Abstract
For most membrane proteins, the transmembrane domain (TMD) is more than just an anchor to the membrane. The TMDs of hepatitis C virus (HCV) envelope proteins E1 and E2 are extreme examples of the multifunctionality of such membrane-spanning sequences. Indeed, they possess a signal sequence function in their C-terminal half, play a major role in endoplasmic reticulum localization of E1 and E2, and are potentially involved in the assembly of these envelope proteins. These multiple functions are supposed to be essential for the formation of the viral envelope. As for the other viruses of the family Flaviviridae, these anchor domains are composed of two stretches of hydrophobic residues separated by a short segment containing at least one fully conserved charged residue. Replacement of these charged residues by an alanine in HCV envelope proteins led to an alteration of all of the functions performed by their TMDs, indicating that these functions are tightly linked together. These data suggest that the charged residues of the TMDs of HCV glycoproteins play a key role in the formation of the viral envelope.
The rough endoplasmic reticulum (ER) is the site of eukaryotic membrane protein synthesis. The majority of the transmembrane (TM) proteins are synthesized on membrane-bound ribosomes and are cotranslationally integrated into the ER membranes. These proteins are endowed with an ER targeting signal. Upon arrival at the ER membrane, such a signal can either be cleaved by a signal peptidase or remain attached and function as a TM anchor (62, 74). A TM protein with a cleavable signal sequence has a second signal, the stop-transfer sequence, which functions as an anchor domain. This membrane-spanning sequence is supposed to fold as an α-helix, which can be considered as an autonomous folding domain (54). Until recently, the membrane-spanning domain was mainly thought of as a stretch of hydrophobic residues, whose only function was to anchor the protein in the lipid bilayer. However, the primary structures of TM domains (TMDs) of various proteins exhibit strikingly high levels of conservation among different species. Such a high degree of conservation makes it likely that membrane-spanning domains contain important information, not only in terms of hydrophobicity. Indeed, the observation that the TMDs of Golgi resident proteins sufficed to localize reporter molecules to the Golgi apparatus (38) suggested other functions for TMDs. This observation was extended to include residents of the ER (53, 68, 75, 76), and together these results argue for a role of the TMD in protein sorting. The presence of one or several hydrophilic residues within the hydrophobic TM sequence of proteins has been reported to ensure their ER retention (2, 36, 58). TMDs can also play a crucial role in oligomerization. For instance, specific interactions between TMDs are essential for the assembly of class II major histocompatibility complex molecules (10), T-cell receptor (11), and glycophorin A molecules (35). TMDs have also been proposed to play a direct role in signal transduction (42). Therefore, structure-function studies of TMDs is of major interest to better understand biological events such as the biogenesis of a viral envelope.
Hepatitis C virus (HCV) is the causal agent of hepatitis C, which is a major health problem worldwide (30). HCV is a positive-stranded RNA virus which belongs to the family Flaviviridae (24). Its genome encodes a single polyprotein of 3,008 to 3,037 amino acid residues (6) that is co- and posttranslationally cleaved to generate at least 10 polypeptides (C, E1, E2, p7, NS2, NS3, NS4A, NS4B, NS5A, and NS5B) (59). The two HCV envelope proteins, E1 and E2, are obtained after cleavage of the polyprotein by host signal peptidase(s) (56). HCV glycoproteins E1 and E2 are heavily modified by N-linked glycosylation and are TM glycoproteins with a large N-terminal ectodomain and a C-terminal hydrophobic anchor (18, 55). The C-terminal halves of the TM sequences of E1 and E2 are supposed to play the role of signal sequences for E2 and the p7 polypeptide, respectively (56).
HCV glycoproteins assemble to form a noncovalent heterodimer which is retained in the ER (14). This heterodimer is believed to be the prebudding form of an HCV glycoprotein complex (18). Recently, the TMDs of E1 and E2 have been shown to retain a reporter protein in the ER compartment, indicating that these domains play a major role in the subcellular localization of HCV glycoprotein complex (7, 8). In addition, as recently demonstrated, the glycans of the reporter protein were not processed by Golgi enzymes, indicating that the TMDs of E1 and E2 are responsible for true retention in the ER, without recycling through the Golgi apparatus (7, 21). The TMDs of HCV glycoproteins have also been suggested to play a role in the assembly of the E1E2 heterodimer. Indeed, deletion of the C-terminal hydrophobic sequence of E2 (47, 63) or its replacement by the membrane anchor of CD4 or a glycosyl phosphatidylinositol moiety (8) has been shown to abolish the formation of E1E2 heterodimer.
The TMDs of HCV envelope proteins are multifunctional. Indeed, besides their role as membrane anchors, the putative TMDs of both HCV glycoproteins possess a signal sequence function and play a major role in subcellular localization and potentially in assembly of these envelope proteins. Here we report that all of these functions can be disrupted by mutating charged residues present in these TMDs. In addition, comparative sequence analysis of TMDs of envelope proteins of other Flaviviridae viruses reveals common features for all of the members of this family.
MATERIALS AND METHODS
Cell culture.
The HepG2, CV-1, and 143B (thymidine kinase deficient) cell lines were obtained from the American Type Culture Collection, Rockville, Md. Cell monolayers were grown in Dulbecco's modified essential medium supplemented with 10% fetal bovine serum.
Construction of mutant and chimeric proteins.
Site-directed mutagenesis was performed by enzymatic inverse PCR as described by Stemmer and Morris (70). HCV sequences were amplified from clones derived from the H strain (22, 23). Amino acid residues modified by site-directed mutagenesis were all replaced by an Ala residue. The following oligonucleotides were used to generate the mutant proteins E2D (5′-TCCTGGTCTCTGCAGCCGCGCGCGTCTGCTCCTGCTTGTGGATG-3′ [Asp728 mutant]), E2R (5′-TCCTGGTCTCTGCAGACGCGGCCGTCTGCTCCTGCTTGTGGATGATG-3′ [Arg730 mutant]), E2DR (5′-TCC TGG TC TC TGCAGCCGCGGCCG TC TGC TCC TGCT TG TGGATGATG-3′ [double mutant Asp728-Arg730]), E1K (5′-TCCTCGTCTCTGGGGAACTGGGCGGCGGTCCTGGTAGTGCTGCTGCTATTTGCCGGC-3′ [Lys370]), and E1NK (5′-TCCTCGTCTCTGGGGGCCTGGGCGGCGGTCCTGGTAGTGCTGCTGCTATTTGCCGGC-3′ [double mutant Asn367-Lys370]). Boldface letters indicate the mutations introduced in the HCV sequence. The fragments containing the mutations in E2 were introduced into pTM1/E2 (a plasmid containing the entire sequence of E2 with its signal sequence, corresponding to amino acids 371 to 746 on the HCV polyprotein) generating pTM1/E2D, pTM1/E2R, and pTM1/E2DR. The fragments containing the mutations in E1 were introduced into pTM1/E1 (a plasmid containing the entire sequence of E1 with its signal sequence, corresponding to amino acids 171 to 383 on HCV polyprotein) and pTM1/E1E2p7 (a plasmid containing the end of the sequence of the capsid protein; the signal sequence of E1; and the entire sequences of E1, E2, and p7, corresponding to amino acids 132 to 809 on HCV polyprotein) (23), generating pTM1/E1NK and pTM1/E1NKE2p7, respectively. The E1 sequence of pTM1/E1NKE2p7 was introduced into pTM1/E2, generating pTM1/E1NKE2. Plasmids pTM1/E2E1 and pTM1/E2DRE1 were obtained by fusing the 3′ end sequence of E2 (wild type or mutated) and the 5′ end sequence of E1 by enzymatic inverse PCR (69). These plasmids contain the sequence of E2 and its signal sequence (residues 370 to 746) in fusion with the sequence of E1 (residues 192 to 383).
Plasmids pTM1/CD4-E1K, pTM1/CD4-E1NK, and pTM1/CD4-E2DR contain the signal sequence and the ectodomain of CD4 (amino acids 1 to 371) in fusion with the mutated TMD of E1 (amino acids 353 to 383) or E2 (amino acids 718 to 746). These plasmids were obtained by introducing the mutated E1 or E2 fragment into pTM1/CD4(1–371) plasmid which contains the sequence of the ectodomain of CD4. pTM1/CD4-E1 and pTM1/CD4-E2 contain the signal sequence of CD4 and its ectodomain (amino acids 1 to 371) in fusion with the C-terminal 31 and 29 amino acids of E1 (amino acids 353 to 383) and E2 (amino acids 718 to 746), respectively. Between these sequences, there is a junction sequence encoding two additional amino acids (Gly and Ser). Plasmid pTM1/CD4-E1370 contains the signal sequence and the ectodomain of CD4 in fusion with the first 18 amino acids of the TMD of E1 (amino acids 353 to 370). Plasmid pTM1/E2730 contains the sequence of E2 from which the last 16 amino acids (amino acids 731 to 746) have been deleted. Sequences of DNA fragments obtained by PCR amplification were verified by sequencing.
Generation and growth of viruses.
