Abstract
Background and aims
Eosinophilic oesophagitis (EoE) is characterised by symptoms of esophageal dysfunction and oesinophil tissue infiltration. The EoE Diagnostic Panel (EDP) can distinguish between active and non-active EoE using a set of 77 genes. Recently, the existence of distinct EoE variants featuring symptoms similar to EoE, such as oesophageal dysfunction but lacking eosinophil infiltration, had been determined.
Methods
We used oesophageal biopsies from patients with histologically active (n=10) and non-active EoE (n=9) as well as from healthy oesophageal controls (n=5) participating in the Swiss Eosinophilic Esophagitis Cohort Study (SEECS) and analysed the gene expression profile in these biopsies by total RNA-sequencing (RNA-seq). Moreover, we employed the publicly accessible RNA-seq dataset (series GSE148381) as reported by Greuter et al, encompassing a comprehensive genomic profile of patients presenting with EoE variants.
Results
A novel, diagnostic gene expression panel that can effectively distinguish patients with histologically active conventional EoE from patients with EoE in histological remission and control individuals, and from three newly discovered EoE variants was identified. Histologically Active EoE Diagnostic Panel (HAEDP) consists of 53 genes that were identified based on differential expression between histologically active EoE, histological remission and controls (p≤0.05). By combining the HAEDP with EDP, we expanded our knowledge about factors that may contribute to the inflammation in EoE and improved our understanding of the underlying mechanisms of the disease. Conversely, we suggested a compact group of genes common to both HAEDP and EDP to create a reliable diagnostic tool that might enhance the accuracy of EoE diagnosis.
Conclusion
We identified a novel set of 53 dysregulated genes that are closely associated with the histological inflammatory activity of EoE. In combination with EDP, our new panel might be a valuable tool for the accurate diagnosis of patients with EoE as well as for monitoring their disease course.
Keywords: oesophageal disorders, oesophagitis, oesophageal disease
WHAT IS ALREADY KNOWN ON THIS TOPIC
Eosinophilic oesophagitis (EoE) is known for oesophageal dysfunction and eosinophil infiltration, and the EoE Diagnostic Panel (EDP) differentiates active from non-active EoE.
WHAT THIS STUDY ADDS
This study introduces the Histologically Active EoE Diagnostic Panel (HAEDP) of 53 genes, enhancing diagnosis by distinguishing between active EoE, remission, controls and new EoE variants.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
The HAEDP’s detailed gene profiling improves the diagnosis of histologically active EoE and offers a new tool for exploring EoE pathogenesis, potentially leading to novel research in understanding and treating EoE.
Introduction
Oeosinophilic esophagitis (EoE) is a chronic, type 2 inflammatory disease which is restricted to the oesophagus.1 2 The diagnosis of EoE is based on the 2018 International Consensus Guidelines1 and relies on symptoms of oesophageal dysfunction as well as dense eosinophilic infiltration of the oesophagus with at least 15 eosinophils per high-power field. Furthermore, EoE diagnosis requests the exclusion of other systemic and local conditions that might contribute to EoE-associated symptoms and/or oesophageal eosinophilia, for example, GERD, eosinophilic gastroenteritis or Crohn’s disease with upper GI tract involvement.1 3
Thus, due to the large number of relevant differential diagnosis, tools that can accurately diagnose EoE are highly needed in clinical practice. A diagnostic tool of this kind is the EoE Diagnostic Panel (EDP),4 which consists of a molecular multiplex quantitative PCR (qPCR)-based assay of 94 genes. The panel design is based on the EoE transcriptome identified by Blanchard et al 5 and subsequent studies.6–9 Only 77 genes out of the 94 genes of the EDP panel were shown to be significantly dysregulated in EoE tissue, whereof 50 genes were upregulated and 27 genes were downregulated.4 6 7 The set of genes covered by the EDP represented the first tissue-based molecular diagnostic tool that accurately differentiates active EoE from non-active EoE in paediatric and adult patients4 10 11 and the EDP score correlated with oesophageal eosinophil counts.4 10 12 Moreover, the EDP score reliably distinguishes EoE from GERD.7 13 Recently, a multicentre cohort study demonstrated the capacity of the EDP to detect oesophageal inflammation in patients with EoE using a single distal or proximal oesophageal biopsy.6
The correlation of the EDP with features of the EoE histological scoring system and the EoE endoscopic reference scoring highlighted the existence of three distinct EoE endotypes, namely EoE1–3.10 Of note, even though all three endotypes revealed comparable eosinophil levels, they clearly differed concerning the extent of their histological, endoscopic and molecular alterations.10 Furthermore, a recent multicentre study by Greuter et al 14 described three distinct EoE variants, namely EoE-like oesophagitis, lymphocytic esophagitis and non-specific oesophagitis. Patients suffering from one of those EoE variants present with clinical symptoms of EoE and oesophageal dysfunction but without intraepithelial esinophilia on histological examination. EoE-like oesophagitis is defined by the presence of <15 esinophils per high-power field, and by typical histological EoE features, particularly dilated intercellular spaces and basal zone hyperplasia.15 Lymphocytic oesophagitis is defined by a high number of intraepithelial lymphocytes (≥30 lymphocytes per hpf), which are gathered mainly in peripapillary fields, peripapillary spongiosis and by the absence of intraepithelial granulocytes.16 Non-specific oesophagitis is defined by histological infiltration of lymphocytes or neutrophils without fulfilling the numerical and distributional criteria of lymphocytic }>esophagitis.15
Here, we aimed to identify a diagnostic panel that can effectively distinguish patients with conventional EoE and active histological inflammation from patients with conventional EoE in histological remission and from control individuals, and from the three newly discovered EoE variants (EoE-like esophagitis, lymphocytic oesophagitis, non-specific oesophagitis). To achieve this, we identified a novel set of 53 dysregulated genes closely associated with the inflammatory activity of EoE. Our new panel might be a valuable tool for accurate diagnosis of patients with EoE as well as for monitoring their disease course.
Methods
Patient samples
Oesophageal biopsies and clinical data were collected from patients participating in the Swiss Eosinophilic Oesophagitis Cohort Study (SEECS).16 Oesophageal biopsies were histologically evaluated by a pathologist. Based on this evaluation, patients were attributed to the following categories: nine patients in histological inflammatory remission (peak eosinophil count <5/hpf, absence of basal cell hyperplasia), thereof four patients with endoscopic/histological findings suggestive of fibrosis (EoE RF+) and five patients without fibrosis (EoE RF−). Ten patients with histologically active inflammation (peak eosinophil count >15/hpf, presence of basal cell hyperplasia), thereof five patients with fibrosis (EoE AF+) and five patients without fibrosis (EoE AF−); five oesophagus-healthy controls. From 5 patients, biopsies were taken from the proximal oesophagus, while from the remaining 19 patients, biopsies were taken from the distal oesophagus. Five biopsies from proximal oesophagus were included: one in a control patient, one in a patient with EoE who was histologically active with fibrosis, one in a patient with EoE who was not histologically active with fibrosis and two patients with EoE in histological remission with fibrosis (online supplemental table 1).
gutjnl-2023-331743supp001.pdf (994.2KB, pdf)
Patient enrolment
Patients included in the study were categorised based on the SEECS criteria.16 First, physicians completed a questionnaire describing the patient’s symptoms. Overall symptom severity in the past 24 hours, in the past 7 days and in the past 30 days were ranked on a Likert scale from 0 (no symptoms) to 10 (most severe) (online supplemental figure 4A). Second, physicians assessed the endoscopic disease activity that included scoring: white exudates (absent, mild, severe), fixed rings (absent, mild, moderate, severe, not available), vascular pattern (normal, decreased), furrows (absent, present), strictures (absent, low-grade, intermediate-grade, high-grade, not available) in both proximal and distal oesophagus. Next, overall endoscopic disease activity was evaluated and included endoscopic inflammatory activity (online supplemental figure 4B) (based on the severity of white exudates, vascular pattern and furrows) and endoscopic fibrotic activity (online supplemental figure 4C) (based on the severity of rings and strictures). Subsequently, the physician assigned an endoscopic overall activity on a scale ranging from 0 (indicating inactive EoE) to 10 (representing the most active EoE) (online supplemental figure 4D).
