Abstract
The incorporation of envelope (Env) glycoproteins into virions is an essential step in the retroviral replication cycle. Lentiviruses, including human immunodeficiency virus type 1 (HIV-1), encode Env glycoproteins with unusually long cytoplasmic tails, the functions of which have not been fully elucidated. In this study, we examine the effects on virus replication of a number of mutations in a helical motif (α-helix 2) located near the center of the HIV-1 gp41 cytoplasmic tail. We find that, in T-cell lines, small deletions in this domain disrupt the incorporation of Env glycoproteins into virions and markedly impair virus infectivity. Through the analysis of viral revertants, we demonstrate that a single amino acid change (34VI) in the matrix domain of Gag reverses the Env incorporation and infectivity defect imposed by a small deletion near the C terminus of α-helix 2. These results provide genetic evidence, in the context of infected T cells, for an interaction between HIV-1 matrix and the gp41 cytoplasmic tail and identify domains of both proteins involved in this putative interaction.
Retroviral envelope (Env) glycoproteins are synthesized in the endoplasmic reticulum as precursor proteins that are proteolytically cleaved by a cellular protease during their transport to the cell surface (11, 39). Cleavage of the Env precursor generates the two components of the mature Env glycoprotein complex: the surface (SU) and transmembrane (TM) Env glycoprotein subunits. In the case of human immunodeficiency virus type 1 (HIV-1), the precursor, SU, and TM Env glycoproteins have been designated gp160, gp120, and gp41, respectively. After reaching the plasma membrane, the Env complex is incorporated into budding virions by a process whose details remain to be elucidated.
A striking feature of lentiviruses is that they encode TM Env glycoproteins with cytoplasmic tails (CTs) that are much longer than those of other retroviruses. The HIV-1 and HIV-2 TM CTs, for example, are generally around 150 residues in length, more than five times longer than those of the avian and murine oncoretroviruses. Previous studies have observed diverse effects of HIV-1 gp41 CT mutations, including defective Env incorporation (6, 9, 46), impaired virus infectivity (6, 9, 13), reduced gp160 processing and Env stability (13), disrupted basolateral targeting of virus release (2, 25), and slower Env internalization from the cell surface (3, 33, 36). The structure of the gp41 CT has not been determined; however, several regions within the tail are likely to adopt a helical folding (8, 40). One of these, termed α-helix 1, is located at the C terminus of gp41; another, referred to as α-helix 2, is located near the center of the CT (9). These helical motifs have also been referred to as lentivirus lytic peptides due to their ability to associate with and disrupt lipid bilayers (20, 28, 29, 38). The periodic spacing of Leu residues in α-helix 2 suggests that it may form a Leu zipper (23).
Mixed results have been reported concerning whether or not the incorporation of lentiviral Env glycoproteins into virions requires a direct interaction with the viral Gag proteins during assembly. Evidence for a direct Gag-Env interaction includes the findings that (i) mutations in the HIV-1 MA can block HIV-1 Env incorporation (5, 9, 12, 45) (this incorporation defect can be reversed by pseudotyping virions with heterologous Env glycoproteins containing short CTs or by removing the gp41 CT [9, 12, 26]), (ii) HIV-1 Env directs basolateral budding of Gag in polarized epithelial cells (25), (iii) a direct binding between HIV-1 MA and gp41 CT peptides has been reported in an in vitro system (4), and (iv) the Gag and Env proteins of simian immunodeficiency virus can be coimmunoprecipitated in virus-expressing cells (41). In contrast, several observations support a more passive mode of Env incorporation: heterologous Env proteins (e.g., those of murine leukemia virus, human T-cell leukemia virus, and vesicular stomatitis virus [VSV]) can be incorporated into HIV-1 virions (22, 26), and in some cases the gp41 CT is dispensable for HIV-1 Env incorporation (9, 12, 13, 42). A limitation of previous studies is that the cell types used in the biochemical analysis of Env incorporation have generally been those that are readily transfectable, rather than those that naturally support productive, spreading HIV-1 infections. We recently reported that truncation of the gp41 CT markedly impaired Env incorporation when virus was produced in most T-cell lines and primary monocyte-derived macrophages but had only a modest effect when virus was derived from HeLa and MT-4 cells (31).
We previously determined that α-helix 2 of the gp41 CT forms the boundary between truncation mutants whose incorporation is blocked by a single amino acid change in HIV-1 MA and truncated Env glycoproteins whose incorporation is independent of MA mutation (9). Furthermore, we observed that truncations that fell within α-helix 2 had profound effects on Env incorporation and virus infectivity (9). These results suggested that α-helix 2 might play a critical role in gp41 CT function. In this report, we examine the effects of a number of α-helix 2 mutations on virus infectivity and Env incorporation. Biochemical analysis of Env incorporation in T-cell lines capable of sustaining a productive HIV-1 infection is made possible by the use of a high-level, transient HIV-1 expression system based on pseudotyping with the VSV G glycoprotein (VSV-G) (31). Through the isolation and characterization of viral revertants which appear spontaneously in infected T-cell line cultures, we demonstrate that a single amino acid change in MA can reverse the Env incorporation defect imposed by a small deletion near the C terminus of α-helix 2. These results provide genetic evidence, in the context of infected T-cell lines, for an interaction between MA and α-helix 2 of the gp41 CT.
