Abstract
Throughout the life span of a host, bifidobacteria have shown superior colonization and glycan abilities. Complex glycans, such as human milk oligosaccharides and plant glycans, that reach the colon are directly internalized by the transport system of bifidobacteria, cleaved into simple structures by extracellular glycosyl hydrolase, and transported to cells for fermentation. The glycan utilization of bifidobacteria introduces cross-feeding activities between bifidobacterial strains and other microbiota, which are influenced by host nutrition and regulate gut homeostasis. This review discusses bifidobacterial glycan utilization strategies, focusing on the cross-feeding involved in bifidobacteria and its potential health benefits. Furthermore, the impact of cross-feeding on the gut trophic niche of bifidobacteria and host health is also highlighted. This review provides novel insights into the interactions between microbe-microbe and host-microbe.
Subject terms: Applied microbiology, Microbial communities
Introduction
Bifidobacteria are abundant in the human gut and other warm-blooded animals, and are present in the rumen of ruminants, vagina1, intestines of honeybees2, oral cavity3, dairy products4, and breast milk5. Gut colonization by bifidobacteria occurs in the early stages of life and is closely associated with health and aging. Bifidobacteria are transmitted vertically from mother to offspring and are commonly found in the gut of healthy breastfed infants, accounting for 60–70% of all gut bacteria6. Aging alters the number and diversity of bifidobacteria in the human gut. In adulthood, the relative abundance of bifidobacteria decreases to approximately 10%, however remains stable, whereas that in aging individuals accounts for approximately 5% of the total gut bacteria6. In infancy, Bifidobacterium breve, B. longum, and B. bifidum are commonly found in feces7, while in adulthood, B. adolescentis, B. longum, and B. pseudocatenulatum are the dominant species8,9. The relative abundance of B. adolescentis decreases with age, whereas that of B. breve and B. longum is dominant in the gut of aging individuals10.
The rich and complex sources of carbon in the gut tract provide a trophic niche for gut microbiota. The differential utilization of glycans influences gut microbiota formation in gut niches, which may explain why certain species of bifidobacteria are more common in the gut tract of infants or adults11,12. Infant breast milk and adult diets contain a high abundance of glycans and oligosaccharides with complex structures that cannot be digested by the human body and hence pass through the large intestine as substrates for the gut microbiota. Bifidobacteria metabolize monosaccharides, disaccharides, and oligosaccharides; however, different bifidobacterial species prefer dietary glycans, including inulin-type fructan (ITF)13,14, resistant starch (RS)15, galactan16,17, xylan18,19, and arabinan20, while others utilize host-derived glycans, such as human milk oligosaccharides (HMOs)21,22 and mucins23.
The metabolism of complex glycans by human gut microbiota is mediated by carbohydrate-active enzymes (CAZymes). Bifidobacteria has been estimated to use approximately 14.64% of the CAZyme gene, including glycosyl hydrolases (GHs) and glycosyl transferases (GTs), for the transport, degradation, and regulation of glycans, which is only slightly less than that of Bacteroides (20%). Gut microbiota employ similar strategies to maintain and break down complex glycans24,25. Bifidobacteria encode a series of modular glucanase complexes that are anchored to the cell surface by transmembrane domains. These contain genes encoding GHs to extracellularly digest long oligosaccharides or glycans and substrate-binding proteins (SBPs) of the adenosine triphosphate (ATP)-binding cassette (ABC) transport system to capture oligosaccharides digested before entering cells26,27. No strain has been identified to utilize all glycans; however, specific types of glycans are utilized. Metabolic specificity exists between different bifidobacteria and other gut microbiota in the range, species, and utilization strategies of available glycans, which induces interactions between bifidobacteria and other flora, including cross-feeding, to promote the gut adaptability of bifidobacteria28.
The utilization of HMOs by bifidobacteria is an example of occupying a specific trophic niche in the gut. B. longum and B. bifidum establish a cross-feeding relationship with other microbes (such as Eubacterium) in early life by sharing their HMO metabolites degraded by extracellular glycosidases, including fucose and short-chain fatty acids (SCFAs)29,30. This cross-feeding strategy can augment the accessibility of glycans to gut microbiota and mediate interspecific interactions through metabolite reuse, directly affecting host gut homeostasis and health31,32.
Understanding glycan utilization and cross-feeding mechanisms will help to facilitate the discovery of new genes, enzymes, and metabolites that are potentially involved in bifidobacteria-microbe associations and host health. Therefore, this review describes the glycan utilization properties of bifidobacteria and examines the glycan-based cross-feeding activities and mechanisms involved in bifidobacteria, as well as their impact on host health.
Glycan preference and metabolic pathway of bifidobacteria
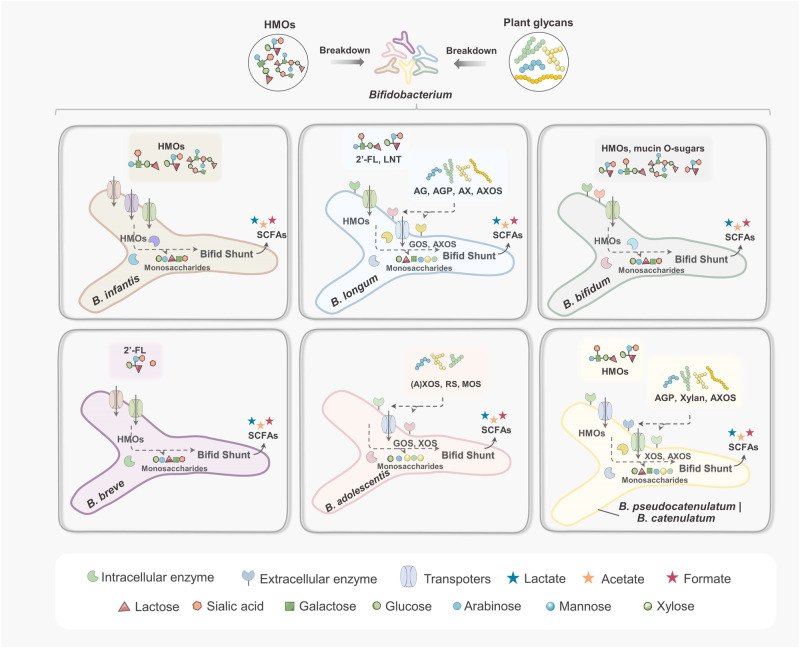
The colonization advantages of bifidobacteria depend on the availability, demand, and consumption rates of specific nutrient resources. The different carbohydrate metabolisms of bifidobacteria in different periods influence their selective adaptation to different gut environments. Due to differences in trophic sources, the intestinal environment of infants differs from that of adults, characterized by bifidobacteria including B. longum subsp. infantis (B. infantis), B. breve, and B. bifidum, which use HMOs as the primary carbon source; however, when introducing complex diets after weaning, the bifidobacteria B. longum subsp. longum (B. longum), B. adolescentis, B. catenulatum, and B. pseudocatenulatum, which have a greater capacity to degrade plant glycans, are more prevalent6,33,34. The breakdown of glycans by bifidobacteria depends on a significant number of GHs located extracellularly or within the cell wall. Bifidobacteria have evolved several homologous enzymes with overlapping, yet different, substrate specificities. Different species have different intestinal adaptations to cope with different glycan structures. Complex glycans are first degraded by extracellular GHs into monosaccharides or oligosaccharides, some of which are transported directly to the cytoplasm by transport proteins, while others are phosphorylated. These monosaccharides or oligosaccharides, and other simple carbohydrates, enter the bifid-shunt pathway (fructose-6-phosphate phosphoketolase central fermentative pathway) of bifidobacteria and are metabolized and used for ATP production35. The metabolic pathways of bifidobacteria are closely associated with the type and concentration of extracellular sugars in the gut. Bifidobacteria normally produce various metabolites, including acetate, ethanol, and formate, depending on the amount of sugar available; when extracellular sugars are abundant, acetate and lactate are generated36. Therefore, bifidobacteria adopt different strategies to colonize, and the capture, degradation, and metabolism of glycans by bifidobacteria can determine their trophic niches in the gut tract (Fig. 1 and Table 1).
Fig. 1. Major glycan utilization strategies of bifidobacteria.
Bifidobacteria complete the degradation by cleaving the glycan by extracellular enzymes or internalizing the glycan directly. B. infantis can internalize most of the HMOs through the transporter system, and B. bifidum degrade the HMOs (including fucosylated glycans) by abundant extracellular enzymes for further internalization and uptake. B. breve utilize limited HMOs (mainly 2ʼ-FL). B. longum excel in the utilization of phytoglycans containing arabinose structures. B. adolescentis is an excellent utilizer of resistant starch. Both B. pseudocatenulatum and B. catenulatum can consume some HMOs and phytoglycans.
Table 1.
