Abstract
The efficient cytosolic delivery of proteins is critical for advancing novel therapeutic strategies. Current delivery methods are severely limited by endosomal entrapment, and detection methods lack sophistication in tracking the fate of delivered protein cargo. HaloTag, a commonly used protein in chemical biology and a challenging delivery target, is an exceptional model system for understanding and exploiting cellular delivery. Here, we employ a combinatorial strategy to direct HaloTag to the cytosol. We established the use of Virginia Orange, a pH-sensitive fluorophore, and JF585, a similar but pH-agnostic fluorophore, in a fluorogenic assay to ascertain protein localization within human cells. Using this assay, we investigated HaloTag delivery upon modification with cell-penetrating peptides, carboxyl group esterification, and co-treatment with an endosomolytic agent. We found efficacious cytosolic entry with two distinct delivery methods. This study expands the toolkit for detecting the cytosolic access of proteins and highlights that multiple intracellular delivery strategies can be used synergistically to effect cytosolic access. Moreover, HaloTag is poised to serve as a platform for the delivery of varied cargo into human cells.
Graphical Abstract
INTRODUCTION
Leveraging the remarkable potency and specificity of proteins has revolutionized the therapeutic landscape. Rationally designed biologics have shown efficacy in a wide range of etiologies, leading to their prevalence in the clinic.1-3 To date, however, these advances have been restricted to extracellular targets, leaving an estimated ¾ of disease-relevant, intracellular human proteins undruggable.4 Unlike small-molecule therapeutics, which can enter cells via diffusion, proteins require adjuvants to mediate cellular uptake. Thus, the efficient intracellular delivery of proteins would both advance basic research and open new therapeutic avenues.
From this perspective, the HaloTag protein is an ideal candidate for investigating cytosolic delivery. HaloTag (HT7) is an engineered bacterial haloalkane dehalogenase, derived from Rhodococcus dehalogenase (DhaA), that has been altered5 (L47V, S58T, D78G, Y87F, L88M, C128F, A155T, E160K, A167V, A172T, K175M, C176G, K195N, A224E, N227D, E257K, T264A, F272N, Y273L, P291S, A292T, Q294E, Q294E, Y295I, S296, G297) to prevent catalytic turnover but enable the O-alkylation of an active-site glutamic acid residue using a haloalkane of choice.6 Depending on the substrate, the labeling kinetics of HaloTag are ~104–107 M−1 s−1, allowing for rapid saturation of the enzyme with a defined ligand.7 Given its attractive features, HaloTag is frequently used for the imaging of fusion proteins, as a handle for protein purification, and as a model protein.5,8-12
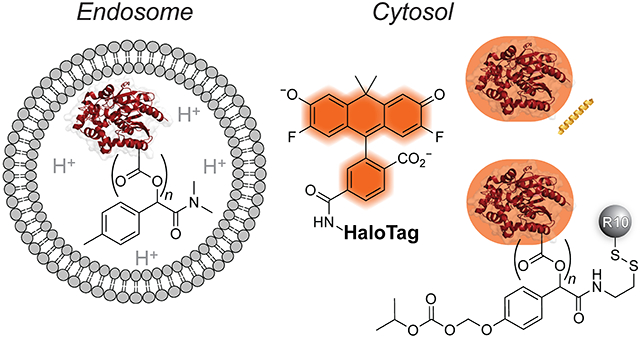
The outer membrane of mammalian cells is highly anionic.13-15 Because of repulsive Coulombic forces, HaloTag, which is a highly anionic protein (pI 4.916), is a special challenge for cellular delivery (Figure S3).17 Several approaches for cytosolic protein delivery have sought to overcome this barrier. Proteins have been translocated to the cytosol of cells via electroporation,18 polymer encapsulation,19 conjugation to transduction domains (e.g., cell-penetrating peptides),4,20-23 supercharging,24-27 co-treatment with endosomolytic agents,28,29 thiol–disulfide exchange,30 and cationic lipids.31,32 Recently, several reversible methods have enabled the traceless delivery of proteins into cells,33-36 thereby addressing an important consideration for clinical translation. The majority of the aforementioned strategies induce endosomal uptake of proteins, followed by endosomal escape to access the cytosol and the therapeutic target of interest. Notably, endosomal escape is frequently a limiting factor in protein delivery experiments, as typically <10% of the cargo reaches the cytosol, and the remainder is trapped in the endosome and trafficked toward lysosomal degradation.37
Several functional assays have been developed to assess the fate of protein cargoes, including protein complementation assays,38-40 corrective splicing assays,41,42 and assays that leverage cytosolic enzymes to label an exogenously delivered tag.43-47 In addition, fluorogenic assays46 and fluorescence correlation spectroscopy48-50 imaging techniques provide complementary approaches to assess protein internalization. Most of these methods, however, rely on enzymatic activity after “tag” recognition, which requires the engineering of new cell lines,51 hampering generality. Alternative techniques require the use of highly specialized instrumentation and advanced modeling to yield experimental results.51
A pH-sensitive fluorophore could be the basis for a general strategy to discern the localization of protein cargo. This approach would leverage the difference in pH between the cytosol, which has a near-neutral pH, and the endosome, which has an acidic pH of 5–6,52,53 as “on” and “off” switches for fluorescence. Naphthofluorescein (ε = 44,000 M−1 cm−1; Φ = 0.14),54,55 in particular, has been used in this context.56-59 Naphthofluorescein, however, contains an extensive aromatic ring system, which is disadvantageous given the propensity for cellular uptake and subsequent localization of small biomolecules to be altered by a pendant dye.60-63 While the size of the fluorophore plays an outsized role in cell penetration of small biomolecules as compared to large ones, like HaloTag (~35 kDa), establishing a smaller pH-based fluorescent reporter than naphthofluorescein is desirable to minimize the effect of the tag in any biomolecule.
