Abstract
Endoplasmic reticulum (ER) stress is a potentially lethal condition that is induced by the abnormal accumulation of unfolded or misfolded secretory proteins in the ER. In eukaryotes, ER stress is managed by the unfolded protein response (UPR) through a tightly regulated, yet highly dynamic, reprogramming of gene transcription. Although the core principles of the UPR are similar across eukaryotes, unique features of the plant UPR reflect the adaptability of plants to their ever-changing environments and the need to balance the demands of growth and development with the response to environmental stressors. The past decades have seen notable progress in understanding the mechanisms underlying ER stress sensing and signalling transduction pathways, implicating the UPR in the effects of physiological and induced ER stress on plant growth and crop yield. Facilitated by sequencing technologies and advances in genetic and genomic resources, recent efforts have driven the discovery of transcriptional regulators and elucidated the mechanisms that mediate the dynamic and precise gene regulation in response to ER stress at the systems level.
Introduction
In his evolution theory, Charles Darwin articulated that “a plant on the edge of a desert is said to struggle for life against the drought, though more properly it should be said to be dependent on the moisture”1, which encapsulates the inherent struggle for survival plants face in their natural habitats. As sessile organisms, plants lack the mobility to evade adverse environmental conditions, relying instead on robust preparation and resilience. Such immobility exposes them to a multitude of environmental stresses, including drought, heat, salinity and pathogen infections, throughout the life cycle. Notably, when confronted with such adverse conditions, the demand for proper folding of secretory proteins – a crucial process orchestrated in the endoplasmic reticulum (ER) – can exceed the ER biosynthetic capacity2,3. This imbalance gives rise to a potentially lethal condition known as ER stress. ER stress is managed by a suite of conserved signalling pathways, collectively termed the unfolded protein response (UPR), which swiftly reprogramme the expression of molecular chaperone and foldase genes, augmenting the protein folding capacity of the ER. Concurrently, the UPR curtails general secretory protein synthesis and promotes protein degradation through ER-associated protein degradation4,5 and ER-phagy6, thereby alleviating the ER burden. When effective, these ER stress management strategies enable plants to adapt to environmental changes, ensuring their survival and growth in dynamic environments. Should these adaptive measures prove inadequate, the UPR can trigger programmed cell death2,7,8.
The fundamental mechanisms of the UPR are highly conserved across eukaryotes2. However, plants have evolved unique UPR features to adapt to environmental challenges and control organ growth in stressful conditions9-13, which involves the addition or omission of regulators and pathways. Unlike the three distinct branches found in metazoans2, only two signalling cascades initiated by the ER membrane protein sensors ribonuclease and kinase inositol requiring 1 (IRE1) and basic leucine zipper 28 (bZIP28) exist in the plant UPR (Fig. 1). Among these, IRE1 stands out as the most highly conserved ER stress sensor2,9, as the bZIP28/ATF6 branch is present in metazoans and plants but absent in yeast2. IRE1 represses a universal growth regulator, target of rapamycin (TOR)10; this repression of TOR function indicates that the UPR governs conserved key developmental factors, albeit through yet unknown mechanisms. Even if components of the UPR are conserved across kingdoms, their mechanism of action can differ in plants. For instance, Bax inhibitor 1 (BI-1) has a crucial role as a highly conserved modulator of cell death in metazoans, in which it interacts with and inhibits IRE1 in response to ER stress14, yet BI-1 does not suppress IRE1 activity during plant ER stress11,15. These nuanced differences underscore the intricate nature of the plant UPR, honed over millions of years of divergent evolution and adaptation to the environment.
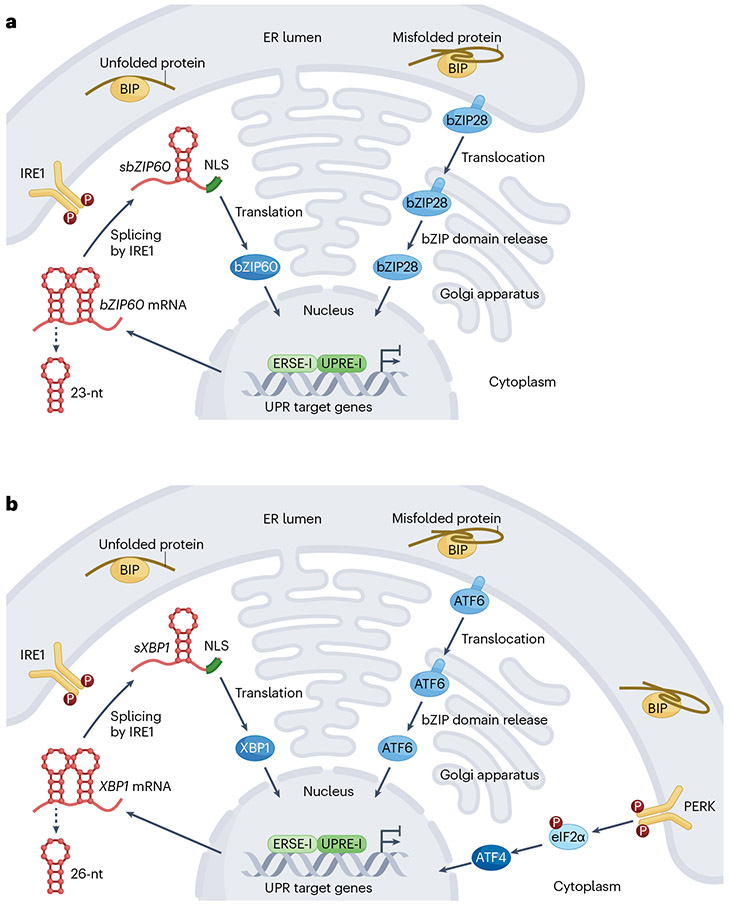
Fig. 1 |. The ER stress response in plants versus metazoans.
a, Endoplasmic reticulum (ER) stress sensing and signalling mechanisms in the plant unfolded protein response (UPR). In response to ER stress, plants activate a protective mechanism called the UPR to maintain the stability of the ER. The plant UPR is primarily controlled by two distinct signalling pathways. The first is a response mediated by bZIP28, a transcription factor (TF) associated with the ER membrane, which, when ER stress occurs, plays a crucial role by undergoing a process of post-translational cleavage. This cleavage takes place in the Golgi apparatus, and, as a result of it, the N-terminal part of bZIP28 is released and moves to the nucleus. Inside the nucleus, the N-terminal domain of bZIP28 regulates the expression of genes involved in responding to ER stress. In the second pathway, a dual-functioning protein kinase and ribonuclease, IRE1, is activated when ER stress occurs. IRE1 splices the mRNA encoding the transcription factor bZIP60. This splicing event leads to a change in the mRNA sequence, generating a transcription factor with a nuclear localization signal (NLS; indicated by a green bar). The newly formed bZIP60 with an NLS enters the nucleus, where it participates in regulating genes related to the ER stress response. IRE1 controls the abundance of transcripts also through regulated IRE1-dependent decay, a process not highlighted in this figure. Additionally, bZIP28 and bZIP60 can form combinations either by pairing with themselves (homodimerization) or with each other (heterodimerization). Together, they collaborate to control the expression of genes that help the plant adapt to ER stress via binding to cis-regulatory elements such as ER stress-responsive element-I (ERSE-I) or unfolded protein response element-I (UPRE-I). b, ER stress sensing and signalling mechanisms in the metazoan UPR. IRE1, bZIP28 (ATF6) and bZIP60 (XBP1) maintain their structural integrity and processing mechanisms in metazoans, reflecting the evolutionary importance of these pathways. However, in the metazoan UPR, a distinctive branch emerges, orchestrated by the protein kinase RNA-like ER kinase (PERK). As an immediate adaptive response to ER stress, PERK phosphorylates eukaryotic translation initiation factor 2 subunit-α (eIF2α), causing a transient reduction of protein synthesis. Simultaneously, the phosphorylated eIF2α triggers a specific translation programme, targeting a select group of mRNAs, including ATF4 mRNA and others harbouring upstream open reading frames within their 5′ untranslated regions. ATF4 is a transcription factor that functions as a master regulator, orchestrating the expression of a multitude of UPR target genes. 23-nt, 23-nucleotide mRNA segment; 26-nt, 26-nucleotide mRNA segment; P, phosphate group; sbZIP60, spliced bZIP60 mRNA; sXBP1, spliced XBP1 mRNA.
A better understanding of the mechanisms involved in ER stress management has the potential to help meet the food demands of a growing world population16, for example, by promoting the development of stress-tolerant crop varieties and improving global agricultural sustainability. Among the various layers that constitute the plant UPR, the dynamics of transcriptional responses stand out as a key regulatory point for restoring ER homeostasis. Recent insights from systems-level approaches have revealed that gene regulation within the plant UPR is a finely orchestrated process, tightly governed by master regulators17,18, their co-regulators19-24, cis-regulatory elements (CREs)17,19 and epigenetic components25, which differentially act depending on the intensity, duration and nature of the environmental challenges leading to ER stress. Of note, although the basic gene regulatory mechanisms of UPR signalling seem to be highly conserved across plant lineages, the UPR has evolved in a species-specific manner (Box 1).
Box 1. Evolution of the UPR across plant lineages.
