Abstract
We constructed human immunodeficiency virus type 1 (HIV-1) vectors that will allow higher levels of gene expression in T cells. Gene expression under the control of an internal cytomegalovirus (CMV) immediate-early promoter in a self-inactivating lentiviral vector (CSCG) is 4- to 15-fold lower in T-cell lines (SUPT1 and CEMX174) than in non-lymphoid-cell lines (HeLa and 293T). This is in contrast to a Moloney murine leukemia virus (MoMLV)-based retrovirus vector (SRαLEGFP). We therefore replaced the internal CMV promoter of CSCG with three different murine oncoretroviral long terminal repeat (LTR) promoters—murine sarcoma virus (MSV), MoMLV (MLV), and the LTR (termed Rh-MLV) that is derived from the ampho-mink cell focus-forming (AMP/MCF) retrovirus in the serum of one rhesus macaque monkey that developed T-cell lymphoma following autologous transplantation of enriched bone marrow stem cells transduced with a retrovirus vector preparation containing replication-competent viruses (E. F. Vanin, M. Kaloss, C. Broscius, and A. W. Nienhuis, J. Virol. 68:4241–4250, 1994). We found that the combination of Rh-MLV LTR and a partial gag sequence of MoMLV (Δgag871–1612) in CS-Rh-MLV-E gave the highest level of enhanced green fluorescent protein (EGFP) gene expression compared with MLV, MSV LTR, phosphoglycerate kinase, and CMV promoters in T-cell lines, as well as activated primary T cells. Interestingly, there was a further two- to threefold increase in EGFP expression (thus, 10-fold-higher expression than with CMV) when the Rh-MLV promoter and Δgag871–1612 were used in a self-inactivating-vector setting that has a further deletion in the U3 region of the HIV-1 LTR. These hybrid vectors should prove useful in gene therapy applications for T cells.
Lentivirus vectors based on human immunodeficiency virus type 1 (HIV-1) can transduce nondividing cells in vitro and in vivo (6, 12, 18, 23, 27). This is an advantage over retrovirus vectors derived from oncoretroviruses, such as Moloney murine leukemia virus (MoMLV) because these retrovirus vectors require proliferation of the target cells for integration (8, 25). Several measures have been taken to develop HIV-1 vectors that are safer for gene therapy. First, accessory genes (vif, vpr, vpu, and nef) that are not essential for transduction were eliminated from a packaging construct (12, 13, 30). Second, the use of a three-plasmid expression system that consists of packaging, envelope, and vector constructs has minimized the possibility of generating replication-competent virus through recombination (21, 22, 23). Third, self-inactivating (SIN) vectors are constructed by deletions in the 3′ long terminal repeats (LTRs) of the HIV-1 vectors (19, 31). Miyoshi et al. (19) have deleted 133 nucleotides in the U3 region of the 3′ LTR, including the TATA box and binding sites for the transcription factors Sp1 and NF-κB, in the CS-G vector. Zufferey et al. (31) have generated a SIN vector (SIN-18) that has a more extensive 400-nucleotide deletion in the U3 region of the 3′ LTR, thus eliminating all of the transcriptional elements located upstream of the SP1 binding sites. These SIN vectors also enable the regulated expression of genes from internal promoters by eliminating any cis-acting effects of the HIV-1 LTR (19, 31).
