Abstract
Open reading frame (ORF) 57 of Kaposi's sarcoma-associated herpesvirus (KSHV) encodes a homolog of known posttranscriptional regulators that are essential for replication in other herpesviruses. Here, we examined the expression of this gene and the function(s) of its product. KSHV ORF 57 is expressed very early in infection from a 1.6-kb spliced RNA bearing several in-frame initiation codons. Its product is a nuclear protein that, in transient assays, has little effect on the expression of luciferase reporter genes driven by a variety of KSHV and heterologous promoters. However, ORF 57 protein enhances the accumulation of several viral transcripts, in a manner suggesting posttranscriptional regulation. These transcripts include not only known cytoplasmic mRNAs (e.g., ORF 59) but also a nuclear RNA (nut-1) that lacks coding potential. Finally, ORF 57 protein can also modulate the effects of the ORF 50 gene product, a classical transactivator known to be required for lytic induction. The expression from some (e.g., nut-1) but not all (e.g., tk) ORF 50-responsive promoters can be synergistically enhanced by coexpression of ORF 50 and ORF 57. This effect is not due to upregulation of ORF 50 expression but rather to a posttranslational enhancement of the transcriptional activity of ORF 50. These data indicate that ORF 57 is a powerful pleiotropic effector that can act on several posttranscriptional levels to modulate the expression of viral genes in infected cells.
Kaposi's sarcoma (KS), an endothelial tumor with neoangiogenic and inflammatory components, is a common neoplasm of AIDS patients. Recent evidence strongly implicates a novel lymphotropic herpesvirus, KS-associated herpesvirus (KSHV; also called human herpesvirus 8), in the pathogenesis of KS (5, 12; for reviews, see references 14 and 41). KSHV DNA is found in virtually all of the spindle (endothelial) cells of clinically apparent KS lesions (8, 46), as well as in tumor-infiltrating monocytes (6) and circulating B cells (1, 27, 50). Consistent with its classification as a lymphotropic gammaherpesvirus, KSHV is also tightly linked to certain B-cell lymphomas, termed primary effusion lymphomas (9, 45). Although KSHV infection of KS spindle cells is predominantly latent, lytic replication is also evident in the tumor (28, 35, 36, 46), and growing evidence suggests that the lytic cycle contributes importantly to tumorigenesis. For example, the incidence of KS is greatly decreased when AIDS patients at risk for KS are treated with ganciclovir, a drug that specifically blocks lytic viral replication (25). Moreover, several lytic-cycle gene products can stimulate inflammatory and angiogenic responses in surrounding cells and tissues (2, 3, 7). Lytic replication has also been posited to be required for KSHV spread from its presumed lymphoreticular reservoir to its endothelial targets (24). Thus, the study of the KSHV lytic cycle (and the switch from latency to lytic growth) is important not only to fully characterize the molecular basis of viral replication but also to further inform our evolving notions of KS pathogenesis.
Lytic herpesviral replication is characterized by a temporally regulated cascade of viral gene expression. Immediate-early (IE) genes, many of which encode activators of gene expression, are expressed first. Their expression leads to upregulation of delayed-early (DE) genes, whose products include proteins involved in viral DNA replication; following replication, the so-called late (L) genes, primarily encoding virion structural proteins, are expressed. Two of the earliest genes to be transcribed in KSHV-infected B cells are open reading frame (ORF) 50 and ORF 57. We (23, 24) and others (47) have recently shown that ORF 50 expression can trigger lytic reactivation of KSHV in infected B cells. ORF 50 is an IE gene (53) whose product is a transcriptional transactivator (23, 24, 47), and this activity is required for viral reactivation by all known chemical inducers (e.g., tetradecanoyl phorbol acetate (TPA) and sodium butyrate) (24). In addition to its classical DE targets (24), the ORF 50 gene product can also upregulate the promoter for ORF 57 (23).
KSHV ORF 57 is homologous to known posttranscriptional regulators in other herpesviruses. One of these, ICP27 of herpes simplex virus (HSV), is a pleiotropic regulator whose functions include downregulation of intron-containing transcripts and upregulation of certain late messages (40, 43). Temperature-sensitive mutations have shown that ICP27 is essential for lytic viral replication and is required for inhibition of host cell splicing, an activity that contributes to host shutoff and to the downregulation of intron-containing genes in transient assays (17, 18). ICP27 has also been shown to shuttle from the nucleus to the cytoplasm and to promote the export of intronless viral RNAs (26, 32, 39, 44). The other gammaherpesviruses, Epstein-Barr virus (EBV) and herpesvirus saimiri (HSV), also encode ICP27 homologs (13, 21, 30) which, while less extensively studied, also appear to modulate gene expression in a posttranscriptional fashion (38, 42, 51).
Here, we have examined the fine structure and expression of KSHV ORF 57 mRNA in BCBL-1 cells, a primary effusion lymphoma cell line harboring latent but inducible KSHV genomes, and present an initial characterization of the activities of its product. Our results show that ORF 57 is a complex pleiotropic effector that can act on several levels to augment viral gene expression.
MATERIALS AND METHODS
Cell lines, plasmids, and probes.
CV-1 cells were propagated and maintained in Dubecco's modified Eagle medium H21 (DME-H21 medium) supplemented with 10% fetal calf serum and penicillin-steptomycin at 37°C in 5% CO2.
To create intron-containing versions (see Fig. 3 and 8) of the KSHV reporter vectors described previously (24), we used PCR to amplify the simian virus 40 (SV40) T antigen intron from the pGL2 promoter vector (Promega) by using the following primers (which introduced terminal XbaI sites): IN 1 (5′ GCGCTCTAGAAGAGATTTAAAGCTC) and IN 2 (5′ GCGCTCTAGACAGTTCCATAGGTTG). The resulting PCR fragment was cut with XbaI and cloned into the XbaI site of each KSHV promoter pGL3 vector at the 3′ end of the luciferase coding region. The nut-1 reporter (pGL3 nut-1 A) (see Fig. 6) was created by PCR amplification of pGL3 nut-1 (24) as template by using an internal pGL3 basic primer containing a BglII site and the primer 5′ GCGAGATCTAGCCAAGGTGACT. The PCR-amplified fragment was digested with BglII and was ligated into the BglII site of pGL3 basic (Promega). To create the C-terminal epitope-tagged version of ORF 57, we performed two cloning steps. First, we digested pcDNA 3.1 ORF 57 (24) with BamHI and XhoI to release the ORF 57 coding region and cloned that fragment into the BamHI and XhoI sites of pcDNA 3.1-V5-His A (Invitrogen). Second, we used PCR to amplify the 3′ end of ORF 57 without its stop codon, so that the C-terminal tag could be expressed in frame with ORF 57. The following primers were used to PCR amplify the ORF 57 3′ end from a genomic clone: primer 57 V1 (5′ GCAGAGCGAGCTAGCACTGATCAAAC) and primer 57 V1 (5′ GCGGGTTCGAAAGAAAGTGGATAAAAG). The PCR fragment was digested with NheI and SfuI and was cloned into the NheI and SfuI sites of the vector created in step 1 to make pcDNA ORF 57-V5.
FIG. 3.
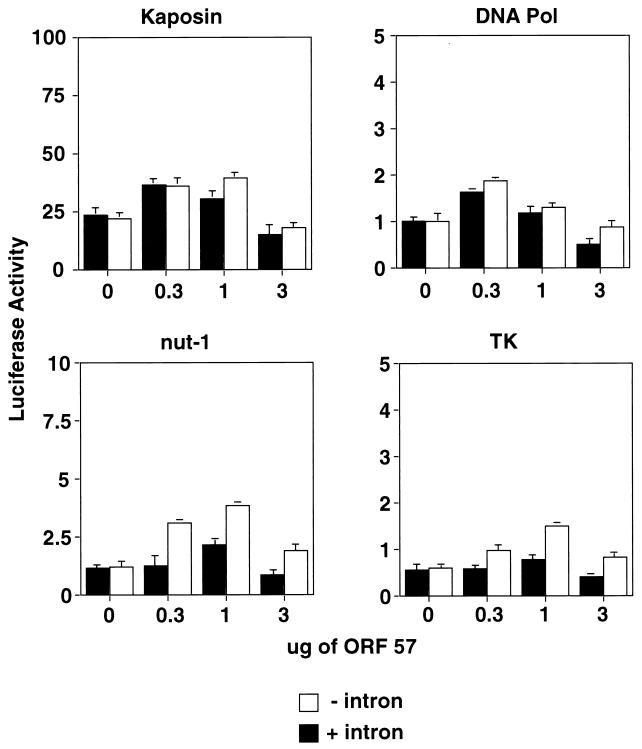
Effect of ORF 57 expression on viral promoter constructs with and without introns. CV-1 cells were cotransfected with increasing amounts of pcDNA 3.1 ORF 57 (0, 0.3, 1, and 3 μg) and fixed amounts of the following KSHV promoter luciferase constructs with or without an intron: DNA Pol, Kaposin, Nut-1, and TK. Luciferase activity was measured 48 h posttransfection. Graphs are representative examples of experiments performed in duplicate. Error bars represent standard deviations.
