Abstract
The present study analyzed B-cell clonality and bovine leukemia virus (BLV) provirus integration sites in cattle with enzootic bovine leukosis (EBL) having BLV proviral copy numbers less or greater than the number of bovine nucleated cells. EBL cattle with BLV copy numbers less than the number of bovine nucleated cells showed monoclonal and biclonal proliferation of B-cells with one BLV provirus integration site. On the other hand, EBL cattle with BLV copy numbers greater than the number of bovine nucleated cells showed monoclonal proliferation of B-cells with two BLV provirus integration sites. These results suggest that superinfection of BLV can occur in EBL cattle.
Keywords: bovine leukemia virus, clonality, enzootic bovine leukosis, superinfection
Bovine leukemia virus (BLV), a member of the Retroviridae family, integrates into the bovine B-cell genome as a provirus [11]. Almost all BLV-infected cattle remain asymptomatic, while 1–5% develop enzootic bovine leukosis (EBL) with monoclonal or oligoclonal B-cell proliferation [6]. The retroviral phenomenon of superinfection resistance (SIR) in infected cells has been known, with one BLV provirus integrated per B cell [10]. However, BLV proviral copy numbers can exceed the number of bovine nucleated cells in EBL cattle [1], and there are few reports on the mechanism underlying BLV proviral copy numbers greater than the number of nucleated cells in EBL cattle.
B-cell clonality and BLV integration sites have been analyzed by B-cell clonality polymerase chain reaction (PCR) and inverse PCR, respectively [8, 9]. In addition, the number of proviral copies per 100,000 nucleated cells has been calculated using the BLV-CoCoMo-qPCR method [12]. In the present study, the number of BLV proviral copies per B cell in EBL cattle was evaluated by B-cell clonality PCR, inverse PCR, and BLV-CoCoMo-qPCR to estimate the number of BLV provirus copies within B-cells in EBL cattle. Moreover, whole genome of BLV in EBL cattle with a few integration sites were detected by sequence.
Blood and several organs from 68 cattle with suspicions of bovine lymphoma were provided by local veterinarians and meat hygiene inspection centers in Chiba, Ibaraki, Iwate, and Hokkaido, Japan. Samples included lymph nodes (mediastinal, superficial, subiliac, medial, and iliac lymph nodes) and solid tumors in several organs (heart, lung, abomasum, liver, kidney, and uterus) (Supplementary Table 1). One sample from each cattle was used. Pathological findings confirmed the presence of B-cell lymphoma in all cattle. Genomic DNA was extracted from the tissues using the QIAamp® DNA Mini Kit (QIAGEN, GmbH, Hilden, Germany) and stored at −30°C.
BLV proviral copy numbers per 100,000 nucleated cells were quantified by the BLV-CoCoMo-qPCR method using the CoCoMo®-BLV Primer/Probe (RikenGenesis, Tokyo, Japan) and TaqMan™ Gene Expression Master Mix (Thermo Fisher Scientific, Foster City, CA, USA) according to the manufacturers’ instructions.
B-cell clonality analysis using PCR for immunoglobulin heavy chain gene rearrangement was performed as described previously [9]. Briefly, PCR was conducted using a HotStarTaq® DNA Polymerase (QIAGEN GmbH) with primer pairs (BoVHF1 and BoVHR1). The amplification program was the same as that reported previously. PCR products were electrophoresed on 2% agarose gels.
Inverse PCR was performed as described previously [8]. Briefly, genomic DNA was digested with Pst I (Takara Bio Inc., Shiga, Japan) and self-ligated using the DNA Ligation Kit Mighty Mix (Takara). PCR was performed using a GoTaq® Master Mix (Promega, Madison, WI, USA) with inverse primers (Pst-F and Pst-R). The amplification program was the same as that reported previously. PCR products from each sample were electrophoresed on 2% agarose gels.