Vaccinia virus recombinants were generated by homologous recombination essentially as described previously (31) and plaque purified twice on 143B cells under bromodeoxyuridine selection (50 μg/ml). Stocks of vaccinia virus recombinants were grown and titrated on CV-1 monolayers. The following vaccinia virus recombinants have been described previously: vTF7-3 (expressing the T7 DNA-dependent RNA polymerase) (25), vE1E2p7 (expressing HCV glycoproteins E1 and E2 and the p7 polypeptide) (23), vCE1 (expressing HCV proteins C and E1) (47), vCD4-E1 (expressing the ectodomain of CD4 in fusion with the sequence of the TMD of E1) (7), vE2717-CD4 (expressing the ectodomain of E2 in fusion with the membrane anchor signal of CD4), vCD4 (expressing the full-length CD4), and vCD4-E2 (expressing the ectodomain of CD4 in fusion with the TMD of E2) (8).
Antibodies.
Rabbit anti-mouse immunoglobulin G (IgG) was purchased from DAKO (Copenhagen, Denmark). Rhodamine-conjugated donkey anti-mouse IgG was obtained from Jackson Immunoresearch (West Grove, Pa.). Monoclonal antibodies (MAbs) A4 and A11 (anti-E1 [19]), H53 (anti-E2 [8]), A11 (anti-E2 [19]), and OKT4 (anti-CD4 [57]) were produced in vitro by using a MiniPerm apparatus (Heraeus) as recommended by the manufacturer.
Metabolic labeling, immunoprecipitation, and endoglycosidase digestions.
Cells expressing HCV proteins were metabolically labeled with 35S-Protein Labeling mix (3.7 × 106 Bq/ml) as described previously (19). Cells were lysed with 0.5% Igepal CA-630 in Tris-buffered saline (TBS; 50 mM Tris-Cl [pH 7.5], 150 mM NaCl). Immunoprecipitations were carried out as described previously (20). For endoglycosidase digestion, immunoprecipitated proteins were eluted from protein A Sepharose in 30 μl of dissociation buffer (0.5% sodium dodecyl sulfate [SDS], 1% 2-mercaptoethanol) by boiling for 10 min. The protein samples were then divided into equal portions for digestion with either endo-β-N-acetylglucosaminidase H (endo H) or peptide–N-glycosidase F (PNGase F) and an undigested control. Digestions were carried out for 1 h at 37°C in the buffer provided by the manufacturer. Digested samples were mixed with an equal volume of 2× Laemmli sample buffer and analyzed by SDS polyacrylamide gel electrophoresis (PAGE).
Immunofluorescence.
Subconfluent HepG2 cells grown on coverslips were infected by the appropriate vaccinia virus recombinants at a multiplicity of infection of 3 PFU/cell. At 8 h postinfection, cells were fixed for 10 min with paraformaldehyde (4% in phosphate-buffered saline). Cells were then permeabilized or not for 30 min at room temperature with TBS containing 0.1% Triton X-100. The expression of the proteins of interest was revealed with MAb H53 (anti-E2; dilution 1/600) or OKT4 (anti-CD4; dilution, 1/100) followed by rabbit anti-mouse (rhodamine conjugated) Ig (dilution, 1/100).
Western blotting.
Proteins bound to nitrocellulose membranes were revealed by enhanced chemiluminescence detection (ECL system) as recommended by the manufacturer (Amersham). Briefly, after separation by SDS-PAGE under reducing conditions, proteins were transferred to nitrocellulose membranes by using a Trans-Blot apparatus (Bio-Rad) and revealed with specific MAbs (A4 or A11; dilution of 1/250) followed by rabbit anti-mouse Ig conjugated to peroxidase (dilution, 1/1,000).
Sequence analysis.
The protein sequence of the putative TMDs of envelope proteins from each virus strain (identified by its EMBL accession number [see the legend to Fig. 9]) was used to search all variants in the EMBL database by using the FASTA homology search program (52). Incomplete TMD sequences were removed from the list of matching sequences. Multiple amino acid sequence alignments and consensus sequences were carried out with CLUSTAL W program (71). Visualization of the most represented amino acid at each position was done with the MPSA program (C. Blanchet et al., unpublished data). All of the analyses were made by using the Network Protein Sequence @nalysis facilities (NPS@) through the IBCP server (http://pbil.ibcp.fr/NPSA). Prediction of minimal membrane segments was deduced from dense alignment surface (DAS) analysis (12) (http://www.biokemi.su.se/∼server/DAS).
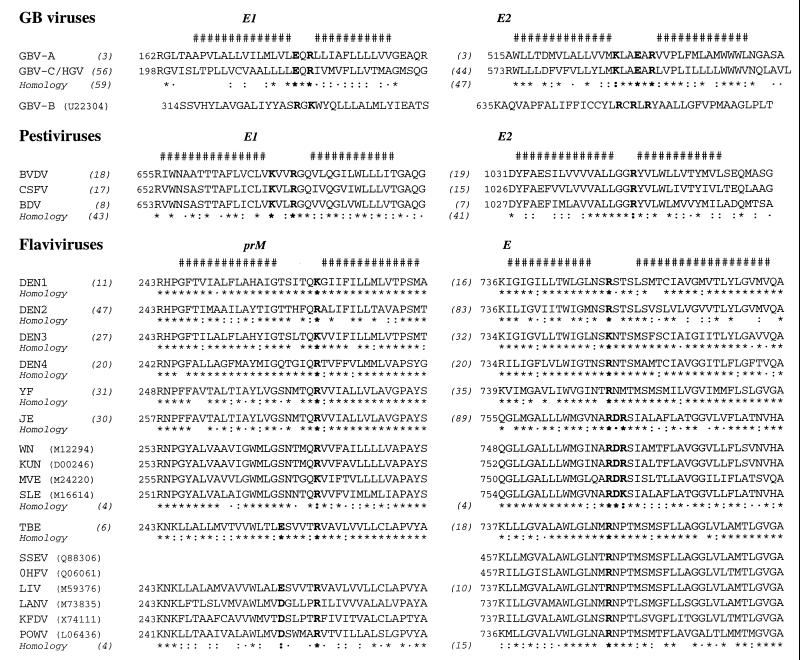
FIG. 9.
Comparison and conservation of amino acid sequences of the putative TMD of envelope proteins in the Flaviviridae family. The consensus sequence of most represented amino acid was deduced from the sequence analysis of natural variants available in the EMBL database as described in Materials and Methods. The number of variants analyzed is indicated in parentheses, and the amino acid homology at each position is symbolized as follows: asterisk, fully conserved; colon, conserved; dot, similar. The consensus of putative minimal TM segments is shown at the top (#). Conserved charge residues located between the two stretches of hydrophobic residues are in boldface. The amino acid numbering refers to the EMBL database sequence accession number when indicated. For consensus sequences, numbering is according to the sequences for which the EMBL accession number is indicated (in parentheses) in the following references: GBV-A and GBV-B viruses (49, 67); GBV-C/HGV virus (U44402) (34, 37); BVDV, bovine viral diarrhea virus (M96687) (15); CSFV, classical swine fever virus (J04358) (61); BDV, border disease virus (AF037405) (1); DEN1, dengue virus type 1 (M23027) (45); DEN2 (M20558) (16); DEN3 (M93130) (51); DEN4 (M14931) (77); YF, yellow fever virus (X03700) (60); JE, Japanese encephalitis virus (M55506) (50); WN, West Nile virus (5); KUN, Kunjin virus (9); MVE, Murray Valley encephalitis virus (13); SLE, St. Louis encephalitis virus (72); TBE, tick-borne encephalitis virus (M27157) (39); SSEV, Spanish sheep encephalitis virus (43); OHFV, Omsk hemorrhagic fever virus (28); LIV, Louping ill virus (M59376) (65); LANV, Langat virus (41); KFDV, Kyasanur Forest disease virus (73); POWV, tick-borne Powassan virus (40). Note that no prM sequence is available for OHFV or SSEV.
RESULTS
Organization of the TMDs of HCV glycoproteins.
Some RNA viruses synthesize their polypeptides as a polyprotein which is cleaved co- or posttranslationally by viral and/or host proteases. As exemplified by the alphaviruses (27), the fact that the envelope proteins of this type of virus are translated from a single coding region implies that internal signal peptides must be used. In the case of HCV glycoproteins, the signal sequences of E1 and E2 are present at the C terminus of the immature form of the capsid protein and in the second half of the TMD of E1, respectively, and a hydrophobic sequence present in the second half of the TMD of E2 is believed to be the signal sequence for a polypeptide called p7 (56) (Fig. 1A).
FIG. 1.
Organization of the TMDs of HCV glycoproteins. (A) Processing of the N-terminal region of HCV polyprotein generating the envelope proteins E1 and E2. The positions of the C-terminal residues of C, E1, and E2 are indicated at the top. (B) Amino acid variability of the putative TMDs of HCV glycoproteins E1 and E2. The consensus patterns of 340 and 133 natural variants of E1 and E2, respectively, are presented. The observed residues at each position are indicated in decreasing order of frequency from top to bottom. The less frequently observed residues (<5%) are not presented. The positions of the amino acid residues indicated above the consensus sequences correspond to their position in the polyprotein. Fully conserved, conserved, and similar residues at each position are symbolized by an asterisk, colon, and dot, respectively. The fully conserved Lys370 residue in E1 and Asp728 and Arg730 residues in E2 which have been mutated in this study are shown in boldface. #, prediction of minimal membrane segments deduced from dense alignment surface (DAS) analysis (12).