In the second part of patient enrolment, the histological activity was evaluated by a pathologist. Peak esinophil count in the epithelium (online supplemental figure 4E), eosinophil abscesses—defined as aggregates of 10 or more cells in a cluster either intraepithelial or on the surface (present, absent, cannot be evaluated), enlargement of the epithelial basal layer (<14%, 15%–33%, 34%–66%, >67%, cannot be evaluated) were determined. Next, a detailed analysis of lamina propria was implemented to determine if lamina propria is present in biopsy (present, can be evaluated; present, cannot be evaluated; absent), and when present and can be evaluated, peak eosinophils in lamina propria were counted. Finally, the fibrosis severity of lamina propria was assessed as absent, mild/moderate and severe. Subsequently, patients were classified as non-fibrotic if fibrosis was absent, while those with mild or severe fibrosis in the lamina propria were classified as fibrotic. In addition, overall histological disease activity was scored (online supplemental figure 4F) between 0 (inactive disease) and 10 (most active disease).
Taking all these factors into account, the treating gastroenterologist completed the physician global assessment of EoE activity, using a Likert scale from 0 (inactive EoE) to 10 (most active EoE), indicating the activity of the patient’s EoE at the time of biopsy collection (online supplemental figure 4G). This assessment reflected the expert opinion on EoE activity, considering symptoms, endoscopy and histology makers at the time of biopsy collection, and is presented as ‘disease activity score’.16
RNA sequencing
One biopsy per patient was collected in RNALater and used for RNA sequencing (RNA-seq) analysis. RNA isolation was performed by Microsynth, Balgach, Switzerland. Library preparations and RNA-seq were performed by the Functional Genomics Center Zurich, Zürich, Switzerland. The sequencing libraries were sequenced using the Illumina Hiseq. FastQC (V.0.11.5) was used to check the data quality. The human genome GRCh38 from Ensemble was used as a reference genome for STAR (V.2.5.4) for mapping the reads. The analysis was performed in R. Bulk RNA-seq of histologically defined samples is accessible on request in the National Center for Biotechnology Information (NCBI) Sequence Read Accessible (SRA) database with the accession code PRJNA1036399.
Differential gene expression analysis
Differential gene expression analysis of experimental groups was performed using the Bioconductor package DESeq2. Each group, namely controls, EoE AF+, EoE AF−, EoE RF+ and EoE RF− was taken as a single factor. The representative genes for histological EoE activity were selected based on the following comparisons: (a) controls to EoE AF−, (b) EoE RF− to EoE AF− and (c) EoE RF+ to EoE AF+. All genes were divided into two lists for upregulated and downregulated genes. The upregulated genes were those with a positive fold-2-change for EoE active, while downregulated genes were those with a negative fold-2-change for EoE active. The lists of upregulated and downregulated genes from those comparisons were compared online to create a Venn diagram. Differential gene expression levels were considered significant when the adjusted p value was ≤0.05.
Heatmaps
Normalised counts of the selected genes were used to create heatmaps. The heatmaps were visualised using the pheatmap function from the pheatmap package V.1.0.12. Rows and columns were hierarchically clustered. Clustering methods were set as ‘complete’, and the clustering distance was set as ‘Euclidean’. Briefly, the sample in each colu mn was considered as an individual cluster. Then, each stage joined similar clusters together until the last cluster was formed. The clustering distance was based on the Euclidean method, that is, the correlation between clusters increased when the distance was smaller.
Publicly available dataset
The publicly available dataset GSE148381 of the genome-wide expression from oesophageal biopsies of subjects with EoE-like inflammatory diseases, EoE, GERD and controls was used.14 This dataset comprised 10 patients with EoE, 6 patients with GERD, 7 controls, 13 patients with EoE-like disease, 5 patients with lymphocytic EoE and 10 patients with non-specific EoE. Fastq files from the Bioproject PRJNA626361 were downloaded and processed similarly to the present dataset.14
Statistical analysis
Data were analysed and graphed using RStudio (V.2023.03.0+386) and GraphPad Prism software (V.8.3.0). For statistical analyses, GraphPad Prism software was used. The analysis was conducted using a one-way analysis of variance with Holm-Šídák’s multiple comparisons test and a single pooled variance. The mean of each column was compared with the mean of every other column. Results are presented as mean±SEM. Differences between the means were considered significant when p value was ≤0.05.
Results
Identification of a distinct EoE transcriptome profile that defines patients with histologically active EoE
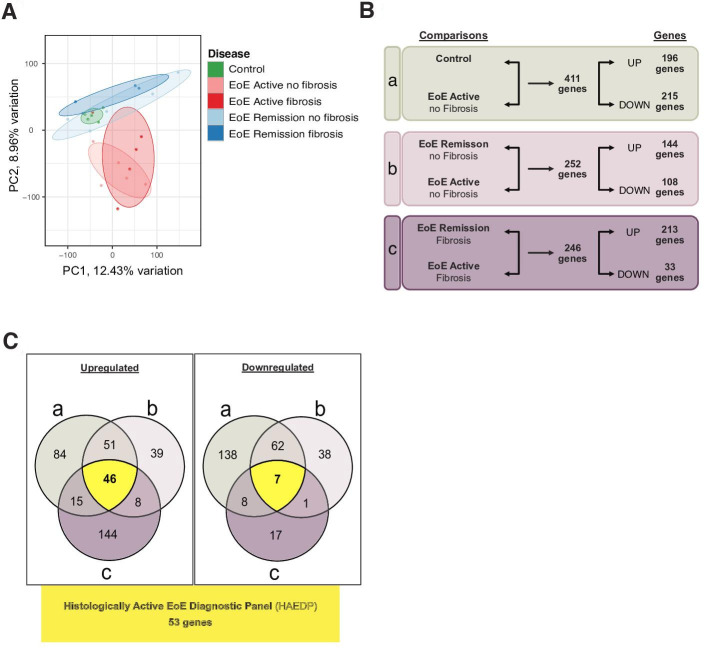
In our present study, the gene expression profiles of 24 oesophageal biopsy samples from 19 patients with EoE and 5 controls were analysed by total RNA-seq. Detailed characteristics of the samples can be found in online supplemental table 1. All subjects were adult patients between 25 and 70 years of age, 55% (n=13) were males. Principal component analysis (PCA) demonstrated significant differences in the gene expression patterns in patients with EoE with histologically active disease that are different from patients with EoE in histological remission and controls. Of note, there was no difference between samples derived from controls and patients with EoE in histological remission and between patients with or without fibrosis, regardless of their histological inflammation status (figure 1A). To identify genes that might contribute to the inflammatory activity of EoE and explain the observed differences between histologically inflamed and uninflamed tissue, we identified differentially expressed genes (DEG) with an adjusted p value ≤0.05 and performed the following comparisons: (a) controls versus histologically active EoE without fibrosis (EoE AF−), (b) EoE in histological remission without fibrosis (EoE RF−) versus active EoE without fibrosis (EoE AF−) and (c) EoE in histological remission with fibrosis (EoE RF+) versus histologically active EoE with fibrosis (EoE AF+). We identified genes that were only associated with the inflammatory activity of the disease but not with the onset of fibrosis. The DEG were divided into genes being either upregulated (log2 fold change (FC) ≥2) or downregulated (log2FC ≤−2) in samples derived from patients with histologically active EoE. As shown in figure 1B, the DEG analysis between controls and EoE AF− patients revealed a total of 411 differently regulated genes, comprising 196 upregulated and 215 downregulated genes (a). Moreover, a comparison between EoE RF− and EoE AF− patients found a total of 252 differentially regulated genes, comprising 144 upregulated and 108 downregulated genes (b). Finally, 246 genes were differentially expressed with 213 upregulated and 33 downregulated genes when comparing EoE RF+ with EoE AF+ patient groups (c). Using Venn diagram representation for either upregulated or downregulated genes, we then identified genes that were differentially expressed specifically associated with the histological inflammatory activity of EoE (figure 1C). By this approach, genes that were either upregulated (46 genes) or downregulated (7 genes) in all three comparisons and we named this set of genes the Histologically Active EoE Diagnostic Panel (HAEDP). The lists of upregulated and downregulated genes extracted from the Venn diagrams and their corresponding intersections are available in online supplemental tables 2 and 3, respectively. In addition, hierarchical heatmaps of the genes upregulated and downregulated in histologically active EoE in all comparisons (figure 1C) are available in online supplemental figure 1. In summary, we identified a novel set of 53 dysregulated genes that are closely associated with the inflammatory activity of EoE and might provide a novel insight into the molecular mechanisms underlying EoE pathogenesis.
