Introduction
The global prevalence of preterm birth (PTB) has remained mostly unchanged in the decade between 2010 (at 9.8%) and 2020 (at 9.9%), with approximately 13.4 million infants being born too early in 2020.1 The rates remained the highest in South Asia and Sub-Saharan Africa where survival, especially for those born extremely preterm, is the lowest.1 In the United States, the rates have also varied but only modestly: from 10.6% in 1990 to its peak of 12.8% in 2006 and back to 10.4% in 2022, resulting in 380,548 infants being born too early in 2020.2,3
Prematurity is an important risk factor for neonatal mortality and adverse infant outcomes such as sepsis, necrotizing enterocolitis, jaundice (hyperbilirubinemia), and neurodevelopmental delay. This persistent threat to a healthy pregnancy could be greatly reduced by the development of prediction tools that can identify – early in pregnancy and with high accuracy – women who are at risk. Once identified, these women could for example be offered a treatment, such as low-dose aspirin known to reduce the risk,4 and be closely followed until the delivery. In low-income settings, where efficient resource utilization is particularly critical, prediction tools could become crucial to save lives, allowing women at risk to be more efficiently triaged and monitored. Furthermore, identified biomarkers could lead to a better understanding of the biological pathways that are critical in the etiology of PTB, thereby guiding and improving prenatal care pathways.
Why the proteome?
The complexity of PTB, both spontaneous and medically induced, and its various etiologies and associated risk factors pose a challenge to developing prediction tools and unveiling the etiologies of PTB.5 High-throughput measurements covering the genome, transcriptome, proteome, metabolome, lipidome, or microbiome have enabled the collection of detailed omics measurements that reflect biological processes in human physiology and capture molecular differences caused by diseases. Various omics approaches have captured the dynamics of pregnancy6,7 and its adverse outcomes, including PTB,8 preeclampsia (PE),9,10 and onset of labor.11
The genome is the fundamental code of DNA that determines an organism’s capacity for expressing myriad of proteins, many of which are further modified after they have been translated from mRNA. These modifications include phosphorylation, glycosylation, ubiquitination, methylation, acetylation, oxidation, and nitrosylation. Protein translation is cell-, time-, and condition-dependent, and post-translational modifications affect protein structure and function. While relevant, these processes are not the focus of this review.
The transcriptome reflects the active expression of genes. However, the biological consequences of gene expression are probably better understood when evaluating proteins, as not all transcripts result in protein synthesis, and post-translational alterations of proteins (alternative splicing which allows a single gene to code for multiple proteins) and abundance can be directly measured. Moreover, transcripts are more transient in the circulation, and their measurements can be more technically challenging.
The metabolome encompasses the complete set of small molecules in a biological system (including substrates, intermediates, and products of cellular metabolism more generally), and constitutes a more complex array of molecules than the proteome. Metabolites can serve as biomarkers and help identify pathways that are contributing to a particular phenotype as they are directly linked to cellular function. However, the complexities and the dynamic nature of the metabolome pose challenges requiring highly standardized collection and processing protocols to ensure reproducibility.
This review focuses on the discovery of proteomics signatures in plasma, sera, urine, or cervicovaginal fluid that might be useful for diagnosing or predicting various adverse pregnancy outcomes, such as spontaneous PTB or PE. The latter is a significant cause of PTB because early delivery may be indicated to protect the mother and/or the baby. The proteome of amniotic fluid has been studied for the same purpose, but now access to this source is limited, as amniocentesis has become less common due to the introduction of noninvasive pregnancy tests (NIPT). Thus, the proteome of amniotic fluid will not be discussed in detail. Importantly, there is ample evince to suggest that the proteome represented in plasma, sera, and urine contains diagnostic and predictive information regarding the risk for PTB and PE. Moreover, combining proteomics approaches with other omics, some of them involving single cell proteomics measures (e.g., single cell mass cytometry by time-of-flight mass spectrometry or CyTOF) does improve its diagnostic and predictive value. The analysis of the proteome can also provide insight into the pathogenesis of PTB and PE, thereby revealing possible targets for intervention.
The proteome reflects both genetic and environmental influences on a biological system and contains information that allows predicting pregnancy outcomes12 including PTB8 with typically better accuracy than other omics. However, the predictive power of proteomics signatures can be enhanced when integrated with other omics datasets (Fig. 1).8,10 Numerous protein biomarker candidates have been reported for PTB.8,13–30 These findings are encouraging and provide the basis for continued efforts aiming at the development of clinically useful tests for the early prediction of PTB and PE using a broadly-available assays. Such efforts will need to address major challenges including the prospective validation of identified protein biomarker candidates and the demonstration of their robustness across diverse populations.
Fig. 1. Predictive Modeling of Preterm Birth.
