Abstract
The entry of vaccinia virus (VV) into the host cell results in the delivery of the double-stranded DNA genome-containing core into the cytoplasm. The core is disassembled, releasing the viral DNA in order to initiate VV cytoplasmic transcription and DNA replication. Core disassembly can be prevented using the VV early transcription inhibitor actinomycin D (actD), since early VV protein synthesis is required for core uncoating. In this study, VV intracellular cores were accumulated in the presence of actD and isolated from infected cells. The content of these cores was analyzed by negative staining EM and by Western blotting using a collection of antibodies to VV core and membrane proteins. By Western blot analyses, intracellular actD cores, as well as cores prepared by NP-40–dithiothreitol treatment of purified virions (NP-40/DTT cores), contained the core proteins p25 (encoded by L4R), 4a (A10L), 4b (A3L), and p39 (A4L) as well as small amounts of the VV membrane proteins p32 (D8L) and p35 (H3L). While NP-40/DTT cores contained the major putative DNA-binding protein p11 (F17R), actD cores entirely lacked this protein. Labeled cryosections of cells infected for different periods of time in the presence or absence of actD were subsequently used to follow the fate of VV core proteins by EM. These EM images confirmed that p11 was lost at the plasma membrane upon core penetration. The cores that accumulated in the presence of actD were labeled with antibodies to 4a, p39, p25, and DNA at all times examined. In the absence of the drug the cores gradually lost their electron-dense inner part, concomitant with the loss of p25 and DNA labeling. The remaining core shell still labeled with antibodies to p39 and 4a/4b, implying that these proteins are part of this structure. These combined data are discussed with respect to the structure of VV as well as core disassembly.
Vaccinia virus (VV), the prototype of the poxvirus family, contains a double-stranded DNA genome of 190 kb encoding over 200 proteins, of which about 100 make up the brick-shaped particle (10). Poxviruses are unique in that both transcription and DNA replication occur in the host cell cytoplasm since the virus encodes for its own enzymes required for these two processes (26).
Assembly of VV starts with the formation at 5 to 6 h postinfection (p.i.) of typical crescent-shaped membranes modified by viral membrane proteins. We and others have shown that these membranes are derived from the smooth endoplasmic reticulum (ER)/intermediate compartment and are composed of two tightly apposed cisternal membranes (32, 39, 46; see reference 12 for a different interpretation). Consistently, we have shown that a number of viral membrane proteins are cotranslationally inserted into the rough ER and transported to and retained in the intermediate compartment in infected cells (21, 34). These crescents then form the completely spherical immature viruses (IVs) composed of the two cisternal membranes and an electron-dense central part that contains the viral core proteins (5, 9, 38, 42). When these spherical IVs take up the DNA, they undergo a complex series of morphological changes resulting in the brick-shaped intracellular mature virus (IMV), the first of the two infectious forms of VV. Morphologically, this transition results in the formation of the (quasi-brick-shaped) core structure, while at the biochemical level it coincides with the cleavage of at least three of the major core proteins (19, 20, 27, 43, 44, 47).
The molecular details that underly the formation as well as the structure of this quasi-brick-shaped core are poorly understood. Recombinant viruses have been generated in which the expression of specific core proteins is regulated by inducible promoters (29, 45, 48). The information obtained from such genetic studies has, however, been limited since in the absence of specific core proteins, assembly was either blocked at the IV stage (29, 48) or generated IMVs that appeared morphologically indistinguishable from wild-type virus (45).
Other approaches to study the core structure have included a systematic disassembly of intact, purified IMV. Treatment of IMVs with a mixture of the detergent NP-40 and a reducing agent has been used to chemically prepare VV cores (8, 37). In fact, VV proteins are classically defined as core proteins when they remain associated with pelletable particles after treatment of the IMV with detergent and reducing agents. The major components of such cores are the gene products of A10L (4a), A3L (4b), A4L (p39), L4R (p25), and F17R (p11) as well as the viral genome and the enzymes required for early transcription (15).
A final approach aimed at understanding the core structure has been to study intermediates of uncoating upon VV infection. Although VV entry is poorly understood (see Discussion), it is generally accepted that its final result is the delivery of the viral core (seemingly indistinguishable from chemically prepared cores) into the cytoplasm (6). Studies by Sarov and Joklik (36) and by Holowczak (13) have shown that several core uncoating intermediates, characterized by different sedimentation properties, different protein content, as well as different morphological appearances, can be distinguished in infected cells. However, these studies were performed before the VV genome was sequenced, and many of the proteins associated with these uncoating intermediates remain therefore unidentified. Other studies have shown that when the synthesis of VV early mRNAs and/or early proteins is blocked, a specific uncoating intermediate accumulates in infected cells (13, 17, 35). In this intermediate the viral genome is still protected from DNase digestion, suggesting that its release from the core requires the synthesis of at least one early viral protein (17, 30).
In this study, we have analyzed cores isolated from infected cells in the presence of actinomycin D (actD), an inhibitor of VV early transcription, and compared them to cores that had been prepared by treatment of the virus with NP-40 and dithiothreitol (NP-40/DTT cores). Moreover, in a detailed time course we have analyzed the fate of several typical core proteins in infected cells by electron microscopy (EM). The combined results have implications not only for VV uncoating but also for the structure of the IMV.
MATERIALS AND METHODS
Preparation of virus stocks.
HeLa cells (CCL2) were obtained from the American Type Culture Collection and grown in Dulbecco's modified Eagle medium (DMEM) containing penicillin and streptomycin and 5% heat-inactivated fetal calf serum (FCS). VV (Western Reserve strain) was propagated in HeLa cells as described before (15). The virus was semipurified by pelleting through a 36% (wt/vol) sucrose cushion in a Beckman SW40-Ti rotor at 24,000 rpm and 4°C for 30 min. Virus pellets were resuspended in 10 mM Tris-HCl (pH 9.0), aliquoted, and stored at −80°C. 35S-labeled IMV was prepared as described by Krijnse Locker and Griffiths (22). To prepare virus whose DNA genome was labeled with [3H]thymidine, cells were infected at a multiplicity of infection (MOI) of 3 to 5. After 2 h, 0.5% FCS and 25 μCi of [3H]thymidine (Amersham) per ml were added to the inoculum, and the virus was isolated 3 days later. All virus stocks were plaque titrated on HeLa cells and were typically between 108 and 5 × 108 PFU per ml. By measuring the optical density at 260 nm (OD260) of a diluted virus preparation, the amount of particles per milliliter was calculated (15). By dividing this number by the PFU per milliliter as measured by plaque assay, the particle/PFU ratio was determined as 50. The incorporated amount of radioactivity was determined by spotting an aliquot of the virus preparation on filter paper, followed by trichloroacetic acid precipitation and liquid scintillation counting.
Antisera.
The antibodies to p16 (A14L), p8 (A13L), p21 (A17L), p11 (F17R), and anticore have been described before (15, 22, 34). Antibodies generously provided by colleagues were as follows: p14 (A27L [31]) and p39 (A4L [24]) (from M. Esteban); p25 (L4R) and 4a/4b (A10L/A3L [43, 44]) (from D. Hruby); and p32 (D8L [28]) (from E. Niles). The antibody recognizing p35 (H3L [4]) was made using a peptide corresponding to amino acids 31 to 43 of the H3L gene. Rabbits were immunized as described by Cudmore et al. (5). The anti-DNA immunoglobulin M (IgM) monoclonal antibody was from Boehringer GmbH (Mannheim, Germany).
Purification of viral and subviral structures from infected cells.