FIG. 8.
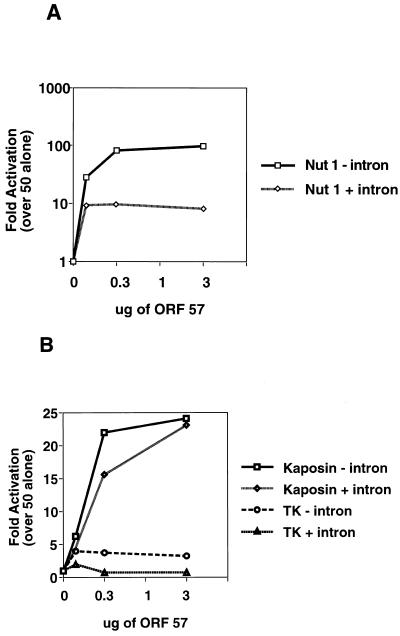
Effect of introns on synergistic activation of different promoters by the ORF 50-ORF 57 combination. CV-1 cells were cotransfected with fixed amounts of the indicated KSHV promoter constructs with or without introns (Nut-1, TK, and Kaposin), a fixed amount of the transcriptional activator, pcDNA 3 ORF 50, and increasing amounts of pcDNA 3.1 ORF 57 (0, 0.3, 1, and 3 μg). Luciferase activity was measured 48 h after transfection. (A) Log scale graph displaying the effects on the Nut-1 promoter with and without an intron. (B) Graph displaying the effects on the Kaposin and TK promoters with and without an intron. Values are plotted as fold luciferase activity over that of ORF 50 alone, which was set to 1. Error bars representing standard deviations are from two experiments performed in duplicate. Note that activation is plotted on an arithmetic scale in B and on a log scale in A.
FIG. 6.
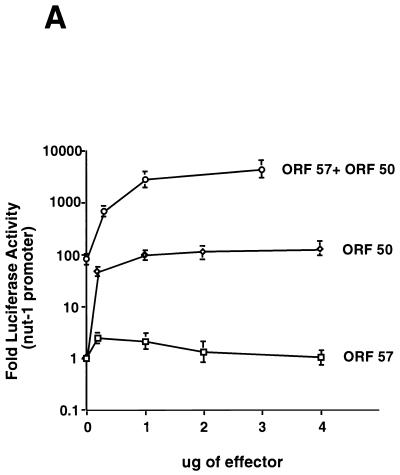
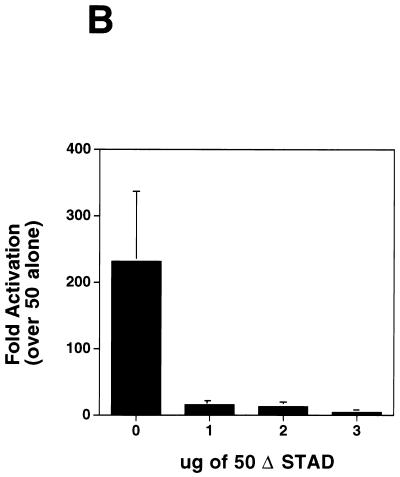
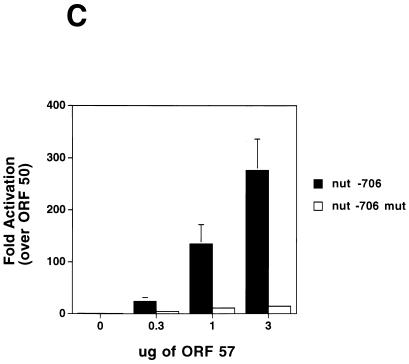
ORF 57 augments the effect of ORF 50 on the Nut-1 promoter. (A) ORF 50-ORF 57 synergy on the nut1-reporter. CV-1 cells were transfected with a fixed amount of the nut-1 promoter reporter A (intronless), and the indicated amounts of either pcDNA 3 ORF 50 or pcDNA 3.1 ORF 57 or 1 μg of pcDNA 3 ORF 50 plus the indicated amounts of pcDNA 3.1 ORF 57. Luciferase activity was measured 48 h posttransfection and was plotted on the graph. Error bars represent standard deviations from four experiments performed in duplicate. (B) Dominant negative ORF 50 inhibits the augmentation of ORF 50 by ORF 57. CV-1 cells were transfected with fixed amounts of pcDNA 3 ORF 50 (1 μg), pcDNA 3.1 ORF 57 (1 μg), and the Nut-1 promoter (0.1 μg) along with increasing amounts of pCMV-myc-nuc-ΔSTAD (0, 1, 2, and 3 μg) (23). Forty-eight hours posttransfection, luciferase activity was measured and values were plotted that represented fold activation relative to that of ORF 50 alone, which was set to 1. (C) Mutations in the nut-1 promoter abolish synergy between ORF 50 and ORF 57. CV-1 cells were transfected with 2 μg of pcDNA 3 ORF 50, increasing amounts of ORF 57, and either 1 μg of the wild-type nut-1-706 reporter or the nut-1-706 mutant reporter. Forty-eight hours posttransfection, luciferase activity was measured and values were plotted that represent fold activation relative to that of ORF 50 alone, which was set to 1.
The ORF 59/58 expression plasmid (pcDNA 3.1 ORF 59/58) expresses the bicistronic ORF 59/58 message encompassing the region from the AUG to 152 bases downstream of the poly(A) signal. The following primers introduced restriction enzyme sites by PCR amplification by using a lambda genomic clone from a KS lung tumor as template: primer 59 (added XbaI site) (5′ GCGCTCTAGAATGCCTGTGGATTTTCAC) and primer 58 (added EcoRV site) (5′ GCGCGATATCCCAATGCAGTTGAACTAC). The 2.4-kb PCR product was cloned into pCR2.1 (Invitrogen). The PCR product was released from pCR2.1 with HindIII, was treated with Klenow, and was then digested with XbaI. This product was cloned into pcDNA 3.1 (Invitrogen) which had been digested with BbsI (to remove the vector-provided poly(A) signal), was treated with Klenow, and was then cut with XbaI, to create pcDNA 3.1 ORF 59/58. To create a probe for the ORF 59/58 Northern blot, we digested pcDNA 3.1 ORF 59/58 with EcoRV to release a 500-bp fragment spanning nucleotide (nt) 94330 to nt 94852.
The nut-1 expression plasmid was created by PCR amplification of a genomic lambda clone by using the following primers: nut-1 L (5′ ACTGGGACTGCCCAGTC) and nut-1 R (5′ ATGGATTAAACATTGCCATTTAT). The resulting product was cloned into pCR 2.1 (Invitrogen), this was digested with HindIII and EcoRV. The resulting fragment was cloned into the HindIII and EcoRV sites of pcDNA 3 (Invitrogen) to create pcDNA 3 nut-1. A double-stranded (ds) DNA probe for nut-1 was created by digesting pcDNA 3 nut-1 with EcoRI. The GCR and K5 expression plasmids were made by PCR amplification of genomic lamda clones with the following primers, and subsequent cloning of the PCR products into pCR 3.1 (Invitrogen): for GCR, GCR 1 (5′ CGATCGCGGCCGCACCTATACTACTTGTT) and GCR 2 (5′ CAGCTTGATCACCGCGGGCTACGTGGTGGC); and for K5, K5 1 (5′ CCAAGTGGTTGTTCAACCGT) and K5 2 (5′ AGCTGCAAAGATGGCGTCTA). DNA probes (ds) for GCR and K5 contained only coding sequences.