The TMD of E1, which has been proposed to start at position 353 (7), is composed of two hydrophobic stretches connected by a short hydrophilic segment (367 to 370) (Fig. 1B). Examination of 133 sequences of natural variants reported in the protein data banks shows that the two hydrophobic stretches exhibit some variability, although most of the changes are conservative and should not modify the conformation of these structural elements. Nevertheless, this variability contrasts with the high conservation of the connecting segment which contains a fully conserved Lys residue at position 370. The presence of an Asn residue at position 367 should also be mentioned, although Ala is observed in 14% of natural variants. The TMD of E2 is 29 amino acids in length (amino acids 718 through 746 on the polyprotein [48]) and presents a sequence organization similar to that of the TMD of E1. It is worth noting that the two charged residues of the connecting segment (Asp728 and Arg730) are fully conserved and the two surrounding hydrophobic stretches are highly conserved over the 340 sequences of natural variants. In conclusion, the full conservation of the charged residues present in the TMDs of E1 and E2 suggests that their presence is essential for the functions played by these TMDs.
Stable membrane integration of HCV glycoproteins necessitates the presence of their whole TMD.
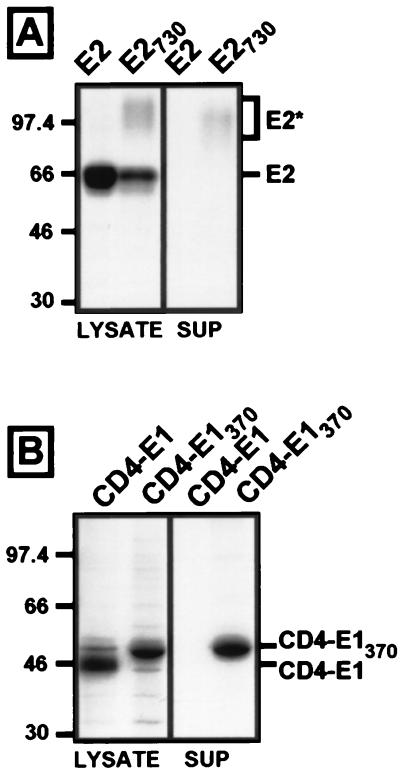
Since the second stretch of hydrophobic residues of the TMDs of E1 and E2 is supposed to act as a signal sequence (56), it is reasonable to think that the first stretch should be a stop-transfer signal. In addition, to play both functions of stop-transfer and signal sequence, the TMDs of E1 and E2 are expected to cross the ER membrane twice, at least transiently. To analyze whether the first stretch of hydrophobic residues can act as a true stop-transfer sequence, proteins with the second stretch deleted were produced. As shown in Fig. 2A, deletion of the second stretch of hydrophobic residues at the C terminus of E2 led to the appearance of a second band (E2*) when expressed by a vaccinia virus recombinant in HepG2 cells (E2730, lysate). In addition, this deletion led to a partial secretion of E2 (Fig. 2A, E2730, supernatant). Indeed, as quantified by phosphorimaging, about 27% of E2730 was detected in the supernatant, whereas no secretion was observed in the absence of any deletion. It is worth noting that the E2* band, detected in the supernatant and the cell lysate (Fig. 2A, E2730), is more diffuse and has a slower electrophoretic mobility. This is due to processing of the glycans, as previously observed after endo H treatment when the TMD of E2 is deleted (data not shown) (47). A similar approach was developed to analyze the role of the second stretch of hydrophobic residues present in the TMD of E1. However, since it has been previously shown that E1 does not fold properly in the absence of E2 (47), deletion of the second hydrophobic stretch was done in the context of a chimeric protein made of the ectodomain of CD4 in fusion with the TMD of E1 (CD4-E1) (7). As shown in Fig. 2B, CD4-E1 with the second stretch of hydrophobic residues (CD4-E1370) deleted and as expressed by a vaccinia virus recombinant was released in the supernatant of HepG2 cells. About 70% of CD4-E1370 was detected in the supernatant, whereas no secretion was detected in the absence of any deletion of CD4-E1. It is worth noting that CD4-E1370 had a slower electrophoretic mobility than CD4-E1 (Fig. 2B). This might be due to processing of the glycans of CD4-E1370. However, the difference in the electrophoretic mobilities of CD4-E1 and CD4-E1370 was already observed after a short pulse (data not shown), suggesting that this might not be the case. Abnormal electrophoretic mobility has been previously reported for E1 molecules with their TMD truncated (23). It is likely that deletions in the TMD of CD4-E1 have the same effect as that observed for E1.
FIG. 2.
The first hydrophobic stretch of the TMDs of HCV glycoproteins is not sufficient for an efficient arrest of translocation. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. At 4.5 h postinfection, cells were pulse-labeled for 10 min and chased for 4 h. Cell lysates and supernatants (sup) were immunoprecipitated with MAb H53 (E2 and E2730) (A) or OKT4 (CD4-E1 and CD4-E1370) (B). Proteins were separated by SDS-PAGE (10% polyacrylamide). The sizes (in kilodaltons) of protein molecular mass markers are indicated on the left. The slow-migrating form of E2 is indicated by an asterisk.
Taken together, these data indicate that the first stretch of hydrophobic residues present at the C terminus of HCV glycoproteins E1 and E2 is not sufficient for an efficient membrane anchorage. This suggests that stable membrane integration of these proteins necessitates the presence of their whole TMD.
Charged residues located between the two stretches of hydrophobic residues play a major role in ER retention.
As described above, charged residues (Lys370 for E1 and Asp728 and Arg730 for E2) present in the TMDs of HCV envelope proteins are fully conserved (Fig. 1B). In addition, their location between two stretches of hydrophobic residues suggests that they might play a major role in some of the functions played by the TMDs of HCV glycoproteins. Since these TM sequences act as ER retention signals (7, 8), we suspected that the charged residues might play an important role in the ER localization of HCV glycoproteins. Indeed, the presence of a charged residue(s) within the hydrophobic TM sequence of proteins has been reported to ensure their ER retention (2).
To test this hypothesis, the charged residues located between the two stretches of hydrophobic residues were mutated into alanine. This residue was chosen because it is well recognized that it has potentially minimal effects on local protein conformation when replacing another residue. Mutants of E2 obtained by replacing Asp728 and/or Arg730 by Ala were named E2D, E2R, and E2DR, respectively. The E2-CD4 chimera, in which the TMD of E2 has been replaced by the membrane anchor of CD4 (8), was used as a control of export out of the ER. Mutated proteins expressed by vaccinia virus recombinants were analyzed by SDS-PAGE. As shown in pulse-chase experiments, two bands were detected for the mutated proteins (E2DR and E2-CD4) after immunoprecipitation with the conformation-sensitive anti-E2 MAb H53 (Fig. 3, four left lanes). The fast-migrating band of the mutated proteins had the same estimated molecular mass as wild-type E2. Due to the slow folding of E2 and because we used a conformation-sensitive MAb, the intensity of E2 was lower during the pulse as previously reported (8, 21). The slow-migrating band (E2*), observed for the mutants and not detected for wild-type E2, started to be detected after 1 h of chase for E2-CD4 and after 2 h of chase for E2DR. The intensity of this band was higher for E2-CD4 and E2DR (Fig. 3) than for E2D and E2R (data not shown). Since the slow-migrating band was observed later during the chase, we suspected that it could correspond to molecules having their glycans processed by Golgi enzymes and which are no longer retained in the ER. Indeed, the slow-migrating bands of E2-CD4 and E2DR showed some resistance to endo H treatment (Fig. 3, E2R right lanes), suggesting that they have moved out of the ER. The E2R band had a lower intensity and was more diffuse than the slow-migrating band observed before endo H treatment (E2*). This might be due to the heterogeneity in the sensitivity to endo H treatment of some glycans. It is worth noting that, even after 4 h of chase, endo H-sensitive forms were still observed (E2S). This is probably due to the slow folding of E2, as previously suggested for E2-CD4 (8). It should be noted that a small portion of E2DR was secreted. This is likely due to the absence of E1, which has been shown to stabilize the membrane insertion of E2 (L. Cocquerel, J.-C. Meunier, A. Op De Beeck, C. Wychowski, and J. Dubuisson, unpublished data). To confirm the role of Asp728 and Arg730 in the TMD of E2, the same mutations were introduced in the context of a chimeric molecule corresponding to the ectodomain of CD4 in fusion with the TMD of E2. This mutant (CD4-E2DR) and its nonmutated form (CD4-E2) and the wild-type CD4 molecule were expressed by vaccinia virus recombinants, and their sensitivity to endo H treatment was analyzed in pulse-chase experiments (Fig. 4). The CD4 protein contains two N-linked glycans, and only one of them becomes endo H resistant (64). The wild-type CD4 molecule acquired endo H resistance after 1 h of chase, whereas CD4-E2 remained endo H sensitive even after 4 h of chase, as previously observed (8). Similarly to what was observed for CD4, the majority of CD4-E2DR molecules became endo H resistant after 1 h of chase. These data confirm that the ER retention is abolished when Asp728 and Arg730 are both replaced by Ala residues.
FIG. 3.