HeLa cells were grown to 80% confluency in 6-cm-diameter dishes (approximately 1.5 × 106 cells). The virus was thawed and sonicated at 35 kHz for 1 min at room temperature in a water bath sonicator. Usually, cells were infected with an MOI of between 50 and 100. Inocula were prepared in serum-free DMEM and were put on the cells for various periods of time (between 0.5 and 3 h), depending on the experiment. When actD (Sigma) was used, the drug was added together with the virus. At 3 h p.i., the inoculum was removed, and the cells were washed once with phosphate-buffered saline (PBS), and DMEM containing 0.5% FCS and 3 μg of actD was added. To isolate disassembled forms of VV, the cells were washed twice with PBS and put on ice. Subsequently, they were scraped from the dish in PBS and pelleted by low-speed centrifugation. Cell pellets were resuspended in 500 μl of 10 mM Tris-HCl (pH 9.0), and the cells were broken with a Dounce homogenizer (10 to 15 strokes). After removal of the nuclear fraction (by centrifugation in a Eppendorf microcentrifuge at 2,500 rpm for 5 min), the cytoplasmic fraction was directly loaded onto a linear 12 to 26% (wt/wt) sucrose gradient in 10 mM Tris-HCl (pH 9.0). Directly prior to loading, samples were sonicated for 1 min at 35 kHz in a water bath sonicator. The gradient was run in a Beckman SW40-Ti rotor at 15,000 rpm and 4°C for 25 min. Gradients were pumped out from the top using a peristaltic pump and fraction collector. The volume of the gradient (14 ml) was divided into 23 to 25 equal fractions. In some sedimentation analyses, NP-40/DTT cores were included for comparison.
ELISA on gradient fractions using an anticore antiserum.
To identify the position of the core peak in gradients loaded with unlabeled virus preparations, we developed an enzyme-linked immunosorbent assay (ELISA) using the anticore antibody. Twenty-five-microliter samples of gradient fractions were diluted twice in 10 mM Tris-HCl (pH 9.0). An ELISA plate was coated with these samples for 30 min at 37°C. After blocking with 0.5% gelatin in PBS, a 1:1,000 dilution of the anticore antiserum was added to the wells. After incubation with goat anti-rabbit coupled to horseradish peroxidase (HRP) (Bio-Rad) as second antibody, the ELISA was developed using a 3,3′,5,5′-tetramethylbenzidine-HRP detection kit (Pierce, KMF Laborchemie Handels GmbH, St. Augustin, Germany) and enhanced chemiluminescence (ECL) according to the manufacturer's instructions, and the OD450 of the wells was determined in an ELISA plate reader.
Immunolabeling of particles from entry experiments and preparation of cryosections.
The presence of various VV proteins on the surface of viral or subviral structures from cell lysates or gradient fractions was analyzed using a panel of VV antibodies. Samples (5 to 10 μl) were put on carbon-coated copper grids and incubated for 15 min at room temperature. After blocking with 10% FCS in PBS, a standard immunogold labeling procedure (9) was carried out, followed by negative staining using 0.3% uranyl acetate or 2% ammonium molybdate. For cryosections, HeLa cells grown to 70% confluency in 6-cm-diameter dishes were infected at an MOI of 200 as described elsewhere (22), using semipurified virus prepared as described above. The cells were fixed at the indicated times after infection by adding an equal volume of fixative to the medium, and cryosections were prepared and labeled essentially as described by van der Meer et al. (41).
Western blot of particles from entry experiments.
Using a Pierce bicinchoninic acid protein assay reagent, the protein concentration of the particles, of purified IMV and of NP-40/DTT cores was determined according to the instructions of the manufacturer. About 800 ng of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 15% gel, transferred to nitrocellulose, and detected as described elsewhere (9).
RESULTS
Biochemical isolation of natural cores.
To study intracellular cores, we designed a method to isolate them from infected cells. For this, we prepared IMV in which either the DNA or proteins had been labeled by using [3H]thymidine or [35S]methionine/cysteine, respectively. The cells were broken in a hypotonic buffer by using a Dounce homogenizer, and only mild sonification was used to dissolve aggregates. Subsequently, the samples were loaded onto a sucrose gradient and analyzed by biochemical and EM methods.
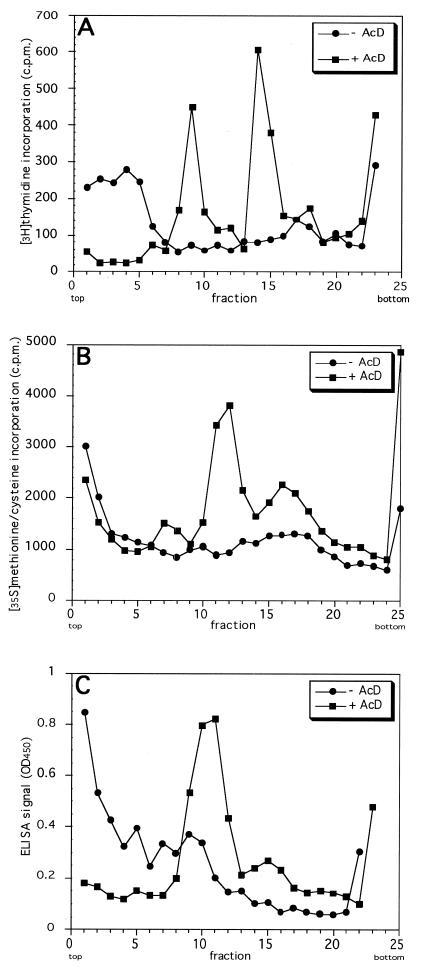
Distributions of the various types of radiolabel in sucrose gradients from a typical experiment are shown in Fig. 1A and B. A large amount of the labeled virus and/or disassembly intermediates remained at the top of the gradient, possibly due in part to aggregation with cellular membranes. Although intact virus particles and cores were seen by EM in fractions from the central part of the gradient, the separation of these two structures and the reproducibility of these experiments were not satisfactory. Therefore, we decided to let disassembly intermediates accumulate by using the drug actD, a transcription inhibitor which is known to block VV uncoating (17, 25).
FIG. 1.
Separation of viral and subviral components from VV-infected cells on sucrose gradients. HeLa cells were infected at an MOI of 50 to 100 with virus that had been metabolically prelabeled with [3H]thymidine (A) or [35S]methionine/cysteine (B) or was unlabeled (C). Cells were either infected for 3 h in the absence of actD (−AcD) or infected in the presence of 3 μg of actD per ml (+AcD), after which the infection was continued for 3 h in the presence of the drug. Cell lysates were prepared as described in Materials and Methods and separated on 12 to 26% (wt/vol) sucrose gradients. When radioactively labeled virus was used (A and B), the location of viral and subviral components in the gradient was determined by liquid scintillation counting. When unlabeled virus was used (C), the location of the viral cores in the gradient was determined by ELISA using the anticore antibody (see Materials and Methods).
Radiolabeled virus was added to HeLa cells in the presence or absence of 3 μg of actD per ml. The cells infected in the absence of the drug were harvested at the end of the absorption period, while those infected in the presence of actD were incubated for another 3 h in the presence of the drug before being harvested and analyzed. The use of actD had a remarkable effect. When either [3H]thymidine- or [35S]methionine/cysteine-labeled virus was used, the large amount of label at the top of the gradient was no longer seen (Fig. 1A and B). Instead, two clear peaks were detected in the central region of the gradients. Comparison with the sedimentation behavior of intact IMV and artificially prepared NP-40/DTT cores revealed that the material in these two peaks cosedimented with complete virus particles (around fraction 15) and core structures (around fraction 10), respectively (data not shown). The relatively long incubation times in the presence of actD (3 h of absorption and 3 h of infection) were required to resolve the distinct (virus- and core-containing) peaks. Shorter or longer times of infection in the absence of actD did not affect the pattern (no detectable peaks with the bulk of the material remaining at the top of the gradient) obtained after 3 h of absorption, while shorter incubations in the presence of actD did not result in discrete core and virion peaks (not shown).
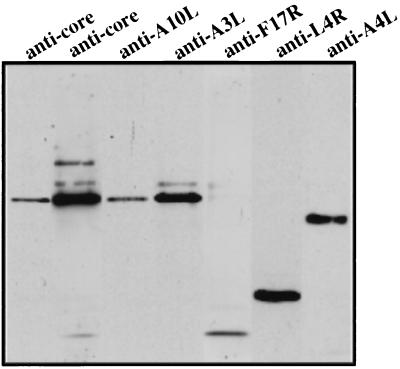
To confirm the nature of the peaks, we developed an ELISA using a rabbit antiserum that had been raised against cores prepared by NP-40/DTT stripping of purified IMV. By Western blotting this antibody recognizes predominantly a 65-kDa band representing the mature forms of 4a and 4b (A10L and A3L, respectively) and weakly p11 (F17R) (Fig. 2), but no other viral proteins. For simplicity, this antibody will further be referred to as anticore antibody. Undenatured material from gradient fractions was used as antigen to coat the wells of an ELISA plate. As expected, the ELISA produced a strong signal only in the wells coated with material from the putative core peak (Fig. 1C), not in wells containing intact virus in which the core antigens should not be accessible to the antiserum.
