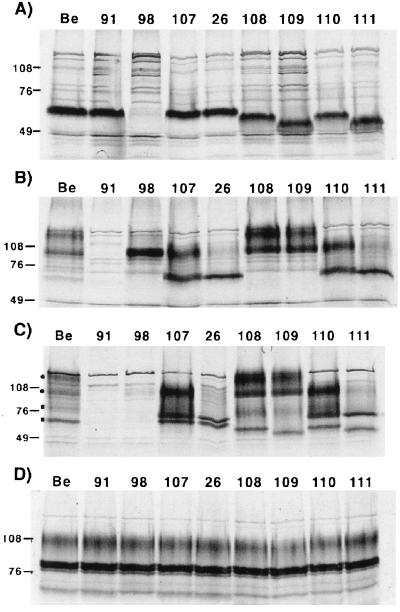
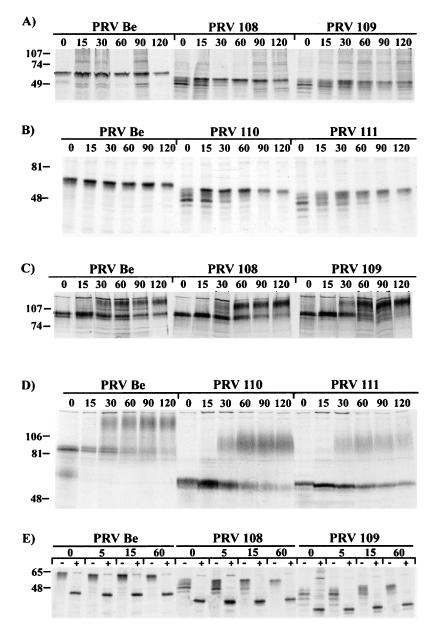
Steady-state levels of the gI and gE proteins were analyzed by immunoprecipitations performed with extracts from PK15-infected cells labeled with [35S]methionine and [35S]cysteine. The results are shown in Fig. 2. The rabbit polyclonal antibody used to immunoprecipitate gI recognized only the immature form of the protein. The immature gI protein in wild-type (PRV Be)-infected cells had a molecular mass of approximately 65 kDa (Fig. 2A). An identical protein was seen in PRV 91 (gE null)-, PRV 107 (anc gE)-, and PRV 26 (sec gE)-infected cells but was absent from PRV 98 (gI null)-infected cells. As predicted, the gI proteins made after infection with PRV 108 (anc gI) and PRV 109 (sec gI) migrated slightly further than wild-type gI protein in SDS-polyacrylamide gels, with apparent molecular masses of approximately 61 and 55 kDa, respectively. As expected, the gI protein made by PRV 110 (anc gI/anc gE) was indistinguishable from that made by PRV 108, and the protein made by PRV 111 (sec gI/sec gE) was similar to the protein produced by PRV 109. The gI and gE proteins produced by the revertant viruses were indistinguishable from wild-type virus as assayed by Western blot analysis (data not shown).
FIG. 2.
Steady-state immunoprecipitation analysis. PK15 cells were infected at an MOI of 10 with either PRV Be (wild type), PRV 91 (gE null), PRV 98 (gI null), PRV 107 (anc gE), PRV 26 (sec gE), PRV 108 (anc gI), PRV 109 (sec gI), PRV 110 (anc gI/anc gE), or PRV 111 (sec gI/sec gE) and labeled for 12 h with [35S]methionine-[35S]cysteine prior to preparing cell lysates. Immunoprecipitations were performed on extracts using either rabbit polyvalent antiserum to the immature form of gI (A), rabbit polyvalent antiserum to gE (B), monoclonal antiserum to gE when it is complexed with gI (C), or goat polyvalent antiserum to gC (D). Panels A, B, and D show immunoprecipitations performed with denatured extracts, while the extracts for panel C were not denatured prior to immunoprecipitation. Circles and squares in panel C denote gE- gI-specific bands, respectively. Positions of apparent molecular mass markers (kilodaltons) are shown on the left.
Rabbit polyclonal antiserum to the gE ectodomain immunoprecipitated immature and mature forms of gE from Be-infected cells, as shown in Fig. 2B. The precursor form of gE had an apparent molecular mass of 93 kDa, and the mature form had an apparent molecular mass of 110 kDa. Identical proteins were made in PRV 108- and PRV 109-infected cells. The gE-specific proteins were absent in PRV 91-infected cells, and the precursor form was the predominant form seen in PRV 98-infected cells, as previously reported (39). As noted previously, the gE proteins expressed by PRV 107 and PRV 26 migrated faster than wild-type gE and had immature forms of 70 and 65 kDa, respectively, and mature forms of 93 and 85 kDa, respectively (36, 38). Importantly, the gE protein produced by PRV 110 was indistinguishable from that produced by PRV 107, and the gE protein made by PRV 111 was indistinguishable from that made by PRV 26. Extracts were also immunoprecipitated for an unrelated glycoprotein, gC, as a loading control (Fig. 2D).
We performed coimmunoprecipitations of gI and gE from undenatured extracts by using MAb 1/14, which specifically recognizes gE complexed with gI (Fig. 2C). MAb 1/14 immunoprecipitated the 93-kDa immature and 110-kDa mature gE proteins, as well as the 65-kDa immature and 80-kDa mature forms of gI from PRV Be-infected cells. These proteins were absent from PRV 91 (gE null)- or PRV 98 (gI null)-infected cells. Both gE and gI could be readily coimmunoprecipitated by the antiserum in PRV 107-, PRV 26-, PRV 108-, PRV 109-, PRV 110-, or PRV 111-infected cells.
Unfortunately, we were unable to demonstrate that PRV 109 expressed a secreted gI protein found in the medium of infected cells, as we do not have an antibody that efficiently recognizes the mature form of the gI protein (data not shown). We have shown previously that PRV 26 encodes a secreted gE protein (38) and have evidence that PRV 111 also encodes a secreted form of gE (data not shown).
Steady-state localization of the gI-gE complex.