Analysis of the endo H sensitivity of E2, E2DR, and E2-CD4 glycoproteins. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. At 4.5 h postinfection, cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb H53 and then treated (+) or not (−) with endo H. Proteins were separated by SDS-PAGE (10% polyacrylamide) under reducing conditions. E2*, slow-migrating form of E2; E2R, endo H-resistant form of E2; E2S, deglycosylated form of E2.
FIG. 4.
Role of charged residues in ER retention mediated by the TMD of HCV glycoproteins. Chimeras made of the ectodomain of CD4 in fusion with wild-type or mutated TMD of HCV glycoproteins were expressed by vaccinia virus recombinants. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 5 PFU/cell. At 4.5 h postinfection, cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb OKT4 and then treated (+) or not (−) with endo H. Proteins were separated by SDS-PAGE (10% polyacrylamide) under reducing conditions. Deglycosylated proteins are indicated by an asterisk.
To evaluate the role of the charged residue located in the TMD of E1, the same strategy as that developed for E2 was used. However, as mentioned above, E1 does not fold properly in the absence of E2 (47), so the mutations were introduced in the context of a chimeric protein made of the ectodomain of CD4 in fusion with the TMD of E1 (CD4-E1) (7). The Lys residue at position 370 was replaced by Ala (CD4-E1K). In addition, in order to analyze the effect of Lys370, the Asn at position 367 was also replaced by an Ala (CD4-E1NK). Indeed, Asn is a polar residue which might also play a role in ER retention, but its replacement by Ala in 14% of E1 HCV variants (Fig. 1B) suggests only a secondary role for Asn367. These mutants (CD4-E1K and CD4-E1NK), as well as CD4-E1, were expressed by vaccinia virus recombinants, and their sensitivity to endo H treatment was analyzed in pulse-chase experiments. As shown previously (7), CD4-E1 remained endo H sensitive even after 4 h of chase (Fig. 4). In contrast, an endo H-resistant form started to be detected after 1 h of chase for CD4-E1NK. Similar results were obtained with CD4-E1K (data not shown), indicating that Lys370 is required for ER retention. It is worth noting that no secretion of CD4-E1NK was observed.
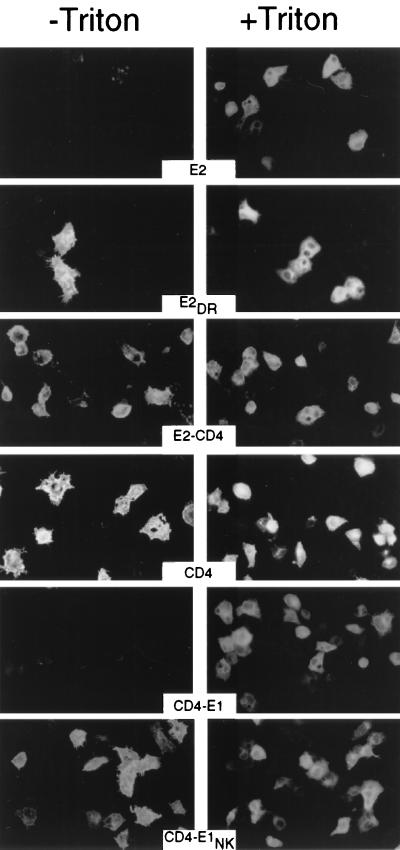
Since our data showed that mutations of charged residues in the TMDs of E1 and E2 abolished the ER retention, we wanted to know whether these mutated molecules would reach the cell surface. For this purpose, nonpermeabilized cells expressing these mutated proteins were analyzed by immunofluorescence. Control Triton X-100-permeabilized cells were analyzed in parallel. In contrast to what was observed for E2, cells expressing E2DR or E2-CD4 were labeled in the absence of permeabilization (Fig. 5). Similarly, cells expressing CD4 or CD4-E1NK were detectable without detergent treatment, whereas those expressing CD4-E1 were only detected after permeabilization. The few spots of fluorescence observed in nonpermeabilized cells expressing E2 or CD4-E1 correspond to labeling of dead cells. These data indicate that mutations of charged residues in the TMDs of E1 and E2 lead to cell surface expression.
FIG. 5.
Cell surface expression of mutant proteins analyzed by indirect immunofluorescence. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinant at a multiplicity of infection of 3 PFU/cell. At 8 h postinfection, cells were fixed with paraformaldehyde, permeabilized or not with Triton X-100, and immunostained with anti-E2 (E2, E2DR, and E2-CD4) or anti-CD4 (CD4, CD4-E1NK, and CD4-E1) MAbs.
Altogether, these data indicate that charged amino acid residues located within the middle of the TMDs of HCV glycoproteins play a major role in ER retention of these proteins.
Charged residues located between the two stretches of hydrophobic residues are necessary for signal sequence function in the polyprotein.
Since the second stretch of hydrophobic residues present in the TMDs of HCV glycoproteins plays the role of a signal sequence, we suspected that mutation of the charged residues would alter this function. To test this hypothesis, the mutations were introduced in a polyprotein containing E1 and E2 (E1E2 or E2E1). The mutations in the TMD of E1 were introduced in the E1E2 polyprotein (E1NKE2), whereas the mutations in E2 were introduced in a polyprotein in which the positions of E1 and E2 have been inverted (E2DRE1). These proteins were expressed by vaccinia virus recombinants and analyzed in pulse-chase experiments followed by immunoprecipitation. As described previously (14, 19, 55), both E1 and E2 were detected after immunoprecipitation with a MAb directed against either E1 or E2 in the absence of any mutation in the E1E2p7 polyprotein, which was used as a control (Fig. 6A). As previously observed (19), the mobility of E1 increased during the chase, presumably as a result of trimming of mannose-rich core glycans. This explains the difference in mobility observed between E1 precipitated by MAb H53 after 4 h of chase and E1 precipitated by MAb A4 during the pulse. It has been shown previously that the cleavage between E1 and E2 and the assembly of these glycoproteins are similar in the presence and absence of p7 (14). In the context of E1NKE2, bands corresponding to E1 and E2 were also detected after immunoprecipitation with anti-E1 or -E2 MAbs, indicating that some signal sequence cleavage had occurred in the E1NKE2 polyprotein. However, the intensity of these bands was lower, and an additional band (labeled E1NKE2* in Fig. 6A) which migrated slightly faster than E2 was detected after immunoprecipitation with the anti-E1 MAb (A4). The intensity of this band was high after the pulse and decreased rapidly during the chase, suggesting a rapid degradation for this protein. The absence of detection of the latter by anti-E2 MAb H53 is probably due to misfolding of this protein, since H53 is a conformation-sensitive MAb (8, 21). We suspected that this new band (E1NKE2*) could be an uncleaved polyprotein with the E2 portion remaining cytosolic. To confirm the absence of cleavage between E1 and E2, E1NKE2 was expressed in HepG2 cells and analyzed by Western blotting. As shown in Fig. 6B, E1NKE2* was detected by anti-E1 as well as anti-E2 MAbs, indicating that it is an uncleaved polyprotein. It is worth noting that the E1NKE2*/E1 and E1NKE2*/E2 ratios were lower than those observed in the early time points of pulse-chase experiments (Fig. 6A). This is probably due to the shorter half-life of E1NKE2*. In addition, the differences in the intensities of E1 and E2 expressed from E1NKE2 or E1E2 observed in Fig. 6B were lower than those observed in Fig. 6A. This is likely due to the accumulation of proteins detected by Western blotting. PNGase F treatment of E1NKE2* indicated that this protein is glycosylated and the size of the deglycosylated form of this protein is compatible with that of an uncleaved deglycosylated E1E2 polyprotein (data not shown). In addition, its faster electrophoretic mobility than that of E2 suggests that only the E1 portion was glycosylated. Together these data support the hypothesis that E1NKE2* is an uncleaved polyprotein. They also indicate that the mutations introduced in the TMD of E1 partially abolish the signal sequence function.
FIG. 6.
Role of charged residues on the signal sequence function of the TMD of HCV glycoprotein E1. HepG2 cells were coinfected with vTF7-3 and a vaccinia virus recombinant expressing E1E2 or E1NKE2 at a multiplicity of infection of 5 PFU/cell. (A) At 4.5 h postinfection, cells were pulse-labeled for 10 min and chased for the indicated times (in hours). Cell lysates were immunoprecipitated with MAb H53 (anti-E2) or A4 (anti-E1). Proteins were separated by SDS-PAGE (10% polyacrylamide) under reducing conditions. (B) Infected cells were lysed at 4.5 h postinfection. After separation by SDS-PAGE, the proteins of interest were revealed by Western blotting with an anti-E1 (A4) or anti-E2 (A11) MAb. The uncleaved E1NKE2 polyprotein is indicated by an asterisk.