Major glycan utilization strategies of bifidobacteria
| Bifidobacterial strains | Types of glycan | Strategies for glycan processing | Metabolites | References |
|---|---|---|---|---|
| B. infantis | ||||
| ATCC 15697 | LNT and LNnT | Hexosaminidase hydrolyzes β 1-3 bonds in LNT and LNnT to release GlcNAc, which is then deacetylated by GlcNAc-6-P deacetylase (nagA) and deaminated by glucosamine-6-P isomerase (nagB). | GlcNAc, acetate, ethanol, formate, and lactate | 149 |
| Bi-26 | 2ʼ-FL | 2ʼ-FL is transported by a special ABC transporter, and the genes encoding fucose peroxidase and ATP transporter are up-regulated during fermentation. | Fucose, acetate, lactate, 1,2-PD, and formate | 150,151 |
| EVC001 | Human milk glycoproteins | Endo-β-N-acetylglucosaminidase releases N-glycans. | Lactate and acetate | 152 |
| B. longum | ||||
| 105-A | Lactulose (contains Gal-β1, 4-Rha structure) | The SBP encoded by the BL105A 0502 gene internalizes lactulose. Gh42β-galactosidase is a candidate enzyme for Gal-β1, 4-RHA degradation. | Acetate and lactate | 153,154 |
| M12 | 2ʼ-FL and LNT | B. longum M12 contains the GH95 gene (α-1-2-L-fucosidase), which can grow on 2ʼ-FL and LNT as the sole carbon source, but lacks GH29 (α-1,3/4-fucosidase) and cannot utilize LNnT. | -- | 46 |
| JCM7052 | Gum arabic AGP | α-l-Rhap-(1 → 4)-β-d-GlcpA-(1 → 6)-β-d-Galp-(1 → 6)-d-Gal tetrasaccharide is produced by the cooperation of three extracellular enzymes including BlArafE, which is a new α-1-arabinofuranosidase (GH43/34) for splitting up the α1,4-Araf linkage. | Oligosaccharides (tet-rasaccharide S4) | 122 |
| JCM7052 | AGP | 3-O-α-D-galactosyl-α-L-arabinofuranosidase (GAfase) (GH39) has responsibility for the release of α-D-Galp-(1 → 3)-L-Ara and β-L-Arap-(1 → 3)-L-Ara. | L-arabinose, and galactose | 54 |
| JCM1217 | Type II AG, and larch wood AG | β-1,6-galactobiose is produced by the combination of three enzymes including GH43_24 exo-β-1,3-galactanase (Bl1,3 Gal), GH30_5 exo-β-1,6-galactobiohydrolase (Bl1,6 Gal) and GH43_22 α-L-arabinofuranosidase (BlArafA). | β-1,6-galactobiose, and arabinofuranose | 16 |
| NCC2705 | High-mannose N-glycan | After cleaving by an endo-β-N-acetylglucosaminidase (GH85), N-glycan is broken down by three GH38 α-mannosidases and a GH125 α-1,6-mannosidase. | Mannose, acetate formate, and ethanol | 56 |
| NCIMB 8809 | Hydroxycinnamic acids (HCAs) | The CaeA esterase in an arabinoxylan/arabinan metabolism cluster can cleave several HCA-containing oligosaccharides. | --- | 155 |
| B. breve | ||||
| UCC2003 | Lacto-N-biose (LNB) | Three transcriptional regulators (LntR, NahR, and NagR1) are involved in regulating LN (n) T/LNB metabolism. | --- | 156 |
| UCC2003 | 4-galactosyl-kojibiose and lactulosucrose | β-galactosidase and the specific gene clusters (Bbr_1551 to Bbr_1553) are used to degrade GOS and lactulose. | -- | 157 |
| DSM 20091 | GOS | GosDEC, GalCDE transporters, and extracellular GH53 enzymes are used to degrade GOS. | -- | 158 |
| JCM1254, JCM7004, TMC3108, and TMC3115 | 2ʼ-FL, 3ʼ-FL, LNnT, and LNFP I | A combination of seven extracellular GH enzymes degrades HMOs, releasing degradants into the extracellular space. | LNB, lactose, galactose, and fucose | 66 |
| B. bifidum | ||||
| JCM1254 | LNT | LNBase (LnbB) is specific for LNT degradation. | lacto-N-biose I and lactose | 45 |
| JCM 1254 | Mucin O-glycans | Degradation of mucin O-glycans by GH 20 sulfoglycosidase (BbhII) and GlcNAc-6S-specific carbohydrate-binding module (CBM) 32. | N-acetylglucosamine-6-sulfate | 71 |
| B. adolescentis | ||||
| P2P3 | High amylose corn starch | RSD1/2/3 and starch-binding modules (CBM25, CBM26 and CBM74) are used for RS degradation. | Maltooligosaccharides | 72 |
| DSMZ 20083 | β-manno-oligosaccharide (MOS) | ABC and MFS transporters facilitate the uptake of linear MOS, while GH1 β-glucosidase and GH32 β-furanoglycosidase catalyze the cleavage of MOS. | Acetate, lactate, and formate | 77 |
| ATCC 15703 | AXOS (DP 2–4 and mono-substituted) | GH43 α-L-arabinofuranosidase is responsible for degradation. | Lactate and acetate | 159 |
| B. pseudocatenulatum | ||||
| MP80 | 2ʼ-FL | A series of gene clusters containing GH29 and GH95 enzymes perform degradation of fucosylated HMOs. | 1, 2-PD | 160 |
| JCM 1200 | Sucrose (Suc) and N-acetyl sucrosamine (SucNAc) | Sucrose phosphorylase is responsible for Suc degradation, and β-fructofuranosidase is for SucNAc. | -- | 85 |
| ED02 | XOS and linear xylan | An extracellular GH10 endo-β-1.4 xylanase exhibits activity against both XOS and xylan. | XOS fractions of the various DP | 161 |
| YIT 4072 T | Arabinoxylan hydrolysate (AXH) | Five GH43 enzymes and three transporters participate in the degradation of AXOS and XOS. | Arabinose and xylose | 162 |
| B. catenulatum subspecies kashiwanohense | ||||
| JCM 15439T | AX, xylan, and XOS | Extracellular xylanase can cleave AX into XOS and AXOS, which are subsequently further catabolized by intracellular arabino-franosidase and xylosidase into arabinose and xylose. | Arabinose and xylose | 87 |
| YIT 13060 | 2ʼ-FL, lacto-N-difucohexaose (LNDFH) | 2ʼ-FL and LNDFH are translocated intracellularly and further degraded in cooperation with fucosidase, β-galactosidase, and Lacto-N-biosidases. | GLcNAC, fucose, galactose, and glucose | 87 |
B. infantis primarily internalizes HMOs
The ability of B. infantis to intracellularly utilize most HMOs provides a competitive advantage over other HMO-consuming species (such as B. breve), resulting in its dominance in the infant gut during breastfeeding37. The processing of 2ʼ-fucosyl lactose (2ʼ-FL) by B. infantis depends on the ABC transporters for intracellular digestion. B. infantis contains several intracellular enzymes including fucosidase, β-galactosidase, LNB phosphorylase, N-acetyl-β-hexosaminidase, and sialidase, to degrade fucosylated and sialylated HMOs38.
Transporter specificity is necessary for the effective uptake of HMOs. Sakanaka et al.26 have reported that the two FL transporters (FL1-BP and FL2-BP) of B. infantis can be used to absorb 2ʼ-FL and 3ʼ-FL, and the expression of FL transporters influences the abundance of B. infantis. Intracellular FL is utilized by α-fucosidase to produce fucose, which results in the secretion of 1,2-propanediol (1,2-PD), acetate, and lactate39. Non-fucose glycosylated neutral HMOs, including lactose-N-tetrasaccharide (LNT), lacto-N-neotetraose (LNnT), lactose-N-bisaccharide (LNB), and N-acetyl glucosamine, stimulate the growth of B. infantis35,40. Duar et al.41 determined that the utilization of LNT and LNnT by B. infantis EVC001 is closely associated with the Blon2175-2177 ABC transporter in its H5 cluster, which contains a complete gene repertoire of HMO utilization, while the H5-negative strains exhibit growth defects on the carbon source.
B. longum can metabolize arabinoxylan
Colonization by B. longum dominates the entire lifespan of the host42. The abundance of B. longum is closely associated with food intake at different growth stages, while the gene pool for glycan acquisition and metabolism can be selectively altered due to changes in certain dietary components43,44. The colonization superiority of B. longum is manifested in its ability to grow on HMOs and metabolize complex arabinoxylan (AX) substrates.
Some B. longum metabolize specific HMOs, including LNT and 2ʼ-FL rather than sialylated HMOs, as well as mucin O-glycans, thereby occupying trophic niches in the gut during early life45. Sakanaka et al.45 identified an extracellular lacto-N-biosidase (LnbX) from B. longum JCM1217 that could consume LNT and release LNB and lactose via the GNB/LNB pathway. Díaz et al.46 found that B. longum M12 with α-1-2-L-fucosidase (GH95) could grow on 2ʼ-FL and LNT.
The transporters and GHs of B. longum are more likely to metabolize plant-derived glycans (dominated by various arabinose-substituted glycans)47. The enrichment of the gene cluster encoding L-arabinofura (-Araf) facilitates B. longum utilization of these glycans and reflects its ecological adaptability and competitiveness in the adult and aging gut environments48. The GH43 family, which includes α-L-arabinofuranosidase, β-xylosidase, arabinosidase, and xylanase, is substrate-specific and degrades insoluble fibers through cooperation. B. longum has an abundant GH43 gene cluster capable of releasing arabinoxylooligosaccharides (AXOS), arabinose and xylose from arabinogalactan (AG), arabinogalactan (AN), and AX16,49,50. Collectively, the ability of B. longum to degrade AX and AXOS is strain-specific and influenced by different priorities for utilizing different monosaccharide compositions and structures (such as degree of polymerization (DP), linkage type, and side-chain space structure)51. For example, B. longum JCM 1217 may prefer natural and partially degraded AX components with higher DP, side chain content, and arabinosyl-monosubstituted structure, possibly due to differences in the transport system specificity and membrane localization of the enzymes14. AG and arabinogalactan protein (AGP) are complex fiber components in plant cell walls that are consumed only by a few sub-species of B. longum; most bifidobacterial species cannot absorb full AGP52. Fujita et al.16 reported a type II AG-degrading enzyme in B. longum JCM1217 that can grow on larch AGP but cannot act on gum arabic AGP with a more complex structure. The high molecular weight arabinose side chain in the highly modified AGP causes steric hindrance, resulting in limited cleavage of β-1,3/1,6 bonds in the galactose skeleton by β-galactosidase and arabinoglycosidase and insufficient galactose and galactooligosaccharides (GOS) release53. Sasaki et al.54 identified a novel arabinofuranosidase (named GAfase) that can cleave the l-Ara structure of gum arabic AGP to remove steric hindrance, a key factor in the growth of B. longum JCM7052 in gum arabic AGP. These relatively limited degradations suggest that B. longum must cooperate with other bacteria to completely consume the AGP and AG complex. Moreover, B. longum selectively utilizes other N-glycans for growth. Specifically, the α-mannosidases, N-acetyl glucosaminidase, and α-glucosidase of B. longum NCC2705 release mannose and N-acetyl glucosamine (GlcNAc) from mannans through cooperation55,56.
B. breve has limited growth on complex HMOs
HMO metabolism in B. breve is highly similar to B. infantis, which absorbs intact oligosaccharides through the ABC transport system and degrades them in cells. However, the complete gene pool required to metabolize complex HMOs, such as fucosyllactose and sialyl-lactose, and the utilization of HMO components by B. breve are limited to N-glycans, including LNT, LNnT, and N-disaccharide (LNB)57,58. The genome of B. breve mostly contains genes involved in the breakdown of lactose, fucose, sialic acid, and amino sulfate59–61. Specifically, the external sialidase (GH33) in B. breve can help to release sialic acid from mucin and grow on free sialic acid in the gut environment29. The N-acetyl glucosamine 6-phosphate deacetylase of B. breve UCC2003 can efficiently break down GlcNAc-6-sulfate residues in O-glycans61. Bottacini et al.62 analyzed the genomes of 20 B. breve strains and determined that α-glucosidases (GH13 and GH31), β-glucosidase (GH1 and GH3), and β-galactosidases (GH2 and GH42) were involved in the metabolism of α-glucans (such as maltose), cellobiose, and galactose, respectively. Notably, most B. breve strains cannot utilize fucosylated HMOs (2ʼ-FL or 3ʼ-FL), but utilize fucose produced by other bacteria63.
B. bifidum extracellularly degrade HMOs and mucin O-sugars
Compared with other bifidobacterial species, B. bifidum has a complete set of extracellular GHs, including α-sialidase, α-fucosidase, N-Acetyl-β-hexosaminidase, β-galactosidase, and LNBase, to assimilate complex HMOs and leave degradation products, such as lactose, fucose, and sialic acid, outside the cell45,64–66. Nishiyama et al.67 identified a sialidase (SiaBb2) involved in the degradation of HMOs and mucin and may promote the adhesion and colonization of B. bifidum on the surface of the intestinal mucosa. Additionally, B. bifidum lacks specific enzymes to degrade N-glycans, however, has a substrate preference for O-glycans, which avoids competition with other bifidobacterial species in the guts of breast-fed infants. B. bifidum encodes N-acetyl galactosidase (GH101) and Lacto-N-biosidase (GH136), which degrade mucin O-glycosidic bonds, and has abundant carbohydrate-binding modules (CBMs) that promote the proximity of GHs to substrates68,69. Takada et al.70 found that the β-N-acetylglucosaminidases of B. bifidum can specifically degrade β-GlcNAc linkages of mucin core structures, while Katoh et al.71 reported that B. bifidum could release GlcNAc-6S from sulfated O-glycans using sulfoglycosidase and may affect the metabolism of other bacteria.
B. adolescentis prefers to degrade starch
The competitive advantage of B. adolescentis is mainly reflected in its utilization of plant glycans, including RS, mannooligosaccharides (MOS), and inulin. B. adolescentis contains α-amylases, glycogen debranching enzyme, pullulanase, and a specific starch-binding module, which facilitate the complete degradation of RS72,73. Enzymes encoding other glycans, including galactosidase, mannosidase, β-xylosidase, and arabinofuranosidase, similarly reflect the preference of B. adolescentis in degrading dietary glycans74. Moreover, Mulualem et al.75 identified an α-galactosidase (BgaC) from B. adolescentis ATCC15703 that produces GOS from lactose through transglycosylation. Notably, B. adolescentis consumes oligosaccharides and is more likely to grow on MOS (DP ≤ 4) and mannose, using β-glucosidase (GH1) and β-fructofuranosidase (GH32), than complex β-mannan76,77. Salas-Veizaga et al.78 reported that B. adolescentis metabolizes glucuronosylated-XOS (GXOs) and XOs (DP 2-6) to produce SCFAs. B. adolescentis prefers (A)XOS to arabinose or xylose, and encodes a gene for endo-1,4-β-xylanase capable of growing on wheat AX9,14,79.
B. pseudocatenulatum and B. catenulatum consume host and plant glycans
B. pseudocatenulatum and B. catenulatum are found in the guts of infants and adults. Their CAZyme genes differ in hosts of different ages6,80, suggesting that both species have a relatively well-developed degradation system for both host and plant sugars.
