Abstract
We previously reported specific interaction of cellular glyceraldehyde 3-phosphate dehydrogenase (GAPDH), the key glycolytic enzyme, and La protein, the RNA polymerase III transcription factor, with the cis-acting RNAs of human parainfluenza virus type 3 (HPIV3) and packaging of these proteins within purified virions (B. P. De, S. Gupta, H. Zhao, J. Z. Drazba, and A. K. Banerjee, J. Biol. Chem. 271:24728–24735, 1996). To gain further insight into these molecular interactions, we analyzed the virion-associated GAPDH and La protein using two-dimensional gel electrophoresis and immunoblotting. The GAPDH was resolved into two major and one minor molecular species migrating in the pI range of 7.6 to 8.3, while the La protein was resolved into five molecular species in the pI range of 6.8 to 7.5. The GAPDH isoforms present in the virions were also detected in the cytoplasmic fraction of CV-1 cell extract, albeit as minor species. On the other hand, the multiple molecular forms of La protein as seen within the virions were readily detected in the total CV-1 cell extract. Further analysis of virion-associated GAPDH by in vivo labeling with [32P]orthophosphate revealed the presence of multiple phosphorylated species. The phosphorylated species were able to bind specifically to the viral cis-acting 3′ genome sense RNA but failed to bind to the leader sense RNA, as determined by gel mobility shift assay. In contrast, the La protein isoforms present within the virions were not phosphorylated and bound to the viral cis-acting RNAs in a phosphorylation-independent manner. The GAPDH isoforms purified from the CV-1 cell cytoplasmic fraction inhibited viral transcription in vitro. Consistent with this, flag-tagged recombinant GAPDH synthesized by using the vaccinia virus expression system also inhibited viral transcription. Together, these data indicate that specific phosphorylated forms of GAPDH associate with HPIV3 and are involved in the regulation of virus gene expression.
Human parainfluenza virus type 3 (HPIV3), a paramyxovirus, is second only to respiratory syncytial virus in causing severe respiratory tract infections in children and young adults (6). The single-stranded RNA genome of HPIV3, 15,461 nucleotides (nt) long, is contained within a helical nucleocapsid (15). Three virus-encoded proteins, the nucleocapsid protein N (68 kDa), the phosphoprotein P (90 kDa), and the RNA polymerase L (257 kDa), are associated with the nucleocapsid to form a transcribing ribonucleoprotein (RNP) complex (1, 15). The N protein enwraps the genome RNA, while the L and P proteins together constitute the RNA-dependent RNA polymerase complex that transcribes and replicates the N-bound genome RNA. During transcription, the RNA polymerase synthesizes a short leader RNA, followed by six capped and polyadenylated mRNAs. During replication, on the other hand, a full-length plus-strand copy of the genome RNA is synthesized which, in turn, serves as the template for synthesis of the minus-strand genome RNA (1, 6, 15). Studies from our laboratory, as well as others, indicate that cellular cytoskeletal proteins such as actin and tubulin are involved in the activation of transcription of several paramyxoviruses (4, 9, 12, 22, 34, 35). Similarly, cellular RNA binding proteins that interact with the viral cis-acting elements are believed to be involved in the regulation of transcription and replication (25, 26, 28).
The minus-sense genomic 3′ end and its complementary leader sequence are the key cis-acting RNA elements that regulate viral transcription and replication. The 3′ end of the genome RNA serves as the site for interaction of the RNA polymerase to initiate RNA synthesis. The leader sequence at the 5′ end of the nascent plus-strand RNA, on the other hand, is involved in the initiation of encapsidation by the N protein during replication. Thus, the N protein, which remains complexed with the P protein, is the viral trans-acting component that modulates the balance between transcription and replication. However, some studies indicate that in addition to N protein, some cellular protein(s) may play a role in modulating the switch from transcription to replication (14). In fact, comparison of the nucleotide sequences of different paramyxovirus 5′- and 3′-terminal RNA sequences identified highly conserved regions which may form the cognate site for binding of cellular proteins (50). In the case of HPIV3, we recently reported that cellular glyceraldehyde 3-phosphate dehydrogenase (GAPDH) and La protein interact with the viral cis-regulatory RNAs (11). Further analysis revealed that these cellular proteins are also packaged within the progeny virions, suggesting a role(s) for these cellular proteins in HPIV3 transcription and replication.
GAPDH (37 kDa), a thoroughly studied key enzyme of the glycolytic pathway, is responsible for the oxidative phosphorylation of glyceraldehyde 3-phosphate by NAD+ and inorganic phosphate. Evidence is emerging that GAPDH is also involved in various other cellular functions, such as DNA binding (39, 52), tRNA export (45), uracil DNA glycosylase activity (3, 32), and association with polyribosomes (42). Furthermore, an RNA-unwinding property has been identified in GAPDH which is believed to facilitate translation of mRNAs in the polyribosomes (23). In accord with these varied activities, GAPDH is distributed both in the cytosol and in the nucleus and has been shown to exist in multiple molecular forms (33). Moreover, certain isoforms of GAPDH have been shown to be involved in specific functions (17, 33). Recently, GAPDH has been shown to interact with the cis-acting RNAs of several viruses, including HPIV3 (11), hepatitis A virus (43), hepatitis B virus (54), rabies virus (A. K. Gupta et al., unpublished data), and measles virus (B. P. De et al., unpublished data). These findings suggest that GAPDH, besides being involved in normal cellular function, may also play a role in the regulation of gene expression of several viruses.
The La protein (50 kDa) is an essential factor in RNA polymerase III transcription (18, 29) and has been shown to be involved in normal cellular functions, as well as in some pathological conditions, such as systemic lupus erythematosus and Sjögren's syndrome (48). In normal cell function, it is required for efficient termination of RNA polymerase III-mediated transcripts and thereby facilitates the reinitiation of RNA chains (13, 29). Specific phosphorylation of La protein by casein kinase II (CKII) at Ser366 has been shown to regulate this process (13). Recent studies indicate that the La protein exists in multiple molecular forms (30) and thus may be involved in diverse activities, as reported for GAPDH (17, 30, 33, 37, 41). Similar to GAPDH, the La protein also interacts with cis-acting RNAs of several viruses, including HPIV3 (11), vesicular stomatitis virus (26, 53), rabies virus (25), Sindbis virus (38), poliovirus (31), and human immunodeficiency virus (5). Although the La protein's role in the virus life cycle remains largely unclear, its interaction with poliovirus RNA has been shown to relieve structural constraint during translation.
To gain further insight into the molecular mechanism of GAPDH and La protein interaction with HPIV3 and their role in the virus life cycle, we have analyzed these virion-associated proteins by two-dimensional gel electrophoresis and immunoblotting. Here we demonstrate that specific phosphorylated forms of GAPDH interact with the viral genomic cis-acting RNA and remain packaged within the virions. Moreover, purified GAPDH containing the phosphorylated isoforms inhibited HPIV3 mRNA synthesis by purified viral RNP in vitro. These data indicate that specific phosphorylated forms of GAPDH play a role in the regulation of virus gene expression.
MATERIALS AND METHODS
Cells and viruses.
CV-1 cells (ATCC CCL 185) were propagated in monolayers as described previously (8). HPIV3 (HA-1; NIH 47885) was grown in CV-1 cells and purified as described previously (8).
HPIV3 transcription in vitro.