The internal cytomegalovirus (CMV) immediate-early promoter has been widely used in lentivirus vectors to control gene expression in mammalian cells. It is expressed efficiently in a variety of cell types (6, 12, 18) but generally less efficiently in lymphoid cells (2). Since dysfunctional T cells are often associated with an inherited or acquired immunodeficiency (11, 16, 26), there are numerous gene therapy applications for such dysfunctional T cells (including HIV-1 disease). We therefore tested other promoters that are expressed well in T cells. We tested the MoMLV (hereafter termed MLV) LTR because overexpression of proto-oncogenes under the control of the MLV LTR has been demonstrated to be one of the common features of MoMLV-induced thymic lymphoma (28). We also tested the LTR of a novel retrovirus, ampho-mink cell focus-forming (AMP/MCF) virus (termed the Rh-MLV LTR), that was identified in the serum of a rhesus monkey that developed rapidly progressive T-cell lymphomas involving the thymus, lymph nodes, liver, spleen, and bone marrow (9, 24, 29). The lymphomas were the result of insertional mutagenesis by the productive retroviral infections in the monkeys that had received autologous transplantation of enriched bone marrow stem cells transduced with a retrovirus vector preparation containing replication-competent viruses (29). The AMP-MCF replication-competent virus arose by two recombination events—first, a recombination between the vector and packaging-encoding sequences in the producer clone used for transduction of bone marrow stem cells (vector-helper recombinant); second, a recombination involving the genome of the vector-helper recombinant and endogenous murine retroviral genomes in the producer clone. The LTR of the AMP-MCF virus also differs from the MLV LTR by a 23-bp insertion (a direct copy of the 23 bp 3′ to the site of insertion) in the enhancer region of the U3 segment.
In the present study, we used the gene for enhanced green fluorescent protein (EGFP) as a reporter gene and analyzed the effects of various murine oncoretrovirus LTR promoters in a SIN lentivirus vector. Our results show that the Rh-MLV LTR is a stronger promoter than the MLV and CMV promoters, and the partial gag sequence of MoMLV in the context of an HIV-1-based vector is essential for the high level of gene expression in human T lymphocytes.
MATERIALS AND METHODS
Construction of vectors.
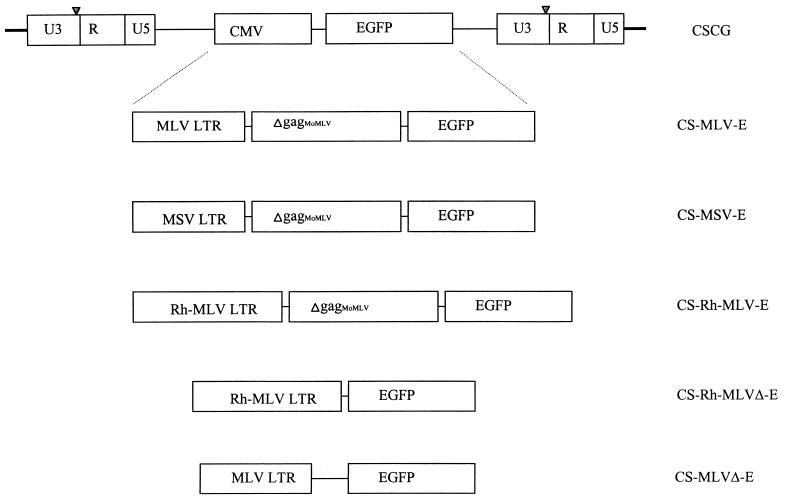
The HIV-1-based vectors CS-MSV-E, CS-MLV-E, CS-MLVΔ-E, CS-Rh-MLV-E, and CS-Rh-MLVΔ-E were derived from pCS or pCSCG (19). CS-MSV-E was constructed from pCS and SRαLEGFP (4). SRαLEGFP was digested with BglII, filled in with Klenow fragments, and then excised by NheI. The resulting 2.2-kb fragment, which contains the Moloney murine sarcoma virus (MSV) LTR, a partial gag sequence of MoMLV from LNL6 (Δgag803–1612; nucleotides 803 to 1612, as described in GenBank accession number M63653), and EGFP, was ligated to a 7-kb HpaI/XbaI fragment of pCS. To make CS-MLV-E, a 2.2-kb fragment of CS-MSV-E that contains the MSV LTR, Δgag803–1612 of MoMLV, and EGFP was subcloned into the XhoI and BstXI sites of the pBlu2SKM vector. The resulting intermediate construct, pBS-MSV-EM, was digested with EcoRI, filled in with Klenow fragments, digested with XmaI to liberate the MSV LTR, and then ligated to a 700-bp fragment of SRαLEGFP (which contains the MLV LTR) to give pBS-MLV-EM′. The 2.2-kb BstXI/XhoI fragment of pBS-MLV-EM′ was then religated to the same sites in CS-MSV-E. To make CS-MLVΔ-E, pCSCG was digested with EcoRI, filled in with Klenow fragments, and digested with NheI to liberate a 500-bp fragment that contains the CMV promoter. The resulting 8-kb fragment was then ligated to an ∼950-bp fragment of pBS-MLV-EM′. The latter fragment was prepared by digesting pBS-MLV-EM′ with BstXI, filling in with Klenow fragments, and digesting with SpeI.