The α-fucosidases GH29 and GH95 induced B. pseudocatenulatum MP80 to exclusively utilize lower-molecular-weight fucosylated HMOs, such as lactodifucotetraose81. B. pseudocatenulatum has an endo-1,4-β-xylanase (GH10) that can thrive on long-chain xylan-derived polysaccharides (XOS with polymerization degrees 2–4), which may become a critical feature of B. pseudocatenulatum, consistent with the finding that bifidobacteria have a limited ability to degrade xylan82. Additionally, endo-1,4-β-xylanase cuts the main chain of xylan to produce XOS and xylose. While some metabolites are absorbed by the intracellular utilization system, others are released into the extracellular environment for use by secondary consumers82. Specifically, a β-L-arabinopyranosidase (AAfase) is characterized in the genome of B. pseudocatenulatum MCC10289 for the assimilation of AGP side chains and L-arabinose83. Hosaka et al.84 determined that B. pseudocatenulatum JCM 1200 expresses sucrose phosphorylase and β-fructofuranosidase to hydrolyze sucrose and analog disaccharide N-acetyl sucrosamine (SucNAc)85.
Similarly, the genome of B. catenulatum contains genes for GHs involved in HMOs and xylan, starch, and their derived oligosaccharides, which can adapt to changes in the host diet86. HMOs, including 2ʼ-FL and lacto-N-difucohexaose, are translocated intracellularly by B. catenulatum and degraded into GLcNAC and monosaccharides in cooperation with fucosidase, β-galactosidase, and Lacto-N-biosidases87. The extracellular xylanase of B. catenulatum can cleave AX into XOS and AXOS, which are subsequently catabolized by intracellular arabino-franosidase and xylosidase into arabinose and xylose87.
Glycan utilization capacity of bifidobacteria shapes cross-feeding interactions with other species
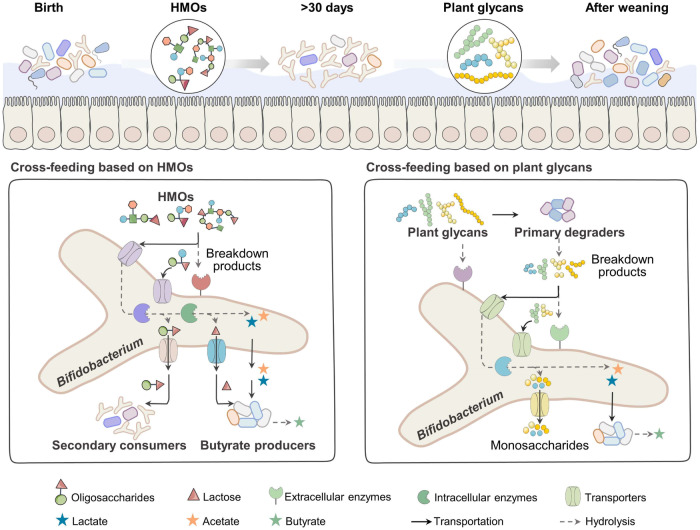
The gut microbiota is mostly auxotrophic, and the assimilation of glycans by different species may be similar (metabolic redundancy). Therefore, there remains a need to compete with other species or obtain nutrition from the gut environment88,89. Relying on other microorganisms that release metabolites into the environment, competing species can coexist in equilibrium; these cost-free metabolites promote microbial interactions90. Cross-feeding exists between species with different metabolisms, specifically those that utilize particular complex compounds to liberate metabolic by-products that are further assimilated by others that cannot grow on those compounds alone28,90,91. Bifidobacteria rely on their enzymatic breakdown and oligosaccharide transport systems to consume glycans from the host, resulting in stable colonization of the gut tract and facilitating cross-feeding relationships within bifidobacterial species or with other bacteria, such as Bacteroides and butyrate producers92 (Fig. 2 and Table 2).
Fig. 2. Cross-feeding strategy of bifidobacteria throughout the life span.
Cross-feeding of bifidobacteria exists throughout the life cycle. The major categories are HMO-based and phytoglycan-based cross-feeding. Specifically, when co-cultured on HMOs, bifidobacteria are more likely to cross-feed HMO degradation products to other non-HMO degradation-dominant bacteria and butyrate producers. When cross-fed on phytoglycans, bifidobacteria are more likely to thrive by relying on the extracellular degradation products of other dominant degrading bacteria.
Table 2.
Cross-feeding strategies of bifidobacteria based on glycan utilization
| Bifidobacterial strains | Cross-feeders | Types of glycan | Secondary and final metabolites | Strategies for cross-feeding | References |
|---|---|---|---|---|---|
| Cross-feeding based on HMO and Mucins | |||||
| B. infantis ATCC15697 | A. caccae L1-92 | HMOs | Lactose, lactate, and acetate | B. infantis releases metabolites from HMOs to help A. caccae produce butyrate. | 93 |
| B. longum BSM11-5, B. breve DSM 20213, and B. infantis DSM 20088 | E. hallii 3353 | Fucosyllactose and L-fucose | Glucose, lactate, acetate, and 1, 2-PD | L-fucose is metabolized to produce lactate, 1, 2-PD, and acetate, which are further converted to butyrate, formate, and propionate by E. hallii. | 30 |
| B. longum LH206 | B. pseudocatenulatum LH657, LH659 | LNnT | 2ʼ-FL and galactose | The products of LNnT are degraded by B. longum and are cross-fed to B. pseudocatenulatum, and the product of 2ʼ-FL is metabolized by B. pseudocatenulatum, to support the growth of B. longum. | 29 |
| B. bifidum ATCC 15696 | B. breve 24b | 2ʼ-O-Fucosyl-Lactose | Lactose and fucose | Lactose and fucose are cross-fed to B. breve 24b for 1, 2-PD production. | 98 |
| B. bifidum CCX 19041 and B. infantis CCX 19042 | B. breve CCX 19061 | Sialylated immunoglobulin G | N-acetylneuraminic acid (Neu5Ac) and galactose | Neu5Ac and galactose degraded by B. bifidum CCX 19041 and B. infantis CCX 19042 are cross-fed to B. breve CCX 19061. | 100 |
| B. bifidum PRL2010 | B. breve UCC2003 | 3ʼ-SL | Sialic acid and lactose | B. breve uses the extracellular degradation products of B. bifidum. | 163 |
| B. bifidum ATCC 15696 | B. breve JCM 7019 | 6ʼ-SL | Sialic acid and lactose | B. breve JCM 7019 consumes the hydrolysates by extracellular sialidase of B. bifidum ATCC 15696. | 99 |
| B. bifidum BSM28-1 | B. breve BRS 26-2, B. infantis DSM 20088, and E. hallii DSM 3353 | Mucins | Lactose, formate, and acetate | B. bifidum BSM28-1 provides lactose to the other three strains, and E. hallii consumes secondary metabolites for butyrate and propionate production. | 94 |
| B. breve UCC2003 | Lm. reuteri ATCC PTA 6475 | Fucose | 1, 2-PD | The pduCDE operon of L. reuteri is responsible for utilizing 1, 2-PD produced by B. breve UCC2003. | 102 |
| Cross-feeding based on plant glycans | |||||
| B. longum LMG 11047 | L. paracasei 8700:2 and Anaerotipes caccae DSM 14662 T, or E. hallii DSM 17630 | ITF | Oligofructose, lactate, and acetate | B. longum LMG 11047 grows on short-chain inulin produced by Lactobacillus and releases acetate and lactate which are further converted to butyrate and gases. | 118 |
| B. longum NCC2705 | E. rectale ATCC 33656 | AXOS | Acetate, arabinose, and xylose | Both bacteria could degrade XOS. E. rectale ATCC 33656 uses acetate produced by B. longum to produce butyrate and xylose; the latter is consumed by B. longum. | 108 |
| B. longum NCC 2705 | B. caccae ATCC 43185 | Larch wood AG | Carbohydrate fragments of AG, lactate | B. longum NCC 2705 depends on the products of AG degradation by B. caccae ATCC 43185, while Bacteroides may utilize lactate produced by B. longum. | 53 |
| B. longum BB536 | A. caccae DSM 14662 and Roseburia intellalis DSM 14610 | FOS | Acetate and fructose | A. caccae DSM 14662 grows on fructose released by B. longum BB536, while Roseburia intellalis DSM 14610 consumes acetate. | 164 |
| B. adolescentis L2-32, and B. adolescentis DSM 20083 | E. hallii L2-7, and R. homini A2-183 | Starch or FOS | Lactate, acetate, and oligosaccharides | Lactate and acetate could be cross-fed to E. hallii, and oligosaccharides could be used as growth substrates for R. hominis. | 104 |
| B. adolescentis ATCC 15703 | R. hominis A2-183 | Linear or galactose-substituted β-mannan-oligosaccharides | Acetate | Acetate helps R. hominis A2-183 produce butyrate and grow on mannan-oligosaccharides. | 109 |
| B. adolescentis DSMZ 20083 | B. ovatus DSMZ 1896 | Galactomannans | β-mannooligosaccharides | B. adolescentis DSMZ 20083 relies on short β-mannooligosaccharides (DP3). | 115 |
| B. adolescentis L2-32 | F. prausnitzii S3/L3 | FOS | Acetate and FOS residues | B. adolescentis cross-feeds acetate to F. prausnitzii and grows better due to FOS residues released by F. prausnitzii. | 165 |
| B. adolescentis LMG10734 | F. prausnitzii DSM 17677(T) | ITF | Acetate | Acetate is used for butyrate production. | 110 |
| B. pseudolongum ST6 | B. animaiis subsp. lactis RG1 | Hi-Maize starch | Maltose | Maltose is released by B. pseudolongum and cross-fed to B. animaiis. | 111 |
| B. breve UCC2003 | Bacteroides cellulosilyticus DSM 14838 (Baccell) | Larch wood AG | β-1,3-GOS | B. breve UCC2003 uses the short β-1,3-GOS released by Baccell, eventually producing succinic acid and acetate. | 166 |
| B. bifidum PRL2010 | B. breve 12 L, B. adolescentis 22 L, or Bacteroides thermophilum JCM1207 | RS2-resistant starch or xylan | Glucose and or maltose | B. bifidum PRL2010 may utilize glucose and or maltose released by other bifidobacterial strains, and the genes involved in glycolysis in these species were upregulated. | 112 |
| B. animalis subsp. lactis DSM-10140 | B. ovatus DSM-1896 or Bacteroides xylanisolvens DSM-18836 | Beechwood and corncob xylans | XOS | B. animalis subsp. lactis grows on XOS produced by Bacteroides, releasing lactate and acetate. | 19 |
HMO-based cross-feeding of bifidobacteria allows for its dominance in the guts of infants
Cross-feeding of bifidobacteria with butyrate producers based on HMOs
B. infantis metabolizes HMOs to produce oligosaccharides and metabolites (such as 1, 2-PD and acetate) that play important roles in the gut tract of breastfed infants by cross-feeding with other bacteria such as Eubacterium hallii and Anaerostipes caccae30,93,94.
Cheng et al.95 found that B. infantis utilizes 6ʼ-sialyllactose (6ʼ-SL) for acetate production and is cross-fed to Faecalibacterium prausnitzii for butyrate conversion, which causes B. infantis to proliferate as a result. This may be associated with the fact that F. prausnitzii secretes extracellular sialidase to promote the expression of sialidase in B. infantis, thereby increasing acetic acid production, and that the occurrence of this cross-feeding activity depends on the molecular structure of HMOs95,96.
Cooperative degradation of HMO between bifidobacterial species
When extracellularly degrading HMOs, the products produced by bifidobacteria (including B. bifidum and B. longum) are partially released into the public environment, inducing cross-feeding with other bifidobacterial species with weaker HMO-utilizing capacity.
