TO THE EDITOR
Hypopigmented mycosis fungoides (HMF) is the most common type of cutaneous T-cell lymphoma in children (Amorim et al., 2018). HMF has a benign course and rarely progresses past localized disease, a striking difference from classic MF, which sometimes progresses from patches to tumors in an average of 12.4 years. On the basis of the identification of dominant clonal T-cell populations, classic MF is seen as a malignancy of CD4+ skin-resident memory cells, whereas HMF is considered to be a malignancy of CD8+ T cells (El-Shabrawi-Caelen et al., 2002). The difference in malignant cell type between these two mycosis fungoides (MF) subtypes does not explain the indolent disease course of HMF because another cutaneous malignancy of activated cytotoxic CD8+ T cells, a primary cutaneous CD8+ aggressive epidermotropic T-cell lymphoma, has rapid tumor dissemination and fatal outcome. Some authors have suggested that HMF may present as a low-grade lymphoproliferative disorder (Martinez-Escala et al., 2017) or some type of T-cell dyscrasia (Magro et al., 2014) rather than a true lymphoma. In addition, Martínez Villarreal et al. (2020) recently hypothesized that CD8+ T cells in HMF are not neoplastic but rather reactive clonal cells, whose activation causes collateral damage, including hypopigmentation from altered melanocyte function.
We hypothesize that HMF is a malignancy of the CD4+ T-cell population that is so small in comparison with that of clonal reactive CD8+ T cells that malignant CD4+ T cells cannot be detected by current methods used in clinical practice (such as PCR-based clonality assessment and standard one-label immunohistochemistry) but can be identified with single-cell RNA sequencing (scRNA-seq). To test our hypothesis, we constructed a reference mapping model utilizing the scRNA-seq data from two patients with classic MF deposited in the National Center for Biotechnology Information Gene Expression Omnibus (accession numbers GSM5280111 and GSM5047045) (Jonak et al., 2021; Rindler et al., 2021a), one normal skin sample (accession number GSM4450726) (Gaydosik et al., 2020), three atopic dermatitis samples (accession number GSE153760) (Rojahn et al., 2020), three psoriasis samples (accession number GSE162183) (Gao et al., 2021), and six vitiligo samples (accession number GSE203262) (Shiu et al., 2022). After reducing the number of genes to match our targeted scRNA-seq, we observed that each patient with classic MF displayed apparent Uniform Manifold Approximation and Projection clustering of malignant cells and other cell types of the tumor microenvironment (Figure 1a and b). The gene expression heatmap identifies genes characteristic to each cell type (Figure 1c), with the top 20 most differentially expressed genes present in the malignant cluster (Figure 1d). We tested whether a reference mapping model could be used to predict malignant cells in other MF datasets. The malignant cell cluster predicted in GSM5047045 by our model exactly corresponded to the clonal population of CD4+ T cells in this dataset with a high area under the curve (Figure 1e). In contrast, reference mapping to the normal skin sample and nonmalignant inflammatory skin diseases detected no malignant cells as expected (Figure 1f and Supplementary Figure S1a–c). To exclude any influence of the CD4 gene on malignant cell prediction, we removed CD4 from our model and repeated reference mapping. Even without the primary marker for CD4+ T cells, our model continued to perform well in predicting malignant cells in GSM5047045 (Figure 1g). The high accuracy of prediction and lack of false positives shows that the reference mapping method is valid for identifying cells with malignant phenotypes in classic MF.
Figure 1. Reference mapping model accurately predicts malignant CD4+ T cells in classic MF.