Figure 1.
The Histologically Active EoE Diagnostic Panel (HAEDP) differentiates patients with histologically active eosinophilic oesophagitis (EoE) from patients with EoE in remission and control individuals. (A) Principal component analysis visualises the geometric distance between samples. Each dot corresponds to a patient sample. (B) Graphical representation of the differentially expressed genes in the analysis comparing (a) controls with EoE AF−, (b) EoE RF− with EoE AF− and (c) EoE RF+ with EoE AF+ (adjusted p≤0.05). (C) Venn diagrams of downregulated (log2FC ≤−2) and upregulated (log2FC ≥2) genes differentially expressed in histologically active EoE compared with controls and EoE in histological remission. Intersections of both Venn diagrams correspond to the HAEDP (controls n=5, EoE AF(+) n=5, EoE AF(−) n=5, EoE RF(+) n=4, EoE RF(−) n=5). FC, fold change.
gutjnl-2023-331743supp002.xlsx (25KB, xlsx)
gutjnl-2023-331743supp003.xlsx (26.8KB, xlsx)
Patients with histologically active EoE identified by the HAEDP can be further divided based on their disease activity score
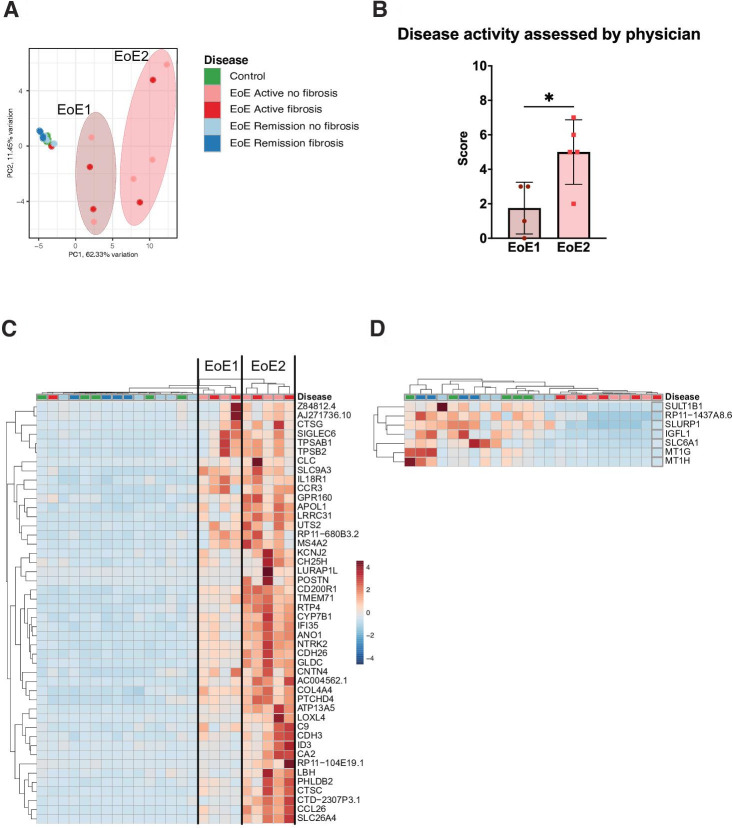
We next illustrated the distribution of the respective patient samples in a PCA that was based on HAEDP and its associated genes (figure 2A). Our data showed that samples from patients with histologically active EoE were clearly separated from both the samples derived from patients with EoE in histological remission and the samples from controls. Of note, the samples from patients with histologically active EoE were further divided into two distinct clusters, which we named EoE1 and EoE2. Initially, we hypothesised that the clustering into EoE1 and EoE2 might be linked to the infiltration of eosinophils and thus the eosinophil counts in the oesophageal epithelium. We expected eosinophil counts to be higher in the EoE2 subgroup compared with the EoE1 subgroup, due to the fact that the expression of CCL26, a key chemical messenger that attracts esinophils, was higher in the EoE2 subgroup. The final distinction between EoE1 and EoE2 was made based on a composite scoring system that supported the distinction between EoE1 and EoE2. This composite scoring system was an overall disease activity score assessed by a physician and is described in detail in the ‘Methods’ section. However, these EoE subtypes did not correlate with the eosinophil counts in the biopsies nor with the location of origin of the histologically active EoE samples (online supplemental figure 2). However, we found that the disease activity score of the EoE2 group was significantly higher than the score of the EoE1 group (p≤0.05), thus indicating a potential clinical relevance of this subgrouping (figure 2B).
Figure 2.
The Histologically Active EoE Diagnostic Panel (HAEDP) assesses disease severity in patients with histologically active eosinophilic oesophagitis (EoE). (A) Principal component analysis visualises the geometric distance between samples after applying the HAEDP gene list. Each dot corresponds to a patient. (B) The overall disease activity score assessed by the physician was determined for the EoE1 and EoE2 subgroups (*p≤0.05 using non-parametric t-test analysis). Hierarchical heatmaps of 46 upregulated (C) and 7 downregulated (D) genes of HAEDP in patients with histologically active EoE samples. Each column represents a patient, and each row represents a gene. Hierarchical clustering was used to analyse the data, and distance methods were set as ‘Euclidean’ (controls n=5, EoE AF(+) n=5, EoE AF(−) n=5, EoE RF(+) n=4, EoE RF(−) n=5).