The mutations introduced in the TMD of E2 were tested in the context of an E2E1 polyprotein (E2DRE1) to evaluate their role in signal sequence function. However, since the positions of HCV glycoproteins have been inverted on the polyprotein, we first tested the expression and processing of the nonmutated E2E1 polyprotein by Western blotting analysis. As shown in Fig. 7, a band corresponding to E2 was revealed by the anti-E2 MAb, and bands corresponding to different glycoforms of E1 (19, 33) were revealed by the anti-E1 MAb. It is worth noting that inversion of the positions of E1 and E2 on the polyprotein has a negative effect on the efficiency of glycosylation of E1 (J. Dubuisson, S. Duvet, J.-C. Meunier, A. Op De Beeck, R. Cacan, C. Wychowski, and L. Cocquerel, unpublished data). Detection of both E1 and E2 indicates that the polyprotein has been cleaved and confirms the presence of a signal sequence function in the C terminus of E2. In contrast, when the mutations were introduced in the TMD of E2, no band corresponding to E1 or E2 was observed, but a band (E2DRE1*) with slower electrophoretic mobility than E2 was detected by both the anti-E1 and anti-E2 MAbs (Fig. 7). It has to be noted that, in addition to E2DRE1*, a second band migrating faster than E2, but slower than E1, was also revealed by the anti-E1 MAb (Fig. 7, E2DRE1). This band might correspond to a nonglycosylated uncleaved E2DRE1 molecule. However, it was not recognized by the anti-E2 MAb. Alternatively, this could be a cleaved form of E2DRE1 with a portion of E2 deleted. Together these data suggest that E2DRE1* is an uncleaved polyprotein. They also indicate that the mutations introduced in the TM sequence of E2 abolish the signal sequence function. It is worth noting that the signal sequence function present at the C terminus of E2 was totally abolished when charged residues were mutated in this protein, whereas mutations at the C terminus of E1 had only a partial effect. Nevertheless, these data indicate that the charged residues located within the middle of the TMDs of HCV glycoproteins are important for the signal sequence function.
FIG. 7.
Role of charged residues in the signal sequence function of the TMD of HCV glycoprotein E2. HepG2 cells were coinfected with vTF7-3 and a vaccinia virus recombinant expressing E2E1 or E2DRE1 at a multiplicity of infection of 5 PFU/cell. Infected cells were lysed at 4.5 h postinfection. After separation by SDS-PAGE (10% polyacrylamide) under reducing conditions, proteins of interest were revealed by Western blotting with an anti-E1 (A4) or anti-E2 (A11) MAb. The uncleaved E2DRE1 polyprotein is indicated by an asterisk.
Charged residues located between the two stretches of hydrophobic residues play a major role in the assembly of E1E2 complex.
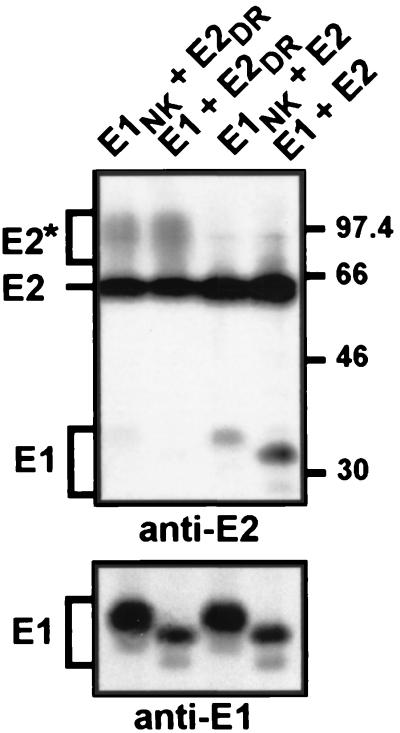
HCV glycoproteins interact to form a noncovalent heterodimer (14), and their TMDs have been suggested to play a role in the assembly of the complex (8, 47, 63). For this reason, we wanted to know whether the point mutations introduced in the TMDs of HCV glycoproteins would have some effect on the assembly of the heterodimer. Since the mutations introduced in the TMD of E1 partially abolish the signal sequence function for E2, the effect of the mutations on HCV glycoprotein assembly could not be analyzed in the context of an E1E2 polyprotein. To circumvent this problem, the mutations were introduced in E1 and E2 proteins expressed individually by vaccinia virus recombinants, and coexpression of HCV glycoproteins was obtained by coinfecting cells with both recombinant viruses. To analyze the association of these glycoproteins, coinfected cells were pulse-labeled for 10 min and chased for 4 h. These conditions have been previously shown to correspond to the peak of detection of the heterodimer (14). The formation of HCV glycoprotein complexes was then analyzed by immunoprecipitation with the conformation-sensitive E2-specific MAb H53 (8, 21) and revealed by SDS-PAGE. As shown in Fig. 8, E1 was coprecipitated with E2 when wild-type HCV glycoproteins were coexpressed in trans, indicating that a coexpression in cis is not necessary for assembly of the heterodimer. When E2DR was coexpressed with E1, only the E2 band was detected by immunoprecipitation with MAb H53. In contrast, a faint band corresponding to E1 was detected when E1NK and E2 were coexpressed, and E1 was barely detectable when the mutations were introduced in both molecules. The expression of E1 or E1NK in these coinfection experiments was confirmed by immunoprecipitation with an anti-E1 MAb (Fig. 8, anti-E1). It is worth noting that the mutated form of E1 migrated slightly more slowly than the wild-type protein. This is probably due to processing of the glycans.
FIG. 8.
Role of charged residues in the assembly of the E1E2 polyprotein complex. HepG2 cells were coinfected with vTF7-3 and the appropriate vaccinia virus recombinants at a multiplicity of infection of 5 PFU/cell. At 4.5 h postinfection, cells were pulse-labeled for 10 min and chased for 4 h. Cell lysates were immunoprecipitated with MAb H53 (anti-E2) or A4 (anti-E1), and proteins were separated by SDS-PAGE (10% polyacrylamide) under reducing conditions. The sizes (in kilodaltons) of protein molecular mass markers are indicated on the right. The slow-migrating form of E2 is indicated by an asterisk.
Together, these data indicate that the charged residues located within the middle of the TMDs of HCV glycoproteins play a major role in assembly of E1E2 heterodimer.
Sequence analysis of the TM sequences of the other members of the family Flaviviridae.
It was of great interest to see whether some structural similarities could be observed in the sequences of the TMDs of envelope proteins of other viruses belonging to the same family. The Flaviviridae family is divided in three genera: Hepacivirus, Flavivirus, and Pestivirus (46). In addition, HCV is related to recently identified tamarin or human viruses known as GBV-A, GBV-B, and GBV-C (the latter being also called hepatitis G virus, or HGV) (37, 66, 67).
Analysis of the C-terminal sequences of the envelope proteins in the family Flaviviridae (Fig. 9) reveals that their putative TMDs have an organization similar to those of HCV glycoproteins (Fig. 1B). Because of the high conservation of the connecting sequences located between the hydrophobic stretches, specific patterns emerge for different virus groups. In all cases, the presence of at least one positively charged residue (Arg or Lys) is observed. The full conservation of this basic residue(s) within each viral group suggests that they are involved in functions similar to those reported above for the TMDs of HCV envelope proteins. Besides the Arg or Lys residues, and depending on the virus group and/or the envelope protein, additional charged residues which are remarkably conserved can be observed. Typically (i) a second basic residue is always present in the TMDs of pestivirus and GBV-B E1 proteins, and a total of three Arg residues are observed in the TMD of GBV-B E2 protein; (ii) an acidic residue (Glu or Asp) is observed in prM of tick-borne encephalitis virus and related viruses, as well as in GBV-A and GBV-C E1 proteins; and (iii) both an acidic residue and an additional basic residue are present in the TMDs of GBV-A and GBV-C E2 proteins, as well as in the TMD of E protein of Japanese encephalitis virus and related viruses. This quite broad variability of connecting segments observed between different viral groups might be related to specific intra- and/or intermolecular recognition for each virus and its closely related species.
It is worth noting that the connecting segments of prM and E in the Flavivirus genus appear longer than their counterparts in the other genera. They contain several well-conserved polar residues (Asn, Gln, Ser, and/or Thr), while only one polar fully conserved residue (Gln) is observed in the TMDs of E1 of the GB viruses and pestiviruses. In addition, the second hydrophobic stretch of the TMD of E in the Flavivirus genus is clearly longer (19 residues) than its counterpart in the other genera (12 to 13 residues). It should be underlined that this hydrophobic stretch is potentially long enough to form an α-helix which spans the membrane entirely, while the short hydrophobic segments in HCV, GB viruses, and pestiviruses do not fit with the classical view of TM α-helices. These differences might reflect some divergence in the functions played by the TMDs of the envelope proteins in the Flavivirus genus.
DISCUSSION
It is well documented that TM sequences can play other roles than simply anchoring a protein in a membrane. HCV glycoproteins are good examples of additional functions performed by TMDs and represent an attractive model with which to understand the multifunctionality of such domains. We show here that mutation of charged residues located in the TMDs of HCV glycoproteins leads to an alteration in the processing, subcellular localization, and assembly of these envelope proteins. These data suggest that these charged residues play a key role in the formation of the viral envelope.