To make CS-Rh-MLVΔ-E, the 5-3BS plasmid was digested with AatII, blunt ended by T4 DNA polymerase, and then digested with SstII to liberate an 820-bp fragment that contained the Rh-MLV LTR. This Rh-MLV LTR-containing fragment was cloned into the pCS vector that was previously digested by EcoRI, filled in by Klenow fragments, and digested with SstII. The resulting construct, pCS-Rh-MLVΔ, was then digested with HpaI and XhoI and ligated to a 700-bp XhoI fragment of pCSCG (which contains the EGFP reporter gene) filled in with NheI to generate CS-Rh-MLVΔ-E. To generate CS-Rh-MLV-E, two intermediate constructs (pCS-IRES-EGFP and pCS-Rh-MLVΔ-IRES-EGFP) were made. For pCS-CMV-IRES-EGFP, pSRαIRES-EGFP was digested with BglII, filled in with Klenow fragments, and digested with EcoRI to liberate an ∼1.4-kb fragment that contained the IRES-EGFP gene sequence. This 1.4-kb fragment was then cloned into the HpaI and EcoRI sites of pCS. For pCS-Rh-MLVΔ-IRES-EGFP, pCS-Rh-MLVΔ was digested with XhoI, filled in with Klenow fragments, and then digested with BamHI to liberate an ∼900-bp fragment that contained the Rh-MLV LTR. This DNA fragment was ligated to the Eco47III and BamHI sites of the pCS-IRES-EGFP vector. CS-Rh-MLV-E was then constructed by cloning a ∼1.7-kb fragment of pBS-MSV-EM (SpeI digested, filled in with Klenow fragments, and XhoI digested) into the HpaI and XhoI sites of pCS-Rh-MLVΔ-IRES-EGFP. To generate SIN-18-Rh-MLV-E, an ∼600-bp fragment (which contains the housekeeping phosphoglycerate kinase [PGK] promoter) of RRL-PGK-EGFP-SIN18 (31) was liberated by EcoRV and AgeI. The resulting vector fragment was then ligated to the ∼1.4-kb fragment of CS-Rh-MLV-E (SacII digested, blunted by T4 polymerase, and AgeI digested) that contains the Rh-MLV promoter and the partial gag sequence of MoMLV.
We have also generated two SIN vectors that contain the partial untranslated gag sequence of LNL6 (Δgag871–1612; nucleotides 871 to 1612 as described in GenBank accession number M63653) of MoMLV in the context of CMV (CS-CMV-ΔgagE) and PGK (SIN18-PGK-ΔgagE) internal promoters. To generate CS-CMV-ΔgagE, the EGFP reporter gene of CSCG was replaced by the cloning of an ∼1.3-kb SpeI-XhoI fragment of pBS-MSV-EM (which contains the partial gag sequence and the EGFP reporter gene) into the NheI and XhoI sites of CSCG. For SIN18-PGK-ΔgagE, an SpeI-NotI fragment of SRαLEGFP (containing the partial gag sequence and the EGFP reporter gene) was first subcloned into the same sites of pBluSK2M (pBS-gagE). The EGFP reporter gene of SIN-18 was then replaced by the cloning of an ∼1.3-kb XmaI-SacII fragment of pBS-gagE (that contains the partial gag sequence and the EGFP reporter gene) into the AgeI and SacII sites of the SIN-18 vector.
Production and determination of titers of VSVG-pseudotyped lentivirus reporter virus.