FIG. 2.
Western blot showing that the anticore antibody predominantly recognizes a 65-kDa band. Proteins of purified IMV were separated by SDS-PAGE on a 10% gel and blotted onto nitrocellulose. Core proteins were detected with antibodies to 4a (anti-A10L), 4b (anti-A3L), p11 (anti-F17R), p25 (anti-L4R), and p39 (anti-A4L), followed by anti-rabbit coupled to HRP and ECL. The two leftmost lanes were probed with the anticore antibody. Upon short exposure (the leftmost lane), the antibody detects a 65-kDa band only. Since the cleaved forms of 4a and 4b cannot be resolved by SDS-PAGE, it is not clear whether the antibody reacts with the mature form of 4a or 4b. Upon longer exposure (second lane from the left), however, the antibody does detect the uncleaved forms of 4a and 4b (of which a variable fraction is always present in purified IMV preparations) as well as the 11-kDa F17R gene product.
Characterization of core structures by immunolabeling.
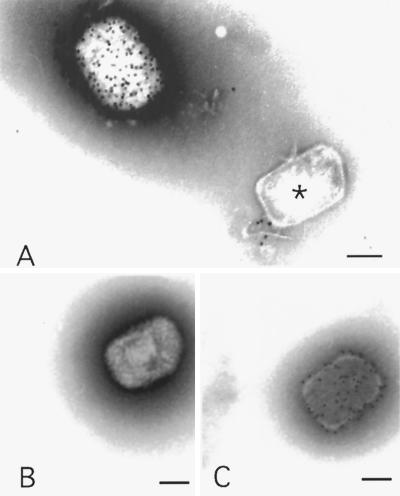
Postnuclear supernatants (PNS) from infected cells treated with actD or gradient fractions were analyzed by negative staining EM. In total PNS we detected only two distinct structures, which appeared to represent intact virus particles and cores (Fig. 3). Later examination of the two peaks in the various gradients revealed some cross-contamination, but in general the material in the virus and core peaks was relatively pure.
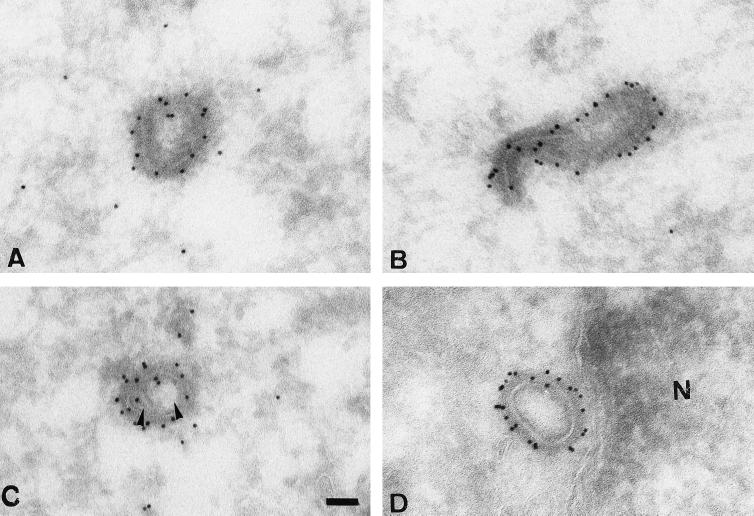
FIG. 3.
Negative staining EM and immunolabeling of intact IMVs and actD cores. (A) Labeling with anti-p14. The upper left shows a heavily labeled IMV; in the lower right an unlabeled core is evident. (B and C) labeling with anticore, showing an unlabeled IMV (B) and a labeled actD core (C). Bars = 100 nm.
To characterize the two structures in more detail, we used a set of specific antisera against typical VV core and membrane proteins to label the surface of virus and cores, respectively, and evaluated labeling by negative staining EM. In this approach, only antigens on the surface of the particle have access to antibodies. The results are summarized in Table 1, while Fig. 3 shows examples of such labeling by negative staining EM. We detected intact IMVs heavily labeled for the surface protein p14 but no labeling with this antibody on intracellular cores (Fig. 3A). Conversely, the anticore antibody detected abundant labeling on the cores (Fig. 3C), while the IMVs were unlabeled (Fig. 3B).
TABLE 1.
Quantitation of immunolabeling with antibodies to VV core and membrane proteins on purified IMVs and isolated intracellular cores
| Antibody (gene) | Classificationa (reference[s]) | Labeling (avg ± SD)b
|
|
|---|---|---|---|
| IMV | Cores | ||
| Anticore | Core | 1 ± 1 | 42 ± 18 |
| p21 (A17L) | Membrane (32) | 24 ± 5 | 0 |
| p16 (A14L) | Membrane (32) | 0 | 0 |
| p32 (D8L) | Membrane (28) | 28 ± 6 | 5 ± 2 |
| P35 (H3L) | Membrane (4) | 0 | 9 ± 4 |
| p8 (A13L) | Membrane (34) | 37 ± 9 | 0 |
| p14 (A27L) | Membrane (31) | 79 ± 19 | 0 |
| p11 (F17R) | Core (15, 18) | 0 | 0 |
| p39 (A4L) | Core + membrane (5) | 0 | 6 ± 2 |
| p25 (L4R) | Core (15, 47) | 0 | 2 ± 2 |
Based on observations made with cores prepared by NP-40/DTT treatment.
Average amount of gold particles per IMV or intracellular core from postnuclear supernatant of HeLa cells infected in the presence of actD (n = 20).
Intact IMVs labeled as expected; the surface labeled significantly with antibodies to the membrane proteins p8 (A13L [34]) and p32 (D8L [40]) and showed moderate levels of labeling with antibodies to the cleaved N terminus of p21 (A17L) (Table 1). No p16 (A14L) labeling was detected, consistent with previous data showing that this membrane protein is part of the inner of the two VV membranes (32, 34). Poor labeling was also detected with antibodies to the IMV membrane protein p35 (H3L [4]), suggesting that the epitope recognized by our antibody may not be exposed on the surface of the IMV. Finally, as expected, no labeling was found on intact IMVs with antibodies to core proteins (Table 1). On the VV cores, no labeling of the membrane proteins p21, p8, and p16 was detected. Surprisingly, low but significant labeling was observed for the putative membrane proteins p32 and p35 (Table 1). The core proteins p39 and 4a/4b (labeled with anticore) were detected on the surface of the cores, but no labeling was seen with antibodies to p11 (F17R) and p25 (L4R), suggesting that these proteins might be buried inside the core (see below).
Characterization of viral and subviral structures by Western blotting.