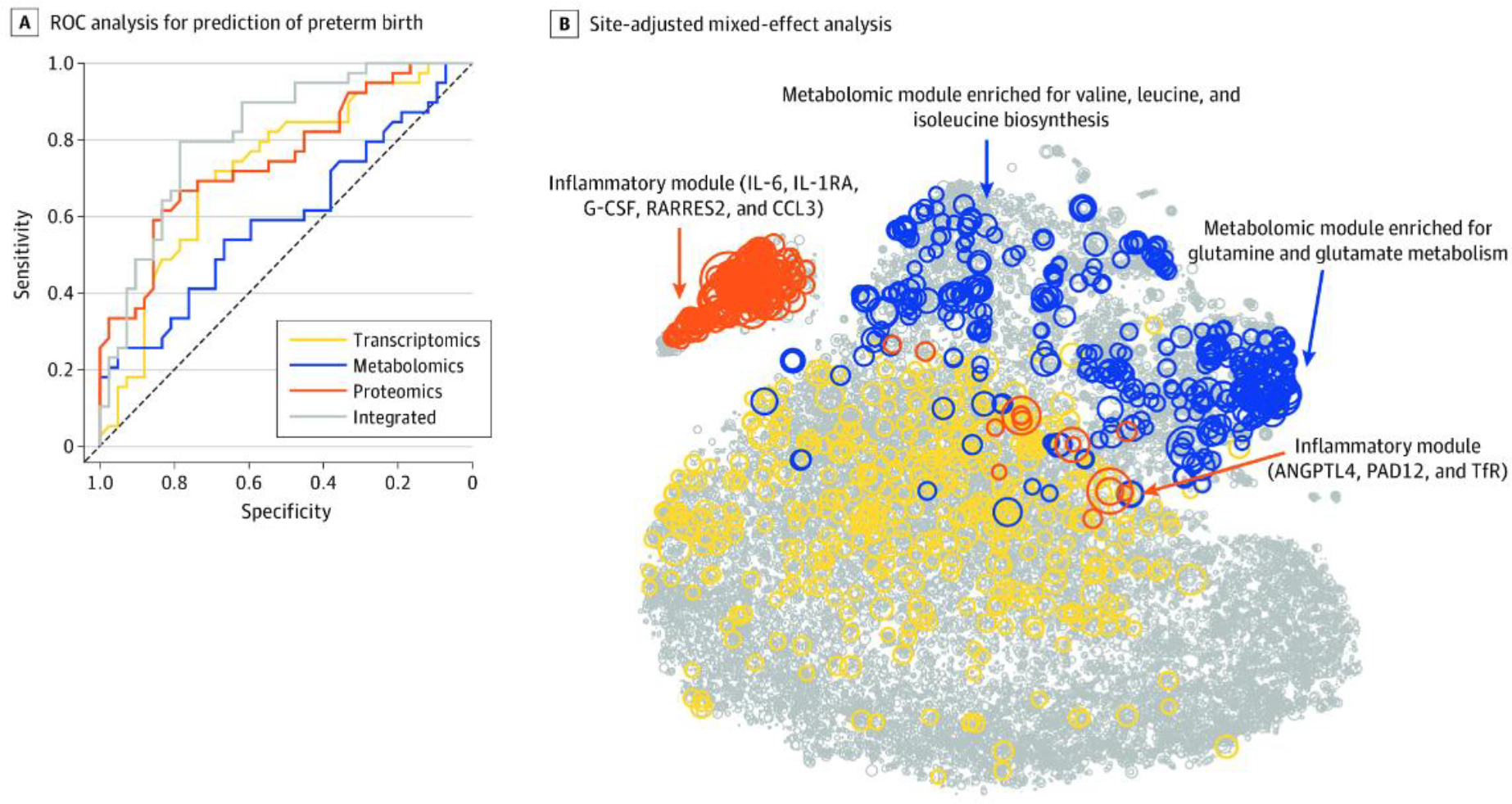
A. This receiver operating characteristic (ROC) curve analysis used each biological modality and the integrated approach. The mean area under the ROC curve and 95% confidence interval (CI) for each modality were as follows: transcriptomics (area under the ROC [AUROC]; 0.73; 95% CI: 0.61, 0.83), metabolomics (AUROC: 0.59; 95% CI: 0.47, 0.72), proteomics (AUROC: 0.75; 95% CI: 0.64, 0.85), and integrated (AUROC: 0.83; 95% CI: 0.72, 0.91). B. Circle size is proportional to −log10 (Wilcoxon) p-value for discrimination between term pregnancies and preterm births. Top features included an inflammatory module (which included interleukin 6 [IL-6]; IL-1 receptor antagonist [IL-1RA], a regulatory member of the IL-1 family whose expression is induced IL-1β under inflammatory conditions; granulocyte colony-stimulating factor [G-CSF]; retinoic acid receptor responder protein 2 [RARRES2]; chemokine ligand 3 [CCL3]; angiopoietin-like 4 [ANGPTL4]; protein-arginine deiminase type II [PADI2]; and transferrin receptor [TfR]) and a metabolomic module (which was enriched for glutamine and glutamate metabolism [Fisher test for pathway enrichment analysis p<4.4×10−9] and valine, leucine, and isoleucine biosynthesis pathways [p<7.3×10−6]). From Jehan F, Sazawal S, Baqui AH, et al. Multiomics characterization of preterm birth in low- and middle-income countries. JAMA Netw Open 2020;3(12):e2029655.
Methods for Proteome Analysis
Analytical methods to assess the proteome are classified as ‘targeted’ and ‘untargeted’. The primary objective of targeted approaches is to identify biomarker candidates in a predetermined set of proteins; whereas, untargeted approaches aim to identify proteins without predefining specific targets.31 The major analytical platforms for targeted approaches either use antibodies or aptamers.32,33 The scope of targeted approaches has dramatically changed during the last decade as current assays can be highly multiplexed, allowing for the simultaneous analysis of over 10,000 proteins in a given specimen. As such, targeted approaches, originally not suitable for relevant discovery work, are now commonly used to derive health-relevant biosignatures. Compared with untargeted approaches, they currently also provide higher sensitivity for the detection of low abundance proteins, particularly in sera and plasma. However, untargeted approaches have limitations. They may provide biased results as antibodies or aptamers against relevant proteins may not be included in the assay.34 Additionally, different antibody- or aptamer-based assays may provide incongruent results as the specificity and binding sites of used antibodies and aptamers can vary.35 As such, validation of targeted proteomic results by an alternative assay is critically important when developing a predictive or diagnostic tool, or inferring relevant biology.
Untargeted approaches are anchored in mass spectrometry (MS), which measures the mass-to-charge ratio (m/z) of ionized analytes, thereby detecting the number of ions at each m/z value and mapping it to the mass spectrum.36 MS methods include surface enhanced laser desorption ionization (SELDI), matrix assisted laser desorption ionization (MALDI) coupled with time-of-flight (TOF), and gas chromatography MS (GC-MS) or liquid chromatography MS (LC-MS).37 Untargeted approaches overcome some of the limitations inherent to targeted approaches. A major advantage is their ability to identify proteins that were not initially thought of in an experimental context. In other words, untargeted approaches can protect against preconceived notions. They can also identify proteins that would be missed by antibodies or aptamers designed for their detection due to the posttranslational modifications of these proteins. As indicated earlier, a major limitation of untargeted approaches is the detection of low abundance proteins as their representation on the mass spectrum may be masked by high abundance proteins.
Considering the advantages and disadvantages of targeted and untargeted approaches, they should be considered complementary methods. The selection of either or both approaches critically depends on a particular study design and rationale.
Biological Compartments
The search for proteomics signatures predictive or diagnostic of PTB spans the analyses of specimens from different sources including tissue (e.g., placenta), plasma, sera, urine, cervicovaginal fluid, amniotic fluid, and exosomes. The choice of the specimen source is ideally driven by a particular study question. However, access to tissue and the feasibility of sample collection often dictates the source. It is therefore not surprising that most investigation heavily rely on the collection of blood specimens. For blood specimens, proteomic analyses can either be performed in sera or plasma, which are processed differently. The question then arises whether the diagnostic or predictive power of proteomics signatures varies when derived in sera or plasma. Addressing this question requires the direct comparison of the predictive power of sera- and plasma-derived biosignatures using simultaneously collected samples. Such a study was recently performed using samples from 73 pregnant women and assaying over a thousand proteins to derive a signature predicting gestational age at the time of sampling.38 The results demonstrated a significantly higher predictive power for a plasma-derived signature compared to a serum-derived signature. A likely explanation for this difference is that serum is subjected to the degradation of proteins while processed.