MATERIALS AND METHODS
Cells, viruses, and plasmids.
CEM (12D-7), H9, HeLa, and MAGI cells were maintained as described previously (18). Human peripheral blood mononuclear cells (PBMC) were stimulated with 1 μg of phytohemagglutinin (PHA) per ml for 3 days and maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS), 2 mM glutamine, antibiotics, and 20 U of recombinant human interleukin-2 (Roche Molecular Biochemicals) per ml. For infection of T-cell lines and PBMC, virus was obtained by transfecting HeLa cells with the T-cell-line-tropic molecular clone pNL4-3 (1) or derivatives containing mutations in α-helix 2 of the gp41 CT. VSV-G was expressed using the plasmid pHCMV-G (44) (kindly provided by J. Burns).
Mutagenesis of the gp41 CT α-helix 2.
A template for oligonucleotide-directed mutagenesis was constructed by cloning the BamHI-KpnI fragment from pNL4-3 (nucleotides [nt] 8465 to 9005) into M13mp18. Mutations in α-helix 2 were introduced by site-directed mutagenesis using methods described by Kunkel et al. (21). The mutagenized fragments were then recloned into pNL4-3 using the unique BamHI (nt 8465) and XhoI (nt 8887) restriction sites and were sequenced in their entireties. α-Helix 2 of the gp41 CT overlaps with the rev open reading frame. Certain mutations (not used in this study) near the N terminus of α-helix 2 caused marked Rev defects (unpublished data). In contrast, the mutations reported here had no significant effect on levels of Gag and Env expression, indicating normal Rev function.
Molecular cloning of viral revertants.
Virus supernatant was harvested at the peak of virus replication from CEM (12D-7) or H9 cultures infected with mutant d5, d7, or d8. Virus stocks were normalized for reverse transcriptase (RT) activity and used to infect fresh CEM (12D-7) or H9 cells in parallel with the wild-type (wt) virus. At the peak of virus replication in the infected cultures, total DNA was purified using a QIAamp blood kit (Qiagen). A 0.98-kbp fragment spanning the gp41 coding region was PCR amplified from the extracted DNA. The primers used were as follows: (+), 5′-GGAATGCTAGTTGGAGTAA-3′, and (−), 5′-TGCCTTGTAAGTCATTGGT-3′. The plus-sense and minus-sense primers bound in the vicinity of pNL4-3 nt positions 8040 and 9020, respectively. For analysis of the d8 revertant, a 1.2-kbp fragment spanning the MA coding region was also PCR amplified as described previously (9). Amplified DNA was purified and sequenced. The PCR products were digested with BamHI and KpnI (nt 8465 and 9005) and cloned into pUC19. Following resequencing, BamHI-XhoI fragments from the pUC19 clones were exchanged for the BamHI-XhoI fragment (nt 8465 to 8887) of pNL4-3 to generate the pNL4-3/275LP/d5 and pNL4-3/271IN/d7 double mutants. To generate pNL4-3/d8/34VI, both pNL4-3/34VI (9) and pNL4-3/d8 were digested with BssHII (nt 711) and EcoRI (nt 5743) and the 5-kbp fragment from pNL4-3/34VI was ligated with the 10-kbp fragment from pNL4-3/d8.
Transfections and infections.
Virus stocks of NL4-3, α-helix 2 mutants, and VSV-G pseudotypes were obtained by transfecting HeLa cells using the calcium phosphate precipitation method (14, 16). Transfected cell supernatants were harvested, filtered (0.45-μm-pore-size filter), normalized for RT activity, and used in infections as described below. Infections of T-cell lines and PBMC were performed as previously described (12, 18). RT assays were performed as reported previously (12). CEM (12D-7) cells (5 × 106) were infected with 1 ml of VSV-G-pseudotyped HIV-1. To block subsequent spread of virions bearing the HIV-1 Env glycoprotein following VSV-G-mediated infection, the CXCR4 inhibitor T22 (32) was added to the infected cultures.
Metabolic labeling and radioimmunoprecipitation.
One day following infection, infected cells were plated in 5 ml of Cys-free RPMI medium supplemented with 10% FBS and 1 μM T22 in 25-cm2 flasks. After addition of [35S]Cys (500 μCi), the cultures were incubated for 16 h at 37°C. Preparation of cell and viral lysates and immunoprecipitation of cell- and virion-associated proteins with AIDS patient sera (HIV-1 neutralizing sera; obtained from the National Institutes of Health AIDS Research and Reference Reagent Program; catalog no. 1983 and 1984) have been described previously (10). Quantitative analysis of bands visualized by radioimmunoprecipitation was performed with a FujiX BAS2000 Bio-Image analyzer.
Western blot analysis.
Cell lysates prepared from HeLa or CEM (12D-7) cells that were infected with VSV-G-pseudotyped HIV-1 were separated by sodium dodecylsulfate–10% polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride membranes (Millipore) using a semidry blotter. Membranes were incubated with the anti-gp41 monoclonal antibody T32 (7) (a kind gift of P. Earl) or AIDS patient sera. Subsequently, membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (Amersham Pharmacia), and the antibody-bound proteins were detected by enhanced chemiluminescence (Amersham Pharmacia).
Flow cytometric analysis of HIV-1 Env surface expression.