Orthologues of the master Arabidopsis unfolded protein response (UPR) regulators have been successfully identified and characterized in rice (Oryza sativa L.)117-121, maize (Zea mays)36,120,122, sorghum (Sorghum bicolor L.)123, potato (Solanum tuberosum L.)124 and tomato (Solanum lycopersicum cv. Moneymaker)125. Interestingly, copy numbers of UPR genes vary among plant species, likely owing to multiple rounds of genome duplication events that occurred independently in each species126. In plants, the duplication of genes and genomes followed by sub-functionalization and neo-functionalization are generally considered as the main contributors to the emergence of new traits, especially in stress resilience127,128. For instance, whereas Arabidopsis and maize possess three and two isoforms of the IRE1 gene, respectively, the rice genome only contains a single isoform121. Furthermore, the Arabidopsis IRE1 gene partially duplicated to generate an isoform known as IRE1c, which encodes a protein that lacks the ER luminal domain12, considered a sensor region129,130. Although IRE1c does not have a major role in sensing ER stress, it functions in gametophyte development with IRE1b12. The maize genome encodes one bZIP transcription factor, in addition to the homologue of bZIP60 that encodes a type II membrane-anchored protein, yet sequence analyses indicate that it matches both bZIP17 and bZIP2836. Additionally, the pro-death factor PHOSPHATASE TYPE 2CA-INTERACTING RING FINGER PROTEIN 1 (PIR1), which acts downstream of IRE1 under ER stress, is found exclusively in dicots (for example, soybean) and not in monocots (for example, maize and sorghum)22. Moreover, the EMS-MUTAGENIZED BRI SUPPRESSOR (EBS7) seems to be a plant-specific component of a broadly conserved ER-associated protein degradation complex131. These observations underscore the dynamic evolution of the UPR throughout plant lineages.
Here, we review research into gene regulation in the plant UPR and its transformative impact on understanding ER stress management in plants. We first provide a historical overview of the application of genetic technologies for understanding gene regulation in the plant UPR and resulting biological insights into underlying mechanisms. Next, we delve into the intricate gene regulation models of the plant UPR built on results gathered from cutting-edge systems-wide approaches. Finally, we consider the potential implementation of emerging technological advances to enhance our insights into the gene regulation of the plant UPR.
A timeline of plant UPR research
Originating from a fortuitous observation and capitalizing on Arabidopsis thaliana as a model plant system (Fig. 2), as well as substantial work in crop species9,26,27 (Box 2), plant UPR research gained momentum with the advent of sequencing technologies and the availability of numerous genetic and genomic tools and resources, including multi-omics data, for this tractable multicellular model species. Upon completion of the Arabidopsis reference genome28, DNA microarray technologies soon enabled measurements of gene expression levels in response to ER stress at genome scale in a single experiment29. In 2003, the first transcriptome-wide studies, covering approximately one-third of Arabidopsis genes, implicated hundreds of genes associated with non-secretory biological pathways in the UPR and identified the existence of ER stress-responsive CREs in the promoters of ER stress-responsive genes30,31. Subsequent research adopted microarray technologies with increased gene coverage and under various conditions32-34. For example, Mishiba et al. revealed that 96% of genes that were previously downregulated in ER stress were not differentially regulated in a double loss-of-function mutant of IRE1a and IRE1b, indicating that the majority of ER stress-inducible transcriptional changes are under the control of IRE1a and IRE1b.34 Although the microarray platforms did not cover the entire collection of Arabidopsis genes and had inevitable technical challenges (for example, high levels of signal-to-noise ratio or the inability to distinguish homologues), they successfully revealed a global landscape of gene expression reprogramming of the plant UPR under ER stress conditions and initiated notable discoveries in the early genomics era.
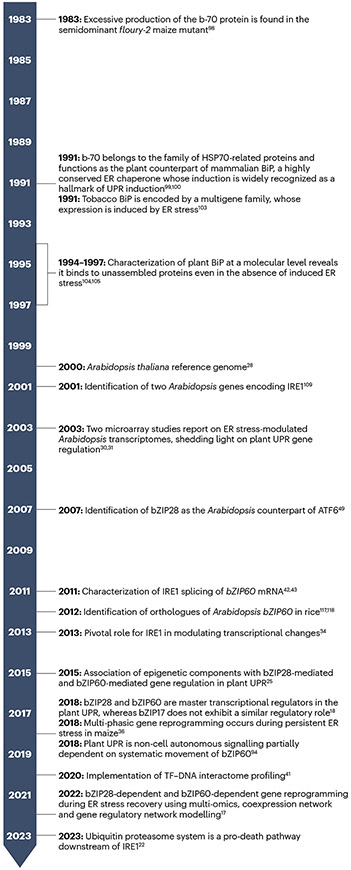
Fig. 2 |. Historical timeline of plant UPR research.
Plant unfolded protein response (UPR) research has evolved along historical milestones, a selection of which is highlighted in the figure. In 1983, Galante et al. observed excessive production of the b-70 protein in the semidominant floury-2 maize mutant, which exhibited reduced levels of zein, a maize endosperm storage protein98. It was not until 1991 that the surplus b-70 production in the maize mutant was fully characterized. It was confirmed that b-70 belongs to the family of HSP70-related proteins and functions as the plant counterpart of the mammalian luminal binding protein (BiP), a critical protein associated with UPR induction99-101. The floury-2 mutation caused a signal peptide defect in an α-zein protein, resulting in the accumulation of abnormal zein proteins that induces BiP overexpression102. By the early 1990s, researchers established that plant BiP, like its metazoan counterpart, aided in protein folding within the endoplasmic reticulum (ER). Using tobacco as a model, they found that BiP bound to assembly-defective proteins even without induced ER stress, demonstrating its role in protein folding and quality control103-105. In the late 1990s, research shifted to Arabidopsis, a model plant with abundant genetic and genomic tools106-108. Research in Arabidopsis enabled the identification of plant counterparts of key UPR components42,43,49,109-115 and provided a platform to study ER stress responses at the molecular level but also at the organismal level94. Microarray technologies116 and later RNA-seq technologies revolutionized genomics, enabling genome-scale analysis of gene expression in response to ER stress. These tools unveiled the roles of UPR-bZIP transcription factors (TFs; bZIP28 and bZIP60) and dynamic transcriptomic changes17,18,30-34. In recent years, chromatin immunoprecipitation followed by sequencing (ChIP–seq)17,36 and enhanced yeast-one hybrid (eY1H) screening21,41 have revealed hundreds of transcription factor-binding targets and upstream UPR biomarker genes, respectively, offering a deeper understanding of plant UPR gene regulation and its broader implications, which has been further expanded with multi-omics approaches17,21,22,41. These discoveries have substantially advanced our understanding of the plant UPR, shedding light on the molecular mechanisms underlying gene regulation in response to ER stress in plants. These large-scale datasets are invaluable resources for future functional research in crops.
Box 2. Insights into gene regulation for crop UPR implementation.
Environmental stresses, including drought, heat, salinity and pathogen infections, can directly or indirectly induce ER stress, leading to substantial agronomic losses3,38,64,71-76,124. This realization has prompted plant researchers to harness knowledge gained from Arabidopsis in investigating unfolded protein response (UPR) gene regulation in crops. These efforts have established valuable connections between the functional roles of UPR gene regulation and crop resilience and productivity3,132. For example, drawing parallels with the heat stress-inducible activation of IRE1 in Arabidopsis42, Li et al. observed a similar response in maize, in which heat stress triggered the unconventional splicing of bZIP6038. Intriguingly, the increased levels of maize spliced bZIP60 (sbZIP60) conferred heat tolerance by directly influencing the expression of a crucial transcription factor (TF), heat shock transcription factor 13, through direct DNA binding via a canonical endoplasmic reticulum (ER) stress-responsive cis-regulatory element (CRE). Tomato also exhibits heat stress-mediated induction of sbZIP60, bZIP28 and BiPs125, indicating conserved gene regulatory mechanisms between monocots and dicots. Notably, overexpressing BiP in soybean enhances drought tolerance, resulting in reduced expression levels of drought-responsive genes133. Knockout Chinese Cabbage lines, which have a disrupted gene encoding ELONGATED HYPOCOTYL 5 (HY5) — a transcriptional repressor in the UPR that competes with bZIP28 on the gene promoters in Arabidopsis20 — show reduced growth inhibition under ER stress compared with the wild type134. Collectively, these results underscore the feasibility of engineering the UPR to generate stress-resilient crops.
The development of high-throughput data collection techniques has enabled the simultaneous exploration of transcriptomes and TF–DNA interactomes in crops, providing foundational resources for further characterizations. For instance, an RNA sequencing dataset has been generated in maize during an extensive time frame of ER stress, along with open chromatin and small RNA profiles36. These multi-omics data revealed the existence of extensive ER stress-inducible waves of transcriptomic changes as well as chromatin accessibility and microRNA abundance changes. Herath and Verchot reported transcriptomic changes induced by ER stress in potato, revealing an extension of the UPR to other signalling pathways135. Furthermore, enhanced yeast-one hybrid screens of 23 promoter fragments from six maize UPR biomarker gene promoters against 600 maize TFs unveiled hundreds of interactions with a wide array of multifunctional TFs, broadening the scope of plant UPR beyond ER stress41. These large-scale datasets are invaluable resources for future functional research in crops.
The advent of RNA sequencing (RNA-seq) technology35 marked a notable leap in understanding transcriptome changes, enabling functional genomics (for example, mutant analyses) and time course analyses at scale. In 2018, Kim et al. applied RNA-seq to perform comparative transcriptome profiling in double loss-of-function mutants of the UPR transcription factors (TFs), bZIP28, bZIP60 (hereafter called UPR-bZIP TFs) and bZIP17, under exposure to multiple ER stress inducers, including tunicamycin18. This study underscored intricate regulatory cohorts of these TFs in the UPR. In the same year, Srivastava et al. performed RNA-seq in maize (Zea mays) with high resolution of time points under persistent ER stress, which spanned from the early pro-survival to mid-phase and the late pro-death phase36. This study revealed multi-modal phases of the transcriptome during ER stress, suggesting that the interplay of transcriptional alterations associated with diverse phase-specific biological pathways has a critical role in the UPR. These studies demonstrate the power of RNA-seq in obtaining a comprehensive landscape of ER stress-inducible transcriptomic changes. Since then, several RNA-seq studies have afforded notable biological insights into gene regulation in the plant UPR17,22,23,37,38. For example, the recent application of coexpression network modelling to transcriptomic datasets revealed the existence of ER stress-responsive gene modules associated with distinct biological pathways and has unveiled dynamic transcriptomic changes when ER stress is resolved17. Through RNA-seq analyses in a maize mutant lacking functional bZIP60, Li et al. revealed a bZIP60-mediated molecular link of the UPR to the heat stress response38.