Vesicular stomatitis virus G (VSVG)-pseudotyped lentivirus vectors were produced by calcium phosphate-mediated transfection of 293T cells as described previously (4). In brief, 20 × 106 293T cells were transfected with 5 μg of pHCMVG, 12.5 μg of pCMVΔR8.2DVPR, and 12.5 μg of the lentivirus reporter vector (CSCG, CS-MLV-E, CS-MLVΔ-E, CS-Rh-MLV-E, or CS-Rh-MLVΔ-E). Culture supernatant that contained viruses was collected on days 2, 3, and 4 posttransfection, pooled, and filtered through a 0.22-μm-pore-size filter. To make VSVG-pseudotyped retrovirus vectors, 20 × 106 293T cells were transfected with 5 μg of pHCMVG, 12.5 μg of SVψ−envMLV (3, 15), and 12.5 μg of SRαLEGFP.
Virus titers were determined by infection of HeLa cells, followed by flow analysis of the percentages of EGFP+ HeLa cells on day 3 after infection. HeLa cells (5 × 104/well) were cultured in a 24-well plate overnight and were infected with VSVG-pseudotyped viruses by incubating the cells with 0.25 ml of virus supernatant (at appropriate dilutions, usually unconcentrated [1×] and 1/10× and 1/50× diluted) in the presence of Polybrene (10 μg/ml; Sigma) at 37°C for 2 h. The virus supernatant was then removed and replaced with 0.5 ml of Dulbecco's modified Eagle medium with 10% calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. The percentages of EGFP+ HeLa cells were analyzed on day 3 after infection. Viruses were diluted in Iscove's modified Dulbecco's medium with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml to various dilutions (usually 1/10× and 1/50×). Unconcentrated viral supernatant of all lentivirus-based vectors generally yielded a titer of 0.1 × 106 to 2 × 106 infectious units (IU)/ml; SRαLEGFP generally yielded a titer of 104 IU/ml.
Activation of primary PBMC cultures.
Peripheral blood mononuclear cells (PBMC) were separated over a Ficoll-Hypaque gradient (Amersham Pharmacia, Uppsala, Sweden). A culture plate was coated with 10 μg of goat anti-mouse immunoglobulin G antibodies (Cappel, Durham, N.C.)/ml at 37°C for 2 h, washed with sterilized phosphate-buffered saline, and recoated with 1 μg of anti-CD3 monoclonal antibody (MAb) (Coulter Corp., Hialeah, Fla.)/ml at 37°C for 2 h. Nonadherent PBMC were obtained after depleting macrophages by 2 h of adherence to plastic and were cultured in a 6-well plate (at 106 per well) that was coated with immobilized anti-CD3 MAb. The cells were cultured in RPMI 1640 supplemented with 10% AB serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and 0.5 μg of soluble anti-CD28 MAb (Pharmingen, San Diego, Calif.)/ml. Activated T cells were collected 48 to 60 h after activation and were used for infection experiments.
Infection of target cells in vitro.
CEMX174 and SUPT1 cells were cultured in Iscove's modified Dulbecco's medium (GIBCO) with 10% fetal bovine serum, 100 U of penicillin/ml, and 100 μg of streptomycin (GIBCO)/ml. HeLa and 293T cells were cultured in Dulbecco's modified Eagle medium with 10% calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Activated T cells were cultured in RPMI 1640 (GIBCO) supplemented with 10% fetal bovine serum, 100 U of penicillin/ml, 100 μg of streptomycin/ml, and interleukin 2 (1 μg/ml). The cells (105) were infected with VSVG-pseudotyped viruses by incubation with 0.25 ml of virus supernatant (at appropriate dilutions) in the presence of Polybrene (10 μg/ml) at 37°C for 2 h. Three days postinfection, the cells were analyzed by flow cytometry for EGFP expression.
Flow cytometric analysis.