HPIV3 transcription was carried out by using purified viral RNP extracted from purified virions and further purified as described previously (8). Briefly, the purified virions (2 mg at 500 μg/ml) were disrupted in 10 mM Tris-HCl (pH 8.0) containing 5% glycerol, 0.4 M NaCl, 1.85% Triton X-100, and 0.6 mM (final concentration) dithiothreitol (DTT) at room temperature for 10 min. The lysate was centrifuged through 30% (vol/vol) glycerol (1.5 ml) in 20 mM HEPES-KOH (pH 7.5) and 1 mM DTT onto a 100% glycerol cushion (0.5 ml) using an SW 50.1 rotor at 40,000 rpm for 2 h. The purified RNP was collected from the top of the 100% glycerol cushion and used in the transcription reaction. The transcription reaction mixture, in a 50-μl final volume, contained 100 mM HEPES (pH 8.0); 100 mM KCl; 5 mM MgCl2; 1 mM DTT; 1 mM each ATP, GTP, CTP, and UTP; 25 U of human placental RNase inhibitor; 2 μg of RNP; and partially purified actin. Incubation was carried out at 30°C for 3 h. The in vitro-synthesized RNAs were purified by phenol extraction and ethanol precipitation and analyzed by primer extension using an N mRNA-specific primer that produces a 91-nt-long primer extension DNA product. The extended DNA was analyzed by 5% polyacrylamide–urea gel electrophoresis, followed by autoradiography.
Plasmid construction and T7 RNA polymerase transcription.
Construction of two plasmids containing the 73-nt 3′ genomic-sense (3′-GS) and leader sense (LS) RNAs, respectively, under control of the T7 promoter was described earlier (11). Radiolabeled RNAs were synthesized using these plasmid DNAs after linearization with HgaI in an in vitro transcription reaction mixture containing 500 μM each ATP, GTP, and CTP; 25 μM UTP; 50 μCi of [α-32P]UTP; and 20 U of T7 RNA polymerase in a 50-μl volume in accordance with the manufacturer's (Boehringer Mannheim) protocol. The 73-nt-long transcript would contain 55-nt from the leader region and 18 nt from the N gene. In vitro-synthesized RNAs were analyzed in a 10% polyacrylamide–urea gel, and the radiolabeled RNAs were excised. The RNAs were then eluted in a buffer containing 0.5 M ammonium acetate, 1 mM EDTA, and 0.1% sodium dodecyl sulfate (SDS), and purified by phenol extraction and ethanol precipitation.
Cell labeling and in vitro phosphorylation of proteins.
CV-1 cells were grown in monolayers in Dulbecco modified Eagle medium supplemented with 10% fetal bovine serum. The cells were grown overnight in phosphate-free medium containing dialyzed serum before addition of [32P]orthophosphate. Cells were infected with HPIV3 at a multiplicity of infection of 5 and labeled at 5 h postinfection with [32P]orthophosphate. The medium containing the progeny virions was collected at 40 h postinfection and centrifuged at 1,500 × g to remove cell debris. The virions were pelleted from the supernatant by further centrifugation at 100,000 × g for 2 h. For two-dimensional protein gel electrophoresis, the viral pellet was directly dissolved in two-dimensional gel electrophoresis sample buffer and analyzed.
In vitro phosphorylation of GAPDH and La protein was performed using rat brain protein kinase (PKC) and recombinant CKII as described previously (2, 10).
Gel mobility shift assay.
Binding of 32P-labeled 3′-GS and LS RNAs to the cellular proteins was performed by following the procedure described previously (11). The reaction mixture (20 μl) contained 15 mM HEPES-KOH (pH 8.0), 15 mM KCl, 0.25 mM EDTA 0.25 mM DTT, 5 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride, 200 μg of yeast tRNA per ml, 10% glycerol, 0.1 ng of radiolabeled RNA, and (unless otherwise indicated) 0.5 μg of purified cellular proteins or 50 ng of commercially available rabbit muscle GAPDH. The reaction mixture was incubated at room temperature for 30 min, and the samples were analyzed in a 6% nondenaturing polyacrylamide gel in 0.5× Tris-borate-EDTA buffer. The gel was run at 150 V and room temperature and then dried and subjected to autoradiography.
Two-dimensional protein gel electrophoresis.
CV-1 cells uninfected or infected with HPIV3 were lysed directly in the two-dimensional gel electrophoresis sample buffer (36). First-dimensional gel electrophoresis was performed using the method described by Harrington et al. (21), with slight modifications. Isoelectrophoresis was performed in a 20-cm-long glass tube for 10 kV-h with a separation pH gradient of 6.8 to 8.5. Second-dimensional electrophoresis was performed using the procedure described by Harrington et al. (21). Proteins were transferred onto nitrocellulose membrane for Western blot analysis (51). The proteins on the membrane were detected using specific antibodies, followed by ECL (Amersham). 32P-labeled samples were exposed to X-ray films after Western blot analysis where indicated.
Purification of cellular and recombinant GAPDH.
Soluble cytoplasmic GAPDH was purified from A549 cells by following a previously published procedure with modifications (11). Cells (108) were harvested in phosphate-buffered saline and pelleted by centrifugation at 1,000 × g for 10 min. The cell pellet was suspended in 2 ml of hypotonic buffer containing 10 mM Tris-HCl (pH 8.0), 10 mM NaCl, and 1 mM DTT. The cells were then lyzed by four cycles of freezing and thawing, and the lysate was clarified by centrifugation at 10,000 × g for 10 min. The clarified supernatant was dialyzed overnight against buffer A (50 mM HEPES-KOH [pH 7.5], 5 mM MgCl2, 10 mM KCl, 3% glycerol, 1 mM DTT). The dialyzed extract was loaded onto a phosphocellulose column (1-ml bed volume) equilibrated with buffer A. The column was washed with 4 column volumes of buffer A, and the bound proteins were eluted with a 0 to 1 M NaCl (5-ml total volume) linear gradient in buffer A. Aliquots of individual fractions were analyzed by Western blotting using anti-GAPDH antibody. These fractions were also analyzed by using anti-actin antibody in the Western blot.
Recombinant flag-tagged GAPDH was expressed using the recombinant VTF7-3 vaccinia virus expression system. Briefly, full-length rat GAPDH cDNA was spliced into vector plasmid pGEM4 under control of the T7 promoter at the BamHI-KpnI site. The plasmid DNA was further engineered such that the C terminus of the expressed protein would contain 8-amino-acid flag (DYKDDDDK). The resulting plasmid DNA was transfected into HeLa cells in a six-well plate that were infected with VTF7-3 using Lipofectin reagent following the manufacturer's (Boehringer Mannheim) protocol. A 12 h postinfection or posttransfection, the cells were labeled with [35S]methionine (20 μCi/ml) in methionine-free medium for 6 h. Cytoplasmic proteins were extracted using luciferase assay kit cell lysis buffer and following the manufacturer's (Boehringer Mannheim) protocol. The radiolabeled, flag-tagged GAPDH in the extract was purified by immunoprecipitation using anti-flag antibody M2 conjugated to Sepharose beads in accordance with the manufacturer's (Sigma) protocol. Immunoprecipitated proteins in the beads were estimated, analyzed by SDS-polyacrylamide gel electrophoresis, and directly used for HPIV3 transcription in vitro.
RESULTS
Packaging of multiple molecular forms of GAPDH and La protein within HPIV3 virions.