In case that a few bands were detected by inverse PCR, BLV provirus integration sites were identified. Products of inverse PCR were purified from the gels using QIAquick Gel Extraction Kit (QIAGEN) and the purified products were submitted to Eurofins Japan Inc. for sequencing using amplification primers. The BLV provirus genome was identified using the National Center for Biotechnology Information (NCBI) BLAST program (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The bovine genome sequence adjacent to BLV provirus genome was compared using Cow BLAT Search (https://genome.ucsc.edu/cgi-bin/hgBlat) against the April 2018 freeze of the cow genome sequence.
Complete genome sequences of BLV in EBL cattle with a few integration sites were BLV strains were identified as described previously [7]. Briefly, PCR for BLV genome was performed using genomic DNA and two primer pairs (BLV 1–17 + BLV 4565–4586, and BLV 4416–4436 + BLV 8703–8720) [7]. DNA libraries were prepared using the QIAGEN® QIAseq FX DNA Library Kit (QIAGEN) and sequenced on an Illumina iSeq system using 2 × 150 bp paired-end reads. Reads mapping against a reference BLV genome (Accession No. EF600696.1) was performed for determination of whole genome sequences of the BLV using CLC Genomics Workbench v. 20.0 (CLC bio, Aarhus, Denmark).
Table 1 and Supplementary Table 1 show the results of BLV-CoCoMo-qPCR, B-cell clonality PCR, and inverse PCR. In 66 of the 68 EBL cattle, BLV proviral copy numbers were 70,200–99,975 copies per 100,000 nucleated cells. In B-cell clonality PCR and inverse PCR, one and two bands were detected in 64 and 2 of the 66 EBL cattle (EBL-31 and EBL-42), respectively (Fig. 1). In the remaining 2 EBL cattle (EBL-33 and EBL-52), BLV proviral copy numbers were 161,315 and 198,243 copies per 100,000 nucleated cells, respectively, and a single band and two bands were observed in B-cell clonality PCR and inverse PCR, respectively (Fig. 1). In each of 4 EBL cattle in which inverse PCR demonstrated 2 bands, 2 different integration sites were identified (chr 5: 46963363-4 and chr 20: 2223266-7 in EBL-31, chr 2: 28090975-6 and chr 10: 63666897–8 in EBL-33, chr.1: 136221663-4 and chr. 14: 31131921-2 in EBL-42, and chr. 2: 71557048-9 and chr. 24: 25455752-3 in EBL-52). Moreover, one complete genome sequence of BLV was identified in each of these 4 EBL cattle.
Table 1. Results of bovine leukemia virus (BLV) CoCoMo-qPCR, B-cell clonality PCR, and inverse PCR.
| Number of enzootic bovine leukosis cases | BLV proviral copy numbers (copies/100,000 cells) | B-cell clonality PCR | Inverse PCR |
|---|---|---|---|
| 64 | 70,200–99,975 | Single band | Single band |
| 2 | 73,125 and 99,315 | Double bands | Double bands |
| 2 | 161,315 and 198,243 | Single band | Double bands |
Fig. 1.
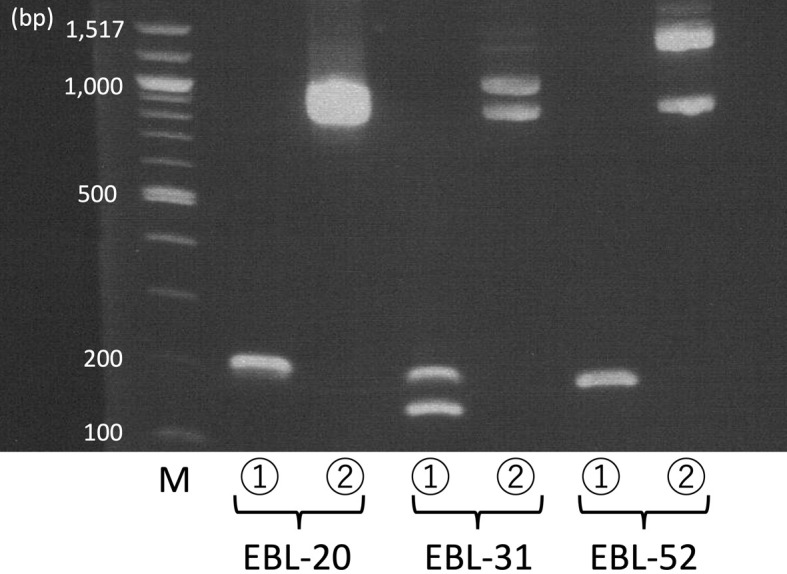
Representative results of B-cell clonality polymerase chain reaction (PCR) and inverse PCR. Peripheral blood from enzootic bovine leukosis (EBL)-20, lymph node from EBL-31, and lymph node from EBL-52 were used. ①: B-cell clonality PCR, ②: inverse PCR, and M: molecular weight marker.
