Abstract
Nucleoside analog chain terminators such as 3′-azido-3′-deoxythymidine (AZT) and 2′,3′-dideoxy-3′-thiacytidine (3TC) represent an important class of drugs that are used in the clinic to inhibit the reverse transcriptase (RT) of human immunodeficiency virus type 1. Recent data have suggested that mutant enzymes associated with AZT resistance are capable of removing the chain-terminating residue with much greater efficiency than wild-type RT and this may, in turn, facilitate rescue of DNA synthesis; these experiments were performed using physiological concentrations of pyrophosphate or nucleoside triphosphates, respectively. The present study demonstrates that the M184V mutation, which confers high-level resistance to 3TC, can severely compromise the removal of chain-terminating nucleotides. Pyrophosphorolysis on 3TC-terminated primer strands was not detectable with M184V-containing, as opposed to wild-type, RT, and rescue of AZT-terminated DNA synthesis was significantly decreased with the former enzyme. Thus, mutated RTs associated with resistance to AZT and 3TC possess opposing, and therefore incompatible, phenotypes in this regard. These results are consistent with tissue culture and clinical data showing sustained antiviral effects of AZT in the context of viruses that contain the M184V mutation in the RT-encoding gene.
Although considerable progress has been made in the treatment of AIDS, the emergence of mutated variants of human immunodeficiency virus type 1 (HIV-1) resistant to antiviral drugs is a major problem. Resistant breakthrough viruses, and also the persistence of reservoirs of latently infected cells, can limit the success of highly active antiretroviral therapy that involves a combination of potent inhibitors of the virus-encoded reverse transcriptase (RT) and protease enzymes (9, 12, 13, 40). The prolonged clinical use of nucleoside analog chain terminators, e.g., 3′-azido-3′-deoxythymidine (AZT or zidovudine) and 2′,3′-dideoxy-3′-thiacytidine (3TC or lamivudine) gives rise to resistant viruses that contain mutations in the RT enzyme (4, 10, 21, 22, 35, 36, 37, 38). This class of inhibitors competes with natural deoxynucleoside triphosphate (dNTP) pools after being phosphorylated by cellular kinases. DNA synthesis is blocked once the chain terminator is incorporated, since the nucleoside analog lacks a 3′-OH group that is required to continue the polymerization process. Mutant enzymes can reduce the probability of incorporation of a chain terminator by lowering the affinity of the latter for the dNTP binding pocket and/or reducing the efficiency of the catalytic step. The crystal structures of HIV-1 RT bound to DNA-DNA with (16) and without (17) an incoming dNTP provide a basis to understand how individual amino acid substitutions might alter the RT structure in the vicinity of the polymerase active site to confer resistance to nucleoside-analog RT inhibitors. Current knowledge with regard to mechanisms of HIV resistance to this class of drugs has recently been reviewed (34).
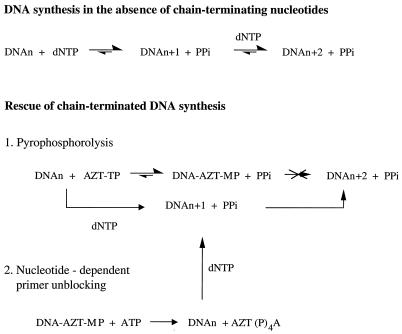
Understanding the mechanisms involved in HIV resistance to 3TC and AZT, particularly in the context of combination treatment, is of enormous practical importance, since both compounds are important components of many currently employed drug regimens. The M184V mutation in RT that confers high-level resistance to 3TC is located in the YMDD motif that constitutes a portion of the polymerase-active site of RT and other polymerases (1). Mutated RT that contains the M184V mutation (RT-M184V) can discriminate between 3TC triphosphate (3TC-TP) and dCTP, such that the incorporation rates of 3TC monophosphate (3TC-MP) are 20- to 100-fold less than with the wild-type (wt) enzyme (7, 8, 19, 28, 30). The mechanism of HIV resistance to AZT appears to be different. Tissue culture and clinical data have revealed that multiple mutations in the “fingers” and “palm” subdomains of RT are required to confer resistance to AZT (22); however, enzymes that contain a combination of amino acid substitutions frequently associated with AZT resistance, i.e., M41L, D67N, K70R, T215Y/F, and K219Q, incorporated AZT-MP at rates almost equal to that of the wt enzyme (6, 18, 19, 20). Recently, it was demonstrated that these AZT-resistant enzymes have increased rates of pyrophosphorolysis (2), the back-reaction of nucleotide incorporation. Thus, chain termination in this case is not irreversible and DNA synthesis can be rescued through removal of the incorporated AZT-MP (Fig. 1). It has further been shown that unblocking of chain-terminated primers can also be achieved with nucleoside triphosphates (24, 25).
FIG. 1.
Rescue of chain-terminated DNA synthesis via pyrophosphorolysis or nucleotide-dependent primer unblocking. Pathways of the forward and back-reactions catalyzed by HIV-1 RT are shown under different reaction conditions. In the absence of chain-terminating nucleotides, DNA synthesis is practically irreversible (14) and, therefore, the back-reaction plays only a minor role at physiological concentrations of PPi. In contrast, when chain-terminating nucleotides, such as AZT-MP, are incorporated into the growing strand, the forward reaction is blocked and pyrophosphorolysis can effectively occur. The addition of normal nucleotides may finally rescue the formerly blocked polymerization process. Removal of terminal nucleotides and rescue of DNA synthesis can, alternatively, be facilitated in the presence of nucleoside triphosphates such as ATP. Mutant enzymes that confer resistance to AZT showed an increase in both pyrophosphorolysis (2) and nucleotide-dependent primer unblocking (25), suggesting a possible rescue of DNA synthesis as a mechanism involved in such resistance.