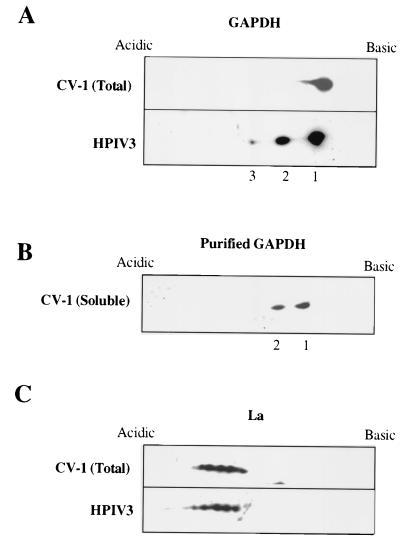
We previously demonstrated that cellular GAPDH and La protein specifically interact with the HPIV3 cis-acting RNAs and also remain packaged within the progeny virions (11). Cellular GAPDH and La protein have been shown to undergo posttranslational modifications generating multiple molecular forms which are involved in various cellular functions (17, 30, 33, 37, 41). In light of these observations, we investigated the involvement of such molecular forms of GAPDH and La protein in the interaction with HPIV3. Virion-associated GAPDH and La protein were analyzed using two-dimensional gel electrophoresis, followed by immunoblotting. As shown in Fig. 1A, three distinct isoforms of GAPDH (two major and one minor) that migrated in the pI range of 7.6 to 8.3 were present within the virions. To determine whether these isoforms were also present in the cellular pool, we similarly analyzed the GAPDH present in the CV-1 cell extract. Surprisingly, in the total cellular pool, GAPDH was identified predominantly as a single species (pI of about 8.4) that was different from the species packaged within the virions. Further analysis by cell fractionation showed that in the soluble cytosolic fraction there are minor species that are similar to the virion-associated GAPDH isoforms (data not shown). These species were subsequently purified from the cytosolic fraction by successive chromatography using DEAE-cellulose and phosphocellulose columns and confirmed to be similar to those present within the virions with respect to pI (Fig. 1B). The particulate fraction, on the other hand, contained the major isoform (>90%), similar to that seen in Fig. 1A. These results indicate that the soluble isoforms of GAPDH in the pI range of 7.6 to 8.3 are selectively packaged within the virions, possibly due to their strong interaction with the viral genomic cis-regulatory RNA.
FIG. 1.
Molecular forms of GAPDH and La within HPIV3 virions and in CV-1 cells. HPIV3 virion-associated GAPDH and La protein were subjected to two-dimensional gel electrophoresis, followed by Western blot analysis using anti-GAPDH and anti-La antibodies. Similarly, the GAPDH and La protein present in CV-1 cell extract were analyzed. Migration patterns in isoelectric focusing gels are shown. The CV-1 cell cytosolic GAPDH was purified by chromatography on DEAE-cellulose and phosphocellulose columns and analyzed by two-dimensional gel electrophoresis. Panels: A, GAPDH present in the virions and CV-1 cell extract; B, GAPDH purified from the CV-1 cell cytosolic fraction; C, La protein present in virions and CV-1 cell extract.
As shown in Fig. 1C, analysis of virion-associated La protein revealed the presence of five distinct molecular forms which were acidic in nature and migrated in the pI range of 6.8 to 7.5. All five isoforms were also detected in the total cellular pool in the same relative amounts as within the virion. These data suggest that all five isoforms of La protein might interact with the cis-regulatory RNA with similar affinity and possibly play some role in the virus's life cycle. We consequently focused primarily on characterization of the specific molecular forms of GAPDH present within the virions.
The virion-associated GAPDH isoforms are highly phosphorylated.
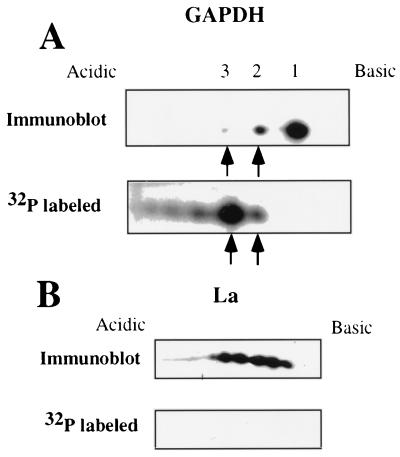
Packaging of specific molecular forms of GAPDH within the HPIV3 virions suggested that a subpopulation of GAPDH is possibly modified e.g., by phosphorylation, and thus interacted with the virions (24, 41). To examine the phosphorylation status of GAPDH isoforms present in the virions, CV-1 cells were infected with HPIV3 at a multiplicity of infection of 5 and labeled at 5 h postinfection with [32P]orthophosphate. At 40 h postinfection, released virions were harvested from the medium and subjected to two-dimensional polyacrylamide gel electrophoresis. The proteins were detected by Western blotting, followed by autoradiography, and 32P-labeled spots were superimposed on the specific spots observed by Western blotting. Similar to that shown in Fig. 1A, two major isoforms were detected by Western blot analysis (Fig. 2A) whereas five 32P-labeled species were identified. Superimposition of these spots showed that the major isoform (spot 1) was not labeled with 32P. However, the same spot was detected by Western blotting when we used antiphosphoserine and antiphosphothreonine antibodies (data not shown), suggesting that this isoform was packaged from the preexisting prephosphorylated pool of cytosolic GAPDH. In contrast, the highly acidic isoforms, spots 2, 3, and several others remained undetected by Western blotting due to their presence in small amounts but were clearly detected in the 32P autoradiogram because of the high level of phosphorylation of these species. These results suggest that specific phosphorylated isoforms of GAPDH are packaged within the virions.
FIG. 2.
Phosphorylation status of virion-associated GAPDH and La. HPIV3 virions harvested from 32P-labeled CV-1 cells as described in Materials and Methods were analyzed by two-dimensional gel electrophoresis. GAPDH and La protein were detected by Western blot analysis using anti-GAPDH and anti-La antibodies. The blot was then exposed to X-ray film to detect 32P-labeled spots. Arrows indicate that these spots were detected by both Western blotting and 32P labeling. The basic isoform (spot 1) is not phosphorylated, whereas the minor species that were not detected in the Western blot were heavily phosphorylated. Panels: A, analysis of GAPDH by immunoblotting and autoradiography; B, analysis of La by immunoblotting and autoradiography.
The virion-associated La protein, on the other hand, was detected in the Western blot in multiple molecular forms but was undetectable in the 32P autoradiogram (Fig. 2B). This indicated that the La protein is packaged within the virions in the unphosphorylated forms. This was also confirmed by the findings that virion-associated La isoforms were not detected by antiphosphoserine and antiphosphothreonine antibodies (data not shown). The microheterogeneity of the La protein present within the virions could therefore be due to some modifications other than phosphorylation.
Involvement of PKC in the phosphorylation of GAPDH.