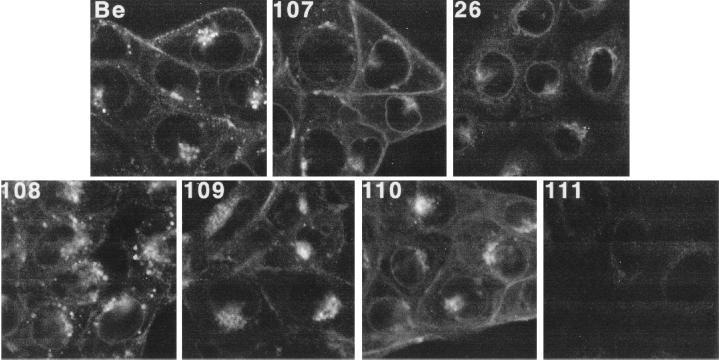
To determine the steady-state localization of the gI-gE complex formed after infection with the various mutants, indirect immunofluorescence was performed with MAb 1/14 on fixed, permeabilized cells at 4 h postinfection. PRV Be-, PRV 107-, and PRV 26-infected cells are shown in Fig. 3 for comparison. The gI-gE complex formed after infection with PRV Be and PRV 107 localized in very similar patterns, with staining evident in the nuclear envelope, the endoplasmic reticulum (ER), Golgi region, and plasma membrane. As reported earlier, cells infected with PRV 107 showed more gI-gE complex on the cell surface than cells infected with wild-type virus, due to a disruption in endocytosis of the complex (36, 37). Cells infected with PRV 26 showed gI-gE complex localization mainly in the ER and Golgi region and very little staining on the plasma membrane, as previously reported (38). The gI-gE complex in cells infected with PRV 108, PRV 109, or PRV 110 localized similarly to wild-type gI-gE complex; however, several differences were noted. In PRV 108-infected cells, there were numerous gI-gE-positive vesicles scattered throughout the cytosol of the infected cells, more so than was seen in wild-type-infected cells. In PRV 110-infected cells, the plasma membrane showed bright gI-gE staining which was very similar to that of cells which had been infected with PRV 107. Finally, it was difficult to detect positive gI-gE staining in PRV 111-infected cells.
FIG. 3.
Steady-state distribution of gI-gE complexes. PK15 cells were infected at an MOI of 10 for 4 h with either PRV Be (wild type), PRV 107 (anc gE), PRV 26 (sec gE), PRV 108 (anc gI), PRV 109 (sec gI), PRV 110 (anc gI/anc gE), or PRV 111 (sec gI/sec gE). The cells were fixed, permeabilized, and reacted with a MAb that specifically recognized gE when it was complexed with gI (MAb 1/14). An Alexa-568-conjugated secondary antibody was used to visualize bound MAb. Confocal sections were taken through the center of the cells.
Endocytosis of the gI-gE complex.
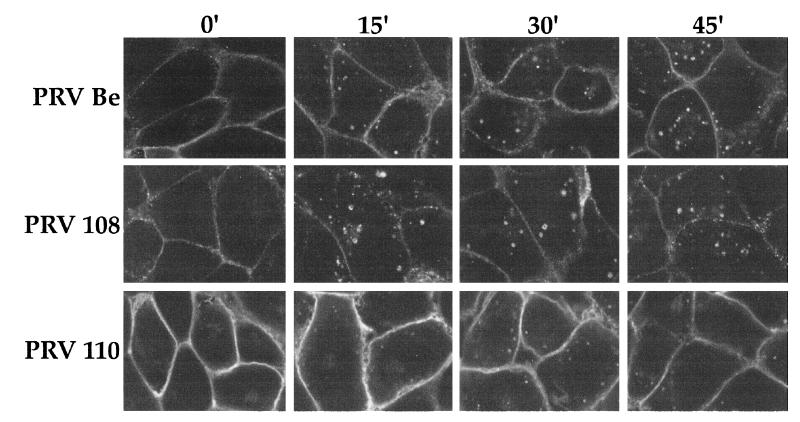
We determined the ability of the PRV gI-gE complexes to internalize at 4 h postinfection using an indirect immunofluorescence endocytosis assay with MAb 1/14. As shown in Fig. 4, the wild-type gI-gE complex was detected on the plasma membrane of infected PK15 cells when the cells were not shifted to 37°C. However, after a temperature shift for the indicated times, the complex accumulated in the interior of the cells in cytoplasmic vesicles which became larger and more numerous with longer incubation at 37°C. The gI-gE complex formed after infection with PRV 108 (anc gI) was internalized similarly to wild-type gI-gE complex. Numerous vesicles accumulated in the interior of the cells after the temperature shift. The gI-gE complex formed after infection with PRV 110 (anc gI/anc gE) was also observed on the plasma membrane of PK15 cells, which were brightly stained at the 0-min time point. However, after a shift to 37°C for up to 45 min, most of the complex remained on the surface of the cells and did not accumulate appreciably in the interior of the cells, as previously reported for PRV 107 (anc gE) (36), although a few gI-gE-positive vesicles were seen in the interior of the cells.
FIG. 4.
Endocytosis of the gI-gE complexes. PK15 cells were infected at an MOI of 10 with either PRV Be (wild type), PRV 108 (anc gI), or PRV 110 (anc gI/anc gE) for 4 h prior to an indirect immunofluorescence endocytosis assay as described in Materials and Methods. Briefly, the cells were incubated at 4°C with MAb 1/14, which specifically recognized gE when it was complexed with gI. The cells were shifted to 37°C for the indicated times to allow internalization of the protein complex, then fixed, permeabilized, and reacted with a secondary antibody to visualize the bound primary antibody. Confocal sections were taken through the center of the cells.
Plaque size in MDBK cells.
Viruses that lack gE and gI form small plaques on MDBK cells (17, 18, 42). It has also been noted that the plaques formed by a gI null virus are slightly larger than plaques formed by a gE null virus (17). We analyzed plaques formed by the viral mutants on MDBK cells at 72 h postinfection (Table 2). Plaques formed by wild-type virus had average diameters of 1.3 mm (set to 100%). Plaques formed after infection with the revertant viruses were not statistically different from wild-type plaques. Plaques formed by PRV 91 (gE null) and PRV 107 (anc gE) were significantly smaller, 70 to 72% of the wild-type size as previously reported (36). PRV 26 (sec gE) made plaques that were 85% of the wild-type size as noted previously (38). A virus lacking gI (PRV 98) also made plaques that were 85% of the wild-type size. Plaques formed by PRV 108 (anc gI) and PRV 109 (sec gI) were slightly larger than plaques formed by PRV 98 and were 92% of the wild-type size. There was no statistical difference between plaques formed by wild-type virus and PRV 108 or PRV 109. PRV 110 (anc gI/anc gE) made plaques that were identical to those made by PRV 91 and PRV 107 (73% of the wild-type size), while PRV 111 (sec gI/sec gE) formed plaques that were indistinguishable from those of PRV 98 and PRV 26 (85% of the wild-type size).
TABLE 2.