In this study, we investigated the rescue of DNA synthesis in the context of the M184V mutation to evaluate the roles of pyrophosphorolysis and of nucleotide-dependent primer unblocking with regard to enzymes that display resistance to 3TC. Our data show that rescue of DNA synthesis was severely impaired with this mutated RT. This effect was strongly pronounced with 3TC-terminated primers and was significant, as well, with AZT-terminated primers. These results help to explain the resensitization to AZT on the part of viruses that contain AZT resistance-conferring mutations, as well as the M184V mutation in the RT gene.
MATERIALS AND METHODS
Enzymes and nucleic acids.
The wt HIV-1 RT, RT-, M184V, and AZT-resistant mutant RT containing the amino acid substitutions D67N, K70R, T215F, and K219Q (RT-AZTr) were prepared and purified as previously described (2, 29). The primer-template substrate used in this study was derived from the polypurine tract of the HIV-1 genome (11). The following oligonucleotides were employed: 5′-CGTTGGGAGTGAATTAGCCCTTCCAGTCCCCCCTTTTCTTTTAAAAAGTGCTAAGA-3′ (termed 57D), which served as a DNA template, and 5′-TTAAAAGAAAAGGGGGG-3′ (termed 17D), 5′-TTAAAAGAAAAGGGGGGA-3′ (termed 18D), and 5′-TTAAAAGAAAAGGGGGGAC-3′ (termed 19D), which were used as primer strands.
Constructs 57D and 17D were used as a primer-template combination to study the rescue of DNA synthesis, while constructs 18D and 19D were used to generate 3TC- and AZT-terminated primers, respectively. All DNA oligonucleotides were chemically synthesized and purchased from GIBCO-BRL Inc., Montreal, Quebec, Canada.
Preparation of AZT- and 3TC-terminated primer strands.
To generate chain-terminated primers, oligonucleotides 18D and 19D were first radiolabeled at the 5′ end and subsequently extended with 3TC-TP or AZT-TP using HIV-1 RT. 5′-end labeling of oligonucleotides was performed with [γ-32P]ATP and T4 polynucleotide kinase (GIBCO-BRL). Reactions were allowed to proceed for 30 min, and then heat inactivation of the kinase was performed at 95°C for 5 min. Next, oligonucleotide 57D (120 nM), which served as a template strand, was added to the solution containing the 32P-labeled primer (100 nM). The primer and template were hybridized by denaturing of the nucleic acids in the mixture at 95°C for 2 min, followed by incubation at 72°C (10 min) and cooling for 20 min to room temperature. The preannealed primer-template substrate (100 nM) was incubated with HIV-1 RT (150 nM) in a buffer containing 50 mM Tris-HCl (pH 7.8) and 50 mM NaCl. AZT-TP or 3TC-TP (100 μM) was added to form ternary RT–primer-template–dNTP complexes. Oligonucleotides 18D and 19D, respectively, permitted incorporation of 3TC-MP and AZT-MP at the next template positions. Reactions were initiated with MgCl2 at a final concentration of 6 mM and allowed to proceed for 60 min at 37°C. Chain-terminated primers were purified on 15% polyacrylamide–7 M urea gels containing 50 mM Tris borate (pH 8) and 1 mM EDTA.
Pyrophosphorolysis.
The specific activities of the three different RT species that were studied differed slightly from one another; this is most likely attributable to differences in amino acid composition (19, 29). To specifically monitor putative differences among RTs with regard to the back-reaction (pyrophosphorolysis), all enzyme preparations had to be studied under conditions that would ensure the same efficiency of polymerization. The polymerase activity was analyzed under steady-state conditions using the same primer-template combination that was employed to study the influence of the various amino acid substitutions on rates of pyrophosphorolysis. For this purpose, the preannealed primer-template substrate (100 nM 17D-57D) was incubated for 5 min at 37°C with either wt or mutated HIV-1 RT (10 to 20 nM) in a buffer containing 50 mM Tris-HCl (pH 7.8), 50 mM NaCl, and a dNTP cocktail that allowed incorporation of the first four nucleotides (dATP, dCTP, dTTP, and ddGTP at a final concentration of 100 μM each). Polymerization was restricted to a limited number of incorporation events to ensure that measurements of specific activity would not be confounded by differences in processivity among these RT species (2, 3). Reactions were initiated by MgCl2 (6 mM) and stopped after 5 min by addition of EDTA to a final concentration of 40 mM. Samples were precipitated with ethanol, resuspended in 95% formamide, and finally resolved on 12% polyacrylamide–7 M urea gels, followed by overnight exposure to Kodak Bio Mat films. Band intensities were analyzed by densitometry or by molecular image analysis. Using this protocol, all enzyme preparations were adjusted to allow extension of 20% of the primer during a 5-min reaction. To monitor pyrophosphorolysis, the preannealed primer-template substrates (100 nM) that contained either AZT- or 3TC-terminated primer strands were preincubated for 5 min at 37°C with HIV-1 RT (50 nM) in a buffer containing 50 mM Tris-HCl (pH 7.8), 50 mM NaCl, and 6 mM MgCl2.
Pyrophosphorolytic cleavages were initiated by adding PPi at a final concentration of 150 μM. Reactions were performed at 37°C and stopped at different times by adding 1-μl aliquots of reaction mixture to 9 μl of 95% formamide containing 40 mM EDTA. Samples were finally heat denatured (5 min at 95°C) and analyzed as described above.
Rescue of DNA synthesis in the presence of PPi and ATP.