In the present study, EBL cattle with BLV proviral copy numbers <100,000 copies per 100,000 nucleated cells showed a single band in B-cell clonality PCR and inverse PCR, indicating monoclonal proliferation of B-cells with one integration site of BLV proviral DNA, as well as two bands in B-cell clonality PCR and inverse PCR, indicating biclonal proliferation of B-cells with one integration site of BLV proviral DNA. On the other hand, EBL cattle with BLV proviral copy numbers >100,000 copies per 100,000 nucleated cells showed a single band and two bands in B-cell clonality PCR and inverse PCR, respectively, indicating monoclonal proliferation of B-cells with two integration sites of BLV proviral DNA. In EBL cattle with BLV proviral copy numbers greater than the number of nucleated cells, a few BLV proviruses might have integrated into the bovine genome.
Integration of a second provirus is typically prevented in retrovirus infection because of SIR, although multiple integrations of human T-cell lymphotropic virus type I, which is related to BLV, have been reported [4, 13]. The main form of SIR is receptor occupancy by viral envelope proteins, which prevents the binding of a second virus [10]. Moreover, receptor down-modulation is a way to prevent a second viral infection in cells with human immunodeficiency virus (HIV), a retrovirus [3, 5]. Furthermore, viral proteins F12-HIV, Gag, Vif, and Nef have a strongly reduced ability to initiate the retrotranscription process in HIV-infected cells, and second viral infection is thus prevented [2]. The preset study identified one complete genome of BLV in EBL cattle with monoclonal proliferation of B-cells and two integration sites of BLV proviral DNA. The results demonstrated identical BLV provirus integrated into 2 different sites. However, there are few reports on SIR in cells infected with BLV, and thus further investigation is necessary to elucidate the mechanism of BLV superinfection in EBL cattle.
The present study found monoclonal proliferation of B-cells with two BLV provirus integration sites in EBL cattle. However, no direct evidence for multiple integrations of BLV provirus was obtained. Further investigation using single cell analysis in EBL with BLV proviral copy numbers exceeding the number of bovine nucleated cells will be necessary.
In conclusion, we analyzed B-cell clonality and BLV provirus integration sites in EBL cattle, and found that EBL cattle with BLV proviral copy numbers less than the number of bovine nucleated cells had monoclonal or biclonal proliferation of B-cells with one integration site of BLV proviral DNA, whereas those with BLV proviral copy numbers greater than the number of bovine nucleated cells had monoclonal proliferation of B-cells with two integration sites of BLV proviral DNA. These results suggest that superinfection of BLV can occur in EBL cattle. However, the mechanism of superinfection of BLV in EBL cattle and differences in the characteristics of tumor cells with one or two integration sites of BLV proviral DNA remain unclear, warranting further investigation.
CONFLICTS OF INTEREST
The Laboratory of OSG Veterinary Science for Global Disease Management is an endowment laboratory, supported by a grant from OSG Corporation.
Supplementary Material
Acknowledgments
We thank veterinarians of the Chiba, Iwate, and Tokachi Agriculture Mutual Aid Associations, Ibaraki Livestock Hygiene Service Center, and Tokachi and Hayakita Meat Hygiene Inspection Centers for sampling. This work was supported by Japan Society for the Promotion of Science (JSPS) KAKENHI Grant Numbers 23K14077 and 23H02378.