CEM (12D-7) cells (106) that were uninfected or infected with VSV-G-pseudotyped wt or mutant HIV-1 were incubated for 1 h at 4°C with 12.5 μg of the T32 anti-gp41 monoclonal antibody per ml in phosphate-buffered saline (PBS) supplemented with 1% FBS. The cells were then washed twice with PBS supplemented with 1% FBS and incubated with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G for 30 min at 4°C. Cells were then washed once with PBS supplemented with 1% FBS and once with PBS and resuspended in 1% formaldehyde in PBS. Cell surface Env expression was analyzed with a FACScan flow cytometer (Becton Dickinson).
Cell surface biotinylation.
Cell surface proteins on VSV-G-pseudotype-infected CEM (12D-7) cells were biotinylated essentially as described previously (37) using 0.25 mg of sulfo-N-hydroxysuccinimide–biotin (Pierce). Biotinylated cell lysates were immunoprecipitated with AIDS patient sera (to detect total intracellular Env expression) or precipitated with neutravidin-agarose beads (Pierce) (to detect cell surface Env) and subjected to Western blotting with rabbit anti-gp120 polyclonal antibody as described above. Brefeldin A treatment was performed for 4 h prior to biotinylation with 5 μg of brefeldin A (Sigma) per ml.
Single-cycle MAGI infectivity assay.
Single-cycle infectivity of HeLa-derived virus was determined by transfecting HeLa cells with wt or mutant molecular clones, harvesting virus stocks, normalizing them for RT activity, and infecting the MAGI cell line (19). To measure the infectivity of T-cell-line-derived virus, HeLa cells were cotransfected with wt or mutant molecular clones and a VSV-G expression vector. HeLa-derived stocks were normalized for RT activity and used to infect CEM (12D-7) cells. Two days postinfection, virus-containing supernatants were harvested from the CEM (12D-7) cells, normalized for RT activity, and used to infect the MAGI cell line. Infected MAGI cells were fixed, stained, and scored 2 days postinfection essentially as described previously (19).
RESULTS
Small deletions in α-helix 2 of the gp41 CT markedly impair virus replication in T-cell lines.
To determine the role of the α-helix 2 domain of the gp41 CT in HIV-1 replication, we introduced a number of single and double amino acid substitutions, and several small deletions, into this region (Fig. 1). These mutations were designed primarily to reduce the predicted helical nature of this sequence or to disrupt its ability to form a leucine zipper. We first examined the effect of these changes on virus replication kinetics in the CEM (12D-7) and H9 T-cell lines. In general, single and double amino acid changes produced minor to moderate effects: 261LW, 273EP, 275LR, and 281EP replicated with near-wt kinetics; 259HP, 260RP, 263DP, 289LR, and 275LR/289LR replicated with a delay of 4 to 6 days relative to wt NL4-3; 263DP/273EP and 263DP/281EP were delayed 8 and 12 days, respectively, relative to wt (data not shown). In CEM (12D-7) cells, the d7 and d8 deletion mutants replicated with 40- and 14-day delays, respectively, whereas d5 failed to replicate (Fig. 2A). In H9 cells, d5, d7, and d8 replicated with delays of 20, 10, and 32 days, respectively, relative to wt (Fig. 2B). In both CEM (12D-7) and H9 cells, the d6 deletion mutant failed to produce any detectable RT activity. We also examined replication of the deletion mutants in PHA-stimulated human PBMC. No replication was observed for any of the mutants (data not shown). Thus, small deletions in gp41 α-helix 2 markedly impaired virus replication in both T-cell lines and primary PBMC.
FIG. 1.
Mutagenesis of α-helix 2 of the gp41 CT. A linear representation of the gp41 CT is shown at the top; the shaded area represents the membrane-spanning domain of gp41 (MSD). Two predicted helical domains within the CT (α-helix 1 and 2) and gp41 amino acid positions are indicated. The wt sequence is shown; residues which could form a Leu zipper are boxed. Below the wt sequence are indicated the positions of the α-helix 2 mutations. Dashes denote amino acid identity with wt; dots represent deleted residues.
FIG. 2.
Replication kinetics of α-helix 2 deletion mutants. Virus stocks, obtained by transfection of HeLa cells with the indicated molecular clones, were normalized for RT activity and used to infect the CEM (12D-7) (A) and H9 (B) T-cell lines. mock, mock infected.
Characterization of α-helix 2 deletion mutants in HeLa cells.
We next tested the infectivity of virus stocks obtained by transfecting HeLa cells with wt or mutant molecular clones in the single-cycle MAGI assay (19). Relative infectivities of d5, d6, d7, and d8, normalized for the RT activity of each input inoculum, were 27 ± 20, 20 ± 19, 89 ± 32, and 51% ± 24% of the wt value (average of at least four assays, ± standard deviation). To examine the effect of the α-helix 2 deletions on Env incorporation, we transfected HeLa cells in parallel with wt or mutant molecular clones, metabolically labeled the transfected cells with [35S]Cys, and immunoprecipitated cell- and virion-associated proteins with AIDS patient sera. The results indicated that the deletion mutations had no significant effect on Env incorporation when virus was produced in HeLa cells; in a typical assay, the amount of gp120 in d5, d6, d7, and d8 virions (normalized for virion p24) was 160, 106, 93, and 83% of that of the wt. These results demonstrated that the deletion mutations, particularly d5 and d6, significantly impaired virus infectivity in the MAGI assay even in the absence of a reduction in Env incorporation.
