Abstract
We aimed to describe the landscape, including molecular, epidemiological, and clinical aspects of CHIKV infections in the Ribeirao Preto region, an area endemic to dengue. We randomly screened 3744 plasma samples that had undergone DENV diagnosis to evaluate CHIKV-RNA using an in-house RT-PCR assay. Positive samples were followed clinically, and RNA samples were submitted to whole genome sequencing. Seventeen cases (0.5 %) were positive for CHIKV-RNA despite being negative for DENV-RNA. Notably, half of the patients experienced prolonged arthralgia lasting more than 90 days. Compared with the healthy control group, leukopenia and thrombocytopenia were observed in all CHIKV-positive individuals with statistically significant P values (P < 0.0001 and P = 0.0003, respectively). The genomic analysis revealed that the CHIKV strains being studied are classified within the East-Central-South-African (ECSA) genotype. This analysis identified new mutations, E1: K211E and E2: V264A, while the previously known mutation E1: A226V was not detected among these strains. This study highlights the need for epidemiological surveillance and preparedness for potential CHIKV epidemics in Brazil, particularly where other arboviruses co-circulate.
Keywords: Chikungunya virus, Molecular epidemiology, Phylogeny, Arbovirus
Introduction
Chikungunya fever is a mosquito-borne disease caused by the Chikungunya virus (CHIKV), a member of the Alphavirus genus within the Togaviridae family. Over the past decade, CHIKV has emerged and has become a significant global health concern due to its rapid spread, considerable mortality, and economic impact. CHIKV is usually associated with acute polyarthralgia, characterized by high fever, rash, and severe joint pain that can last months after infection [1]. The disease is primarily transmitted by Aedes mosquitoes, with four distinct genotypes: the West African, Asian, East/Central/South African (ECSA), and Indian Ocean Lineage genotype [2], contributing to its global epidemiology.
In the Americas, there has been an observed increase in CHIKV cases and deaths in the early months of 2023, with 30,707 reported cases and 14 deaths, surpassing the numbers reported in 2022 (271,176 cases/95 deaths) and 2021 (137,025 cases/12 deaths) [3]. Brazil accounted for 64 % (174,517) of the cases in 2022 [4], with the Northeast region having the highest incidence rate of 257.4 cases per 100,000 inhabitants and 41 % of death notifications. São Paulo reported 5155 and 3945 cases in the Southeast region in 2022 and 2023, respectively.
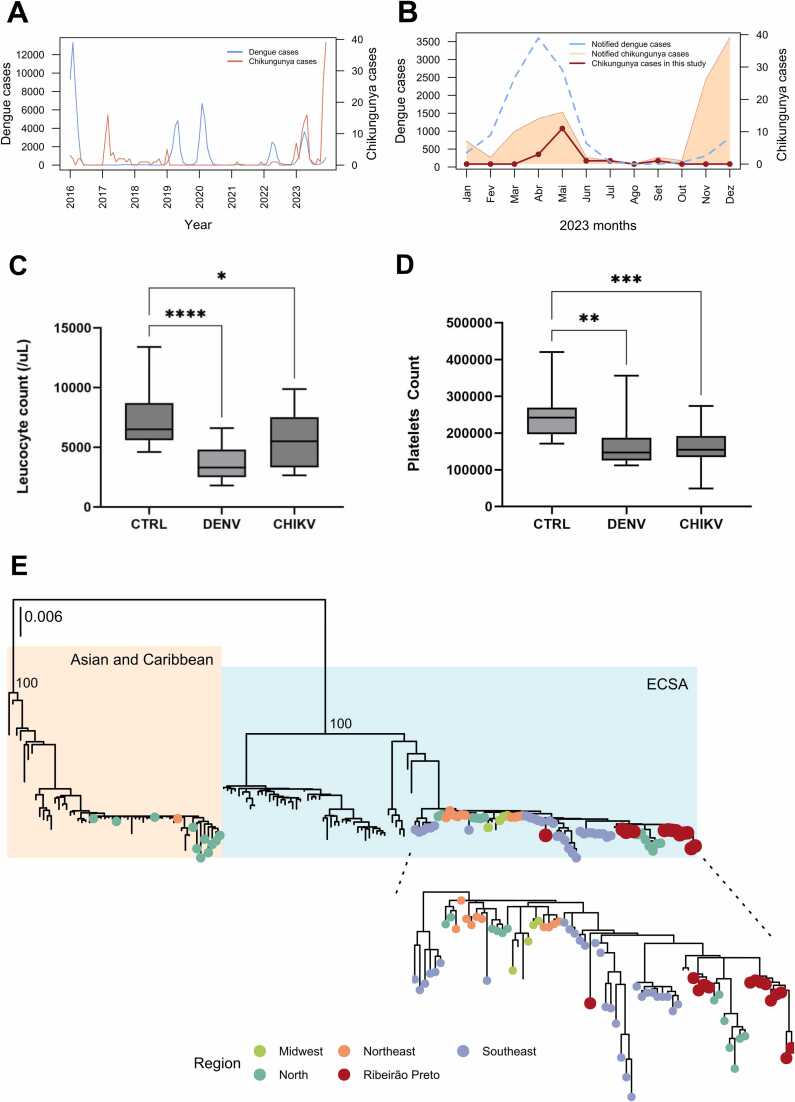
Although the CHIKV epidemic has not yet reached Ribeirão Preto, there has been a notable increase in CHIKV cases since the beginning of the 2023 seasonal period compared to previous years, as depicted in Fig. 1A. Notably, 75 % of these cases are autochthonous. Concurrently, DENV outbreaks have occurred in multiple years (2016, 2019, 2020, 2022, and 2023) as shown in Fig. 1A. Therefore, this situation raises concerns due to the favorable conditions for other arboviruses, such as CHIKV, and consequently for coinfections.