Infected cells were harvested, run on a FACScan flow cytometer (Becton-Dickinson & Co., San Jose, Calif.), and analyzed by the Cell Quest program (Becton-Dickinson) for the percentage of EGFP+ cells and the level of EGFP expression. Five thousand to 10,000 events were acquired for each sample. To determine the expression of CD4 and CD8 molecules on activated T cells, 5 × 105 infected cells were costained with anti-CD4-PE (Coulter) and anti-CD8-PerPC (Becton-Dickinson) MAbs.
RESULTS
Construction and generation of modified SIN vectors.
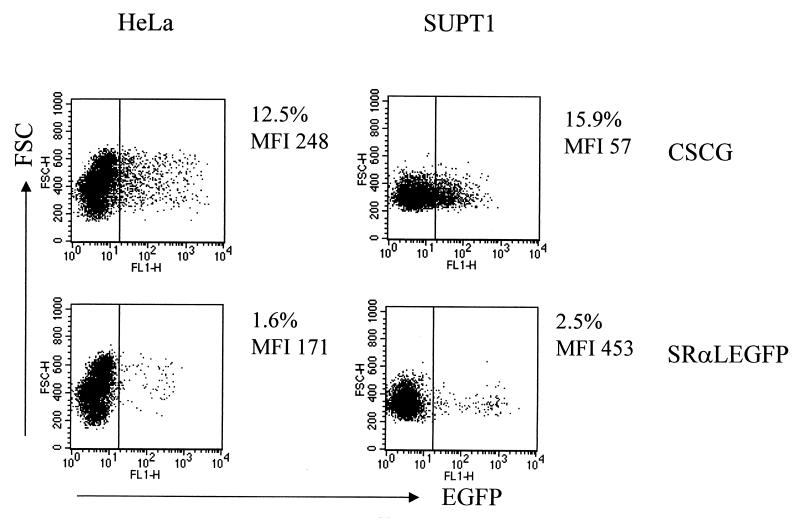
The lentivirus-based SIN vector CSCG has a 133-bp deletion in the U3 region of the 3′ LTR that eliminates the transcriptional interference of gene expression from the internal CMV promoter in the provirus. EGFP expression under the control of the CMV promoter in CSCG is about four times higher in HeLa cells (mean fluorescence intensity [MFI], 248) than in SUPT1 cells (MFI, 57) (Fig. 1). We constructed modified lentivirus vectors that allow higher levels of gene expression in T cells. SRαLEGFP is a MoMLV-based vector that contains the MLV LTR, a partial untranslated gag sequence of MoMLV, and the EGFP reporter gene in its proviral form. The 809-bp MoMLV partial gag sequence (Δgag803–1612, derived from LNL6 nucleotides 803 to 1612 as described in GenBank accession number M63653) of SRαLEGFP was derived from the retrovirus vector LNSX (5, 17), which contains the “extended packaging sequence” essential for the increase in retrovirus vector titers and gene transfer efficiency. In addition, a stop codon was inserted in place of the Pr65 gag start codon to prevent synthesis of Pr65 gag (17). We compared CSCG with SRαLEGFP in infected HeLa and SUPT1 cells and found that, in contrast to CSCG, SRαLEGFP has a higher EGFP expression in SUPT1 than in HeLa cells. EGFP expression under SRαLEGFP in SUPT1 cells is generally 8- to 10-fold higher than that of CSCG (Fig. 1 and Table 1), suggesting that MLV LTR is a stronger promoter than CMV in SUPT1 cells. We therefore tested the promoter activity of various oncoretrovirus LTRs in a SIN vector. The internal CMV promoter of CSCG was replaced by an oncoretrovirus LTR derived either from MLV (CS-MLV-E) or MSV (CS-MSV-E) (Fig. 2). We also tested a novel LTR that has been identified in the AMP-MCF retrovirus found in the serum of a monkey with lymphoma (CS-Rh-MLV-E) (9, 29) (Fig. 2). We hypothesize that since this LTR was derived from a rhesus macaque T-cell tumor it should be expressed efficiently in primate T cells. All three vectors also contain the untranslated partial gag sequence of MoMLV (from the SRαLEGFP vector) and the EGFP gene as a reporter. We have also constructed CS-MLVΔ-E and CS-Rh-MLVΔ-E vectors that are devoid of the partial gag sequence. VSVG-pseudotyped vectors were generated by transient cotransfection of each lentivirus vector construct with a packaging construct and a VSVG expression construct in 293T cells. The culture supernatant of the transfectant cells was collected, and the titer was determined by quantitation of the number of EGFP-positive HeLa cells in flow cytometry. The unconcentrated virus supernatants of all vectors, including the CSCG vector, generally yielded a titer of 0.1 × 106 to 2 × 106 IU/ml (see figures below). Thus, the use of murine oncoretrovirus LTR internal promoters in the context of a SIN vector provides vector titers comparable to those of CSCG. SRαLEGFP generally yielded a titer of 104 IU/ml.