All experiments were performed with oligonucleotide 57D as the template strand and the 17D primer. 5′-end labeling of the primer and the formation of duplex DNA were performed as described above. The duplex (100 nM) was preincubated with wt or mutant RT (50 nM), followed by the addition of dATP (50 μM), dCTP (50 μM), and AZT-TP (10 μM) to generate an AZT-terminated primer strand. Under these conditions, about 90% of the primer was found to be extended after 2 min. The reaction was then allowed to proceed for 20 min to convert the 17D primer completely into the AZT-terminated form. After 20 min, a cocktail of dTTP (100 μM), ddGTP (100 μM), and PPi (150 μM) or ATP (3.5 mM) was added to the reaction mixture to assay rescue of DNA synthesis at the next template position. (A 35 mM solution of ATP was pretreated with 1 U of inorganic pyrophosphatase [Sigma], which liberated 1 μmol of putative contaminating pyrophosphate per min.) Aliquots were removed at different time points and analyzed as described above. Experiments that monitored the rescue of 3TC-terminated DNA synthesis followed the same procedure, except that a cocktail of dATP (50 μM) and 3TC-TP (50 μM) was used to generate the terminated primer and a mixture of dCTP (100 μM) and ddTTP (100 μM) was added to follow rescue of DNA synthesis.
RESULTS
Pyrophosphorolysis on AZT- and 3TC-terminated primer strands.
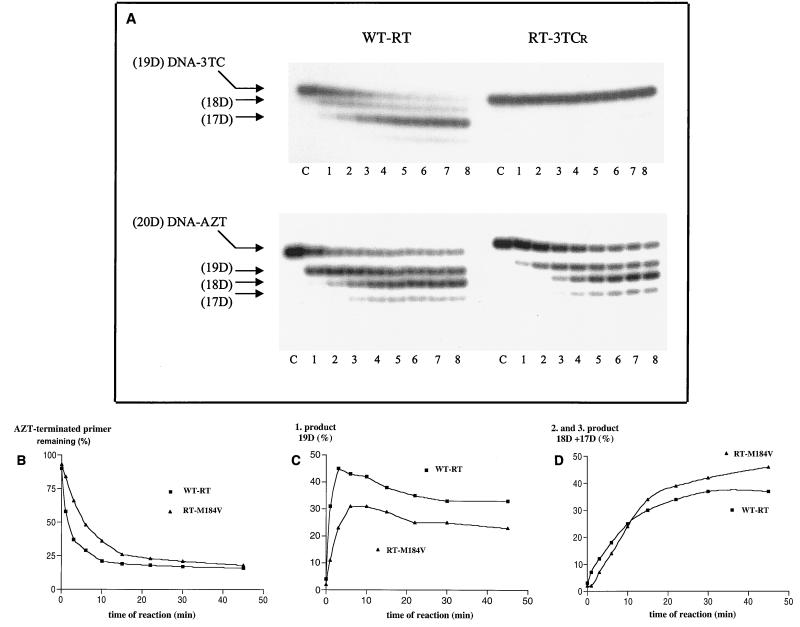
Diminished rates of 3TC-MP incorporation by RT-M184V have been well documented (7, 8, 19, 28, 30). Whether the reverse reaction, i.e., pyrophosphorolysis, has the same tendency has not been studied. Yet, this question is important in evaluating the impact of the M184V substitution on the efficiency with which chain-terminated DNA synthesis can be rescued. To address this issue, we first compared the rates of pyrophosphorolytic cleavage on a 3TC-terminated primer using wt HIV-1 RT and RT-M184V. We employed a well-established DNA-DNA primer-template system that was used to study the initiation of HIV-1 plus-strand DNA synthesis (11). Results obtained with this substrate may contribute insight additional to that obtained on RNA-dependent pyrophosphorolysis with the wt and AZT-resistant enzymes (2). The time course experiment of Fig. 2A (left, top) shows that wt RT removed 3′-terminal 3TC-MP with considerable efficiency; the terminal residue was almost completely cleaved within the first 10 min of the reaction (lane 4). In contrast, RT-M184V was virtually unable to complete this back-reaction and pyrophosphorolytic cleavages were almost nondetectable with this mutant enzyme, as shown in Fig. 2A (right, top). The 3TC-terminated primer remained uncleaved, even when relatively high concentrations of RT were used, e.g., 300 nM HIV-1 RT and 100 nM primer-template. Thus, RT-M184V possesses a phenotype that is the opposite of that of RT that contains amino acid substitutions associated with AZT resistance, i.e., increased rates of pyrophosphorolysis (2).
FIG. 2.
Pyrophosphorolysis catalyzed by wt RT and RT-M184V. The time course of pyrophosphorolytic cleavage was analyzed with the same DNA primer-template substrate that was recently employed to study mechanistic aspects of initiation of HIV plus-strand synthesis (11). (A) Time course of removal of 3TC-MP (top) and AZT-MP (bottom). Chain-terminated primers are labeled (20D) DNA-AZT and (19D) DNA-3TC. Reaction products are indicated by (19D), (18D), and (17D), with numbers referring to the nucleotide length of the primer and D designating DNA. RT–primer-template complexes were incubated with 150 μM PPi as described in Materials and Methods. Reactions were stopped at 1, 3, 6, 10, 15, 22, 30, and 45 min and analyzed on 15% polyacrylamide gels. Lanes 1 to 8 represent these reactions, and lane C is a control performed in the absence of PPi. (B, C, and D) Graphic representation of the data shown at the bottom of panel A. Panel B shows the decrease in the amount of the uncleaved primer, while C and D show the formation of the primary product and subsequent reaction products, respectively.