Simple substitutions within xanthene dyes enable the tuning of their spectroscopic properties.64-73 For example, substitutions can bias the dye toward a closed, lactone form, which has negligible fluorescence or an open, zwitterionic form, which is highly fluorescent. This equilibrium can also be affected by the microenvironment of the fluorophore.74
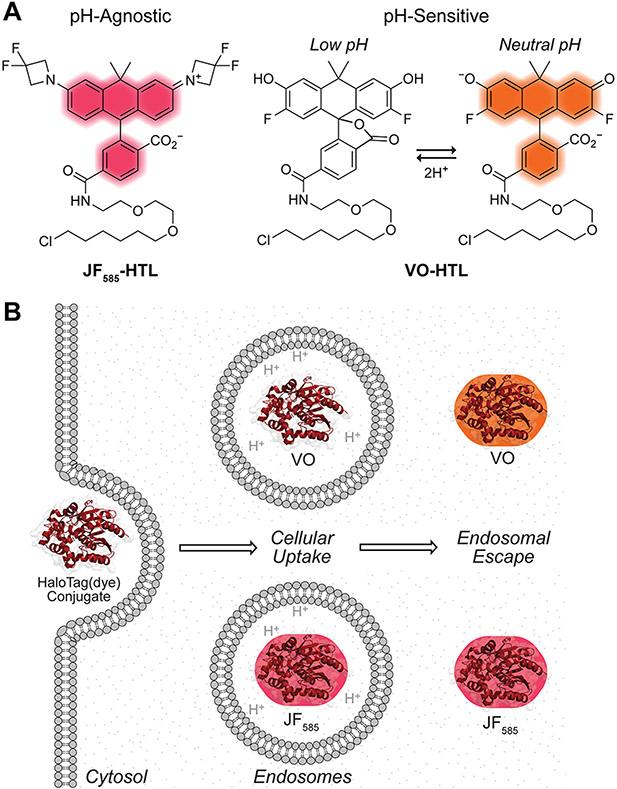
Two complementary carbaxanthene-based dyes are ideal for our purpose (Figure 1A). One is Janelia Fluor 585 (JF585; εmax = 156,000 M−1 cm−1; Φ = 0.78), which is ~19-fold brighter than naphthofluorescein, has λmax/λem = 585 nm/609 nm, is designed to fluoresce only after binding within the HaloTag active site and does not have a proton that is titratable under physiological conditions.67,69 The other dye is Virginia Orange (VO; ε = 90,900 M−1 cm−1; Φ = 0.40), which is ~6-fold brighter than naphthofluorescein (ε = 90,900 M−1 cm−1; Φ = 0.40), has λmax/λem 555 nm/581 nm, and has highly pH-sensitive fluorescence (h = 1.46) with a transition at pH 6.75.75,76 As early endosomes mature to late endosomes, their pH drops from ~6.5 to ~5.5.77 Lysosomes, their ultimate locale, have pH ~4.5.77 This acidity means that VO is virtually non-fluorescent in endosomes and lysosomes.76 Hence, we hypothesized that the conjugation of JF585 or VO to HaloTag would enable the monitoring of its endosomal escape during protein delivery experiments and the quantification of the efficiency of its cytosolic entry (Figure 1B).
Figure 1.
(A) Structures of JF585 and VO with a pendant HTL. (B) The low pH of endosomes and the pH-sensitive fluorescence of Virginia Orange (but not JF585) can be used to ascertain the subcellular localization of HaloTag(dye) conjugates.
We interrogated three distinct strategies for the cytosolic delivery of HaloTag(dye) constructs. Specifically, we explored conjugation with a cell-penetrating peptide, bioreversible esterification, and co-treatment with an endosomolytic agent to deliver HaloTag when conjugated to either JF585 or VO. We observed varying cellular localizations that depend upon the delivery strategy.
RESULTS
HaloTag(dye) Conjugates Can Inform Cellular Localization.
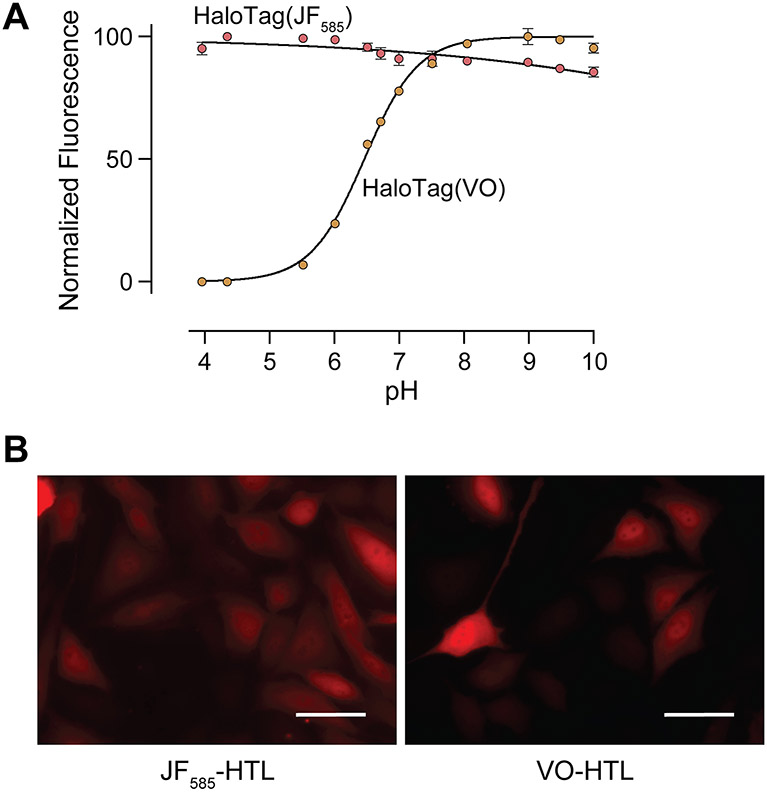
Our first aim was to validate that JF585 and VO would be capable of distinguishing between endosomal and cytosolic compartments as a function of pH. We have reported the pH-dependence of the fluorescence of JF585 and VO for the dyes alone67,69,75,76 but not for HaloTag(dye) conjugates. To do so, we produced HaloTag in Escherichia coli with a TEV protease-cleavable poly-histidine tag for ease of purification. After purification and proteolytic cleavage (Figure S1), we conjugated HaloTag to either JF585 or VO equipped with a pendant HaloTag ligand (HTL) (Figure 1A). Analysis with mass spectrometry validated the identity of the HaloTag(JF585) and HaloTag(VO) constructs (Figure S2). Next, we measured the fluorescence of each HaloTag(dye) as a function of pH. We found that the fluorescence of HaloTag(JF585) is pH-agnostic (Figure 2A), as expected.67,69 In contrast, we found that the fluorescence of HaloTag(VO) has a transition at pH 6.46 ± 0.01. This value is slightly lower than that of the dye alone (pH = 6.75),75,76 which seems surprising given the anionic microenvironment that the dye occupies on the surface of HaloTag (Figures S3 and S4). However, the cooperativity of the pH transition is also smaller somewhat diminished (h = 1.05) compared to that of the free dye (h = 1.46), suggesting stabilization of the open form of the dye by the polar environment of the protein surface and perhaps explaining the lower pKa value. Taken together, this dye pair seems capable of distinguishing total protein uptake from that localized in the cytosol.