Gene regulation is mediated mainly by dynamic interactions between TFs and DNA39. Therefore, identifying global binding targets of TFs is a straightforward approach to understanding gene regulatory mechanisms in the plant UPR. To decipher the regulatory hierarchy under ER stress, chromatin immunoprecipitation followed by sequencing (ChIP–seq) has been applied in multiple ER stress studies in plants, including Arabidopsis17,21 and maize36, revealing hundreds of TF binding targets and temporal binding intensity changes of, for example, UPR-bZIP TFs. In maize, RNA-seq data were captured along with open chromatin and small RNA profiles during an extensive period of ER stress36; these multi-omics data revealed extensive waves of transcriptomic changes as well as chromatin accessibility and microRNA abundance changes induced by ER stress36. Enhanced yeast-one hybrid (Y1H) screening, a semi-automatic version of Y1H40, identified numerous TFs upstream of UPR biomarker genes in Arabidopsis21 and maize41, facilitating the identification of new regulators in the UPR. Specifically, enhanced Y1H screens of 23 promoter fragments from six maize UPR biomarker genes against 500 maize TFs unveiled hundreds of interactions with a wide array of multifunctional TFs, broadening the scope of plant UPR beyond ER stress41. By integrating transcriptomes and protein–DNA interactomes into gene regulatory networks17,21,22,41, multi-omics approaches have greatly advanced our understanding of plant UPR gene regulation in recent years, providing foundational resources for further characterization.
Mechanisms of UPR gene regulation
Eukaryotic cells consistently monitor the levels of unfolded or misfolded proteins in the ER, enabling communication of ER homeostasis status with the nucleus and cytoplasm to prompt rapid transcriptional changes if ER stress occurs, that is, when ER protein-folding capacity is insufficient2,9. In this section, we delve into the fundamental aspects of plant UPR signalling and how dynamic gene expression changes ultimately influence cell fate under ER stress. We focus on Arabidopsis as most of the advances have been gained in this model species.
ER stress sensing and cytoplasmic signalling
The ER membrane-associated protein sensors IRE1 and bZIP28 possess an ER luminal domain for sensing unfolded proteins and cytosolic regions that facilitate transcriptional regulation (Fig. 1). Upon ER stress, the type I transmembrane protein IRE1 oligomerizes and autophosphorylates to activate its RNase activity, splicing a 23-nucleotide intron in the bZIP60-encoding mRNA42,43 (Fig. 1). This frameshift results in a spliced mRNA encoding a TF with a nuclear localization signal, known as spliced bZIP60 (sbZIP60). Direct IRE1 phosphorylation targets other than itself have yet to be identified in vivo in any organism. In addition to controlling gene expression through the splicing of bZIP60 mRNAs, IRE1 can exert post-transcriptional gene regulation through a conserved process dubbed regulated IRE1-dependent decay34,44,45. The GTP-binding protein β1 (AGB1) seems to participate in UPR gene regulation mediated by IRE1, as evidenced by dramatic transcriptional reprogramming of the UPR biomarker genes ERdj3A and ERdj3B, and higher growth suppression in response to ER stress in a triple-mutant of IRE1a, IRE1b and AGB1, relative to a double-mutant of IRE1a and IRE1b46,47.
In the other UPR branch, bZIP28, a type II transmembrane protein, is typically retained in the ER through its association with BiP48. Upon ER stress, it dissociates from BiP and translocates to the Golgi apparatus, where it is cleaved by site-1 protease (S1P) and subsequently by S2P49. The released N-terminal fragment contains a bZIP domain and relocates to the nucleus (Fig. 1). However, a later genetic study showed that bZIP28 undergoes proteolytic cleavage by S2P and as-yet-unidentified protease(s), bypassing the S1P cleavage in the Golgi apparatus50. Therefore, further investigation is needed to establish the mechanisms underlying the sequential cleavage of bZIP28.
Because the main regulatory UPR branch initiated by the protein kinase RNA-like ER kinase (PERK) in metazoans is missing in plants9 – although PERK-like activity exists in plants through a stress-inducible orthologue of the eIF2α kinase, general control non-repressible 2 (GCN2)51 – the plant UPR seems to centralize its transcriptional regulation around bZIP60 and bZIP28 (Fig. 1). Overall, the plant UPR utilizes a combination of these two signalling pathways to relay sensing signals to the nucleus, where dynamic gene expression changes are orchestrated to maintain ER proteostasis and uphold cellular functions under ER stress.
Other ER membrane-anchored proteins respond to ER stress and participate in UPR gene regulation. For example, two TFs of the plant-unique NAC (No apical meristem, Arabidopsis transcription activation factor and Cup-shaped cotyledon) family, NAC08952 and NAC06253, are anchored to the ER membrane, cleaved and transported to the nucleus under ER stress and abiotic stresses, modulating programmed cell death. B cell lymphoma 2-associated athanogene 7 (BAG7), another ER membrane-associated protein, is proteolytically processed in response to heat stress, which could trigger ER stress, and is translocated into the nucleus to regulate gene expression changes necessary for stress tolerance54. Currently, the extent of gene regulation by these membrane-associated proteins compared with bZIP28 and bZIP60 remains largely unknown.
Nuclear signalling
In the nucleus, the UPR-bZIP TFs bZIP28 and bZIP60 can heterodimerize and homodimerize19, and they can thus independently or jointly regulate hundreds or thousands of genes involved in a broad array of biological pathways, including ER protein translocation, folding and secretion, as well as the degradation of misfolded proteins17,18,25. In transactivation assays using tobacco infiltration, introducing agrobacterium cells containing sbZIP60 alone induced a 2.5-fold increase in luciferase gene expression driven by a BiP3 promoter, whereas introducing both bZIP28 and sbZIP60 effector cells resulted in a 14.7-fold increase22. These findings suggest that bZIP28 and bZIP60 work synergistically to coordinate gene expression changes in the UPR. Notably, integrating RNA-seq and ChIP–seq profiles revealed that over 90% of the genes differentially regulated by bZIP28 and bZIP60 under ER stress and recovery are not directly bound by these TFs17, hinting to indirect regulation via transcriptional signal cascades in the UPR.
When they work together, bZIP28 and bZIP60 regulate their target genes by directly binding to the promoters via canonical ER stress-responsive CREs19,21,22. In plants, two canonical ER stress-responsive CREs are predominantly enriched in the promoters of ER stress-responsive genes: ER stress-responsive element-I (ERSE-I; 5′-CCAAT-N10-CACG-3′) and unfolded protein response element-I (UPRE-I; 5′-TGACGTG-G/A-3′)17,19,30,55. Mutations in these motifs significantly reduce the binding intensity and transcriptional activity of bZIP28 and bZIP6019,21. ChIP–seq analyses of bZIP28 and bZIP60 confirmed that ERSE-I is the most highly enriched motif in the centre of global bZIP28-binding and bZIP60-binding peaks17, supporting the idea that UPR-bZIP TFs regulate UPR target genes by binding to promoters via ERSE-I in vivo17. Plant UPR gene regulation also involves ERSE-I and UPRE-I variants, such as ERSE-II (5′-ATTGG-N2-CACG-3′)17,56 UPRE-II (5′-GATGACGCGTAC-3′)17,57 and UPRE-III (5′-TCATCG-3′)17,58.
The observation that some transcription of UPR genes occurs in a triple-mutant lacking bZIP28, bZIP60 and their repressor, G-class bZIP TF2 (GBF2)21, raises the question of whether UPR gene expression may be modulated by yet another pathway working redundantly but much less prominently than bZIP28 and bZIP60. Such a pathway might be governed by bZIP17, an ER membrane protein sensor that is activated by a proteolytic machinery similar to bZIP28 (that is, sequential cleavage by S1P and S2P)59,60. Although bZIP17 has been demonstrated to interact with bZIP28 or bZIP60 in yeast using a yeast two-hybrid system19, heterodimerization has yet to be substantiated through other means, and, importantly, in vivo under conditions of ER stress. Consequently, the functional role of bZIP17 in regulating the transcriptional activities of bZIP28 and bZIP60 in the UPR remains to be explored. Genetic mutant and transcriptome analyses have revealed significant contributions of bZIP17 to plant growth under physiological conditions and salt stress13,18. However, it is noteworthy that bZIP17 is not considered a primary regulator in the UPR. Indeed, one RNA-seq study18 highlighted bZIP28 and bZIP60, but not bZIP17, as major modulators in transcriptional responses to ER stress. Particularly in ER stress conditions, the expression of UPR biomarker genes was not induced in a bzip28/bzip60 double mutant, whereas it was significantly induced in bzip17/bzip28 and bzip17/bzip60 double mutants18, which supports the hypothesis that bZIP17 is dispensable for UPR gene regulation in ER stress. The challenge of isolating a viable triple-mutant involving bZIP17, bZIP28 and bZIP60 has impeded the exploration of potential functional redundancies among these bZIP TFs.
Gene expression dynamics upon ER stress
During ER stress, the plant UPR swiftly enacts an adaptive phase to help cells survive by enhancing the quality control machinery for maintaining ER homeostasis. This encompasses maintaining optimal rates of protein production and folding as well as rapid responses to diverse stimuli such as plant hormones, nutrient levels, growth factors, energy status and redox balance17,30-33,36. However, when the UPR capacity to maintain ER homeostasis is overwhelmed, cells initiate cell death programmes such as autophagy and metabolic changes36. This transition coincides with a decline in sbZIP60 expression36, a proxy for IRE1 activity42,43, suggesting that reduced IRE1 splicing activity may contribute to cell death. BI-1, a modulator of cell death in both plants and animals61,62, has a role in this process, with its expression peaking in the middle–late phase of the adaptive UPR36 and being modulated by the IRE1-bZIP60 branch11. These results support a model in which IRE1 helps make transitional decisions for cellular fates. Emerging evidence increasingly points to transcriptional reprogramming in the plant UPR extending beyond the adaptive–apoptotic transition, encompassing various biological pathways, such as organ development, metabolism, hormone regulation and stress responses17,36. Interestingly, the expression of bZIP28 and unspliced bZIP60 is highly co-induced upon ER stress17,36 and upon abiotic and biotic stresses63,64 (Fig. 3). This finding suggests the existence of transcriptional regulatory mechanisms for bZIP60 and bZIP28 that are independent of IRE1-mediated post-transcriptional modification and proteolysis cleavage, respectively. Further observations suggest that the expression of unspliced bZIP60 is intricately regulated by multiple TFs, with its promoter having an ERSE-I-like sequence that can be bound by multiple TFs in vivo, including bZIP60 itself17, bZIP28 (ref. 17) and Abscisic acid insensitive 5 (ABI5)22. Furthermore, enhanced Y1H screen studies21,41 revealed that 120 Arabidopsis TFs and 107 maize TFs bind to the 1-kb promoter of Arabidopsis bZIP60 and maize bZIP60, respectively21,41. These findings suggest complex transcriptional regulation of unspliced bZIP60, potentially involving feedback loops between UPR-bZIP TFs. Overall, at the transcriptional level, the plant UPR coordinates multi-phase gene reprogramming, which has an impact on not only the adaptive–apoptotic transition but also a wide array of biological pathways.