To create the truncated version of the nut-1 reporter, pGL3 nut-1-706, pGL3 nut-1 (24) was digested with SmaI and TthIII followed by blunt-end ligation. The mutant version of this reporter, pGL3nut-1-706 mut, was created by linker scanning and resulted in substitution of the ORF 50 response element with a 12-bp linker containing a NdeI site and three nucleotides on either side. (J. Chang, D. Lukac, and D. Ganem, unpublished data).
cDNA cloning.
The cDNA clones containing ORF 57 were isolated from a poly(dT)-primed library made by R. Renne from induced BCBL-1 cells in Lambda Zap (Stratagene). Screening was performed with a 436-bp ds DNA fragment spanning the region from nt 82839 to nt 83278.
RNase protection and primer extension.
RNase protection was performed with the RPA II kit from Ambion (Austin, Tex.) according to the manufacturer's instructions with the following modifications. The following plasmid was constructed to make the single-stranded RNA probe for RNase protection: a 400-bp KpnI genomic fragment containing the genomic sequence from nt 81948 to nt 82346 was cloned into the KpnI site of pSP72 (Promega). The plasmid was linearized with XbaI and was transcribed with T7 RNA polymerase in the presence of [32P]UTP to create an antisense probe. Approximately 106 cpm of probe was hybridized to 7.5 μg of RNA from either uninduced or TPA-induced BCBL-1 cells overnight at 42°C. The samples were then digested with a 1:100 dilution of the kit-provided RNase A-RNase T1 solution plus a 1:100 dilution of the kit-provided RNase T1 solution for 1 h at 37°C. Following precipitation of the digested RNA, the samples were separated on an 8% denaturing acrylamide gel. The gel was exposed to Kodak XAR-5 film for 3 days.
Primer extension was performed essentially as described by Zhong et al. (52). Briefly, 10 μg of RNA from TPA-induced BCBL-1 cells was hybridized with the primer 57 PE (5′ CTCTAGGATGCCCTTCATAATGTC). The samples were separated on an 8% denaturing acrylamide gel and were exposed to Kodak XAR 5 film for 3 days. A sequencing ladder created by sequencing a genomic clone with the primer 57 PE was loaded on the gel adjacent to the primer extension products for size comparison.
Immunofluorescence.
CV-1 cells were plated to 60% confluency on glass cover slips and were transfected with either pcDNA 3.1-V5-HisA or pcDNA 3.1-V5-HisA ORF 57 (Invitrogen). Forty-eight hours after transfection with Lipofectamine (Gibco BRL), the cells were washed according to the manufacturer's instructions and were then fixed for 30 min in fresh 4% paraformaldehyde in phosphate-buffered saline (PBS). After washing, the cells were permeabilized in 1× PBS–0.1% Triton X–0.1% sodium citrate for 10 min at 4°C. Subsequently, the cells were blocked in 1× PBS–1.0% Triton X–0.5% Tween–3.0% bovine serum albumin for 30 min at room temperature. The cells were then incubated with a 1:300 dilution of the mouse anti-V5 antibody (Invitrogen), were diluted into the blocking solution at room temperature for 1 h, and were washed three times with 1× PBS. Finally, the cells were incubated with tetramethyl rhodamine isothiocyanate (TRITC)-conjugated goat anti-mouse F(ab′)2 fragments at a dilution of 1:100 for 1 h at room temperature and then washed as described above.
Transfections.
CV-1 cells were plated at 105 cells per well in a six-well tissue culture dish the day prior to transfection. In all transfections, pcDNA 3 was used as a filler plasmid to normalize total DNA in each transfection. For the luciferase assays, 4 to 5 μg of total of DNA was diluted into 100 μl of serum-free DME-H21 medium, and 10 μl of Superfect transfection reagent (Qiagen) was added. For amounts of effector plasmids used, see the figure legends. After a 10-min incubation during which the cells were washed once in 1× PBS, 700 μl of complete medium was mixed with the DNA-Superfect media mixture and was added to each well.
After 3 h of incubation, the cells were washed once with 1× PBS, and 2 ml of fresh complete medium was added. Following a 48-h incubation, the cells were washed twice with 1× PBS and were scraped into 150 μl of 1× reporter lysis buffer (Promega). The cell extracts were vortexed for 30 s, and the debris was removed by centrifugation for 15 s at 16,000 × g in a microcentrifuge. The supernatant was transferred to a new tube, and 20-μl aliquots were analyzed by luciferase assays according to the manufacturer's instructions (Promega). All graphs of luciferase activity represent the results of experiments performed at least three times, in duplicate, unless otherwise noted. Because all of the reporters we attempted to use as internal standards were affected by ORF 57 expression, we chose to perform multiple replicates of each experiment and to indicate standard deviations of the replicates as error bars. In some experiments, the standard deviation is so small that the error bars are not visible.
Northern blotting.
For Northern blotting, CV-1 cells were plated at 5 × 105 cells per 100-mm-diameter dish the day prior to transfection. For the ORF 50 Northern blotting, cells were transfected with 10 μg of total plasmid DNA consisting of 2.5 μg of pcDNA ORF 50, and 0, 0.75, 2.5, or 7.5 μg of pcDNA ORF 57. For the ORF 59/58, nut-1, GCR, and K5 Northern analyses, cells were transfected as above with 10 μg of total plasmid DNA consisting of 5 μg of the expression vector for the target gene and either 0 or 1 μg of pcDNA 3 ORF 57. Superfect transfection reagent (Qiagen) was used according to the manufacturer's instructions. Forty-eight hours after transfection, total RNA was harvested by using RNAzol B (Tel-Test, Inc., Friendswood, Tex.) as recommended by the manufacturer. Total RNA (10 μg) was separated on a 1% agarose–17% formaldehyde gel and was transferred to Hybond-N membrane for 12 h in 10× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The blots were UV cross-linked and hybridized with either the ORF 50 antisense riboprobe (described in Lukac et al. [23]), as in Kirshner et al. (19), or with ds DNA probes for ORF 59/58, GCR, K5, and nut-1 (as in Lagunoff et al. [20]). The blots were exposed to Kodak XAR5 film for 2 days.
Western blotting.
For Western blotting, cells were transfected as described above. Forty-eight hours after transfection, the cells were scraped into 10-S buffer (23), were incubated on ice for 10 min, then were centrifuged for 5 min at 16,000 × g in a microcentrifuge, whereupon the supernatant was removed. The protein concentration of the extracts was determined by Bradford assay, and equal amounts of protein (10 μg) were separated either by sodium dodecyl sulfate–8% polyacrylamide gel electrophoresis (ORF 50) or by sodium dodecyl sulfate–12.5% polyacrylamide gel electrophoresis (ORF 59/58). Following electrophoresis, proteins were transferred and detected as described by Lukac et al. (23). The primary antibody to ORF 59/58 (24) was used at a 1:500 dilution.
RESULTS
Expression of KSHV ORF 57.