Fig. 1.
Epidemiological and Clinical Characteristics of CHIKV Infections in Ribeirão Preto, São Paulo, Brazil. A) Annual distribution of reported CHIKV and DENV cases in Ribeirão Preto from 2016 to 2023. B) Distribution of CHIKV-positive cases, highlighting the notable increase in CHIKV cases in 2023, with 75 % autochthonous. C) Comparative analysis of leukocyte counts in CHIKV-positive individuals versus healthy controls, indicating significant leukopenia (P < 0.0001). D) Comparative analysis of platelet counts in CHIKV-positive individuals versus healthy controls, showing significant thrombocytopenia (P = 0.0003). E) Phylogenetic tree of CHIKV sequences from Ribeirão Preto, clustering with the ECSA genotype reference strains from Brazilian and South American isolates. The red dots represent the 14 complete genomes sequenced in this study.
Methods
During the peak of the DENV outbreak in May 2023, a random screening was conducted on 3744 plasma samples from individuals presenting with febrile syndrome related to arbovirus infections at the Basic Health Units of Ribeirão Preto, located in the Northeast part of São Paulo State, Brazil. CHIKV-RNA was detected using an in-house RT-PCR assay based on the method described by Lanciotti et al., 2006 [5]. Whole genome sequencing of CHIKV was performed utilizing the Illumina COVIDSeq protocol (Illumina), with adaptation employing CHIKV-specific primers [6]. The sequencing protocol is accessible at protocols.io under "Pathogen Whole Genome Sequencing - Multiplexed amplicon sequencing" [7]. Sample selection was based on the cycle threshold (Ct) value (<30, mean 26.01 ± 8.15). Subsequently, the Epidemiological Service of the municipality was informed, and patients were subject to clinical monitoring until their symptoms were resolved.
Results
The initial selection of 3744 plasma samples for this study was based on samples that had undergone DENV diagnosis, as they exhibited symptoms indicative of arbovirus infections. Simultaneously, all samples were tested for CHIKV using RT-PCR. Out of the 3744 plasma samples analyzed, 17 (0.5 %) tested positive for CHIKV-RNA while being negative for DENV-RNA. All cases were autochthonous, with 70.6 % male and an average age of 49 (ranging from 0 to 76 years). We highlight the positivity in a newborn of 4 months whose mother tested negative for CHIKV RT-PCR. The infant had ceased to receive breastfeeding one month prior to testing. Interestingly, it was found that one of the infant's siblings also tested positive for CHIKV RT-PCR during the timeframe. These findings suggest that the mode of transmission in this instance is more likely vector-borne rather than maternal.