This result is also interesting in the light of earlier findings that viruses containing the M184V substitution in RT alongside relevant AZT resistance-conferring mutations became resensitized to AZT (23). Therefore, we next analyzed the efficiency of pyrophosphorolysis with an AZT-terminated primer in the context of the 3TC-resistant mutant enzyme. As shown in Fig. 2A (bottom) and B, pyrophosphorolytic cleavages were still diminished with RT-M184V, although this effect was less pronounced than that observed with the 3TC-terminated primer. Quantification of these data revealed that a two- to threefold reduction in pyrophosphorolytic removal of AZT had occurred with RT-M184V. These differences were restricted to the first cleavage event that required removal of the terminal AZT-MP, while subsequent cleavage events, that involved pyrophosphorolysis of “normal” nucleoside monophosphates, occurred at rates similar to those seen with wt RT (Fig. 2C and D). At the start of the reaction (less than 10 min), these ensuing cleavages, that yielded DNA fragments with 18 and 17 nucleotides, appeared to be delayed with RT-M184V. This is a consequence of the diminished rates of pyrophosphorolysis of the AZT-terminated primer. However, as the concentration of the initial cleavage product increased, these shorter products appeared to be stronger with the mutated RT than with wt RT. This represents an “internal control,” indicating that the diminished rate of pyrophosphorolysis of AZT-terminated primers with the 3TC-resistant mutant enzyme is a specific effect attributable to the amino acid change at residue 184. Were the decrease in pyrophosphorolysis nonspecific and, as such, attributable to putative differences in active-site concentrations of the enzyme preparations used, then diminished rates of pyrophosphorolysis on the AZT-terminated primer with “normal” nucleoside monophosphates would also have been observed.
Rescue of chain-terminated DNA synthesis.
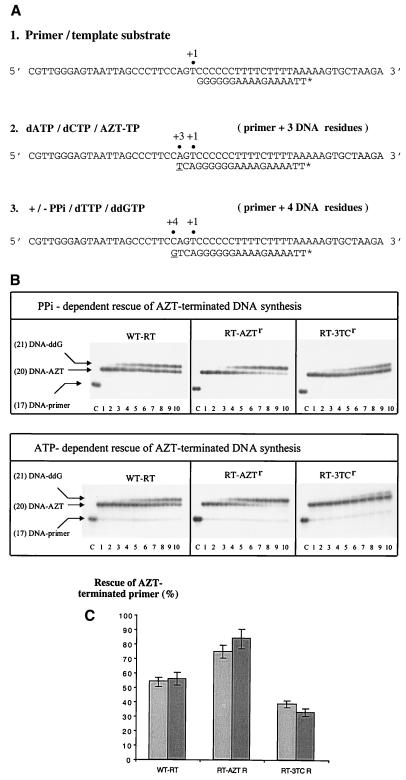
We next analyzed whether differences in rates of the back-reaction would translate into corresponding alterations with regard to the ability of the various RTs to rescue chain-terminated DNA synthesis. In this context, we compared wt RT, RT-M184V, and RT-AZTr, which showed increased rates of rescue of AZT-terminated DNA synthesis with both PPi (2) and ATP (25). In order to reduce the complexity of the reaction to a minimum, we devised an assay that allows us to monitor the rescue of DNA synthesis at a single template position (Fig. 3A); this involves the quantitative termination of DNA synthesis by AZT-TP and the rescue of the reaction using a mixture of dTTP and PPi. Removal of AZT-MP and incorporation of dTMP is then monitored by the addition of ddGTP at the next template position.
FIG. 3.
Comparison of pyrophosphate- and nucleotide-dependent rescue of DNA synthesis. (A) Schematic representation of the assay used to study these events. RT–primer-template complexes were incubated with dATP, dCTP, and AZT-TP to generate an AZT-terminated primer (step 1). Reactions were allowed to proceed for 20 min to quantitatively convert the primer strand into an AZT-terminated product (step 2). Rescue of DNA synthesis requires removal of AZT-MP and incorporation of dTMP and at least an additional nucleotide. The AZT-terminated complex was therefore incubated simultaneously with dTTP, ddGTP, and PPi or ATP (step 3). The extent of rescued DNA synthesis is thus reflected by the efficiency with which ddGTP is incorporated at position +4. (B) Rescue of AZT-terminated DNA synthesis with wt RT, RT-AZTr, and RT-M184V. Reactions were performed with 150 μM PPi (top) and 3.5 mM ATP (bottom). Lane C shows the unextended primer, and lane 1 shows the AZT-terminated primer. Lanes 2 to 10 show reactions performed for 1, 3, 6, 10, 15, 22, 30, 45, and 60 min, respectively. (C) Graphic representation of data shown in panel B, including standard deviations. Rescue of AZT-terminated DNA synthesis was compared after a 60-min reaction with PPi (light bar) and ATP (dark bar), respectively.
Figure 3B shows a time course of the rescue of DNA synthesis performed with wt RT, RT-AZTr, and RT-M184V using physiological concentrations of PPi. In agreement with previous results performed with RNA templates (2), the use of RT-AZTr resulted in rescue of DNA synthesis to levels higher than those seen with wt RT. The opposite effect was seen with RT-M184V, in which case rescue of DNA synthesis was seen to a lesser extent than with the wt enzyme. The same pattern was seen when ATP was used to remove the terminal residue. Thus, regardless of whether ATP or PPi was employed, the M184V mutation in RT resulted in diminished rescue of DNA synthesis while substitutions in RT associated with resistance to AZT increased the strength of the reaction.