Plaque size on MDBK cells
| Virus | Size (mm)a | % of wild-type level |
Pb with respect to:
|
|
|---|---|---|---|---|
| PRV Be | PRV 98 | |||
| PRV Be | 1.3 ± 0.16 | 100 | NA | <0.005 |
| PRV 91 | 0.93 ± 0.10 | 72 | <0.005 | <0.005 |
| PRV 98 | 1.1 ± 0.06 | 85 | <0.005 | NA |
| PRV 107 | 0.94 ± 0.09 | 72 | <0.005 | <0.005 |
| PRV 26 | 1.1 ± 0.06 | 85 | <0.005 | 0.40 |
| PRV 108 | 1.3 ± 0.08 | 100 | 0.20 | <0.005 |
| PRV 109 | 1.2 ± 0.08 | 92 | 0.07 | <0.005 |
| PRV 110 | 0.96 ± 0.09 | 74 | <0.005 | <0.005 |
| PRV 111 | 1.1 ± 0.07 | 85 | <0.005 | 0.63 |
| PRV 108R | 1.3 ± 0.09 | 100 | 0.82 | <0.005 |
| PRV 109R | 1.3 ± 0.11 | 100 | 0.60 | <0.005 |
| PRV 110R | 1.3 ± 0.07 | 100 | 0.11 | <0.005 |
Plaque size averages were taken from 20 isolated plaques. The standard deviations are indicated.
Determined by Student's t test. NA, not applicable (this is the virus to which others are compared).
Pulse-chase analysis.
We used pulse-chase analysis to determine the rate of processing of either the gI or gE protein after infections with the various viral mutants. As shown in Fig. 5A, the gI antiserum immunoprecipitated the 65-kDa immature form of the protein at the 0-min time point from wild-type (PRV Be)-infected cell lysates. This protein remained stable up to 120 min of chase. As this antibody does not recognize the mature form of the gI protein, the conversion of immature to mature gI protein could not be monitored. It is important to note, however, that the amount and electrophoretic mobility of this protein were identical at all time points when wild-type virus was used. In the latest time point, 120 min, some of the immature gI protein began to disappear, presumably due to its conversion to the mature form. The pulse-chase analyses of gI after infections with PRV 108 (anc gI) or PRV 109 (sec gI) showed results similar to one another. At the 0- and 15-min time points, multiple gI species could be detected. These proteins resolved to one form after a 30-min chase. However, even after a 30-min chase, this band was diffuse and continued to become more distinct until 90 to 120 min of chase. The results of immunoprecipitations from PRV 110- or PRV 111-infected cells are shown in Fig. 5B. The results obtained with PRV 110 were indistinguishable from those obtained with PRV 108, and those with PRV 111 were indistinguishable from those of PRV 109.
FIG. 5.
Pulse-chase analysis. PK15 cells were infected at an MOI of 10 with either PRV Be (wild type), PRV 108 (anc gI), PRV 109 (sec gI), PRV 110 (anc gI/anc gE), or PRV 111 (sec gI/sec gE). The cells were pulse labeled for 7 min with [35S]methionine-[35S]cysteine and chased for the times indicated with nonradioactive medium before collection of cell lysates. Cell lysates were denatured and immunoprecipitated with polyclonal antiserum to the immature form of gI (A, B, and E) or polyclonal antiserum to gE (C and D). In panel E, half of the samples were digested with endo H after immunoprecipitation, indicated with +. Apparent molecular mass markers (kilodaltons) are indicated on the left. Samples shown in panels B, D, and E were run on larger gels to help discriminate between the different forms of the proteins.
The results of immunoprecipitations using the rabbit polyclonal antiserum directed against the gE cytoplasmic domain are shown in Fig. 5C. In PRV Be-infected cells, only the immature gE protein was detected at 0 and 15 min of chase. The mature form of the protein was first immunoprecipitated at the 30 min time point. The half-time for conversion of the wild-type gE precursor to the mature species was between 30 and 60 min. In contrast to results obtained with the gI antiserum, pulse-chase analysis of gE after infections with PRV 108 and PRV 109 gave results identical to those for wild-type virus. The appropriate gE-specific proteins were immunoprecipitated, and the half-time to conversion to the mature form was identical. The gE pulse-chase from PRV 110- or PRV 111-infected cells is shown in Fig. 5D. Immunoprecipitations were performed with a polyclonal antibody that recognizes the ectodomain of the protein. The gE protein made by PRV 110 was converted to the mature form with similar kinetics as wild-type gE protein. The gE protein produced after infection with PRV 111 however, showed a delay in processing to the mature form and was not completely processed after 120 min of chase, as shown previously for PRV 26 (sec gE) (38).
To further define the processing defect of the gI proteins, we repeated the pulse-chase analysis using shorter chase periods and treated half of the immunoprecipitates with endo H. Endo H digestion cleaves high-mannose N-linked sugar modifications received in the ER or cis-Golgi from the peptide backbone of glycoproteins. The gI protein contains five potential N-linked glycosylation sites as determined by protein sequence. The results are shown in Fig. 5E. At all time points analyzed, PRV Be-infected cells showed a single protein of 65 kDa that was sensitive to endo H digestion and was digested to a faster-migrating form of approximately 45 kDa. These proteins were indistinguishable from one another at all time points. In lysates from PRV 108-infected cells, at least five proteins were immunoprecipitated with gI antiserum at the 0-min time point. These proteins resolved to one slower-migrating form of 60 kDa between 15 and 60 min of chase, as shown in Fig. 5A. All forms of the protein were completely sensitive to endo H digestion. The results of experiments with PRV 109 were similar to those done with PRV 108. At least five endo H-sensitive proteins were immunoprecipitated at 0 min, and these proteins were resolved to a single slower-migrating form.
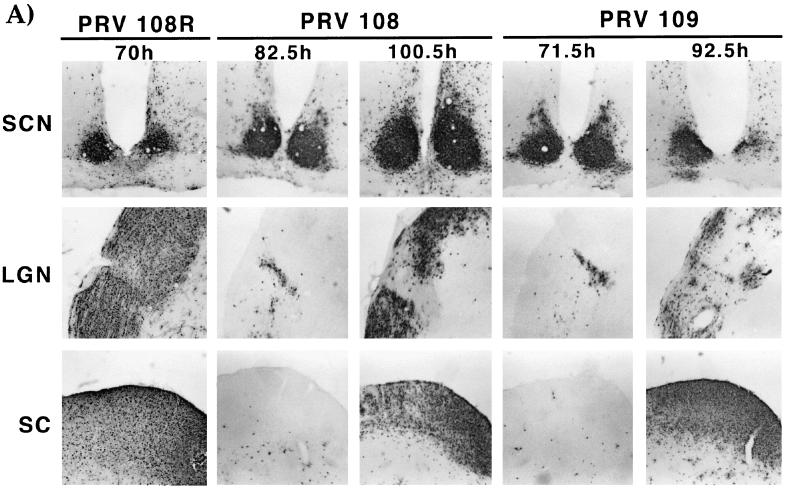
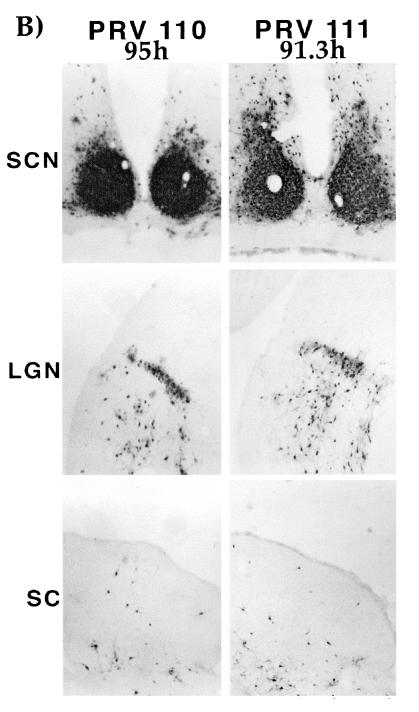
Spread in the rat CNS.