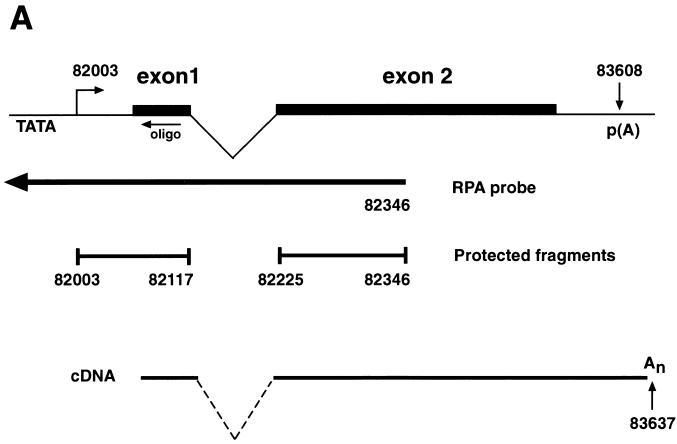
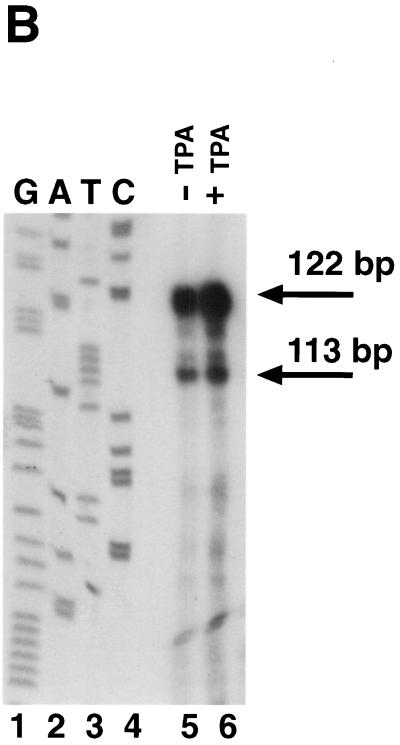
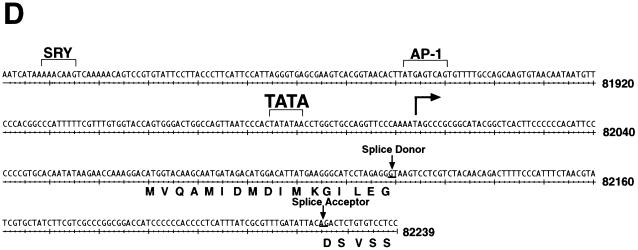
We have previously shown that KSHV ORF 57 is a lytic gene expressed between 2 and 4 h after TPA induction of BCBL-1 cells, immediately following the appearance of ORF 50 transcripts but prior to most DE mRNAs (23). To determine the fine structure of ORF 57 mRNA, we first screened an oligo(dT)-primed cDNA library from TPA-induced BCBL-1 cells with a probe for ORF 57. Six clones were obtained, none of which were full length. The longest (schematically depicted in Fig. 1A) contained a 3′ end at nt 83637, 29 nt downstream of a canonical poly(A) signal, and a 5′ end at nt 82100. An intron of 108 bp was located at the 5′ end of this clone, with consensus splice donor and acceptor sites (Fig. 1A). To locate the transcriptional start site, we performed RNase protection and primer extension analyses. The antisense riboprobe used for RNase protection extended from the beginning of the second exon to 400 bp upstream (Fig. 1A). After hybridization to RNA from either TPA-induced or uninduced BCBL-1 cells, the samples were treated with RNases A and T1 for 1 h at 37°C and were separated on a 6% denaturing gel. Two protected fragments were generated (Fig. 1B). These protected fragments are more abundant in the induced lane but are also present in the uninduced lane due to the 1 to 4% of BCBL-1 cells that undergo spontaneous reactivation. These results indicate a transcription start site at nt 82003. To confirm these results, we performed primer extension with an end-labeled oligonucleotide that hybridized to the start of exon 1 (Fig. 1A). After annealing of this primer to RNA from TPA-induced BCBL-1 cells and extension with reverse transcriptase, a single product of 116 nt was detected (Fig. 1C). By comparing the size of this product to a sequencing ladder generated by using the same primer, we were able to confirm the start site at nt 82003. Analysis of the region 5′ to the start site revealed a TATA box 24 bp upstream of the transcriptional start site as well as several consensus transcription factor binding sites (Fig. 1D). Scanning the region downstream of the start site revealed the existence of four methionine (ATG) codons in frame with exon 1. As we do not yet know which of the potential start codons is/are utilized, in constructing an ORF 57 expression vector, a genomic fragment containing all potential start codons was employed. Although the genomic sequence originally predicted a size of 275 amino acids (aa) for ORF 57, our cDNA cloning and transcript mapping reveals that ORF 57 actually has a larger coding region (potentially 456 aa) in vivo.
FIG. 1.
(A) Diagram of KSHV ORF 57 mRNA structure. ORF 57 is spliced, and the transcriptional start sites and poly(A) location are shown. The position of the antisense riboprobe used for RNase protection and sizes of the resulting protected fragments are depicted, as well as the oligonucleotide used for primer extension. The longest cDNA clone is also shown below. (B) RNase protection to identify the transcriptional start site of ORF 57 mRNA. Autoradiogram of a 6% denaturing acrylamide gel containing the protected species. Total RNA from either untreated (lane 5) or TPA-treated (lane 6) BCBL-1 cells was hybridized to the riboprobe shown in panel A and was digested with RNase, denatured, and electrophoresed. A sequencing ladder was run on the same gel and used for sizing (lanes 1 to 4). (C) Primer extension to confirm the start site was performed with labeled oligonucleotide (diagrammed in panel A) annealed to TPA-treated BCBL-1 RNA (lane 5). A sequencing ladder generated by the same primer on genomic DNA was run on the gel (lanes 1 to 4) for size comparison. The arrow indicates the extension product and its corresponding nucleotide position, also indicated by the asterisk on the left. Nucleotide positions are according to the sequence of Russo et al. (37). (D) Nucleotide sequence surrounding the transcription start site (arrow). Translated sequence from the first AUG in mRNA is shown below the DNA sequence.
Subcellular localization of ORF 57 protein.
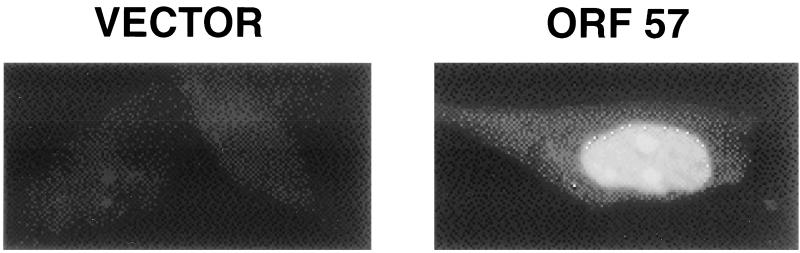
Analysis of the predicted aa sequence of ORF 57 reveals several arginine-rich potential nuclear localization signals. To determine the cellular localization of ORF 57, we first constructed an expression vector for ORF 57 with an epitope tag (pcDNA 3.1-V5; Invitrogen) fused to the C terminus. After verification that this expression vector generated a functional ORF 57 product (as defined by its ability to synergize with ORF 50) (see below), we examined CV-1 cells transiently transfected with this construct by immunofluorescence with anti-V5 monoclonal antibodies (MAbs). The left panel of Fig. 2 shows the control-vector-transfected cells 48 h posttransfection; no staining is observed. When transfected with the ORF 57 expression vector, intense staining is observed only in the nuclei of CV-1 cells (Fig. 2, right panel).
FIG. 2.
The C-terminally tagged version of ORF 57 is localized to the nucleus. The panels show results of immunofluorescence staining of CV-1 cells 48 h after transfection with either empty pcDNA 3.1-V5 vector (left panel) or pcDNA 3.1-V5 ORF 57 (right panel). Reactivity to anti-V5 antibody (Invitrogen) was detected with a TRITC-conjugated rabbit anti-mouse antibody.
Effect of ORF 57 on KSHV-promoter-driven reporter genes.
To begin the analysis of ORF 57 function, we examined the ability of the protein to regulate expression of luciferase reporter genes driven by a variety of KSHV DE and latent promoters. The tested promoters included those from the following KSHV genes: nut-1 (also called PAN), tk (ORF 21), DBP (ORF 6), DNA polymerase (Pol) (ORF 9), and Kaposin. CV-1 cells were cotransfected with pcDNA 3.1 ORF 57 and the reporter constructs, and luciferase activity was measured. Each target was examined over a 10-fold range of concentrations of the ORF 57 expression vector; most were unaffected, and in no case did we observe more than a twofold effect over the basal level of expression in the absence of ORF 57. Figure 3 shows representative results for four promoters: DNA Pol, Kaposin, nut-1, and tk (white bars). Several heterologous promoters were also examined in this fashion (e.g., cytomegalovirus (CMV) IE and SV40 E) with similar results (J. R. Kirshner, unpublished data; see also Fig. 7). These data indicate that ORF 57 is not a broad-spectrum transcriptional activator such as adenovirus E1A or HSV ICP0. Because HSV-1 ICP27 regulation of reporter expression is influenced by the presence or absence of introns in the body of the transcript, we constructed isogenic, intron-containing versions of these luciferase reporters (driven by the promoters for DNA Pol, Kaposin, nut-1, and tk) and tested them alongside their intronless counterparts. We used the SV40 T antigen intron, which has been shown to be a target of regulation by ICP27 (40). Figure 3 (black bars) shows the results obtained for each of the 4 constructs, again tested over a 10-fold range of concentration of cotransfected ORF 57 plasmid. We saw no consistent effect of the presence of intronic sequences for each KSHV reporter tested. Some constructs (e.g., that driven by the DNA Pol promoter) were completely unaffected, while others showed small (twofold) effects at selected concentrations of ORF 57 (see Fig. 3 for representative examples). Interestingly, this finding was not limited to the SV40 T intron: an SV40 early-promoter-driven luciferase construct bearing a synthetic intron derived from β-globin likewise failed to display regulation by ORF 57, as did its intronless counterpart (data not shown). Moreover, an authentic KSHV gene (ORF 50) bearing its native intron was not downregulated by ORF 57 coexpression (see below and Fig. 7). These results argue against the existence of an ORF-57-encoded activity that globally impairs splicing or actively represses expression from intron-containing genes. However, they do not exclude the possibility that some viral genes might display intron-dependent responses to ORF 57, and examples of this phenomenon are presented below (see Fig. 8).