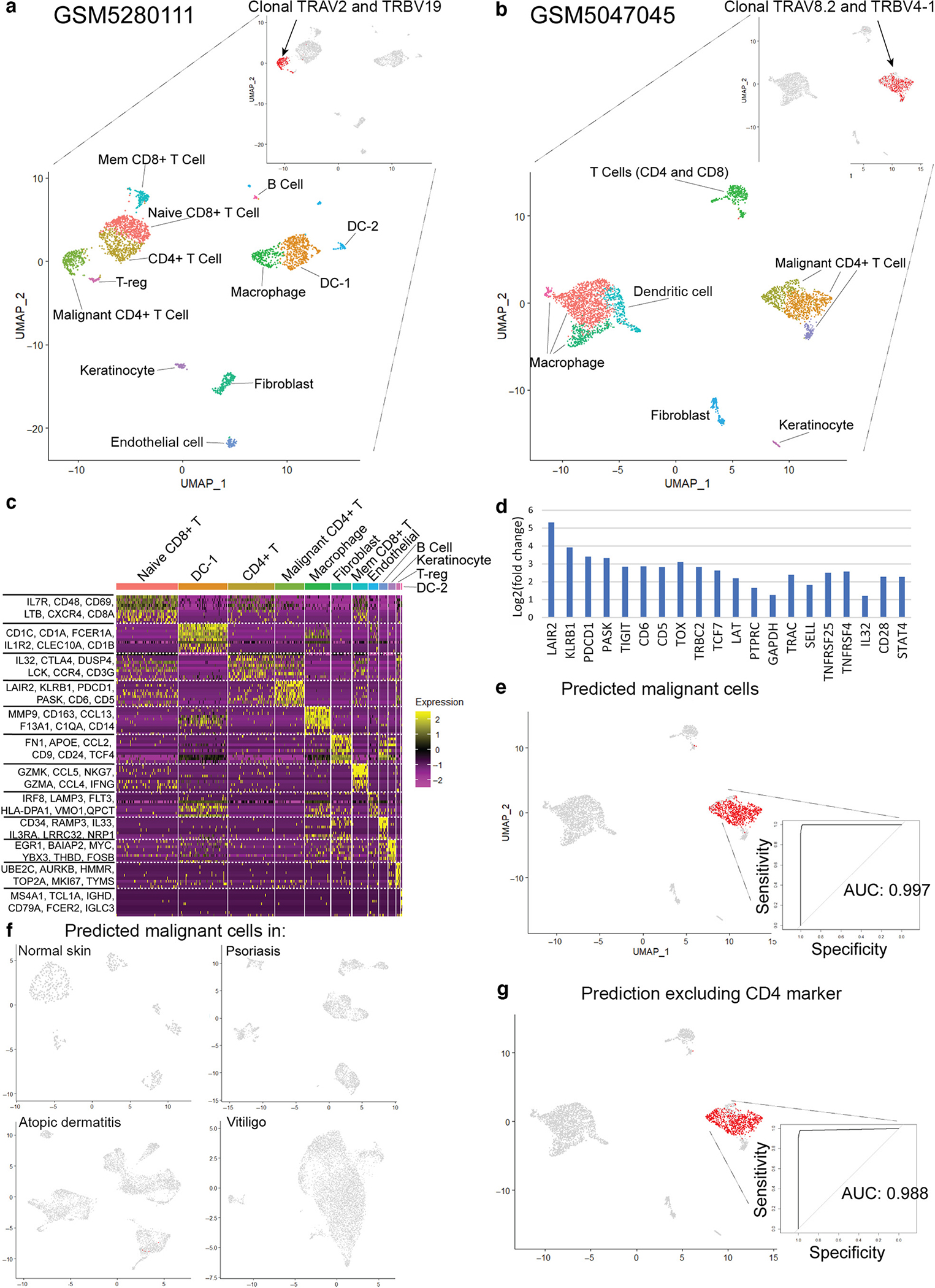
(a) UMAP of targeted scRNA-seq in one patient with classic MF (GEO accession number GSM5280111) and identification of malignant and nonmalignant cell types of the tumor microenvironment. (b) UMAP of scRNA-seq in a second patient with classic MF (GEO accession number GSM5047045). (c) Heatmap of cell clusters in GSM5280111. Genes with the greatest differential expression in each region are listed. (d) Identification of the top 20 differentially expressed genes of the malignant CD4+ T-cell population in GSM5280111. (e) Reference mapping prediction of malignant cells in GSM5047045 using a model built on GSM5280111. (f) Reference mapping prediction of malignant cells in normal skin, psoriasis, atopic dermatitis, and vitiligo samples using the model. (g) Reference mapping prediction of malignant cells in GSM5047045, excluding CD4 from the model. AUC, area under the curve; DC, dendritic cell; GEO, Gene Expression Omnibus; MF, mycosis fungoides; scRNA-seq, single-cell RNA sequencing; T-reg, regulatory T cell; UMAP, Uniform Manifold Approximation and Projection.