In the rat retina infection paradigm, retinal ganglion cells lining the retina are the first order neurons that become infected. After infection of these cell bodies, virus travels down the axon terminals and infects second-order, synaptically connected neurons in retinorecipient areas. Wild-type virus (PRV Be) can travel to all retinorecipient areas, including the suprachiasmatic nucleus (SCN), the lateral geniculate nucleus (LGN), including the dorsal and ventral aspects (dLGN and vLGN) and intergeniculate leaflet (IGL), as well as the SC (8). The SCN and the IGL are broadly defined as circadian rhythm centers while the dLGN, vLGN, and SC are visual centers. gE or gI null viruses can infect only the SCN and the IGL (39). We determined the ability of the various mutant viruses to infect these areas after intraocular infection to test the role of the gI cytoplasmic domain in gI-facilitated spread of virus. Animals infected with PRV 108 (anc gI) and PRV 109 (sec gI) survived until 67 to 103 h postinfection, while those infected with PRV 110 and PRV 111 survived until 71 to 95 h postinfection. Animals infected with PRV Be or revertant viruses never survived beyond 71.5 h postinfection (n = 13). We analyzed the brains of animals that survived to both early and late times after infection; the results are illustrated in Fig. 6 and tabulated in Table 3. Figure 6A shows a brain from an animal infected with PRV 108R as an example of a wild-type infection. This virus established a robust infection in all retinorecipient regions. The patterns of infection in the brains of animals infected with PRV 108 and PRV 109 were indistinguishable from one another and showed striking features. If an animal died at an earlier time point after infection, the staining pattern in the brain was indistinguishable from that of an animal infected with a gI null virus (39). Staining was observed only in the SCN and the IGL (the restricted infection pattern of gE and gI null mutants). If the animals survived to late times postinfection (>90 h postinfection), all areas normally infected by wild-type virus were also infected by the viruses. Most notably, the SC was heavily infected in animals with longer survival times. This kinetic effect was not observed with the single gE anchored or secreted mutant (36, 38). The brains of animals infected with either PRV 110 or PRV 111 are shown in Fig. 6B. The infection pattern in these brains was not dependent on the time at which the animal died. All brains had a staining pattern that was identical to that for a gI null virus, with strong staining evident only in the SCN and the IGL. The staining pattern in brains of animals infected with the revertant viruses was identical to the pattern for animals infected with wild-type virus (data not shown).
FIG. 6.
Localization of viral antigen in brain sections. Animals were infected with the indicated viruses by intraocular injection as described in Materials and Methods. Upon signs of imminent death, the animals were sacrificed and the brains were removed and analyzed for viral antigen with a polyvalent rabbit antiserum generated against whole virus particles (Rb133). Serial sections (35 μm) through the coronal plane were cut, processed, and mounted on slides. Representative sections containing the SCN, LGN, including the dorsal and ventral aspects as well as the IGL and the SC are pictured. (A) Brain slices from an animal infected with PRV 108R taken at 70 h postinfection, shown as an example of a wild-type infection pattern. Representative brains from animals that were sacrificed at early (82.5 and 71.5 h) and late (100.5 and 92.5 h) times after infection with PRV 108 and PRV 109 are also shown. (B) Brain slices from animals that were sacrificed at the longest survival time after infection with PRV 110 (95 h) and PRV 111 (91 h).
TABLE 3.
Detection of viral antigen
| Virus | Timeb (h) | Infectiona
|
||||
|---|---|---|---|---|---|---|
| SCN | dLGN | IGL | vLGN | SC | ||
| PRV Be | 63.1ce | ++++ | ++++ | ++++ | ++++ | ++++ |
| PRV 108R | 67.2c | ++++ | ++++ | ++++ | ++++ | ++++ |
| PRV 109R | 66.8c | ++++ | ++++ | ++++ | ++++ | ++++ |
| PRV 110R | 58c | ++++ | ++++ | ++++ | ++++ | ++++ |
| PRV 98 | 83–88d | ++++ | − | ++++ | +/− | − |
| PRV 107 | 74–108d | ++++ | ++ | ++++ | ++ | +++ |
| PRV 26 | 74–123d | ++++ | ++ | ++++ | ++ | +++ |
| PRV 108 | 76.8 | ++++ | − | ++++ | +/− | +/− |
| 82.5 | ++++ | − | ++++ | +/− | +/− | |
| 84.3 | ++++ | − | ++++ | ++ | +/− | |
| 93 | ++++ | +++ | ++++ | +/− | +++ | |
| 100.5 | ++++ | ++++ | ++++ | ++++ | ++++ | |
| 103 | +++ | +++ | ++++ | ++++ | ++++ | |
| PRV 109 | 67 | ++++ | − | ++++ | − | − |
| 70.5 | ++++ | − | ++++ | +/− | − | |
| 71.5 | ++++ | − | ++++ | +/− | +/− | |
| 73.5 | ++++ | − | ++++ | +/− | + | |
| 75 | ++++ | − | ++++ | ++ | + | |
| 92.5 | ++++ | +++ | ++++ | +++ | ++++ | |
| PRV 110 | 71 | ++++ | − | ++++ | +/− | − |
| 83.8 | ++++ | + | ++++ | +/− | − | |
| 95 | ++++ | − | ++++ | +/− | − | |
| PRV 111 | 81.8 | ++++ | − | ++++ | ++ | − |
| 83.5 | ++++ | − | ++++ | +/− | − | |
| 83.8 | ++++ | − | ++++ | +/− | − | |
| 84 | ++++ | − | ++++ | +/− | − | |
| 91.3 | ++++ | − | ++++ | ++ | − | |
Scored from ++++ (extensive infection of all areas) to − (little to no infection of the area).
Time postinfection that the animal showed signs of imminent death and was sacrificed.
Average time to signs of imminent death.
Range in which animals showed signs of imminent death.
From reference 36.
Virulence of truncation mutants.