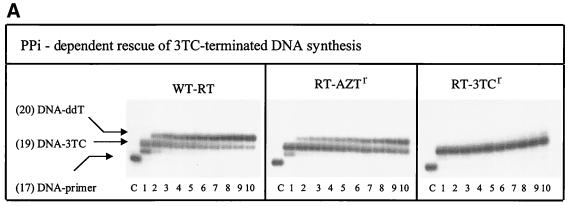
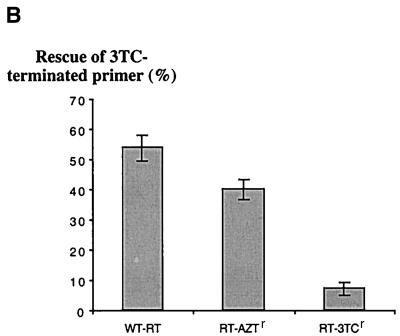
The increased rescue of DNA synthesis with the RT-AZTr enzyme was specifically seen with the AZT-terminated primers, while 3TC-terminated DNA synthesis was rescued at levels lower than those seen with wt RT (Fig. 4). Moreover, and in accordance with the severely restricted rates of pyrophosphorolysis shown in Fig. 2A, RT-M184V was able to rescue 3TC-terminated DNA synthesis neither in the presence of PPi (Fig. 4A and B) nor in the presence of ATP (data not shown). In this context, it is interesting that neither wt RT nor RT-AZTr was able to quantitatively incorporate 3TC-MP while RT-M184V did this effectively over longer reaction times, i.e., >15 min using a 1:2 molar ratio of RT and the nucleic acid substrate. This result may reflect a poor affinity of RT-M184V for the 3TC-terminated primers and, consequently, does not contradict previous data that showed diminished rates of incorporation of 3TC-MP. Indeed, since this enzyme was used at concentrations lower than that of the primer-template, the 3TC-terminated primer should have been expected to compete with the unextended fraction. A relatively poor affinity for the 3TC-terminated primer likely results in preferential binding of the M184V mutated enzyme to the unmodified 3′-OH terminus and might help to explain the fact that the primer was quantitatively extended. Such diminution in affinity for 3TC-terminated primers on the part of RT-M184V is consistent with the severely compromised pyrophosphorolysis associated with this enzyme.
FIG. 4.
Rescue of 3TC-terminated DNA synthesis. (A) Reactions performed in the presence of 150 μM pyrophosphate with wt RT, RT-AZTr, and RT-M184V. The reaction conditions were similar to those described in the legend to Fig. 3, except that chain-terminating 3TC-MP was incorporated at template position +2 and rescue of DNA synthesis was monitored by the addition of ddTMP at position +3. It is noteworthy that incorporation of 3TC-MP was not quantitatively achieved with wt RT and RT-AZTr (see Results). (B) Graphic representation of the data shown in panel A with standard deviations. Rescue of 3TC-terminated DNA synthesis was compared after a 60-min reaction.
DISCUSSION
We have demonstrated that HIV-1 RT-M184V is virtually unable to remove 3′-terminal 3TC-MP via pyrophosphorolytic cleavage. This mutated enzyme is also known to possess diminished processivity (3), increased fidelity or diminished nucleotide misincorporation with regard to both the RNA-dependent DNA polymerase and DNA-dependent DNA polymerase steps of reverse transcription (8, 15, 26, 27, 31, 39), and diminished incorporation of 3TC-MP (7, 8, 19, 28, 30) in comparison with wt RT. However, the present demonstration of compromised pyrophosphorolytic cleavage is the strongest effect of this mutation on RT function seen to date. Crystal structure analysis of HIV-1 RT bound to a DNA-DNA primer-template substrate, with (16) and without bound dNTP (17), as well as the structures of M184V and M184I mutated enzymes (23), may provide an explanation for our observation. Modeling of the inhibitor at the active site of these structures suggested a steric clash between the sugar moiety of 3TC-TP and mutation at position 184, which is believed to account for the diminished incorporation rate of 3TC-MP. Since pyrophosphorolysis represents the back-reaction of nucleotide incorporation, it follows that such unfavorable interactions are involved during the removal of terminal nucleoside monophosphates as well. However, as corroborated by our data, the back-reaction should be more sensitive to this structural hindrance than the forward reaction, since rates of pyrophosphorolytic cleavage are normally significantly slower than rates of nucleotide incorporation (14).
Our finding that pyrophosphorolysis on 3TC-terminated primer strands is virtually eliminated in the case of RT-M184V seems paradoxical, since one could argue that increased rates of pyrophosphorolysis should lead to augmented rescue of chain-terminated DNA synthesis and therefore contribute to higher levels of drug resistance. Indeed, precisely such mechanisms have been proposed to explain RT resistance to AZT (2). Thus, any decrease in rates of pyrophosphorolytic cleavage might be expected to increase drug sensitivity. However, the 200- to 1,000-fold increase in the 50% inhibitory concentration that has been reported for 3TC in association with viruses containing the M184V mutation is extremely high (4, 10, 35, 36, 37). Therefore, the diminished rate of incorporation of 3TC-MP by RT-M184V is likely both necessary and sufficient to ensure that viral replication can continue in the presence of the drug and the rescue of DNA synthesis appears to be irrelevant with regard to mechanisms that confer resistance to 3TC. However, impaired pyrophosphorolysis may be one of the factors that contribute to the reduced replication fitness of M184V-containing variants of HIV-1; this may relate to the possibility that a single nucleotide addition is sufficient to severely compromise reverse transcription. Given the scenario that 3TC-MP is eventually incorporated, DNA synthesis would most likely be efficiently blocked, due to a lack of pyrophosphorolytic cleavage by RT-M184V. Kinetics studies have indicated that the incorporation efficiency (kpol/Kd) of 3TC-MP is as high as the formation of G-T mismatches (2.4 × 10−3 versus 1.1 × 10−3 μM−1 s−1) and significantly higher than those of more unfavorable mismatch combinations (8). Thus, the probability that 3TC-MP is incorporated by RT-M184V in vivo is not negligible, which helps to explain previous observations that the viral burden in the presence of 3TC does not rebound to baseline levels, despite the presence of the M184V mutation (36).