Second, we applied our reference mapping model built on the patients with classic MF reported previously to our patients with HMF. We utilize a fresh skin biopsy from an African American male aged 32 years with a 7-year history of persistent hypopigmented patches in the bathing-trunk distribution (Figure 2a) after written, informed consent was obtained as a part of NCT00177268 (University of Pittsburgh Institutional Review Board number 0411029). A skin biopsy showed a sparse infiltrate of atypical CD8+ epidermotropic lymphocytes (Figure 2b). Targeted scRNA-seq analysis allowed us to identify critical components of the HMF microenvironment (Figure 2c). Indeed, we identified a clonal population of CD8+ T cells using TCRα CDR3 amino acid sequences (Figure 2d and e). However, when we applied our reference malignant mapping model to this patient with HMF, we identified a distinct group of CD4+ T cells completely overlapping with malignant transcriptome separately from clonal CD8+ T cells (Figure 2d). Examining the differentially expressed genes identified in malignant CD4+ T cells from GSM5280111, we observe similar gene expression patterns between predicted malignant cells in our HMF and malignant CD4+ T cells from GSM5280111. In contrast, gene expression in CD8+ T cells in our patient with HMF did not overlap with the malignant transcriptome (Figure 2g). In fact, clonal CD8+ T cells had more similar expression patterns than polyclonal CD8+ T cells compared with predicted malignant cells (Figure 2e and f). These data highlight the presence of a small clonal CD4+ T-cell population carrying malignant transcriptome in a patient with HMF.
Figure 2. Targeted scRNA-seq and reference mapping analysis of one patient with HMF indicates a malignant CD4+ T-cell population separate from clonal CD8+ T cells.
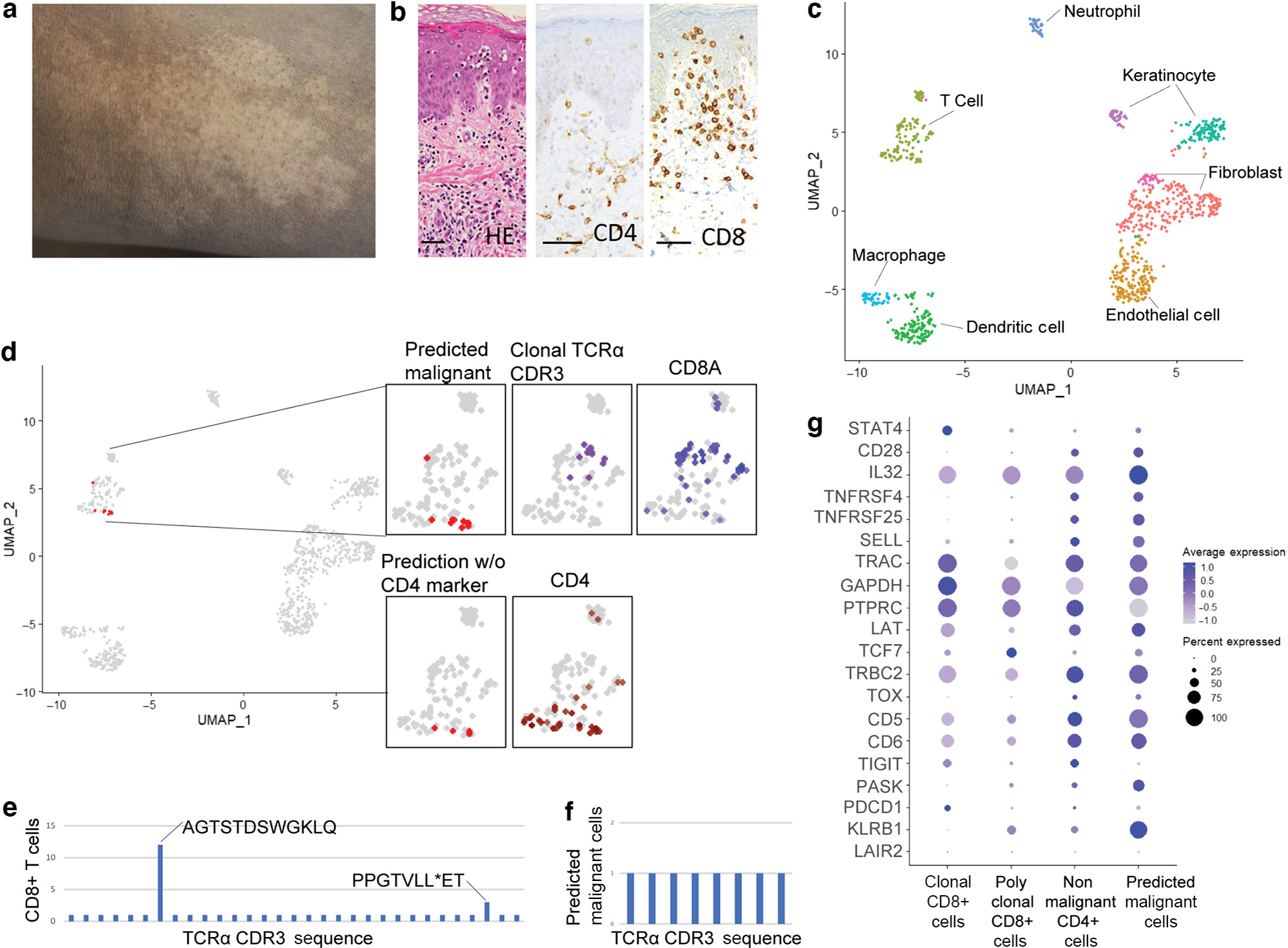
(a) Hypopigmented patch. (b) Atypical CD8+ epidermotropic lymphocytes on skin biopsy (H&E, CD4, and CD8 staining; bar = 50 μm). (c) UMAP projection of scRNA-seq from a patient with HMF. (d) Reference mapping prediction of malignant cells in a patient with HMF using model built on GSM5280111. Clustering of clonal T cells, CD4+ T cells, and CD8+ T cells are also shown. (e) TCR-α CDR3 amino acid sequences in CD8+ T cells. (f) TCR-α CDR3 amino acid sequences in predicted malignant T cells. (g) Dot plot of the top 20 differentially expressed genes used in the reference mapping model showing gene expression in clonal CD8+ T cells, polyclonal CD8+ T cells, nonmalignant CD4+ T cells, and predicted malignant cells in the patient with HMF. HMF, hypopigmented mycosis fungoides; scRNA-seq, single-cell RNA sequencing; STAT4, singal transducer and activator of transcription 4; UMAP, Uniform Manifold Approximation and Projection.