For example, the products of LNnT degradation by B. longum could be cross-fed to B. pseudocatenulatum, and the oligosaccharides from 2ʼ-FL degradation by B. pseudocatenulatum support the growth of B. longum29. B. bifidum is not the dominant species in the intestinal tract of infants; however, as an extracellular degrader of HMOs, it cross-feeds with other species by releasing HMO derivatives, thus affecting the composition of the gut microbiota97. HMO metabolism between B. bifidum and B. breve is complementary, which also contributes to cross-feeding between the two strains61. B. bifidum uses fucosidase and FL transporters to release lactose and fucose extracellularly, which cross-feeds to B. breve. This indicates that B. bifidum is easy to cross-feed with other fucose consumers who lack extracellular fucosidase. However, B. bifidum prefers to grow on lactose rather than fucose, suggesting altruism as its primary function98. In addition to cooperation, B. breve can compete with B. bifidum for lactose; the key to coexistence is that B. bifidum releases more lactose by upregulating the gene expression of the enzymes and transporters involved98. A similar phenomenon was observed in the co-culture of mucin and sialylated glycan59,99. Chen et al.100 demonstrated that B. bifidum released sialic acid and lactose from sialylated glycans, which supported the growth of B. infantis and B. breve as secondary metabolites of cross-feeding. B. bifidum does not assimilate fucose, galactose, NeuAc, and GlcNAc-6S; therefore, the degradation of mucin O-glycan may be accomplished in cooperation with other species, such as B. breve, that utilizes these mucin-derived carbohydrate fragments66,101.
HMO-based cross-feeding of Bifidobacteria with other gut bacteria
Cross-feeding relationships established between bifidobacteria and other gut bacteria can help to refine the network of interactions within the gut microbiota during early life. Cheng et al.102 demonstrated that the metabolite 1,2-PD produced by B. breve through fermenting fucose could promote the colonization of Limosilactobacillus reuteri in the gut of gnotobiotic mice. Nogacka et al.103 investigated the 2-ʼFL-based metabolic interactions between B. bifidum and Lactobacillus gasseri and found that B. bifidum IPLA20048 promoted the proliferation of L. gasseri IPLA20136 by cross-feeding the extracellular degradation products (galactose, fucose, and lactose) and that genes encoding α-fucosidase involved in carbohydrate transport are upregulated in B. bifidum thereby increasing carbohydrate production.
The gut microbiota of breastfed infants undergoes significant changes in response to diet before and after weaning, thereby facilitating more complex microbial interactions and inducing gradual stabilization and maturation of the structure and composition of the gut microbiota.
Phytoglycan-based cross-feeding of bifidobacteria after weaning enables its persistence in the gut
Cross-feeding of bifidobacteria with butyrate producers based on plant glycans
Acetic and lactic acids are the end products of oligosaccharides metabolism by bifidobacteria and are important substrates of cross-feeding between bifidobacteria and butyric acid bacteria104–106. When grown on AXOS, except for the conversion of acetic acid to butyric acid, butyrate producers consume AXOS to produce arabinose and xylose, which can be consumed as substrates by B. longum107,108. Bhattacharya et al.109 investigated the cross-feeding between B. adolescentis and Roseburia based on galactose-substituted β-mannan-oligosaccharides.
Moens et al.110 confirmed that bifidobacteria can easily cross-feed on FOS and inulin-type fructan to produce the bifidogenic and butyrogenic effects and that the conversion degree of acetic acid to butyric acid was closely associated with the differences in the glycan-degrading ability of bifidobacterial species. Specifically, when co-cultured with F. prausnitzii, B. breve can only utilize fructose, cannot consume FOS and ITF, and relies on the monosaccharides produced by F. prausnitzii to degrade fructan for growth. B. adolescentis prefers short-chain FOS over ITF, allowing it to cross-feed acetate to F. prausnitzii and utilize its FOS for continued growth. In contrast, B. longum or B. angulatum competes with F. prausnitzii on ITF, resulting in the lowest efficiency of butyric acid synthesis in the co-culture system110.
Phytoglycan-based cross-feeding between bifidobacterial species expands carbon source availability
When co-cultured, bifidobacterial species that cannot degrade plant glycans rely on other species that can degrade. For example, Centanni et al.111 reported that when grown on Hi-Maize, B. pseudolongum extracellularly produced type 1 pullulanase and alpha-amylase for the release of glucose, maltose, and maltotriose, which are cross-fed to B. animalis. More importantly, bifidobacterial species with similar nutrient metabolism can expand their trophic niches by cross-feeding the same substrates rather than competing. For example, B. bifidum PRL2010 lacks a degradation system that utilizes plant glycans, such as starch and xylan; however, when co-cultured with B. adolescentis 22 L, B. breve 12 L, and B. thermophilum JCM1207, it can grow on simple carbohydrates released by other bifidobacteria and promote its sugar utilization gene expression112. When co-cultivated with other bifidobacteria, B. adolescentis exhibits the most significant alterations in genes involved in xylose metabolism compared with other species, reflecting the enhanced genetic adaptability of these strains, which may help hosts to expand the utilization potential of carbohydrates during specific dietary changes113.
Bifidobacteria cooperates with other gut bacteria to degrade complex plant glycans
Cross-feeding of breakdown products of complex glycans among the gut microbiota depends on the complexity of the target glycan. Due to the lack of a complete degradation and transport system, bifidobacteria have a limited capacity for recalcitrant plant glycans, including xylan, long-chain inulin, and mannan; therefore, they rely on other bacteria to extracellularly release soluble oligosaccharides as cross-feeding substrates, such as AXOS, XOS, FOS, and mannose76,114. Some members of the genera Bacteroides and Lactobacillus are dominant glycan-degrading bacteria that can cross-feed with Bifidobacterium, including B. longum, B. animalis, and B. adolescentis19,115. When grown on simple xylans, such as glucuronoxylan and wheat arabinoxylan, B. adolescentis ATCC 15703 relies on AXOS produced by Bacteroides ovatus ATCC 8483 for growth in the absence of a degradation system containing xylanase (GH98)116. The cross-feeding of fructans is closely associated with the substituents and length of the sugar chain, and complex fructans (i.e., those containing sucrose units) are degraded by primary degraders to produce short-chain compounds that are utilized by secondary consumers117. B. longum tends to use short-chain inulin, leading to cooperation with Lactobacillus paracasei to completely degrade long-chain inulin and produce fructose, lactate, and acetate118.
B. longum has a cross-feeding relationship with Bacteroides based on xlyan, type II AG, and AGP. Wang et al.53 reported that when co-cultured with Bacteroides caccae ATCC 43185, B. longum NCC 2705 thrived on the carbohydrate fragments of larch wood AG degraded by B. caccae. Vega-Sagardía et al.119 reported that B. ovatus HM222 degraded xylan and promoted the growth of B. longum PT4 by releasing XOS and extracellular enzymes and that the secretion of α-L-arabinoglycosidase, L-arabinose isomerase, and xylulose isomerase increased in B. longum PT4. However, due to the poor growth of Bacteroides at pH <5.5, with the production of acetate and lactate by B. longum fermentation, the substrate conversion (from succinic acid to propionate) efficiency of Bacteroides decreases, resulting in succinic acid accumulation53. Therefore, cross-feeding with Bacteroides is pH-limited, and the combined bacteria used to degrade complex AG or xylan require the participation of other species, such as Prevotella and Roseburia intestinalis120,121. Notably, the cross-feeding relationship between B. longum and Bacteroides is mutual, and the secondary products of plant glycan metabolism from B. longum can be cross-fed to some Bacteroides with a poor glycan metabolic system. For example, AGP-related oligosaccharides degraded by B. longum can be cross-fed with Bacteroides vulgatus, which promotes xylosidase expression in the latter strain122. Sonnenburg et al.123 determined that B. longum promoted the functions of mannosidase and xylosidase in Bacteroides thetaiotaomicron when co-cultivated on plant glycans. Given the limited data, whether the cross-feeding interactions involving bifidobacteria contribute positively to the balance of carbohydrate utilization within the entire intestinal community remains to be determined.
Availability of glycans and cross-feeding activities shape the critical position of Bifidobacterium during life span
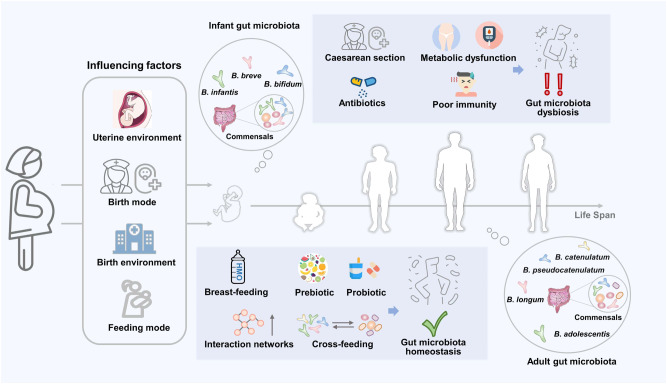
The cross-feeding activities produced by bifidobacteria underscore their key ecological role in acquiring or providing substrates from or for other bacteria. This dependent interaction can also enhance the metabolic function of the participants, thus expanding their original trophic niche. The supplementation with bifidobacteria-containing multistrain synbiotics to construct a network of interactions between different members of the gut can help the gut microbiota to mature and stabilize during early life and increase the secretion of beneficial metabolites (such as butyrate), ultimately preventing chronic diseases associated with an imbalanced gut microbiota44,124. This synergistic combination of strains synergistically regulates gut microbiota and health homeostasis and is less affected by endogenous microorganisms than a single strain125,126. However, current research on the regulation of health by multi-strain synbiotics is only at the phenomenal description level, and clear explanations of the underlying reasons for the combination, the basis of its formulation, and the mechanism of its effectiveness remain scarce. Therefore, the possibility and necessity of such multi-strain synbiotics should be explored based on the preference of probiotics for glycans and the cross-feeding their relationships127,128. Gut microbiota is often affected by age, dietary habits, and antibiotics, necessitating tailored nutrition based on the needs of different hosts using appropriate and individualized strategies to regulate flora homeostasis34,129 (Fig. 3).
Fig. 3. Benefits of cross-feeding of bifidobacteria throughout life span.
The bifidobacteria of the host are influenced by several factors from birth. Breastfeeding and the use of probiotics and prebiotics contribute to the establishment of a network of interactions of the gut microbiota, including cross-feeding. The cross-feeding relationship can facilitate the formulation of multi-strain synbiotics, which are effective in improving intestinal homeostasis and mitigating related disorders including dysbiosis, metabolic syndrome, and immunodeficiency, due to cesarean section and antibiotic use.
Beneficial effects of cross-feeding involved in bifidobacteria during early life
Ameliorating the adverse consequences of microbial colonization caused by cesarean section and antibiotic use
Mode of birth (cesarean section) and antibiotic treatment negatively impact gut microbiota during infancy, as evidenced by the accumulation of pathogenic bacteria, decrease in Bifidobacterium and Bacteroides, and increase the risk of metabolic, inflammatory, and immune disorders in infants127,130,131. Supplementation with Bifidobacterium strains with extensive HMO metabolism (such as B. infantis), probiotics, and prebiotics that can stimulate the growth of Bifidobacterium have been used to reverse gut microbial dysbiosis induced by childbirth132,133.
B. breve has been reported to grow well on 2ʼ-FL by coexisting with other species encoding an extracellular fucosidase (GH95). Lou et al.132 found that supplementation with R. gnavus can promote the proliferation of B. breve, which utilizes lactose released by the former from the 2ʼ-FL, and contribute to the shift of the preterm microbiome to a bifidobacteria-rich community. Korpela et al.127 developed a mixture of Lactobacillaceae rhamnoides, B. breve, and Propionibacterium freundenreichii combined with FOS, which promoted the colonization of Bifidobacterium and reduced the abundance of Enterococcaceae, Clostridiaceae, and Veillonellaceae, ultimately correcting adverse changes in the gut microbiome due to cesarean section and antibiotic induction. A metaproteome analysis revealed that the enzymes used for HMO degradation in Bifidobacterium (including β-galactosidase and β-galactosyl N-acetyl hexosaminephosphorylase) were significantly expressed and Lactobacillus rhamnosus and might benefit from the monosaccharides released from HMO degradation127. These studies suggest that the administration of probiotics and/or prebiotics with gut-resident species (especially Bifidobacterium) as allies is effective in restoring gut microbiota disorders in infants; such multi-strain and prebiotic combinations with synergistic effects can be used as supplements in infant formulas.