The α-helix 2 deletion mutants show Env incorporation defects in a T-cell line.
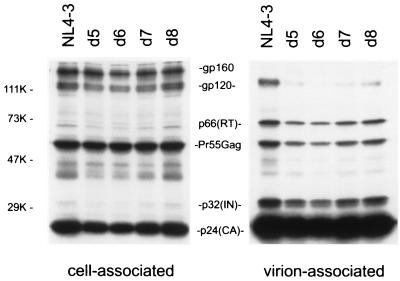
We recently observed (31) that truncation of the gp41 CT had little effect on Env incorporation when virus was produced from HeLa cells but drastically reduced levels of virion gp120 and gp41 when virus was produced by most T-cell lines. We were therefore interested in determining the effect of the d5 to d8 deletions on Env incorporation in T-cell lines. Since levels of virus expression suitable for biochemical analyses are generally not observed immediately following transfection of T-cell lines, we utilized a high-level, transient HIV-1 expression system based on pseudotyping with VSV-G (31). HeLa cells were cotransfected with a VSV-G expression vector (44) and either wt pNL4-3 or derivatives expressing the gp41 mutants. VSV-G-pseudotyped virus stocks were harvested and used to infect CEM (12D-7) cells. One day postinfection, cells were metabolically labeled overnight with [35S]Cys, virions were pelleted by ultracentrifugation, and cell and virion lysates were prepared and immunoprecipitated with AIDS patient sera. Examination of cell-associated material demonstrated that all mutants showed essentially wt levels of Env expression and processing (Fig. 3). However, the deletion mutations significantly reduced Env incorporation; in an average of three independent assays, virion gp120 levels relative to p24 were reduced to approximately 35 (d5), 10 (d6), or 20% (d7 and d8) of those present on wt virions.
FIG. 3.
Radioimmunoprecipitation analysis of α-helix 2 deletion mutants. CEM (12D-7) cells were infected with wt or mutant NL4-3 virions pseudotyped with VSV-G. Infected cells were metabolically labeled overnight with [35S]Cys. Virion-associated material was obtained by pelleting the infected cell supernatant in an ultracentrifuge; lysates derived from cell- and virion-associated material were immunoprecipitated with AIDS patient sera (see Materials and Methods). The positions of the Env precursor gp160, the mature surface glycoprotein gp120, p66 (RT), the Gag precursor Pr55Gag, p32 (IN), and p24 (CA) are indicated; the sizes of molecular mass markers are shown in kilodaltons (K). The results are representative of two independent experiments.
The virion gp120 data presented in Fig. 3 demonstrate that the deletion mutations caused modest (3-fold) to severe (10-fold) defects in Env incorporation in the CEM (12D-7) T-cell line. To investigate whether this incorporation defect might result from reduced cell surface Env expression, the levels of wt and mutant Env on the cell surface were measured. Cell surface Env expression was analyzed by flow cytometry using CEM (12D-7) cells that were uninfected or infected with VSV-G-pseudotyped HIV-1. All mutants showed essentially wt levels of Env at the cell surface (Fig. 4). These results suggest that the Env incorporation defect imposed by small deletions in gp41 α-helix 2 does not result from reduced cell surface Env expression. This conclusion is supported by the observation that the d8 mutation did not affect the levels of gp120 detected in cell surface biotinylation experiments using infected CEM (12D-7) cells (see below).
FIG. 4.
FACS analysis of cell surface Env expression. Cell surface immunostaining with an anti-gp41 monoclonal antibody (T32) was performed using CEM (12D-7) cells infected with wt or mutant NL4-3 virions pseudotyped with VSV-G. Averages of duplicate experiments are presented ± standard deviations. Solid bars represent mean fluorescence intensities determined by FACScan; open bars indicate percent Env-positive cells. Approximately 40% of cultures infected with the wt were Env positive.
Isolation of revertants of α-helix 2 deletion mutants.
To gain further insights into the mechanism by which the α-helix 2 deletion mutations impaired virus replication, we sought to determine whether the RT peak detected at delayed time points (Fig. 2) was due to the emergence of viral revertants. Supernatants were harvested at the peak of virus replication, normalized with wt for RT activity, and used to reinfect fresh CEM (12D-7) or H9 cells. In all cases, the repassaged viruses replicated with accelerated kinetics relative to the original mutant (data not shown), suggesting the presence of viral revertants in the infected cultures.
To examine the possibility that the putative revertants harbored a second-site compensatory mutation(s), viral DNA was prepared from infected cultures at the peak of RT production and PCR amplification was performed using primers specific for the gp41 CT coding region. The repassaged d5 contained second-site changes at gp41 amino acid 205 (S→L) and 275 (L→P); the putative d7 revertant acquired a change at gp41 residue 271 (I→N). Both putative revertants retained the original (d5 and d7) mutations. By constructing and analyzing double mutants containing second-site changes in combination with the original deletions, we determined that the 275LP and 271IN substitutions were sufficient to repair the virus replication defect imposed by the d5 and d7 deletions, respectively (data not shown).