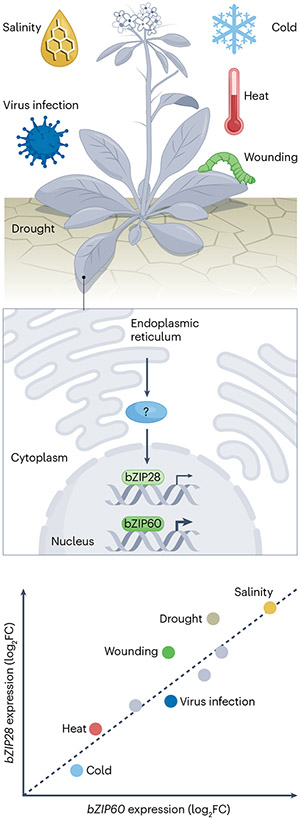
Fig. 3 |. Transcriptional coregulation of bZIP28 and bZIP60 in response to various stress conditions.
The plant unfolded protein response is responsive to diverse environmental stresses, including abiotic stresses, such as heat, drought and cold, and biotic stresses, such as pathogen infections or tissue damage by herbivores. Of note, the expression of bZIP28 and unspliced bZIP60 exhibits high inducibility in response to endoplasmic reticulum stress, albeit with bZIP28 showing lower transcriptional amplitude than bZIP60. For instance, analysis of expression data from 82 Arabidopsis transcriptome datasets sourced from the EMBL-EBI Expression Atlas revealed Pearson correlation coefficients of 0.66 (P = 1.31 × 10−7) and 0.59 (P = 4.15 × 10−4) under abiotic and biotic stress conditions, respectively, which supports a high degree of correlation of the expression of the two master regulators during stress (see the graph). log2FC, log-transformed fold change (stress/control).
Context dependency of gene reprogramming
The remarkable plasticity in growth and development65,66, coupled with quantifiable responses to ER stress inducers11,21,22,67, renders plants an ideal system for elucidating the dynamics and mechanisms of ER stress-responsive gene regulation through phenotypic screening (Fig. 4). Three distinct experimental approaches to ER stress conditions discussed here have played a pivotal role in the molecular characterization of the plant UPR.
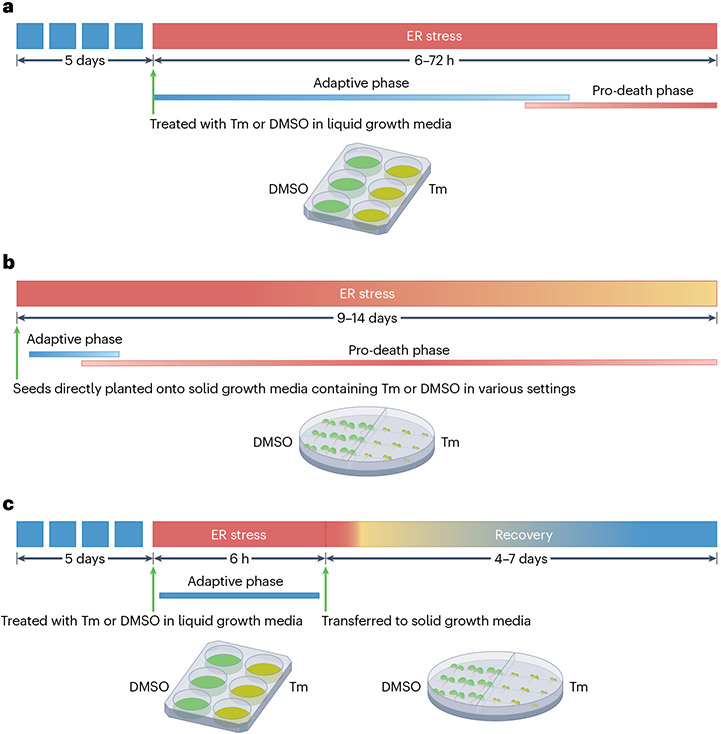
Fig. 4 |. Experimental designs for analysing responses to ER stress in Arabidopsis.
a, Adaptive endoplasmic reticulum (ER) stress. In this approach, 5-day-old seedlings are subjected to a high dose of tunicamycin (Tm) (for example, 500 ng ml−1) in liquid growth media using multi-well plates for a duration determined by the experimental objectives. The Tm solvent dimethyl sulfoxide (DMSO) is used as mock control. b, Chronic ER stress. Seeds are directly sown onto solid growth media containing a low dose of Tm (for example, 25 ng ml−1) or DMSO as control. Different doses of Tm can induce various cellular environments (for example, low dose for pro-survival environment; high dose for pro-death environment) that researchers can utilize to investigate their biological questions. Bi-plates featuring Tm-containing media on one side and DMSO-containing media on the other are advantageous for this method. Phenotypic assessments are typically conducted between 9 and 14 days after planting. c, ER stress recovery. After exposure to adaptive ER stress, seedlings are transferred to plates containing solid growth media for recovery. Growth restoration is evaluated between 5 and 7 days after the transfer. Collectively, these experimental systems offer powerful means to analyse molecular and physiological responses to ER stress in a highly reproducible manner.
Adaptive ER stress
In the context of adaptive ER stress, seedlings grown in physiological conditions are transferred to growth media containing an ER stress inducer, allowing for the timely controlled exploration of gene regulatory responses in a dose-dependent manner17,68 (Fig. 4a). ER stress induced by the drug treatment triggers a rapid and massive wave of transcriptional changes as an adaptive response to maintain ER homeostasis17,30,36. For instance, the expression of the UPR biomarker genes BiP3 and ERdj3B sees exponential increases exclusively during early phases (for example, 2 h or 4 h) of adaptive ER stress30,36. A double loss-of-function mutant of bZIP28 and bZIP60 showed almost no signs of BiP3 transcriptional induction11. 1ntriguingly, the TF single mutants bzip28-2 and bzip60-2 exhibit varying degrees of attenuation in the transcriptional induction of BiP3 and ERdj3B11. Therefore, although functional redundancy between bZIP28 and bZIP60 is evident, the extent to which they contribute to the UPR gene regulation seems to be distinct, depending on target genes. However, diversified functions of UPR-bZIP TFs during adaptive ER stress are confined to a small set of genes. Indeed, RNA-seq data have revealed that transcriptome profiles are largely similar among wild type (Col-0), bzip28-2 and bzip60-2, with only 59 and 62 differentially expressed genes (DEGs) in a bZIP28-dependent and bZIP60-dependent manner, respectively17. Therefore, UPR-bZIP TFs predominantly act as functionally redundant master regulators during adaptive ER stress. Recent research has further highlighted that during the adaptive phase of ER stress, pathways related to growth and development are downregulated, whereas those associated with plant hormones are upregulated17,22,36. Adaptive phases witness dynamic alterations in the expression of genes tied to a wide range of significant biological pathways18,22,30-33,36, supporting a broad integration of the UPR with cellular homeostasis.
Chronic (unresolved) ER stress
Chronic ER stress assays generally involve germination and seedling growth on media containing low levels of ER stress inducer to mimic conditions of unresolved ER stress and/or insufficient UPR (Fig. 4b). Although some studies have explored gene expression changes during chronic ER stress69,70, the nature of the gene regulatory mechanism in this condition remains speculative. For instance, the expression of BiP3 at 5 days of chronic ER stress diminished by 29.5% compared with that at 3 days, remaining stable thereafter for 10 days (D.K.K. and F.B., unpublished observations). This observation suggests that the functional role of BiP3 as a critical adaptive strategy component is most likely confined to the early stages of the UPR, and, thus, that samples taken after 3 days of planting may not offer biologically relevant insights into BiP3-specific functions. Therefore, understanding whether molecular snapshots reflect genuine molecular changes remains a challenge. Nevertheless, chronic ER stress set-ups, being easy to establish and specifically designed for observing effects accumulated over an extended period, prove ideal for investigating growth phenotypes under ER stress, particularly in mutant screenings in which qualitative rather than quantitative phenotypes are of interest (for example, life or death)12,22,43,69,71.
ER stress recovery
Because environmental stresses that plants experience can trigger ER stress64,72-76, the recovery of ER function is essential for re-establishing cellular homeostasis and resume growth, which is hampered during ER stress22,67,76 (Fig. 4c). It was shown recently that, in addition to making critical cell fate decisions under ER stress, the plant UPR contributes to the coordination of gene expression changes and restores growth when ER stress is resolved11,17. A transcriptional profiling of Arabidopsis seedlings exposed to ER stress for 6 h and then set in conditions of ER stress recovery upon ER stress inducer washout showed that the numbers of DEGs at 12 h and 24 h of ER stress recovery were significantly higher (2,484 and 2,930, respectively) with respect to the period just before recovery (576 DEGs at 6 h of adaptive ER stress)17. This drastic expansion in transcriptional programming encompasses not only gene quantity but also diverse functions, involving cellular, molecular, physiological and metabolic changes necessary for growth re-establishment. A pressing question is how systematic transcriptional coordination is achieved in a relatively short time frame. Increasing evidence suggests that the diversified roles of bZIP28 and bZIP60 play an important part. The single knockout mutants of bZIP28 and bZIP60 (bzip28-2 and bzip60-2, respectively) have no visible phenotype under ER stress48, leading to the classification of these regulators as functionally redundant under ER stress. However, in recovery from ER stress, bzip28-2 exhibits a more pronounced delay in root growth re-establishment than bzip60-211,17. Notably, the time course RNA-seq analyses during ER stress recovery reveal almost twice as many DEGs in bzip28-2 or bzip60-2 as there are in Col-0 at 12 h and 24 h of recovery, whereas the number of DEGs was similar at the onset of recovery (0 h, which is 6 h of ER stress). The phenotype and transcriptomic data collectively suggest that bZIP28 and bZIP60 have diversified roles alongside shared functions specifically when ER stress is resolved, influencing a wide range of biological pathways, including root growth, photosynthesis and primary metabolism.