We indeed identified a patient with active EoE appearing in the inactive group. However, we currently lack a specific explanation for this discrepancy, as patient enrolment in this category was determined by both a physician and a pathologist. Nevertheless, the complexity of clinical interpretations and potential variations in disease presentation may contribute to such instances.
Since the aim of our study was to identify genes that can differentiate patients with histologically active EoE from controls and patients with EoE in histological remission, the HAEDP genes were plotted as heatmaps demonstrating the genes upregulated (figure 2C) or downregulated (figure 2D) in patients with histologically active EoE when compared with samples from controls or patients with EoE in histological remission. Interestingly, no difference in sample clustering based on the fibrotic status was observed (data not shown). Our data suggest that the HAEDP can be effectively employed to detect histologically active EOE regardless of the region of origin of the biopsy or the fibrotic tissue status. Moreover, the HEADP differentiates between patients with active EoE and controls or patients with EoE in remission and can identify the severity of the disease in patients with active EoE.
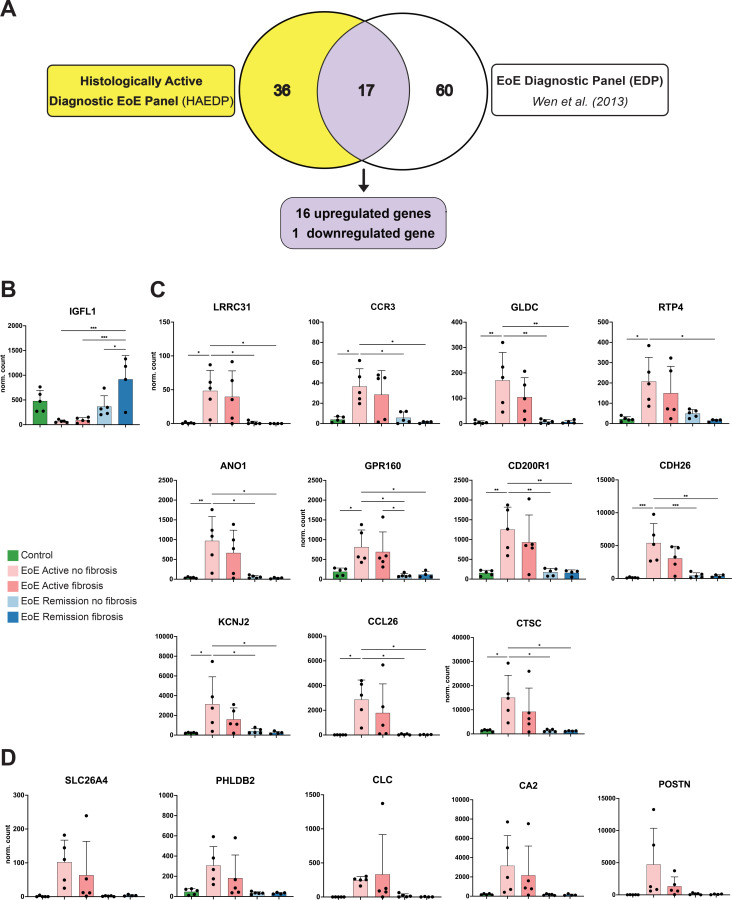
Seventeen genes are common between the HAEDP and the EDP and might serve as markers for histologically active EoE
Our discovered HAEDP included 53 genes in total, of which 46 were upregulated and 7 were downregulated in histologically active EoE tissues. To further optimise the diagnostic accuracy of our HAEDP, we combined the pattern of our HAEDP with the pattern of the EDP described previously by Wen et al.4 The original EDP consisted of 77 genes, of which 50 were upregulated and 27 were downregulated in EoE tissue. As shown in figure 3A, the overlap of both gene lists revealed that the two panels share only 17 genes while the additional 36 genes of our HAEDP were newly discovered. Thus, we anticipated that the panel of 17 genes common to HAEDP and EDP might be most suitable to characterise the histological inflammatory activity of EoE. The specific messenger RNA expression patterns of all of those 17 genes, as observed in our 5 patient groups, are plotted in figure 3B–D. IGF-like family member 1 (IGFL1) was the only downregulated gene in the samples from patients with histologically active EoE. However, normalised counts of the IGFL1 gene were not statistically different between histologically active EoE and control samples. At the same time, there was a significant difference between histologically active EoE when compared with EoE RF+ samples (figure 3B). Of note, the normalised gene counts of LRRC31, CCR3, GLDC, RTP4, anoctamin 1 (ANO1), GPR160, CD200R1, CDH26, KCNJ2, CCL26 and CTSC were significantly higher in the samples derived from patients of the EoE AF− group than in the control group or in the EoE in histological remission group (figure 3C).
Figure 3.
The Histologically Active EoE Diagnostic Panel (HAEDP) and EoE Diagnostic Panel (EDP) share a common gene list. (A) Graphical representations of the number of genes overlapping between the EDP identified by Wen et al 4 and the HAEDP. Seventeen genes were common to both panels and visualised as significantly downregulated in histologically active eosinophilic oesophagitis (EoE) (B), significantly upregulated in histologically active EoE (C) and upregulated but significantly not different in histologically active EoE (D). In addition, 36 genes were uniquely found in HAEDP, and 60 genes were uniquely found in EDP. Data are represented as the normalised counts mean±SEM. *P≤0.05, **p≤0.01, ***p≤0.001 using one-way analysis of variance test followed by Holm-Šídák’s multiple comparisons test (controls n=5, EoE AF(+) n=5, EoE AF(−) n=5, EoE RF(+) n=4, EoE RF(−) n=5). IGFL1, IGF-like family member 1.
Additionally, five genes, namely SLC26A4, PHLDB2, CLC, CA2 and periostin (POSTN), showed a trend towards an increase in the samples derived from patients with histologically active EoE when compared with samples from either controls or patients with EoE in histological remission, although this trend was not statistically significant (figure 3D). Thus, we suggest a compact group of 17 genes common to the EDP and the HEADP to create a reliable diagnostic tool. By combining HEADP with the existing EDP, we might further enhance the accuracy of EoE diagnosis (figure 3).