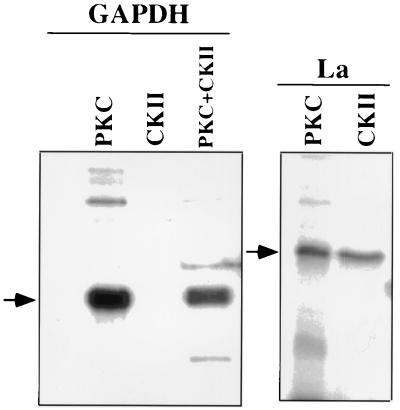
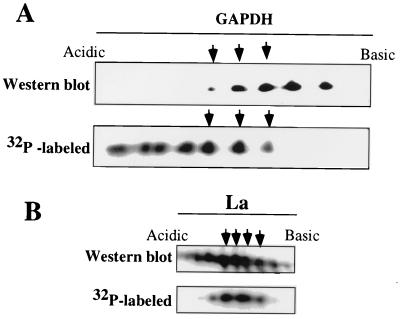
Next, we investigated whether GAPDH and the La protein were phosphorylated in vitro and phosphorylation had similar effects on the migration of these proteins, as analyzed by two-dimensional polyacrylamide gel electrophoresis. We used commercially available purified rabbit muscle GAPDH and bacterially expressed human La protein. Phosphorylation of these proteins in vitro was performed using rat brain PKC and recombinant CKII. As shown in Fig. 3, GAPDH was specifically phosphorylated by PKC but not by CKII. The La protein, on the other hand, was phosphorylated by both PKC and CKII. The phosphorylated GAPDH and La protein and their unphosphorylated counterparts were then analyzed by two-dimensional polyacrylamide gel electrophoresis. As shown in Fig. 4A, unphosphorylated rabbit muscle GAPDH, unlike CV-1 cell GAPDH (Fig. 1), was resolved into five distinct species. Such cell type-specific differences in GAPDH have previously been shown (17, 33, 49). Nonetheless, phosphorylation of GAPDH in vitro resulted in the migration of these species toward a more acidic pI, similar to the migration pattern of virion-associated phosphorylated GAPDH isoforms (Fig. 2A). Thus, it seems that the observed migration of virion-associated GAPDH following phosphorylation (Fig. 2A) toward an acidic region was indeed due to the increased degree of phosphorylation. The La protein isoforms, unlike GAPDH, displayed a minor shift in their migration pattern toward the acidic region following phosphorylation, indicating that phosphorylation also contributes, to some extent, to the microheterogeneity of La protein. Since multiple unphosphorylated forms of La protein are found within the virions (Fig. 2B), it appears that differently modified forms of La protein are selectively packaged by HPIV3 virions.
FIG. 3.
In vitro phosphorylation of rabbit muscle GAPDH and bacterially expressed La. Rabbit muscle GAPDH and bacterially expressed La protein were phosphorylated using [γ-32P]ATP and commercially available PKC and CKII. Phosphorylated proteins were resolved by SDS–10% polyacrylamide gel electrophoresis, dried, and exposed to X-ray film. The migration positions of GAPDH and La protein are indicated by the arrows.
FIG. 4.
Migration patterns of in vitro-phosphorylated GAPDH and La protein in two-dimensional gel electrophoresis. Rabbit muscle GAPDH and bacterially expressed La protein were phosphorylated in vitro with PKC and CKII, respectively, and analyzed by two-dimensional gel electeophoresis. The proteins were detected by Western blotting using anti-GAPDH and anti-La antibodies, followed by autoradiography, for the detection of 32P-labeled proteins. The arrows indicate that these same spots were detected by Western blotting, as well as autoradiography. Several 32P-labeled spots were not visible in the Western blot. The two very basic spots detected in the Western blot are not labeled with 32P.
Phosphorylation-mediated regulation of the interaction of GAPDH with viral cis-acting RNAs.
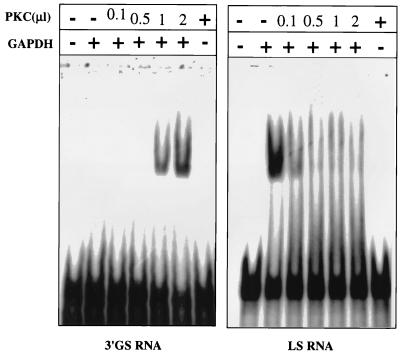
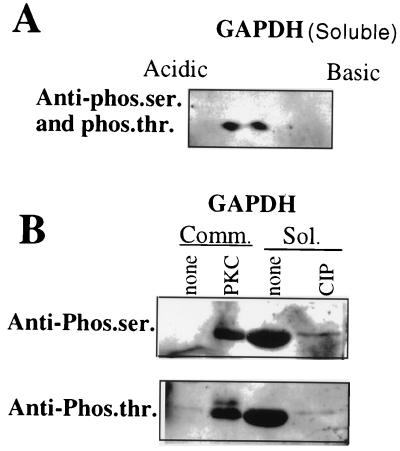
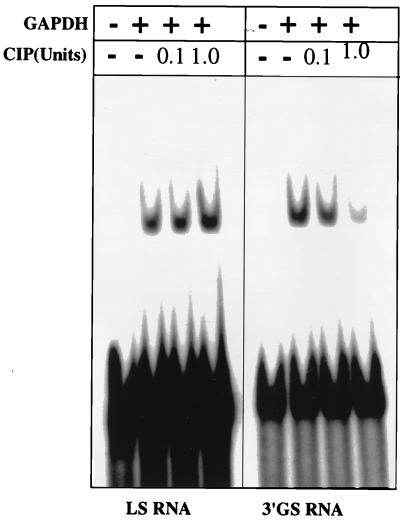
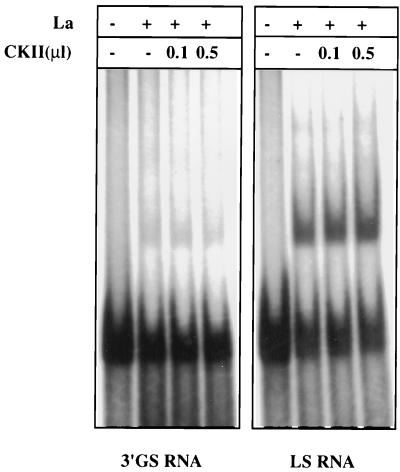
Our previous studies indicated that purified cellular cytosolic GAPDH binds to 3′-GS RNA with about threefold higher affinity than to LS RNA (11). This raised the possibility that the isoforms that bind to the 3′-GS RNA with higher affinity perhaps represent the phosphorylated forms of GAPDH, as seen within the virions. To investigate this, we phosphorylated commercially available rabbit muscle GAPDH in vitro with PKC and studied its interaction with 3′-GS and LS RNAs by gel mobility shift assay. As shown in Fig. 5, phosphorylation of GAPDH with increasing concentrations of PKC showed progressively increased binding of GAPDH to the 3′-GS RNA. By contrast, phosphorylation resulted in a dramatic decrease in binding of GAPDH to the LS RNA, indicating a phosphorylation-mediated switch of the binding property of GAPDH. To further investigate the role of phosphorylation in binding of GAPDH to 3′-GS and LS RNAs, we used GAPDH purified from the CV-1 cell cytosolic fraction (11). Purified GAPDH was subjected to phosphorylation in vitro using PKC and [γ-32P]ATP. However, the purified cytosolic GAPDH could not be phosphorylated in vitro, suggesting that it is present in fully phosphorylated form (data not shown). This was confirmed when purified cytosolic GAPDH was analyzed by two-dimensional gel electrophoresis and immunoblotting with anti-GAPDH, as well as antiphosphoserine and antiphosphothreonine, antibodies. As shown in Fig. 6A, the molecular species of purified cellular GAPDH (pI range of 7.6 to 8.3) that reacted with anti-GAPDH antibody (Fig. 1B) were also recognized by antiphosphoserine and antiphosphothreonine antibodies. This indicated that the cellular GAPDH is phosphorylated at Ser/Thr residues, presumably by PKC. The cellular cytosolic GAPDH was then analyzed by Western blotting after dephosphorylation using calf intestinal alkaline phosphatase. As shown in Fig. 6B, the cytosolic GAPDH did not react with either phosphoserine- or phosphothreonine-specific antibody following dephosphorylation. Some reactivity of antiphosphoserine and antiphosphothreonine antibodies with the band most likely represents a small amount of GAPDH that was not completely dephosphorylated by the phosphatase. The unphosphorylated state of rabbit muscle GAPDH was confirmed by its inability to react with the antibodies. However, it efficiently reacted following phosphorylation by PKC. Having confirmed the phosphorylated state of cytosolic GAPDH, we treated the GAPDH with increasing concentrations of calf intestinal alkaline phosphatase and analyzed its binding to the 3′-GS and LS RNAs in vitro. As shown in Fig. 7, dephosphorylation of GAPDH resulted in increased binding to the LS RNA with a concomitant decrease in binding to the 3′-GS RNA. These results confirm that phosphorylation of GAPDH regulates its interaction with the viral cis-acting RNAs in vitro. The role of phosphorylation in the interaction of La protein with viral cis-acting RNAs was similarly investigated. Bacterially expressed La protein was used for binding to LS and 3′-GS RNAs in gel mobility shift assays following phosphorylation by CKII in vitro. As shown in Fig. 8, La protein specifically bound to the LS RNA and CKII-mediated phosphorylation had no effect on the binding. These data indicate that the specific interaction of La protein with LS RNA, unlike that of GAPDH, does not depend upon the phosphorylation state of the protein.