Coordination of complex UPR gene regulation
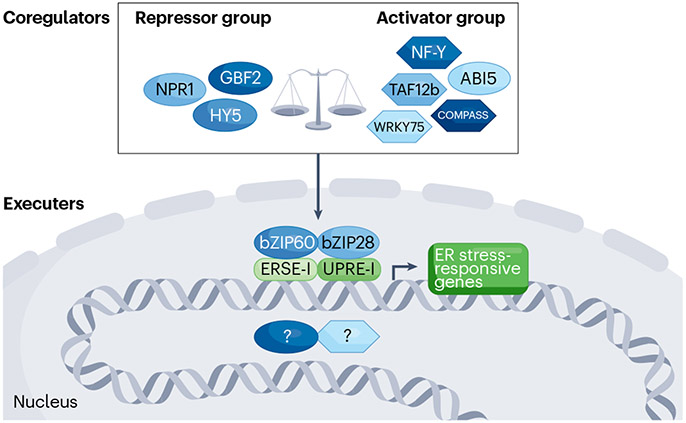
In various biological processes, such as developmental transitions, cell fate specification and responses to environmental stresses, gene expression changes are often orchestrated through the concerted action of master regulators77,78. These regulators dynamically coordinate their activities by interacting with coregulators in a highly interconnected network79. Although this mechanism enables efficient and rapid temporal changes to take place, it necessitates dominant master regulators capable of orchestrating molecular events, along with numerous coregulators that customize transcriptional activities based on cellular status and available resources. In the context of plant UPR gene regulation, identifying coregulators of UPR-bZIP TFs has been challenging due to functional redundancy within the UPR and the multifunctionality of TFs. Recent studies, however, have revealed that the scope of UPR gene regulatory machinery is more extensive than previously thought and that the said machinery is finely controlled by diverse coregulators that adapt to ER stress progression. This maximizes regulatory impact while minimizing unnecessary energy expenditure during critical cellular conditions. In this section, we highlight coregulatory factors working alongside UPR-bZIP TFs and the underlying mechanisms.
Transcriptional co-regulators
The first coregulators identified and functionally characterized in association with UPR-bZIP TFs were NF-Y subunits19, also known as CCAAT box binding factors80. Through physical interactions, NF-Y subunits, which bind to the first subunit (5′-CCAAT-3′) of ERSE-I, form a transcriptional complex with bZIP28 to promote BiP3 expression. This co-binding offers gene regulatory specificity given that the second subunit (5′-CACG-3′) of ERSE-I is nearly identical to the G-box (5′-CACGTG-3′) recognized by bZIP or bHLH TFs81. These observations imply that other bZIP TFs can function as heteromeric protein complexes to increase possible regulatory combinations and are potentially involved in UPR-bZIP TFs-mediated gene regulation during the onset, progression or recovery of ER stress. However, identifying coregulators among the 78 bZIP TFs82 and 162 bHLH TFs83 encoded in the Arabidopsis genome, along with understanding their working mechanisms, is akin to finding a needle in a haystack. To address this fundamental challenge, the plant UPR research community has recently adopted genetic, genomic and multi-omics approaches. For instance, the bZIP TF HY5, which functions as a positive regulator of light signalling84, represses the transcriptional activities of bZIP28 under ER stress by competing for ERSE-I binding20. GBF2, a crucial component of abscisic acid signalling85, competes with UPR-bZIP TFs for binding to the BiP3 promoter via the G-box, thereby maintaining appropriate gene expression levels21. Interestingly, in a triple mutant of bZIP28, bZIP60 and GBF2, the expression of BiP3 and ERdj3B was de-repressed, suggesting the presence of unknown transcriptional executer(s) independent of UPR-bZIP TFs. ABI5, a bZIP TF involved in stress responses86, positively regulates UPR biomarker genes by binding to the bZIP60 promoter via the G-box22. There are also a few examples of non-bZIP TF coregulators. For example, NON EXPRESSOR OF PR1 (NPR1), a critical redox-regulated master regulator of salicylic acid-dependent responses87, antagonizes transcriptional activities of UPR-bZIP TFs through a physical interaction in the nucleus24. WRKY75 positively regulates the transcriptional activity of bZIP28 through physical interactions, yet WRKY75 alone has no activity on the promoters of UPR biomarker genes88. Importantly, the functions of GBF2, ABI5 and NPR1 in the UPR seem independent of their originally reported roles in the hormone signalling pathways, as both abscisic acid and salicylic acid levels remain unchanged during ER stress22,24. TBP-ASSOCIATED FACTOR 12b (TAF12b), a core factor of TF subcomplexes within the transcription preinitiation complex89, acts as a cofactor of bZIP60 in the UPR, enhancing the transcriptional activity of bZIP60 through physical interactions23. How the physical interactions between TAF12b and bZIP60 positively affect bZIP60’s transcriptional activity remains unknown. Taken together, in this model (Fig. 5), the transcriptional activities of bZIP28 and bZIP60 are finely tuned by coactivators and co-repressors to elicit rapid but precise transcriptional reprogramming in a dynamically changing environment.
Fig. 5 |. A schematic model for gene regulation in plant UPR.
The transcriptional activities of unfolded protein response (UPR)-bZIP transcription factors (TFs) are meticulously orchestrated by the collaborative interplay of coactivators and corepressors. The plant UPR transcriptional system can be broadly categorized into two key components: ‘executer’ and ‘coregulators’. The coregulators, in turn, are further classified into ‘activator’ and ‘repressor’, based on their functional relationship with UPR-bZIP TFs. These coregulators are pivotal in fine-tuning the UPR response, ensuring the fidelity of gene expression. The executer group predominantly comprises UPR-bZIP TFs; however, it is important to note the potential existence of as-yet-uncharacterized executer(s). Maintaining a delicate balance between the activities of repressors and activators is crucial for the swift execution of gene expression changes. This equilibrium is important, especially in the context of limited energy resources and challenging environmental conditions, in which efficient resource allocation is essential for plant survival and adaptation. bZIP TFs are represented by elliptical shapes, whereas non-bZIP TFs are denoted by hexagons. ERSE-I and UPRE-I are canonical ER stress-responsive cis-regulatory elements. COMPASS, complex proteins associated with Set1; ER, endoplasmic reticulum.
Epigenetic regulators
The packaging of DNA with histone octamers into chromatin provides a sophisticated regulatory module for diverse gene regulation patterns in plant growth, development and adaptation to the environment90,91. Posttranslational modifications of histones directly or indirectly govern chromatin states, making them crucial for gene expression92. Given their critical role in gene regulation, histone modifications have been extensively studied in the context of abiotic stress-induced gene expression changes90,93. Concerning the UPR, Song et al. reported that COMPASS (complex proteins associated with Set1)-like components interact with bZIP28 and bZIP6025, directing the deposition of an activation mark H3K4me3 on the promoters of the UPR biomarker genes. Although these results underscore an aspect of the intricately regulated nature of gene regulation within the plant UPR, COMPASS may be the only epigenetic regulatory machinery in the UPR.
CREs
Canonical CREs such as ERSE-I and UPRE-I have crucial roles in mediating TF binding to the promoters of ER stress-responsive genes, as described above. However, it is worth noting that de novo motif analyses in RNA-seq studies have identified numerous other TF-binding motifs that could function as CREs. For instance, the promoters of DEGs regulated in a bZIP28-dependent or bZIP60-dependent manner during ER stress recovery showed significant enrichments of C2H2 and NAC TF-binding motifs17. Similarly, ChIP–seq analyses of bZIP28 and bZIP60 revealed significant enrichments of MYB96-binding, CRABS CLAW (CRC)-binding, WRKY75-binding and PHL4-binding motifs in UPR-specific bZIP28-binding and bZIP60-binding peaks (that is, presence only in tunicamycin-treated samples but not in DMSO-treated ones). These findings suggest that TFs other than bZIPs can participate in UPR-bZIP TF-mediated gene regulation. Further investigation is needed to understand whether and how these potential CREs function in mediating UPR gene regulation.
Conclusions and future perspectives
Rapid and precise transcriptional responses are at the heart of the UPR, ultimately determining the fate of a cell under ER stress. Yet, the mechanisms orchestrating this transcriptional coordination remain largely enigmatic. Adding to this complexity, the plant UPR has a systemic component enabled by the movement of bZIP60 through plasmodesmata94, supporting the idea that the coordination of the UPR assumes a non-cell autonomous component. The transcriptional machinery orchestrated by UPR-bZIP TFs is exquisitely tailored to meet the demands of dynamically changing ER stress environments. It comprises a wide array of transcriptional regulators, permitting fine-tuning with activators and repressors, all within a highly interconnected network. Although this intricate mechanism shares commonalities with responses to other environmental stresses, such as heat, drought and pathogen infection78,95, the uniqueness of the plant UPR system lies mainly in the dominance of two master regulators, bZIP28 and bZIP60, that coordinate a multitude of gene expression changes. Such streamlined centralization, although seemingly risky, seems to be highly efficient and effective for timely decisions.