EDP-specific and HAEDP-specific genes differentiate histologically active EoE
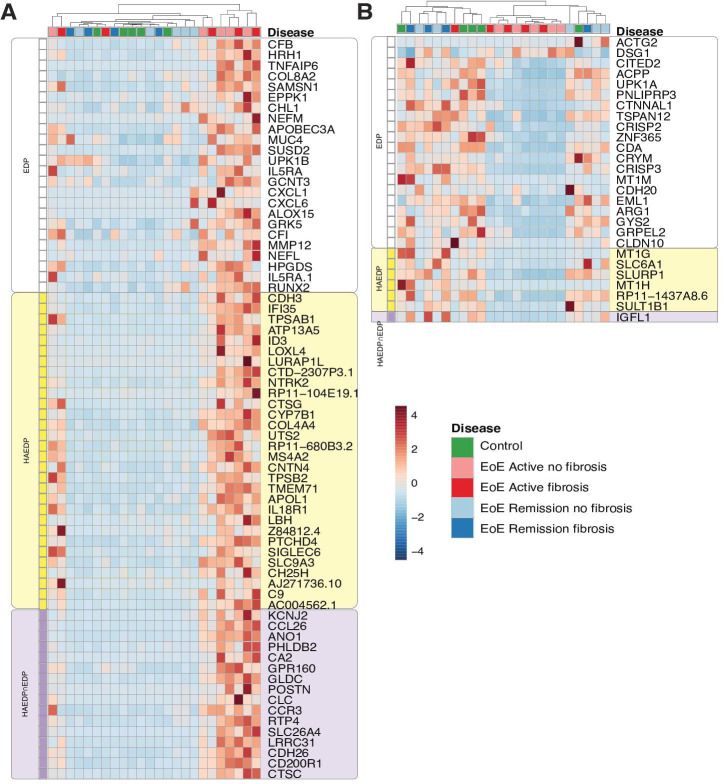
We next investigated the gene expression profile of all 113 genes that are either part of the HAEDP (53 genes) and/or the EDP (77 genes) in our patient cohort. As mentioned above, 17 genes are common to both the HAEDP as well as the EDP (figure 4). Genes classified as upregulated in histologically active EoE are visualised in figure 4A, while those categorised as downregulated in histologically active EoE are depicted in figure 4B. In our analysis, we identified 24 upregulated genes that are unique to the EDP, 36 upregulated genes that are unique to the HAEDP as well as an additional 16 upregulated genes that are common to both the EDP and the HAEDP. Conversely, we identified 20 downregulated genes specific to EDP, 6 downregulated genes exclusive to HAEDP and only 1 downregulated gene common to both panels. These findings underscore the complementary nature of both panels.
Figure 4.
Combined EoE Diagnostic Panel (EDP) and Histologically Active EoE Diagnostic Panel (HAEDP) precisely differentiated histologically active eosinophilic oesophagitis (EoE). Hierarchical heat diagram of genes of the EDP and HAEDP that are upregulated (A) and downregulated (B) in patients with EoE. Each column in the diagram represents a patient, and each row represents a gene. The genes are clustered by panels: white represents genes exclusive to EDP only, yellow represents genes exclusive to HAEDP only and purple represents genes common to EDP and HAEDP (controls n=5, EoE AF(+) n=5, EoE AF(−) n=5, EoE RF(+) n=4, EoE RF(−) n=5).
To identify more genes that can distinguish between histologically active EoE, EoE in remission and controls, we analysed genes that are unique to either the EDP only, the HAEDP only or that overlap between the EDP and the HAEDP. By doing so, we aimed to expand our knowledge of factors that may contribute to the inflammation in EoE and improve our understanding of the underlying mechanisms of the disease (figure 4).
The combination of the HAEDP and the EDP allows differentiation of patients with histologically active EoE from patients with EoE variants
In the next step, we aimed to investigate whether the combined gene expression pattern of the HAEDP and the EDP might be useful to distinguish conventional EoE from other EoE variants. For this purpose, we used the recent publicly available dataset (series GSE148381) from Greuter et al 14 describing the three EoE variants. This dataset contained the gene expression profiles of oesophageal biopsies derived from 13 patients with EoE-like oesophagitis, 5 patients with lymphocytic oesophagitis, 10 patients with non-specific oesophagitis, 10 patients with EoE, 6 patients with GERD as well as 7 controls.
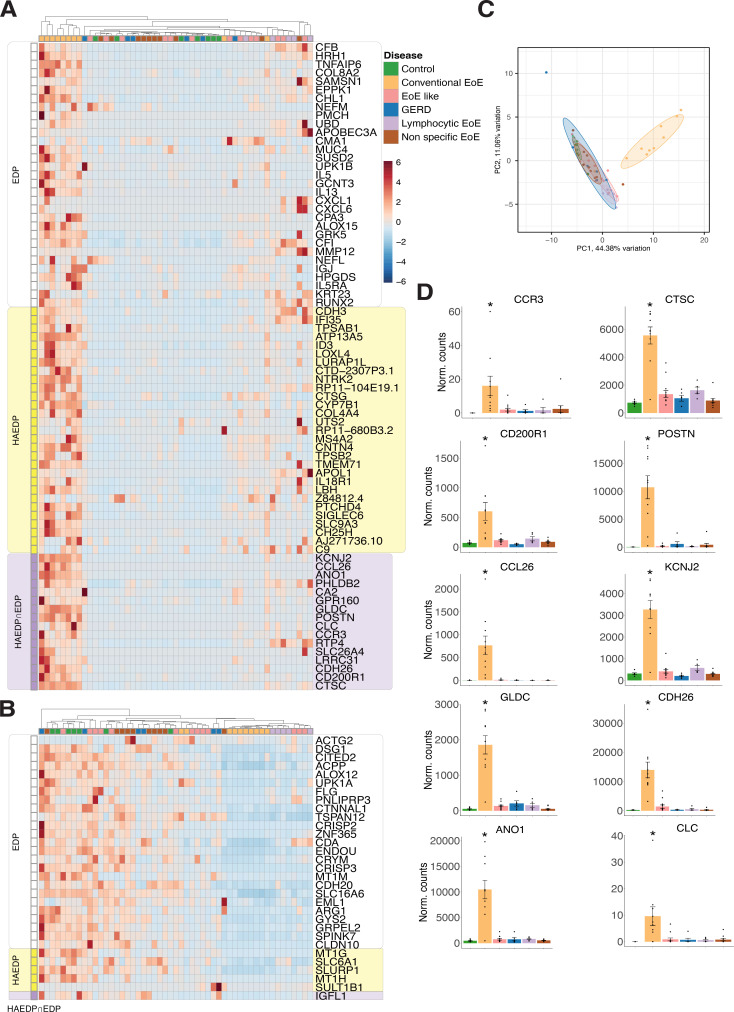
The genes that were either specific for the EDP or the HAEDP or present in both panels were compared with the dataset from Greuter et al. Only the genes present in the EDP and/or the HAEDP, as well as in the dataset from Greuter et al were visualised as heatmaps (figure 5A,B). In the heatmaps, the patients from the dataset from Greuter et al were hierarchically clustered based on the expression profiles of upregulated and downregulated genes described above in the EDP and/or the HAEDP (figure 5A,B). We found that 31 upregulated and 25 downregulated genes exclusively belonged to the EDP, 29 upregulated and 5 downregulated genes exclusively belonged to the HAEDP and 16 upregulated and 1 downregulated genes were common to the two panels and also present in the dataset from Greuter et al.
Figure 5.
Combined EoE Diagnostic Panel (EDP) and Histologically Active EoE Diagnostic Panel (HAEDP) detect conventional eosinophilic oesophagitis (EoE) among EoE variants. Hierarchical heatmap of genes of the EDP and HAEDP applied on the RNA-sequencing dataset from Greuter et al 14: (A) upregulated genes and (B) downregulated genes (C) principal component analysis after applying HAEP and EDP panels to visualise the distribution of the samples. (D) List of genes upregulated only in conventional EoE as compared with other EoE variants and control. Data are the normalised counts mean±SEM. *P≤0.001 using one-way analysis of variance test followed by Holm-Šídák’s multiple comparisons test (controls n=7, conventional EoE n=10, GERD n=6, EoE-like disease n=13, lymphocytic EoE n=5, non-specific EoE n=10).
Next, to investigate the variation of the gene expression in different EoE variants included in the data repository from Greuter et al, we applied a list of genes derived from the EDP as well as the HAEDP and performed a PCA (figure 5C). The results demonstrated that the specific gene expression profiles allows discrimination of oesophageal tissue samples derived from patients with conventional EoE from patients with GERD, EoE-like disease, lymphocytic EoE and non-specific EoE and controls.