FIG. 5.
Effect of phosphorylation on the binding of GAPDH to cis-acting RNAs. Rabbit muscle GAPDH was phosphorylated in vitro using unlabeled ATP and increasing amounts of commercially available PKC (0.08 mU/μl). About 100 ng of GAPDH was used in a gel mobility shift assay with in vitro-transcribed [32P]UTP-labeled cis-acting RNAs (3′-GS and LS RNAs), as indicated. PKC alone was used as a gel shift control.
FIG. 6.
Analysis of GAPDH using antiphosphoserine (Anti-phos.ser.) and antiphosphothreonine (Anti-phos.thr.) antibodies. Commercial (Comm.) and purified cellular soluble (Sol.) GAPDH were subjected to two-dimensional gel electrophoresis or SDS–10% polyacrylamide gel electrophoresis, followed by Western blot using anti-GAPDH and antiphosphoserine or antiphosphothreonine antibodies. The proteins were either phosphorylated by PKC or dephosphorylated by calf intestinal alkaline phosphatase (CIP), as indicated, before gel electrophoresis. Panels: A, GAPDH analyzed by two-dimensional gel electrophoresis; B, GAPDH analyzed by SDS–10% polyacrylamide gel electrophoresis.
FIG. 7.
Effect of dephosphorylation on the binding of cytoplasmic GAPDH to cis-acting RNAs. GAPDH was purified from the soluble fraction (S100) of CV-1 cells by column chromatography as described in Materials and Methods. Purified GAPDH (30 ng) was treated with increasing amounts of phosphatase and then used in gel shift assays with 32P-labeled 3′-GS and LS RNAs. Mock-treated GAPDH served as a control. Phosphatase alone did not bind to either of the probes (data not shown). Treatment of GAPDH with increasing amounts (0.1 and 1.0 U) of calf intestinal alkaline phosphatase (CIP; Boehringer Mannheim) and binding of the phosphatase-treated GAPDH to 3′-GS and LS RNAs are shown.
FIG. 8.
Effect of phosphorylation on the binding of La protein to cis-acting RNAs. Bacterially expressed La protein was phosphorylated in vitro using unlabeled ATP and increasing amounts of commercially available CKII (0.2 mU/μl). About 100 ng of La protein was used in a gel mobility shift assay with in vitro-transcribed [32P]UTP-labeled cis-acting RNAs (3′-GS and LS RNAs), as indicated. The faster-migrating band represents the free probe.
Inhibition of HPIV3 transcription in vitro by purified GAPDH.
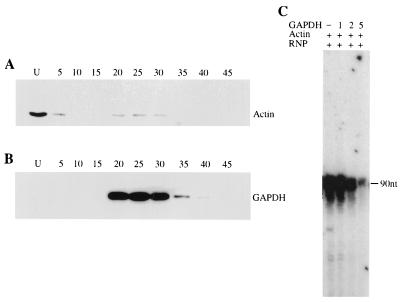
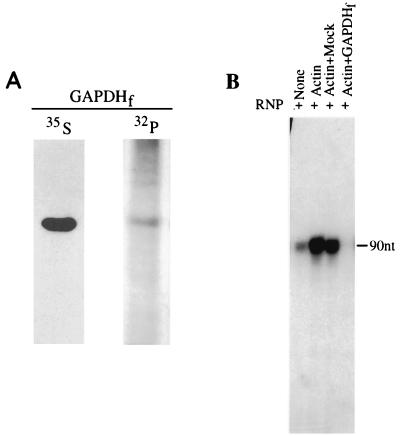
Specific interaction of GAPDH isoforms with viral cis-acting RNAs and packaging within the progeny virions suggest a role for GAPDH in the regulation of viral transcription and replication. To investigate this, we partially purified CV-1 cell cytoplasmic phosphorylated GAPDH. As shown in Fig. 9A and B, the cytoplasmic GAPDH was separated from actin in the phosphocellulose column in which GAPDH was eluted in the bound fraction whereas actin was eluted in the unbound fraction of the column. The actin, purified free of GAPDH, was used to activate transcription by purified viral RNP for mRNA synthesis in vitro. The partially purified GAPDH was then examined for its effect on viral mRNA synthesis in vitro. As shown in Fig. 9C, increasing amounts of GAPDH inhibited viral transcription efficiently in a dose-dependent manner, as determined by primer extension analysis. The commercial unphosphorylated form of GAPDH, on the other hand, had no effect on transcription but gained significant inhibitory activity following phosphorylation by PKC in vitro (data not shown). To further confirm the inhibitory role of the phosphorylated form of GAPDH in viral transcription, we used recombinant GAPDH. The GAPDH was expressed in the recombinant vaccinia virus expression system, and the plasmid DNA was engineered such that the expressed recombinant GAPDH would be flag tagged at the C terminus. The protein was metabolically labeled with [35S]methionine as well as [32P]orthophosphate. The radiolabeled, flag-tagged recombinant GAPDH was purified by immunoprecipitation with anti-flag antibody conjugated to Sepharose beads. As shown in Fig. 10A, the flag-tagged recombinant GAPDH was highly purified and was present in the phosphorylated form. The beads containing immunoprecipitated GAPDH were then directly used in the transcription reaction. Mock-treated cell extract was similarly used for immunoprecipitation with antiflag antibody-conjugated Sepharose beads and used as a control. As shown in Fig. 10B, purified, flag-tagged recombinant GAPDH, like cellular cytoplasmic GAPDH, strongly inhibited viral transcription in vitro. Together, these data indicate that phosphorylated forms of GAPDH associate with viral RNP to inhibit mRNA synthesis by the viral RNA polymerase.
FIG. 9.
Inhibition of HPIV3 transcription in vitro by GAPDH. Cellular GAPDH was purified from HeLa cells as described in Materials and Methods. Fractions (numbered in panels A and B) were analyzed for GAPDH and actin by Western blotting. U, the unbound fraction. Actin was eluted in the unbound fraction (A), and the protein concentration was estimated to be 1 mg/ml. The GAPDH was eluted from the column in 20 to 30 fractions (B). These GAPDH fractions were pooled, and the protein concentration was estimated to be 0.4 mg/ml. In vitro transcription (C) was carried out as described in Materials and Methods in a transcription reaction mixture containing purified RNP (1 μg) and partially purified actin (5 μg). Increasing amounts of GAPDH (the values above the lanes are volumes in microliters) was added to the transcription reaction mixture. Viral transcription was measured by primer extension analysis as described in Materials and Methods. The migration position of the 90-nt primer extension product, as confirmed by using a sequencing ladder, is indicated.