By measuring the mean time to signs of imminent death after infection by various mutants, we assessed the contribution of the gI cytoplasmic domain to virulence (6, 7). As shown in Table 4, we found that animals infected with all of the revertant viruses (PRV 108R, PRV 109R, and PRV 110R) were identical to animals infected with wild-type virus, with times to signs of imminent death of 67.3, 64.6, and 58.0 h, respectively. By contrast, animals infected with PRV 98 (gI null) or PRV 91 (gE null) lived to extended times postinfection (78.7 and 79.5 h). We observed that animals infected with mutants expressing anchored gI (PRV 108) or secreted gI (PRV 109) showed mean times to signs of imminent death at 89.9 and 75.0 h. More animals will need to be analyzed to determine if these numbers are statistically different from one another. In addition, we found that infections with the double-truncation mutants PRV 110 (anc gE/anc gI) and PRV 111 (sec gE/sec gI) were similar to those obtained with the single-truncation mutants (83.3 and 84.7 h). We observed a large standard deviation in these measurements with some viruses, as shown in Table 3. We have noted this previously with various attenuated viruses (36). Not only did animals infected with all of these mutant viruses live to extended times after infection, they also displayed mild symptoms typical of infection with an attenuated virus.
TABLE 4.
Virulence of truncation mutantsa
| Virus | No. of animals | Mean time to symptoms (h) ± SD | Symptoms |
Pb with respect to:
|
||
|---|---|---|---|---|---|---|
| PRV Be | PRV 91 | PRV 98 | ||||
| PRV Be | 3 | 63.3 ± 1.3c | Severed | NAe | <0.005 | <0.005 |
| PRV 108R | 3 | 67.3 ± 6.4 | Severe | 0.344 | 0.007 | 0.017 |
| PRV 109R | 4 | 64.6 ± 4.5 | Severe | 0.634 | <0.005 | <0.005 |
| PRV 110R | 3 | 58.0 ± 5.3 | Severe | 0.173 | <0.005 | <0.005 |
| PRV 91 | 6 | 79.5 ± 3.9 | Attenuated | <0.005 | NA | 0.761 |
| PRV 98 | 6 | 78.7 ± 4.5 | Attenuated | <0.005 | 0.761 | NA |
| PRV 108 | 6 | 89.9 ± 10.5 | Attenuated | <0.005 | 0.044 | 0.017 |
| PRV 109 | 6 | 75 ± 9.0 | Attenuated | 0.065 | 0.286 | 0.389 |
| PRV 110 | 3 | 83.3 ± 12.0 | Attenuated | 0.045 | 0.485 | 0.424 |
| PRV 111 | 5 | 84.9 ± 3.7 | Attenuated | <0.005 | 0.045 | 0.042 |
After infection, animals were monitored 24 h a day for signs of imminent death, at which point they were sacrificed and the time was recorded.
Determined by Student's t test.
From reference 36.
Virulence (degree of pathogenesis after rat eye infection) was measured in terms of mean time to death or appearance of symptoms. Typical severe symptoms of PRV disease in the rat include ruffling of the fur, nasal discharge, hunched posture, labored breathing, uncoordinated movement, and scratching or rubbing of face, particularly in the area surrounding the injected eye. Not only do these symptoms appear more rapidly after wild-type virus infection, but also they are markedly severe in intensity. Animals infected with mutant viruses lacking gE or gI live longer and exhibit milder symptoms that are delayed in onset (attenuated). However, all infected animals ultimately die.
NA, not applicable.
DISCUSSION
Previous studies have shown that expression of the gE ectodomain with wild-type gI is sufficient to mediate spread to areas not infected by a gE or gI null virus (36, 38). The experiments in this report were initiated to determine if the gI cytoplasmic domain is dispensable for anterograde transmission of virus in the rat CNS. To test this idea, we constructed viral mutants that expressed the ectodomain of gI (anchored or secreted gI) in combination with wild-type gE. We also constructed double mutants that expressed either anchored gI and anchored gE or secreted gI and secreted gE. We hypothesized that if the gE and gI ectodomains functioned in concert, then the cytoplasmic domains of both proteins would be dispensable for spread. Consequently, all of these viruses would have the same phenotype as a virus expressing either anchored or secreted gE in combination with wild-type gI. The results that we obtained, however, suggest that the actions of gE and gI are more complicated than just the action of a single complex. These results are summarized in Table 5. Our data suggest that gE and gI may play different roles in mediating spread in the CNS. In addition, we discovered that the cytoplasmic domain of gI is required for full virulence and that it is responsible for efficient posttranslational N-linked glycosylation of the gI ectodomain.
TABLE 5.
Summary of gE and gI mutantsa
| Virus | Expressed protein
|
Spread MDBK | Spread animals
|
Virulence | Endocytosis | ||
|---|---|---|---|---|---|---|---|
| gE | gI | Early | Late | ||||
| PRV Be | WT | WT | +++ | +++ | +++ | +++ | +++ |
| PRV 91 | Null | WT | − | − | − | − | ND |
| PRV 98 | WT | Null | − | − | − | − | ND |
| PRV 107 | Anc | WT | − | ++ | ++ | − | − |
| PRV 25 | FS | WT | − | ++ | ++ | − | − |
| PRV 26 | Sec | WT | + | ++ | ++ | − | − |
| PRV 108 | WT | Anc | ++ | − | ++ | − | +++ |
| PRV 109 | WT | Sec | ++ | − | ++ | − | ND |
| PRV 110 | Anc | Anc | − | − | − | − | − |
| PRV 111 | Sec | Sec | + | − | − | − | ND |
Phenotypes are scored based on similarity to either a wild-type (WT) virus (+++) or a gE or gI null virus (−). ND, not determined; Anc, anchored; FS, Sec, secreted.
Our in vitro analysis showed that the truncated gI mutants were expressed to high levels, maintained the ability to complex with gE, and were not significantly altered in their steady-state localization within the infected cell. This finding proves that interaction between PRV gE and gI does not require the cytoplasmic domain of either protein but requires only the ectodomains of both of the proteins, as has been suggested for other alphaherpesviruses (2, 29). In addition, we observed no difference in endocytosis of the complex in cells infected with PRV 108 (anc gI) compared to wild-type virus. Endocytosis in cells infected with PRV 110 (anc gI/anc gE) was indistinguishable from that for cells infected with the single-anchored gE mutant (PRV 107) (36). Clearly the gI cytoplasmic domain has no effect on directing endocytosis of the complex and that all of the signals necessary for directing endocytosis of the complex are encoded in the gE cytoplasmic domain.