FIG. 1.
Comparison of the control of EGFP gene expression in CSCG and SRαLEGFP vectors. HeLa and SUPT1 cells were infected by unconcentrated virus supernatant of CSCG (virus titer, 0.5 × 105 IU/ml; MOI, 0.125) and SRαLEGFP (virus titer, 0.64 × 104 IU/ml; MOI, 0.016). The percentage of EGFP+ cells and the MFI of EGFP expression of the infected cells were analyzed by flow cytometry 3 days after infection. FSC, forward scatter; FL, fluorescence.
TABLE 1.
Summary of EGFP expression of various vectors in different cell lines
| Construct | Relative EGFP expressiona (± SD)
|
|||
|---|---|---|---|---|
| SUPT1 | CEMX174 | HeLa | 293T | |
| CSCG | 1 | 1 | 1 | 1 |
| CS-MLV-E | 2.9 ± 0.5 | 2.2 ± 0.5 | 0.35 ± 0.08 | 0.13 ± 0.02 |
| CS-Rh-MLV-E | 4.9 ± 1.0 | 2.5 ± 1.0 | 0.38 ± 0.05 | 0.13 |
| CS-MSV-E | 0.98 ± 0.09 | 1.1 ± 0.2 | 0.32 ± 0.02 | 0.09 ± 0.005 |
| CS-Rh-MLVΔ-E | 2.5 ± 0.5 | 1.0 ± 0.2 | 0.25 ± 0.05 | 0.09 ± 0.005 |
| CS-MLVΔ-E | 2.3 ± 0.6 | 1.1 ± 0.3 | 0.26 ± 0.06 | 0.1 ± 0.006 |
| SRαLEGFP | 10.9 ± 2.7 | 6.2 ± 1.0 | 0.99 ± 0.19 | 0.21 ± 0.05 |
Relative to the EGFP mean fluorescence level of the CSCG construct.
FIG. 2.
Maps of various lentivirus-retrovirus hybrid vectors developed from a SIN HIV-1-based CSCG vector. The internal CMV immediate-early promoter was removed from CSCG and replaced with an oncoretrovirus LTR (MLV, Rh-MLV, or MSV) with or without a partial gag sequence of MoMLV.
CS-MLV-E has higher EGFP expression in T cell lines than CSCG.