Positive individuals were symptomatic, suspected of having arbovirus-febrile syndrome, and sought medical attention approximately three days after the onset of symptoms. Predominant reported symptoms included fever (90 %), myalgia (90 %), headache (65 %), nausea (60 %), vomiting (35 %), rash (30 %), retro-orbital pain (24 %), inappetence (18 %), and petechiae (6 %). Arthralgia was observed in only 53 % of the cases, with 60 % experiencing intense joint pain. Primary treatments included analgesics, corticoids, and anti-inflammatory drugs. Arthralgia persisted for more than 90 days in fifty percent of the patients. Interestingly, leukopenia and thrombocytopenia were observed in all CHIKV-positive individuals, showing significant P values (P < 0.0001 and P = 0.0003, respectively) compared to healthy individuals (controls), as depicted in Fig. 1C. The most prevalent comorbidities among these individuals were hypertension and diabetes. Patients were monitored until the resolution of acute symptoms, and none exhibited severe manifestations. (Supplementary Data, Table S1).
All samples met the specific criterion for viral whole genome sequencing, which requires a cycle threshold (Ct) value of less than 30. However, only 14 out of the 17 samples analyzed achieved a coverage greater than 90 % and a depth exceeding 1,000X, accounting for 82.3 % of the total. All complete genomes of CHIKV sequences were deposited in the GISAID database (Supplementary Data, Table S2). Multiple alignments were performed using 166 complete genome sequences from NCBI (Supplementary Data, Table S2). Phylogenetic analysis revealed that the CHIKV sequences belong to the ECSA genotype and cluster with reference strains from Brazilian and South American isolates (Fig. 1D, red dots), specifically from Brazil's North and Southeast regions. Our findings align with the data reported by Xavier et al. (2023), underscoring the crucial role of Brazil's Northeast region as the primary source for the dispersion of CHIKV. We did not detect the E1-A226V mutation associated with CHIKV outbreaks in Asia, which is linked to an enhanced transmission potential for Aedes albopictus mosquitoes [8]. Additionally, there is an absence of novel mutations, namely E1: K211E and E2: V264A, which are associated with enhanced fitness in Aedes aegypti [9]. The E1-T98A mutation is present in all sequences compared to the S27-African prototype (NCBI Reference Sequence: NC_004162.2) [10].
Discussion
In Brazil, the diagnosis of CHIKV relies initially on clinical evaluation, with confirmation mainly through IgM serology [4]. However, it is essential to note that while RT-PCR is considered the gold standard and the most reliable verification method, its availability in the public health network is limited. Interestingly, Thiboutot and colleagues (2010) demonstrated that cell counts of leukocytes and platelets are not commonly reduced in CHIKV [11]. However, our findings demonstrated a decrease in these cell counts, further allowing for diagnostic confusion with other arboviruses, such as dengue fever.
Despite São Paulo's low incidence rate for CHIKV (Fig. 1A), areas like Ribeirão Preto, with endemic DENV, present all the conditions that favor the emergence of a new arbovirus, such as CHIKV. Our data provides a local context, but its significance extends beyond that, as it can aid in comprehending the spread of CHIKV nationwide and forecasting potential epidemics.
The current scenario is an alert for future outbreaks, given that this region has not yet been affected. Hence, it is imperative to conduct epidemiological surveillance studies to investigate clinical events and closely monitor the evolving situation [12]. Despite the frequent DENV epidemics in the Ribeirão Preto region, there remain uncertainties about the sporadic and imported cases of CHIKV in this area. CHIKV strains identified in our study are similar to those endemic to Brazil, suggesting a predominately domestic circulation rather than introduction from neighboring countries. The presence of the primary vector species, Ae. aegypti and Ae. albopictus, in the state of São Paulo, including Ribeirão Preto, is a well-documented fact [13]. Moreover, the increasing average temperatures correlate with the geographic spread of Ae. aegypti. Notably, in terms of larval density, Ae. aegypti tends to surpass Ae. albopictus. It is essential to recognize that factors such as transmission efficiency, susceptibility to viruses, and adaptability can vary significantly between these vector species [14], [15].
Considering Ribeirão Preto an endemic area for DENV, co-circulation of DENV and CHIKV poses complex challenges for public health strategies, as well as for the diagnosis and treatment protocols [16]. This complexity arises from the fact that immunity to one virus does not confer immunity to the other, as they have distinct immunological profiles. Furthermore, the transmission dynamics in such regions could be significantly affected by factors like population susceptibility and previous immunity levels. However, it is crucial to consider the potential biases in reporting and surveillance data, given that CHIKV screening is still not a standard requirement for arbovirus-febrile onset cases.