We measured cell-to-cell spread in vitro by analyzing plaques formed by the various viruses. In this report, we found that PRV 108 (anchored gI) and PRV 109 (secreted gI) formed plaques that were not statistically different in size from plaques formed by wild-type virus, while a gI null virus formed plaques that were 85% of the wild-type size. Although the gE cytoplasmic domain is important for spread in this assay, the gI cytoplasmic domain must play only a minor role in promoting cell-to-cell spread in MDBK cells. When double mutants were analyzed, we found that the double-anchored mutant made plaques identical in size to those made by a gE null virus, while a double-secreted mutant made plaques indistinguishable from those made by a gI null virus. The combination of the mutations resulted in viruses that were more defective than viruses with single gE or gI truncation mutants. Therefore, gE and gI must not play equivalent roles in directing cell-to-cell spread.
The kinetics of gI processing was affected by loss of the cytoplasmic domain. This result seen in PRV 108-, PRV 109-, PRV 110-, or PRV 111-infected cells was striking and novel. It is difficult to imagine how the loss of the gI cytoplasmic domain could affect the rate of glycosylation of the ectodomains of the proteins. If the proteins were cotranslationally glycosylated, glycosylation would be just as efficient as seen with wild-type protein. A single stop codon after the sequences encoding the transmembrane domain of the protein should not affect cotranslational processes. The available data suggest, then, that wild-type gI protein must be “posttranslationally” glycosylated after complete translation and insertion of the protein into the ER. As the protein encoded by PRV 108 is lacking only the cytoplasmic domain of gI, the data suggest that the cytoplasmic domain of the protein must be translated or the rate of glycosylation of the ectodomain is reduced. Therefore, without the cytoplasmic domain, anchored and secreted gI proteins are not glycosylated as efficiently as wild-type protein and all five glycosylation events can be observed during the pulse-chase as a ladder of gI-specific proteins. The C-terminal domain of gI could affect the glycosylation rate of the ectodomain by altering the conformation of the protein or by binding to another protein.
Like the MDBK plaque size analysis, the in vivo data also show differences between the gI and gE truncation mutants. Previous experiments demonstrated that the N-terminal 428 amino acids of gE are sufficient to sponsor viral infection of all retinorecipient areas of the brain that wild-type virus infects (36, 38). However, unlike viruses expressing truncated proteins due to nonsense mutations in gE, PRV 108 (anc gI) and PRV 109 (sec gI) were not wild type in neuroinvasiveness to visual centers after rat retina infections. These mutants displayed a kinetic defect in infection of these areas. In contrast to PRV 107, 25, and 26, PRV 108 and 109 were not able to infect these regions unless the animals survived beyond 90 h postinfection. We conclude that the gI ectodomain is not sufficient for wild-type neuroinvasiveness and that the gI cytoplasmic domain also plays a modulatory role in mediating anterograde transneuronal transfer of virus. In addition to data obtained with the single-truncation mutants, we discovered that double-truncation mutants [PRV 110 (anc gI/anc gE) and PRV 111 (sec gI/sec gE)] were indistinguishable from gE or gI null mutants in infecting the visual centers. Together, these results suggest that neuroinvasiveness requires not only the gE and gI ectodomains but also one of the cytoplasmic domains of the proteins. We suggest that gE and gI are not equivalent in providing this function. The gI cytoplasmic domain provides better function in promoting spread to visual centers because mutants lacking the gI cytoplasmic domain spread slower while those lacking the gE cytoplasmic domain spread like wild-type virus. An argument could be made that the rate defect seen with PRV 108 and PRV 109 infections reflects the gI processing defect observed in vitro. However, this argument is unlikely based on infections with PRV 110 and PRV 111. Unlike PRV 108 and PRV 109, these viruses were defective in spread regardless of the time to death of the animal, yet the gI processing defect was identical to that seen with PRV 108 or PRV 109. If the in vivo phenotype reflected the tissue culture phenotype, then animal infections with PRV 110 and PRV 111 should have been identical to those obtained with PRV 108 and PRV 109.
We are currently considering two models for transsynaptic spread by which both ectodomains and at least one of the cytoplasmic domains are required. These models are similar to the models presented by Dingwell and Johnson (9) and Brideau et al. (4). In model 1, the gE and gI ectodomains must be targeted to the appropriate site to achieve efficient spread. While targeting information would reside in either of the cytoplasmic domains, the gI cytoplasmic domain would be more efficient at directing the complex. In support of this, we found that the gE and gI transmembrane and cytoplasmic domains could direct GFP to similar intracellular locations during a viral infection (data not shown). Targeting could be achieved through signals encoded by these domains or through the interaction of these domains with another protein (i.e., Us9). These signals would be redundant, encoded by both of the domains, and defects in targeting would not be seen until mutations of both were made. One drawback of this model is the fact that the gE and gI cytoplasmic domains share virtually no sequence or motif homologies. It is unlikely that these two domains could encode the same targeting motifs or ability to interact with the same protein. The second model is similar to the first in that the ectodomains of the proteins are still responsible for mediating spread. However, the ectodomains must be activated to mediate spread. Activation would be achieved primarily through the gI cytoplasmic domain. For example, the ectodomain of the gE-gI complex could change conformation due to the presence of the gI cytoplasmic domain. In the absence of the gE cytoplasmic domain (PRV 107, PRV 25, and PRV 26), the ectodomains are active and spread occurs. In the absence of the gI cytoplasmic domain (PRV 108 and PRV 109), activation occurs slowly and there is a rate defect in infecting visual centers. In the absence of both domains (PRV 110 and PRV 111), activation does not occur and spread is identical to that of the null mutants. We are currently exploring both of these possibilities.
Although neuroinvasiveness of PRV relies primarily on the gE and gI ectodomains and partly on the gI cytoplasmic domain, virulence of PRV clearly requires both the gE and the gI cytoplasmic domains. The gE and gI cytoplasmic domains are both required for an infected animal to exhibit severe symptoms and rapid death characteristic of infections with a wild-type virus. We suggest that both the gE and gI domains act in concert to mediate this expression of virulence. The mechanism by which the gE and gI cytoplasmic domains promote virulence remains to be elucidated. One intriguing possibility involves phosphorylation of the gE cytoplasmic domain. Perhaps phosphorylation of gE results in a signal transduction cascade culminating in new gene expression that is responsible for the induction of the symptoms that we score as virulence. The presence of the gI cytoplasmic domain may be required to control phosphorylation/dephosphorylation of the gE cytoplasmic domain. We know that in PK15 cells, gE is phosphorylated just as efficiently after infections with PRV 108 (anc gI) and PRV 109 (sec gI) or even after infection with a gI null (data not shown). It is not clear though, whether phosphorylation in these cultured cells reflects the phosphorylation that would occur in infected animals. Another possibility is that the gE and gI cytoplasmic domains cooperatively bind a cellular or viral protein. Binding of the protein by gE and gI would lead to the expression of the virulence phenotype. We are in the process of testing these ideas.