Statistical Analyses
Statistical and computational analyses of proteomic data sets include both classical statistics methods and machine learning approaches. Hypothesis-driven analyses testing associations between a few proteins and PTB can be addressed with classical hypothesis testing,39 along with an adjustment for multiple comparisons to control for false discovery.40 In contrast, finding the most predictive biomarkers among thousands of proteins requires the use of machine-learning methods. Most suitable for the analysis of these high-dimensional data sets, characterized by numerous measurements (features) and typically a smaller number of samples, are sparsity-promoting regression methods that select a small subset of the most informative features from all features. In principle, such analysis requires evaluating the predictive power of all possible feature subsets. However, in the case of high-dimensional data, the number of generated subsets is too large to allow for such evaluation. This challenge is addressed by introducing penalization schemes that remove features with poor predictive power, thereby selectively considering features with the highest predictive power.41 Another challenge in a clinical setting is that computational algorithms for feature selection have to be trained in data sets obtained in relatively small cohorts. A consequence of a limited cohort size is that small perturbations in the data set can yield different model features. The limited reliability of feature selection is a well-recognized problem of stability in high-dimensional statistical inference.42,43 In other words, there remains significant uncertainty regarding the choice of model features, if such models are derived in small cohorts. Advanced algorithms specifically addressing this problem include Stability selection44, Knockoffs-based 45–46, bootstrap-enhanced Lasso (Bolasso),47 and Stabl.48 These novel approaches result in a sparse and reliable set of biomarkers. The stability selection method improves model robustness by using bootstrapping and selecting features that are chosen with the highest frequency, whereas the approach by Barber and Candès45 and Candès et al.46 introduces artificial features to separate random feature selection from the selection of truly informative features (i.e., knockoffs). Stabl is the latest implementation of such an algorithm that integrates both approaches. Specifically, Stabl determines a feature frequency selection threshold based on data, by adding random features to the data set and allowing separating noise from signal. Truly informative features are selected more frequently during bootstrapping then the added random features. By using this approach, Stabl creates a sparse set of highly reliable features. The above approaches provide a high-impact advancement, as the cohort sizes of many clinical studies are too small to allow for meaningful analyses with deep-learning (DL) algorithms. However, when large datasets are available, complex patterns could potentially be discovered by DL algorithms, resulting in prediction with higher accuracy. A few studies have used DL for the discovery of predictive biosignatures in proteomics49 and a multiomics data set.50
Clinical Studies
Numerous studies have examined the association between proteins in different biological compartments, most commonly sera or plasma, and PTB as well as PE resulting in PTB. These studies reported an array of proteins. Sentinel results are reviewed in this section and are listed in Table 1. A comprehensive list of omics biomarkers, including the proteome, has been published in a recent systematic review.51
Table 1.
Overview of studies on proteomic biomarkers for prediction of preterm birth (PTB).
| Study | Identified Biomarkers | Matrix | Size | When | Method |
|---|---|---|---|---|---|
| Targeted studies | |||||
| Massaro et al. (2009)24 | - IL-6 | - Amniotic fluid | - 8 PTB - 92 Term |
- 16–22 wks | - ELISA - p<0.05 |
| Manning et al. (2019)22 | - IL-1β - IL-8 - IL-6 |
- Cervical fluid | - 44 PTB - 90 Term - All with history of PTB or cervical surgery |
- 22–24 wks | - Multiplex platform - ELISA - PCR - Bonferroni corr. (p<0.01) |
| Goepfert et al. (2001)16 | - IL-6 | - Cervical fluid | - 125 PTB - 125 Term |
- 22–246/7 wks | - ELISA |
| Sorokin et al. (2010)28 | - IL-6 - CRP |
- Sera | - 47 PTB (<32 wks) - 423 Term |
- 24–32 wks | - ELISA - p<0.05 |
| Ghezzi et al. (2002)15 | - CRP | - Amniotic fluid | |||
| Wallenstein et al. (2016)30 | - sVEGFR-3 - sIL-2Rα - sTNFR1 |
- Sera | - 34 PTB - 34 Term - BMI>30 kg/m2 |
- 15–20 wks | - Random forest: classification and regression trees (CART) |
| Saade et al. (2016)27 | - IBP4/SHBG ratio | - Sera | - 248 Spontaneous PTB | - 19–21 wks | |
| Markenson et al. (2020)23 | - IBP4/SHBG ratio | ||||
| Khanam et al. (2022)20 | - IBP4/SHBG ratio | - 300 Women | |||
| Untargeted studies | |||||
| D’ Silva et al. (2018)13 | - 30, including 9 phosphoproteins, 11 glycoproteins - Pathways: blood coagulation, plasminogen activation, vitamin D metabolism, and angiogenesis |
- Sera | - 10 PTB - 10 Term |
- 11–13+6/7 wks | - 2DE and MS |
| Gunko et al. (2016)18 | - 25 including IL-6, VEGFA - Pathways: angiogenesis, proteolysis, transcription, inflammation processes, binding, and transportation of various ligands |
- Serum | - 10 PTB - 10 Term |
- 16–17 wks | - MS - p<0.05 |
| Pereira et al. (2010)25 | - Pathways: complement /coagulation cascade; inflammation/immune response; fetal-placental development; extracellular matrix proteins | - Serum (glycoproteome and peptidome) | - 48 Spontaneous PTB ≤33 wks - 62 PTB with GA≥34 wks |
- 20–336/7 wks | - p<0.05 |
| Jehan et al. (2020)8 |
- An inflammatory module | - Plasma | - 39 PTB - 42 term |
- Early pregnancy (median 13.6 wks) |
- targeted proteomics (1,002) AUC: 0.75 95% CI: 0.64, 0.85 |
| Tarca et al. (2021)29 | - PDE11A - ITGA2B - IL-6 - ANGPT1 - MMP7 - ITGA2B - Vascular wall pathways, nervous system development, developmental biology, focal adhesion, VEGFA/VEGFR-2 signaling, and membrane trafficking pathways |
- Plasma | - 62 Spontaneous PTB - 4 PPROM - 39 Term |
- 17–23 wks - 27–33 wks |
- targeted plasma proteomics (1,125) AUC: 0.76 95% CI: 0.72, 0.8 |
| Lynch et al. (2016)21 | - Complement factors B and H - Coagulation factors IX and IX ab - Pathways: complement cascade, the immune system, and the clotting cascade |
- Plasma | - 41 PTB - 88 Term |
- 10–15 wks |
- targeted proteomics (1,129) |
| Gudicha et al. (2022)17 | - SNAP25 GPI - PTPN11 - OLR1 - ENO1 - GAPDH - CHI3L1 - RETN - CSF3 - LCN2 - CXCL1 - CXCL8 - PGLYRP1 - LDHB - IL-6 - MMP8 - PRTN3 |
- Amniotic fluid | - 90 Women with a short cervix | - 1,310 proteins measured using aptamer-based multiplex platform (SOMAmer) | |
| Romero et al. (2008)26 | - 39 features identified | - Amniotic fluid | - 60 PTB - 59 Term |
- SELDI | |
| Hong et al. (2020)19 | - VEGFR-1 - Lipocalin-2 - Fc fragment of IgG binding protein |
- Amniotic fluid | - 139 | - 24–326/7 wks | - LC-MS - validation using ELISA |