The first stretch of hydrophobic residues present in the TMDs of HCV glycoproteins E1 and E2 is not sufficient for an efficient arrest of translocation. A similar observation has been reported for a glycoprotein (prM) of another member of the family Flaviviridae (dengue virus type 4) (44). Natural stop-transfer sequences generally consist of more than 18 mainly hydrophobic amino acid residues and are followed by positive charges (62). They have two functions: to interrupt the ongoing protein translocation and to anchor the final protein in the membrane. The inefficiency in membrane insertion in the absence of the second hydrophobic stretch (this work and reference 44) might be due to the short length of the first hydrophobic sequence of the TMDs of the envelope proteins in the Flaviviridae family. However, it has been shown experimentally that an artificial hydrophobic sequence as short as 8 leucine residues can cause an efficient stop of translocation (32). In contrast, when a membrane-spanning sequence was made of alanine residues, up to 19 residues were necessary to obtain the same stop-transfer efficiency. With a 50:50 mixture of leucine and alanine, at least 11 residues were required. The first hydrophobic stretch of E2 contains 6 leucine residues which could compensate for its short length (11 residues). However, the charged residues located after this hydrophobic segment can also have some effect on the efficiency of a stop-transfer sequence. Indeed, positive charges after the hydrophobic segment are more favorable than negative charges (32). For instance, in the case of prM of dengue virus type 4, an additional arginine residue introduced after the first stretch of hydrophobic residues has been shown to increase the efficiency of its membrane insertion (44). It is likely that the short length of the first stretch of hydrophobic residues in the TMD of HCV glycoproteins and the lack of reinforcement by a positive charge make the first stretch of hydrophobic sequence an inefficient stop-transfer signal.
We demonstrate here that charged amino acid residues located in the segments connecting the two hydrophobic stretches of HCV glycoprotein TMDs play a major role in ER retention of these proteins. ER retention by TMDs has been reported for several TM proteins (2, 3, 36, 76), and a usual feature of membrane determinants for ER retention is the presence of one or several hydrophilic residues within the hydrophobic TMD. The introduction of charged residues in the hydrophobic segment of a plasma membrane protein is sufficient to cause its retention in the ER, and their effect is strongest when they are localized toward the middle of the TMD (2). For HCV glycoproteins, there is no clear evidence that the charged residues are located in the middle of a single membrane-spanning segment. In the case in which the double-membrane-spanning topology is maintained, these charged residues should be exposed on the cytosolic face of the membrane. However, it cannot be excluded that a reorientation of the second stretch of hydrophobic residues occurs immediately after the signal sequence cleavage at its C terminus. If there is such a flipping of the second hydrophobic segment, the TMDs of HCV glycoproteins could form a single TM segment with the charged residues in the middle of the membrane-spanning sequence. The mechanism of ER localization by such a signal is poorly understood, but it has been proposed to be due to interactions with proteins involved in the ER retrieval machinery (36). However, this does not explain how some proteins, like HCV glycoproteins (21), are strictly retained in the ER by their TMD.
In the absence of any reorientation in the TMDs of HCV glycoproteins, the two membrane-spanning segments are expected to be short. Hydrophobic sequences which are shorter than the average of those of plasma membrane proteins have been shown to be involved in Golgi or ER retention (4, 29, 53, 76), and a lipid-based mechanism has been proposed for TMD-mediated retention (4). The mixed lipid populations in the intracellular membrane would separate into lipid microdomains with distinct compositions, thicknesses, and degrees of structural perturbability. Proteins would selectively partition into one domain and so be prevented from entering transport vesicles comprising the other domain by virtue of physical properties of their TMDs. Whether the TM sequences of HCV glycoproteins fit into this model remains to be proven.
The charged amino acid residues located in the middle of the TMDs of HCV glycoproteins also play a major role in the assembly of HCV glycoproteins. These data suggest a direct interaction between the TMDs of HCV glycoproteins. This interaction might be due to a direct involvement of the charged residues of the TMDs of E1 and E2. This might explain the lack of interaction when these charged residues are replaced by an alanine. However, it is very likely that the interaction between the TMDs involves a direct contact between the hydrophobic segments of E1 and E2. Therefore, the lack of interaction after mutation of the charged residue(s) might be due to a conformational change in the structure of the TMDs of HCV envelope proteins.
Sequence analysis of the TMDs of envelope proteins of other Flaviviridae viruses revealed a similar organization for all the members of this viral family. Despite some differences observed for the members of the Flavivirus genus, the presence of at least one positively charged residue was systematically observed in the short segment connecting the two hydrophobic stretches. One can expect that the TMDs of the envelope proteins of the other Flaviviridae viruses should play functions similar to those reported here for HCV glycoproteins and that this charged residue might play a crucial role in these functions. However, when other charged or polar residues are present in the connecting segment, their concomitant mutation might also be required to modify the functions of these TMDs.
In addition to the functions described above, the TMDs of HCV glycoproteins might also play a direct role in the assembly of the particle. Viral envelope proteins can play a major role in virus budding (26), which is a late stage of virus assembly corresponding to the acquisition of a membrane that surrounds the nucleocapsid (17). Virus budding can occur at the plasma membrane or at an intracellular membrane along the secretory pathway. For the Flaviviridae viruses, ultrastructural studies of virus-infected cells indicate that virion morphogenesis occurs in association with intracellular membranes believed to be derived from the ER (59). It has generally been thought that budding is driven by interactions between the viral TM proteins and the nucleocapsid. This model fits very well for the alphaviruses (70). However, it is now evident that enveloped viruses use various kinds of proteins for budding. Indeed, virus budding can also be driven by capsid or core protein only (retrovirus), by membrane protein only (coronavirus), or by matrix protein with the assistance of spikes and ribonucleoprotein (rhabdovirus and possibly paramyxovirus and orthomyxovirus) (26). The mechanism of budding has not been specifically studied for members of the family Flaviviridae, but flavivirus envelope proteins can exhibit an independent budding activity. Indeed, they can form capsid-free membrane particles by themselves (59). This observation suggests that, at least for the genus Flavivirus, the envelope proteins play a major role in budding. However, this does not exclude the possibility of a role played by the capsid. In this case, we would expect to have some interaction between the nucleocapsid and cytosolic residues of the envelope proteins. The charged residues which were shown to play an important role for the functions of the TMDs of HCV glycoproteins would be potential candidates for such interactions.
As shown in this work, the various functions played by the TMDs of HCV glycoproteins can be disrupted by mutating charged residues present in the membrane anchor of these proteins, indicating that these functions are tightly linked together. These charged residues probably play a crucial role in the structure of these domains. The analysis of the three-dimensional structure of these domains should allow further understanding of their multifunctionality. Such a knowledge would also help our understanding of some crucial events in the biogenesis of the envelope of HCV and other viruses of the Flaviviridae family.
ACKNOWLEDGMENTS
We thank François Letourneur and Françoise Jacob-Dubuisson for critical reading of the manuscript, André Pillez and Sophana Ung for excellent technical assistance, C. Blanchet for the MPSA program, and G. Deléage for Network Protein Sequence @nalysis (http://phil.ibcp.fr/NPSA).
This work was supported by the CNRS, the Institut Pasteur de Lille, a PRFMMIP grant from the French Ministry of Research, and grant 9736 from the ARC.