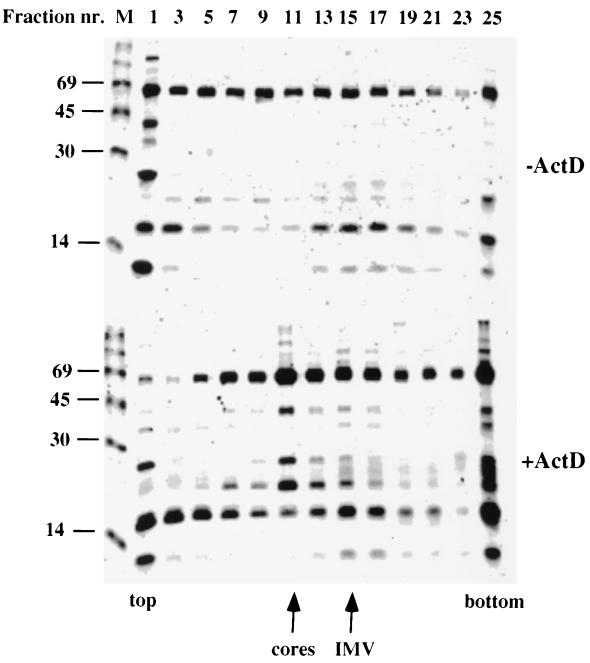
To investigate the protein content of the isolated core fractions, we performed an SDS-PAGE analysis of the gradient fractions from Fig. 1B in which 35S-labeled virus was used. Again, when analyzing the fractions of cells infected in the absence of actD, we could detect no clear pattern. In the presence of actD, however, a number of viral proteins were specifically concentrated in the core peak compared to the virus-containing fractions (Fig. 4). Among those was the characteristic doublet of 65 kDa, most likely representing the major core proteins 4a and 4b. Four other abundant proteins were clearly present in the core peak fractions, one migrating at about 40 kDa (most likely p39) and three with molecular masses between 15 and 28 kDa. Surprisingly, a ≈11-kDa band, that most likely corresponded to the major core protein p11, was depleted from the core fraction while clearly present in the virus-containing peak (Fig. 4; see below).
FIG. 4.
35S-labeled pattern of subviral components isolated from VV-infected cells separated on sucrose gradients. Gradient fractions from Fig. 1B were separated by SDS-PAGE on a 15% gel, which was then processed for autoradiography. The upper panel represents gradient fractions from untreated (-ActD) cells, while the lower panel is from treated (+ActD) cells. The top and bottom of the gradients, as well as positions of the core and IMV peak, are indicated. In each case, fractions with even and odd numbers were pooled such that, e.g., fraction 1 denotes the combined radioactive pattern of fractions 1 and 2. M, 14C-labeled markers of 14, 30, 45, and 69 kDa.
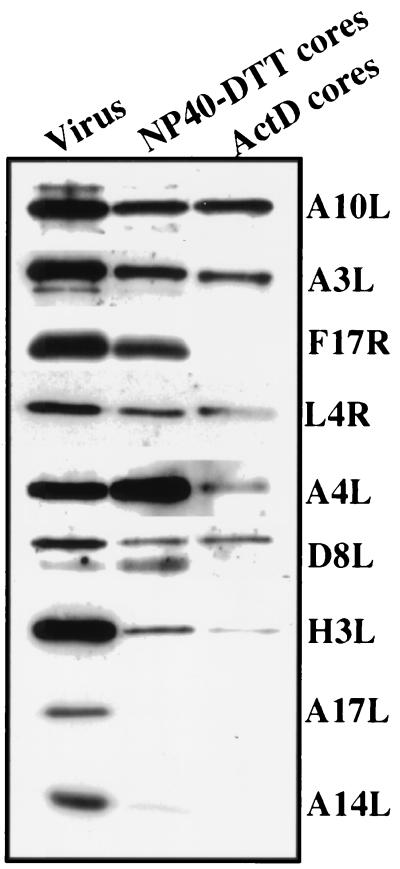
To more firmly establish the protein content of the subviral structures, we subjected them to Western blotting using a variety of antibodies to membrane and core proteins of the IMV. The protein pattern was compared to those of pure IMV preparations and of NP-40/DTT cores. The latter cores behaved as expected, with a total absence of the typical membrane proteins p16 and p21, while p39, 4a, and 4b as well as p25 and p11 were clearly present (Fig. 5). The subviral particles isolated from infected cells also contained the typical core proteins 4a, 4b, p39, and p25 but not p16 and p21 (Fig. 5). Consistent with the SDS-PAGE analysis, these structures appeared to lack the major putative DNA-binding protein p11 (18). Finally, in agreement with the negative staining results, small amounts of the IMV membrane proteins p35 and p32 were found on both the NP-40/DTT and actD cores (Fig. 5; see Discussion).
FIG. 5.
Detection by Western blotting of several VV core and membrane proteins in purified IMV (Virus), NP-40/DTT cores, and actD cores. In each lane of an SDS–15% polyacrylamide gel, 800 ng of purified IMV, NP-40/DTT cores, or actD cores was loaded and after electrophoresis blotted onto nitrocellulose. VV core and membrane proteins were detected by Western blotting using ECL.
Characterization of VV cores on cryosections.
The experiments described above suggested that the cores that had accumulated in the presence of actD were devoid of one of the most abundant core proteins. This could be explained in three possible ways: p11 was (i) lost upon VV entry, (ii) lost from the core intracellularly upon exposure to the cytoplasm, or (iii) artifactually lost during the isolation procedure. To address this issue, we immunolabeled cryosections prepared from cells infected for different periods of time in either the presence or absence of actD. Using antibodies to selected core proteins, labeling was quantified on IMVs that had remained outside at the plasma membrane and on intracellular cores. The data (Table 2) show several surprising results.
TABLE 2.
Quantitation of immunolabeling on IMVs and intracellular cores with antibodies to VV core proteins and to DNA on cryosections
| Treatment | Time of infection (min)a | Labeling (avg ± SD)b
|
||||
|---|---|---|---|---|---|---|
| Anticorec | p39 | p11 | p25 | Anti-DNA | ||
| −ActD | Cores | |||||
| 60 | 13 ± 5 | 12 ± 3 | 0 | 5 ± 2 | 14 ± 8 | |
| 120 | 13 ± 5 | 8 ± 3 | 0 | 5 ± 3 | 4 ± 4 | |
| 180 | 5 ± 3 | 5 ± 3 | 0 | 1 ± 1 | 1 ± 1 | |
| IMV | 18 ± 6 | 12 ± 4 | 12 ± 3 | 6 ± 2 | 9 ± 3 | |
| +ActD | Cores | |||||
| 60 | 16 ± 5 | 10 ± 3 | 0 | 7 ± 2 | 21 ± 6 | |
| 120 | 12 ± 4 | 8 ± 3 | 0 | 6 ± 2 | 17 ± 7 | |
| 180 | 16 ± 5 | 9 ± 4 | 0 | 4 ± 3 | 18 ± 6 | |
| IMV | 19 ± 6 | 10 ± 3 | 9 ± 3 | 5 ± 3 | 11 ± 5 | |
Labeling on cryosections from cells infected in the absence (−ActD) or presence (+ActD) of actD.
Quantitation of labeling with antibodies to core proteins on intracellular cores accumulated for the indicated times of infection and on extracellular IMVs at 60 min of infection.
Average amount of labeling on 20 intracellular cores or on IMVs.