IL, interleukin; ELISA, enzyme-linked immunosorbent assay; PCR, polymerase chain reaction; CRP, C-reactive protein; BMI, body mass index; 2DE, 2-dimensional gel electrophoresis; MS, mass spectrometry; AUC, area under the curve; CI, confidence interval; PPROM, preterm premature rupture of membranes; SELDI, surface enhanced laser desorption ionization; LC, liquid chromatography
The strong association between intrauterine infection and PTB suggests that the resulting inflammatory response is a main driver of PTB.52 A hallmark of inflammation is the increase of cytokines including interleukin (IL)-1, IL-6, IL-8 and C-reactive protein (CRP).53,54 Associations between different pro-inflammatory cytokines with PTB have been observed in multiple studies. For example, associations between IL-6 and PTB were shown in different biological compartments including amniotic fluid,24 cervical fluid16,22 and sera.18,28 Consistent with these reports is a recently published proteomics profile derived with aid of a machine-learning analysis and considering 1,125 simultaneously measured plasma proteins. This profile included IL-6.29 Further evidence highlighting the importance of inflammation in PTB are increased levels of C-reactive protein in amniotic fluid15 and plasma28, and increased levels of IL-1β and IL-8, next to IL-6, in cervical fluid.22
A remarkably large study including 248 women with spontaneous PTB pregnancies examined the ratio of insulin-like growth factor-binding protein 4 (IBP4) and sex hormone-binding globulin (SHBG) in serum.27 The cohort was split into discovery, verification, and validation sub-cohorts. In the validation cohort, the IBP4/SHBG ratio predicted spontaneous PTB with modest accuracy (area under the curve [AUC]=0.67) when measured between 19 and 21 weeks of gestation, and with an increased accuracy (AUC=0.75) after stratification by the body-mass-index (BMI). These findings were subsequently validated with an AUC=0.67 considering the ratio, and an AUC=0.71 after BMI stratification.23 A subsequent study in a cohort of 300 women from Bangladesh, Pakistan, and Tanzania validated this biomarker with lower accuracy (AUC=0.64), which could be increased (AUC=0.79) by adding endoglin, prolactin, and tetranectin to the prediction model.20 These studies resulted in the development of a commercially available test for predicting the risk of PTB based on the IBP4/SHBG ratio.
Studies using an untargeted approach identified a number of proteins in sera13,18,25 and plasma that were significantly associated with PTB.8,21,29 For example, a study reporting a readout of 628 proteins in serum of 20 women using two-dimensional gel electrophoresis (2DE) and MS identified 30 proteins involved in immunological, developmental, and metabolic processes.13 Another study, using a targeted approach to measure 1,012 proteins in plasma from 81 women8 built a multivariate model to predict the risk of PTB. However, the model had moderate predictive power as evidenced by an AUC=0.75.
Comparisons among studies8,13,18,21,25,29 examining and reporting a wide array of protein candidates reveals limited overlap with some exceptions, one being IL-6. While divergent findings may partially be due to methodological differences, inconsistent findings likely reflect the heterogeneity of the studied cohorts and pathophysiologies underlaying PTB. This view is supported by a recent investigation measuring over 1,000 plasma proteins with the same analytical platform to predict the risk of PE in two demographically distinct cohorts of pregnant women.14 Multivariate models derived separately in each cohort predicted the risk of PE with good accuracy in the respective cohort. However, either model failed to predict the risk of PE in the alternative cohort, emphasizing the need to study large and diverse cohorts to develop generalizable prediction models. While the proteins associated with PTB vary across studies,8,13,18,21,25,29 pathway analysis points to important biology that likely drives the development of PTB including pathways relevant to inflammation8,25,29 and angiogenesis.13,18
From an interventional perspective the established association between IL-1 and PTB is interesting, as it may offer a therapeutic approach by targeting IL-1. While several IL-1 antagonists have been approved for clinical use, none have been approved for the prevention of PTB.55 One hindrance is that IL-1 antagonists have failed to prevent PTB in animal models.56 Furthermore, there is a relevant risk that straight antagonism at the IL-receptor to decrease IL-1 binding, which has a role during labor, may interfere with normal delivery.55 These obstacles may be overcome by further examining an IL-1R allosteric modulator, which has proven effective in mice.57 However, from a drug development perspective, a significant challenge is the difficulty in conducting randomized clinical trials to evaluate therapeutic candidates, as the incidence of PTB is about 10%. Developing a reliable test for the early prediction of PTB would allow for an enriched trial design, greatly enhancing the feasibility of conducting interventional clinical studies.
Preeclampsia
While the pathophysiology of PE is not fully understood, placental ischemia likely plays a causal role.58 Ischemia changes the level of circulating angiogenic and antiangiogenic factors, including decreases of angiogenic factors such as vascular endothelial growth factor (VEGF) and placental growth factor (PlGF), and increases of antiangiogenic factors such as soluble VEGF receptor-1 (sVEGFR-1) or sFlt-1 and soluble endoglin (sEng).59 Investigations focusing on biomarker discovery to predict PE indeed revealed changes in circulating angiogenic factors60 with multiple studies showing increased sFlt-1 and decreased of PIGF levels.61 However, neither of these biomarkers had shown sufficient predictive power when examined alone.62,63 Importantly, examining the sFlt-1/PIGF ratio increased the prediction accuracy and is now used in clinical practice.64–66 The use of the sFlt-1/PIGF ratio is recommended by the current National Institute for Health and Care Excellence (NICE) guidelines.67 They specifically suggest that a ratio >38 is indicative for the short-term PE risk during the 24–366/7 weeks gestational period. The sFlt-1/PIGF ratio can be used in conjunction with clinical factors and uterine artery Doppler results to increase accuracy.64 However, determining the sFlt-1/PIGF ratio is particularly useful in pregnancy after the 24th week, while clinical features suggestive of PE may already have manifested.68 As such, the sFlt-1/PIGF ratio can viewed as diagnostic rather than a predictive tool. The search for proteomics signatures that can help predict PE early during pregnancy rather than diagnose it later during pregnancy remains a high-yield objective.10 Table 2 summarized the results of untargeted studies, which predominantly revealed angiogenic and inflammatory proteins associated with PE early and late in pregnancy.69,70
Table 2.