Sequencing of d8-derived PCR fragments indicated that no changes were present in the gp41 CT coding region, apart from the original d8 deletion. To assess the possibility that a d8 compensatory change might map to MA, we sequenced PCR products obtained by amplification of the MA coding region. Intriguingly, the putative d8 revertant that emerged independently in both CEM (12D-7) and H9 cells acquired the same second-site change in MA: a V→I substitution at MA residue 34.
The Env incorporation defect imposed by the d8 deletion is reversed by the 34VI change in MA.
To evaluate whether the MA 34VI change was responsible for the improved replication kinetics observed upon repassage of d8, a 34VI/d8 double mutant was constructed. CEM (12D-7) cells were transfected in parallel with wt pNL4-3 or derivatives containing the d8 or 34VI/d8 mutation (Fig. 5A). In the pNL4-3-transfected culture, virus replication peaked on day 8 posttransfection; peak virus production in d8-transfected cells occurred 12 days later. In contrast, the replication kinetics of the 34VI/d8 double mutnat were delayed only 4 days relative to wt. We also observed that the 34VI/d8 mutant showed wt replication kinetics in H9 cells (Fig. 5B). To examine the effect of the 34VI change in natural target cells, the replication kinetics of d8 and 34VI/d8 in PHA-stimulated human PBMC were examined. As shown in Fig. 5C, the 34VI/d8 double mutant displayed a marked improvement in replication relative to d8. These results indicate that the MA 34VI change is sufficient to largely reverse the replication defect imposed by d8 in T-cell lines and primary PBMC.
FIG. 5.
Replication kinetics of wt, d8, 34VI, and 34VI/d8 in CEM (12D-7) (A), H9 (B), and PHA-stimulated human PBMC (C). CEM (12D-7) cells were transfected with the indicated molecular clones; H9 and PBMC were infected with HeLa-derived virus stocks.
We next examined whether the 34VI change improved d8 infectivity in a single-cycle assay. CEM (12D-7) cells were infected with VSV-G-pseudotyped wt, d8, 34VI/d8, and KFS (env minus [12]) virions; virus stocks were harvested, and their infectivities were measured by MAGI assay (Materials and Methods) (Fig. 6). d8 showed an infectivity approximately 25% of that of wt, whereas 34VI/d8 showed near-wt infectivity. These results demonstrate that, using virus stocks obtained from CEM (12D-7) cells, the d8 infectivity defect is repaired by the MA 34VI substitution.
FIG. 6.
MAGI infectivities of virus stocks produced from CEM (12D-7) cells infected with VSV-G pseudotypes. HeLa cells were cotransfected with wt, KFS (env-minus), d8, 34VI, or 34VI/d8 molecular clones and a VSV-G expression vector. The VSV-G-pseudotyped virus stocks were normalized for RT activity and used to infect CEM (12D-7) cells. Two days postinfection, virus-containing supernatants were harvested, normalized for RT activity, and used to infect the MAGI cell line (19). Infected MAGI cells were fixed, stained, and scored 2 days postinfection. The infectivity of wt virus stocks was approximately 3 × 104 infectious units/ml. Data presented are averages of at least two assays, ± standard deviations.
As indicated in Fig. 3, the d8 mutation caused a significant defect in Env incorporation in the CEM (12D-7) T-cell line. To determine whether the improved replication kinetics and single-cycle infectivity observed with the 34VI/d8 revertant were due to an increase in Env incorporation relative to the original d8 mutant, we performed radioimmunoprecipitation analysis of cell- and virion-associated material derived from CEM (12D-7) cells (Fig. 7A). 34VI/d8 virions contained approximately 70% of wt levels of gp120 relative to p24, a substantial improvement over the fivefold-reduced gp120 levels observed in d8 virions. The 34VI single mutant displayed wt levels of Env incorporation. To determine whether the reduced levels of gp120 present on d8 virions are the result of increased gp120 shedding from virions following Env incorporation, we measured the levels of gp41 on wt, d8, 34VI, and 34VI/d8 virions. Consistent with the gp120 data (Fig. 7A), d8 showed a significant reduction in virion gp41 which was restored to near-wt levels in 34VI/d8 double mutant virions (Fig. 7B). These results support the conclusion drawn from the gp120 data that d8 reduces Env incorporation and that this defect is reversed by the 34VI change in MA.
FIG. 7.
Analysis of wt and mutant Env incorporation into virions. (A) Radioimmunoprecipitation analysis of wt, d8, 34VI, and 34VI/d8. CEM (12D-7) cells were infected with wt or mutant virions pseudotyped with VSV-G. Labeling and immunoprecipitation were performed as indicated in the Fig. 3 legend. (B) Analysis of gp41 by Western blotting. Cell and virion lysates were prepared from CEM (12D-7) cells infected with VSV-G-pseudotyped KFS (Env-minus), NL4-3, d8, 34VI, or 34VI/d8. Samples were transferred to polyvinylidene difluoride membranes, blotted with an anti-gp41 monoclonal antibody (T32), and reprobed with AIDS patient sera to detect p24 (CA) (Materials and Methods). These results are representative of duplicate experiments.