Figure 2.
(A) pH-Dependence of the fluorescence of HaloTag(dye) constructs. Values are the mean ± SE (n = 3). HaloTag(VO) has an apparent pKa of 6.46 ± 0.01 (h = 1.05). (B) Microscopy images of HeLa cells expressing cytosolically localized HaloTag and treated with either JF585-HTL or VO-HTL. Images were normalized to untransfected cells treated with either fluorophore. Images are representative of uptake profiles of at least two independent experiments, each performed with three technical replicates. Scale bars: 50 μM. Additional images are shown in Figure S12.
To validate this difference in a cellular context, we used a transfection-based method to confirm the fluorescence of each dye in the cytosol. HeLa cells were transiently transfected with mRNA that encodes cytosolic HaloTag. Cells were then treated with JF585-HTL or VO-HTL via pulse-chase epifluorescence imaging, followed by flow cytometry, to identify cells that express HaloTag (Figure 2B, S12-S14). We anticipated that when treated with either fluorescent ligand, cells containing HaloTag would fluoresce and that un-transfected cells would not. Indeed, images of transfected cells treated with either fluorophore revealed a cytosolic HaloTag population, unlike images of un-transfected cells. Additionally, we observed no background fluorescence in un-transfected cells following the same treatment and washout period as transfected cells, indicating that the observed fluorescent signal is specifically associated with transfection (S14). These data indicate that both dyes fluoresce when bound to HaloTag in the cytosol, enabling unambiguous interpretation of protein delivery experiments.
Modification of HaloTag(dye) Conjugates.
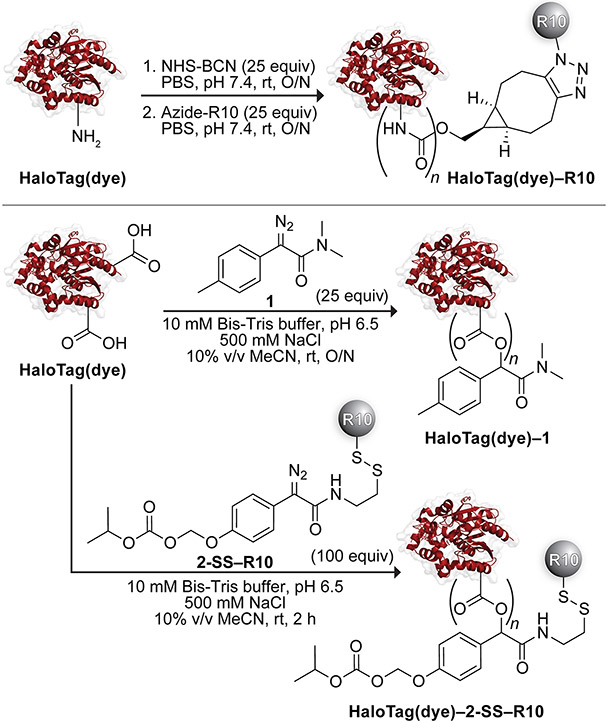
With a validated assay for cytosolic localization in hand, we sought to investigate the effect of different delivery agents on the uptake of HaloTag (Scheme 1). First, we appended deca-arginine (R10) to protein amino groups on each HaloTag(dye) construct. To do so, we irreversibly modified HaloTag using lysine amidation with NHS-BCN. Then, we clicked an azide-containing R10 peptide onto the protein. The extent of labeling (typically, 0–1 R10 moieties) was determined by mass spectrometry (Figures S5 and S6).
Scheme 1.
Semisynthesis of HaloTag(dye) Conjugates
Next, we masked HaloTag carboxyl groups by esterification with a tuned diazo compound.33,35 Upon cellular entry, endogenous intracellular esterases cleave these esters, unveiling the nascent protein. Previously, we reported that esterification using diazo compound 1 directed both the green fluorescent protein33 and human ribonuclease 135 to the cytosol. Each of these proteins has, however, a pI value that is higher than that of HaloTag. To assess the cellular localization of HaloTag with diazo compound 1, we modified the surface of HaloTag(JF585) or HaloTag(VO) with this compound in an acidic buffer containing 500 mM NaCl to mediate labeling with minimal protein precipitation. We investigated two ranges of ester labels. The identity of the conjugates was confirmed using mass spectrometry and found to contain either 0–3 labels (low labeling) or 2–8 (high labeling) (Figures S7 and S8). Installation of more labels on the surface of HaloTag led to the precipitation of the protein, consistent with previous reports correlating the number of labels with protein insolubility as the pI of the protein approaches the pH of the solution.78
Recently, we deployed a novel diazo compound, 2-SSpy, that enables a late-stage modification with a ligand of choice.36 To modify HaloTag with this compound, 2-SSpy was briefly pre-mixed with the thiol-equipped R10 to form a disulfide linkage prior to reaction with the protein. The typical extent of labeling was found to be 0–1 (Figure S9). Based on a report of the solvent accessibility of cysteine residues in HaloTag,79 we explored disulfide-linked HaloTag conjugates as another means for delivery but found the ensuing linkages to be unstable (Figure S11).
Finally, we used the L17E peptide in a co-treatment strategy to effect cellular entry. L17E is an engineered version of the toxic peptide M-lycotoxin from Wolf spider venom.29 L17E was optimized to adopt an α-helical structure that selectively disrupts endosomal membranes, thus mediating endosomal escape of the co-treated protein or other cargo into the cytosol. Altogether, we evaluated four distinct uptake strategies using the complementary dye pair.
Cellular Uptake Experiments with Microscopy.