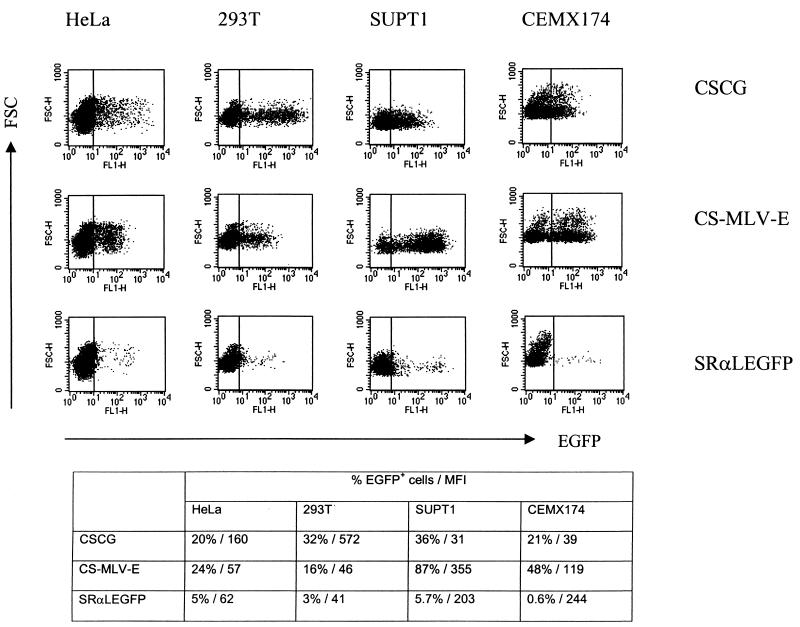
The CS-MLV-E vector differs from CSCG only in the internal promoter (MLV LTR instead of CMV) and the inclusion of the Δgag803–1612 of MoMLV. Thus, this lentivirus-based vector contains a transcriptional promoter-enhancer closely resembling that of the SRαLEGFP provirus. CS-MLV-E was used to infect HeLa, 293T, and T-cell lines (SUPT1 and CEMX174) as target cells in order to analyze the promoter activity of the MLV LTR in the SIN vector. CSCG and SRαLEGFP were used as controls. The percentage of EGFP-positive cells and the MFI of EGFP expression of the infected cells were analyzed 3 days after infection. The CSCG vector has a greater MFI of EGFP expression in HeLa and 293T (MFIs, 160 and 572, respectively) than in the CSCG-infected T-cell lines (Fig. 3). The MFIs of EGFP in the CSCG-infected SUPT1 and CEMX174 cells were usually 4- to 20-fold lower than those in HeLa or 293T cells (Fig. 1 and 3). In contrast, the MFI of EGFP expression in SRαLEGFP-infected SUPT1 or CEMX174 cells is always two- to sixfold higher than that in HeLa or 293T cells (Fig. 1 and 3). The EGFP expression of SRαLEGFP in SUPT1 or CEMX174 is generally 6- to 10-fold higher than that of CSCG, despite the fact that SRαLEGFP has a lower virus titer than CSCG (Fig. 3 and Table 1). We compared the EGFP expression of our CS-MLV-E vector to that of CSCG and SRαLEGFP. The EGFP expression of our CS-MLV-E vector in these four cell lines resembles that of the SRαLEGFP vector, i.e., the CS-MLV-E vector has a higher EGFP expression in T-cell lines (SUPT1 and CEMX174) than in HeLa and 293T cells. The MFI of EGFP expression of the SUPT1 or CEMX174 cells which were infected by unconcentrated virus supernatant of CS-MLV-E is 3- to 10-fold higher than that of the CSCG-infected cells (Fig. 3).
FIG. 3.
EGFP expression of CS-MLV-E in lymphoid- and non-lymphoid-cell lines. Lymphoid cells (SUPT1 and CEMX174) and non-lymphoid cells (HeLa and 293T) were infected by unconcentrated virus supernatant of CSCG (virus titer, 0.8 × 105 IU/ml; MOI, 0.2), CS-MLV-E (virus titer, 0.96 × 105 IU/ml, MOI, 0.24), and SRαLEGFP (virus titer, 2 × 104 IU/ml; MOI, 0.05). The percentage of EGFP+ cells and the MFI of EGFP expression of the infected cells were analyzed by flow cytometry 3 days after infection. FSC, forward scatter; FL, fluorescence.