ACKNOWLEDGMENTS
We thank J. Goodhouse for help and advice with the confocal images. We also thank K. Bienkowska-Szewczyk and T. Ben-Porat for gE antisera, and we thank F. Hughson for reagents and protocols. Many thanks go to members of the Enquist lab and to B. Banfield for support and critical reading of the manuscript. R.S.T. also sincerely acknowledges P. Husak for providing pPH3.
This work was supported by NINDS grant 1RO133506 to L.W.E. and NIH grant 5T32GM07388 to R.S.T.
REFERENCES
- 1.Babic N, Klupp B, Brack A, Mettenleiter T C, Ugolini G, Flamand A. Deletion of glycoprotein gE reduces the propagation of pseudorabies virus in the nervous system of mice after intranasal inoculation. Virology. 1996;219:279–284. doi: 10.1006/viro.1996.0247. [DOI] [PubMed] [Google Scholar]
- 2.Basu S, Dubin G, Basu M, Nguyen V, Friedman H M. Characterization of regions of herpes simplex virus type 1 glycoprotein E involved in binding the Fc domain of monomeric IgG and in forming a complex with glycoprotein I. J Immunol. 1995;154:260–267. [PubMed] [Google Scholar]
- 3.Ben-Porat T, Kaplan A S. Molecular biology of pseudorabies virus. In: Roizman B, editor. The herpesviruses. New York, N.Y: Plenum Publishing Corp.; 1985. pp. 105–173. [Google Scholar]
- 4.Brideau A D, Card J P, Enquist L W. Role of pseudorabies virus Us9, a type II membrane protein, in infection of tissue culture cells and the rat central nervous system. J Virol. 2000;74:843–845. doi: 10.1128/jvi.74.2.834-845.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Card J P, Levitt P, Enquist L W. Different patterns of neuronal infection after intracerebral injection of two strains of pseudorabies virus. J Virol. 1998;72:4434–4441. doi: 10.1128/jvi.72.5.4434-4441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Card J P, Enquist L W. Neurovirulence of pseudorabies virus. Crit Rev Neurobiol. 1995;9:137–162. . (Erratum, 9:preceding 311.) [PubMed] [Google Scholar]
- 7.Card J P, Whealy M E, Robbins A K, Enquist L W. Pseudorabies virus envelope glycoprotein gI influences both neurotropism and virulence during infection of the rat visual system. J Virol. 1992;66:3032–3041. doi: 10.1128/jvi.66.5.3032-3041.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Card J P, Whealy M E, Robbins A K, Moore R Y, Enquist L W. Two alpha-herpesvirus strains are transported differentially in the rodent visual system. Neuron. 1991;6:957–969. doi: 10.1016/0896-6273(91)90236-s. [DOI] [PubMed] [Google Scholar]
- 9.Dingwell K S, Johnson D C. The herpes simplex virus gE-gI complex facilitates cell-to-cell spread and binds to components of cell junctions. J Virol. 1998;72:8933–8942. doi: 10.1128/jvi.72.11.8933-8942.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dingwell K S, Brunetti C R, Hendricks R L, Tang Q, Tang M, Rainbow A J, Johnson D C. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J Virol. 1994;68:834–845. doi: 10.1128/jvi.68.2.834-845.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dubin G, Frank I, Friedman H M. Herpes simplex virus type 1 encodes two Fc receptors which have different binding characteristics for monomeric immunoglobulin G (IgG) and IgG complexes. J Virol. 1990;64:2725–2731. doi: 10.1128/jvi.64.6.2725-2731.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubin G, Basu S, Mallory D L, Basu M, Tal-Singer R, Friedman H M. Characterization of domains of herpes simplex virus type 1 glycoprotein E involved in Fc binding activity for immunoglobulin G aggregates. J Virol. 1994;68:2478–2485. doi: 10.1128/jvi.68.4.2478-2485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enquist L W, Card J P. Pseudorabies virus: a tool for tracing neuronal connections. In: Lowenstein P R, Enquist L W, editors. Protocols for gene transfer in neuroscience: towards gene therapy of neurological disorders. New York, N.Y: John Wiley & Sons, Inc.; 1996. pp. 333–348. [Google Scholar]
- 14.Enquist L W, Dubin J, Whealy M E, Card J P. Complementation analysis of pseudorabies virus gE and gI mutants in retinal ganglion cell neurotropism. J Virol. 1994;68:5275–5279. doi: 10.1128/jvi.68.8.5275-5279.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Favoreel H W, Nauwynck H J, Van Oostveldt P, Mettenleiter T C, Pensaert M B. Antibody-induced and cytoskeleton-mediated redistribution and shedding of viral glycoproteins, expressed on pseudorabies virus-infected cells. J Virol. 1997;71:8254–8261. doi: 10.1128/jvi.71.11.8254-8261.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fuchs W, Rziha H J, Lukacs N, Braunschweiger I, Visser N, Lutticken D, Schreurs C S, Thiel H J, Mettenleiter T C. Pseudorabies virus glycoprotein gI: in vitro and in vivo analysis of immunorelevant epitopes. J Gen Virol. 1990;71:1141–1151. doi: 10.1099/0022-1317-71-5-1141. [DOI] [PubMed] [Google Scholar]
- 17.Jacobs L. Glycoprotein E of pseudorabies virus and homologous proteins in other alphaherpesvirinae. Arch Virol. 1994;137:209–228. doi: 10.1007/BF01309470. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs L, Mulder W A, Priem J, Pol J M, Kimman T G. Glycoprotein I of pseudorabies virus (Aujeszky's disease virus) determines virulence and facilitates penetration of the virus into the central nervous system of pigs. Acta Vet Hung. 1994;42:289–300. [PubMed] [Google Scholar]
- 19.Jacobs L, Mulder W A, Van Oirschot J T, Gielkens A L, Kimman T G. Deleting two amino acids in glycoprotein gI of pseudorabies virus decreases virulence and neurotropism for pigs, but does not affect immunogenicity. J Gen Virol. 1993;74:2201–2206. doi: 10.1099/0022-1317-74-10-2201. [DOI] [PubMed] [Google Scholar]