FIG. 7.
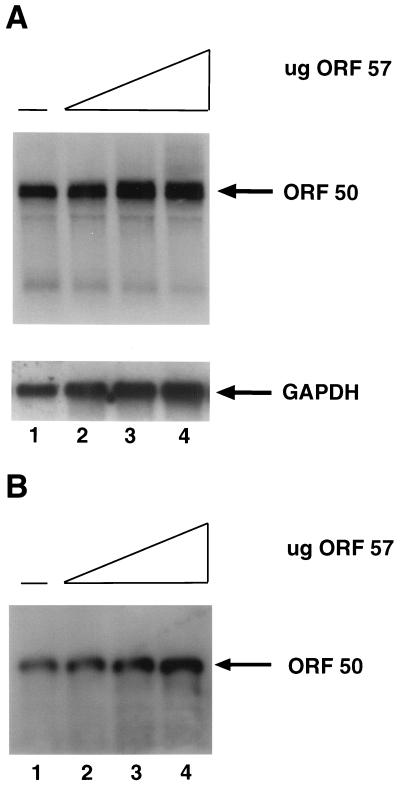
ORF 50 mRNA and protein levels are not significantly increased by ORF 57 expression. (A) Top, autoradiogram of a Northern blot containing total RNA from CV-1 cells transfected with a fixed amount of pcDNA 3 ORF 50 and the following amounts of pcDNA 3.1 ORF 57: 0 μg (lane 1), 0.75 μg (lane 2), 2.5 μg (lane 3), and 7.5 μg (lane 4). An antisense riboprobe for ORF 50 was hybridized to the blot. Bottom, the blot was reprobed with a GAPDH-specific probe as a loading control. (B) Western blot analysis of protein extracts from CV-1 cells transfected with a fixed amount of pcDNA 3.1 ORF 50 and the following amounts of pcDNA 3.1 ORF 57: 0 μg (lane 1), 0.5 μg (lane 2), 2.5 μg (lane 3), and 5 μg (lane 4). The ORF 50 protein was detected with a polyclonal antibody and then with a secondary rabbit anti-mouse antibody conjugated to horseradish peroxidase and ECL (Amersham). Equal amount of extracts as determined by Bradford assay from each transfection were loaded onto the gel.
ORF 57 alone can regulate KSHV gene expression.
Thus far, in the context of artificial reporter constructs, ORF 57 expression was observed to have little effect on gene expression on its own. However, such chimeric reporters bear little resemblance to the natural targets of ORF 57 regulation in vivo. We therefore turned to the examination of transcription units containing authentic viral sequences to more closely mimic the targets found in a viral infection. In scanning for KSHV genes that might be subject to ORF 57 regulation, we focused on the subclass of DE genes involved in DNA replication, as these genes have been implicated in such regulation in both HSV (49) and EBV (42). ORF 59 is a KSHV homolog of EBV BMRF1, a DNA Pol accessory factor whose expression has been shown to be enhanced by EBV M protein expression (42). This gene was a particularly attractive target for our work because of the availability of MAbs to the ORF 59 protein (24). ORF 59 is expressed as the 5′ gene in an unspliced bicistronic transcript with ORF 58, and polyadenylation occurs 3′ to ORF 58 (10; R. Renne and D. Ganem, unpublished data). Accordingly, we cloned the genomic region spanning ORFs 58 and 59, including the relevant KSHV poly(A) signal, downstream of the CMV IE promoter, which we have shown is not substantially regulated by ORF 57 (see Fig. 7). This construct was transfected into CV-1 cells in the presence or absence of pcDNA 3.1 ORF 57, and the levels of RNA and protein were measured by Northern blotting and immunoblotting, respectively. Figure 4A shows that in total RNA from whole-cell extracts of such transfectants, ORF 59 mRNA levels in the presence of ORF 57 were nearly 20-fold higher than those produced in its absence. This stimulation was also reflected in the levels of ORF 59 protein (Fig. 4B). Since the CMV promoter is not significantly upregulated by ORF 57, the bulk of this upregulation is posttranscriptional. Similar induction was observed when the KSHV poly(A) signal was replaced by the poly(A) signal from a heterologous bovine growth hormone gene.
FIG. 4.
ORF 57 increases ORF 59/58 mRNA and protein levels. (A) Top, autoradiogram of a Northern blot containing total RNA from CV-1 cells transfected with a fixed amount of pcDNA 3.1 ORF 59/58 in the presence (+) or absence (−) of 0.1 μg of pcDNA 3.1 ORF 57. A ds DNA probe specific for ORF 59 was used; bottom, the blot was reprobed with a glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-specific probe as a loading control. (B) Western blot analysis of protein extracts from CV-1 cells transfected with a fixed amount of pcDNA 3.1 ORF 59/58 in the presence (+) or absence (−) of 1 μg of pcDNA 3.1 ORF 57. The ORF 59 protein was detected with a KSHV MAb, and then with a secondary goat anti-mouse conjugated to horseradish peroxidase and ECL (Amersham). Equal amounts (10 μg) of extracts as determined by Bradford assay from each transfection were loaded onto the gel.
Similar screens of other early KSHV genes revealed that this effect is not limited to ORF 59 (Fig. 5). A particularly interesting case is that of nut-1 RNA (Fig. 5A). In this experiment, full-length nut-1 sequences bearing their native poly(A) signals were cloned downstream of the CMV promoter; this allows examination of ORF 57 effects on the nut-1 transcript independent of any possible effects on the nut-1 promoter. A nearly 20-fold enhancement in the accumulation of nut-1 RNA is observed in total RNA from whole-cell lysates (Fig. 5A). However, other KSHV-containing transcripts (e.g., GCR and K5) driven by the CMV promoter [and employing the bovine growth hormone poly(A) signal] were unaffected (Fig. 5B), indicating that posttranscriptional upregulation by ORF 57 is transcript specific and confirming that ORF 57 does not upregulate the CMV promoter.
FIG. 5.
Upregulation of nut-1 RNA accumulation by ORF 57 expression. The indicated constructs (bottom) were transfected into CV-1 cells in the presence (+) or absence (−) of 1 μg of pcDNA 3.1 ORF 57. Forty-eight hours later, total RNA was prepared from whole-cell lysates, electrophoresed through 1% agarose-formaldehyde gels, transferred to Hybond-N+ membranes, and hybridized to probes specific for nut-1 (A) or ORF 74/GCR (B, left panel) or ORF K5 (B, right panel). Below each panel is shown rRNA bands in ethidium bromide-stained lanes as a loading control.
Augmentation of ORF 50 activity by ORF 57 protein.
In an authentic KSHV infection, ORF 57 is not expressed in isolation; rather, it is produced with other powerful regulatory proteins, of which the best characterized is the IE transactivator encoded by ORF 50. Accordingly, we examined the effects of ORF 57 expression on the (intronless) nut-1-driven luciferase reporter in the presence or absence of the ORF 50 protein, which we have previously established can strongly upregulate this promoter (24). As shown in Fig. 6A, while ORF 57 alone had little effect on nut-1-promoted luciferase expression, ORF 50 alone, as expected, activated reporter expression 80-fold. We then selected a level of ORF 50 expression vector that produced maximal activation (1 μg/transfection), and to this we added increasing amounts of ORF 57 expression vector and assayed luciferase expression. The addition of even low levels of ORF 57 to ORF 50 resulted in a striking (40- to 50-fold) further upregulation of expression over that generated by ORF 50 alone.