Enzymes associated with resistance to AZT possessed the opposite phenotype; i.e., they manifested increased rates of rescue of DNA synthesis with either PPi (2) or ATP (25). The mechanisms involved in the two reactions may be similar. Presumably, the β- and γ-phosphates of nucleoside triphosphates can occupy the same binding site as PPi as pyrophosphorolysis proceeds. Further studies are required to determine whether there is any preference for PPi or for nucleoside triphosphates or whether both are employed with similar efficiencies in vivo. Identification of reaction intermediates in HIV-infected cells may shed some light on this issue. However, data that are available from cell-free assays indicate that both PPi and nucleoside triphosphates need to be considered as cellular components capable of removing chain-terminating stop nucleotides. In addition, although removal of the chain terminator is definitely a prerequisite for rescue of DNA synthesis, other factors can influence this process as well. These include increased affinity of mutated RT-AZTr for the AZT-terminated primer (2, 5) and increased enzyme processivity (2), both factors that can facilitate rescue of DNA synthesis. Conversely, efficient binding of the dNTP that is complementary to the next template position can diminish the removal of the chain terminator and, in turn, diminish such rescue (25).
Our results also show that amino acid substitutions in the RT enzyme and the chemical nature of the inhibitor are important parameters that can critically affect the efficiency with which the terminal residue that blocks DNA synthesis is removed. An increase in rescue of DNA synthesis with RT-AZTr was specifically seen for AZT-terminated primer strands, while 3TC-terminated DNA synthesis was rescued even at lower levels than those seen with wt RT. This result is in accordance with tissue culture and clinical data that show that AZT-resistant viruses are not cross-resistant to 3TC. Conversely, AZT-resistant viruses can be resensitized in the context of AZT-3TC combination therapy alongside the appearance of the M184V mutation (23). These observations are consistent with our current results showing that RT-M184V possesses a diminished rescue capacity for AZT-terminated DNA synthesis. Recent experiments performed with mutant enzymes containing the M184V mutation in a background of amino acid substitutions associated with resistance to AZT further support these observations (M. Götte, D. Arion, M. A. Parniak, and M. A. Wainberg, unpublished data).
The rescue of chain-terminated DNA synthesis, regardless of whether it is mediated by PPi or ATP, must be considered an important mechanism to explain HIV resistance to AZT. It will be of interest to study such reactions in the context of other mutated RTs and nucleoside analog inhibitors. However, our data make it clear that unblocking of the primer does not represent a unifying mechanism to explain all HIV resistance to nucleoside reverse transcriptase inhibitors. Indeed, our findings with both 3TC and AZT and the corresponding drug-resistant RT species point to the involvement of diametrically opposed mechanisms. The diminished chain terminator incorporation rate is important with regard to resistance to 3TC but not AZT; in contrast, rescue of DNA synthesis is important with regard to AZT resistance but may be irrelevant in the case of 3TC and RT-M184V.
ACKNOWLEDGMENTS
This study was supported by grants to M.A.W. and M.A.P. from the Medical Research Council of Canada.
REFERENCES
- 1.Argos P. A sequence motif in many polymerases. Nucleic Acids Res. 1988;16:9909–9916. doi: 10.1093/nar/16.21.9909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arion D, Kausshik N, McCormick S, Borkow G, Parniak M A. Phenotypic mechanism of HIV-1 resistance to 3′-azido-3′-deoxythymidine (AZT): increased polymerization processivity and enhanced sensitivity to pyrophosphate of the mutant viral reverse transcriptase. Biochemistry. 1998;37:15908–15917. doi: 10.1021/bi981200e. [DOI] [PubMed] [Google Scholar]
- 3.Back N K T, Nijhuis M, Keulen W, Boucher C A B, Essnik B B O, van Kuilenburg A B P, van Gennip A H, Berkhout B. Reduced replication of 3TC-resistant HIV-1 variants in primary cells due to a processivity defect of the reverse transcriptase enzyme. EMBO J. 1996;15:4040–4049. [PMC free article] [PubMed] [Google Scholar]
- 4.Boucher C A B, Cammack N, Schipper P, Schuurman R, Rouse P, Wainberg M A, Cameron J M. High level resistance to (−) enantiomeric 2′-deoxy-3′-thiacytidine in vitro is due to one amino acid substitution in the catalytic site of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother. 1993;37:2231–2234. doi: 10.1128/aac.37.10.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canard B, Sarfati S R, Richardson C C. Enhanced binding of azidothymidine-resistant human immunodeficiency virus 1 reverse transcriptase to the 3′-azido-3′-deoxythymidine 5′-mono-phosphate-terminated primer. J Biol Chem. 1998;273:14596–14604. doi: 10.1074/jbc.273.23.14596. [DOI] [PubMed] [Google Scholar]