REFERENCES
- 1.Becher P, Shannon A D, Tautz N, Thiel H J. Molecular characterization of border disease virus, a pestivirus from sheep. Virology. 1994;198:542–551. doi: 10.1006/viro.1994.1065. [DOI] [PubMed] [Google Scholar]
- 2.Bonifacino J S, Cosson P, Shah N, Klausner R D. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 1991;10:2783–2793. doi: 10.1002/j.1460-2075.1991.tb07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bonifacino J S, Suzuki C K, Klausner R D. A peptide sequence confers retention and rapid degradation in the endoplasmic reticulum. Science. 1990;247:79–82. doi: 10.1126/science.2294595. [DOI] [PubMed] [Google Scholar]
- 4.Bretcher M S, Munro S. Cholesterol and Golgi apparatus. Science. 1993;261:1280–1281. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 5.Castle E, Nowak T, Leidner U, Wengler G. Sequence analysis of the viral core protein and the membrane-associated proteins V1 and NV2 of the flavivirus West Nile virus and of the genome sequence for these proteins. Virology. 1985;145:227–236. doi: 10.1016/0042-6822(85)90156-4. [DOI] [PubMed] [Google Scholar]
- 6.Chamberlain R W, Adams M, Saeed A A, Simmonds P, Elliott R M. Complete nucleotide sequence of a type 4 hepatitis C virus variant, the predominant genotype in the Middle East. J Gen Virol. 1997;78:1341–1347. doi: 10.1099/0022-1317-78-6-1341. [DOI] [PubMed] [Google Scholar]
- 7.Cocquerel L, Duvet S, Meunier J-C, Pillez A, Cacan R, Wychowski C, Dubuisson J. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J Virol. 1999;73:2641–2649. doi: 10.1128/jvi.73.4.2641-2649.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cocquerel L, Meunier J-C, Pillez A, Wychowski C, Dubuisson J. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J Virol. 1998;72:2183–2191. doi: 10.1128/jvi.72.3.2183-2191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coia G, Parker M D, Speight G, Byrne M E, Westaway E G. Nucleotide and complete amino acid sequences of Kunjin virus: definitive gene order and characteristics of the virus-specific proteins. J Gen Virol. 1988;69:1–21. doi: 10.1099/0022-1317-69-1-1. [DOI] [PubMed] [Google Scholar]
- 10.Cosson P, Bonifacino J. Role of transmembrane domain interactions in the assembly of class II MHC molecules. Science. 1992;258:659–662. doi: 10.1126/science.1329208. [DOI] [PubMed] [Google Scholar]
- 11.Cosson P, Lankford S P, Bonifacino J, Klausner R D. Membrane protein association by potential intramembrane charge pairs. Nature. 1991;351:414–416. doi: 10.1038/351414a0. [DOI] [PubMed] [Google Scholar]
- 12.Cserzo M, Wallin E, Simon I, von Heijne G, Elofsson A. Prediction of transmembrane alpha-helices in procaryotic membrane proteins: the dense alignment surface method. Protein Eng. 1997;10:673–676. doi: 10.1093/protein/10.6.673. [DOI] [PubMed] [Google Scholar]
- 13.Dalgarno L, Trent D W, Strauss J H, Rice C M. Partial nucleotide sequence of the Murray Valley encephalitis virus genome—comparison of the encoded polypeptides with yellow fever virus structural and non-structural proteins. J Mol Biol. 1986;187:309–323. doi: 10.1016/0022-2836(86)90435-3. [DOI] [PubMed] [Google Scholar]
- 14.Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, Hahn Y S, Rice C M, Dubuisson J. Formation of native hepatitis C virus glycoprotein complexes. J Virol. 1997;71:697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Moerlooze L, Lecomte C M, Brown-Shimer S L, Schmetz D, Guiot C, Vandenbergh D J, Allaer D, Rossius M, Chappuis G, Dina D, Renard A, Martial J A. Nucleotide sequence of the bovine viral diarrhea virus Osloss strain: comparison with related viruses and identification of specific DNA probes in the 5′ untranslated region. J Gen Virol. 1993;74:1433–1438. doi: 10.1099/0022-1317-74-7-1433. [DOI] [PubMed] [Google Scholar]
- 16.Deubel V, Kinney R M, Trent D W. Nucleotide sequence and deduced amino acid sequence of the nonstructural proteins of dengue type 2 virus, Jamaica genotype: comparative analysis of the full-length genome. Virology. 1988;165:234–244. doi: 10.1016/0042-6822(88)90677-0. [DOI] [PubMed] [Google Scholar]
- 17.Dubois-Dalcq M, Holmes K V, Rentier B. Assembly of enveloped RNA viruses. Vienna, Austria: Springer-Verlag KG; 1984. [Google Scholar]
- 18.Dubuisson J. Folding, assembly and subcellular localization of HCV glycoproteins. Curr Top Microbiol Immunol. 1999;242:135–148. doi: 10.1007/978-3-642-59605-6_7. [DOI] [PubMed] [Google Scholar]
- 19.Dubuisson J, Hsu H H, Cheung R C, Greenberg H B, Russell D G, Rice C M. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dubuisson J, Rice C M. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J Virol. 1996;70:778–786. doi: 10.1128/jvi.70.2.778-786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duvet S, Cocquerel L, Pillez A, Cacan R, Verbert A, Moradpour D, Wychowski C, Dubuisson J. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J Biol Chem. 1998;273:32088–32095. doi: 10.1074/jbc.273.48.32088. [DOI] [PubMed] [Google Scholar]
- 22.Feinstone S, Alter H J, Dienes H P, Shimizu Y, Popper H, Blackmore D, Sly D, London W T, Purcell R H. Non-A, non-B hepatitis in chimpanzees and marmosets. J Infect Dis. 1981;144:588–598. doi: 10.1093/infdis/144.6.588. [DOI] [PubMed] [Google Scholar]
- 23.Fournillier-Jacob A, Cahour A, Escriou N, Girard M, Wychowski C. Processing of the E1 glycoprotein of hepatitis C virus expressed in mammalian cells. J Gen Virol. 1996;77:1055–1064. doi: 10.1099/0022-1317-77-5-1055. [DOI] [PubMed] [Google Scholar]
- 24.Francki R I B, Fauquet C M, Knudson D L, Brown F. Classification and nomenclature of viruses. Fifth Report of the International Committee on Taxonomy of Viruses. Arch Virol. 1991;2(Suppl.):223. [Google Scholar]
- 25.Fuerst T R, Niles E G, Studier F W, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122–8126. doi: 10.1073/pnas.83.21.8122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garoff H, Hewson R, Opstelten D-J E. Virus maturation by budding. Microbiol Mol Biol Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garoff H, Liljeström P, Metsikkö K, Lobigs M, Wahlberg J. Formation and function of the Semliki Forest virus membrane. In: Brinton M A, Heinz F X, editors. New aspects of positive-strand RNA viruses. Washington, D.C.: American Society for Microbiology; 1990. pp. 166–172. [Google Scholar]
- 28.Gritsun T S, Lashkevich V A, Gould E A. Nucleotide and deduced amino acid sequence of the envelope glycoprotein of Omsk haemorrhagic fever virus; comparison with other flaviviruses. J Gen Virol. 1993;74:287–291. doi: 10.1099/0022-1317-74-2-287. [DOI] [PubMed] [Google Scholar]
- 29.Honsho M, Mitoma J Y, Ito A. Retention of cytochrome b5 in the endoplasmic reticulum is transmembrane and luminal domain-dependent. J Biol Chem. 1998;273:20860–20866. doi: 10.1074/jbc.273.33.20860. [DOI] [PubMed] [Google Scholar]
- 30.Houghton M. Hepatitis C viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1035–1058. [Google Scholar]
- 31.Kieny M-P, Lathe R, Drillien R, Spehner D, Skory S, Schmitt D, Wiktor T, Koprowski H, Lecocq J-P. Expression of rabies virus glycoprotein from a recombinant vaccinia virus. Nature. 1984;312:163–166. doi: 10.1038/312163a0. [DOI] [PubMed] [Google Scholar]
- 32.Kuroiwa T, Sakaguchi M, Mihara K, Omura T. Systematic analysis of stop-transfer sequence for microsomal membrane. J Biol Chem. 1991;266:9251–9255. [PubMed] [Google Scholar]
- 33.Lanford R E, Notvall L, Chavez D, White R, Frenzel G, Simonsen C, Kim J. Analysis of hepatitis C virus capsid, E1, and E2/NS1 proteins expressed in insect cells. Virology. 1993;197:225–235. doi: 10.1006/viro.1993.1583. [DOI] [PubMed] [Google Scholar]
- 34.Leary T P, Muerhoff A S, Simons J N, Pilot-Matias T J, Erker J C, Chalmers M L, Schlauder G G, Dawson G J, Desai S M, Mushahwar I K. Sequence and genomic organization of GBV-C: a novel member of the flaviviridae associated with human non-A-E hepatitis. J Med Virol. 1996;48:60–67. doi: 10.1002/(SICI)1096-9071(199601)48:1<60::AID-JMV10>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 35.Lemmon M A, Flanagan J M, Hunt J F, Adair B, Bormann B J, Dempsey C, Engelman D. Glycophorin A dimerization is driven by specific interactions between transmembrane α-helices. J Biol Chem. 1992;267:7683–7689. [PubMed] [Google Scholar]
- 36.Letourneur F, Cosson P. Targeting to the endoplasmic reticulum in yeast cells by determinants present in transmembrane domains. J Biol Chem. 1998;273:33273–33278. doi: 10.1074/jbc.273.50.33273. [DOI] [PubMed] [Google Scholar]
- 37.Linnen J, Wages J, Zhang-Keck Z Y, Fry K E, Krawczynski K, Alter H, Koonin E, Gallagher M, Alter M, Hadziyannis S, Karayiannis P, Fung K, Nakatsuji Y, Shih W K, Young L, Piatak M, Hoover C, Fernandez J, Chen S, Zou J C, Morris T, Hyams K C, Ismay S, Lifson J D, Hess G, Foung S K H, Thomas H, Bradley D, Margolis H, Kim J P. Molecular cloning and disease association of hepatitis G virus: a transfusion-transmissible agent. Science. 1996;271:505–508. doi: 10.1126/science.271.5248.505. [DOI] [PubMed] [Google Scholar]