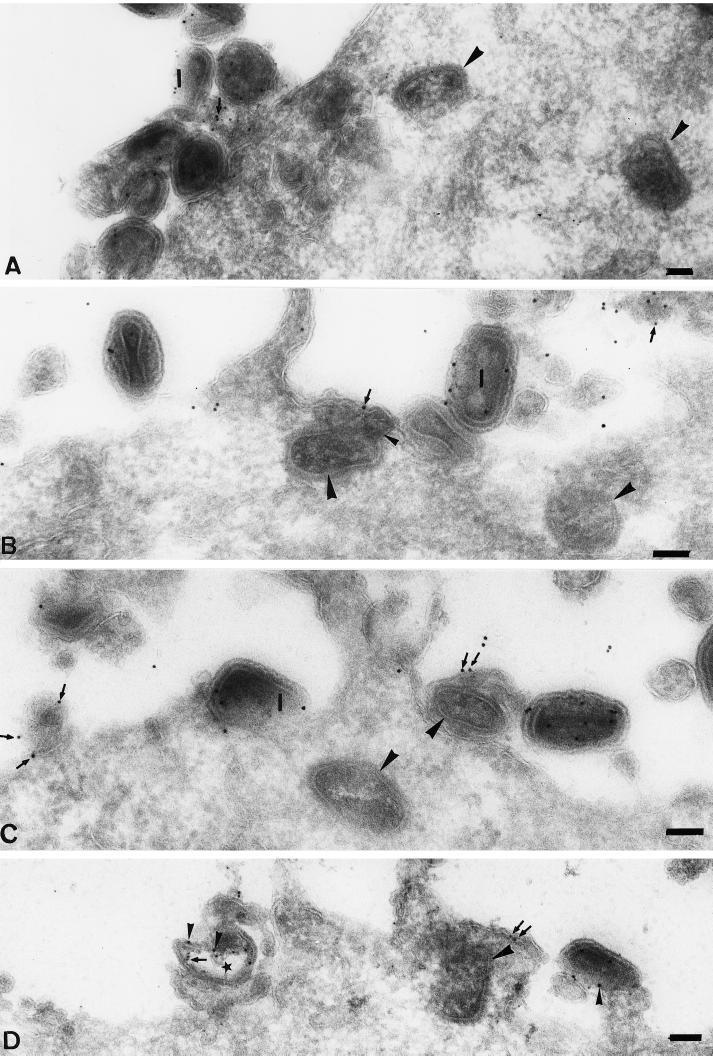
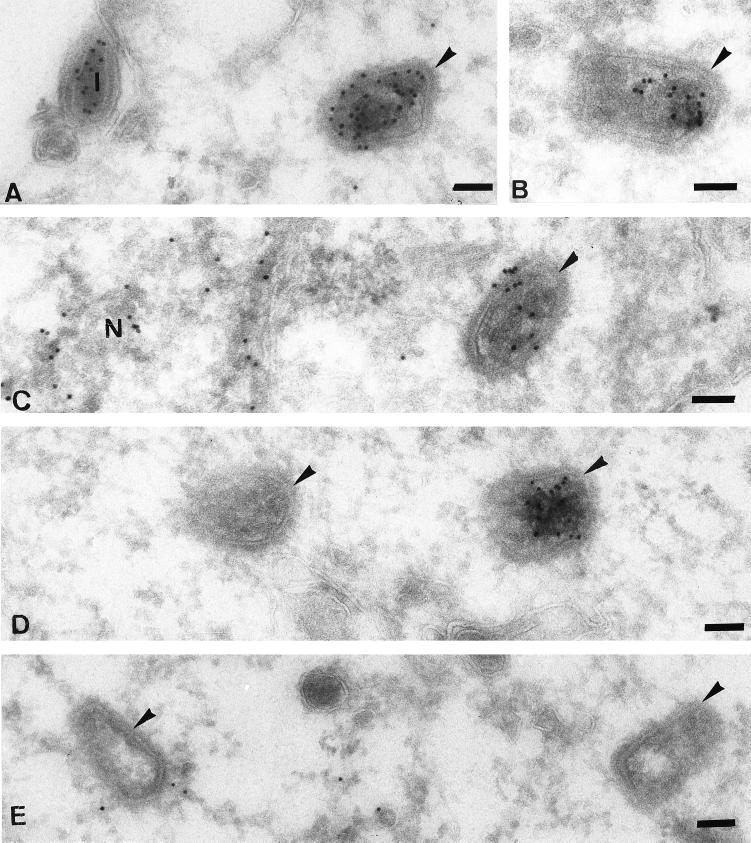
First, we observed that, in agreement with the Western blot results, the intracellular cores were entirely devoid of p11 labeling (Fig. 6B to D). Consistently, on extracellular IMVs the labeling for p11 appeared to be associated more with the viral membranes than with the core (Fig. 6B and C). Many images suggested that p11 separated from the core at the plasma membrane (Fig. 6B to D), leaving what appeared to be p11-positive viral membranes at the cell surface (Fig. 6D). The images shown in Fig. 6 also revealed a number of striking features of the entry process and the fate of the viral core. First, we found that many of the virions at the plasma membrane looked significantly different from normal IMVs (Fig. 6C, 7B and C, and 9A). Although the significance of these morphological changes is not clear at present, by negative staining EM of our purified virus preparations such particles were never observed, making it unlikely that they represented damaged virions contained in the inoculum (not shown). Second, Fig. 6B and D show what we believe to be putative entry intermediates, supporting the idea that the viral membranes as well as the nonmembrane protein p11 remain outside while the core crosses the membrane.
FIG. 6.
P11 is absent from intracellular cores. The cryosections, all from HeLa cells infected for 30 min in the absence of actD, show intracellular cores (large arrowheads) that are devoid of p11 labeling; extracellular IMVs (I) are labeled. Note that the bulk of the labeling on the IMVs is excluded from the interior, nucleoid region. (B to D) Putative entry intermediates in which the cores are separated from the outer membranes and p11 remains extracellularly. The small arrowhead in panel B shows a possible connection between the viral membranes and the core. (B and C) Membrane fragments (small arrows) at the plasma membrane that label for p11; (B and D) such fragments adjacent to an incoming core; (D) double labeled for p11 (10-nm gold; small arrowheads) and p16 (5-nm gold; small arrows), showing a viral membrane fragment (star) labeled for both markers that is outside the cell. Bars = 100 nm.
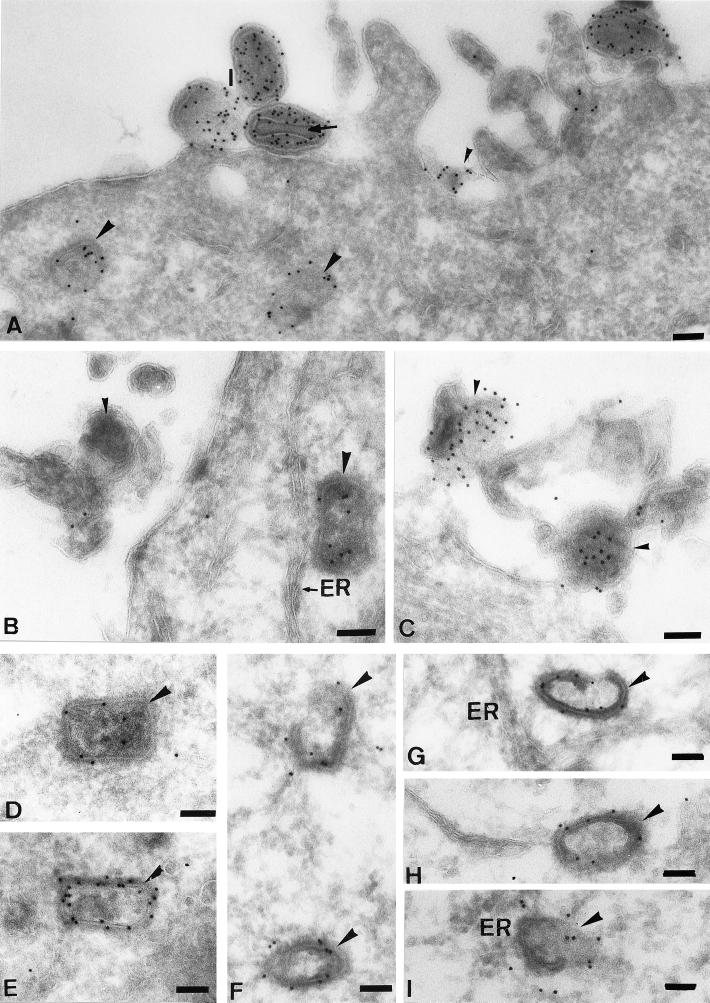
FIG. 7.
Labeling of intracellular cores with the anticore antibody. In all images, the intracellular cores are indicated by large arrowheads. (A) From 60-min infection in the presence of actD, showing labeling of two cores and four extracellular virions (I). The small arrowhead shows an extracellular membrane fragment labeled for anticore. Note that in the IMVs the labeling is excluded from the central nucleoid region (arrow in panel A). (B and C) From 90-min infection in the absence of actD. Small arrowheads show extracellular virions that have dramatically changed their shape upon encountering the plasma membrane. The intracellular cores are often close to the ER (B, G, and I). (D and E) From 120- and 60-min infections, respectively, in the presence of actD. (F through I) At 90 min postinfection in the absence of the drug, showing the appearance of full (D and E) versus electron-lucent (F to I) cores that all label with anticore. The images in panels F and G also show what appears to be an opening in the core shell. Bars = 100 nm.