Promoting a dominant position of bifidobacteria and perfecting intestinal community assembly
HMO-based metabolic interactions can shape the composition of infant gut microbiota. Bifidobacterium (especially B. infantis) is prioritized for gut colonization of breastfed infants, as determined by their superior ability to assimilate HMOs21,134. Bifidobacteria exert a priority effect based on HMO metabolism shortly after birth and consume most of the carbon sources in the gut, which prevents the colonization of later species, stimulates the establishment of infant intestinal homeostasis, and inhibits the growth of pathogens (such as Clostridium difficile and Salmonella)135. Intestinal colonization of B. breve is an example of priority effects. Considering the limited utilization ability of B. breve toward HMO, the formation of its dominant position is not simply related to the phenotype of HMO consumption but depends on cross-feeding with other bifidobacteria61,66.
The cross-feeding activity involved in bifidobacteria plays a key role in the assembly of gut microbiota. Chang et al.136 found that B. infantis Bg2D9 promoted the colonization of Prevotella copri Bg131 in the gut of malnourished mice and facilitated the release of arabinose from diets containing arabinoglycans, which contributed to the proliferation of other species in the gut, including B. catenulatum and Blautia obeum, by cross-feeding. This could be used as a dietary intervention in severely acutely malnourished children.
In summary, the gut microbiota is highly malleable after birth and it is possible to promote gut homeostasis by introducing diets with appropriate synbiotics. Determining the processes and outcomes of these interactions can be used for dietary interventions to achieve personalized nutrition137.
Beneficial effects of cross-feeding involved in bifidobacteria during adulthood
Reestablishing gut microbiota structure and gut homeostasis
The prevalence of endogenous bifidobacteria is lower in adulthood than early life; however, due to the increase in dietary diversity, cross-feeding activities involved in bifidobacteria facilitate more interactions with the gut microbiota and host34. Dietary supplementation with multi-strain synbiotics helps to promote the proliferation of bifidobacteria and to reestablish a stable gut community138,139. Exposure to antibiotics can disrupt the abundance and diversity of the gut microbiota, and the introduction of deleted core microbiota can help to rebuild the gut immune homeostasis through syntrophic relationships125. For example, Button et al.140 developed a synbiotic of HMOs and B. infantis to improve the imbalance in the gut microbiota and affect the growth of Enterobacteriaceae by promoting the production of acetate and butyrate. A follow-up study by Button et al.141 found that this HMO-dependent bifidobacterial strategy could increase the abundance of lactate consumers (Veillonella spp.) and alleviate antibiotic-induced dysbiosis of the gut microbiota by decreasing the pro-inflammatory metabolite p-cresol sulfate.
Stimulating secretion of beneficial metabolites and improving metabolic status
Due to the cross-feeding relationships with butyrate producers, changes in the abundance of Bifidobacterium are strongly correlated with fecal acetate and butyrate concentrations, and their metabolic interactions can modulate metabolic syndromes such as type 2 diabetes (T2D) and obesity142,143. Butyrate plays an important role in the regulation of blood glucose levels144. Perraudeau et al.145 designed a synbiotic containing Akkermansia muciniphila, B. infantis, three butyrate producers, and inulin to improve T2D and determined that the cross-feeding activities of A. muciniphila and B. infantis with butyrate producers promoted butyrate production, which warrants in vivo validation. Multistrain synbiotics have also been used to improve energy metabolism, immune function, and gut microbiota in individuals with obesity146,147. Kanazawa et al.142 introduced a synbiotic composed of GOS, B. longum, and B. bifidum to regulate obesity, and determined that an increase in Ruminococcus can be used for butyrate production. Nguyen et al.148 used arabinoxylan to treat obesity through co-occurrence network analysis and found that B. longum released oligosaccharides through the degradation of arabinoxylan for synergistic and mutual interactions with Bacteroides, Phascolarctobacterium, and Subdoligranulum to promote the production of acetate and mitigate obesity. Dietary changes and synbiotic interventions can be used to increase the abundance of bifidobacteria in the gut tract and promote the secretion of beneficial metabolites, thereby mitigating metabolic disorders; however, the clinical significance remains to be determined.
Future perspectives
Nutrition changes, such as those from breast milk in early life to complex glycans in adulthood and beyond, directly determine the evolution of bifidobacteria in the gut. The glycan preference of bifidobacteria is useful to adapt to changes at different ages and dietary stages and introduce cross-feeding relationships between bifidobacterial strains and other gut microbiota, providing positive feedback to the host. Therefore, the glycan utilization strategies of Bifidobacterium and their cross-feeding networks must be explored. However, unlike Bacteroides, evidence of the intracellular and extracellular GHs and transport systems of Bifidobacterium remains scarce.
The ability of the transport system to recognize specific substrates may influence cross-feeding, which warrants further investigation. In addition, cross-feeding between Bacteroides and Bifidobacterium based on AX and AG has been reported; however, the evidence remains inadequate, and the syntrophic relationship with other glycans, such as β-glucans and fructans, remains to be elucidated. Future studies should further investigate the cross-feeding of bifidobacterial species (such as B. longum and B. pseudocatenulatum) and that between bifidobacteria and gut microbiota (such as Lactobacillus).
The prevalence of bifidobacteria decreases with age. The cross-feeding activities of bifidobacteria are beneficial to the host; therefore, dietary intervention strategies must be adopted to regulate health status. However, most existing studies on the health benefits of multistrain synbiotics have focused on the outcome of the intervention rather than the process and lack the scientific basis for such synergistic combinations. This necessitates the development of individual formulas and consideration of intervention times based on the gut microbiota of individuals at different ages or nutritional stages. The beneficial effects of bifidobacteria involved in multistrain synbiotics and strategies to alter the relationship between gut microbiota and host must be determined. The targeted modulation of bifidobacteria through cross-feeding strategies can shift the classic nutritional studies of flora to molecular nutrition, which will be the future trend of microbiome therapy.
Funding
This work was supported by National Natural Science Foundation of China Key Program (32372296), Natural Science Foundation of Jiangsu Province (BK20220155, BE2021623), the Key Scientific and Technological Research Projects in the Key Areas of the Xinjiang Production and Construction Corps (2018AB010).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Leilei Yu, Email: edyulei@126.com.
Fengwei Tian, Email: fwtian@jiangnan.edu.cn.
References
- 1.Lundgren SN, et al. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome. 2018;6:109–120. doi: 10.1186/s40168-018-0490-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vernier, C. L. et al. Gut microbiota contribute to variations in honey bee foraging intensity. ISME J. wrae030 10.1093/ismejo/wrae030 (2024). [DOI] [PMC free article] [PubMed]
- 3.Manome A, et al. Acidogenic potential of oral Bifidobacterium and its high fluoride tolerance. Front. Microbiol. 2019;10:1099. doi: 10.3389/fmicb.2019.01099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Le Barz M, et al. In vivo screening of multiple bacterial strains identifies Lactobacillus rhamnosus Lb102 and Bifidobacterium animalis ssp. lactis Bf141 as probiotics that improve metabolic disorders in a mouse model of obesity. FASEB J. 2019;33:4921–4935. doi: 10.1096/fj.201801672R. [DOI] [PubMed] [Google Scholar]
- 5.Duranti S, et al. Maternal inheritance of bifidobacterial communities and bifidophages in infants through vertical transmission. Microbiome. 2017;5:66–79. doi: 10.1186/s40168-017-0282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arboleya S, Watkins C, Stanton C, Ross RP. Gut bifidobacteria populations in human health and aging. Front. Microbiol. 2016;7:1204–1213. doi: 10.3389/fmicb.2016.01204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sims IM, Tannock GW. Galacto- and fructo-oligosaccharides utilized for growth by cocultures of bifidobacterial species characteristic of the infant gut. Appl Environ. Microbiol. 2020;86:e00214–e00220. doi: 10.1128/AEM.00214-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmidt V, Enav H, Spector TD, Youngblut ND, Ley RE. Strain-level analysis of Bifidobacterium spp. from gut microbiomes of adults with differing lactase persistence genotypes. mSystems. 2020;5:e00911–e00920. doi: 10.1128/mSystems.00911-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu J, et al. Population-level variation in gut bifidobacterial composition and association with geography, age, ethnicity, and staple food. NPJ Biofilms Microbiomes. 2023;9:98. doi: 10.1038/s41522-023-00467-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma T, et al. The diversity and composition of the human gut lactic acid bacteria and bifidobacterial microbiota vary depending on age. Appl Microbiol Biotechnol. 2021;105:8427–8440. doi: 10.1007/s00253-021-11625-z. [DOI] [PubMed] [Google Scholar]
- 11.Kelly SM, Munoz-Munoz J, van Sinderen D. Plant glycan metabolism by Bifidobacteria. Front Microbiol. 2021;12:609418. doi: 10.3389/fmicb.2021.609418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cockburn DW, Koropatkin NM. Polysaccharide degradation by the intestinal microbiota and its influence on human health and disease. J. Mol. Biol. 2016;428:3230–3252. doi: 10.1016/j.jmb.2016.06.021. [DOI] [PubMed] [Google Scholar]
- 13.Nagy, D. U. et al. Effect of chicory-derived inulin-type fructans on abundance of Bifidobacterium and on bowel function: A systematic review with meta-analyses. Crit. Rev. Food Sci. Nutr. 1–18 10.1080/10408398.2022.2098246 (2022). [DOI] [PubMed]
- 14.Rivière A, Selak M, Geirnaert A, Van den Abbeele P, De Vuyst L. Complementary mechanisms for degradation of inulin-type fructans and arabinoxylan oligosaccharides among bifidobacterial strains suggest bacterial cooperation. Appl. Environ. Microbiol. 2018;84:e02893–17. doi: 10.1128/AEM.02893-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Selak M, et al. Inulin-type fructan fermentation by bifidobacteria depends on the strain rather than the species and region in the human intestine. Appl. Microbiol Biotechnol. 2016;100:4097–4107. doi: 10.1007/s00253-016-7351-9. [DOI] [PubMed] [Google Scholar]
- 16.Fujita K, et al. Degradative enzymes for type II arabinogalactan side chains in Bifidobacterium longum subsp. longum. Appl. Microbiol. Biotechnol. 2019;103:1299–1310. doi: 10.1007/s00253-018-9566-4. [DOI] [PubMed] [Google Scholar]
- 17.Li, S., Hu, J., Yao, H., Geng, F. & Nie, S. Interaction between four galactans with different structural characteristics and gut microbiota. Crit. Rev. Food Sci. Nutr. 1–11 10.1080/10408398.2021.1992605 (2021). [DOI] [PubMed]
- 18.Wang Z, et al. Xylan alleviates dietary fiber deprivation-induced dysbiosis by selectively promoting Bifidobacterium pseudocatenulatum in pigs. Microbiome. 2021;9:227. doi: 10.1186/s40168-021-01175-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zeybek N, Rastall RA, Buyukkileci AO. Utilization of xylan-type polysaccharides in co-culture fermentations of Bifidobacterium and Bacteroides species. Carbohydr. Polym. 2020;236:116076. doi: 10.1016/j.carbpol.2020.116076. [DOI] [PubMed] [Google Scholar]