- 20.Johnson D C, Feenstra V. Identification of a novel herpes simplex virus type 1-induced glycoprotein which complexes with gE and binds immunoglobulin. J Virol. 1987;61:2208–2216. doi: 10.1128/jvi.61.7.2208-2216.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson D C, Frame M C, Ligas M W, Cross A M, Stow N D. Herpes simplex virus immunoglobulin G Fc receptor activity depends on a complex of two viral glycoproteins, gE and gI. J Virol. 1988;62:1347–1354. doi: 10.1128/jvi.62.4.1347-1354.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knapp A C, Enquist L W. Pseudorabies virus recombinants expressing functional virulence determinants gE and gI from bovine herpesvirus 1.1. J Virol. 1997;71:2731–2739. doi: 10.1128/jvi.71.4.2731-2739.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knapp A C, Husak P J, Enquist L W. The gE and gI homologs from two alphaherpesviruses have conserved and divergent neuroinvasive properties. J Virol. 1997;71:5820–5827. doi: 10.1128/jvi.71.8.5820-5827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kritas S K, Nauwynck H J, Pensaert M B. Dissemination of wild-type and gC-, gE-and gI-deleted mutants of Aujeszky's disease virus in the maxillary nerve and trigeminal ganglion of pigs after intranasal inoculation. J Gen Virol. 1995;76:2063–2066. doi: 10.1099/0022-1317-76-8-2063. [DOI] [PubMed] [Google Scholar]
- 25.Kritas S K, Pensaert M B, Mettenleiter T C. Invasion and spread of single glycoprotein deleted mutants of Aujeszky's disease virus (ADV) in the trigeminal nervous pathway of pigs after intranasal inoculation. Vet Microbiol. 1994;40:323–334. doi: 10.1016/0378-1135(94)90120-1. [DOI] [PubMed] [Google Scholar]
- 26.Kritas S K, Pensaert M B, Mettenleiter T C. Role of envelope glycoproteins gI, gp63 and gIII in the invasion and spread of Aujeszky's disease virus in the olfactory nervous pathway of the pig. J Gen Virol. 1994;75:2319–2327. doi: 10.1099/0022-1317-75-9-2319. [DOI] [PubMed] [Google Scholar]
- 27.Litwin V, Jackson W, Grose C. Receptor properties of two varicella-zoster virus glycoproteins, gpI and gpIV, homologous to herpes simplex virus gE and gI. J Virol. 1992;66:3643–3651. doi: 10.1128/jvi.66.6.3643-3651.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mettenleiter T C, Zsak L, Kaplan A S, Ben-Porat T, Lomniczi B. Role of a structural glycoprotein of pseudorabies in virus virulence. J Virol. 1987;61:4030–4032. doi: 10.1128/jvi.61.12.4030-4032.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mijnes J D, Lutters B C, Vlot A C, van Anken E, Horzinek M C, Rottier P J, de Groot R J. Structure-function analysis of the gE-gI complex of feline herpesvirus: mapping of gI domains required for gE-gI interaction, intracellular transport, and cell-to-cell spread. J Virol. 1997;71:8397–8404. doi: 10.1128/jvi.71.11.8397-8404.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mulder W, Pol J, Kimman T, Kok G, Priem J, Peeters B. Glycoprotein D-negative pseudorabies virus can spread transneuronally via direct neuron-to-neuron transmission in its natural host, the pig, but not after additional inactivation of gE or gI. J Virol. 1996;70:2191–2200. doi: 10.1128/jvi.70.4.2191-2200.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mulder W A, Jacobs L, Priem J, Kok G L, Wagenaar F, Kimman T G, Pol J M. Glycoprotein gE-negative pseudorabies virus has a reduced capability to infect second- and third-order neurons of the olfactory and trigeminal routes in the porcine central nervous system. J Gen Virol. 1994;75:3095–3106. doi: 10.1099/0022-1317-75-11-3095. [DOI] [PubMed] [Google Scholar]
- 32.Roizman B. Herpesviridae: a brief introduction. In: Knipe D M, Fields B N, editors. Fundamental virology. 2nd ed. New York, N.Y: Raven Press, Ltd.; 1991. pp. 841–847. [Google Scholar]
- 33.Ryan J P, Whealy M E, Robbins A K, Enquist L W. Analysis of pseudorabies virus glycoprotein gIII localization and modification by using novel infectious viral mutants carrying unique EcoRI sites. J Virol. 1987;61:2251–2257. doi: 10.1128/jvi.61.10.2962-2972.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rziha H J, Mettenleiter T C, Ohlinger V, Wittmann G. Herpesvirus (pseudorabies virus) latency in swine: occurrence and physical state of viral DNA in neural tissues. Virology. 1986;155:600–613. doi: 10.1016/0042-6822(86)90220-5. [DOI] [PubMed] [Google Scholar]
- 35.Strebel K, Beck E, Strohmaier K, Schaller H. Characterization of foot-and-mouth disease virus gene products with antisera against bacterially synthesized fusion proteins. J Virol. 1986;57:983–991. doi: 10.1128/jvi.57.3.983-991.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tirabassi R S, Enquist L W. Mutation of the YXXL endocytosis motif in the cytoplasmic tail of pseudorabies virus gE. J Virol. 1999;73:2717–2728. doi: 10.1128/jvi.73.4.2717-2728.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tirabassi R S, Enquist L W. Role of envelope protein gE endocytosis in the pseudorabies virus life cycle. J Virol. 1998;72:4571–4579. doi: 10.1128/jvi.72.6.4571-4579.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tirabassi R S, Townley R A, Eldridge M G, Enquist L W. Characterization of pseudorabies virus mutants expressing carboxy-terminal truncations of gE: evidence for envelope incorporation, virulence, and neurotropism domains. J Virol. 1997;71:6455–6455. doi: 10.1128/jvi.71.9.6455-6464.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whealy M E, Card J P, Robbins A K, Dubin J R, Rziha H J, Enquist L W. Specific pseudorabies virus infection of the rat visual system requires both gI and gp63 glycoproteins. J Virol. 1993;67:3786–3797. doi: 10.1128/jvi.67.7.3786-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wittmann G, Rziha H J. Aujeszky's disease (pseudorabies) in pigs. In: Wittmann G, editor. Herpesvirus diseases of cattle, horses and pigs. Boston, Mass: Kluwer; 1989. pp. 230–325. [Google Scholar]
- 41.Yang M, Card J P, Tirabassi R S, Miselis R R, Enquist L W. Retrograde, transneuronal spread of pseudorabies virus in defined neuronal circuitry of the rat brain is facilitated by gE mutations that reduce virulence. J Virol. 1999;73:4350–4359. doi: 10.1128/jvi.73.5.4350-4359.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zsak L, Zuckermann F, Sugg N, Ben-Porat T. Glycoprotein gI of pseudorabies virus promotes cell fusion and virus spread via direct cell-to-cell transmission. J Virol. 1992;66:2316–2325. doi: 10.1128/jvi.66.4.2316-2325.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuckermann F A, Mettenleiter T C, Schreurs C, Sugg N, Ben-Porat T. Complex between glycoproteins gI and gp63 of pseudorabies virus: its effect on virus replication. J Virol. 1988;62:4622–4626. doi: 10.1128/jvi.62.12.4622-4626.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]