Further analysis revealed that 10 out of the 16 upregulated genes that are common to both the EDP as well as the HAEDP were significantly upregulated in samples from patients with conventional EoE when compared with samples from controls and patients with GERD or EoE variants (p≤0.001, figure 5D). Thus, we propose that the expression of these genes, for example, CCR3, CTSC, CD200R1, POSTN, CCL26, KCNJ2, GLDC, CDH26, ANO1 and CLC, might help to differentiate conventional EoE from controls, patients with GERD and more importantly from the EoE variants, namely EoE-like disease, lymphocytic EoE and non-specific EoE. Similarly, online supplemental figure 3A shows that 8 out of 31 EDP-specific genes are significantly upregulated in conventional EoE relative to controls, GERD and EoE variants. In addition, online supplemental figure 3B indicates that 17 out of 29 upregulated HAEDP-specific genes exhibit significant upregulation in conventional EoE.
In summary, these data demonstrate that the combined EDP and HAEDP allow differentiation of conventional, histologically active EoE from controls, EoE in histological remission and different EoE variants.
Discussion
In our study, we have identified a panel of 53 genes that are dysregulated in the oesophageal tissue of patients with histologically active EoE and can separate histologically active EoE from EoE in remission and control individuals. Thus, we named this newly discovered panel the HAEDP.
By applying DEG analysis, we were able to demonstrate that non-inflamed patients, which include both control individuals as well as patients in histological remission, can be distinguished from the patients with histologically active EoE based on their gene expression profile. This signifies that by examining the genes common to the HAEDP and EDP, it is possible to differentiate between control oesophageal tissue and histologically inflamed tissue in patients with EoE. Therefore, the 53 dysregulated genes proposed within the HAEDP can be considered as a panel for the identification of a histologically active EoE disease. This can be achieved by qPCR and immunohistological staining, which are both widely available diagnostic tools for routine laboratories.
This might be highly relevant for patient care since EoE diagnosis relies on clinical, histological and endoscopic disease assessment.2 In light of recent discoveries of EoE endotypes and emerging EoE variants, there is a growing need to identify biomarkers that can contribute to a more effective diagnosis of EoE.14 15 17
A decade ago, microarrays5 and RNA-seq18 were used to establish EoE transcriptomic profiles from samples obtained from the US EoE centres. These transcriptomic profiles were generated by comparing oesophageal tissue from patients with EoE with that of control individuals. In our study, we analysed the EoE transcriptome using well-defined histologically active and inflamed EoE biopsies from patients of the SEECS.16 Compared with 1607 significantly dysregulated transcripts from previously published RNA-seq analysis,18 our approach has identified 658 dysregulated transcripts (sum of all genes presented in figure 1C). This difference might be attributed to the stringent use of histologically strictly defined biopsies and the limited number of analysed patients. Additionally, as proposed with HAEDP, an EoE transcriptome profile was created irrespective of the onset of fibrosis.
We further observed a separation within the histologically active EoE patient group into two distinct subgroups, named EoE1 and EoE2. However, on thorough analysis, we did not uncover any significant correlation between the eosinophil counts, basal zone hyperplasia and the clustering into EoE1 and EoE2. On the contrary, we observed an upregulation of KCNJ2, POSTN, ANO1, CDH26 and GLDC genes in the EoE2 patients cluster. Interestingly, potassium channel KCNJ2 was found to be expressed at high levels in the patients with EoE,15 ANO1 was identified as a key driver of epithelial cell proliferation during the onset of EoE19 and POSTN expression is found to be altered in EoE and normalise on treatment.20
Furthermore, the overall disease activity score assigned by physicians was significantly higher in EoE2 compared with EoE1, indicating greater disease activity when considering a combination of clinical, endoscopic and histological diagnostic features.
The EDP is known for distinguishing patients with active EoE from control individuals or patients with EoE in remission.4 In contrast, we have demonstrated that the HAEDP can also assist in distinguishing patients with histologically active EoE from those who are histologically defined as in remission and those who are control individuals. As described previously, EoE variants were defined as disease entities sharing clinical characteristics but not histological features, such as intraepithelial eosinophilia, with conventional EoE.15 We hypothesised that the EDP and the HAEDP could differentiate EoE variants based on the gene expression profiles. We found that both panels can differentiate conventional EoE from controls and patients with EoE variants. Furthermore, we have identified 10 genes common to both the EDP and the HAEDP that are significantly upregulated exclusively in EoE tissue when compared with controls and each of the EoE variants.
On a molecular level, while the role of CLC26 and its receptor CCR3 is well-known in the pathogenesis of EoE,21 the mechanistic role of the ion channels KCNJ2, ANO1, the classical hallmark of eosinophilic inflammation CLC,22 immune modulating CD200R123 and of cell attachment inducing POSTN24 is still unknown and requires further investigation. We also found that CDH26 is upregulated in patients with histologically active EoE and can discriminate conventional EoE from the EoE variants. This is particularly interesting since CDH26 might be a promising target in future drug development in EoE. CDH26 regulates epithelial cell polarisation and cytoskeletal structure.25 By interacting with integrin alpha 4 and integrin alpha E, CDH26 may impact leucocyte migration, localisation and activation during inflammation.26 In the murine model of allergic airway disease, CDH26 deficiency reduced airway mucus production, airway hyper-responsiveness and airway eosinophilia.27 In in vitro cell culture, CDH26 knockdown inhibited interleukin (IL)-13-induced IL-4R alpha and IL-13R alpha upregulation and suppressed downstream JAK1 and STAT6 phosphorylation.27 Current research indicates that CDH26 regulates IL-4 signalling in epithelial cells, making it a potential therapeutic target for IL-4R-mediated allergic diseases, with EoE being one of them.28
It should be noted that we used a single oesophageal biopsy frozen in RNALater buffer for bulk RNA-seq. Our results and results published by others11 indicate that a single biopsy might be enough for molecular diagnosis of histologically active EoE. By using the HEADP analysis on a single biopsy sample, we can differentiate active EoE from control individuals and patients with EoE in remission and from other EoE variants. Given the patchy nature of EoE, it is advisable to obtain biopsies from both the proximal and distal oesophagus29 and to investigate whether the location of the biopsy sampling influences the readout. In the present study, the selection of oesophageal biopsies and their location of origin was based on the availability of biopsies accessible within the framework of the SEECS. From each patient enrolled in the SEECS, both proximal and distal biopsies were initially collected. However, after very detailed endoscopic and histological characterisation, only biopsies that fulfilled the criteria of a given group were included for further molecular analysis. Obtaining oesophageal biopsies that contain both the epithelial compartment as well as the lamina propria is technically challenging but crucial for assessing fibrosis.30 Therefore, the selection of biopsies for further analysis was limited to those where sufficient lamina propria material was available. It is important to acknowledge this limitation, as the variability in biopsy locations may impact the generalisability of our findings. To address this, future investigations should strive to include a more comprehensive sampling strategy that encompasses both proximal and distal oesophageal regions, preferably performed with biopsy forceps models that have a higher yield for subepithelial tissue when compared with standard biopsy forceps models.30 This approach will enable a more nuanced understanding of the disease’s patchy nature. Additionally, to validate the reliability and accuracy of HEADP, it is necessary to assess it in a larger population. The patients studied in our work were on different treatment regimens while the samples were collected (online supplemental table 4). It remains possible that such treatments affect the oesophageal transcriptome. Furthermore, it must be noted that the population in which the HAEDP was developed only included patients on a proton pump inhibitors (PPIs) (and excluded those treated with swallowed steroids), so this difference might at least partially account for some of the differences observed in gene expression pattern observed in the HAEDP.