- 20.Arzamasov AA, van Sinderen D, Rodionov DA. Comparative genomics reveals the regulatory complexity of bifidobacterial arabinose and arabino-oligosaccharide utilization. Front Microbiol. 2018;9:776. doi: 10.3389/fmicb.2018.00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ojima MN, et al. Priority effects shape the structure of infant-type Bifidobacterium communities on human milk oligosaccharides. ISME J. 2022;16:2265–2279. doi: 10.1038/s41396-022-01270-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thomson P, Medina DA, Garrido D. Human milk oligosaccharides and infant gut bifidobacteria: Molecular strategies for their utilization. Food Microbiol. 2018;75:37–46. doi: 10.1016/j.fm.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 23.Zúñiga M, Monedero V, Yebra MJ. Utilization of host-derived glycans by intestinal Lactobacillus and Bifidobacterium species. Front Microbiol. 2018;9:1917. doi: 10.3389/fmicb.2018.01917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwalm ND, Groisman EA. Navigating the gut buffet: Control of polysaccharide utilization in Bacteroides spp. Trends Microbiol. 2017;25:1005–1015. doi: 10.1016/j.tim.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 25.Milani C, et al. Genomics of the genus Bifidobacterium reveals species-specific adaptation to the glycan-rich gut environment. Appl Environ. Microbiol. 2016;82:980–991. doi: 10.1128/AEM.03500-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sakanaka M, et al. Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci. Adv. 2019;5:eaaw7696. doi: 10.1126/sciadv.aaw7696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Singh RP. Glycan utilisation system in Bacteroides and Bifidobacteria and their roles in gut stability and health. Appl. Microbiol. Biotechnol. 2019;103:7287–7315. doi: 10.1007/s00253-019-10012-z. [DOI] [PubMed] [Google Scholar]
- 28.Giri S, et al. Metabolic dissimilarity determines the establishment of cross-feeding interactions in bacteria. Curr. Biol. 2021;31:5547–5557.e6. doi: 10.1016/j.cub.2021.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Lawson MAE, et al. Breast milk-derived human milk oligosaccharides promote Bifidobacterium interactions within a single ecosystem. ISME J. 2020;14:635–648. doi: 10.1038/s41396-019-0553-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwab C, et al. Trophic interactions of infant bifidobacteria and Eubacterium hallii during L-fucose and fucosyllactose degradation. Front Microbiol. 2017;8:95. doi: 10.3389/fmicb.2017.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oña L, et al. Obligate cross-feeding expands the metabolic niche of bacteria. Nat. Ecol. Evol. 2021;5:1224–1232. doi: 10.1038/s41559-021-01505-0. [DOI] [PubMed] [Google Scholar]
- 32.Douglas AE. The microbial exometabolome: Ecological resource and architect of microbial communities. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375:20190250. doi: 10.1098/rstb.2019.0250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kan Z, et al. Genotyping and plant-derived glycan utilization analysis of Bifidobacterium strains from mother-infant pairs. BMC Microbiol. 2020;20:277. doi: 10.1186/s12866-020-01962-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Derrien M, Turroni F, Ventura M, van Sinderen D. Insights into endogenous Bifidobacterium species in the human gut microbiota during adulthood. Trends Microbiol. 2022;30:940–947. doi: 10.1016/j.tim.2022.04.004. [DOI] [PubMed] [Google Scholar]
- 35.Li S, You X, Rani A, Özcan E, Sela DA. Bifidobacterium infantis utilizes N-acetylglucosamine-containing human milk oligosaccharides as a nitrogen source. Gut Microbes. 2023;15:2244721. doi: 10.1080/19490976.2023.2244721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Versluis DM, et al. A multiscale spatiotemporal model including a switch from aerobic to anaerobic metabolism reproduces succession in the early infant gut microbiota. mSystems. 2022;7:e0044622. doi: 10.1128/msystems.00446-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matsuki T, et al. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat. Commun. 2016;7:11939. doi: 10.1038/ncomms11939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sela DA, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl Acad. Sci. USA. 2008;105:18964–18969. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dedon LR, Özcan E, Rani A, Sela DA. Bifidobacterium infantis metabolizes 2’fucosyllactose-derived and free fucose through a common catabolic pathway resulting in 1,2-propanediol secretion. Front Nutr. 2020;7:583397. doi: 10.3389/fnut.2020.583397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Arzamasov AA, et al. Human milk oligosaccharide utilization in intestinal bifidobacteria is governed by global transcriptional regulator NagR. mSystems. 2022;7:e0034322. doi: 10.1128/msystems.00343-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Duar RM, et al. Comparative genome analysis of Bifidobacterium longum subsp. infantis strains reveals variation in human milk oligosaccharide utilization genes among commercial probiotics. Nutrients. 2020;12:3247. doi: 10.3390/nu12113247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vatanen T, et al. Genomic variation and strain-specific functional adaptation in the human gut microbiome during early life. Nat. Microbiol. 2019;4:470–479. doi: 10.1038/s41564-018-0321-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blanco G, et al. Revisiting the metabolic capabilities of Bifidobacterium longum subsp. longum and Bifidobacterium longum subsp. infantis from a glycoside hydrolase perspective. Microorganisms. 2020;8:723. doi: 10.3390/microorganisms8050723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vatanen T, et al. A distinct clade of Bifidobacterium longum in the gut of Bangladeshi children thrives during weaning. Cell. 2022;185:4280–4297.e12. doi: 10.1016/j.cell.2022.10.011. [DOI] [PubMed] [Google Scholar]
- 45.Yamada C, et al. Molecular insight into evolution of symbiosis between breast-fed infants and a member of the human gut microbiome Bifidobacterium longum. Cell Chem. Biol. 2017;24:515–524.e5. doi: 10.1016/j.chembiol.2017.03.012. [DOI] [PubMed] [Google Scholar]
- 46.Díaz R, Torres-Miranda A, Orellana G, Garrido D. Comparative genomic analysis of novel Bifidobacterium longum subsp. longum strains reveals functional divergence in the human gut microbiota. Microorganisms. 2021;9:1906. doi: 10.3390/microorganisms9091906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arboleya S, et al. Gene-trait matching across the Bifidobacterium longum pan-genome reveals considerable diversity in carbohydrate catabolism among human infant strains. BMC Genomics. 2018;19:33. doi: 10.1186/s12864-017-4388-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Odamaki T, et al. Genomic diversity and distribution of Bifidobacterium longum subsp. longum across the human lifespan. Sci. Rep. 2018;8:85. doi: 10.1038/s41598-017-18391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Komeno M, Hayamizu H, Fujita K, Ashida H. Two novel α-l-arabinofuranosidases from Bifidobacterium longum subsp. longum belonging to glycoside hydrolase family 43 cooperatively degrade arabinan. Appl Environ. Microbiol. 2019;85:e02582–18. doi: 10.1128/AEM.02582-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komeno M, et al. Two α-L-arabinofuranosidases from Bifidobacterium longum subsp. longum are involved in arabinoxylan utilization. Appl. Microbiol Biotechnol. 2022;106:1957–1965. doi: 10.1007/s00253-022-11845-x. [DOI] [PubMed] [Google Scholar]
- 51.Song A-X, Li L-Q, Yin J-Y, Chiou J-C, Wu J-Y. Mechanistic insights into the structure-dependant and strain-specific utilization of wheat arabinoxylan by Bifidobacterium longum. Carbohydr. Polym. 2020;249:116886. doi: 10.1016/j.carbpol.2020.116886. [DOI] [PubMed] [Google Scholar]
- 52.Sasaki Y, Uchimura Y, Kitahara K, Fujita K. Characterization of a GH36 α-D-galactosidase associated with assimilation of gum arabic in Bifidobacterium longum subsp. longum JCM7052. J. Appl Glycosci. (1999) 2021;68:47–52. doi: 10.5458/jag.jag.JAG-2021_0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang Y, LaPointe G. Arabinogalactan utilization by Bifidobacterium longum subsp. longum NCC 2705 and Bacteroides caccae ATCC 43185 in monoculture and coculture. Microorganisms. 2020;8:1703. doi: 10.3390/microorganisms8111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sasaki Y, et al. Novel 3-O-α-d-Galactosyl-α-l-arabinofuranosidase for the assimilation of gum arabic arabinogalactan protein in Bifidobacterium longum subsp. longum. Appl Environ. Microbiol. 2021;87:e02690–20. doi: 10.1128/AEM.02690-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cordeiro RL, et al. N-glycan utilization by Bifidobacterium gut symbionts involves a specialist β-mannosidase. J. Mol. Biol. 2019;431:732–747. doi: 10.1016/j.jmb.2018.12.017. [DOI] [PubMed] [Google Scholar]
- 56.Cordeiro RL, et al. Mechanism of high-mannose N-glycan breakdown and metabolism by Bifidobacterium longum. Nat. Chem. Biol. 2022;19:281–229. doi: 10.1038/s41589-022-01202-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ojima MN, et al. Ecological and molecular perspectives on responders and non-responders to probiotics and prebiotics. Curr. Opin. Biotechnol. 2022;73:108–120. doi: 10.1016/j.copbio.2021.06.023. [DOI] [PubMed] [Google Scholar]
- 58.Walsh C, Lane JA, van Sinderen D, Hickey RM. Human milk oligosaccharide-sharing by a consortium of infant derived Bifidobacterium species. Sci. Rep. 2022;12:4143. doi: 10.1038/s41598-022-07904-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yokoi T, et al. O-acetylesterase activity of Bifidobacterium bifidum sialidase facilities the liberation of sialic acid and encourages the proliferation of sialic acid scavenging Bifidobacterium breve. Environ. Microbiol Rep. 2022;14:637–645. doi: 10.1111/1758-2229.13083. [DOI] [PubMed] [Google Scholar]
- 60.James K, et al. Metabolism of the predominant human milk oligosaccharide fucosyllactose by an infant gut commensal. Sci. Rep. 2019;9:15427. doi: 10.1038/s41598-019-51901-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Connell Motherway M, et al. Carbohydrate syntrophy enhances the establishment of Bifidobacterium breve UCC2003 in the neonatal gut. Sci. Rep. 2018;8:10627. doi: 10.1038/s41598-018-29034-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bottacini F, et al. Comparative genomics and genotype-phenotype associations in Bifidobacterium breve. Sci. Rep. 2018;8:10633. doi: 10.1038/s41598-018-28919-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ruiz-Moyano S, et al. Variation in consumption of human milk oligosaccharides by infant gut-associated strains of Bifidobacterium breve. Appl Environ. Microbiol. 2013;79:6040–6049. doi: 10.1128/AEM.01843-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sabater C, Ruiz L, Margolles A. A machine learning approach to study glycosidase activities from Bifidobacterium. Microorganisms. 2021;9:1034. doi: 10.3390/microorganisms9051034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Martín R, et al. The infant-derived Bifidobacterium bifidum strain CNCM I-4319 strengthens gut functionality. Microorganisms. 2020;8:1313. doi: 10.3390/microorganisms8091313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gotoh A, et al. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci. Rep. 2018;8:13958. doi: 10.1038/s41598-018-32080-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nishiyama K, et al. Bifidobacterium bifidum extracellular sialidase enhances adhesion to the mucosal surface and supports carbohydrate assimilation. mBio. 2017;8:e00928–17. doi: 10.1128/mBio.00928-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Katoh T, et al. Enzymatic adaptation of Bifidobacterium bifidum to host glycans, viewed from glycoside hydrolyases and carbohydrate-binding modules. Microorganisms. 2020;8:481. doi: 10.3390/microorganisms8040481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tarracchini C, et al. Genetic strategies for sex-biased persistence of gut microbes across human life. Nat. Commun. 2023;14:4220. doi: 10.1038/s41467-023-39931-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Takada H, Katoh T, Sakanaka M, Odamaki T, Katayama T. GH20 and GH84 β-N-acetylglucosaminidases with different linkage specificities underpin mucin O-glycan breakdown capability of Bifidobacterium bifidum. J. Biol. Chem. 2023;299:104781. doi: 10.1016/j.jbc.2023.104781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Katoh T, et al. A bacterial sulfoglycosidase highlights mucin O-glycan breakdown in the gut ecosystem. Nat. Chem. Biol. 2023;19:778–789. doi: 10.1038/s41589-023-01272-y. [DOI] [PubMed] [Google Scholar]