Overview of studies on proteomic biomarkers for prediction of preeclampsia (PE)
| Study | Identified Biomarkers | Matrix | Size | When | Method |
|---|---|---|---|---|---|
| Targeted studies | |||||
| Honigberg et al. (2016)61 | - sFlt-1 - PIGF |
- 2,355 Women | - 10 wks - 18 wks - 26 wks - 5 wks |
- AUROC curve | |
| Zeisler et al (2016)66 | - sFlt-1/PIGF ratio | - Sera | - 500 Discovery - 550 Validation |
- 24–366/7 wks | - Elecsys assays for sFlt-1 - electrochemiluminescence immunoassay platform for PIGF |
| Levine et al. (2006)65 | - sEng - sFlt-1/PIGF ratio |
- Sera | - 72 PTB PE - 120 Term PE - 120 Gestational hypertension - 120 SGA - 120 Controls |
- 21–32 wks - 33–42 wks |
- ELISA |
| Herraiz et al. (2018)64 | - sFlt-1/PIGF ratio | - 24–28 wks | |||
| Taylor et al. (2015)72 | - Leptin | - Sera | - 430 PE - 316 Controls |
- 9–26 wks | - Generalized linear model |
| Untargeted studies | |||||
| Ouyang et al. (2007)71 |
- Leptin | - Plasma | - 53 PE - 20 Controls |
||
| Chen et al. (2022)70 | - IGFBP4 - ITIH2–4 |
- Plasma | - 17 Early-onset PE - 18 Late-onset PE - 18 Controls |
- At hospital admission for delivery | - Untargeted MS; 370 proteins - Targeted for validation |
| Maric et al. (2022)10 | - Leptin - VEGFA - SEL-L - SEL-E |
- Plasma | - 17 PE - 16 Controls |
- longitudinally | - Untargeted LC-MS (1,305 proteins) |
| Beernik et al. (2022)69 | - ApoD - SEL-L - Ficolin-2 - Serum amyloid A-1 - Fibrinogen beta chain - Cartilage acidic protein 1 - Mannan-binding lectin serine protease 1 |
- Sera | - 23 PE - 23 Controls |
- 11 wks - 14 wks |
- Untargeted LC-MS |
AUROC, area under the receiver operating characteristics curve; SGA, small-for-gestational-age; ELISA, enzyme-linked immunosorbent assay; MS, mass spectrometry; LC, liquid chromatography
Increased levels of leptin in early and mid-pregnancy have been observed in pregnant women developing clinically overt PE later in pregnancy, if adjusted for maternal BMI.10,71,72 Associations of PE with other potential biomarkers include plasma protein-A (PAPP-A)73 and uric acid.6 However, the predictive power of reported isolated proteins is poor. Instead, integrating these proteins in composite signatures can significantly improve predictive accuracy.74,75 Specifically, first trimester screening information including maternal clinical factors, the uterine artery pulsatility index, the mean arterial pressure (MAP), and the maternal serum proteins PAPP-A and PIGF resulted in a 95% detection rate with a 10% false-positive rate.74 A large subsequent study including over 35,000 women as a training cohort, and over 25,000 women as a validation cohort confirmed the utility of the composite signature approach. First trimester screening of maternal factors, MAP, uterine artery pulsatility index, and PIGF resulted in an AUC>95% for early-onset PE and >80% for PE in general.75 While yielding high accuracy, these composite signatures require tests that are not conducted during routine prenatal care. As such, proteomic signatures composed of multiple proteins and derived by interrogating a large array of proteins with machine-learning methods do have the potential to advance the development of sufficiently accurate prediction models that may not require results of demanding clinical tests.10
Future Directions
Significant research efforts aiming to develop tests that predict preterm birth and PE before they become clinically manifest and point to underlaying mechanisms have identified an array of biomarker candidates including genes, proteins, and metabolites. Yet, the development of predictive tools that are sufficiently accurate to be of clinical utility has been hindered by limited predictive power, missing or incomplete validation, restricted generalizability, and constrained clinical use as pointed out for the sFlt-1/PIGF ratio. The development of highly multiplexed and sensitive platforms allowing for the simultaneous analysis of over 10,000 proteins holds the promise that predictive tests with higher accuracy will be developed in the future. Importantly, the derivation of predictive proteomics signatures will rely on advanced computational algorithms that adequately address the highly intercorrelated and redundant nature of the proteome. As important is the derivation of such signatures in large and diverse populations using designs that allow for cross and independent validation. It is conceivable that these investigations will reveal that accurate proteomic signatures vary for different demographic groups, sub-groups of PTB and PE with different or only partially congruent underlying pathophysiology, and a particular clinical context including gestational age at the time of specimen sampling. Large scale proteomics studies in diverse populations will also shed more light on the biology and pathways driving PTB and PE, which are incompletely understood. Such knowledge will facilitate the development of preventive and therapeutic interventions.
A highly promising approach to derive predictive tests of sufficient accuracy is the combination of proteomics with other omics, i.e., the conduct of multiomics studies. The integration of information from various biological layers, including the genome, transcriptome, proteome, and immunome in mid-sized study cohorts has already revealed that multiomics models improve predictive power.8 They also provide a more complete view of the biological process underlying PTB and PE, and as such, may be a powerful approach to improve disease diagnostics,76 predictive accuracy,8 and the understanding of interrelationships between different omics, thereby pointing to important pathophysiologic drivers of PTB and PE.10 Various advanced computational methods exist for the integration of omics datasets, generally classified as early-, late-, or hybrid- fusions.77 Early fusion approaches concatenate and use features from all omic sources to train and validate a predictive model. Late fusion approaches first build predictive models for each omics dataset, and then combine these predictions to build an integrated model. As noted previously, including the proteome in an integrative approach can greatly enhance the accuracy of an integrated model, as it offers superior accuracy on its one when compared with other omics.8,10 A common approach when deriving integrated models is to assign more weight to the omics dataset that are particularly informative.78 As such the proteome, likely being quite informative, will weigh heavily on the accuracy of the final model. Inclusion of the proteome should therefore be strongly considered when engaging in a multiomics approach.
One important limitation of multiomics models is that some relevant multiomics data may be difficult to obtain in a clinical setting. Multiomics biosignatures may therefore not directly translate into a simple predictive test. They may require substitution of parameters that cannot readily be measured, and such substitution will require the simultaneous consideration of all available biological, demographic, and clinical data. The promise of this approach resides in the inherently redundant nature of these datasets.