In cells treated with actD, the typical core proteins p39, p25, 4a/4b (labeled using the anticore antibody) as well as the DNA (labeled with a monoclonal antibody to DNA) remained mostly associated with the cores (Table 2). In general anti-p39 and anticore tended to label primarily the outer rim of the cores (Fig. 7A, 7E to H, and 8), while anti-DNA and p25 labeled mostly the inner, central part (Fig. 9A to E and 10A to D). In some sections this inner part occasionally revealed what appeared to be the viral nucleoid (Fig. 9B to E and 10A to D). Cores that accumulated in the absence or presence of actD often tended to be located close to the ER or, occasionally, the nuclear envelope (Fig. 7B, 7G, 7I, 8D, 9H, and 10D).
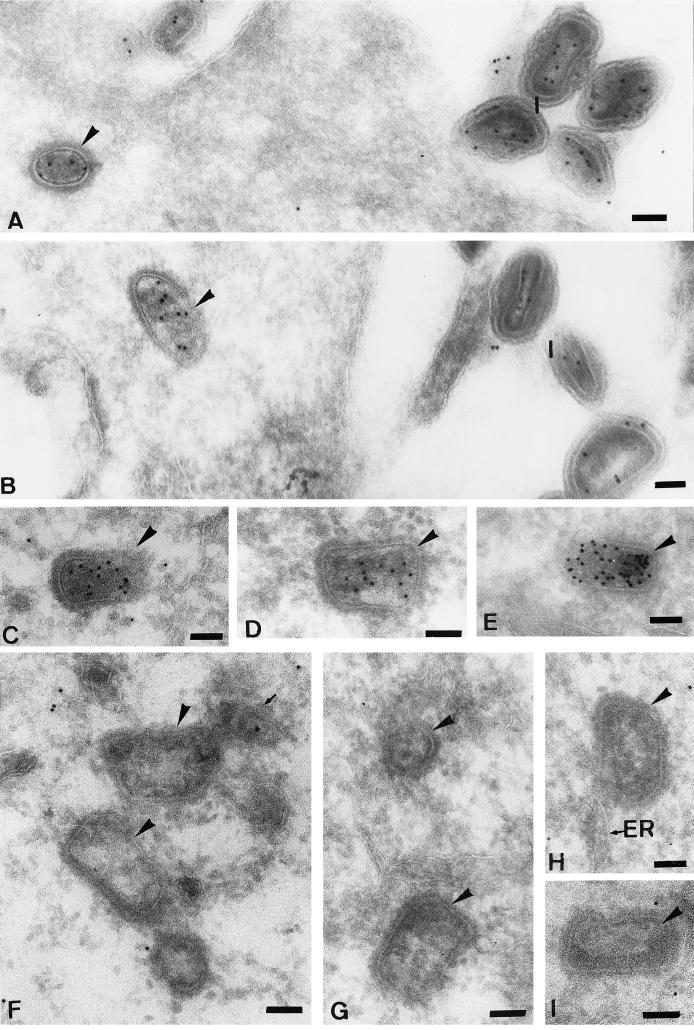
FIG. 9.
P25 labeling of intracellular cores and IMVs. (A and E) From cells at 30 min after infection; (B) 60 min of infection (all in the absence of actD) (C) 180 min in the presence of actD; (D and F through I) from cells infected for 90 min in the absence of the drug. (A through E) typical p25 localization to intracellular cores, in which the inner electron-dense part is heavily labeled. In the extracellular virions (I) in panels A and B, p25 also mostly labels the most central nucleoid part. At later times of infection in the absence of actD (F to I), the intracellular cores are devoid of p25 labeling, while this labeling is not lost in the presence of actD (C). Some of the images in panels F through I show that while the innermost part as well as the p25 labeling of these cores is lost, some stain-excluding material that lines the core shell persists. The arrow in panel F denotes a structure that seems to have left the adjacent core (large arrowhead) and which is labeled for p25. The core in panel H is close to ER membranes. Bars = 100 nm.
FIG. 8.
Localization of p39 to intracellular cores in the absence of actD. (A, C, and D) Cells infected for 120 min; (B) after a 60-minute infection. Panels A through D clearly show that p39 is mostly localized to the surface of the outer shell of the cores. Panel D clearly shows electron-dense material surrounding the core shell, which we believe to be a spike-like structure (5, 33), with which the p39 labeling is predominantly associated. (B) Rare image of a putative core uncoating intermediate. The core in panel C appears to have two compartments (arrowheads) separated by a piece of core shell, giving the impression of this core being bilobed. The core in panel D is close to the nuclear envelope (N, nucleus). Bars = 100 nm.
FIG. 10.
Cryosections labeled with anti-DNA. (A to C and E) From cells infected for 30 min; (D and E) from cells at 90 min postinfection (all in the absence of actD). Anti-DNA labels mostly the central part of the cores (arrowheads) as well as of the IMV (I) in panel A. (D and E) Cores devoid of labeling for anti-DNA. In panel C, the nucleus (N) also labels for DNA. Bars = 100 nm.
In the absence of actD, labeling for p25, p39, 4a/4b (labeled with anticore) and DNA was gradually lost from the core (Table 2). Instead, at the later time points, we often observed labeling for these antigens in the cytoplasm, but we do not know whether this labeling was associated with any particular (cellular or viral) structure (not shown). Quantitation showed that the labeling for DNA and p25 was lost more rapidly from the core structure than that for p39 and anticore. These quantitative data appeared consistent with our morphological observations, whereas the actD cores always retained a full appearance; in the absence of the drug, the cores appeared to gradually lose their electron-dense, nucleoid material. Such electron-lucent cores still labeled for anticore and p39 (Fig. 7F to I and 8) but were devoid of p25 and anti-DNA labeling (Fig. 9F to I, 10D, and 10E), suggesting that under uncoating conditions these latter components were able to dissociate from the remaining (p39 and anticore-positive) core structure. These cores that had lost the DNA and p25 also revealed several interesting characteristics. First, anticore labeled mainly the membrane-like shell of the core (Fig. 7E to H), while the anti-p39 labeling tended to be some distance away from this structure, being more associated with what we believe to be spike-like protrusions of the core (5, 33) (Fig. 8A, C, and D). Second, the empty core shells often appeared to have acquired what looked like an opening on one side of the core (Fig. 7F to H and 9I). Finally, although p25 and DNA had left these cores, some electron-dense material lining the inner part of the core shell was still apparent (Fig. 7F and H, 8A and D, 9F to I, and 10E show clear examples); we did not analyze the composition of this material.
DISCUSSION
In this study we performed a detailed analysis of VV intracellular cores. Since we were unable to repeat the results obtained by Sarov and Joklik (35) and Holowczak (13), in which distinct uncoating intermediates were separated on sucrose gradients, we accumulated cores in the presence of actD and compared these to chemically prepared cores. Uncoating beyond the actD-sensitive step was nevertheless studied at the ultrastructural level.
The composition of intracellular cores.
Our data shed more light on the structure of the intracellular VV core. VV cores with a fuller appearance and empty cores seen in this study were described before, obtained either by systematic chemically disintegration of the IMVs (8, 14) or by separating uncoating intermediates upon viral infection (13). Our combined results strongly suggest that p39 and 4a/4b may be part of the outer shell of the core, while p25 and the genome are located in the inner part. This was demonstrated by negative staining EM, showing that p39 and 4a/4b were exposed on the surface of the core, while p25 and DNA (not shown) were not. Moreover, in cryosections, antibodies to p25 and DNA labeled mostly the central part, while p39 and 4a labeled the rims of the cores. In addition, when the central part of the cores was lost, the electron-lucent cores were devoid of p25 and DNA, but the remaining shell labeled with p39 and 4a/4b. Since p25 has been proposed to be a double-stranded and single-stranded DNA (and RNA)-binding protein (2, 47), its location inside the core, close to DNA, appears logical.
We have shown before that p39 is located between the core and the viral membranes and have suggested that the protein may be part of the spike-like structure on the core surface (5, 33). The present data clearly corroborate these previous findings, showing that p39 is also on the surface of intracellular cores, while inspection of some sections suggested their localization to the core spikes. Although we have not attempted to quantify our Western blot data, this technique suggested that the actD cores contained less p39 than NP-40/DTT cores. This may imply that p39, like p11, is either partially lost at the plasma membrane or lost upon isolation of the actD core from infected cells. This latter explanation appears most plausible considering the EM data; no labeling for p39 was seen at the plasma membrane (except in extracellular virions), while quantitation showed that the early cores (60 min after infection) labeled as much for anti-p39 as did the extracellular IMVs.