Despite significant progress over the past decades, uncovering the molecular mechanisms driving dynamic transcriptional changes in plant UPR remains a formidable challenge, in particular owing to technical limitations. For instance, mutant suppressor screening has proven powerful in identifying qualitative phenotypes such as life versus death22,23, but additional assessments are required for quantitative traits (for example, root length), potentially causing delays in the selection processes. This limitation becomes even more apparent when dealing with molecular components exhibiting a high degree of functional redundancy, a characteristic often found in the plant UPR9. Furthermore, analysis of regulatory complexity has been conducted on bulk populations of millions of highly differentiated input cells, complicating the identification of functionally redundant transcriptional regulators. Consequently, UPR research needs to embrace state-of-the-art approaches that dissect this regulatory complexity at higher resolution, both temporally and spatially. The integration of single-cell omics technologies for transcriptome profiling96 and machine learning-based network models97 being trained with accumulated datasets presents an auspicious starting point for this journey.
Identifying the molecular components and their regulation within the UPR using plants as models will not only aid in the development of climate-resilient crops (Box 2) but also offer fresh perspectives on the UPR of other species, potentially uncovering novel mechanisms for the management of ER stress in intact organisms.
Acknowledgements
The authors’ work is supported by the National Institutes of Health (R35GM136637) with contributing support from the Great Lakes Bioenergy Research Center, U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research (DE-SC0018409), Chemical Sciences, Geoscience and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (DE-FG02-91ER20021), and MSU AgBioResearch (MICL02598).
Glossary
- Abscisic acid
A plant hormone that regulates numerous biological pathways, including growth, development and responses to environmental stress.
- Chaperone
A group of functionally related proteins that facilitate and regulate polypeptide folding within cells, including the highly conserved heat shock protein 70 (HSP70), which is present in all cellular compartments.
- Chromatin immunoprecipitation followed by sequencing
(ChIP–seq). The most popular in vivo technique to identify DNA sequences bound by a transcription factor of interest at genome scale.
- DNA microarray
A technology utilizing over 20,000 DNA probes attached to a surface, which are hybridized with cDNA (reverse-transcribed from mRNA using reverse transcriptase), enabling simultaneous measurement of expression levels of numerous genes.
- Endoplasmic reticulum
(ER). A cellular organelle composed of a membranous tubule network involved in the synthesis of protein and lipids and the export of secreted proteins.
- ER-associated protein degradation
A cellular pathway that eliminates misfolded proteins in the ER via ubiquitination followed by degradation by a protein-degradation complex known as the proteasome.
- ER-phagy
A mechanism that degrades portions of the ER in response to an excessive accumulation of misfolded proteins.
- Foldase
A particular set of molecular chaperones that facilitate the non-covalent folding of proteins in an ATP-dependent manner.
- Golgi apparatus
A cellular organelle where proteins and lipids are modified, packaged and transported to target destinations.
- Nuclear localization signal
An amino acid sequence that serves as a ‘tag’ for directing a protein into the nucleus through nuclear transport.
- Plasmodesmata
Small channels that connect plant cells to each other, providing living tubes between cells.
- Salicylic acid
A plant hormone that is crucial for triggering plant defence against both biotic and abiotic stresses, using morphological, physiological and biochemical mechanisms.
- Tunicamycin
A mixture of four homologous nucleoside antibiotics that inhibits N-linked glycosylation in the ER, thus disrupting protein folding and maturation.
Related links
Expression Atlas: https://www.ebi.ac.uk/gxa/experiments
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Darwin C. On the Origin of Species (John Murray, 1859). [Google Scholar]
- 2.Walter P & Ron D The unfolded protein response: from stress pathway to homeostatic regulation. Science 334, 1081–1086 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Park CJ & Park JM Endoplasmic reticulum plays a critical role in integrating signals generated by both biotic and abiotic stress in plants. Front. Plant Sci 10, 399 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christianson JC, Jarosch E & Sommer T Mechanisms of substrate processing during ER-associated protein degradation. Nat. Rev. Mol. Cell Biol 24, 777–796 (2023). [DOI] [PubMed] [Google Scholar]
- 5.Smith MH, Ploegh HL & Weissman JS Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science 334, 1086–1090 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang M. et al. ER-phagy: a new regulator of ER homeostasis. Front. Cell Dev. Biol 9, 684526 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crosti P, Malerba M & Bianchetti R Tunicamycin and brefeldin A induce in plant cells a programmed cell death showing apoptotic features. Protoplasma 216, 31–38 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Zuppini A, Navazio L & Mariani P Endoplasmic reticulum stress-induced programmed cell death in soybean cells. J. Cell Sci 117, 2591–2598 (2004). [DOI] [PubMed] [Google Scholar]
- 9.Pastor-Cantizano N, Ko DK, Angelos E, Pu Y & Brandizzi F Functional diversification of ER stress responses in Arabidopsis. Trends Biochem. Sci 45, 123–136 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Angelos E & Brandizzi F The UPR regulator IRE1 promotes balanced organ development by restricting TOR-dependent control of cellular differentiation in Arabidopsis. Plant J. 109, 1229–1248 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruberti C, Lai Y & Brandizzi F Recovery from temporary endoplasmic reticulum stress in plants relies on the tissue-specific and largely independent roles of bZIP28 and bZIP60, as well as an antagonizing function of BAX-Inhibitor 1 upon the pro-adaptive signaling mediated by bZIP28. Plant J. 93, 155–165 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pu Y, Ruberti C, Angelos ER & Brandizzi F AtIRE1C, an unconventional isoform of the UPR master regulator AtIRE1, is functionally associated with AtIRE1B in Arabidopsis gametogenesis. Plant Direct 3, e00187 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao Y, Bassham DC & Howell SH A functional unfolded protein response is required for normal vegetative development. Plant Physiol. 179, 1834–1843 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lisbona F. et al. BAX inhibitor-1 is a negative regulator of the ER stress sensor IRE1alpha. Mol. Cell 33, 679–691 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nagano M. et al. Arabidopsis Bax inhibitor-1 promotes sphingolipid synthesis during cold stress by interacting with ceramide-modifying enzymes. Planta 240, 77–89 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Godfray HC et al. Food security: the challenge of feeding 9 billion people. Science 327, 812–818 (2010). [DOI] [PubMed] [Google Scholar]
- 17. Ko DK & Brandizzi F Advanced genomics identifies growth effectors for proteotoxic ER stress recovery in Arabidopsis thaliana. Commun. Biol 5, 16 (2022). Time-course RNA-seq analyses in UPR mutants under ER stress recovery reveal distinct transcriptional activities of UPR-bZIP TFs through coexpression network analyses.
- 18. Kim JS, Yamaguchi-Shinozaki K & Shinozaki K ER-anchored transcription factors bZIP17 and bZIP28 regulate root elongation. Plant Physiol. 176, 2221–2230 (2018). RNA-seq analyses in genetic mutants demonstrate that bZIP17 does not serve as a core regulator in plant UPR.
- 19. Liu JX & Howell SH bZIP28 and NF-Y transcription factors are activated by ER stress and assemble into a transcriptional complex to regulate stress response genes in Arabidopsis. Plant Cell 22, 782–796 (2010). This study illustrates that NF-Y factors amplify the transcriptional activities of bZIP28, marking NF-Y factors as the first reported coregulators of UPR-bZIP TFs.
- 20.Nawkar GM et al. HY5, a positive regulator of light signaling, negatively controls the unfolded protein response in Arabidopsis. Proc. Natl Acad. Sci. USA 114, 2084–2089 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ko DK & Brandizzi F Transcriptional competition shapes proteotoxic ER stress resolution. Nat. Plants 8, 481–490 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ko DK, Kim JY, Thibault EA & Brandizzi F An IRE1-proteasome system signalling cohort controls cell fate determination in unresolved proteotoxic stress of the plant endoplasmic reticulum. Nat. Plants 9, 1333–1346 (2023). An E3 ligase, PIR1, is identified as a pro-death effector downstream of IRE1, providing new insights in the mechanisms for cell-fate control in ER stress in plants.
- 23.Kim JS et al. Arabidopsis TBP-ASSOCIATED FACTOR 12 ortholog NOBIRO6 controls root elongation with unfolded protein response cofactor activity. Proc. Natl Acad. Sci. USA 119, e2120219119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai YS et al. Salicylic acid-independent role of NPR1 is required for protection from proteotoxic stress in the plant endoplasmic reticulum. Proc. Natl Acad. Sci. USA 115, E5203–E5212 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song ZT et al. Transcription factor interaction with COMPASS-like complex regulates histone H3K4 trimethylation for specific gene expression in plants. Proc. Natl Acad. Sci. USA 112, 2900–2905 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Howell SH Endoplasmic reticulum stress responses in plants. Annu. Rev. Plant Biol 64, 477–499 (2013). [DOI] [PubMed] [Google Scholar]
- 27.Herath V. et al. The plant endoplasmic reticulum UPRome: a repository and pathway browser for genes involved in signaling networks linked to the endoplasmic reticulum. Plant Direct 6, e431 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Arabidopsis Genome Initiative. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408, 796–815 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Yamada K. et al. Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302, 842–846 (2003). [DOI] [PubMed] [Google Scholar]
- 30. Martínez IM & Chrispeels MJ Genomic analysis of the unfolded protein response in Arabidopsis shows its connection to important cellular processes. Plant Cell 15, 561–576 (2003). Together with Noh et al. (2003), the authors pioneer the documentation of ER stress-inducible transcriptomic alterations in Arabidopsis.
- 31. Noh S-J, Kwon CS, Oh D-H, Moon JS & Chung W-I Expression of an evolutionarily distinct novel BiP gene during the unfolded protein response in Arabidopsis thaliana. Gene 311, 81–91 (2003). Alongside Martínez and Chrispeels (2003), the authors contribute to the initial reporting of ER stress-inducible transcriptomic alterations in Arabidopsis.
- 32.Iwata Y, Sakiyama M, Lee M-H & Koizumi N Transcriptomic response of Arabidopsis thaliana to tunicamycin-induced endoplasmic reticulum stress. Plant Biotechnol. 27, 161–171 (2010). [Google Scholar]
- 33.Kamauchi S, Nakatani H, Nakano C & Urade R Gene expression in response to endoplasmic reticulum stress in Arabidopsis thaliana. FEBS J. 272, 3461–3476 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Mishiba K. et al. Defects in IRE1 enhance cell death and fail to degrade mRNAs encoding secretory pathway proteins in the Arabidopsis unfolded protein response. Proc. Natl Acad. Sci. USA 110, 5713–5718 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z, Gerstein M & Snyder M RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet 10, 57–63 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Srivastava R. et al. Response to persistent ER stress in plants: a multiphasic process that transitions cells from prosurvival activities to cell death. Plant Cell 30, 1220–1242 (2018). Conducting time-course RNA-seq analyses in maize, the authors unveil multi-phasic transcriptomic waves during persistent ER stress.