- 72.Jung DH, et al. The presence of resistant starch-degrading amylases in Bifidobacterium adolescentis of the human gut. Int J. Biol. Macromol. 2020;161:389–397. doi: 10.1016/j.ijbiomac.2020.05.235. [DOI] [PubMed] [Google Scholar]
- 73.Kim SY, et al. Enzymatic analysis of truncation mutants of a type II pullulanase from Bifidobacterium adolescentis P2P3, a resistant starch-degrading gut bacterium. Int J. Biol. Macromol. 2021;193:1340–1349. doi: 10.1016/j.ijbiomac.2021.10.193. [DOI] [PubMed] [Google Scholar]
- 74.Argentini C, et al. Ecology- and genome-based identification of the Bifidobacterium adolescentis prototype of the healthy human gut microbiota. Appl Environ. Microbiol. 2024;90:e0201423. doi: 10.1128/aem.02014-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Mulualem DM, et al. Metagenomic identification, purification and characterisation of the Bifidobacterium adolescentis BgaC β-galactosidase. Appl Microbiol Biotechnol. 2021;105:1063–1078. doi: 10.1007/s00253-020-11084-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gao G, et al. BdPUL12 depolymerizes β-mannan-like glycans into mannooligosaccharides and mannose, which serve as carbon sources for Bacteroides dorei and gut probiotics. Int J. Biol. Macromol. 2021;187:664–674. doi: 10.1016/j.ijbiomac.2021.07.172. [DOI] [PubMed] [Google Scholar]
- 77.Mary PR, Monica P, Kapoor M. Insights into β-manno-oligosaccharide uptake and metabolism in Bifidobacterium adolescentis DSMZ 20083 from whole-genome microarray analysis. Microbiol Res. 2023;266:127215. doi: 10.1016/j.micres.2022.127215. [DOI] [PubMed] [Google Scholar]
- 78.Salas-Veizaga DM, Bhattacharya A, Adlercreutz P, Stålbrand H, Karlsson EN. Glucuronosylated and linear xylooligosaccharides from Quinoa stalks xylan as potential prebiotic source for growth of Bifidobacterium adolescentis and Weissella cibaria. LWT. 2021;152:112348. doi: 10.1016/j.lwt.2021.112348. [DOI] [Google Scholar]
- 79.Yang J, et al. Combining of transcriptome and metabolome analyses for understanding the utilization and metabolic pathways of Xylo-oligosaccharide in Bifidobacterium adolescentis ATCC 15703. Food Sci. Nutr. 2019;7:3480–3493. doi: 10.1002/fsn3.1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lin G, et al. The comparative analysis of genomic diversity and genes involved in carbohydrate metabolism of eighty-eight bifidobacterium pseudocatenulatum isolates from different niches of China. Nutrients. 2022;14:2347. doi: 10.3390/nu14112347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shani G, et al. Fucosylated human milk oligosaccharide foraging within the species Bifidobacterium pseudocatenulatum is driven by glycosyl hydrolase content and specificity. Appl Environ. Microbiol. 2022;88:e0170721. doi: 10.1128/AEM.01707-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Watanabe Y, et al. Xylan utilisation promotes adaptation of Bifidobacterium pseudocatenulatum to the human gastrointestinal tract. ISME Commun. 2021;1:62. doi: 10.1038/s43705-021-00066-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sasaki Y, et al. Assimilation of arabinogalactan side chains with novel 3-O-β-L-arabinopyranosyl-α-L-arabinofuranosidase in Bifidobacterium pseudocatenulatum. Microbiome Res Rep. 2023;2:12. doi: 10.20517/mrr.2023.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hosaka H, Kawamura M, Hirano T, Hakamata W, Nishio T. Utilization of sucrose and analog disaccharides by human intestinal bifidobacteria and lactobacilli: Search of the bifidobacteria enzymes involved in the degradation of these disaccharides. Microbiol Res. 2020;240:126558. doi: 10.1016/j.micres.2020.126558. [DOI] [PubMed] [Google Scholar]
- 85.Nagashima M, et al. Growth of Bifidobacterium pseudocatenulatum in medium containing N-acetylsucrosamine: enzyme that induces the growth of this bacterium via degradation of this disaccharide. Glycobiology. 2022;32:540–549. doi: 10.1093/glycob/cwac001. [DOI] [PubMed] [Google Scholar]
- 86.Liu J, Li W, Yao C, Yu J, Zhang H. Comparative genomic analysis revealed genetic divergence between Bifidobacterium catenulatum subspecies present in infant versus adult guts. BMC Microbiol. 2022;22:158. doi: 10.1186/s12866-022-02573-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Orihara K, et al. Characterization of Bifidobacterium kashiwanohense that utilizes both milk- and plant-derived oligosaccharides. Gut Microbes. 2023;15:2207455. doi: 10.1080/19490976.2023.2207455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zengler K, Zaramela LS. The social network of microorganisms - how auxotrophies shape complex communities. Nat. Rev. Microbiol. 2018;16:383–390. doi: 10.1038/s41579-018-0004-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vieira-Silva S, et al. Species-function relationships shape ecological properties of the human gut microbiome. Nat. Microbiol. 2016;1:16088. doi: 10.1038/nmicrobiol.2016.88. [DOI] [PubMed] [Google Scholar]
- 90.Ar P, M, M, D, S Costless metabolic secretions as drivers of interspecies interactions in microbial ecosystems. Nat. commun. 2019;10:103–115. doi: 10.1038/s41467-018-07946-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Fritts RK, McCully AL, McKinlay JB. Extracellular metabolism sets the table for microbial cross-feeding. Microbiol Mol. Biol. Rev. 2021;85:e00135–20. doi: 10.1128/MMBR.00135-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Turroni F, et al. Glycan utilization and cross-feeding activities by Bifidobacteria. Trends Microbiol. 2018;26:339–350. doi: 10.1016/j.tim.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 93.Chia LW, et al. Cross-feeding between Bifidobacterium infantis and Anaerostipes caccae on lactose and human milk oligosaccharides. Benef. Microbes. 2021;12:69–83. doi: 10.3920/BM2020.0005. [DOI] [PubMed] [Google Scholar]
- 94.Bunesova V, Lacroix C, Schwab C. Mucin cross-feeding of infant Bifidobacteria and Eubacterium hallii. Micro Ecol. 2018;75:228–238. doi: 10.1007/s00248-017-1037-4. [DOI] [PubMed] [Google Scholar]
- 95.Cheng L, et al. Effects of different human milk oligosaccharides on growth of bifidobacteria in monoculture and co-culture with Faecalibacterium prausnitzii. Front Microbiol. 2020;11:569700. doi: 10.3389/fmicb.2020.569700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dedon LR, et al. Fucosylated human milk oligosaccharides drive structure-specific syntrophy between bifidobacterium infantis and eubacterium hallii within a modeled infant gut microbiome. Mol. Nutr. Food Res. 2023;67:e2200851. doi: 10.1002/mnfr.202200851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Duranti S, et al. Bifidobacterium bifidum and the infant gut microbiota: An intriguing case of microbe-host co-evolution. Environ. Microbiol. 2019;21:3683–3695. doi: 10.1111/1462-2920.14705. [DOI] [PubMed] [Google Scholar]
- 98.Centanni M, Ferguson SA, Sims IM, Biswas A, Tannock GW. Bifidobacterium bifidum ATCC 15696 and Bifidobacterium breve 24b metabolic interaction based on 2’-O-fucosyl-lactose studied in steady-state cultures in a freter-style chemostat. Appl Environ. Microbiol. 2019;85:e02783–18. doi: 10.1128/AEM.02783-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Nishiyama K, et al. Two extracellular sialidases from Bifidobacterium bifidum promote the degradation of sialyl-oligosaccharides and support the growth of Bifidobacterium breve. Anaerobe. 2018;52:22–28. doi: 10.1016/j.anaerobe.2018.05.007. [DOI] [PubMed] [Google Scholar]
- 100.Chen C, et al. Commensal relationship of three bifidobacterial species leads to increase of Bifidobacterium in vitro fermentation of sialylated immunoglobulin G by human gut microbiota. J. Agric. Food Chem. 2020;68:9110–9119. doi: 10.1021/acs.jafc.0c03628. [DOI] [PubMed] [Google Scholar]
- 101.Egan M, et al. Cross-feeding by Bifidobacterium breve UCC2003 during co-cultivation with Bifidobacterium bifidum PRL2010 in a mucin-based medium. BMC Microbiol. 2014;14:282. doi: 10.1186/s12866-014-0282-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Cheng CC, et al. Ecological importance of cross-feeding of the intermediate metabolite 1,2-propanediol between bacterial gut symbionts. Appl. Environ. Microbiol. 2020;86:e00190–20. doi: 10.1128/AEM.00190-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nogacka AM, Cuesta I, Gueimonde M, de Los Reyes-Gavilán CG. 2-fucosyllactose metabolism by bifidobacteria promotes lactobacilli growth in co-culture. Microorganisms. 2023;11:2659. doi: 10.3390/microorganisms11112659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Belenguer A, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ. Microbiol. 2006;72:3593–3599. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zhao S, Lau R, Zhong Y, Chen M-H. Lactate cross-feeding between Bifidobacterium species and Megasphaera indica contributes to butyrate formation in the human colonic environment. Appl. Environ. Microbiol. 2024;90:e0101923. doi: 10.1128/aem.01019-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kim H, Jeong Y, Kang S, You HJ, Ji GE. Co-culture with bifidobacterium catenulatum improves the growth, gut colonization, and butyrate production of faecalibacterium prausnitzii: In vitro and in vivo studies. Microorganisms. 2020;8:788. doi: 10.3390/microorganisms8050788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Rivière A, Selak M, Lantin D, Leroy F, De Vuyst L. Bifidobacteria and butyrate-producing colon bacteria: Importance and strategies for their stimulation in the human gut. Front Microbiol. 2016;7:979. doi: 10.3389/fmicb.2016.00979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rivière A, Gagnon M, Weckx S, Roy D, De Vuyst L. Mutual cross-feeding interactions between Bifidobacterium longum subsp. longum NCC2705 and Eubacterium rectale ATCC 33656 explain the bifidogenic and butyrogenic effects of arabinoxylan oligosaccharides. Appl. Environ. Microbiol. 2015;81:7767–7781. doi: 10.1128/AEM.02089-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Bhattacharya A, et al. Cross-feeding and enzymatic catabolism for mannan-oligosaccharide utilization by the butyrate-producing gut bacterium Roseburia hominis A2-183. Microorganisms. 2022;10:2496. doi: 10.3390/microorganisms10122496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Moens F, Weckx S, De Vuyst L. Bifidobacterial inulin-type fructan degradation capacity determines cross-feeding interactions between bifidobacteria and Faecalibacterium prausnitzii. Int. J. Food Microbiol. 2016;231:76–85. doi: 10.1016/j.ijfoodmicro.2016.05.015. [DOI] [PubMed] [Google Scholar]
- 111.Centanni M, et al. Bifidobacterium pseudolongum in the ceca of rats fed Hi-Maize starch has characteristics of a keystone species in bifidobacterial blooms. Appl Environ. Microbiol. 2018;84:e00547–18. doi: 10.1128/AEM.00547-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Turroni F, et al. Glycan cross-feeding activities between bifidobacteria under in vitro conditions. Front Microbiol. 2015;6:1030. doi: 10.3389/fmicb.2015.01030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Turroni F, et al. Deciphering bifidobacterial-mediated metabolic interactions and their impact on gut microbiota by a multi-omics approach. ISME J. 2016;10:1656–1668. doi: 10.1038/ismej.2015.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cartmell A, et al. A surface endogalactanase in Bacteroides thetaiotaomicron confers keystone status for arabinogalactan degradation. Nat. Microbiol. 2018;3:1314–1326. doi: 10.1038/s41564-018-0258-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mary PR, Kapoor M. Co-culture fermentations suggest cross-feeding among Bacteroides ovatus DSMZ 1896, Lactiplantibacillus plantarum WCFS1 and Bifidobacterium adolescentis DSMZ 20083 for utilizing dietary galactomannans. Food Res. Int. 2022;162:111942. doi: 10.1016/j.foodres.2022.111942. [DOI] [PubMed] [Google Scholar]
- 116.Rogowski A, et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nat. Commun. 2015;6:7481. doi: 10.1038/ncomms8481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Boger MCL, Lammerts van Bueren A, Dijkhuizen L. Cross-feeding among probiotic bacterial strains on prebiotic inulin involves the extracellular exo-inulinase of Lactobacillus paracasei strain W20. Appl. Environ. Microb. 2018;84:e01539–18. doi: 10.1128/AEM.01539-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Moens F, Verce M, De Vuyst L. Lactate- and acetate-based cross-feeding interactions between selected strains of lactobacilli, bifidobacteria and colon bacteria in the presence of inulin-type fructans. Int. J. Food Microbiol. 2017;241:225–236. doi: 10.1016/j.ijfoodmicro.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 119.Vega-Sagardía M, Delgado J, Ruiz-Moyano S, Garrido D. Proteomic analyses of Bacteroides ovatus and Bifidobacterium longum in xylan bidirectional culture shows sugar cross-feeding interactions. Food Res. Int. 2023;170:113025. doi: 10.1016/j.foodres.2023.113025. [DOI] [PubMed] [Google Scholar]