In summary, PTB is a complex and diverse clinical syndrome influenced by a combination of genetic, biological, and environmental factors. The risk of PTB has been associated with maternal characteristics (e.g., BMI),79 comorbidities and medical history (e.g., previous PTB, and family history of PTB),80 and the interpregnancy interval.81 Similarly, risks factors for PE include a history of PE, chronic hypertension, diabetes, kidney disease, obesity, and nulliparity.82 These data are typically available from the electronic health records, an important source for improving risk assessment.83 Integration of these clinical data with omics measurements, while challenging due to data heterogeneity and high-dimensionality, will be important to improve the accuracy of predicting and diagnosing PTB and PE.10,84
Synopsis:
The complexity of preterm birth (PTB), both spontaneous and medically indicated, and its various etiologies and associated risk factors pose a significant challenge for developing tools to accurately predict risk. This review focuses on the discovery of proteomics signatures that might be useful for predicting spontaneous PTB or preeclampsia (PE), which often results in PTB. We describe methods for proteomics analyses, proteomics biomarker candidates that have so far been identified, obstacles for discovering biomarkers that are sufficiently accurate for clinical use, and the derivation of composite signatures including clinical parameters to increase predictive power. We conclude with an outlook including the derivation of biosignatures with highly multiplexed and sensitive proteomics platforms and the integration of proteomics results with other omics with aid of advanced computational algorithms. Finally, we point to the importance of conducting future biosignature and composite signature studies in large and diverse populations to derive accurate, reliable, generalizable, and clinically useful prediction tools.
Key Points:
Numerous studies have revealed that proteins present in prenatal maternal samples are significantly associated with preterm birth (PTB) and preeclampsia (PE), thereby providing a compelling rationale for advanced proteomics research to derive sufficiently accurate biosignatures to support clinical decision-making in women at risk for PTB or PE.
Current protein biomarker candidates possess insufficient or too restricted predictive power as they lack sufficient accuracy, reliability, and generalizability across diverse populations.
The advancement of highly multiplexed and sensitive platforms simultaneously assessing a large array of proteins, advanced computational methods, and protocols combining proteomics data with other omics and clinical datasets, holds considerable promise for uncovering predictive biosignatures with sufficient accuracy for their clinical use.
Best Practices.
What is the current practice for preterm birth?
Currently, there is no best practice for the prevention for preterm birth (PTB).
What changes in current practice are likely to improve outcomes?
Further studies are needed to establish a set of robust proteomic biomarkers and develop a predictive tool to identify women who would benefit from frequent clinical follow-up and medical interventions, including the preemptive treatment with low-dose aspirin, which reduces the risk of PTB.
Bibliographic Sources:
Magee LA, von Dadelszen P. Aspirin from early pregnancy to reduce preterm birth. Lancet Glob Health 2023;11(3):e314–e5.
Footnotes
Disclosure: The authors have nothing to disclose.
References
- 1.Ohuma EO, Moller AB, Bradley E, et al. National, regional, and global estimates of preterm birth in 2020, with trends from 2010: a systematic analysis. Lancet 2023;402(10409):1261–71. [DOI] [PubMed] [Google Scholar]
- 2.Centers for Disease Control. Percentage of preterm births in the United States from 1990 to 2021 [Graph]. https://www.statista.com/statistics/276075/us-preterm-birth-percentage/. Published 2023. Accessed January 9, 2024.
- 3.March of Dimes. The 2023 March of Dimes Report Card. 2023.
- 4.Magee LA, von Dadelszen P. Aspirin from early pregnancy to reduce preterm birth. Lancet Glob Health 2023;11(3):e314–e5. [DOI] [PubMed] [Google Scholar]
- 5.Romero R, Dey SK, Fisher SJ. Preterm labor: one syndrome, many causes. Science 2014;345(6198):760–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aghaeepour N, Ganio EA, McIlwain D, et al. An immune clock of human pregnancy. Sci Immunol 2017;2(15):eaan2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liang L, Rasmussen MH, Piening B, et al. Metabolic dynamics and prediction of gestational age and time to delivery in pregnant women. Cell 2020;181(7):1680–92 e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jehan F, Sazawal S, Baqui AH, et al. Multiomics characterization of preterm birth in low- and middle-income countries. JAMA Netw Open 2020;3(12):e2029655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han X, Ghaemi MS, Ando K, et al. Differential dynamics of the maternal immune system in healthy pregnancy and preeclampsia. Front Immunol 2019;10:1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marić I, Contrepois K, Moufarrej MN, et al. Early prediction and longitudinal modeling of preeclampsia from multiomics. Patterns (NY) 2022;3(12):100655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stelzer IA, Ghaemi MS, Han X, et al. Integrated trajectories of the maternal metabolome, proteome, and immunome predict labor onset. Sci Transl Med 2021;13(592):eabd9898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aghaeepour N, Lehallier B, Baca Q, et al. A proteomic clock of human pregnancy. Am J Obstet Gynecol 2018;218(3):347 e1–e14. [DOI] [PubMed] [Google Scholar]
- 13.D'Silva AM, Hyett JA, Coorssen JR. Proteomic analysis of first trimester maternal serum to identify candidate biomarkers potentially predictive of spontaneous preterm birth. J Proteomics 2018;178:31–42. [DOI] [PubMed] [Google Scholar]
- 14.Ghaemi MS, Tarca AL, Romero R, et al. Proteomic signatures predict preeclampsia in individual cohorts but not across cohorts - implications for clinical biomarker studies. J Matern Fetal Neonatal Med 2022;35(25):5621–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghezzi F, Franchi M, Raio L, et al. Elevated amniotic fluid C-reactive protein at the time of genetic amniocentesis is a marker for preterm delivery. Am J Obstet Gynecol 2002;186(2):268–73. [DOI] [PubMed] [Google Scholar]
- 16.Goepfert AR, Goldenberg RL, Andrews WW, et al. The Preterm Prediction Study: association between cervical interleukin 6 concentration and spontaneous preterm birth. National Institute of Child Health and Human Development Maternal-Fetal Medicine Units Network. Am J Obstet Gynecol 2001;184(3):483–8. [DOI] [PubMed] [Google Scholar]