FIG. 10.
Purified, flag-tagged recombinant GAPDH (GAPDHf) inhibits HPIV3 transcription in vitro. Flag-tagged GAPDH was expressed in HeLa cells using the recombinant vaccinia virus expression system and was purified by immunoprecipitation as described in Materials and Methods. The expressed protein in parallel experiments was labeled with [35S]methionine and [32P]orthophosphate, immunoprecipitated, and analyzed in SDS-polyacrylamide gels (A). Purified GAPDH (0.5 μg) was used in a transcription reaction (B) as described in the legend to Fig. 9.
DISCUSSION
Cellular proteins have been shown to interact with the cis-acting RNAs of several RNA viruses and are believed to play a role in the regulation of viral transcription and replication (7, 27, 46). We previously reported that cellular GAPDH and La protein interact with the HPIV3 cis-acting RNAs in vitro and, more importantly, that these proteins remain packaged within the progeny virions (11). In this study, we demonstrated by two-dimensional gel electrophoresis that the virion-associated GAPDH exists in several isoforms that are heavily phosphorylated. Consistent with these findings, we observed that cellular cytosolic GAPDH also exists in various phosphorylated forms which appear to be selectively packaged by the virions. By phosphorylation and dephosphorylation of cytosolic GAPDH, we demonstrated that phosphorylation plays a regulatory role in the interaction of GAPDH with the viral cis-acting RNAs (Fig. 2A, 5, and 6). The La protein, on the other hand, was present within the virions in unphosphorylated form (Fig. 2B), and phosphorylation had no effect on its interaction with the cis-acting RNAs (data not shown). In vitro phosphorylation studies indicate that GAPDH is phosphorylated specifically by PKC, whereas La protein is phosphorylated by both PKC and CKII. Thus, it seems that phosphorylated forms of GAPDH specifically interact with the viral genomic cis-acting RNA (3′-GS RNA) and thereby remain packaged within the progeny virions. Moreover, purified GAPDH containing the phosphorylated isoforms inhibited viral transcription in vitro. These data suggest that phosphorylated forms of GAPDH may play a role in the virus's life cycle.
Identification of multiple molecular forms of GAPDH is consistent with the multifunctional nature of the protein. Different molecular forms are believed to be involved in specific biological activities, including DNA replication, DNA repair, translational control of gene expression, and apoptosis (46). The microheterogeneity appears to arise from the variation in the GAPDH primary sequence, as well as posttranslational modification of the protein (17, 30, 33, 37, 40, 41). Although only one gene coding for GAPDH is functional in humans, there are 10 to 30 copies of pseudogenes which are expressed, depending on the types of tissues or state of differentiation (40). Similarly, posttranslational modification of GAPDH, such as phosphorylation, covalent linkage with NAD+, ADP-ribosylation, and S-thiolation, have been reported (30, 37, 41, 49). However, a specific function of these molecular forms has not yet been demonstrated. In this regard, interaction of phosphorylated forms of GAPDH with HPIV3 cis-acting RNAs is the first demonstration of involvement of a specific molecular form of GAPDH in the virus life cycle. Furthermore, distribution of phosphorylated forms of GAPDH in the cytosolic fraction indicates that HPIV3 selectively packages phosphorylated GAPDH from the cytosolic pool. These molecular forms are further phosphorylated within the virions, possibly by a virion-associated protein kinase. Since HPIV3 packages PKCζ, it is conceivable that PKCζ hyperphosphorylates GAPDH within the virions. This notion is supported by the findings that PKCζ is able to phosphorylate GAPDH in vitro (data not shown). The degree of phosphorylation of GAPDH (hypo- or hyperphosphorylation) thus plays a regulatory role in the association of GAPDH with the HPIV3 virions.
Our data demonstrate for the first time that cellular GAPDH which binds to the HPIV3 cis-acting RNA is directly involved in the inhibition of viral transcription. Since HPIV3 utilizes cellular actin for its transcription activation, it appears that both positive and negative transcription factors are involved in the regulation of viral gene expression. This is similar to the findings obtained with Sendai virus, where tubulin was shown to act as a positive transcription factor and another protein, as yet uncharacterized, was shown to act as a negative factor (47). This raises the question of whether interaction of these cellular factors with viral RNP is temporally regulated in infected cells. Our findings indicate that actin interacts with HPIV3 RNP during the early stage of infection, which is consistent with its role in transcription activation (19). The GAPDH, on the other hand, associates with HPIV3 RNP in infected cells during the late stage of the virus replicative cycle, i.e., 24 h postinfection (11). Furthermore, our data indicate that specific phosphorylated forms of GAPDH bind to the 3′-GS RNA and remain packaged within the virion. Thus, it remains to be seen whether phosphorylation of GAPDH is induced in the late stage of the virus replicative cycle. However, our attempts to detect such phosphorylation in infected cells by metabolic labeling with [32P]orthophosphate, followed by immunoprecipitation, failed, perhaps due to interference by a large excess of unphosphorylated GAPDH species at any given time of labeling and immunoprecipitation (data not shown). Regarding physiological significance of the interaction of GAPDH with HPIV3, it is tempting to speculate that the GAPDH-mediated transcription shutoff plays a role in the switch from transcription to replication or is involved in the assembly and budding of virions. In the former case, interaction of GAPDH may favor virus replication in the presence of soluble N protein, although detection of GAPDH in the RNP only at a very late stage of infection, i.e., 24 h postinfection (11), argues against this. However, the possibility remains that a small amount of a highly phosphorylated form, undetected by Western blotting, associates at an early stage of virus replication to mediate this process. This notion is supported by the findings that cellular cytosolic GAPDH and recombinant flag-tagged GAPDH both contain phosphorylated isoforms and strongly inhibit viral transcription. However, we have not been able to establish that the phosphorylated isoforms are the only species that inhibit transcription, because these isoforms could not be purified to homogeneity in sufficient quantity free from the bulk of unphosphorylated GAPDH for use in the transcription reaction. It is noteworthy that a minor subpopulation of GAPDH has been shown to function as an activator of cellular transcription in neuronal cells (33). The GAPDH-mediated inhibition of viral transcription may, alternatively, be involved in virus assembly and budding. In that case, HPIV3 may utilize cellular GAPDH for complete shutoff of viral RNA synthesis during maturation and budding. Given that nonsegmented negative-strand RNA virus M protein is similarly involved in transcription shutoff during budding (16), it remains to be seen whether GAPDH aids in this process. Further studies are therefore needed to investigate these possibilities.
With regard to the La protein, it was also detected in multiple molecular forms within HPIV3 virions. However, unlike that of GAPDH, phosphorylation was found to play no role in the microheterogeneity of virion-associated La protein. This suggests that different modifications of La protein are involved in the regulation of its function in the virus's life cycle. Specific binding of La protein to the LS RNA (11) suggests that the La protein is involved, like RNA polymerase III transcription, in the termination of leader RNA, which has many features in common with RNA polymerase III transcripts (18, 26, 29).