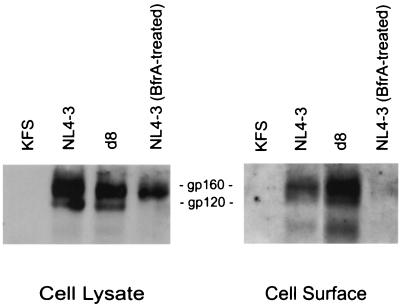
The fluorescence-activated cell sorting (FACS) data presented in Fig. 4 strongly suggest that levels of d5 to d8 Env expression at the cell surface are similar to those of wt. However, since the d8 mutant showed a small reduction in surface expression (Fig. 4), we wished to measure the amount of d8 Env at the cell surface using a different technique. To this end, we performed cell surface biotinylation of CEM (12D-7) cells infected with VSV-G-pseudotyped NL4-3 (wt) and the d8 derivative. As a negative control for Env expression, we again used the env-minus KFS mutant. The left panel of Fig. 8 shows total Env expression; the right side shows cell surface protein (Materials and Methods) (31). To confirm that only cell surface Env was biotinylated, we treated cells expressing wt Env with brefeldin A, which traps glycoproteins in the endoplasmic reticulum (24). The data indicate that the d8 deletion mutant expresses gp160 and gp120 at the cell surface at levels comparable to wt. As expected, gp160 was expressed abundantly in brefeldin A-treated cells but was not efficiently expressed at the cell surface. These results confirm those presented in Fig. 4 and indicate that the reduction in d8 Env incorporation into T-cell-line-derived virus is not the result of impaired cell surface Env expression.
FIG. 8.
Analysis of cell surface d8 Env expression by biotinylation. CEM (12D-7) cells were infected with VSV-G-pseudotyped NL4-3 KFS (env minus), wt NL4-3, or NL4-3/d8. Cells were biotinylated with sulfo-N-hydroxysuccinimide–biotin; cell lysates were immunoprecipitated with AIDS patient sera (to detect total Env expression) or precipitated with neutravidin-agarose beads (to detect cell surface Env) and were subjected to Western blotting with rabbit anti-gp120 polyclonal antibody. To confirm that only cell surface Env was biotinylated, we treated cells infected with VSV-G-pseudotyped NL4-3 with brefeldin A (BfrA) (Materials and Methods).
DISCUSSION
No clear consensus has emerged from previous studies concerning the role of the gp41 CT in HIV-1 replication. In general, previous biochemical analyses were performed in cell lines that can be readily transfected (e.g., COS, CV-1, or HeLa), rather than in cells that are targets for productive HIV-1 infection. In this study, the use of a high-level, transient HIV-1 expression system based on pseudotyping with VSV-G allowed us to determine both biological and biochemical properties of gp41 CT mutants in T-cell-line cultures. We examined the effects of mutations in α-helix 2 of the gp41 CT on virus infectivity and Env incorporation. Small deletions in α-helix 2 markedly impaired virus replication in both T-cell lines (Fig. 2) and primary PBMC, at least in part as a result of a defect in Env incorporation into virions (Fig. 3 and 7). Through the isolation and characterization of viral revertants which appeared spontaneously in infected T-cell-line cultures, we found that second-site changes at gp41 amino acids 275 (L→P) and 271 (I→N) reversed the replication defects imposed by the d5 and d7 deletions, respectively. Most interestingly, we demonstrate that a single amino acid change in MA residue 34 (34VI) reverses the Env incorporation defect imposed by the d8 mutation (Fig. 7). The Env incorporation defect imposed by d8 is not caused by reduced cell surface Env expression, as FACS analysis demonstrated wt levels of gp160-gp41 (Fig. 4) and biotinylation assays (Fig. 8) showed wt levels of gp160 and gp120 on the surface of d8-expressing cells.
Data obtained in the course of this study suggest that, in addition to their effects on Env incorporation, some of the α-helix 2 mutations disrupt virus infectivity even in the absence of an Env incorporation defect. When virus is produced from HeLa cells, the gp41 α-helix 2 deletion mutants show essentially wt Env incorporation, yet HeLa-derived virions display substantially reduced infectivity in the MAGI assay. However, since in some contexts truncation of the entire gp41 CT has little effect on virus infectivity (9, 12, 31, 35), these defects are likely due to conformational changes induced by the mutations rather than to a direct role of α-helix 2 residues in promoting virus infectivity. These small deletions may therefore be functionally analogous to previously reported gp41 CT mutations that impair virus infectivity without disrupting Env incorporation (9, 13, 46). The observation that the α-helix 2 mutations markedly reduce Env incorporation in T-cell lines but not in HeLa cells is consistent with our recent finding that HeLa cells, but not most T-cell lines or primary cell types, are permissive for the incorporation of HIV-1 Env lacking the gp41 CT (31).