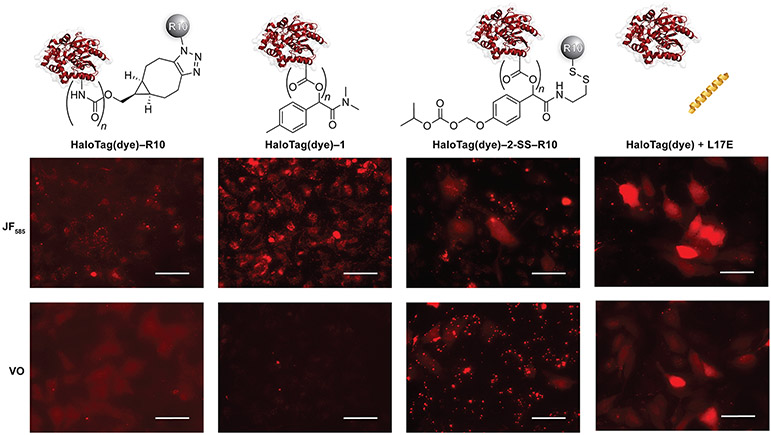
We next assessed the uptake profiles of the protein constructs into live HeLa cells. Cells were treated in serum-free DMEM medium at concentrations and times that were optimized for each HaloTag(JF585) conjugate (Table S1). Subsequent paired experiments were conducted side-by-side with HaloTag(JF585) and HaloTag(VO) conjugates. Epifluorescence microscopy was used to assess cellular localization. We found that all bioconjugates were taken up by cells, with differences in cytosolic access based on the delivery method (Figure 3).
Figure 3.
Microscopy images of HaloTag(dye) conjugates in HeLa cells normalized to cells treated with HaloTag(dye) only. For experimental conditions, see Table S1. Standardized laser intensities were normalized within each treatment condition. Images are representative of uptake profiles of at least two independent experiments performed with three technical replicates. Scale bars: 50 μM. Additional images are shown in Figures S15-S18.
HaloTag(dye)–R10.
HaloTag(JF585)–R10 demonstrated exclusively punctate staining, indicative of endosomal entrapment (Figure 3). When taken together with HaloTag(VO)–R10, however, we observed low fluorescent staining of the cytosol, indicative of a small degree of endosomal escape. Given the anionic nature of the HaloTag surface (Figure S3), we were not surprised that a cationic cell-penetrating peptide does not mediate robust cellular entry.
HaloTag(dye)–1.
Surface esterification with diazo compound 1 was used to investigate the effect of the extent of labeling on HaloTag uptake. We observed successful cellular uptake, but only with a high level of esterification. Imaging revealed endosomal entrapment, evidenced by the punctate staining in cells treated with HaloTag(JF585)–1 and the absence of signal in cells treated with HaloTag(VO)–1 (Figure 3). We attribute the endosomal entrapment of esterified HaloTag to the high anionicity of HaloTag, which cannot be overcome by esterification with diazo compound 1, which ultimately serves to increase the hydrophobicity while only modestly decreasing the anionicity of the protein surface. We did not observe any cellular uptake with a low level of esterification (Figures S20 and S21).
HaloTag(dye)–2-SS–R10.
HeLa cells treated with a 2-SS–R10 ester conjugate showed mixed intracellular localization (Figure 3). Many clusters of cells exhibited cytosolic localization, with diffuse intracellular staining throughout the entire compartment. When cells treated with HaloTag(JF585)–2-SS–R10 were imaged at a higher laser intensity, punctate staining could be observed that was absent in cells treated with HaloTag(VO)–2-SS–R10, indicating endosomal entrapment. We also noted evidence of protein aggregation on the surface of cells treated with HaloTag(VO)–2-SS–R10, which was not observed in other treatment conditions. Nonetheless, the combination of a cell-penetrating peptide with esterification yielded cytosolic access.
HaloTag(dye) plus L17E.
Finally, we cotreated HeLa cells with HaloTag(dye) constructs + L17E. Fluorescence microscopy showed that both constructs localized robustly in the cytosol of cells, especially concentrated in clusters, as evidenced by diffuse staining throughout the cytosol and nucleus (Figure 3). With HaloTag(JF585), we did not observe the punctate staining indicative of endosomal entrapment. Although L17E has been used with other proteins and biomolecules,29,80-83 the demonstration of its efficacy in delivering highly anionic protein cargo is, to our knowledge, without precedent.
Cellular Uptake Experiments with Flow Cytometry.
An advantage of the JF585/VO dye pair is its ability to differentiate between the bulk cellular uptake and the cytosolic localization of conjugates. Having observed uptake patterns that varied between conjugates with fluorescence microscopy, we used flow cytometry to assess delivery efficiencies, which describe the relationship between the total amount of protein cargo taken up by the cell and the amount that ultimately reaches the cytosol. Because of the heterogeneity between each batch of prepared conjugates, as a result of stochastic surface labeling, we sought an analysis that would provide meaningful comparisons. Accordingly, we established a workflow in which cells were first imaged via microscopy and then analyzed by flow cytometry.
Gratifyingly, the flow cytometry results were in agreement with the cellular uptake observed with microscopy. In cells treated with HaloTag(dye)–R10, we observed a modest shift in the bulk population, demonstrating a low level of endosomal escape (Figures S22-S23). When modified with 2–8 ester labels, HaloTag(dye)–1, was efficiently taken up into endosomes but did not exhibit the VO signal that would have been indicative of endosomal escape (Figure S24). By contrast, microscopy revealed a subset of cells co-treated with HaloTag(dye) and L17E exhibited high cellular uptake with both dyes and the absence of punctate staining with HaloTag(JF585), indicating highly efficient cytosolic entry. Consistent with the robust cytosolic delivery of HaloTag(dye) in the presence of L17E that we observed via microscopy, we also observed a subset of highly fluorescent cells and little change to the bulk population via flow cytometry (Figure S25). The percent of cells in each sample treated with HaloTag(dye) + L17E were the same, reinforcing the high efficiency of cytosolic entry. Although we observed the cellular uptake and cytosolic entry of HaloTag(dye)–2-SS–R10 conjugates with both fluorophores via microscopy, the VO conjugate formed aggregates or coacervates on the surface of cells that were resistant to removal by washing and precluded analysis by flow cytometry (Figure S17).