- 17.Gudicha DW, Romero R, Gomez-Lopez N, et al. The amniotic fluid proteome predicts imminent preterm delivery in asymptomatic women with a short cervix. Sci Rep 2022;12(1):11781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunko VO, Pogorelova TN, Linde VA. Proteomic profiling of the blood serum for prediction of premature delivery. Bull Exp Biol Med 2016;161(6):829–32. [DOI] [PubMed] [Google Scholar]
- 19.Hong S, Lee JE, Kim YM, et al. Identifying potential biomarkers related to pre-term delivery by proteomic analysis of amniotic fluid. Sci Rep 2020;10(1):19648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khanam R, Fleischer TC, Boghossian NS, et al. Performance of a validated spontaneous preterm delivery predictor in South Asian and Sub-Saharan African women: a nested case control study. J Matern Fetal Neonatal Med 2022;35(25):8878–86. [DOI] [PubMed] [Google Scholar]
- 21.Lynch AM, Wagner BD, Deterding RR, et al. The relationship of circulating proteins in early pregnancy with preterm birth. Am J Obstet Gynecol 2016;214(4):517 e1–e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Manning R, James CP, Smith MC, et al. Predictive value of cervical cytokine, antimicrobial and microflora levels for pre-term birth in high-risk women. Sci Rep 2019;9(1):11246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Markenson GR, Saade GR, Laurent LC, et al. Performance of a proteomic preterm delivery predictor in a large independent prospective cohort. Am J Obstet Gynecol MFM 2020;2(3):100140. [DOI] [PubMed] [Google Scholar]
- 24.Massaro G, Scaravilli G, Simeone S, et al. Interleukin-6 and Mycoplasma hominis as markers of preterm birth and related brain damage: our experience. J Matern Fetal Neonatal Med 2009;22(11):1063–7. [DOI] [PubMed] [Google Scholar]
- 25.Pereira L, Reddy AP, Alexander AL, et al. Insights into the multifactorial nature of preterm birth: proteomic profiling of the maternal serum glycoproteome and maternal serum peptidome among women in preterm labor. Am J Obstet Gynecol 2010;202(6):555 e1-10. [DOI] [PubMed] [Google Scholar]
- 26.Romero R, Espinoza J, Rogers WT, et al. Proteomic analysis of amniotic fluid to identify women with preterm labor and intra-amniotic inflammation/infection: the use of a novel computational method to analyze mass spectrometric profiling. J Matern Fetal Neonatal Med 2008;21(6):367–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saade GR, Boggess KA, Sullivan SA, et al. Development and validation of a spontaneous preterm delivery predictor in asymptomatic women. Am J Obstet Gynecol 2016;214(5):633 e1–e24. [DOI] [PubMed] [Google Scholar]
- 28.Sorokin Y, Romero R, Mele L, et al. Maternal serum interleukin-6, C-reactive protein, and matrix metalloproteinase-9 concentrations as risk factors for preterm birth <32 weeks and adverse neonatal outcomes. Am J Perinatol 2010;27(8):631–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tarca AL, Pataki BA, Romero R, et al. Crowdsourcing assessment of maternal blood multi-omics for predicting gestational age and preterm birth. Cell Rep Med 2021;2(6):100323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallenstein MB, Jelliffe-Pawlowski LL, Yang W, et al. Inflammatory biomarkers and spontaneous preterm birth among obese women. J Matern Fetal Neonatal Med 2016;29(20):3317–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sobsey CA, Ibrahim S, Richard VR, et al. Targeted and untargeted proteomics approaches in biomarker development. Proteomics 2020;20(9):e1900029. [DOI] [PubMed] [Google Scholar]
- 32.Sun BB, Maranville JC, Peters JE, et al. Genomic atlas of the human plasma proteome. Nature 2018;558(7708):73–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eldjarn GH, Ferkingstad E, Lund SH, et al. Large-scale plasma proteomics comparisons through genetics and disease associations. Nature 2023;622(7982):348–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edwards AM, Isserlin R, Bader GD, et al. Too many roads not taken. Nature 2011;470(7333):163–5. [DOI] [PubMed] [Google Scholar]
- 35.Method of the Year 2012. Nat Methods 2013;10(1):1. [DOI] [PubMed] [Google Scholar]
- 36.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature 2003;422(6928):198–207. [DOI] [PubMed] [Google Scholar]
- 37.Meuleman W, Engwegen JY, Gast MC, et al. Comparison of normalisation methods for surface-enhanced laser desorption and ionisation (SELDI) time-of-flight (TOF) mass spectrometry data. BMC Bioinformatics 2008;9:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Espinosa C, Ali SM, Khan W, et al. Comparative predictive power of serum vs plasma proteomic signatures in feto-maternal medicine. AJOG Glob Rep 2023;3(3):100244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rice JA. Mathematical Statistics and Data Analysis. 3rd ed. Belmont: Brooks/Cole; 2007. [Google Scholar]
- 40.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B Stat Methodol 1995;57(1):289–300. [Google Scholar]
- 41.Tibshirani R, Wainwright M, Hastie T. Statistical learning with sparsity: the Lasso and generalizations. New York: Chapman and Hall/CRC; 2015. [Google Scholar]
- 42.Stability Yu B.. Bernoulli 2013;19(4):1484–500. [Google Scholar]
- 43.Huan X, Caramanis C, Mannor S. Sparse algorithms are not stable: a no-free-lunch theorem. IEEE Trans Pattern Anal Mach Intell 2012;34(1):187–93. [DOI] [PubMed] [Google Scholar]
- 44.Meinshausen N, Bühlmann P. Stability selection. J R Stat Soc Series B Stat Methodol 2010;72(4):417–73. [Google Scholar]
- 45.Barber RF, Candès EJ. Controlling the false discovery rate via knockoffs. Ann Statist 2015;43(5):2055–85. [Google Scholar]
- 46.Candès EJ, Fan Y, Janson L, et al. Panning for gold: ‘Model-X’ knockoffs for high dimensional controlled variable selection. J R Stat Soc Series B Stat Methodol 2018;80(3):551–77. [Google Scholar]
- 47.Bach F Bolasso: model consistent Lasso estimation through the bootstrap. arXiv:08041302 2008. [Google Scholar]
- 48.Hedou J, Marić I, Bellan G, et al. Discovery of sparse, reliable omic biomarkers with Stabl. Nat Biotechnol 2024. doi: 10.1038/s41587-023-02033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hartman E, Scott AM, Karlsson C, et al. Interpreting biologically informed neural networks for enhanced proteomic biomarker discovery and pathway analysis. Nat Commun 2023;14(1):5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leng D, Zheng L, Wen Y, et al. A benchmark study of deep learning-based multi-omics data fusion methods for cancer. Genome Biol 2022;23(1):171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gupta JK, Alfirevic A. Systematic review of preterm birth multi-omic biomarker studies. Expert Rev Mol Med 2022;24:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Romero R, Gomez R, Chaiworapongsa T, et al. The role of infection in preterm labour and delivery. Paediatr Perinat Epidemiol 2001;15 Suppl 2:41–56. [DOI] [PubMed] [Google Scholar]