Interaction of GAPDH and La protein with the cis-acting regulatory RNAs of viruses of different families suggests that these viruses utilize cellular proteins for the same function, i.e., alteration of the structure of template RNA during gene expression. Besides GAPDH and La protein, other RNA binding proteins, such as heterogeneous nuclear RNP particle U and calreticulin, have also been shown to interact with the RNAs of vesicular stomatitis virus and rubella virus, respectively (20, 44). Although the function of these cellular proteins in the virus life cycle remains largely unclear, their role in some viral system is emerging (7, 27, 46). Thus, characterization of specific molecular forms of GAPDH and La protein would certainly help elucidate their functions in the gene expression of HPIV3 and perhaps in other viral systems.
ACKNOWLEDGMENTS
We thank Ranjit Ray for providing anti-HPIV3 antibody.
This work was supported by U.S. Public Health Services grant AI32027 (A.K.B.).
REFERENCES
- 1.Banerjee A K, Barik S, De B P. Gene expression of negative strand RNA viruses. Pharmacol Ther. 1991;51:47–70. doi: 10.1016/0163-7258(91)90041-j. [DOI] [PubMed] [Google Scholar]
- 2.Barik S, Banerjee A K. Cloning and expression of the vesicular stomatitis virus phosphoprotein gene in Escherichia coli: analysis of phosphorylation status versus transcriptional activity. J Virol. 1991;65:1719–1726. doi: 10.1128/jvi.65.4.1719-1726.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baxi M D, Vishvanatha K. Uracil DNA glycosylase/glyceraldehyde 3-phosphate dehydrogenase is an Ap4A binding protein. Biochemistry. 1995;34:9700–9707. doi: 10.1021/bi00030a007. [DOI] [PubMed] [Google Scholar]
- 4.Burke E, Dupuy L, Wall C, Barik S. Role of cellular actin in the gene expression and morphogenesis of human respiratory syncytial virus. Virology. 1998;252:137–148. doi: 10.1006/viro.1998.9471. [DOI] [PubMed] [Google Scholar]
- 5.Chang Y N, Kenan D J, Keene J D, Gatignol A, Jeang K T. Direct interactions between autoantigen La and human immunodeficiency virus leader RNA. J Virol. 1994;68:7008–7020. doi: 10.1128/jvi.68.11.7008-7020.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Collins P L, Chanock R M, Mackintosh K. Parainfluenza viruses. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1205–1241. [Google Scholar]
- 7.De B P, Banerjee A K. Role of host proteins in gene expression of nonsegmented negative strand RNA viruses. Adv Virus Res. 1997;48:169–204. doi: 10.1016/s0065-3527(08)60288-2. [DOI] [PubMed] [Google Scholar]
- 8.De B P, Burdsall A L, Banerjee A K. Role of cellular actin in human parainfluenza virus type 3 genome transcription. J Biol Chem. 1993;268:5703–5710. [PubMed] [Google Scholar]
- 9.De B P, Galinski M S, Banerjee A K. Characterization of an in vitro system for the synthesis of mRNA from human parainfluenza virus type 3. J Virol. 1990;64:1135–1142. doi: 10.1128/jvi.64.3.1135-1142.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De B P, Gupta S, Gupta S, Banerjee A K. Cellular protein kinase C ζ regulates human parainfluenza virus type 3 replication. Proc Natl Acad Sci USA. 1995;92:5204–5208. doi: 10.1073/pnas.92.11.5204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De B P, Gupta S, Zhao H, Drazba J A, Banerjee A K. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase and La protein with cis-acting RNAs of human parainfluenza virus type 3. J Biol Chem. 1996;271:24728–24735. doi: 10.1074/jbc.271.40.24728. [DOI] [PubMed] [Google Scholar]
- 12.De B P, Lesoon A, Banerjee A K. Human parainfluenza virus type 3 transcription in vitro: role of cellular actin in mRNA synthesis. J Virol. 1991;65:3268–3275. doi: 10.1128/jvi.65.6.3268-3275.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan H, Sakulich A L, Goodier J L, Zhang X, Qin J, Maraia R J. Phosphorylation of the human La antigen on serine 366 can regulate recycling of RNA polymerase III transcription complexes. Cell. 1997;88:707–715. doi: 10.1016/s0092-8674(00)81913-3. [DOI] [PubMed] [Google Scholar]
- 14.Fearns R M, Peeles E, Collins P L. Increased expression of the N protein of respiratory syncytial virus stimulates minigenome replication but does not alter the balance between the synthesis of mRNA and antigenome. Virology. 1997;236:188–201. doi: 10.1006/viro.1997.8734. [DOI] [PubMed] [Google Scholar]
- 15.Galinski M S, Wechsler S L. Molecular biology of the paramyxovirus genus. In: Kingsbury D W, editor. Paramyxoviruses. New York, N.Y: Plenum Press; 1991. pp. 41–82. [Google Scholar]
- 16.Garoff H, Hewson R, Opstelten D J E. Virus maturation by budding. Microbiol Mol Biol Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glaser P E, Gross R W. Rapid plasmenylethanolamine-selective fusion of membrane bilayers catalyzed by an isoform of glyceraldehyde 3-phosphate dehydrogenase. Biochemistry. 1995;34:12193–12203. doi: 10.1021/bi00038a013. [DOI] [PubMed] [Google Scholar]
- 18.Gottlieb E, Steiz J A. The RNA binding protein La influences both the accuracy and the efficiency of RNA polymerase III transcription in vitro. EMBO J. 1989;8:841–850. doi: 10.1002/j.1460-2075.1989.tb03445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta S, De B P, Banerjee A K. Involvement of actin microfilaments in the replication of human parainfluenza virus type 3. J Virol. 1998;72:2655–2662. doi: 10.1128/jvi.72.4.2655-2662.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta A K, Drazba J A, Banerjee A K. Specific interaction of heterogeneous nuclear ribonucleoprotein particle U with the leader RNA sequence of vesicular stomatitis virus. J Virol. 1998;72:8532–8540. doi: 10.1128/jvi.72.11.8532-8540.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harrington M G, Gudeman D, Zewert T, Yun M, Hood L. Analytical and micropreparative two-dimensional electrophoresis of proteins. Methods Companion Methods Enzymol. 1991;3:98–108. [Google Scholar]
- 22.Huang Y T, Romito R R, De B P, Banerjee A K. Characterization of the in vitro system for the synthesis of mRNA from human respiratory syncytial virus. Virology. 1993;193:862–867. doi: 10.1006/viro.1993.1195. [DOI] [PubMed] [Google Scholar]
- 23.Karpel R L, Burchard A C. A basic isozyme of yeast glyceraldehyde-3-phosphate dehydrogenase with nucleic acid helix-destabilizing activity. Biochim Biophys Acta. 1981;654:256–267. doi: 10.1016/0005-2787(81)90180-5. [DOI] [PubMed] [Google Scholar]
- 24.Kawamoto R M, Caswell A H. Autophosphorylation of glyceraldehyde-phosphate dehydrogenase and phosphorylation of protein from skeletal muscle microsomes. Biochemistry. 1986;25:656–661. doi: 10.1021/bi00351a022. [DOI] [PubMed] [Google Scholar]