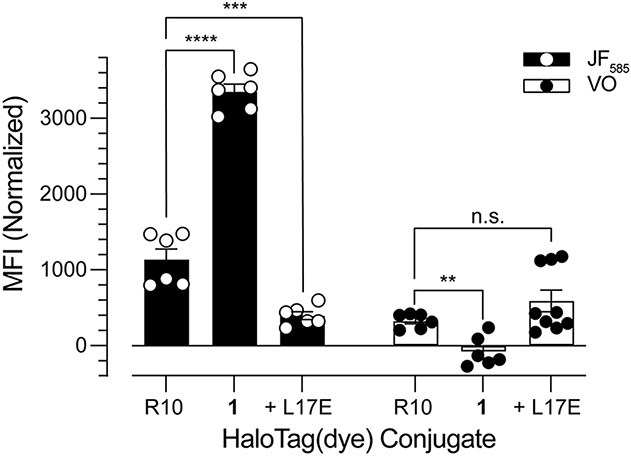
From our flow cytometry analysis, we calculated the mean fluorescence intensity (MFI) of cells treated with HaloTag(dye) conjugates (Figure 4). When cells were treated with HaloTag(JF585) conjugates, we observed MFI values of 1138, 3354, and 396 AU for HaloTag(JF585)–R10, HaloTag(JF585)–1, and HaloTag(JF585) + L17E, respectively. These values highlight that HaloTag(JF585)–1 is ~3-fold more effective at being taken up by cells than HaloTag(JF585)–R10, and that HaloTag(JF585) + L17E is only ~⅓ as effective at enabling cellular uptake when compared to HaloTag(JF585)–R10. Yet, when cells were treated with HaloTag(VO) conjugates, we observed MFI values of 328, −80, and 591 AU for HaloTag(VO)–R10, HaloTag(VO)–1, and HaloTag(VO) + L17E, respectively. This demonstrates that HaloTag(VO) + L17E is ~2-fold more effective at cytosolic delivery than HaloTag(VO)–R10, and that HaloTag(VO)–1 is not detected in the cytosol of cells. These data are summarized in Table 1.
Figure 4.
Mean fluorescence intensity (MFI) of HaloTag(dye) conjugates in HeLa cells. For experimental conditions, see Table S1. Values are the mean ± SE from at least two independent experiments, each performed with three technical replicates; ****p < 0.0001, ***p < 0.0005, **p < 0.0011. The average fluorescence background, calculated from cells treated with HaloTag(dye) alone, was subtracted from all treatment conditions. Standardized laser intensities were used across all experiments.
Table 1.
Summary of Cellular Delivery Efficiencies
| Delivery Strategy |
Total Cellular Uptake (MFIJF585)a |
Cytosolic Delivery (MFIVO)a |
Delivery Efficiency ()a |
|---|---|---|---|
| R10 | 1.0 | 1.0 | 1.0 |
| 1 | 3.0 | 0.0 | 0.0 |
| L17E | 0.35 | 1.8 | 5.2 |
Values are normalized to the uptake profiles of HaloTag(dye)–R10, which is defined as 1.0. MFIJF585 and MFIVO represent the ratio of the MFI of each delivery strategy to the MFI of R10.
With flow cytometry data sets in hand, we calculated the delivery efficiency () by using an equation56 that deploys the MFI of the JF585 and VO conjugates:
| (1) |
Eq 1 relates the total cellular uptake (JF585) and the cytosolic uptake (VO) of a given delivery strategy (denoted as “vehicle”) to one defined as a reference. This normalization ultimately quantifies the efficiency of cytosolic delivery as a fold-change compared to the reference. Because R10 is a well-known cell-penetrating peptide, we set HaloTag(dye)–R10 as our reference and normalized other delivery strategies to that conjugate (Table 1).
Our analysis reveals that while the esterification of HaloTag(dye) with diazo compound 1 provides highly effective entrance into cells via endocytosis, this modification does not afford cytosolic access. This underscores the idiosyncratic nature of protein delivery into cells and highlights the challenge of providing cytosolic access to an extremely anionic payload like HaloTag. In contrast, we observed that the endosomolytic peptide L17E is less efficient at providing cellular entry than R10, but the fraction of HaloTag that reaches the cytosol is ~2-fold greater than R10, representing an ~5-fold increase in delivery efficiency. Thus, L17E is highly efficient at circumventing endosomal entrapment and lysosomal degradation and, ultimately, delivering HaloTag to the cytosol.
CONCLUSIONS
Current methods to assess the cytosolic delivery of proteins are time-consuming to establish, require specialized instrumentation, or introduce enzymatic “tag” recognition sequences that limit generality. Herein, we describe a dye pair with the ability to assess protein delivery efficiencies, enabling a nuanced understanding of how protein delivery agents affect cellular uptake and cytosolic access. Using this novel method, we report the unprecedented (to our knowledge) delivery of HaloTag, a highly anionic and, thus, challenging protein target, to the cytosol of mammalian cells. We anticipate that our results will facilitate the further development of HaloTag as a useful cellular delivery platform to mediate the delivery of other proteins and payloads.
Supplementary Material
ACKNOWLEDGMENTS
The authors are grateful to J. Yang, E. C. Wralstad, N. S. Abularrage, R. L. McPherson, and S. D. Brucks for helpful scientific discussions, and to R. L. McPherson, S. D. Brucks, V. M. Marando, M. C. Hoffman, and K. J. Hetrick for their assistance in reviewing the manuscript. They are grateful to the Koch Institute’s Robert A. Swanson (1969) Biotechnology Center for technical support, specifically the High Throughput Sciences Core for providing the HeLa cell line and performing mycoplasma testing.
Funding
J.V.J. was supported by a Life Sciences Research Foundation fellowship sponsored by the Shurl and Kay Curci Foundation. Y.D.P. was supported by NSF Graduate Research Fellowship 4000143422. This work was supported by Grants R35 GM148220 and P30 CA014051 (NIH) and the Howard Hughes Medical Institute (HHMI).
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/xxxx.xxxxxxx.
Experimental procedures, Table S1, Figures S1-S25, and compound characterization (PDF)
The authors declare no competing financial interest.