- 6.Carroll S S, Geib J, Olsen D B, Stahlhut M, Shafer J A, Kuo L C. Sensitivity of HIV-1 reverse transcriptase and its mutants to inhibition by azidothymidine triphosphate. Biochemistry. 1994;33:2113–2120. doi: 10.1021/bi00174a018. [DOI] [PubMed] [Google Scholar]
- 7.Feng J Y, Anderson K S. Mechanistic studies comparing the incorporation of (+) and (−) isomers of 3TCTP by HIV-1 reverse transcriptase. Biochemistry. 1999;38:55–63. doi: 10.1021/bi982340r. [DOI] [PubMed] [Google Scholar]
- 8.Feng J Y, Anderson K S. Mechanistic studies examining the efficiency and fidelity of DNA synthesis by the 3TC-resistant mutant (184V) of HIV-1 reverse transcriptase. Biochemistry. 1999;38:9440–9448. doi: 10.1021/bi990709m. [DOI] [PubMed] [Google Scholar]
- 9.Finzi D, Blankson J, Siliciano J D, Margolick J B, Chadwick K, Pierson T, Smith K, Lisziewicz J, Lori F, Flexner C, Quinn T C, Chaisson R E, Rosenberg E, Walker B, Gange S, Gallant J, Siliciano R F. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1997;5:512–517. doi: 10.1038/8394. [DOI] [PubMed] [Google Scholar]
- 10.Gao Q, Gu Z, Parniak M A, Cameron J, Cammack N, Boucher C, Wainberg M A. The same mutation that encodes low-level human immunodeficiency virus type 1 resistance to 2′,3′-dideoxyinosine and 2′,3′-dideoxycytidine confers high-level resistance to the (−) enantiomer of 2′,3′-dideoxy-3′-thiacytidine. Antimicrob Agents Chemother. 1993;37:1390–1392. doi: 10.1128/aac.37.6.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Götte M, Maier G, Mochi Onori A, Cellai L, Wainberg M A, Heumann H. Temporal coordination between initiation of HIV (+)-strand DNA synthesis and primer removal. J Biol Chem. 1999;274:11159–11169. doi: 10.1074/jbc.274.16.11159. [DOI] [PubMed] [Google Scholar]
- 12.Gulick R M, Mellors J W, Havlir D, Eron J J, Gonzalez C, McMahon D, Richman D D, Valentine F T, Jonas L, Meibohm A, Emini E A, Chodakewitz J A. Treatment with indinavir, zidovudine, and lamivudine in adults with human immunodeficiency virus infection and prior antiretroviral therapy. N Engl J Med. 1997;337:734–739. doi: 10.1056/NEJM199709113371102. [DOI] [PubMed] [Google Scholar]
- 13.Günthard H F, Wong J K, Ignacio C C, Guatelli J C, Riggs N L, Havlir D V, Richman D D. Human immunodeficiency virus replication and genotypic resistance in blood and lymph nodes after a year of potent antiretroviral therapy. J Virol. 1998;72:2422–2428. doi: 10.1128/jvi.72.3.2422-2428.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh J-C, Zinnen S, Modrich P. Kinetic mechanism of the DNA-dependent DNA polymerase activity of human immunodeficiency virus reverse transcriptase. J Biol Chem. 1993;268:24607–24613. [PubMed] [Google Scholar]
- 15.Hsu M, Inouye P, Rezende L, Richard N, Li Z, Prasad V R, Wainberg M A. Higher fidelity of RNA-dependent DNA mispair extension by M184V drug-resistant that wild-type reverse transcriptase of human immunodeficiency virus type 1. Nucleic Acids Res. 1997;25:4532–4536. doi: 10.1093/nar/25.22.4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang H, Chopra R, Verdine G L, Harrison S C. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- 17.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 Å resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerr S G, Anderson K S. Pre-steady-state kinetic characterization of wild type and 3′-azido-3′deoxythymidine (AZT) resistant human immunodeficiency virus type 1 reverse transcriptase: implication of RNA directed DNA polymerization in the mechanism of AZT resistance. Biochemistry. 1997;36:14064–14070. doi: 10.1021/bi9713862. [DOI] [PubMed] [Google Scholar]
- 19.Krebs R, Immendörfer U, Thrall S H, Wöhrl B M, Goody R S. Single-step kinetics of HIV-1 reverse transcriptase mutants responsible for virus resistance to nucloside inhibitors zidovudine and 3-TC. Biochemistry. 1997;36:10292–10300. doi: 10.1021/bi970512z. [DOI] [PubMed] [Google Scholar]
- 20.Lacey S F, Reardon J E, Furfine E S, Kunkel T A, Bebenek K, Eckert K A, Kemp S D, Larder B A. Chemical studies on the reverse transcriptase and RNase H activities from human immunodeficiency virus strains resistant to 3′-azido-3′deoxythymidine. J Biol Chem. 1992;267:15789–15794. [PubMed] [Google Scholar]
- 21.Larder B A, Darby G, Richman D D. HIV with reduced sensitivity to zidovudine (AZT) isolated during prolonged therapy. Science. 1989;243:1731–1734. doi: 10.1126/science.2467383. [DOI] [PubMed] [Google Scholar]
- 22.Larder B A, Kemp S D. Multiple mutations in HIV-1 reverse transcriptase confer high-level resistance to zidovudine (AZT) Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 23.Larder B A, Kemp S D, Harrigan P R. Potential mechanism for sustained antiretroviral efficacy of AZT-3TC combination therapy. Science. 1995;269:969–699. doi: 10.1126/science.7542804. [DOI] [PubMed] [Google Scholar]