To further investigate the nature of this synergistic effect, we used a dominant negative mutant of ORF 50 termed 50 ΔSTAD. This mutant, whose transcriptional activation domain is deleted, retains dimerization capacity and has been shown to specifically inhibit the ability of wild-type ORF 50 to transactivate reporter genes and to induce lytic replication of KSHV in BCBL-1 cells (23). Increasing amounts of an ORF 50 ΔSTAD expression vector were added to our cotransfection assay containing ORF 50, ORF 57, and the nut-1 reporter. Figure 6B shows that the addition of 50 ΔSTAD inhibits the stimulation of gene expression by the ORF 50-ORF 57 combination in a dose-dependent fashion. Similarly, we found that synergistic upregulation of the nut-1 promoter by the ORF 50-ORF 57 combination is abolished by mutations in the promoter that ablate ORF 50 responsiveness. In studies to be reported elsewhere, mutagenesis of the nut-1 promoter identified a small region required for ORF 50 responsiveness; mutations in this element do not effect basal transcription, but completely abrogate upregulation by ORF 50 (J. Chang, D. Lukac, and D. Ganem, unpublished data). Figure 6C shows that such mutations also abrogate synergy between ORF 50 and ORF 57 proteins. These results indicate that synergy between ORF 50 and ORF 57 on this target requires the specific transactivation activity of ORF 50.
Two independent lines of evidence support the idea that ORF 57 is not simply increasing the amount of the ORF 50 transactivator and thereby further activating the nut-1 promoter. First, the observed effect of ORF 57 operates at saturating levels of the ORF 50 protein; under these conditions, increasing concentrations of ORF 50 have no effect on transactivation (Fig. 6A). Second, direct examination of ORF 50 mRNA and protein levels shows no significant augmentation by ORF 57 expression (Fig. 7). CV-1 cells were cotransfected with pcDNA 3 ORF 50 and increasing amounts of pcDNA 3.1 ORF 57; 48 h after transfection, total RNA was isolated. Equal amounts of this RNA was separated on a 1.2% agarose–17% formaldehyde gel, was transferred to a Hybond-N membrane, and was hybridized to an antisense riboprobe for ORF 50. Figure 5A shows that with increasing amounts of ORF 57, the ORF 50 mRNA levels do not significantly increase. PhosphorImager (Molecular Dynamics) quantitation of the bands in the Northern blot reveals that there is, at most, a threefold increase in the amount of mRNA at the highest concentration of ORF 57, but this ORF 57 level is well above the concentration needed for the superinduction. Extracts of the same transfected cells were also examined by immunoblotting with a rabbit antiserum to ORF 50 (Fig. 7B); no increase was detected in ORF 50 protein levels at concentrations of ORF 57 at which marked synergy was observed by luciferase assay. (At high concentrations of ORF 57, no more than a threefold increase in ORF 50 protein levels was observed.) The possibility of ORF 57 toxicity to host protein synthesis can be excluded for two reasons: (i) basal luciferase levels are not decreased by ORF 57 expression in reporters whose transcripts are not upregulated (Fig. 3), and (ii) levels of ORF 59 protein rise in concert with RNA levels in response to ORF 57 (Fig. 4).
Reexamination of intron effects in the presence of ORF 50 and ORF 57.
In the HSV system, the repressive effect of ICP27 on intron-containing reporters was best seen when expression of the reporters was activated by coexpression of the ICP4 and ICP0 IE transactivators (40). This fact, together with the fact that in natural infection ORF 57 expression is always accompanied by ORF 50 production, led us to reexamine the effects of introns in this context. We tested three pairs of isogenic luciferase reporters containing or lacking the SV40 T intron, driven by the promoters of the nut-1, Kaposin, and tk genes, respectively. Each pair of reporters was examined for luciferase expression in the presence of a fixed saturating dose of ORF 50 and increasing amounts of the ORF 57 expression vector (Fig. 8). Several interesting facts emerged from this analysis. First, the synergistic ORF 50-ORF 57 interaction is promoter specific. While all three promoters are ORF 50 responsive (23), the tk promoter is not synergistically up regulated by the coexpression of ORF 57, while the Kaposin promoter is superinduced by at most 25-fold (versus 80- to 100-fold for the nut-1 promoter) (compare Fig. 8A and B). Second, significant effects of introns were observed in some but not all of these contexts. Specifically, the presence of an intron in the nut-1 construct still allowed upregulation by ORF 57, but this synergy was reduced to 10-fold (versus nearly 100-fold for the intronless reporter [Fig. 8A]). By contrast, little effect of the intron was observed on the Kaposin reporter (Fig. 8B). Notably, we did not observe actual repression of gene expression, i.e., a decrease in luciferase activity below the level observed in the absence of ORF 57 expression. Since the basal level of luciferase was typically quite high in these assays (due to ORF 50 activation), this represents a sensitive assay for detecting repression.
DISCUSSION
These results demonstrate that ORF 57 is a nuclear protein expressed from a spliced lytic mRNA. In transient reporter gene assays, ORF 57 expression has little effect on a wide variety of promoters, suggesting that it is not a broad-spectrum transcriptional activator. Despite extensive searches, we also observed little consistent effect of the presence or absence of introns in such chimeric constructs. Nonetheless, the accumulation of some viral RNAs (e.g., those for ORF 59 and nut-1) can be strongly augmented in the presence of ORF 57, in a manner that suggests posttranscriptional regulation. Moreover, expression from some promoters (especially nut-1) that are upregulated by ORF 50 can be synergistically enhanced by coexpression with ORF 57. This synergy results from a posttranslational enhancement of the transcriptional activity of ORF 50. These data reveal ORF 57 to be a complex pleiotropic effector that can act on several levels to augment viral gene expression.
Although homologous to ICP27, the KSHV ORF 57 protein differs in some important respects from its HSV-1 counterpart. In HSV, ICP27 expression leads to a strong inhibition of RNA splicing, an effect that is thought to be one important contributor to the shutoff of host macromolecular synthesis. (Accordingly, most DE and L mRNAs of HSV are unspliced). Coexpression of ORF 57 with ORF 50 did not impair the accumulation of ORF 50 mRNA or full-length protein (Fig. 7). Since ORF 50 bears an intron in its coding region, this result clearly indicates that ORF 57 does not impair the splicing of this intron (and notably, functional ORF 57 is itself the product of RNA splicing). Moreover, even though research on KSHV gene expression is at an early stage, numerous examples of spliced viral mRNAs have already been documented, including the DE genes for K14/74 (19, 48) and KbZIP (16, 22), and the L genes encoding K8.1 (11, 33) and ORF 29 (34). Recent studies (15) also show that multiply spliced transcripts for K15/LAMP, while expressed at low levels in latency, are strongly up regulated during lytic growth. These findings are inconsistent with a substantial block to RNA splicing or to expression of intron-containing genes throughout the KSHV lytic cycle.
However, although the above considerations argue strongly against a general downregulation of intron-bearing sequences, they do not exclude the possibility that in selected genes activated by ORF 57 such activation might operate preferentially or exclusively on the unspliced version of the RNA. That is, in such genes, ORF 57 might be relatively indifferent to intron-containing sequences but actively up regulate those lacking introns. In other herpesviruses, evidence is growing that the homologs of ORF 57 can shuttle from nucleus to cytoplasm and can mediate the preferential cytoplasmic accumulation of unspliced mRNAs (26, 39, 42, 44). Nothing in the present work excludes this as one potential mode of action of KSHV ORF 57. However, our data indicate that this cannot be the sole mode of action of ORF 57, since (i) its augmentation of ORF 50 activity operates at a posttranslational level (Fig. 7), and (ii) nut-1 RNA, which is localized entirely to the nucleus, is also strikingly up regulated by ORF 57 protein (Fig. 5).
The effects of the intron in the nut-1-promoted reporter are interesting in this regard. Despite the fact that the ORF 50-ORF 57 synergy is expressed primarily by effects at the nut-1 promoter, expression of the intronless version of the gene is upregulated 10-fold more efficiently than its intron-bearing derivative (Fig. 8A). This is consistent with the notion that some intronless mRNAs might be preferentially stabilized or exported for translation in the cytosol. Interestingly, the fact that identical mRNAs do not behave similarly when directed by the Kaposin promoter (Fig. 8B) suggests that events at the promoter can also influence processing and transport of the RNAs. While incompletely understood, such effects have been observed in other systems—for example, 3′ processing of some cellular RNAs is strongly influenced by the promoter used to drive expression of the RNA (29). Clearly, transcriptional and posttranscriptional events are often coupled (4), and we should not be surprised that regulators acting principally by posttranscriptional mechanisms can sometimes be influenced by earlier events in the biogenesis of the target RNA.