REFERENCES
- 1.Aida Y, Murakami H, Takahashi M, Takeshima SN. 2013. Mechanisms of pathogenesis induced by bovine leukemia virus as a model for human T-cell leukemia virus. Front Microbiol 4: 328. doi: 10.3389/fmicb.2013.00328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.D’Aloja P, Olivetta E, Bona R, Nappi F, Pedacchia D, Pugliese K, Ferrari G, Verani P, Federico M. 1998. gag, vif, and nef genes contribute to the homologous viral interference induced by a nonproducer human immunodeficiency virus type 1 (HIV-1) variant: identification of novel HIV-1-inhibiting viral protein mutants. J Virol 72: 4308–4319. doi: 10.1128/JVI.72.5.4308-4319.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoxie JA, Alpers JD, Rackowski JL, Huebner K, Haggarty BS, Cedarbaum AJ, Reed JC. 1986. Alterations in T4 (CD4) protein and mRNA synthesis in cells infected with HIV. Science 234: 1123–1127. doi: 10.1126/science.3095925 [DOI] [PubMed] [Google Scholar]
- 4.Kamihira S, Dateki N, Sugahara K, Yamada Y, Tomonaga M, Maeda T, Tahara M. 2000. Real-time polymerase chain reaction for quantification of HTLV-1 proviral load: application for analyzing aberrant integration of the proviral DNA in adult T-cell leukemia. Int J Hematol 72: 79–84. [PubMed] [Google Scholar]
- 5.Lama J. 2003. The physiological relevance of CD4 receptor down-modulation during HIV infection. Curr HIV Res 1: 167–184. doi: 10.2174/1570162033485276 [DOI] [PubMed] [Google Scholar]
- 6.Murakami H, Yamada T, Suzuki M, Nakahara Y, Suzuki K, Sentsui H. 2011. Bovine leukemia virus integration site selection in cattle that develop leukemia. Virus Res 156: 107–112. doi: 10.1016/j.virusres.2011.01.004 [DOI] [PubMed] [Google Scholar]
- 7.Maezawa M, Fujii Y, Akagami M, Kawakami J, Inokuma H. 2023. Phylogenetic analysis based on whole genome sequence of bovine leukemia virus in cattle under 3 years old with enzootic bovine leukosis. PLoS One 18: e0279756. doi: 10.1371/journal.pone.0279756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maezawa M, Sakaguchi K, Tanaka Y, Watanabe K, Kobayashi Y, Inokuma H. 2021. Detection of monoclonal or oligoclonal integration of bovine leukemia virus proviral DNA by inverse polymerase chain reaction for diagnosis of enzootic bovine leukosis. Comp Clin Pathol 30: 711–714. doi: 10.1007/s00580-021-03265-6 [DOI] [Google Scholar]
- 9.Maezawa M, Watanabe K, Horiuchi N, Matsumoto K, Kobayashi Y, Inokuma H. 2020. Molecular diagnosis of bovine B-cell lymphoma using PCR for immuno- globulin heavy chain gene. J Vet Med Sci 82: 61–63. doi: 10.1292/jvms.19-0418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nethe M, Berkhout B, van der Kuyl AC. 2005. Retroviral superinfection resistance. Retrovirology 2: 52. doi: 10.1186/1742-4690-2-52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schwartz I, Lévy D. 1994. Pathobiology of bovine leukemia virus. Vet Res 25: 521–536. [PubMed] [Google Scholar]
- 12.Takeshima SN, Kitamura-Muramatsu Y, Yuan Y, Polat M, Saito S, Aida Y. 2015. BLV-CoCoMo-qPCR-2: improvements to the BLV-CoCoMo-qPCR assay for bovine leukemia virus by reducing primer degeneracy and constructing an optimal standard curve. Arch Virol 160: 1325–1332. doi: 10.1007/s00705-015-2377-3 [DOI] [PubMed] [Google Scholar]
- 13.Yamamoto I, Takajo I, Umeki K, Morishita K, Hatakeyama K, Kataoka H, Nomura H, Okayama A. 2010. Multiple integrations of human T-lymphotropic virus type 1 proviruses in the engrafted cells from the asymptomatic carriers in NOD/SCID/gammacnull mice. Intervirology 53: 229–239. doi: 10.1159/000302760 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