We were unfortunately unable to detect any of the proteins involved in early transcription either by Western blotting or by EM, most likely because their abundance was too low. Our data therefore allows no conclusions as to their distribution in the VV cores.
Core uncoating, VV entry, and role of p11.
The present data show that the abundant core protein p11 did not enter with the core but remained outside, apparently together with the viral membranes. From its sequence p11 is not predicted to be a membrane protein, and accordingly it does not behave like a membrane protein during assembly (15). Its loss at the plasma membrane is thus most easily explained by a tight interaction with viral membrane proteins. We believe that the loss of p11 is compatible with our recent results on VV entry (J. Krijnse Locker et al., submitted for publication and unpublished data). The accepted view is that VV infects cells via a fusion process at the plasma membrane (1, 3, 7). Although we have also observed images similar to those obtained by Chang and Metz (3) and Armstrong et al. (1) suggesting continuity of the viral membranes with the cell surface, none of the viral membrane proteins that we have analyzed are implanted in the plasma membrane, arguing against fusion. Instead, while attached to the plasma membrane, both the IMV and the extracellular enveloped viral membranes appear to separate from the core. The core then penetrates into the cytoplasm in a manner that is not understood, leaving viral membrane remnants behind, attached to the plasma membrane but not merged with it. Thus, while the loss of the core protein p11 from the IMV (as well as the extracellular enveloped virion [not shown]) during entry appears incompatible with viral membrane fusion, its loss at the cell surface does fit into an elusive nonfusion process of entry.
While the mechanism of VV entry is still under investigation, our data also help define the role of this putative DNA-binding protein (18) in the VV life cycle. Although a functional p11 is essential for IMV assembly (48), we observed that the protein is not colocalized with the DNA-containing nucleoid (as is the other DNA-binding protein p25) but is localized to the space between the viral outer membrane and the core. Our data therefore raise the possibility that in addition to its role in assembly, p11 may perform some function during the entry process as well.
Does the intracellular core contain a membrane?
Cores accumulated in actD-treated cells or prepared after NP-40/DTT treatment contained small amounts of the IMV membrane proteins p32 and p35. This raises the question of whether the core contains any lipid bilayer with which these proteins could associate. On cryosections, the outer shell of the core indeed very much appears as a membrane, in agreement with observations made by Dales (6) and Holowczak (13). Moreover, in a separate study on the structure of the IMV, we have acquired extensive data strongly suggesting that the newly assembled core is indeed surrounded by a membrane that is continuous with the IMV outer cisternal membranes (G. Griffiths et al., unpublished data). To determine whether the intracellular core contained lipids, we ran gradients of isolated actD cores comprising lipids prelabeled with [3H]choline. The results were, however, inconclusive (not shown). An option to consider is that the putative core membrane is rapidly lost during entry. Indeed, studies by Joklik (16) strongly suggested that viral phospholipids dissociated immediately from the incoming particle. After penetration, the organization of the proteins around the previously present bilayer remains, giving the impression of a membrane, while trace amounts of membrane proteins like p32 and p35 remain attached to it. Such a mechanism is not unprecedented in virology; during rotavirus assembly, the virion transiently acquires a membrane derived from the ER. Subsequently, the membrane is lost, leaving the major membrane protein VP7 behind in the lipid-free membrane structure that surrounds the mature particle (reviewed in reference 11).
ACKNOWLEDGMENTS
E.J.S. was supported by an EMBL travel grant from the Netherlands Organization for Scientific Research. Part of this work was supported by an EU TMR grant (ERB4061PL970064) to G.G., K.P., and N.R. and by Norwegian National Research Council grant 131033/300 to N.R. and K.P.
The following people are kindly acknowledged for providing us with the antibodies: Mariano Esteban, Ed Niles, and Denis Hruby.
REFERENCES
- 1.Armstrong J A, Metz D H, Young M R. The mode of entry of vaccinia virus into L cells. J Gen Virol. 1973;21:533–537. doi: 10.1099/0022-1317-21-3-533. [DOI] [PubMed] [Google Scholar]
- 2.Bayliss C D, Smith G L. Vaccinia virion protein VP8, the 25 kDa product of the L4R gene, binds single-stranded DNA and RNA with similar affinity. Nucleic Acids Res. 1997;25:3984–3990. doi: 10.1093/nar/25.20.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chang A, Metz D H. Further investigations on the mode of entry of vaccinia virus into cells. J Gen Virol. 1976;32:275–282. doi: 10.1099/0022-1317-32-2-275. [DOI] [PubMed] [Google Scholar]
- 4.Chertov O Y, Telezhinskaya I N, Zaitseva E V, Golubeva T B, Zinov'ev V V, Ovechkina L G, Mazkova L B, Malygin E G. Amino acid sequence determination of vaccinia virus immunodominant protein p35 and identification of the gene. Biomed Sci. 1991;2:151–154. [PubMed] [Google Scholar]
- 5.Cudmore S, Blasco R, Vincentelli R, Esteban M, Sodeik B, Griffiths G, Krijnse Locker J. A vaccinia virus core protein, p39, is membrane associated. J Virol. 1996;70:6909–6921. doi: 10.1128/jvi.70.10.6909-6921.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dales S. The uptake and development of vaccinia virus in strain L cells followed by labelled viral deoxyribonucleic acid. J Cell Biol. 1963;18:51–72. doi: 10.1083/jcb.18.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doms R W, Blumenthal R, Moss B. Fusion of intra- and extracellular forms of vaccinia virus with the cell membrane. J Virol. 1990;64:4884–4892. doi: 10.1128/jvi.64.10.4884-4892.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Easterbrook K B. Controlled degradation of vaccinia virions in vitro: an electron microscopic study. J Ultrastruct Res. 1966;14:484–496. doi: 10.1016/s0022-5320(66)80077-1. [DOI] [PubMed] [Google Scholar]
- 9.Ericsson M, Cudmore S, Shuman S, Condit R C, Griffiths G, Krijnse Locker J. Characterization of ts16, a temperature-sensitive mutant of vaccinia virus. J Virol. 1995;69:7072–7086. doi: 10.1128/jvi.69.11.7072-7086.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Essani K, Dales S. Biogenesis of vaccinia: evidence for more than 100 polypeptides in the virion. Virology. 1979;95:385–394. doi: 10.1016/0042-6822(79)90493-8. [DOI] [PubMed] [Google Scholar]
- 11.Griffiths G, Rottier P J M. Cell biology of viruses that assemble along the biosynthetic pathway. Semin Cell Biol. 1992;3:367–381. doi: 10.1016/1043-4682(92)90022-N. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hollinshead M, Vanderplasschen A, Smith G L, Vaux D J. Vaccinia virus intracellular mature virions contain only one lipid membrane. J Virol. 1999;73:1503–1517. doi: 10.1128/jvi.73.2.1503-1517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holowczak J A. Uncoating of poxviruses. I. Detection and characterization of subviral particles in the uncoating process. Virology. 1972;50:216–232. doi: 10.1016/0042-6822(72)90362-5. [DOI] [PubMed] [Google Scholar]