We do not yet understand the basis for the remarkable synergy between the product(s) of ORF 50 and ORF 57, nor why they are so promoter selective. In part, this reflects our incomplete understanding of the mechanism of activation by ORF 50. In other studies, we have genetically identified ORF 50 response elements in several different KSHV promoters and have found that such elements display considerable sequence heterogeneity. We note with interest that the two promoters (nut-1 and Kaposin) that are subject to ORF 50-ORF 57 synergy share ORF 50 response elements that are identical in sequence and in their position relative to the start site. By contrast, the ORF 50 response element in the TK promoter is completely divergent from that in nut-1 and Kaposin (J. Chang, D. Lukac, and D. Ganem, unpublished data). This heterogeneity might be one factor contributing to the promoter selectivity of the effect. For example, if ORF 57 expression were to modify the DNA binding activity of ORF 50, such a change might lead to enhanced binding to some but not all recognition sites. Clearly, fuller understanding of this process must await a clearer definition of the biochemistry of ORF 50 DNA binding and its interactions with the basal transcription machinery.
Although synergy with IE regulators has not previously been observed in the gammaherpesvirus homologs of ORF 57, HSV ICP27 has been demonstrated to complex with ICP4, to alter its subcellular location, and to enhance its regulatory actions (31, 54). In the case of the ORF 50-ORF 57 interaction, both proteins are localized to the nucleus independently, and we have no evidence that nuclear subdomains to which ORF 50 is addressed are altered by the presence of ORF 57. We have extensively searched for evidence of a protein-protein interaction between the products of ORF 50 and ORF 57 by using both coimmunoprecipitation assays (in vivo and in vitro) and a mammalian two-hybrid system. To date, however, these studies have been negative.
The effects of ORF 57 on nut-1 expression described herein are dramatic and are likely to play an important role in vivo. Nut-1 RNA is not expressed in latency, as judged by sensitive in situ hybridization analyses (43), but is expressed with enormous efficiency during lytic growth, generating over 105 molecules of nut-1 RNA/cell. We suggest that this remarkable accumulation is due to at least two effects: (i) the ORF 50-ORF 57 synergy that upregulates the primary transcription of the locus and (ii) the ORF-57-mediated posttranscriptional upregulation of nuclear transcript levels. Current work is now focused upon definition of the cis-acting elements responsible for the posttranscriptional effects (both in nut-1 and ORF 59) with the goal of elucidating the molecular mechanism(s) involved in these processes.
ACKNOWLEDGMENTS
We thank Rolf Renne for cDNA cloning and Laurent Coscoy and Andy Polson for expression vectors.
We thank the Howard Hughes Medical Institute for support. D.M.L. is a postdoctoral fellow of the Irvington Institute for Immunological Research.
ADDENDUM
While this manuscript was under review, a paper by A. K. Gupta et al. (16a) was published that supports the role of ORF 57 as a posttranscriptional activator. The authors used nuclear run-on transcription assays to show that the ORF 57 has no effect on transcript initiation rates from a CMV promoter regulating expression of the CAT gene. This data accords well with our findings.
Footnotes
This paper is dedicated to the memory of Rob Sadler, our colleague and friend.
REFERENCES
- 1.Ambroziak J, Blackbourn D, Herndier B, Glogan R, Gullet J, McDonald A, Lennette E, Levy J. Herpesvirus-like sequences in HIV-infected and uninfected Kaposi's sarcoma patients. Science. 1995;268:582–583. doi: 10.1126/science.7725108. [DOI] [PubMed] [Google Scholar]
- 2.Arvanitakis L, Geras-Raaka E, Varma A, Gershengorn M C, Cesarman E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature. 1997;385:347–350. doi: 10.1038/385347a0. [DOI] [PubMed] [Google Scholar]
- 3.Bais C, Santomasso B, Coso O, Arvanitakis L, Raaka E G, Gutkind J S, Asch A S, Cesarman E, Gershengorn M C, Mesri E A, Gerhengorn M C. G-protein-coupled receptor of Kaposi's sarcoma-associated herpesvirus is a viral oncogene and angiogenesis activator. Nature. 1998;391:86–89. doi: 10.1038/34193. . (Erratum, 392:210, 1998.) [DOI] [PubMed] [Google Scholar]
- 4.Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 5.Beral V, Peterman T A, Berkelman R L, Jaffe H W. Kaposi's sarcoma among persons with AIDS: a sexually transmitted infection? Lancet. 1990;335:123–128. doi: 10.1016/0140-6736(90)90001-l. [DOI] [PubMed] [Google Scholar]
- 6.Blasig C, Zietz C, Haar B, Neipel F, Esser S, Brockmeyer N H, Tschacler E, Colombini S, Ensoli B, Sturzl M. Monocytes in Kaposi's sarcoma lesions are productively infected by human herpesvirus 8. J Virol. 1997;71:7963–7968. doi: 10.1128/jvi.71.10.7963-7968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boshoff C, Endo Y, Collins P D, Takeuchi Y, Reeves J D, Schweickart V L, Siani M A, Sasaki T, Williams T J, Gray P W, Moore P S, Chang Y, Weiss R A. Angiogenic and HIV-inhibitory functions of KSHV-encoded chemokines. Science. 1997;278:290–294. doi: 10.1126/science.278.5336.290. [DOI] [PubMed] [Google Scholar]
- 8.Boshoff C, Schulz T F, Kennedy M M, Graham A K, Fisher C, Thomas A, McGee J O, Weiss R A, O'Leary J J. Kaposi's sarcoma-associated herpesvirus infects endothelial and spindle cells. Nat Med. 1995;1:1274–1278. doi: 10.1038/nm1295-1274. [DOI] [PubMed] [Google Scholar]
- 9.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 10.Chan S R, Bloomer C, Chandran B. Identification and characterization of human herpesvirus-8 lytic cycle-associated ORF 59 protein and the encoding cDNA by monoclonal antibody. Virology. 1998;240:118–126. doi: 10.1006/viro.1997.8911. [DOI] [PubMed] [Google Scholar]
- 11.Chandran B, Koelle D M, Corey L, Horvat R, Goldstein E. Reactivities of human sera with human herpesvirus-8-infected BCBL-1 cells and identification of HHV-8-specific proteins and glycoproteins and the encoding cDNAs. Virology. 1998;243:208–217. doi: 10.1006/viro.1998.9055. [DOI] [PubMed] [Google Scholar]
- 12.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 13.Chevallier-Greco A, Manet E, Chavrier P, Mosnier C, Daillie J, Sergeant A. Both Epstein-Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J. 1986;5:3243–3249. doi: 10.1002/j.1460-2075.1986.tb04635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ganem D. KSHV and Kaposi's sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 15.Glenn M, Rainbow L, Aurad F, Davison A, Schulz T F. Identification of a spliced gene from Kaposi's sarcoma-associated herpesvirus encoding a protein with similarities to latent membrane proteins 1 and 2A of Epstein-Barr virus. J Virol. 1999;73:6953–6963. doi: 10.1128/jvi.73.8.6953-6963.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gruffat H, Portes-Sentis S, Sergeant A, Manet E. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) encodes a homologue of the Epstein-Barr virus bZip protein EB1. J Gen Virol. 1999;80:557–561. doi: 10.1099/0022-1317-80-3-557. [DOI] [PubMed] [Google Scholar]
- 16a.Gupta A K, Ruvolo V, Patterson C, Swaminathan S. The human herpesvirus 8 homolog of Epstein-Barr virus SM protein (KS-SM) is a posttranscriptional activator of gene expression. J Virol. 2000;74:1038–1044. doi: 10.1128/jvi.74.2.1038-1044.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardwicke M A, Sandri-Goldin R M. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirshner J R, Staskus K, Haase A, Lagunoff M, Ganem D. Expression of the open reading frame 74 (G-protein-coupled receptor) gene of Kaposi's sarcoma (KS)-associated herpesvirus: implications for KS pathogenesis. J Virol. 1999;73:6006–6014. doi: 10.1128/jvi.73.7.6006-6014.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagunoff M, Ganem D. The structure and coding organization of the genomic termini of Kaposi's sarcoma-associated herpesvirus. Virology. 1997;236:147–154. doi: 10.1006/viro.1997.8713. [DOI] [PubMed] [Google Scholar]