Overall, these data show that reference mapping is, to our knowledge, a previously unreported method that can accurately predict malignant cells in patients with MF among various scRNA-seq datasets. Despite the known variability of scRNA-seq between machines, experiments, and individuals diagnosed with MF (Rindler et al., 2021b), our data showed reference mapping as a robust malignant cell prediction method. Applying the reference mapping to a patient with HMF, we discovered that in fact HMF might be a malignancy of CD4+ T cells. In contrast, dominant clonal CD8+ T cells that did not show malignant alternation in their transcriptome might be reactive.
Designed as a pilot study, our study population is small, involving two patients with classic MF: one patient with HMF and one patient with normal skin. Furthermore, these data are derived from several different sources, and our HMF scRNA-seq dataset had a relatively low total number of cells. Future studies using a larger sample of classic MF, HMF, and normal skin biopsies are needed to confirm the findings in this letter. Nevertheless, we provide data toward understanding the mechanism behind HMF, providing paradigm-shifting results.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by Martin and Dorothy Spatz Foundation (OEA).
Abbreviations:
- HMF
hypopigmented mycosis fungoides
- MF
mycosis fungoides
- scRNA-seq
single-cell RNA sequencing
Footnotes
CONFLICT OF INTEREST
OEA has received research funding/grant support from Actelion, Adaptive Biotechnology, Trillium Therapeutics, Pfizer, Kyowa Kirin, and Malinkrodt; serves as a principal investigator on current clinical trials: Trillium, Eisai, Affimed, Innate Pharma, Tellomak, and Corvus; OEA is a consultant for Castle Biomarkers, SkinJet, Bioniz, Kyowa-Hakka-Kirin, and Allmiral; and OEA is a speaker for Kyowa Kirin and Helsin. The remaining authors state no conflict of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.jid.2022.08.047
Data availability statement
The following datasets related to this article can be found at https://www-ncbi-nlm-nih-gov.pitt.idm.oclc.org/geo/. Samples of the two patients with mycosis fungoides can be found at accession numbers GSM5280111 and GSM5047045, the sample of one normal skin can be found at accession number GSM4450726, three atopic dermatitis samples can be found at accession number GSE153760, three psoriasis samples can be found at accession number GSE162183, and six vitiligo samples can be found at accession number GSE203262. The hypopigmented mycosis fungoides samples were uploaded to the Gene Expression Omnibus database under accession number 212146.