- 120.Aakko J, et al. A carbohydrate-active enzyme (CAZy) profile links successful metabolic specialization of Prevotella to its abundance in gut microbiota. Sci. Rep. 2020;10:12411. doi: 10.1038/s41598-020-69241-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Leth ML, et al. Differential bacterial capture and transport preferences facilitate co-growth on dietary xylan in the human gut. Nat. Microbiol. 2018;3:570–580. doi: 10.1038/s41564-018-0132-8. [DOI] [PubMed] [Google Scholar]
- 122.Sasaki Y, et al. Mechanism of cooperative degradation of gum arabic arabinogalactan protein by Bifidobacterium longum surface enzymes. Appl Environ. Microbiol. 2022;88:e0218721. doi: 10.1128/aem.02187-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sonnenburg JL, Chen CTL, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biol. 2006;4:2213–2226. doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ioannou A, Knol J, Belzer C. Microbial glycoside hydrolases in the first year of life: An analysis review on their presence and importance in infant gut. Front. Microbiol. 2021;12:1–13. doi: 10.3389/fmicb.2021.631282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chng KR, et al. Metagenome-wide association analysis identifies microbial determinants of post-antibiotic ecological recovery in the gut. Nat. Ecol. Evol. 2020;4:1256–1267. doi: 10.1038/s41559-020-1236-0. [DOI] [PubMed] [Google Scholar]
- 126.Ferrari S, et al. The Role of Bifidobacterium bifidum novaBBF7, Bifidobacterium longum novaBLG2 and Lactobacillus paracasei TJB8 to Improve Mechanisms Linked to Neuronal Cells Protection against Oxidative Condition in a Gut-Brain Axis Model. Int. J. Mol. Sci. 2023;24:12281. doi: 10.3390/ijms241512281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Korpela K, et al. Probiotic supplementation restores normal microbiota composition and function in antibiotic-treated and in caesarean-born infants. Microbiome. 2018;6:182. doi: 10.1186/s40168-018-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Nogacka AM, et al. In vitro probiotic modulation of the intestinal microbiota and 2’fucosyllactose consumption in fecal cultures from infants at two months of age. Microorganisms. 2022;10:318. doi: 10.3390/microorganisms10020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Asnicar F, et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat. Med. 2021;27:321–332. doi: 10.1038/s41591-020-01183-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Shao Y, et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature. 2019;574:117–121. doi: 10.1038/s41586-019-1560-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Henrick BM, et al. Bifidobacteria-mediated immune system imprinting early in life. Cell. 2021;184:3884–3898.e11. doi: 10.1016/j.cell.2021.05.030. [DOI] [PubMed] [Google Scholar]
- 132.Lou YC, et al. Infant microbiome cultivation and metagenomic analysis reveal Bifidobacterium 2’-fucosyllactose utilization can be facilitated by coexisting species. Nat. Commun. 2023;14:7417. doi: 10.1038/s41467-023-43279-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Samara J, et al. Supplementation with a probiotic mixture accelerates gut microbiome maturation and reduces intestinal inflammation in extremely preterm infants. Cell Host Microbe. 2022;30:696–711.e5. doi: 10.1016/j.chom.2022.04.005. [DOI] [PubMed] [Google Scholar]
- 134.Laursen MF, Roager HM. Human milk oligosaccharides modify the strength of priority effects in the Bifidobacterium community assembly during infancy. ISME J. 2023;17:2452–2457. doi: 10.1038/s41396-023-01525-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sprockett D, Fukami T, Relman DA. Role of priority effects in the early-life assembly of the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 2018;15:197–205. doi: 10.1038/nrgastro.2017.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Chang, H.-W. et al. Prevotella copri-related effects of a therapeutic food for malnutrition. bioRxiv 2023.08.11.553030 10.1101/2023.08.11.553030 (2023).
- 137.Michelini S, et al. A reverse metabolic approach to weaning: In silico identification of immune-beneficial infant gut bacteria, mining their metabolism for prebiotic feeds and sourcing these feeds in the natural product space. Microbiome. 2018;6:1–18. doi: 10.1186/s40168-018-0545-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hong L, et al. Synbiotics containing nanoprebiotics: A novel therapeutic strategy to restore gut dysbiosis. Front Microbiol. 2021;12:715241. doi: 10.3389/fmicb.2021.715241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Rubin IMC, et al. Synbiotic intervention with Lactobacilli, Bifidobacteria, and inulin in healthy volunteers increases the abundance of Bifidobacteria but does not alter microbial diversity. Appl. Environ. Microbiol. 2022;88:e0108722. doi: 10.1128/aem.01087-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Button JE, et al. Dosing a synbiotic of human milk oligosaccharides and B. infantis leads to reversible engraftment in healthy adult microbiomes without antibiotics. Cell Host Microbe. 2022;30:712–725. doi: 10.1016/j.chom.2022.04.001. [DOI] [PubMed] [Google Scholar]
- 141.Button JE, et al. Precision modulation of dysbiotic adult microbiomes with a human-milk-derived synbiotic reshapes gut microbial composition and metabolites. Cell Host Microbe. 2023;31:1523–1538.e10. doi: 10.1016/j.chom.2023.08.004. [DOI] [PubMed] [Google Scholar]
- 142.Kanazawa A, et al. Effects of synbiotic supplementation on chronic inflammation and the gut microbiota in obese patients with type 2 diabetes mellitus: A randomized controlled study. Nutrients. 2021;13:558. doi: 10.3390/nu13020558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Gupta VK, et al. A predictive index for health status using species-level gut microbiome profiling. Nat. Commun. 2020;11:4635. doi: 10.1038/s41467-020-18476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wu H, et al. The gut microbiota in prediabetes and diabetes: A population-based cross-sectional study. Cell Metab. 2020;32:379–390.e3. doi: 10.1016/j.cmet.2020.06.011. [DOI] [PubMed] [Google Scholar]
- 145.Perraudeau F, et al. Improvements to postprandial glucose control in subjects with type 2 diabetes: A multicenter, double blind, randomized placebo-controlled trial of a novel probiotic formulation. BMJ Open Diab Res Care. 2020;8:e001319. doi: 10.1136/bmjdrc-2020-001319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sergeev IN, Aljutaily T, Walton G, Huarte E. Effects of synbiotic supplement on human gut microbiota, body composition and weight loss in obesity. Nutrients. 2020;12:222. doi: 10.3390/nu12010222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Chaiyasut C, et al. Synbiotic supplementation improves obesity index and metabolic biomarkers in thai obese adults: A randomized clinical trial. Foods. 2021;10:1580. doi: 10.3390/foods10071580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Nguyen NK, et al. Gut microbiota modulation with long-chain corn bran arabinoxylan in adults with overweight and obesity is linked to an individualized temporal increase in fecal propionate. Microbiome. 2020;8:118. doi: 10.1186/s40168-020-00887-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Özcan E, Sela DA. Inefficient metabolism of the human milk oligosaccharides lacto-n-tetraose and lacto-n-neotetraose shifts Bifidobacterium longum subsp. infantis physiology. Front Nutr. 2018;5:46. doi: 10.3389/fnut.2018.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zabel BE, et al. Strain-specific strategies of 2’-fucosyllactose, 3-fucosyllactose, and difucosyllactose assimilation by Bifidobacterium longum subsp. infantis Bi-26 and ATCC 15697. Sci. Rep. 2020;10:15919. doi: 10.1038/s41598-020-72792-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Zabel B, et al. Novel genes and metabolite trends in Bifidobacterium longum subsp. infantis Bi-26 metabolism of human milk oligosaccharide 2’-fucosyllactose. Sci. Rep. 2019;9:7983. doi: 10.1038/s41598-019-43780-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Frese SA, et al. N-glycans from human milk glycoproteins are selectively released by B. infantis EVC001 in vivo. Pediatrics. 2020;146:140. doi: 10.1542/peds.146.1MA2.140a. [DOI] [Google Scholar]
- 153.Hirano R, et al. Next-generation prebiotic promotes selective growth of bifidobacteria, suppressing Clostridioides difficile. Gut Microbes. 2021;13:1973835. doi: 10.1080/19490976.2021.1973835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Yoshida K, et al. Bifidobacterium response to lactulose ingestion in the gut relies on a solute-binding protein-dependent ABC transporter. Commun. Biol. 2021;4:541. doi: 10.1038/s42003-021-02072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kelly SM, O’Callaghan J, Kinsella M, van Sinderen D. Characterisation of a hydroxycinnamic acid esterase from the Bifidobacterium longum subsp. longum taxon. Front Microbiol. 2018;9:2690. doi: 10.3389/fmicb.2018.02690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.James K, O’Connell Motherway M, Penno C, O’Brien RL, van Sinderen D. Bifidobacterium breve UCC2003 employs multiple transcriptional regulators to control metabolism of particular human milk oligosaccharides. Appl Environ. Microbiol. 2018;84:e02774–17. doi: 10.1128/AEM.02774-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ruiz-Aceituno L, Esteban-Torres M, James K, Moreno FJ, van Sinderen D. Metabolism of biosynthetic oligosaccharides by human-derived Bifidobacterium breve UCC2003 and Bifidobacterium longum NCIMB 8809. Int J. Food Microbiol. 2020;316:108476. doi: 10.1016/j.ijfoodmicro.2019.108476. [DOI] [PubMed] [Google Scholar]
- 158.Böger M, van Leeuwen SS, Lammerts van Bueren A, Dijkhuizen L. Structural identity of galactooligosaccharide molecules selectively utilized by single cultures of probiotic bacterial strains. J. Agric. Food Chem. 2019;67:13969–13977. doi: 10.1021/acs.jafc.9b05968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bhattacharya A, et al. Enzyme synergy for the production of arabinoxylo-oligosaccharides from highly substituted arabinoxylan and evaluation of their prebiotic potential. LWT. 2020;131:109762. doi: 10.1016/j.lwt.2020.109762. [DOI] [Google Scholar]
- 160.Heiss BE, et al. Bifidobacterium catabolism of human milk oligosaccharides overrides endogenous competitive exclusion driving colonization and protection. Gut Microbes. 2021;13:1986666. doi: 10.1080/19490976.2021.1986666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Drey E, Kok CR, Hutkins R. Role of Bifidobacterium pseudocatenulatum in degradation and consumption of xylan-derived carbohydrates. Appl. Environ. Microbiol. 2022;88:e0129922. doi: 10.1128/aem.01299-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Saito Y, et al. Multiple transporters and glycoside hydrolases are involved in arabinoxylan-derived oligosaccharide utilization in Bifidobacterium pseudocatenulatum. Appl. Environ. Microbiol. 2020;86:e01782–20. doi: 10.1128/AEM.01782-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Egan M, O’Connell Motherway M, Ventura M, van Sinderen D. Metabolism of sialic acid by Bifidobacterium breve UCC2003. Appl. Environ. Microbiol. 2014;80:4414–4426. doi: 10.1128/AEM.01114-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Falony G, Vlachou A, Verbrugghe K, Vuyst LD. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl. Environ. Microbiol. 2006;72:7835–7841. doi: 10.1128/AEM.01296-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Rios-Covian D, Gueimonde M, Duncan SH, Flint HJ, de los Reyes-Gavilan CG. Enhanced butyrate formation by cross-feeding between Faecalibacterium prausnitzii and Bifidobacterium adolescentis. FEMS Microbiol Lett. 2015;362:fnv176. doi: 10.1093/femsle/fnv176. [DOI] [PubMed] [Google Scholar]
- 166.Munoz J, James K, Bottacini F, Van Sinderen D. Biochemical analysis of cross-feeding behaviour between two common gut commensals when cultivated on plant-derived arabinogalactan. Micro Biotechnol. 2020;13:1733–1747. doi: 10.1111/1751-7915.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]