The results presented here provide genetic evidence, in the context of infected T-cell lines and primary PBMC, for an interaction between MA and the gp41 CT. How might the region of MA encompassing residue 34 interact with the gp41 CT? X-ray crystallography data for both HIV-1 and simian immunodeficiency virus MA suggest that MA has a propensity to trimerize and in so doing forms a lattice containing an array of holes large enough to accommodate a gp41 CT trimer (15, 34). These structural findings raise the possibility that portions of the long gp41 CT fit into the holes present in the MA lattice and that this configuration is stabilized by specific MA-Env interactions. Indeed, the single amino acid MA mutations that we previously observed to disrupt Env incorporation (9, 12) lie in a rim surrounding the large lattice hole (34). However, several aspects of this model remain uncertain. Since Env incorporation precedes and does not require Gag processing (17), the MA domain of Pr55Gag, rather than MA itself, drives the recruitment of Env into virions. While MA has been reported to form trimers in solution (30), it is not clear whether trimerization driven by MA is relevant in the context of Pr55Gag or whether it occurs in virus-expressing cells. Structural data for the intact CT of gp41 are currently lacking, and a variety of folding models have been proposed, several of which predict that gp41 CT α-helices interact with the inner face of the lipid bilayer (20, 28, 40, 43). In any case, particularly considering the very subtle nature of the 34VI substitution, it is interesting to note that this change reverses the Env incorporation defect imposed not only by the gp41 d8 deletion (as shown here) but also by the 12LE MA mutation (9). These results suggest an important role for MA residues 12 and 34, and gp41 CT α-helix 2, in promoting Env incorporation.
ACKNOWLEDGMENTS
We acknowledge A. Ono, D. Demirov, and R. Willey for critical review of the manuscript and for helpful discussions. We thank P. Earl for the T32 anti-gp41 antibody and J. Burns for pHCMV-G. The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program: HIV-1 neutralizing sera (from L. Vujcic) and MAGI cells (from M. Emerman).
REFERENCES
- 1.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ball J M, Mulligan M J, Compans R W. Basolateral sorting of the HIV type 2 and SIV envelope glycoproteins in polarized epithelial cells: role of the cytoplasmic domain. AIDS Res Hum Retrovir. 1997;13:665–675. doi: 10.1089/aid.1997.13.665. [DOI] [PubMed] [Google Scholar]
- 3.Boge M, Wyss S, Bonifacino J S, Thali M. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J Biol Chem. 1998;273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 4.Cosson P. Direct interaction between the envelope and matrix proteins of HIV-1. EMBO J. 1996;15:5783–5788. [PMC free article] [PubMed] [Google Scholar]
- 5.Dorfman T, Mammano F, Haseltine W A, Gottlinger H G. Role of the matrix protein in the virion association of the human immunodeficiency virus type 1 envelope glycoprotein. J Virol. 1994;68:1689–1696. doi: 10.1128/jvi.68.3.1689-1696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Earl P L, Broder C C, Doms R W, Moss B. Epitope map of human immunodeficiency virus type 1 gp41 derived from 47 monoclonal antibodies produced by immunization with oligomeric envelope protein. J Virol. 1997;71:2674–2684. doi: 10.1128/jvi.71.4.2674-2684.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisenberg D, Wesson M. The most highly amphiphilic alpha-helices include two amino acid segments in human immunodeficiency virus glycoprotein 41. Biopolymers. 1990;29:171–177. doi: 10.1002/bip.360290122. [DOI] [PubMed] [Google Scholar]
- 9.Freed E O, Martin M A. Domains of the human immunodeficiency virus type 1 matrix and gp41 cytoplasmic tail required for envelope incorporation into virions. J Virol. 1996;70:341–351. doi: 10.1128/jvi.70.1.341-351.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freed E O, Martin M A. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J Virol. 1994;68:2503–2512. doi: 10.1128/jvi.68.4.2503-2512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed E O, Martin M A. The role of human immunodeficiency virus type 1 envelope glycoproteins in virus infection. J Biol Chem. 1995;270:23883–23886. doi: 10.1074/jbc.270.41.23883. [DOI] [PubMed] [Google Scholar]
- 12.Freed E O, Martin M A. Virion incorporation of envelope glycoproteins with long but not short cytoplasmic tails is blocked by specific, single amino acid substitutions in the human immunodeficiency virus type 1 matrix. J Virol. 1995;69:1984–1989. doi: 10.1128/jvi.69.3.1984-1989.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gabuzda D H, Lever A, Terwilliger E, Sodroski J. Effects of deletions in the cytoplasmic domain on biological functions of human immunodeficiency virus type 1 envelope glycoproteins. J Virol. 1992;66:3306–3315. doi: 10.1128/jvi.66.6.3306-3315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 15.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopkins N, Besmer P, DeLeo A B, Law L W. High-frequency cotransfer of the transformed phenotype and a tumor-specific transplantation antigen by DNA from the 3-methylcholanthrene-induced Meth A sarcoma of BALB/c mice. Proc Natl Acad Sci USA. 1981;78:7555–7559. doi: 10.1073/pnas.78.12.7555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiernan R E, Ono A, Englund G, Freed E O. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J Virol. 1998;72:4116–4126. doi: 10.1128/jvi.72.5.4116-4126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimpton J, Emerman M. Detection of replication-competent and pseudotyped human immunodeficiency virus with a sensitive cell line on the basis of activation of an integrated beta-galactosidase gene. J Virol. 1992;66:2232–2239. doi: 10.1128/jvi.66.4.2232-2239.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kliger Y, Shai Y. A leucine zipper-like sequence from the cytoplasmic tail of the HIV-1 envelope glycoprotein binds and perturbs lipid bilayers. Biochemistry. 1997;36:5157–5169. doi: 10.1021/bi962935r. [DOI] [PubMed] [Google Scholar]