- 21.Lieberman P M, O'Hare P, Hayward G S, Hayward S D. Promiscuous trans-activation of gene expression by an Epstein-Barr virus-encoded early nuclear protein. J Virol. 1986;60:140–148. doi: 10.1128/jvi.60.1.140-148.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin S F, Robinson D R, Miller G, Kung H J. Kaposi's sarcoma-associated herpesvirus encodes a bZIP protein with homology to BZLF1 of Epstein-Barr virus. J Virol. 1999;73:1909–1917. doi: 10.1128/jvi.73.3.1909-1917.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukac D M, Kirshner J R, Ganem D. Transcriptional activation by the open reading frame 50 protein of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J Virol. 1999;73:9348–9361. doi: 10.1128/jvi.73.11.9348-9361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lukac D M, Renne R, Kirshner J R, Ganem D. Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology. 1998;252:304–312. doi: 10.1006/viro.1998.9486. [DOI] [PubMed] [Google Scholar]
- 25.Martin D F, Kuppermann B D, Wolitz R A, Palestine A G, Li H, Robinson C A. Oral ganciclovir for patients with cytomegalovirus retinitis treated with a ganciclovir implant. Roche Ganciclovir Study Group. N Engl J Med. 1999;340:1063–1070. doi: 10.1056/NEJM199904083401402. [DOI] [PubMed] [Google Scholar]
- 26.Mears W E, Rice S A. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology. 1998;242:128–137. doi: 10.1006/viro.1997.9006. [DOI] [PubMed] [Google Scholar]
- 27.Mesri E A, Cesarman E, Arvanitakis L, Rafii S, Moore M A, Posnett D N, Knowles D M, Asch A S. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J Exp Med. 1996;183:2385–2390. doi: 10.1084/jem.183.5.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller G, Heston L, Grogan E, Gradoville L, Rigsby M, Sun R, Shedd D, Kushnaryov V M, Grossberg S, Chang Y. Selective switch between latency and lytic replication of Kaposi's sarcoma herpesvirus and Epstein-Barr virus in dually infected body cavity lymphoma cells. J Virol. 1997;71:314–324. doi: 10.1128/jvi.71.1.314-324.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Neuman de Vegvar H E, Lund E, Dahlberg J E. 3′ End formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell. 1986;47:259–266. doi: 10.1016/0092-8674(86)90448-4. [DOI] [PubMed] [Google Scholar]
- 30.Nicholas J, Gompels U A, Craxton M A, Honess R W. Conservation of sequence and function between the product of the 52-kilodalton immediate-early gene of herpesvirus saimiri and the BMLF1-encoded transcriptional effector (EB2) of Epstein-Barr virus. J Virol. 1988;62:3250–3257. doi: 10.1128/jvi.62.9.3250-3257.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panagiotidis C A, Lium E K, Silverstein S J. Physical and functional interactions between herpes simplex virus immediate-early proteins ICP4 and ICP27. J Virol. 1997;71:1547–1557. doi: 10.1128/jvi.71.2.1547-1557.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phelan A, Dunlop J, Clements J B. Herpes simplex virus type 1 protein IE63 affects the nuclear export of virus intron-containing transcripts. J Virol. 1996;70:5255–5265. doi: 10.1128/jvi.70.8.5255-5265.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raab M S, Albrecht J C, Birkmann A, Yaguboglu S, Lang D, Fleckenstein B, Neipel F. The immunogenic glycoprotein gp35-37 of human herpesvirus 8 is encoded by open reading frame K8.1. J Virol. 1998;72:6725–6731. doi: 10.1128/jvi.72.8.6725-6731.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renne R, Blackbourn D, Whitby D, Levy J, Ganem D. Limited transmission of Kaposi's sarcoma-associated herpesvirus in cultured cells. J Virol. 1998;72:5182–5188. doi: 10.1128/jvi.72.6.5182-5188.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Renne R, Lagunoff M, Zhong W, Ganem D. The size and conformation of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) DNA in infected cells and virions. J Virol. 1996;70:8151–8154. doi: 10.1128/jvi.70.11.8151-8154.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Kedes D, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 37.Russo J J, Bohenzky R A, Chien M C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruvolo V, Wang E, Boyle S, Swaminathan S. The Epstein-Barr virus nuclear protein SM is both a post-transcriptional inhibitor and activator of gene expression. Proc Natl Acad Sci USA. 1998;95:8852–8857. doi: 10.1073/pnas.95.15.8852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sandri-Goldin R M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sandri-Goldin R M, Mendoza G E. A herpesvirus regulatory protein appears to act post-transcriptionally by affecting mRNA processing. Genes Dev. 1992;6:848–863. doi: 10.1101/gad.6.5.848. [DOI] [PubMed] [Google Scholar]
- 41.Schulz T F. Kaposi's sarcoma-associated herpesvirus (human herpesvirus-8) J Gen Virol. 1998;79:1573–1591. doi: 10.1099/0022-1317-79-7-1573. [DOI] [PubMed] [Google Scholar]
- 42.Semmes O J, Chen L, Sarisky R T, Gao Z, Zhong L, Hayward S D. Mta has properties of an RNA export protein and increases cytoplasmic accumulation of Epstein-Barr virus replication gene mRNA. J Virol. 1998;72:9526–9534. doi: 10.1128/jvi.72.12.9526-9534.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith I L, Hardwicke M A, Sandri-Goldin R M. Evidence that the herpes simplex virus immediate early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- 44.Soliman T M, Sandri-Goldin R M, Silverstein S J. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and cytoplasm mediates the expression of late proteins. J Virol. 1997;71:9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Soulier J, Grollet L, Oksenhendler E, Cacoub P, Cazals-Hatem D, Babinet P, d'Agay M F, Clauvel J P, Raphael M, Degos L, et al. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman's disease. Blood. 1995;86:1276–1280. [PubMed] [Google Scholar]
- 46.Staskus K A, Zhong W, Gebhard K, Herndier B, Wang H, Renne R, Beneke J, Pudney J, Anderson D J, Ganem D, Haase A T. Kaposi's sarcoma-associated herpesvirus gene expression in endothelial (spindle) tumor cells. J Virol. 1997;71:715–719. doi: 10.1128/jvi.71.1.715-719.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun R, Lin S F, Gradoville L, Yuan Y, Zhu F, Miller G. A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus. Proc Natl Acad Sci USA. 1998;95:10866–10871. doi: 10.1073/pnas.95.18.10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Talbot S J, Weiss R A, Kellam P, Boshoff C. Transcriptional analysis of human herpesvirus-8 open reading frames 71, 72, 73, K14, and 74 in a primary effusion lymphoma cell line. Virology. 1999;257:84–94. doi: 10.1006/viro.1999.9672. [DOI] [PubMed] [Google Scholar]
- 49.Uprichard S L, Knipe D M. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J Virol. 1996;70:1969–1980. doi: 10.1128/jvi.70.3.1969-1980.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitby D, Howard M R, Tenant-Flowers M, Brink N S, Copas A, Boshoff C, Hatzioannou T, Suggett F E, Aldam D M, Denton A S, et al. Detection of Kaposi sarcoma associated herpesvirus in peripheral blood of HIV-infected individuals and progression to Kaposi's sarcoma. Lancet. 1995;346:799–802. doi: 10.1016/s0140-6736(95)91619-9. [DOI] [PubMed] [Google Scholar]
- 51.Whitehouse A, Cooper M, Meredith D M. The immediate-early gene product encoded by open reading frame 57 of herpesvirus saimiri modulates gene expression at a posttranscriptional level. J Virol. 1998;72:857–861. doi: 10.1128/jvi.72.1.857-861.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong W, Wang H, Herndier B, Ganem D. Restricted expression of Kaposi sarcoma-associated herpesvirus (human herpesvirus 8) genes in Kaposi sarcoma. Proc Natl Acad Sci USA. 1996;93:6641–6646. doi: 10.1073/pnas.93.13.6641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu F X, Cusano T, Yuan Y. Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus. J Virol. 1999;73:5556–5567. doi: 10.1128/jvi.73.7.5556-5567.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhu Z, Schaffer P A. Intracellular localization of the herpes simplex virus type 1 major transcriptional regulatory protein, ICP4, is affected by ICP27. J Virol. 1995;69:49–59. doi: 10.1128/jvi.69.1.49-59.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]