REFERENCES
- Amorim GM, Niemeyer-Corbellini JP, Quintella DC, Cuzzi T, Ramos-E-Silva M. Hypopigmented mycosis fungoides: a 20-case retrospective series. Int J Dermatol 2018;57:306–12. [DOI] [PubMed] [Google Scholar]
- El-Shabrawi-Caelen L, Cerroni L, Medeiros LJ, McCalmont TH. Hypopigmented mycosis fungoides: frequent expression of a CD8 T-cell phenotype. Am J Surg Pathol 2002;26:450–7. [DOI] [PubMed] [Google Scholar]
- Gao Y, Yao X, Zhai Y, Li L, Li H, Sun X, et al. Single cell transcriptional zonation of human psoriasis skin identifies an alternative immunoregulatory axis conducted by skin resident cells. Cell Death Dis 2021;12:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaydosik AM, Queen DS, Trager MH, Akilov OE, Geskin LJ, Fuschiotti P. Genome-wide transcriptome analysis of the STAT6-regulated genes in advanced-stage cutaneous T-cell lymphoma. Blood 2020;136:1748–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonak C, Alkon N, Rindler K, Rojahn TB, Shaw LE, Porkert S, et al. Single-cell RNA sequencing profiling in a patient with discordant primary cutaneous B-cell and T-cell lymphoma reveals micromilieu-driven immune skewing. Br J Dermatol 2021;185:1013–25. [DOI] [PubMed] [Google Scholar]
- Magro CM, Hagen JW, Crowson AN, Liu YC, Mihm M, Drucker NM, et al. Hypopigmented interface T-cell dyscrasia: a form of cutaneous T-cell dyscrasia distinct from hypopigmented mycosis fungoides. J Dermatol 2014;41:609–17. [DOI] [PubMed] [Google Scholar]
- Martínez Villarreal A, Gantchev J, Lagacé F, Barolet A, Sasseville D, Ødum N, et al. Hypopigmented mycosis fungoides: loss of pigmentation reflects antitumor immune response in young patients. Cancers (Basel) 2020;12:2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Escala ME, Kantor RW, Cices A, Zhou XA, Kaplan JB, Pro B, et al. CD8+ mycosis fungoides: a low-grade lymphoproliferative disorder. J Am Acad Dermatol 2017;77:489–96. [DOI] [PubMed] [Google Scholar]
- Rindler K, Bauer WM, Jonak C, Wielscher M, Shaw LE, Rojahn TB, et al. Single-cell RNA sequencing reveals tissue compartment-specific plasticity of mycosis fungoides tumor cells. Front Immunol 2021a;12:666935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rindler K, Jonak C, Alkon N, Thaler FM, Kurz H, Shaw LE, et al. Single-cell RNA sequencing reveals markers of disease progression in primary cutaneous T-cell lymphoma. Mol Cancer 2021b;20:124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojahn TB, Vorstandlechner V, Krausgruber T, Bauer WM, Alkon N, Bangert C, et al. Single-cell transcriptomics combined with interstitial fluid proteomics defines cell type-specific immune regulation in atopic dermatitis. J Allergy Clin Immunol 2020;146:1056–69. [DOI] [PubMed] [Google Scholar]
- Shiu J, Zhang L, Lentsch G, Flesher JL, Jin S, Polleys C, et al. Multimodal analyses of vitiligo skin identify tissue characteristics of stable disease. JCI Insight 2022;7:e154585. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The following datasets related to this article can be found at https://www-ncbi-nlm-nih-gov.pitt.idm.oclc.org/geo/. Samples of the two patients with mycosis fungoides can be found at accession numbers GSM5280111 and GSM5047045, the sample of one normal skin can be found at accession number GSM4450726, three atopic dermatitis samples can be found at accession number GSE153760, three psoriasis samples can be found at accession number GSE162183, and six vitiligo samples can be found at accession number GSE203262. The hypopigmented mycosis fungoides samples were uploaded to the Gene Expression Omnibus database under accession number 212146.