- 25.Kurilla M G, Cabradilla C D, Holloway B P, Keene J D. Nucleotide sequence and host La protein interactions of rabies virus leader RNA. J Virol. 1984;50:773–778. doi: 10.1128/jvi.50.3.773-778.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurilla M G, Keene J D. The leader RNA of vesicular stomatitis virus is bound by a cellular protein reactive with anti-La lupus antibodies. Cell. 1983;34:837–845. doi: 10.1016/0092-8674(83)90541-x. [DOI] [PubMed] [Google Scholar]
- 27.Lai M M C. Cellular factors in the transcription and replication of viral RNA genomes: a parallel to DNA-dependent RNA transcription. Virology. 1998;244:1–12. doi: 10.1006/viro.1998.9098. [DOI] [PubMed] [Google Scholar]
- 28.Leopardi R, Hukkanen V, Vainionpää R, Salmi A A. Cell proteins bind to sites within the 3′ noncoding region and the positive-strand leader sequence of measles virus RNA. J Virol. 1993;67:785–790. doi: 10.1128/jvi.67.2.785-790.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maraia R J, Kenan D J, Keene J D. Eukaryotic transcription termination factor La mediates transcript release and facilitates reinitiation by RNA polymerase III. Mol Cell Biol. 1994;14:2147–2158. doi: 10.1128/mcb.14.3.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDonald L J, Moss J. Stimulation by nitric oxide of an NAD linkage to glyceraldehyde 3-phosphate dehydrogenase. Proc Natl Acad Sci USA. 1993;90:6238–6241. doi: 10.1073/pnas.90.13.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meerovitch K, Sveetkin Y V, Lee H S, Lejbkowicz F, Kenan D J, Chan E K L, Agol V I, Keene J D, Sonenberg N. La autoantigen enhances and corrects aberrant translation of poliovirus RNA in reticulocyte lysate. J Virol. 1993;67:3798–3807. doi: 10.1128/jvi.67.7.3798-3807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyer-Siegler K, Mauro D J, Seal G, Wurzer J C, del Riel J K, Sirover M A. A human nuclear uracil DNA glycosylase is the 37 kDa subunit of glyceraldehyde 3-phosphate dehydrogenase. Proc Natl Acad Sci USA. 1991;88:8460–8464. doi: 10.1073/pnas.88.19.8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Morgenegg G, Winkler G C, Hubscher U, Heizmann C W, Mouis J, Kuenzle C C. Glyceraldehyde 3-phosphate dehydrogenase is a nonhistone protein and a possible activator of transcription in neurons. J Neurochem. 1986;47:54–62. doi: 10.1111/j.1471-4159.1986.tb02830.x. [DOI] [PubMed] [Google Scholar]
- 34.Moyer S A, Baker S C, Horikami S M. Host cell proteins required for measles virus reproduction. J Gen Virol. 1990;71:775–783. doi: 10.1099/0022-1317-71-4-775. [DOI] [PubMed] [Google Scholar]
- 35.Moyer S A, Baker S C, Lessard J L. Tubulin: a factor necessary for the synthesis of both Sendai virus and vesicular stomatitis virus RNAs. Proc Natl Acad Sci USA. 1986;83:5405–5409. doi: 10.1073/pnas.83.15.5405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.O'Farrell P H. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 37.Pancholi V, Fischetti V A. Glyceraldehyde 3-phosphate dehydrogenase on the surface of group A streptococci is also an ADP-ribosylating enzyme. Proc Natl Acad Sci USA. 1993;90:8154–8158. doi: 10.1073/pnas.90.17.8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pardigon N, Strauss J H. Mosquito homolog of the La autoantigen binds to Sindbis virus RNA. J Virol. 1996;70:1173–1181. doi: 10.1128/jvi.70.2.1173-1181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perucho M, Salas J, Salas M L. Identification of the mammalian DNA binding protein p8 as glyceraldehyde 3-phosphate dehydrogenase. Eur J Biochem. 1997;81:557–562. doi: 10.1111/j.1432-1033.1977.tb11982.x. [DOI] [PubMed] [Google Scholar]
- 40.Piechaczyk M, Blanchard J M, Riaad-El Sabouty S, Dani C, Marty L, Jeanteur P. Unusual abundance of vertebrate 3-phosphate dehydrogenase pseudogenes. Nature. 1984;312:469–471. doi: 10.1038/312469a0. [DOI] [PubMed] [Google Scholar]
- 41.Ravichandran V, Seres T, Moriguchi T, Thomas J A, Johnston R B., Jr S-thiolation of glyceraldehyde 3-phosphate dehydrogenase induced by the phagocytosis-associated respiratory burst in blood monocytes. J Biol Chem. 1994;269:25010–25015. [PubMed] [Google Scholar]
- 42.Ryazanov A G, Ashmarina L I, Muronetz V I. Association of glyceraldehyde-3-phosphate dehydrogenase with mono- and poly-ribosomes of rabbit reticulocytes. Eur J Biochem. 1988;171:301–305. doi: 10.1111/j.1432-1033.1988.tb13790.x. [DOI] [PubMed] [Google Scholar]
- 43.Schultz D E, Hardin C C, Lemon S M. Specific interaction of glyceraldehyde 3-phosphate dehydrogenase with the 5′-nontranslated RNA of hepatitis A virus. J Biol Chem. 1996;271:14134–14142. doi: 10.1074/jbc.271.24.14134. [DOI] [PubMed] [Google Scholar]
- 44.Singh N K, Atreya C D, Nakhasi H L. Identification of calreticulin as a rubella virus RNA binding protein. Proc Natl Acad Sci USA. 1994;91:12770–12774. doi: 10.1073/pnas.91.26.12770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh R, Green M R. Sequence-specific binding of transfer RNA by glyceraldehyde-3-phosphate dehydrogenase. Science. 1993;259:365–368. doi: 10.1126/science.8420004. [DOI] [PubMed] [Google Scholar]
- 46.Sirover M A. Role of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in normal cell function and in cell pathology. J Cell Biochem. 1997;66:133–140. [PubMed] [Google Scholar]
- 47.Takagi T, Iwama M, Seta K, Kanda T, Tsukamoto T, Tominaga S, Mizumoto K. Positive and negative host factors for Sendai virus transcription and their organ distribution in rat. Arch Virol. 1996;141:1623–1635. doi: 10.1007/BF01718287. [DOI] [PubMed] [Google Scholar]
- 48.Tan E M. Antinuclear antibodies: diagnostic markers for autoimmune diseases and probes for cell biology. Adv Immunol. 1989;44:93–151. doi: 10.1016/s0065-2776(08)60641-0. [DOI] [PubMed] [Google Scholar]
- 49.Tao Y, Howlett A C, Klein C. Endogenous ADP-ribosylation of glyceraldehyde-3-phosphate dehydrogenase that is not regulated by nitric oxide in Dictyostelium. Cell Signal. 1993;5:763–775. doi: 10.1016/0898-6568(93)90037-m. [DOI] [PubMed] [Google Scholar]
- 50.Tapparel C, Maurice D, Roux L. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J Virol. 1998;72:3117–3128. doi: 10.1128/jvi.72.4.3117-3128.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsai R I, Green H. Studies on a mammalian cell protein (p8) with affinity for DNA in vitro. J Mol Biol. 1973;73:307–316. doi: 10.1016/0022-2836(73)90344-6. [DOI] [PubMed] [Google Scholar]
- 53.Wilusz J, Kurilla M G, Keene J D. A host protein (La) binds to a unique species of minus-sense leader RNA during replication of vesicular stomatitis virus. Proc Natl Acad Sci USA. 1983;80:5827–5831. doi: 10.1073/pnas.80.19.5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zang W-Q, Angela M F, Grant R A, Yen T S B. Identification of glyceraldehyde 3-phosphate dehydrogenase as a cellular protein that binds to the hepatitis B virus posttranscriptional regulatory element. Virology. 1998;248:46–52. doi: 10.1006/viro.1998.9255. [DOI] [PubMed] [Google Scholar]