Building upon the complexity of managing co-circulating DENV and CHIKV in endemic areas like Ribeirão Preto, an additional layer of intricacy emerges when considering the potential for viral transmission through blood transfusion and cell therapy. While transmission of CHIKV through transfusion or cell therapy has not been confirmed, the existence of a viremia period warrants caution regarding this possible route of transmission. This scenario underscores the need for region-specific approaches in managing and understanding the co-circulation of these viruses, emphasizing the need for comprehensive strategies that span from epidemiological monitoring to clinical intervention.
On November 10, 2023, the FDA approved the first live-attenuated vaccine, IXCHIQ® (Valneva) [17], which uses the ECSA genotype with deletion for attenuation. Additionally, exploring treatment strategies to alleviate symptoms should be further studied.
In conclusion, our findings underscore the importance of implementing prompt actions, mainly focusing on vector control measures and monitoring symptomatic cases. Consequently, the spread to new locations encourages further evaluation of the disease’s pathogenesis to facilitate the development of new preventive strategies, diagnostic tools, and therapeutic options. This approach is critical to preventing and controlling emerging infectious disease outbreaks to mitigate the impact on public health.
Author contributions
Methodology: DGLLR, ME, EVS, PNMDC, and LRP. Patient recruitment and editing the article: ECM, DBG, PMAT, AYY, DCDG, and LMRP; Phylogenetic analysis: MG, VF, and DGLLR. Review and editing: SNS, AJM, RTC, DTC, MCE, LCA, and SCS. Data curation, writing, - review, and editing: SK, MG, and DGLLR. Conceptualization: SK and DGLLR. Supervision and Funding acquisition: SCS, DTC, SK, and LCA.
Transparency declaration
The authors declare that they have no conflicts of interest.
Funding
This study was supported by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) through Center for Cell-based Therapy (CTC) (13/08135-2) and Centro para Vigilância Viral e Avaliação Sorológica (CeVIVAS) (21/11944-6), Instituto Nacional de Ciência e Tecnologia em Células-Tronco e Terapia Celular no Câncer (INCTC-CNPq) (465539/2014-9); National Institutes of Health USA grant U01 AI151698 for the United World Arbovirus Research Network (UWARN) and the Collaborative Research Programme - International Center of Genetic Enginering and Biotechnology (CRP-ICGEB) RESEARCH GRANT 2020 Project CRP/BRA20-03, Contract CRP/20/03. M. Giovanetti's funding is provided by Programma Operativo Nazionale Ricerca e Innovazione 2017-2020 (PON) "Ricerca e Innovazione'' 2014–2020. D.G.L.L.R's funding is provided by FAPESP - 22/16349-1.
Ethical statement
This study was approved by the Institutional Ethics Committee of the Faculty of Medicine of Ribeirão Preto (Process CAAE: 59073722.0.0000.5440 registered at Plataforma Brasil).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.jiph.2024.04.026.
Appendix A. Supplementary material
Supplementary material
.
Data availability
Genomes generated in the present study are publicly available in the GISAID database at accession number: EPI_ISL_18699314, EPI_ISL_18699317, EPI_ISL_18699308, EPI_ISL_18699311, EPI_ISL_18699318, EPI_ISL_18699316, EPI_ISL_18699321, EPI_ISL_18699310, EPI_ISL_18699313, EPI_ISL_18699312, EPI_ISL_18699319, EPI_ISL_18699309, EPI_ISL_18699315, EPI_ISL_18699320.
References
- 1.Suhrbier A. Rheumatic manifestations of chikungunya: emerging concepts and interventions. Nat Rev Rheuma. 2019;15:597–611. doi: 10.1038/s41584-019-0276-9. [DOI] [PubMed] [Google Scholar]
- 2.Weaver S.C., Forrester N.L. Chikungunya: evolutionary history and recent epidemic spread. Antivir Res. 2015;120:32–39. doi: 10.1016/j.antiviral.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 3.PAHO/WHO. Epidemiological Alert: Chikungunya increase in the Region of the Americas. 13 Feb 2023. 〈https://www.paho.org/en/documents/epidemiological-alert-chikungunya-increase-region-americas〉.