- 21.Kunkel T A, Roberts J D, Zakour R A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- 22.Landau N R, Page K A, Littman D R. Pseudotyping with human T-cell leukemia virus type I broadens the human immunodeficiency virus host range. J Virol. 1991;65:162–169. doi: 10.1128/jvi.65.1.162-169.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Landschulz W H, Johnson P F, McKnight S L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 24.Lippincott-Schwartz J, Yuan L C, Bonifacino J S, Klausner R D. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lodge R, Gottlinger H, Gabuzda D, Cohen E A, Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68:4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lusso P, di Marzo Veronese F, Ensoli B, Franchini G, Jemma C, DeRocco S E, Kalyanaraman V S, Gallo R C. Expanded HIV-1 cellular tropism by phenotypic mixing with murine endogenous retroviruses. Science. 1990;247:848–852. doi: 10.1126/science.2305256. [DOI] [PubMed] [Google Scholar]
- 27.Mammano F, Kondo E, Sodroski J, Bukovsky A, Gottlinger H G. Rescue of human immunodeficiency virus type 1 matrix protein mutants by envelope glycoproteins with short cytoplasmic domains. J Virol. 1995;69:3824–3830. doi: 10.1128/jvi.69.6.3824-3830.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller M A, Cloyd M W, Liebmann J, Rinaldo C R, Jr, Islam K R, Wang S Z, Mietzner T A, Montelaro R C. Alterations in cell membrane permeability by the lentivirus lytic peptide (LLP-1) of HIV-1 transmembrane protein. Virology. 1993;196:89–100. doi: 10.1006/viro.1993.1457. [DOI] [PubMed] [Google Scholar]
- 29.Miller M A, Garry R F, Jaynes J M, Montelaro R C. A structural correlation between lentivirus transmembrane proteins and natural cytolytic peptides. AIDS Res Hum Retrovir. 1991;7:511–519. doi: 10.1089/aid.1991.7.511. [DOI] [PubMed] [Google Scholar]
- 30.Morikawa Y, Zhang W H, Hockley D J, Nermut M V, Jones I M. Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein, p17. J Virol. 1998;72:7659–7663. doi: 10.1128/jvi.72.9.7659-7663.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami T, Freed E O. The long cytoplasmic tail of gp41 is required in a cell type-dependent manner for HIV-1 envelope glycoprotein incorporation into virions. Proc Natl Acad Sci USA. 2000;97:343–348. doi: 10.1073/pnas.97.1.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, Kishimoto T, Yamamoto N, Nagasawa T. A small molecule CXCR4 inhibitor that blocks T cell line-tropic HIV-1 infection. J Exp Med. 1997;186:1389–1393. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohno H, Aguilar R C, Fournier M C, Hennecke S, Cosson P, Bonifacino J S. Interaction of endocytic signals from the HIV-1 envelope glycoprotein complex with members of the adaptor medium chain family. Virology. 1997;238:305–315. doi: 10.1006/viro.1997.8839. [DOI] [PubMed] [Google Scholar]
- 34.Rao Z, Belyaev A S, Fry E, Roy P, Jones I M, Stuart D I. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 35.Reil H, Bukovsky A A, Gelderblom H R, Gottlinger H G. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rowell J F, Stanhope P E, Siliciano R F. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 37.Salzwedel K, West J T, Hunter E. A conserved tryptophan-rich motif in the membrane-proximal region of the human immunodeficiency virus type 1 gp41 ectodomain is important for Env-mediated fusion and virus infectivity. J Virol. 1999;73:2469–2480. doi: 10.1128/jvi.73.3.2469-2480.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Srinivas S K, Srinivas R V, Anantharamaiah G M, Segrest J P, Compans R W. Membrane interactions of synthetic peptides corresponding to amphipathic helical segments of the human immunodeficiency virus type-1 envelope glycoprotein. J Biol Chem. 1992;267:7121–7127. [PubMed] [Google Scholar]
- 39.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. Plainview, N.Y: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 40.Venable R M, Pastor R W, Brooks B R, Carson F W. Theoretically determined three-dimensional structures for amphipathic segments of the HIV-1 gp41 envelope protein. AIDS Res Hum Retrovir. 1989;5:7–22. doi: 10.1089/aid.1989.5.7. [DOI] [PubMed] [Google Scholar]
- 41.Vincent M J, Melsen L R, Martin A S, Compans R W. Intracellular interaction of simian immunodeficiency virus gag and env proteins. J Virol. 1999;73:8138–8144. doi: 10.1128/jvi.73.10.8138-8144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wilk T, Pfeiffer T, Bosch V. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology. 1992;189:167–177. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- 43.Yang C, Spies C P, Compans R W. The human and simian immunodeficiency virus envelope glycoprotein transmembrane subunits are palmitoylated. Proc Natl Acad Sci USA. 1995;92:9871–9875. doi: 10.1073/pnas.92.21.9871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yee J K, Friedmann T, Burns J C. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol. 1994;43:99–112. doi: 10.1016/s0091-679x(08)60600-7. [DOI] [PubMed] [Google Scholar]
- 45.Yu X, Yuan X, Matsuda Z, Lee T H, Essex M. The matrix protein of human immunodeficiency virus type 1 is required for incorporation of viral envelope protein into mature virions. J Virol. 1992;66:4966–4971. doi: 10.1128/jvi.66.8.4966-4971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yu X, Yuan X, McLane M F, Lee T H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]