REFERENCES
- (1).Deshaies RJ Multispecific drugs herald a new era of biopharmaceutical innovation. Nature 2020, 580, 329–338. [DOI] [PubMed] [Google Scholar]
- (2).Urquhart L. Top product forecasts for 2021. Nat. Rev. Drug Discovery 2021, 20, 10. [DOI] [PubMed] [Google Scholar]
- (3).Urquhart L. Top product forecasts for 2022. Nat. Rev. Drug Discovery 2022, 21, 11. [DOI] [PubMed] [Google Scholar]
- (4).Dougherty PG; Sahni A; Pei D Understanding cell penetration of cyclic peptides. Chem. Rev 2019, 119, 10241–10287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Los GV; Encell LP; McDougall MG; Hartzell DD; Karassina N; Zimprich C; Wood MG; Learish R; Ohana RF; Urh M; Simpson D; Mendez J; Zimmerman K; Otto P; Vidugiris G; Zhu J; Darzins A; Klaubert DH; Bulleit RF; Wood KV HaloTag: A novel protein labeling technology for cell imaging and protein analysis. ACS Chem. Biol 2008, 3, 373–382. [DOI] [PubMed] [Google Scholar]
- (6).Encell LP; Ohana RF; Zimmerman K; Otto P; Vidugiris G; Wood MG; Los GV; McDougall MG; Zimprich C; Karassina N; Learish RD; Hurst R; Hartnett J; Wheeler S; Stecha P; English J; Zhao K; Mendez J; Benink HA; Murphy N; Daniels DL; Slater MR; Urh M; Darzins A; Klaubert DH; Bulleit RF; Wood KV Development of a dehalogenase-based protein fusion tag capable of rapid, selective and covalent attachment to customizable ligands. Curr. Chem. Genomics 2013, 6, 55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Wilhelm J; Kühn S; Tarnawski M; Gotthard G; Tünnermann J; Tänzer T; Karpenko J; Mertes N; Xue L; Uhrig U; Reinstein J; Hiblot J; Johnsson K Kinetic and structural characterization of the self-labeling protein tags HaloTag7, SNAP-tag, and CLIP-tag. Biochemistry 2021, 60, 2560–2575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).England CG; Luo H; Cai W HaloTag technology: A versatile platform for biomedical applications. Bioconjugate Chem. 2015, 26, 975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Buckley DL; Raina K; Darricarrere N; Hines J; Gustafson JL; Smith IE; Miah AH; Harling JD; Crews CM HaloPROTACS: Use of small molecule PROTACs to induce degradation of HaloTag fusion proteins. ACS Chem. Biol 2015, 10, 1831–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Erdmann RS; Baguley SW; Richens JH; Wissner RF; Xi Z; Allgeyer ES; Zhong S; Thompson AD; Lowe N; Butler R; Bewersdorf J; Rothman JE; St Johnston D; Schepartz A; Toomre D Labeling strategies matter for super-resolution microscopy: A comparison between HaloTags and SNAP-tags. Cell Chem. Biol 2019, 26, 584–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Straková K; Lopez-Andarias J; Jimenez-Rojo N; Chambers JE; Marciniak SJ; Riezman H; Sakai N; Matile S Haloflippers: A general tool for the fluorescence imaging of precisely localized membrane tension changes in living cells. ACS Cent. Sci 2020, 6, 1376–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Chen W; Younis MH; Zhao Z; Cai W Recent biomedical advances enabled by HaloTag technology. Biocell 2022, 46, 1789–1801. [PMC free article] [PubMed] [Google Scholar]
- (13).Coulomb CA, Collection de Mémoires Relatifs a la Physique. Gauthier–Villars: Paris, 1884. [Google Scholar]
- (14).Gillmor CS, Coulomb and the Evolution of Physics and Engineering in Eighteenth-Century France. Princeton University Press: Princeton, NJ, 1971. [Google Scholar]
- (15).Varki A; Cummings RD; Esko JD; Freeze HH; Stanley P; Bertozzi CR; Hart GW; Etzler ME, Essentials of Glycobiology. Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, 2009. [PubMed] [Google Scholar]
- (16).Gasteiger E; Hoogland C; Gattiker A; Duvaud S; Wilkins MR; Appel RD; Bairoch A, Protein identification and analysis tools on the ExPASy server. In The Proteomics Protocols Handbook, Walker JM, Ed. Humana Press: Totowa, NJ, 2005; pp 571–607. [Google Scholar]
- (17).Palte MJ; Raines RT Interaction of nucleic acids with the glycocalyx. J. Am. Chem. Soc 2012, 134, 6218–6223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Kim S; Kim D; Cho SW; Kim J; Kim JS Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Lee Y-W; Luther DC; Goswami R; Jeon T; Clark V; Elia J; Gopalakrishnan S; Rotello VM Direct cytosolic delivery of proteins through coengineering of proteins and polymeric delivery vehicles. J. Am. Chem. Soc 2020, 142, 4349–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Schwarze SR; Ho A; Vocero-Akbani A; Dowdy SF In vivo protein transduction: Delivery of a biologically active protein into the mouse. Science 1999, 285, 1569–1572. [DOI] [PubMed] [Google Scholar]
- (21).Nagel YA; Raschle PS; Wennemers H Effect of preorganized charge-display on the cell-penetrating properties of cationic peptides. Angew. Chem., Int. Ed 2017, 56, 122–126. [DOI] [PubMed] [Google Scholar]
- (22).Schneider AFL; Kithil M; Cardoso MC; Lehmann M; Hackenberger CPR Cellular uptake of large biomolecules enabled by cell-surface-reactive cell-penetrating peptide additives. Nat. Chem 2021, 13, 530–539. [DOI] [PubMed] [Google Scholar]
- (23).Zhang X; Cattoglio C; Zoltek M; Vetralla C; Mozumdar D; Schepartz A Dose-dependent nuclear delivery and transcriptional repression with a cell-penetrant MeCP2. ACS Cent. Sci 2023, 9, 277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Fuchs SM; Raines RT Arginine grafting to endow cell permeability. ACS Chem. Biol 2007, 2, 167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Fuchs SM; Rutkoski TJ; Kung VM; Groeschl RT; Raines RT Increasing the potency of a cytotoxin with an arginine graft. Protein Eng. Des. Sel 2007, 20, 505–509. [DOI] [PubMed] [Google Scholar]
- (26).McNaughton BR; Cronican JJ; Thompson DB; Liu DR Mammalian cell penetration, siRNA transfection, and DNA transfection by supercharged proteins. Proc. Natl. Acad. Sci. U. S. A 2009, 106, 6111–6116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Thompson DB; Cronican JJ; Liu DR Engineering and identifying supercharged proteins for macromolecule delivery into mammalian cells. Methods Enzymol. 2012, 503, 293–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Li W; Nicol F; Szoka FC Jr. GALA: A designed synthetic pH-responsive amphipathic peptide with applications in drug and gene delivery. Adv. Drug Del. Rev 2004, 56, 967–985. [DOI] [PubMed] [Google Scholar]