- 37.Herath V & Verchot J Comprehensive transcriptome analysis reveals genome-wide changes associated with endoplasmic reticulum (ER) stress in potato (Solanum tuberosum L.). Int. J. Mol. Sci 23, 13795 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Z, Tang J, Srivastava R, Bassham DC & Howell SH The transcription factor bZIP60 links the unfolded protein response to the heat stress response in maize. Plant Cell 32, 3559–3575 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lemon B & Tjian R Orchestrated response: a symphony of transcription factors for gene control. Genes Dev. 14, 2551–2569 (2000). [DOI] [PubMed] [Google Scholar]
- 40.Gaudinier A. et al. Enhanced Y1H assays for Arabidopsis. Nat. Methods 8, 1053–1055 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ko DK & Brandizzi F A temporal hierarchy underpins the transcription factor-DNA interactome of the maize UPR. Plant J. 105, 254–270 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Deng Y. et al. Heat induces the splicing by IRE1 of a mRNA encoding a transcription factor involved in the unfolded protein response in Arabidopsis. Proc. Natl Acad. Sci. USA 108, 7247–7252 (2011). Together with Nagashima et al. (2011), the authors reveal the unconventional splicing activity of IRE1 on bZIP60 mRNA under ER stress.
- 43. Nagashima Y. et al. Arabidopsis IRE1 catalyses unconventional splicing of bZIP60 mRNA to produce the active transcription factor. Sci. Rep 1, 29 (2011). Together with Deng et al. (2011), the authors demonstrate the unconventional splicing activity of IRE1 on bZIP60 mRNA under ER stress.
- 44.Diwan D, Liu X, Andrews CF & Pajerowska-Mukhtar KM A quantitative Arabidopsis IRE1a ribonuclease-dependent in vitro mRNA cleavage assay for functional studies of substrate splicing and decay activities. Front. Plant Sci 12, 707378 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bao Y. et al. IRE1B degrades RNAs encoding proteins that interfere with the induction of autophagy by ER stress in Arabidopsis thaliana. Autophagy 14, 1562–1573 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen Y & Brandizzi F AtIRE1A/AtIRE1B and AGB1 independently control two essential unfolded protein response pathways in Arabidopsis. Plant J. 69, 266–277 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Afrin T, Costello CN, Monella AN, Kørner CJ & Pajerowska-Mukhtar KM The interplay of GTP-binding protein AGB1 with ER stress sensors IRE1a and IRE1b modulates Arabidopsis unfolded protein response and bacterial immunity. Plant Signal. Behav 17, 2018857 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun L. et al. The lumen-facing domain is important for the biological function and organelle-to-organelle movement of bZIP28 during ER stress in Arabidopsis. Mol. Plant 6, 1605–1615 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Liu JX, Srivastava R, Che P & Howell SH An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. Plant Cell 19, 4111–4119 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Iwata Y. et al. Activation of the Arabidopsis membrane-bound transcription factor bZIP28 is mediated by site-2 protease, but not site-1 protease. Plant J. 91, 408–415 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Lageix S. et al. Arabidopsis eIF2α kinase GCN2 is essential for growth in stress conditions and is activated by wounding. BMC Plant Biol. 8, 134 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang ZT et al. The membrane-associated transcription factor NAC089 controls ER-stress-induced programmed cell death in plants. PLoS Genet. 10, e1004243 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang ZT et al. A plasma membrane-tethered transcription factor, NAC 062/ANAC 062/NTL 6, mediates the unfolded protein response in Arabidopsis. Plant J. 79, 1033–1043 (2014). [DOI] [PubMed] [Google Scholar]
- 54.Li Y, Williams B & Dickman M Arabidopsis B-cell lymphoma2 (Bcl-2)-associated athanogene 7 (BAG7)-mediated heat tolerance requires translocation, sumoylation and binding to WRKY29. New Phytol. 214, 695–705 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Iwata Y, Fedoroff NV & Koizumi N Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. Plant Cell 20, 3107–3121 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kokame K, Kato H & Miyata T Identification of ERSE-II, a new cis-acting element responsible for the ATF6-dependent mammalian unfolded protein response. J. Biol. Chem 276, 9199–9205 (2001). [DOI] [PubMed] [Google Scholar]
- 57.Hayashi S, Takahashi H, Wakasa Y, Kawakatsu T & Takaiwa F Identification of a cis-element that mediates multiple pathways of the endoplasmic reticulum stress response in rice. Plant J. 74, 248–257 (2013). [DOI] [PubMed] [Google Scholar]
- 58.Sun L. et al. The plant-specific transcription factor gene NAC103 is induced by bZIP60 through a new cis-regulatory element to modulate the unfolded protein response in Arabidopsis. Plant J. 76, 274–286 (2013). [DOI] [PubMed] [Google Scholar]
- 59.Liu JX, Srivastava R, Che P & Howell SH Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. Plant J. 51, 897–909 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liu JX, Srivastava R & Howell SH Stress-induced expression of an activated form of AtbZIP17 provides protection from salt stress in Arabidopsis. Plant Cell Environ. 31, 1735–1743 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Ishikawa T, Watanabe N, Nagano M, Kawai-Yamada M & Lam E Bax inhibitor-1: a highly conserved endoplasmic reticulum-resident cell death suppressor. Cell Death Differ. 18, 1271–1278 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watanabe N & Lam E BAX inhibitor-1 modulates endoplasmic reticulum stress-mediated programmed cell death in Arabidopsis. J. Biol. Chem 283, 3200–3210 (2008). [DOI] [PubMed] [Google Scholar]
- 63.Zhang L, Chen H, Brandizzi F, Verchot J & Wang A The UPR branch IRE1-bZIP60 in plants plays an essential role in viral infection and is complementary to the only UPR pathway in yeast. PLoS Genet. 11, e1005164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Henriquez-Valencia C. et al. bZIP17 and bZIP60 regulate the expression of BiP3 and other salt stress responsive genes in an UPR-independent manner in Arabidopsis thaliana. J. Cell. Biochem 116, 1638–1645 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Sparks E, Wachsman G & Benfey PN Spatiotemporal signalling in plant development. Nat. Rev. Genet 14, 631–644 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bradshaw AD Evolutionary significance of phenotypic plasticity in plants. Adv. Genet 13, 115–155 (1965). [Google Scholar]
- 67.Deng Y, Srivastava R & Howell SH Protein kinase and ribonuclease domains of IRE1 confer stress tolerance, vegetative growth, and reproductive development in Arabidopsis. Proc. Natl Acad. Sci. USA 110, 19633–19638 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ruberti C & Brandizzi F Unfolded protein response in Arabidopsis. Methods Mol. Biol 1691, 231–238 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu CY & Nakamura Y Smaller Trichomes With Variable Branches (SVB) and its homolog SVBL act downstream of transcription factor NAC089 and function redundantly in Arabidopsis unfolded protein response. J. Exp. Bot 74, 5870–5880 (2023). [DOI] [PubMed] [Google Scholar]
- 70.Yang Y et al. Stress response proteins NRP1 and NRP2 are pro-survival factors that inhibit cell death during ER stress. Plant Physiol. 187, 1414–1427 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nagashima Y, Iwata Y, Ashida M, Mishiba K & Koizumi N Exogenous salicylic acid activates two signaling arms of the unfolded protein response in Arabidopsis. Plant Cell Physiol. 55, 1772–1778 (2014). [DOI] [PubMed] [Google Scholar]
- 72.Moreno AA et al. IRE1/bZIP60-mediated unfolded protein response plays distinct roles in plant immunity and abiotic stress responses. PLoS ONE 7, e31944 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang SS et al. Tissue-specific transcriptomics reveals an important role of the unfolded protein response in maintaining fertility upon heat stress in Arabidopsis. Plant Cell 29, 1007–1023 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li B. et al. Ufmylation reconciles salt stress-induced unfolded protein responses via ER-phagy in Arabidopsis. Proc. Natl Acad. Sci. USA 120, e2208351120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Reyes-Impellizzeri S & Moreno AA The endoplasmic reticulum role in the plant response to abiotic stress. Front. Plant Sci 12, 755447 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao H, Brandizzi F, Benning C & Larkin RM A membrane-tethered transcription factor defines a branch of the heat stress response in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA 105, 16398–16403 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaufmann K, Pajoro A & Angenent GC Regulation of transcription in plants: mechanisms controlling developmental switches. Nat. Rev. Genet 11, 830–842 (2010). [DOI] [PubMed] [Google Scholar]
- 78.Zhang H, Zhu J, Gong Z & Zhu JK Abiotic stress responses in plants. Nat. Rev. Genet 23, 104–119 (2022). [DOI] [PubMed] [Google Scholar]
- 79.Alvarez JM, Brooks MD, Swift J & Coruzzi GM Time-based systems biology approaches to capture and model dynamic gene regulatory networks. Annu. Rev. Plant Biol 72, 105–131 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Roy B & Lee AS Transduction of calcium stress through interaction of the human transcription factor CBF with the proximal CCAAT regulatory element of the grp78/BiP promoter. Mol. Cell. Biol 15, 2263–2274 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ezer D. et al. The G-box transcriptional regulatory code in Arabidopsis. Plant Physiol. 175, 628–640 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Dröge-Laser W, Snoek BL, Snel B & Weiste C The Arabidopsis bZIP transcription factor family—an update. Curr. Opin. Plant Biol 45, 36–49 (2018). [DOI] [PubMed] [Google Scholar]
- 83.Bailey PC et al. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell 15, 2497–2502 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Osterlund MT, Hardtke CS, Wei N & Deng XW Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466 (2000). [DOI] [PubMed] [Google Scholar]
- 85.Song L. et al. A transcription factor hierarchy defines an environmental stress response network. Science 354, eaag1550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Skubacz A, Daszkowska-Golec A & Szarejko I The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci 7, 1884 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Cao H, Bowling SA, Gordon AS & Dong X Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang LY et al. Orosomucoid proteins limit endoplasmic reticulum stress in plants. New Phytol. 240, 1134–1148 (2023). [DOI] [PubMed] [Google Scholar]
- 89.Robles LM, Wampole JS, Christians MJ & Larsen PB Arabidopsis enhanced ethylene response 4 encodes an EIN3-interacting TFIID transcription factor required for proper ethylene response, including ERF1 induction. J. Exp. Bot 58, 2627–2639 (2007). [DOI] [PubMed] [Google Scholar]
- 90.Lämke J & Bäurle I Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 18, 124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Quint M. et al. Molecular and genetic control of plant thermomorphogenesis. Nat. Plants 2, 15190 (2016). [DOI] [PubMed] [Google Scholar]
- 92.Henikoff S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nat. Rev. Genet 9, 15–26 (2008). [DOI] [PubMed] [Google Scholar]
- 93.Oberkofler V, Pratx L & Bäurle I Epigenetic regulation of abiotic stress memory: maintaining the good things while they last. Curr. Opin. Plant Biol 61, 102007 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Lai YS et al. Systemic signaling contributes to the unfolded protein response of the plant endoplasmic reticulum. Nat. Commun 9, 3918 (2018). This study highlights that plant UPR exhibits non-cell autonomous signalling partially reliant on the systematic movement of bZIP60.