- 53.Pandey M, Chauhan M, Awasthi S. Interplay of cytokines in preterm birth. Indian J Med Res 2017;146(3):316–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Prairie E, Cote F, Tsakpinoglou M, et al. The determinant role of IL-6 in the establishment of inflammation leading to spontaneous preterm birth. Cytokine Growth Factor Rev 2021;59:118–30. [DOI] [PubMed] [Google Scholar]
- 55.Nadeau-Vallee M, Obari D, Quiniou C, et al. A critical role of interleukin-1 in preterm labor. Cytokine Growth Factor Rev 2016;28:37–51. [DOI] [PubMed] [Google Scholar]
- 56.Leitner K, Al Shammary M, McLane M, et al. IL-1 receptor blockade prevents fetal cortical brain injury but not preterm birth in a mouse model of inflammation-induced preterm birth and perinatal brain injury. Am J Reprod Immunol 2014;71(5):418–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dabouz R, Cheng CWH, Abram P, et al. An allosteric interleukin-1 receptor modulator mitigates inflammation and photoreceptor toxicity in a model of retinal degeneration. J Neuroinflammation 2020;17(1):359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dimitriadis E, Rolnik DL, Zhou W, et al. Pre-eclampsia. Nat Rev Dis Primers 2023;9(1):8. [DOI] [PubMed] [Google Scholar]
- 59.Romero R, Chaiworapongsa T. Preeclampsia: a link between trophoblast dysregulation and an antiangiogenic state. J Clin Invest 2013;123(7):2775–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Karumanchi SA. Angiogenic factors in preeclampsia: from diagnosis to therapy. Hypertension 2016;67(6):1072–9. [DOI] [PubMed] [Google Scholar]
- 61.Honigberg MC, Cantonwine DE, Thomas AM, et al. Analysis of changes in maternal circulating angiogenic factors throughout pregnancy for the prediction of preeclampsia. J Perinatol 2016;36(3):172–7. [DOI] [PubMed] [Google Scholar]
- 62.Danielli M, Thomas RC, Gillies CL, et al. Blood biomarkers to predict the onset of pre-eclampsia: A systematic review and meta-analysis. Heliyon 2022;8(11):e11226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Widmer M, Cuesta C, Khan KS, et al. Accuracy of angiogenic biomarkers at ⩽20weeks' gestation in predicting the risk of pre-eclampsia: A WHO multicentre study. Pregnancy Hypertens 2015;5(4):330–8. [DOI] [PubMed] [Google Scholar]
- 64.Herraiz I, Simon E, Gomez-Arriaga PI, et al. Clinical implementation of the sFlt-1/PlGF ratio to identify preeclampsia and fetal growth restriction: A prospective cohort study. Pregnancy Hypertens 2018;13:279–85. [DOI] [PubMed] [Google Scholar]
- 65.Levine RJ, Lam C, Qian C, et al. Soluble endoglin and other circulating antiangiogenic factors in preeclampsia. N Engl J Med 2006;355(10):992–1005. [DOI] [PubMed] [Google Scholar]
- 66.Zeisler H, Llurba E, Chantraine F, et al. Predictive Value of the sFlt-1:PlGF Ratio in Women with Suspected Preeclampsia. N Engl J Med 2016;374(1):13–22. [DOI] [PubMed] [Google Scholar]
- 67.National Institute for Health and Care Excellence (NICE). Diagnostics guidance [DG49]. 2022. [PubMed] [Google Scholar]
- 68.Verlohren S, Herraiz I, Lapaire O, et al. New gestational phase-specific cutoff values for the use of the soluble fms-like tyrosine kinase-1/placental growth factor ratio as a diagnostic test for preeclampsia. Hypertension 2014;63(2):346–52. [DOI] [PubMed] [Google Scholar]
- 69.Beernink RHJ, Zwertbroek EF, Schuitemaker JHN, et al. First trimester serum biomarker discovery study for early onset, preterm onset and preeclampsia at term. Placenta 2022;128:39–48. [DOI] [PubMed] [Google Scholar]
- 70.Chen H, Aneman I, Nikolic V, et al. Maternal plasma proteome profiling of biomarkers and pathogenic mechanisms of early-onset and late-onset preeclampsia. Sci Rep 2022;12(1):19099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ouyang Y, Chen H, Chen H. Reduced plasma adiponectin and elevated leptin in pre-eclampsia. Int J Gynaecol Obstet 2007;98(2):110–4. [DOI] [PubMed] [Google Scholar]
- 72.Taylor BD, Ness RB, Olsen J, et al. Serum leptin measured in early pregnancy is higher in women with preeclampsia compared with normotensive pregnant women. Hypertension 2015;65(3):594–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Luewan S, Teja-Intr M, Sirichotiyakul S, et al. Low maternal serum pregnancy-associated plasma protein-A as a risk factor of preeclampsia. Singapore Med J 2018;59(1):55–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Poon LC, Nicolaides KH. Early prediction of preeclampsia. Obstet Gynecol Int 2014;2014:297397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wright D, Tan MY, O'Gorman N, et al. Predictive performance of the competing risk model in screening for preeclampsia. Am J Obstet Gynecol 2019;220(2):199 e1–e13. [DOI] [PubMed] [Google Scholar]
- 76.Lunke S, Bouffler SE, Patel CV, et al. Integrated multi-omics for rapid rare disease diagnosis on a national scale. Nat Med 2023;29(7):1681–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baltrusaitis T, Ahuja C, Morency LP. Multimodal machine learning: a survey and taxonomy. IEEE Trans Pattern Anal Mach Intell 2019;41(2):423–43. [DOI] [PubMed] [Google Scholar]
- 78.Breiman L Stacked regressions. Mach Learn 1996;24:49–64. [Google Scholar]
- 79.Cnattingius S, Villamor E, Johansson S, et al. Maternal obesity and risk of preterm delivery. JAMA 2013;309(22):2362–70. [DOI] [PubMed] [Google Scholar]
- 80.Koire A, Chu DM, Aagaard K. Family history is a predictor of current preterm birth. Am J Obstet Gynecol MFM 2021;3(1):100277. [DOI] [PubMed] [Google Scholar]
- 81.Schummers L, Hutcheon JA, Hernandez-Diaz S, et al. Association of short interpregnancy interval with pregnancy outcomes according to maternal age. JAMA Intern Med 2018;178(12):1661–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.American College of Obstetrics and Gynecology. Low-dose aspirin use for the prevention of preeclampsia and related morbidity and mortality. Practice Advisory. 2021. [Google Scholar]
- 83.Marić I, Tsur A, Aghaeepour N, et al. Early prediction of preeclampsia via machine learning. Am J Obstet Gynecol MFM 2020;2(2):100100. [DOI] [PubMed] [Google Scholar]
- 84.Higdon R, Earl RK, Stanberry L, et al. The promise of multi-omics and clinical data integration to identify and target personalized healthcare approaches in autism spectrum disorders. OMICS 2015;19(4):197–208. [DOI] [PMC free article] [PubMed] [Google Scholar]