Conclusion
In conclusion, this study introduces a novel set of genes called HAEDP, composed of 53 dysregulated genes that accurately reflect the inflammatory activity of EoE disease. Despite the fact that both EDP and HAEDP are unable to differentiate between patients with fibrostenotic EoE from those without fibrosis, they differentiate between active EoE and EoE variants such as EoE-like, non-specific and lymphocytic EoE. Identifying EoE variants can be challenging as clinical manifestations characteristic of EoE may appear even in the absence of significant eosinophilic infiltration. The proposed HAEDP provides a specific gene expression profile that accurately differentiates EoE variants from classical EoE without the need to differentiate among its three distinct forms and represents a potential improvement in the field of EoE diagnosis. In summary, this study presents new insights into the molecular diagnostics of EoE and highlights the identification of a novel biomarker panel to accurately diagnose histologically active EoE in clinical practice.
Transcript profiling
The RNA-seq data from Greuter et al can be found in the NCBI SRA database with the accession code PRJNA626361.
Bulk RNA-seq of histologically defined samples is accessible on request in the NCBI SRA database with the accession code PRJNA1036399.
Data transparency statement
The demographic information of patients is available in online supplemental table 1. Bulk RNA-seq of histologically defined samples is accessible on request in the NCBI SRA database with the accession code PRJNA1036399.
The RNA-seq from Greuter et al can be found in the NCBI SRA database with the accession code PRJNA626361.
Footnotes
MS and MW contributed equally.
Correction notice: This article has been corrected since it published Online First. Oesinophilic has been corrected to eosinophilic throughout the article, including the title.
Collaborators: Members of the Swiss EoE Cohort Study Group (SEECS): Patrick Aepli, Luc Biedermann, Ruggero Biral, Carine Blanchard, Lorenzo Botteselle, Jan Borovicka, Simon Buetikofer, Emanuel Burri, Joachim Diebold, Annika Eckhold, Annett Franke, Michèle Frei, Thomas Greuter, Chantal Hasler, Wolfram Jochum, Tanja Kilchmann, Seraina Koller, Andrea Kreienbühl, Peter Netzer, Christiane Pelzer, Gabrielle Reichhart, Elodie Ristorcelli, Gerhard Rogler, Jean-Benoit Rossel, Ekaterina Safroneeva, Catherine Saner, Jon-Duri Senn, Alain Schoepfer, Philipp Schreiner, Christine Sempoux, Dagmar Simon, Hans-Uwe Simon, Alex Straumann, Sven Trelle, Achim Weber, Niels Willi, Marcel Zwahlen.
Contributors: EG: generation, collection, assembly, analysis and interpretation of data, drafting the manuscript. YM: conception and design of study, generation, collection, assembly analysis and data interpretation, revision of the manuscript. CM: data generation, collection and assembly. ASc: data collection, revision of manuscript and approval of the final version. CS: data collection, revision of manuscript and approval of the final version. LB: data collection, revision of manuscript and approval of the final version. ASt: data collection, revision of manuscript and approval of the final version. AK: data collection, revision of manuscript and approval of the final version. MS: conception and design of the study, data interpretation, revision of the manuscript and approval of the final version of the manuscript and is guarantor. MW: conception and design of the study, analysis and data interpretation, revision of the manuscript and approval of the final version of the manuscript.
Funding: This study was supported by grants from the EoE Foundation Switzerland (grant no. 2021-09) to MS, grants from the Swiss National Science Foundation (grant nos. 320030_184753 and 320030E_190969) to MS and by the Swiss EoE Cohort Study, supported by Swiss National Science Foundation (grant nos 32003B_204751/1) to ASc.
Competing interests: MS has shares and is a co-founder of Recolony, Zurich, and has shares in PharmaBiome, Zurich. MS served as advisor for AbbVie, Gilead, Fresenius, Topadur, Takeda, Roche and Celltrion. MS received speaker’s honoraria from Janssen, Falk Pharma, Vifor Pharma, Pileje and Bromatech. MS received research grants from AbbVie, Takeda, Gilead, Gnubiotics, Roche, Axalbion, Pharmabiome, Topadur, Basilea, MBiomics, Storm Therapeutics, LimmatTech, Zealand Pharma, NodThera, Calypso Biotech, Pileje, Herbodee, Vifor. ASc received speaker/advisor fees from AbbVie, AstraZeneca, Janssen, Receptos-Celgene-BMS, Dr Falk Pharma, GSK, MSD, Pfizer, Sanofi-Regeneron, Takeda and Vifor. ASc received research grants from AbbVie, AstraZeneca, Receptos-Celgene-BMS, Dr Falk Pharma, GSK, MSD, Pfizer, Sanofi-Regeneron and Vifor. ASt has consultant contracts with AstraZeneca, Receptos-Celene-BMS, Calypso, EsoCap, Dr Falk Pharma, GSK, Pfizer, Sanofi-Regeneron and Shire.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Contributor Information
Collaborators: Swiss EoE Cohort Study Group, Patrick Aepli, Luc Biedermann, Ruggero Biral, Carine Blanchard, Lorenzo Botteselle, Jan Borovicka, Simon Buetikofer, Emanuel Burri, Joachim Diebold, Annika Eckhold, Annett Franke, Michèle Frei, Thomas Greuter, Chantal Hasler, Wolfram Jochum, Tanja Kilchmann, Seraina Koller, Andrea Kreienbühl, Peter Netzer, Christiane Pelzer, Gabrielle Reichhart, Elodie Ristorcelli, Gerhard Rogler, Jean-Benoit Rossel, Ekaterina Safroneeva, Catherine Saner, Jon-Duri Senn, Alain Schoepfer, Philipp Schreiner, Christine Sempoux, Dagmar Simon, Hans-Uwe Simon, Alex Straumann, Sven Trelle, Achim Weber, Niels Willi, and Marcel Zwahlen
Data availability statement
Data are available on reasonable request. Data are accessible on request in the National Center for Biotechnology Information (NCBI) Sequence Read Accessible (SRA) database with the accession code PRJNA1036399.
Ethics statements
Patient consent for publication
Consent obtained directly from patient(s).
Ethics approval
The Swiss eosinophilic oesophagitis Cohort Study was approved by the cantonal ethics committee of the Canton Vaud (PB_2016-01962). Our substudy was approved by the local ethics committee of the Canton Zurich (KEK-ZH 2023-00921). Participants gave informed consent to participate in the study before taking part.