- 95.Eulgem T. Regulation of the Arabidopsis defense transcriptome. Trends Plant Sci. 10, 71–78 (2005). [DOI] [PubMed] [Google Scholar]
- 96.Seyfferth C. et al. Advances and opportunities in single-cell transcriptomics for plant research. Annu. Rev. Plant Biol 72, 847–866 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Ko DK & Brandizzi F Network-based approaches for understanding gene regulation and function in plants. Plant J. 104, 302–317 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Galante E, Vitale A, Manzocchi L, Soave C & Salamini F Genetic control of a membrane component and zein deposition in maize endosperm. Mol. Genet. Genom 192, 316–321 (1983). [Google Scholar]
- 99.Boston RS, Fontes EB, Shank BB & Wrobel RL Increased expression of the maize immunoglobulin binding protein homolog b-70 in three zein regulatory mutants. Plant Cell 3, 497–505 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Fontes EB et al. Characterization of an immunoglobulin binding protein homolog in the maize floury-2 endosperm mutant. Plant Cell 3, 483–496 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Gething MJ & Sambrook J Protein folding in the cell. Nature 355, 33–45 (1992). [DOI] [PubMed] [Google Scholar]
- 102.Coleman CE, Lopes MA, Gillikin JW, Boston RS & Larkins BA A defective signal peptide in the maize high-lysine mutant floury 2. Proc. Natl Acad. Sci. USA 92, 6828–6831 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Denecke J, Goldman MH, Demolder J, Seurinck J & Botterman J The tobacco luminal binding protein is encoded by a multigene family. Plant Cell 3, 1025–1035 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pedrazzini E, Giovinazzo G, Bollini R, Ceriotti A & Vitale A Binding of BiP to an assembly-defective protein in plant cells. Plant J. 5, 103–110 (1994). [Google Scholar]
- 105.Pedrazzini E. et al. Protein quality control along the route to the plant vacuole. Plant Cell 9, 1869–1880 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Page DR & Grossniklaus U The art and design of genetic screens: Arabidopsis thaliana. Nat. Rev. Genet 3, 124–136 (2002). [DOI] [PubMed] [Google Scholar]
- 107.Somerville C & Koornneef M A fortunate choice: the history of Arabidopsis as a model plant. Nat. Rev. Genet 3, 883–889 (2002). [DOI] [PubMed] [Google Scholar]
- 108.O’Malley RC & Ecker JR Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J. 61, 928–940 (2010). [DOI] [PubMed] [Google Scholar]
- 109. Koizumi N. et al. Molecular characterization of two Arabidopsis Ire1 homologs, endoplasmic reticulum-located transmembrane protein kinases. Plant Physiol. 127, 949–962 (2001). This study identifies two genes, IRE1a and IRE1b, encoding IRE1 in the Arabidopsis genome.
- 110.Yoshida H, Haze K, Yanagi H, Yura T & Mori K Identification of the cis-acting endoplasmic reticulum stress response element responsible for transcriptional induction of mammalian glucose-regulated proteins. Involvement of basic leucine zipper transcription factors. J. Biol. Chem 273, 33741–33749 (1998). [DOI] [PubMed] [Google Scholar]
- 111. Iwata Y & Koizumi N An Arabidopsis transcription factor, AtbZIP60, regulates the endoplasmic reticulum stress response in a manner unique to plants. Proc. Natl Acad. Sci. USA 102, 5280–5285 (2005). This study was the first to demonstrate that bZIP60 induces the expression of ER stress-responsive genes.
- 112.Parra-Rojas J, Moreno AA, Mitina I & Orellana A The dynamic of the splicing of bZIP60 and the proteins encoded by the spliced and unspliced mRNAs reveals some unique features during the activation of UPR in Arabidopsis thaliana. PLoS ONE 10, e0122936 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Liou H-C et al. A new member of the leucine zipper class of proteins that binds to the HLA DRα promoter. Science 247, 1581–1584 (1990). [DOI] [PubMed] [Google Scholar]
- 114.Calfon M. et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature 415, 92–96 (2002). [DOI] [PubMed] [Google Scholar]
- 115.Yoshida H. et al. Endoplasmic reticulum stress-induced formation of transcription factor complex ERSF including NF-Y (CBF) and activating transcription factors 6α and 6β that activates the mammalian unfolded protein response. Mol. Cell. Biol 21, 1239–1248 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Shoemaker DD et al. Experimental annotation of the human genome using microarray technology. Nature 409, 922–927 (2001). [DOI] [PubMed] [Google Scholar]
- 117.Hayashi S, Wakasa Y, Takahashi H, Kawakatsu T & Takaiwa F Signal transduction by IRE1-mediated splicing of bZIP50 and other stress sensors in the endoplasmic reticulum stress response of rice. Plant J. 69, 946–956 (2012). [DOI] [PubMed] [Google Scholar]
- 118.Lu SJ, Yang ZT, Sun L, Song ZT & Liu JX Conservation of IRE1-regulated bZIP74 mRNA unconventional splicing in rice (Oryza sativa L.) involved in ER stress responses. Mol. Plant 5, 504–514 (2012). [DOI] [PubMed] [Google Scholar]
- 119.Wang QL, Sun AZ, Chen ST, Chen LS & Guo FQ SPL6 represses signalling outputs of ER stress in control of panicle cell death in rice. Nat. Plants 4, 280–288 (2018). [DOI] [PubMed] [Google Scholar]
- 120.Okushima Y. et al. Isolation and characterization of a putative transducer of endoplasmic reticulum stress in Oryza sativa. Plant Cell Physiol. 43, 532–539 (2002). [DOI] [PubMed] [Google Scholar]
- 121.Wakasa Y, Hayashi S, Ozawa K & Takaiwa F Multiple roles of the ER stress sensor IRE1 demonstrated by gene targeting in rice. Sci. Rep 2, 944 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li Y, Humbert S & Howell SH ZmbZIP60 mRNA is spliced in maize in response to ER stress. BMC Res. Notes 5, 144 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cavalcante FLP et al. Unveiling a differential metabolite modulation of sorghum varieties under increasing tunicamycin-induced endoplasmic reticulum stress. Cell Stress Chaperones 28, 889–907 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ye C, Dickman MB, Whitham SA, Payton M & Verchot J The unfolded protein response is triggered by a plant viral movement protein. Plant Physiol. 156, 741–755 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Löchli K. et al. Crosstalk between endoplasmic reticulum and cytosolic unfolded protein response in tomato. Cell Stress Chaperones 28, 511–528 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Michael TP & VanBuren R Progress, challenges and the future of crop genomes. Curr. Opin. Plant Biol 24, 71–81 (2015). [DOI] [PubMed] [Google Scholar]
- 127.Zou C, Lehti-Shiu MD, Thomashow M & Shiu SH Evolution of stress-regulated gene expression in duplicate genes of Arabidopsis thaliana. PLoS Genet. 5, e1000581 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Van de Peer Y, Maere S & Meyer A The evolutionary significance of ancient genome duplications. Nat. Rev. Genet 10, 725–732 (2009). [DOI] [PubMed] [Google Scholar]
- 129.Bertolotti A, Zhang Y, Hendershot LM, Harding HP & Ron D Dynamic interaction of BiP and ER stress transducers in the unfolded-protein response. Nat. Cell Biol 2, 326–332 (2000). [DOI] [PubMed] [Google Scholar]
- 130.Kimata Y. et al. Genetic evidence for a role of BiP/Kar2 that regulates Ire1 in response to accumulation of unfolded proteins. Mol. Biol. Cell 14, 2559–2569 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Liu Y. et al. EBS7 is a plant-specific component of a highly conserved endoplasmic reticulum-associated degradation system in Arabidopsis. Proc. Natl Acad. Sci. USA 112, 12205–12210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhou Y-F, Qing T, Shu X-L & Liu J-X Unfolded protein response and storage product accumulation in rice grains. Seed Biol. 1, 4 (2022). [Google Scholar]
- 133.Valente MA et al. The ER luminal binding protein (BiP) mediates an increase in drought tolerance in soybean and delays drought-induced leaf senescence in soybean and tobacco. J. Exp. Bot 60, 533–546 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee YR et al. CRISPR/Cas9-mediated HY5 gene editing reduces growth inhibition in Chinese cabbage (Brassica rapa) under ER stress. Int. J. Mol. Sci 24, 13105 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Herath V & Verchot J Comprehensive transcriptome analysis reveals genome-wide changes associated with endoplasmic reticulum (ER) stress in potato. Int. J. Mol. Sci 23, 13795 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]