- (29).Akishiba M; Takeuchi T; Kawaguchi Y; Sakamoto K; Yu HH; Nakase I; Takatani-Nakase T; Madani F; Gräslund A; Futaki S Cytosolic antibody delivery by lipid-sensitive endosomolytic peptide. Nat. Chem 2017, 9, 751–761. [DOI] [PubMed] [Google Scholar]
- (30).Laurent Q; Martinent R; Moreau D; Winssinger N; Sakai N; Matile S Oligonucleotide phosphorothioates enter cells by thiol-mediated uptake. Angew. Chem., Int. Ed 2021, 60, 19102–19106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Futaki S; Ohashi W; Suzuki T; Niwa M; Tanaka S; Ueda K; Harashima H; Sugiura Y Stearylated arginine-rich peptides: A new class of transfection systems. Bioconjugate Chem. 2001, 12, 1005–1011. [DOI] [PubMed] [Google Scholar]
- (32).Sakamoto K; Michibata J; Hirai Y; Ide A; Ikitoh A; Takatani-Nakase T; Futaki S Potentiating the membrane interaction of an attenuated cationic amphiphilic lytic peptide for intracellular protein delivery by anchoring with pyrene moiety. Bioconjugate Chem. 2021, 32, 950–957. [DOI] [PubMed] [Google Scholar]
- (33).Mix KA; Lomax JE; Raines RT Cytosolic delivery of proteins by bioreversible esterification. J. Am. Chem. Soc 2017, 139, 14396–14398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Schneider AFL; Wallabregue ALD; Franz L; Hackenberger CPR Targeted subcellular protein delivery using cleavable cyclic cell-penetrating peptides. Bioconjugate Chem. 2019, 30, 400–404. [DOI] [PubMed] [Google Scholar]
- (35).Ressler VT; Mix KA; Raines RT Esterification delivers a functional enzyme into a human cell. ACS Chem. Biol 2019, 14, 599–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Jun JV; Petri YD; Erickson LW; Raines RT Modular diazo compound for the bioreversible late-stage modification of proteins. J. Am. Chem. Soc 2023, 145, 6615–6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Chao T-Y; Raines RT Fluorogenic label to quantify the cytosolic delivery of macromolecules. Mol. Biosyst 2013, 9, 339–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Johnsson N; Varshavsky A Split ubiquitin as a sensor of protein interactions in vivo. Proc. Natl. Acad. Sci. U. S. A 1994, 91, 10340–10344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Cabantous S; Terwilliger TC; Waldo GS Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol 2005, 23, 102–107. [DOI] [PubMed] [Google Scholar]
- (40).Kato N; Jones J The split luciferase complementation assay. Methods Mol. Biol 2010, 655, 359–376. [DOI] [PubMed] [Google Scholar]
- (41).Dominski Z; Kole R Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl. Acad. Sci. U. S. A 1993, 90, 8673–8677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Kang SH; Cho MJ; Kole R Up-regulation of luciferase gene expression with antisense oligonucleotides: Implications and applications in functional assay development. Biochemistry 1998, 37, 6235–6239. [DOI] [PubMed] [Google Scholar]
- (43).Loison F; Nizard P; Sourisseau T; Le Goff P; Debure L; Le Drean Y; Michel D A ubiquitin-based assay for the cytosolic uptake of protein transduction domains. Mol. Ther 2005, 11, 205–214. [DOI] [PubMed] [Google Scholar]
- (44).Stanford SM; Krishnamurthy D; Kulkarni RA; Karver CE; Bruenger E; Walker LM; Ma CT; Chung TDY; Sergienko E; Bottini N; Barrios AM PCAP-based peptide substrates: The new tool in the box of tyrosine phosphatase assays. Methods 2014, 65, 165–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Verdurmen WPR; Luginbühl M; Honegger A; Plückthun A Efficient cell-specific uptake of binding proteins into the cytoplasm through engineered modular transport systems. J. Controlled Release 2015, 200, 13–22. [DOI] [PubMed] [Google Scholar]
- (46).Chyan W; Raines RT Enzyme-activated fluorogenic probes for live-cell and in vivo imaging. ACS Chem. Biol 2018, 13, 1810–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).Peraro L; Deprey KL; Moser MK; Zou Z; Ball HL; Levine B; Kritzer JA Cell penetration profiling using the chloroalkane penetration assay. J. Am. Chem. Soc 2018, 140, 11360–11369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Larochelle JR; Cobb GB; Steinauer A; Rhoades E; Schepartz A Fluorescence correlation spectroscopy reveals highly efficient cytosolic delivery of certain penta-arg proteins and stapled peptides. J. Am. Chem. Soc 2015, 137, 2536–2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (49).Illien F; Rodriguez N; Amoura M; Joliot A; Pallerla M; Cribier S; Burlina F; Sagan S Quantitative fluorescence spectroscopy and flow cytometry analyses of cell-penetrating peptides internalization pathways: Optimization, pitfalls, comparison with mass spectrometry quantification. Sci. Rep 2016, 6, 36938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (50).Rezgui R; Blumer K; Yeoh-Tan G; Trexler AJ; Magzoub M Precise quantification of cellular uptake of cell-penetrating peptides using fluorescence-activated cell sorting and fluorescence correlation spectroscopy. Biochim. Biophys. Acta—Biomembr 2016, 1858, 1499–1506. [DOI] [PubMed] [Google Scholar]
- (51).Deprey K; Becker L; Kritzer J; Plückthun A Trapped! A critical evaluation of methods for measuring total cellular uptake versus cytosolic localization. Bioconjugate Chem. 2019, 30, 1006–1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Tycko B; Keith CH; Maxfield FR Rapid acidification of endocytic vesicles containing asialoglycoprotein in cells of a human hepatoma line. J. Cell Biol 1983, 97, 1762–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Murphy RF; Powers S; Cantor CR Endosome pH measured in single cells by dual fluorescence flow cytometry: Rapid acidification of insulin to pH 6. J. Cell Biol 1984, 98, 1757–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (54).Lee LG; Berry GM; Chen C-H Vita Blue: A new 633-nm excitable fluorescent dye for cell analysis. Cytometry 1989, 10, 151–164. [DOI] [PubMed] [Google Scholar]
- (55).Wainwright M. The use of dyes in modern biomedicine. Biotechnol. Histochem 2003, 78, 147–155. [DOI] [PubMed] [Google Scholar]