References
- 1. Dellon ES, Liacouras CA, Molina-Infante J, et al. Updated International consensus diagnostic criteria for eosinophilic esophagitis. Gastroenterology 2018;155:1022–33. 10.1053/j.gastro.2018.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sperry SLW, Shaheen NJ, Dellon ES. Toward uniformity in the diagnosis of eosinophilic oesophagitis (EoE): the effect of guidelines on variability of diagnostic criteria for EoE. Am J Gastroenterol 2011;106:824–32. 10.1038/ajg.2011.10 [DOI] [PubMed] [Google Scholar]
- 3. Lucendo AJ, Molina-Infante J, Arias Á, et al. Guidelines on eosinophilic oesophagitis: evidence-based statements and recommendations for diagnosis and management in children and adults. United European Gastroenterol J 2017;5:335–58. 10.1177/2050640616689525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wen T, Stucke EM, Grotjan TM, et al. Molecular diagnosis of eosinophilic oesophagitis by gene expression profiling. Gastroenterology 2013;145:1289–99. 10.1053/j.gastro.2013.08.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blanchard C, Wang N, Stringer KF, et al. Eotaxin-3 and a uniquely conserved gene-expression profile in eosinophilic esophagitis. J Clin Invest 2006;116:536–47. 10.1172/JCI26679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Min S, Shoda T, Wen T, et al. Diagnostic merits of the eosinophilic oesophagitis diagnostic panel from a single esophageal biopsy. J Allergy Clin Immunol 2022;149:782–7. 10.1016/j.jaci.2021.07.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wen T, Rothenberg ME. Clinical applications of the eosinophilic oesophagitis diagnostic panel. Front Med (Lausanne) 2017;4. 10.3389/fmed.2017.00108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sherrill JD, Blanchard C. Genetics of eosinophilic oesophagitis. Dig Dis 2014;32:22–9. 10.1159/000357005 [DOI] [PubMed] [Google Scholar]
- 9. Blanchard C, Mingler MK, Vicario M, et al. IL-13 involvement in eosinophilic oesophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol 2007;120:1292–300. 10.1016/j.jaci.2007.10.024 [DOI] [PubMed] [Google Scholar]
- 10. Shoda T, Wen T, Aceves SS, et al. Eosinophilic oesophagitis endotype classification by molecular, clinical, and histopathological analyses: a cross-sectional study. Lancet Gastroenterol Hepatol 2018;3:477–88. 10.1016/S2468-1253(18)30096-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Dellon ES, Yellore V, Andreatta M, et al. A single biopsy is valid for genetic diagnosis of eosinophilic oesophagitis regardless of tissue preservation or location in the esophagus. J Gastrointestin Liver Dis 2015;24:151–7. 10.15403/jgld.2014.1121.242.bsy [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dellon ES, Veerappan R, Selitsky SR, et al. A gene expression panel is accurate for diagnosis and monitoring treatment of eosinophilic esophagitis in adults. Clin Transl Gastroenterol 2017;8:e74. 10.1038/ctg.2017.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Butz BK, Wen T, Gleich GJ, et al. Efficacy, dose reduction, and resistance to high-dose fluticasone in patients with eosinophilic oesophagitis. Gastroenterology 2014;147:324–33. 10.1053/j.gastro.2014.04.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Greuter T, Straumann A, Fernandez-Marrero Y, et al. Characterization of eosinophilic oesophagitis variants by clinical, histological, and molecular analyses: a cross-sectional multi-center study. Allergy 2022;77:2520–33. 10.1111/all.15233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Salvador Nunes VS, Straumann A, Salvador Nunes L, et al. Eosinophilic oesphagitis beyond eosinophils - an emerging phenomenon overlapping with eosinophilic esophagitis: Collegium Internationale Allergologicum (CIA) update 2023. Int Arch Allergy Immunol 2023;184:411–20. 10.1159/000529910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Safroneeva E, Saner C, Rossel J-B, et al. Cohort profile: the Swiss Eosinophilic Esophagitis Cohort Study (SEECS). Inflamm Intest Dis 2018;2:163–70. 10.1159/000486131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Straumann A, Blanchard C, Radonjic-Hoesli S, et al. A new eosinophilic oesphagitis (EoE)-Like disease without tissue eosinophilia found in EoE families. Allergy 2016;71:889–900. 10.1111/all.12879 [DOI] [PubMed] [Google Scholar]
- 18. Sherrill JD, Kiran KC, Blanchard C, et al. Analysis and expansion of the eosinophilic oesophagitis transcriptome by RNA sequencing. Genes Immun 2014;15:361–9. 10.1038/gene.2014.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Vanoni S, Zeng C, Marella S, et al. Identification of anoctamin 1 (ANO1) as a key driver of oesophageal epithelial proliferation in eosinophilic esophagitis. J Allergy Clin Immunol 2020;145:239–54. 10.1016/j.jaci.2019.07.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Politi E, Angelakopoulou A, Grapsa D, et al. Filaggrin and periostin expression is altered in eosinophilic oesphagitis and normalized with treatment. J Pediatr Gastroenterol Nutr 2017;65:47–52. 10.1097/MPG.0000000000001419 [DOI] [PubMed] [Google Scholar]
- 21. Dunn JLM, Shoda T, Caldwell JM, et al. Esophageal type 2 cytokine expression heterogeneity in eosinophilic oesophagitis in a multisite cohort. J Allergy Clin Immunol 2020;145:1629–40. 10.1016/j.jaci.2020.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ueki S, Miyabe Y, Yamamoto Y, et al. Correction to: Charcot-Leyden crystals in eosinophilic inflammation: active cytolysis leads to crystal formation. Curr Allergy Asthma Rep 2019;19:38. 10.1007/s11882-019-0875-1 [DOI] [PubMed] [Google Scholar]
- 23. Kotwica-Mojzych K, Jodłowska-Jędrych B, Mojzych M. CD200:CD200R interactions and their importance in immunoregulation. Int J Mol Sci 2021;22:1602. 10.3390/ijms22041602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cobo T, Viloria CG, Solares L, et al. Role of Periostin in adhesion and migration of bone remodeling cells. PLoS One 2016;11:e0147837. 10.1371/journal.pone.0147837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lachowicz-Scroggins ME, Gordon ED, Wesolowska-Andersen A, et al. Cadherin-26 (CDH26) regulates airway epithelial cell cytoskeletal structure and polarity. Cell Discov 2018;4:7. 10.1038/s41421-017-0006-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Caldwell JM, Collins MH, Kemme KA, et al. Cadherin 26 is an alpha integrin-binding epithelial receptor regulated during allergic inflammation. Mucosal Immunol 2017;10:1190–201. 10.1038/mi.2016.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Feng Y, Chen S, Xiong L, et al. Cadherin-26 amplifies airway epithelial IL-4 receptor signaling in asthma. Am J Respir Cell Mol Biol 2022;67:539–49. 10.1165/rcmb.2021-0109OC [DOI] [PubMed] [Google Scholar]
- 28. Avlas S, Shani G, Rhone N, et al. Epithelial cell-expressed type II IL-4 receptor mediates eosinophilic oesophagitis. Allergy 2023;78:464–76. 10.1111/all.15510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kumar S, Choi SS, Gupta SK. Eosinophilic oesophagitis: current status and future directions. Pediatr Res 2020;88:345–7. 10.1038/s41390-020-0770-4 [DOI] [PubMed] [Google Scholar]
- 30. Bussmann C, Schoepfer AM, Safroneeva E, et al. Comparison of different biopsy forceps models for tissue sampling in eosinophilic oesophagitis. Endoscopy 2016;48:1069–75. 10.1055/s-0042-117274 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2023-331743supp001.pdf (994.2KB, pdf)
gutjnl-2023-331743supp002.xlsx (25KB, xlsx)
gutjnl-2023-331743supp003.xlsx (26.8KB, xlsx)
Data Availability Statement
Data are available on reasonable request. Data are accessible on request in the National Center for Biotechnology Information (NCBI) Sequence Read Accessible (SRA) database with the accession code PRJNA1036399.