- 24.Meyer P R, Matsuura S E, So A G, Scott W A. Unblocking of chain-terminated primer by HIV-1 reverse transcriptase through a nucleotide-dependent mechanism. Proc Natl Acad Sci USA. 1998;95:13471–13476. doi: 10.1073/pnas.95.23.13471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meyer P R, Matsuura S E, Mohsin Mian A, So A G, Scott W A. A mechanism of AZT resistance: an increase in nucleotide-dependent primer unblocking by mutant HIV-1 reverse transcriptase. Mol Cell. 1999;4:35–43. doi: 10.1016/s1097-2765(00)80185-9. [DOI] [PubMed] [Google Scholar]
- 26.Oude Essnik B B, Back N K, Berkhout B. Increased polymerase fidelity of the 3TC-resistant variants of HIV-1 reverse transcriptase. Nucleic Acids Res. 1997;25:3212–3217. doi: 10.1093/nar/25.16.3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pandey V, Kaushik N, Rege N, Sarafianos S G, Yadav P N S, Modak M J. Role of methionine 184 of human immunodeficiency virus type-1 reverse transcriptase in the polymerase function and fidelity of DNA synthesis. Biochemistry. 1996;35:2168–2179. doi: 10.1021/bi9516642. [DOI] [PubMed] [Google Scholar]
- 28.Quan Y, Gu Z, Li X, Li Z, Morrow C D, Wainberg M A. Endogenous reverse transcription assays reveal high-level resistance to the triphosphate of (−)2′-dideoxy-3′-thiacytidine by mutated M184V human immunodeficiency virus type 1. J Virol. 1996;70:5642–5645. doi: 10.1128/jvi.70.8.5642-5645.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quan Y, Inouye P, Rong L, Götte M, Wainberg M A. Dominance of the E89G substitution in HIV-1 reverse transcriptase in regard to increased polymerase processivity and patterns of pausing. J Biol Chem. 1998;273:21918–21925. doi: 10.1074/jbc.273.34.21918. [DOI] [PubMed] [Google Scholar]
- 30.Quan Y, Gu Z, Liang X C, Parniak M A, Wainberg M A. Endogenous reverse transcriptase assays reveal synergy between combinations of the M184V and other drug resistance-conferring mutations in interactions with nucleoside analog triphosphates. J Mol Biol. 1998;227:237–247. doi: 10.1006/jmbi.1997.1592. [DOI] [PubMed] [Google Scholar]
- 31.Rezende L F, Drosopoulos W C, Prasad V R. The influence of 3TC resistance mutation M184I on the fidelity and error specificity of human immunodeficiency virus type 1 reverse transcriptase. Nucleic Acids Res. 1998;26:3066–3072. doi: 10.1093/nar/26.12.3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rooke R, Tremblay M, Soudeyns H, DeStephano L, Yao X-J, Fanning M, Montaner J S G, O'Shaugnessy M, Gelmon K, Tsoukas C, Ruedy J, Wainberg M A. Isolation of drug-resistant variants of HIV-1 from patients on long-term zidovudine (AZT) therapy. AIDS. 1989;3:411–415. doi: 10.1097/00002030-198907000-00001. [DOI] [PubMed] [Google Scholar]
- 33.Sarafianos S G, Das K, Clark A D, Jr, Ding J, Boyer P L, Hughes S H, Arnold E. Lamivudine (3TC) resistance in HIV-1 reverse transcriptase involves steric hindrance with beta-branched amino acids. Proc Natl Acad Sci USA. 1999;96:10027–10032. doi: 10.1073/pnas.96.18.10027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sarafianos S G, Das K, Ding J, Boyer P L, Hughes S H, Arnold E. Touching the heart of HIV-1 drug resistance: the fingers close down on the dNTP at the polymerase active site. Chem Biol. 1999;6:137–146. doi: 10.1016/s1074-5521(99)80071-4. [DOI] [PubMed] [Google Scholar]
- 35.Schinazi R F, Lloyd R M, Jr, Nguyen M-H, Cannon D L, McMillan A, Ilksoy N, Chu C K, Liotta D C, Bazmi H Z, Mellors J W. Characterization of human immunodeficiency virus resistant to oxathiolane-cytosine nucleosides. Antimicrob Agents Chemother. 1993;37:875–881. doi: 10.1128/aac.37.4.875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schuurman R, Nijhuis M, van Leeuwen R, Shipper P, de Jong D, Collis P, Danner S A, Mulder J, Loveday C, Christopherson C, Boucher C A B. Rapid changes in human immunodeficiency virus type 1 RNA load and appearance of drug-resistant virus populations in persons treated with lamivudine (3TC) J Infect Dis. 1995;171:1411–1419. doi: 10.1093/infdis/171.6.1411. [DOI] [PubMed] [Google Scholar]
- 37.Tisdale M, Kemp S D, Parry N R, Larder B A. Rapid in vitro selection of human immunodeficiency virus type 1 resistant to 3′-thiacytidine inhibitors due to a mutation in the YMDD region of reverse transcriptase. Proc Natl Acad Sci USA. 1993;90:5653–5656. doi: 10.1073/pnas.90.12.5653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wainberg M A. HIV resistance to antagonists of viral reverse transcriptase. In: Dalgleish A G, Weiss R A, editors. HIV and the new viruses. London, United Kingdom: Academic Press, Inc.; 1999. pp. 223–249. [Google Scholar]
- 39.Wainberg M A, Drosopoulos W C, Salomon H, Hsu M, Borkow G, Parniak M A, Gu Z, Song Q, Manne J, Islam S, Castriota G, Prasad V R. Enhanced fidelity of 3TC-selected mutant HIV-1 reverse transcriptase. Science. 1996;217:1282–1285. doi: 10.1126/science.271.5253.1282. [DOI] [PubMed] [Google Scholar]
- 40.Wong J K, Hezareh M, Gunthard H F, Havlir D V, Ignacio C C, Spina C A, Richman D D. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science. 1997;278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]