- 38.Machamer C E. Targeting and retention of Golgi membrane proteins. Curr Opin Cell Biol. 1993;5:606–612. doi: 10.1016/0955-0674(93)90129-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mandl C W, Heinz F X, Kunz C. Sequence of the structural proteins of tick-borne encephalitis virus (western subtype) and comparative analysis with other flaviviruses. Virology. 1988;166:197–205. doi: 10.1016/0042-6822(88)90161-4. [DOI] [PubMed] [Google Scholar]
- 40.Mandl C W, Holzmann H, Kunz C, Heinz F X. Complete genomic sequence of Powassan virus: evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology. 1993;194:173–184. doi: 10.1006/viro.1993.1247. [DOI] [PubMed] [Google Scholar]
- 41.Mandl C W, Iacono-Connors L, Wallner G, Holzmann H, Kunz C, Heinz F X. Sequence of the genes encoding the structural proteins of the low-virulence tick-borne flaviviruses langat TP21 and yelantsev. Virology. 1991;185:891–895. doi: 10.1016/0042-6822(91)90567-u. [DOI] [PubMed] [Google Scholar]
- 42.Marcantonio E E, David F S. Integrin receptor signaling: the propagation of an alpha-helix model. Matrix Biol. 1997;16:179–184. doi: 10.1016/s0945-053x(97)90006-8. [DOI] [PubMed] [Google Scholar]
- 43.Marin M S, Makenzie J, Gao G F, Reid H W, Antoniadis A, Gould E A. The virus causing sheep encephalomyelitis in sheep in Spain is a new member of the tick-borne encephalitis group. Res Vet Sci. 1995;58:11–13. doi: 10.1016/0034-5288(95)90081-0. [DOI] [PubMed] [Google Scholar]
- 44.Markoff L, Chang A, Falgout B. Processing of flavivirus structural glycoproteins: stable membrane insertion of premembrane requires the envelope signal peptide. Virology. 1994;204:526–540. doi: 10.1006/viro.1994.1566. [DOI] [PubMed] [Google Scholar]
- 45.Mason P W, McAda P C, Mason T L, Fournier M J. Sequence of the Dengue-1 virus genome in the region encoding the three structural proteins and the major nonstructural protein NS1. Virology. 1987;161:262–267. doi: 10.1016/0042-6822(87)90196-6. [DOI] [PubMed] [Google Scholar]
- 46.Mayo M A, Pringle C R. Virus taxonomy—1997. J Gen Virol. 1998;79:649–657. doi: 10.1099/0022-1317-79-4-649. [DOI] [PubMed] [Google Scholar]
- 47.Michalak J-P, Wychowski C, Choukhi A, Meunier J-C, Ung S, Rice C M, Dubuisson J. Characterization of truncated forms of hepatitis C virus glycoproteins. J Gen Virol. 1997;78:2299–2306. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- 48.Mizushima H, Hijikata M, Asabe S-I, Hirota M, Kimura K, Shimotohno K. Two hepatitis C virus glycoprotein E2 products with different C termini. J Virol. 1994;68:6215–6222. doi: 10.1128/jvi.68.10.6215-6222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Muerhoff A S, Leary T P, Simons J N, Pilot-Matias T J, Dawson G J, Erker J C, Chalmers M L, Schlauder G G, Desai S M, Mushahwar I K. Genomic organization of GB viruses A and B: two new members of the Flaviviridae associated with GB agent hepatitis. J Virol. 1995;69:5621–5630. doi: 10.1128/jvi.69.9.5621-5630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nitayaphan S, Grant J A, Chang G-J, Trent D. Nucleotide sequence of the virulent SA-14 strain of Japanese encephalitis virus and its attenuated vaccine derivative, SA-14-14-2. Virology. 1990;177:541–552. doi: 10.1016/0042-6822(90)90519-w. [DOI] [PubMed] [Google Scholar]
- 51.Osatomi K, Sumiyoshi H. Complete nucleotide sequence of dengue type 3 virus genome RNA. Virology. 1990;176:643–647. doi: 10.1016/0042-6822(90)90037-r. [DOI] [PubMed] [Google Scholar]
- 52.Pearson W R, Lipman D J. Improved tools for biological sequence comparison. Proc Natl Acad Sci USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pedrazzini E, Villa A, Borgese N. A mutant cytochrome b5 with a lengthened membrane anchor escapes from the endoplasmic reticulum and reaches the plasma membrane. Proc Natl Acad Sci USA. 1996;93:4207–4212. doi: 10.1073/pnas.93.9.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Popot J-L. Integral membrane protein structure: transmembrane α-helices as autonomous folding domains. Curr Opin Struct Biol. 1993;3:532–540. [Google Scholar]
- 55.Ralston R, Thudium K, Berger K, Kuo C, Gervase B, Hall J, Selby M, Kuo G, Houghton M, Choo Q-L. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J Virol. 1993;67:6753–6761. doi: 10.1128/jvi.67.11.6753-6761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Reed K E, Rice C M. Molecular characterization of hepatitis C virus. Curr Stud Hematol Blood Transfus. 1998;62:1–37. doi: 10.1159/000060472. [DOI] [PubMed] [Google Scholar]
- 57.Reinherz E L, Kung P C, Goldstein G, Schlossman S F. Separation of functional subsets of human T cells by a monoclonal antibody. Proc Natl Acad Sci USA. 1979;76:4061–4065. doi: 10.1073/pnas.76.8.4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reth M, Hombach J, Wienands J, Campbell K S, Chien N, Justement L B, Cambier J C. The B-cell antigen receptor complex. Immunol Today. 1991;12:196–201. doi: 10.1016/0167-5699(91)90053-V. [DOI] [PubMed] [Google Scholar]
- 59.Rice C M. Flaviviridae: viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 931–959. [Google Scholar]
- 60.Rice C M, Lenches E M, Eddy S R, Shin S J, Sheets R L, Strauss J H. Nucleotide sequence of yellow fever virus: implications for flavivirus gene expression and evolution. Science. 1985;229:726–733. doi: 10.1126/science.4023707. [DOI] [PubMed] [Google Scholar]
- 61.Ruemenapf T, Meyers G, Stark R, Thiel H J. Hog cholera virus-characterization of specific antiserum and identification of cDNA clones. Virology. 1989;171:18–27. doi: 10.1016/0042-6822(89)90506-0. [DOI] [PubMed] [Google Scholar]
- 62.Sakaguchi M. Mutational analysis of signal-anchor and stop-transfer sequences in membrane proteins. In: von Heijne G, editor. Membrane protein assembly. R. G. Georgetown, Tex: Landes Company; 1997. pp. 135–150. [Google Scholar]
- 63.Selby M J, Glazer E, Masiarz F, Houghton M. Complex processing and protein:protein interactions in the E2:NS2 region of HCV. Virology. 1994;204:114–122. doi: 10.1006/viro.1994.1515. [DOI] [PubMed] [Google Scholar]
- 64.Shin J, Dunbrack R L, Lee S, Strominger J L. Signals for retention of transmembrane proteins in the endoplasmic reticulum studied with CD4 truncation mutants. Proc Natl Acad Sci USA. 1991;88:1918–1922. doi: 10.1073/pnas.88.5.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shiu S Y, Ayres M D, Gould E A. Genomic sequence of the structural proteins of Louping ill virus: comparative analysis with tick-borne encephalitis virus. Virology. 1991;180:411–415. doi: 10.1016/0042-6822(91)90048-g. [DOI] [PubMed] [Google Scholar]
- 66.Simons J N, Leary T P, Dawson G J, Pilot-Matias T J, Muerhoff A S, Schlauder G G, Desai S M, Mushahwar I K. Isolation of novel virus-like sequences associated with human hepatitis. Nat Med. 1995;1:564–569. doi: 10.1038/nm0695-564. [DOI] [PubMed] [Google Scholar]
- 67.Simons J N, Pilot-Matias T J, Leary T P, Dawson G J, Desai S M, Schlauder G G, Muerhoff A S, Erker J C, Buijk S L, Chalmers M L, van Sant C L, Mushahwar I K. Identification of two flavivirus-like genomes in the GB hepatitis agent. Proc Natl Acad Sci USA. 1995;92:3401–3405. doi: 10.1073/pnas.92.8.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith S, Blobel G. The first membrane spanning region of the lamin B receptor is sufficient for sorting to the inner nuclear membrane. J Cell Biol. 1993;120:631–637. doi: 10.1083/jcb.120.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stemmer W P C, Morris S K. Enzymatic inverse PCR: a restriction site independent, single-fragment method for high-efficiency, site directed mutagenesis. BioTechniques. 1992;13:215–220. [PubMed] [Google Scholar]
- 70.Suomalainen M, Liljeström P, Garoff H. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J Virol. 1992;66:4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Trent D W, Kinney R M, Johnson B J B, Vorndam A V, Grant J A, Deubel V, Rice C M, Hahn C. Partial nucleotide sequence of St. Louis encephalitis virus RNA: structural proteins, NS1, NS2a, and NS2b. Virology. 1987;156:293–304. doi: 10.1016/0042-6822(87)90409-0. [DOI] [PubMed] [Google Scholar]
- 73.Venugopal K, Gritsun T, Lashkevich V A, Gould E A. Analysis of the structural protein gene sequence shows Kyasanur Forest disease virus as a distinct member in the tick-borne encephalitis virus serocomplex. J Gen Virol. 1994;75:227–232. doi: 10.1099/0022-1317-75-1-227. [DOI] [PubMed] [Google Scholar]
- 74.von Heijne G. Transcending the impenetrable: how proteins come to terms with membranes. Biochim Biophys Acta. 1988;947:307–333. doi: 10.1016/0304-4157(88)90013-5. [DOI] [PubMed] [Google Scholar]
- 75.Wozniak R W, Blobel G. The single transmembrane segment of gp210 is sufficient for sorting to the pore membrane domain of the nuclear envelope. J Cell Biol. 1992;119:1441–1449. doi: 10.1083/jcb.119.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang M, Ellenberg J, Bonifacino J S, Weissman A M. The transmembrane domain of a carboxy-terminal anchored protein determines localization to the endoplasmic reticulum. J Biol Chem. 1997;272:1970–1975. doi: 10.1074/jbc.272.3.1970. [DOI] [PubMed] [Google Scholar]
- 77.Zhao B, Mackow E R, Buckler-White A J, Markoff L, Chancock R M, Lai C-J, Makino Y. Cloning full-length dengue type 4 viral DNA sequences: analysis of genes coding for structural proteins. Virology. 1986;155:77–88. doi: 10.1016/0042-6822(86)90169-8. [DOI] [PubMed] [Google Scholar]