- 4.Ministério da Saúde. Boletim Epidemiológico 1 2023. 〈https://www.gov.br/saude/pt-br/centrais-de-conteudo/publicacoes/boletins/epidemiologicos/edicoes/2023/boletim-epidemiologico-volume-54-no-01/view〉.
- 5.Lanciotti R.S., Kosoy O.L., Laven J.J., Panella A.J., Velez J.O., Lambert A.J., et al. Chikungunya Virus in US Travelers Returning from India, 2006. Emerg Infect Dis. 2007;13:764–767. doi: 10.3201/eid1305.070015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Quick J., Grubaugh N.D., Pullan S.T., Claro I.M., Smith A.D., Gangavarapu K., et al. Multiplex PCR method for MinION and Illumina sequencing of Zika and other virus genomes directly from clinical samples. Nat Protoc. 2017;12:1261–1276. doi: 10.1038/nprot.2017.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de La-Roque, Debora, Vieira, Elaine S., Kashima S. 2022. Pathogen whole genome sequencing - multiplexed amplicon sequencing.〈https://www.protocols.io/edit/pathogen-whole-genome-sequencing-multiplexed-ampli-cgwbtxan/description〉 [Google Scholar]
- 8.Khongwichit S., Chansaenroj J., Chirathaworn C., Poovorawan Y. Chikungunya virus infection: molecular biology, clinical characteristics, and epidemiology in Asian countries. J Biomed Sci. 2021;28:84. doi: 10.1186/s12929-021-00778-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Familiar-Macedo D., Gama B.E., Emmel V.E., Vera-Lozada G., Abdelhay E., Martins I.S., et al. Molecular aspects of Chikungunya virus infections in cancer patients. Mem Inst Oswaldo Cruz. 2022;117 doi: 10.1590/0074-02760210383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsetsarkin K.A., Chen R., Leal G., Forrester N., Higgs S., Huang J., et al. Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc Natl Acad Sci USA. 2011;108:7872–7877. doi: 10.1073/pnas.1018344108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thiboutot M.M., Kannan S., Kawalekar O.U., Shedlock D.J., Khan A.S., Sarangan G., et al. Chikungunya: a potentially emerging epidemic? PLoS Negl Trop Dis. 2010;4 doi: 10.1371/journal.pntd.0000623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xavier J., Alcantara L.C.J., Fonseca V., Lima M., Castro E., Fritsch H., et al. Increased interregional virus exchange and nucleotide diversity outline the expansion of chikungunya virus in Brazil. Nat Commun. 2023;14:4413. doi: 10.1038/s41467-023-40099-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fonseca Júnior D.P. da, Serpa L.L.N., Barbosa G.L., Pereira M., Holcmam M.M., Voltolini J.C., et al. Vectors of arboviruses in the state of São Paulo: 30 years of Aedes aegypti and Aedes albopictus. Rev Saude Publica. 2019;53:84. doi: 10.11606/s1518-8787.2019053001264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Souza-Neto J.A., Powell J.R., Bonizzoni M. Aedes aegypti vector competence studies: a review. Infect, Genet Evol. 2019;67:191–209. doi: 10.1016/j.meegid.2018.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laporta G.Z., Potter A.M., Oliveira J.F.A., Bourke B.P., Pecor D.B., Linton Y.-M. Global distribution of aedes aegypti and aedes albopictus in a climate change scenario of regional rivalry. Insects. 2023;14:49. doi: 10.3390/insects14010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joob B., Wiwanitkit V. Co-circulation of dengue, chikungunya, and Zika viruses and cross-protection. Rev Panam De Salud Pública. 2019;43:1. doi: 10.26633/RPSP.2019.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schneider M., Narciso-Abraham M., Hadl S., McMahon R., Toepfer S., Fuchs U., et al. Safety and immunogenicity of a single-shot live-attenuated chikungunya vaccine: a double-blind, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2023;401:2138–2147. doi: 10.1016/S0140-6736(23)00641-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Genomes generated in the present study are publicly available in the GISAID database at accession number: EPI_ISL_18699314, EPI_ISL_18699317, EPI_ISL_18699308, EPI_ISL_18699311, EPI_ISL_18699318, EPI_ISL_18699316, EPI_ISL_18699321, EPI_ISL_18699310, EPI_ISL_18699313, EPI_ISL_18699312, EPI_ISL_18699319, EPI_ISL_18699309, EPI_ISL_18699315, EPI_ISL_18699320.