- 14.Ichihashi Y, Oie M, Tsuruhara T. Location of DNA-binding proteins and disulfide-linked proteins in vaccinia virus structural elements. J Virol. 1984;50:929–938. doi: 10.1128/jvi.50.3.929-938.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jensen O N, Houthaeve T, Shevchenko A, Cudmore S, Mann M, Griffiths G, Krijnse Locker J. Identification of the major membrane and core proteins of vaccinia virus by two-dimensional electrophoresis. J Virol. 1996;70:7485–7497. doi: 10.1128/jvi.70.11.7485-7497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joklik W K. The intracellular uncoating of poxvirus DNA. I. The fate of radioactively-labeled rabbitpox virus. J Mol Biol. 1964;8:263–276. doi: 10.1016/s0022-2836(64)80136-4. [DOI] [PubMed] [Google Scholar]
- 17.Joklik W K. The intracellular uncoating of poxvirus DNA. II. The molecular basis of the uncoating process. J Mol Biol. 1964;8:277–288. doi: 10.1016/s0022-2836(64)80137-6. [DOI] [PubMed] [Google Scholar]
- 18.Kao S Y, Bauer W R. Biosynthesis and phosphorylation of vaccinia virus structural protein VP11. Virology. 1987;159:399–407. doi: 10.1016/0042-6822(87)90479-x. [DOI] [PubMed] [Google Scholar]
- 19.Katz E, Moss B. Formation of a vaccinia virus structural polypeptide from a high molecular weight precursor: inhibition of rifampicin. Proc Natl Acad Sci USA. 1970;66:677–684. doi: 10.1073/pnas.66.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Katz E, Moss B. Vaccinia virus structural polypeptide derived from a high molecular weight precursor: formation and integration into virus particles. J Virol. 1970;6:717–726. doi: 10.1128/jvi.6.6.717-726.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krijnse Locker J, Schleich S, Rodriguez D, Goud B, Snijder E J, Griffiths G. The role of a 21-kDa viral membrane protein in the assembly of vaccinia virus from the intermediate compartment. J Biol Chem. 1996;271:14950–14958. doi: 10.1074/jbc.271.25.14950. [DOI] [PubMed] [Google Scholar]
- 22.Krijnse Locker J, Griffiths G. An unconventional role for cytoplasmic disulfide bonds in vaccinia virus proteins. J Cell Biol. 1999;144:267–279. doi: 10.1083/jcb.144.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutay U, Hartmann E, Rapoport T A. A class of membrane proteins with a C-terminal anchor. Trends Biochem Sci. 1993;3:72–75. doi: 10.1016/0962-8924(93)90066-a. [DOI] [PubMed] [Google Scholar]
- 24.Maa J-S, Esteban M. Structural and functional studies of a 39,000-Mr immunodominant protein of vaccinia virus. J Virol. 1987;61:3910–3919. doi: 10.1128/jvi.61.12.3910-3919.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Magee W E, Miller O V. Initiation of vaccinia virus infection in actinomycin D-pretreated cells. J Virol. 1968;2:678–685. doi: 10.1128/jvi.2.7.678-685.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moss B. Poxviridae and their replication. In: Fields B N, Knipe D M, Chanock R M, Hirsch M S, Melnick J L, Monath T P, Roizman B, editors. Fields virology. New York, N.Y: Raven Press; 1990. [Google Scholar]
- 27.Moss B, Rosenblum E N. Protein cleavage and poxvirus morphogenesis: tryptic peptide analysis of core precursors accumulated by blocking assembly with rifampicin. J Mol Biol. 1973;81:267–269. doi: 10.1016/0022-2836(73)90195-2. [DOI] [PubMed] [Google Scholar]
- 28.Niles E G, Seto J. Vaccinia virus gene D8 codes for a virion transmembrane protein. J Virol. 1988;62:3772–3778. doi: 10.1128/jvi.62.10.3772-3778.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ollie W, Wolffe E J, Weisberg A S, Merchlinsky M. Vaccinia virus WR gene A5L is required for morphogenesis of mature virions. J Virol. 1999;73:4590–4599. doi: 10.1128/jvi.73.6.4590-4599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pedley C B, Copper R J. The assay, purification and properties of vaccinia virus-induced uncoating protein. J Gen Virol. 1987;68:1021–1028. doi: 10.1099/0022-1317-68-4-1021. [DOI] [PubMed] [Google Scholar]
- 31.Rodriguez J F, Janeczko R, Esteban M. Isolation and characterization of neutralizing monoclonal antibodies to vaccinia virus. J Virol. 1985;56:482–488. doi: 10.1128/jvi.56.2.482-488.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodriguez J R, Risco C, Carrascosa J L, Esteban M, Rodriguez D. Characterization of early stages in vaccinia virus membrane biogenesis: implication of the 21-kilodalton and a newly identified 15-kilodalton envelope protein. J Virol. 1997;71:1821–1833. doi: 10.1128/jvi.71.3.1821-1833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roos N, Cyrklaff M, Cudmore S, Blasco R, Krijnse Locker J, Griffiths G. A novel immunogold cryoelectron microscopic approach to investigate the structure of the intracellular and extracellular forms of vaccinia virus. EMBO J. 1996;15:2343–2355. [PMC free article] [PubMed] [Google Scholar]
- 34.Salmons T, Kuhn A, Wylie F, Schleich S, Rodriguez J R, Rodriguez D, Esteban M, Griffiths G, Krijnse Locker J. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J Virol. 1997;71:7404–7420. doi: 10.1128/jvi.71.10.7404-7420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarov I, Joklik W K. Characterization of intermediates in the uncoating of vaccinia virus DNA. Virology. 1972;50:593–602. doi: 10.1016/0042-6822(72)90410-2. [DOI] [PubMed] [Google Scholar]
- 36.Sarov I, Joklik W K. Isolation and characterization of intermediates in vaccinia virus morphogenesis. Virology. 1972;52:223–233. doi: 10.1016/0042-6822(73)90411-x. [DOI] [PubMed] [Google Scholar]
- 37.Sarov I, Joklik W K. Studies on the nature and location of capsid polypeptides of vaccinia virions. Virology. 1972;50:579–592. doi: 10.1016/0042-6822(72)90409-6. [DOI] [PubMed] [Google Scholar]
- 38.Sodeik B, Griffiths G, Ericsson M, Moss B, Doms R W. Assembly of vaccinia virus: effects of rifampin on the intracellular distribution of viral protein p65. J Virol. 1994;68:1103–1114. doi: 10.1128/jvi.68.2.1103-1114.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van't Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sodeik B, Cudmore S, Ericsson M, Esteban M, Niles E G, Griffiths G. Incorporation of p14 and p32 into the membrane of the intracellular mature virus. J Virol. 1995;69:3560–3574. doi: 10.1128/jvi.69.6.3560-3574.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.van der Meer Y, Snijder E J, Dobbe J C, Schleich S, Denison M R, Spaan W J M, Krijnse Locker J. The localization of mouse hepatitis virus nonstructural proteins and RNA synthesis indicates a role for late endosomes in viral replication. J Virol. 1999;73:7641–7657. doi: 10.1128/jvi.73.9.7641-7657.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van Slyke J K, Hruby D E. Immunolocalization of vaccinia virus structural proteins during virion formation. Virology. 1994;198:624–635. doi: 10.1006/viro.1994.1074. [DOI] [PubMed] [Google Scholar]
- 43.Van Slyke J K, Whitehead S S, Wilson E M, Hruby D E. The multistep proteolytic maturation pathway utilized by vaccinia virus P4a protein: a degenerate conserved cleavage motif within core proteins. Virology. 1991;183:467–478. doi: 10.1016/0042-6822(91)90976-i. [DOI] [PubMed] [Google Scholar]
- 44.Van Slyke J K, Franke C A, Hruby D E. Proteolytic maturation of vaccinia virus core proteins: identification of a consensus motif at the N termini of the 4b and 25K virion proteins. J Gen Virol. 1991;72:411–416. doi: 10.1099/0022-1317-72-2-411. [DOI] [PubMed] [Google Scholar]
- 45.Wilcock D, Smith G L. Vaccinia virus core protein VP8 is required for virus infectivity, but not for core protein processing or for INV and EEV formation. Virology. 1994;202:294–304. doi: 10.1006/viro.1994.1346. [DOI] [PubMed] [Google Scholar]
- 46.Wolffe E J, Moore D M, Peters P J, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang W P, Bauer W R. Purification and characterization of vaccinia virus structural protein VP8. Virology. 1988;167:578–584. [PubMed] [Google Scholar]
- 48.Zhang Y, Moss B. Vaccinia virus morphogenesis is interrupted when expression of the gene encoding an 11-kilodalton phosphorylated protein is prevented by the Escherichia coli lac repressor. J Virol. 1991;65:6101–6110. doi: 10.1128/jvi.65.11.6101-6110.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]