- (56).Qian Z; Dougherty PG; Pei D Monitoring the cytosolic entry of cell-penetrating peptides using a pH-sensitive fluorophore. Chem. Commun 2015, 51, 2162–2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Qian Z; Martyna A; Hard RL; Wang J; Appiah-Kubi G; Coss C; Phelps MA; Rossman JS; Pei D Discovery and mechanism of highly efficient cyclic cell-penetrating peptides. Biochemistry 2016, 55, 2601–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (58).Stolle AS; Norkowski S; Körner B; Schmitz J; Lüken L; Frankenberg M; Rüter C; Schmidt MA T3SS-independent uptake of the short-trip toxin-related recombinant NleC effector of enteropathogenic Escherichia coli leads to NF-κB p65 cleavage. Front. Cell. Infect. Microbiol 2017, 7, 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (59).Chen K; Pei D Engineering cell-permeable proteins through insertion of cell-penetrating motifs into surface loops. ACS Chem. Biol 2020, 15, 2568–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (60).Fischer R; Waizenegger T; Köhler K; Brock R A quantitative validation of fluorophore-labelled cell-permeable peptide conjugates: Fluorophore and cargo dependence of import. Biochim. Biophys. Acta—Biomembr 2002, 1564, 365–374. [DOI] [PubMed] [Google Scholar]
- (61).Puckett CA; Barton JK Fluorescein redirects a ruthenium-octaarginine conjugate to the nucleus. J. Am. Chem. Soc 2009, 131, 8738–8739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (62).Walrant A; Matheron L; Cribier S; Chaignepain S; Jobin ML; Sagan S; Alves ID Direct translocation of cell-penetrating peptides in liposomes: A combined mass spectrometry quantification and fluorescence detection study. Anal. Biochem 2013, 438, 1–10. [DOI] [PubMed] [Google Scholar]
- (63).Birch D; Christensen MV; Staerk D; Franzyk H; Nielsen HM Fluorophore labeling of a cell-penetrating peptide induces differential effects on its cellular distribution and affects cell viability. Biochim. Biophys. Acta—Biomembr 2017, 1859, 2483–2494. [DOI] [PubMed] [Google Scholar]
- (64).Lavis LD; Raines RT Bright ideas for chemical biology. ACS Chem. Biol 2008, 3, 142–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (65).Grimm JB; Sung AJ; Legant WR; Hulamm P; Matlosz SM; Betzig E; Lavis LD Carbofluoresceins and carborhodamines as scaffolds for high-contrast fluorogenic probes. ACS Chem. Biol 2013, 8, 1303–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (66).Lavis LD; Raines RT Bright building blocks for chemical biology. ACS Chem. Biol 2014, 9, 855–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (67).Grimm JB; English BP; Chen J; Slaughter JP; Zhang Z; Revyakin A; Patel R; Macklin JJ; Normanno D; Singer RH; Lionnet T; Lavis LD A general method to improve fluorophores for live-cell and single-molecule microscopy. Nat. Methods 2015, 12, 244–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (68).Zhou X; Lai R; Beck JR; Li H; Stains CI Nebraska Red: A phosphinate-based near-infrared fluorophore scaffold for chemical biology applications. Chem. Commun 2016, 52, 12290–12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (69).Grimm JB; Muthusamy AK; Liang Y; Brown TA; Lemon WC; Patel R; Lu R; Macklin JJ; Keller PJ; Ji N; Lavis LD A general method to fine-tune fluorophores for live-cell and in vivo imaging. Nat. Methods 2017, 14, 987–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (70).Zhou X; Lesiak L; Lai R; Beck JR; Zhao J; Elowsky CG; Li H; Stains CI Chemoselective alteration of fluorophore scaffolds as a strategy for the development of ratiometric chemodosimeters. Angew. Chem., Int. Ed 2017, 56, 4197–4200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (71).Fang Y; Good GN; Zhou X; Stains CI Phosphinate-containing rhodol and fluorescein scaffolds for the development of bioprobes. Chem. Commun 2019, 55, 5962–5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (72).Brøndsted F; Stains CI Heteroatom-substituted xanthene fluorophores enter the shortwave-infrared region. Photochem. Photobiol 2022, 98, 400–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (73).Brøndsted F; Fang Y; Li L; Zhou X; Grant S; Stains CI Single atom stabilization of phosphinate ester-containing rhodamines yields cell permeable probes for turn-on photoacoustic imaging. Chem.—Eur. J 2024, 30, e202303038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (74).Lavis LD; Rutkoski TJ; Raines RT Tuning the pKa of fluorescein to optimize binding assays. Anal. Chem 2007, 79, 6775–6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (75).Grimm JB; Gruber TD; Ortiz G; Brown TA; Lavis LD Virginia Orange: A versatile, red-shifted fluorescein scaffold for single- and dual-input fluorogenic probes. Bioconjugate Chem. 2016, 27, 474–480. [DOI] [PubMed] [Google Scholar]
- (76).Martineau M; Somasundaram A; Grimm JB; Gruber TD; Choquet D; Taraska JW; Lavis LD; Perrais D Semisynthetic fluorescent pH sensors for imaging exocytosis and endocytosis. Nat. Commun 2017, 8, 1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (77).Hu Y-B; Dammer EB; Ren R-J; Wang G The endosomal–lysosomal system: From acidification and cargo sorting to neurodegeneration. Transl. Neurodegener 2015, 4, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (78).Cheah KM; Jun JV; Wittrup KD; Raines RT Host–guest complexation by β-cyclodextrin enhances the solubility of an esterified protein. Mol. Pharmaceutics 2022, 19, 3869–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (79).Deprey K; Kritzer JA HaloTag forms an intramolecular disulfide. Bioconjugate Chem. 2021, 32, 964–970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (80).Akishiba M; Futaki S Inducible membrane permeabilization by attenuated lytic peptides: A new concept for accessing cell interiors through ruffled membranes. Mol. Pharmaceutics 2019, 16, 2540–2548. [DOI] [PubMed] [Google Scholar]
- (81).Nomura Y; Sakamoto K; Akishiba M; Iwata T; Hirose H; Futaki S Improved cytosolic delivery of macromolecules through dimerization of attenuated lytic peptides. Bioorg. Med. Chem. Lett 2020, 30, 127362. [DOI] [PubMed] [Google Scholar]
- (82).Becker B; Englert S; Schneider H; Yanakieva D; Hofmann S; Dombrowsky C; Macarrón Palacios A; Bitsch S; Elter A; Meckel T; Kugler B; Schirmacher A; Avrutina O; Diederichsen U; Kolmar H Multivalent dextran hybrids for efficient cytosolic delivery of biomolecular cargoes. J. Pept. Sci 2021, 27, e3298. [DOI] [PubMed] [Google Scholar]
- (83).Feng R; Ni R; Chau Y Fusogenic peptide modification to enhance gene delivery by peptide–DNA nano-coassemblies. Biomat. Sci 2022, 10, 5116–5120. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.