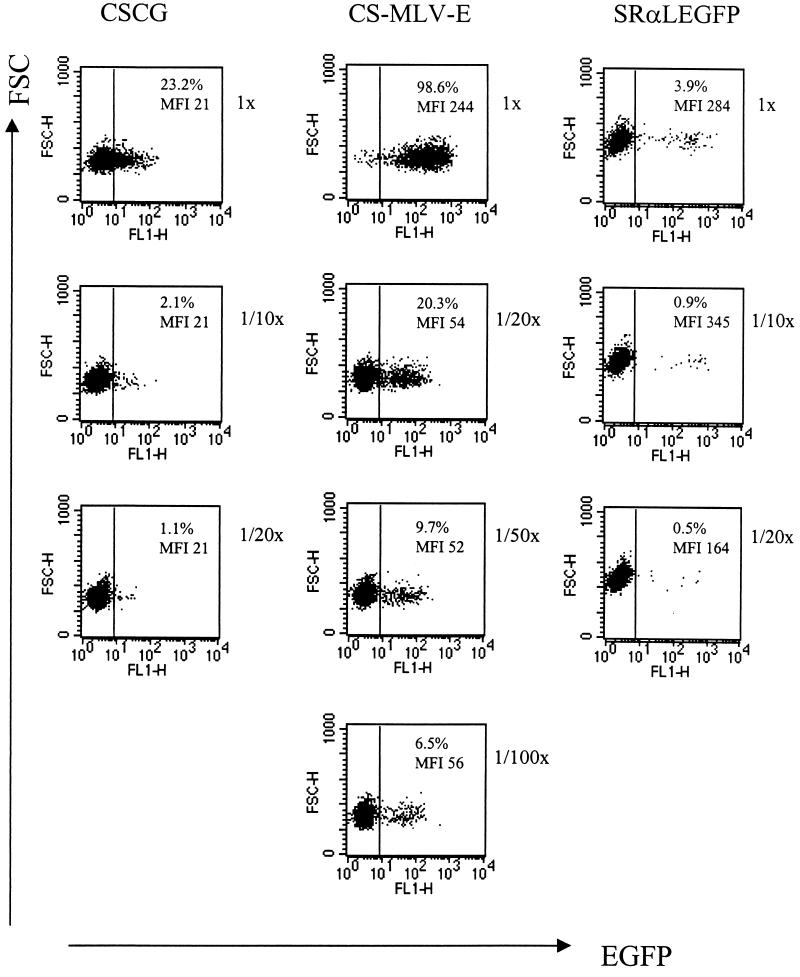
We tested whether the levels of expression of different vectors was independent of the multiplicity of infection (MOI) by titrating unconcentrated virus supernatant of CS-MLV-E and control vectors (CSCG and SRαLEGFP) on SUPT1 cells in an infection experiment. As shown in Fig. 4, a 20-fold dilution of a virus stock of CS-MLV-E that gives 98% EGFP+ cells resulted in a decrease in both the percentage of EGFP+ cells and the MFI of EGFP. Thus, it is likely that more than one virus per cell resulted in a greater MFI at a high MOI. Further dilution of CS-MLV-E resulted in a drop only in the percentage of EGFP-positive cells but not in the MFI of the EGFP-positive infected cells. It is therefore important to note that an accurate comparison of the MFI of the EGFP-positive infected targets requires a low concentration of virus and a low MOI. The MFI of EGFP of the infected cells (at <30% infection) should be a better representation of the promoter activity of the vector constructs. We compared the MFI of EGFP in the CSCG-, CS-MLV-E-, and SRαLEGFP-infected SUPT1 cells at an MOI of less than 0.1. It was found that SRαLEGFP has the highest EGFP expression (MFI, ∼300). CS-MLV-E has a two- to threefold-higher EGFP expression than CSCG (MFIs, approximately 50 and 20, respectively) (Fig. 4 and Table 1).
FIG. 4.
EGFP expression of CS-MLV-E in SUPT1 cells is higher than that of CSCG at low MOIs. SUPT1 cells (0.1 × 106) were infected by virus supernatant of CSCG (virus titer, 3 × 105 IU/ml), CS-MLV-E (virus titer, 1.2 × 106 IU/ml), and SRαLEGFP (virus titer, 0.7 × 105 IU/ml) at the indicated dilutions (undiluted [1×, MOI = 0.8] and 1/10× and 1/20× dilutions for CSCG; 1× (MOI = 3.0), 1/20×, 1/50×, and 1/100× dilutions for CS-MLV-E; 1× (MOI = 0.18), 1/10×, and 1/20× dilutions for SRαLEGFP). The percentage of EGFP+ cells and the MFI of EGFP expression of the infected cells were analyzed by flow cytometry 3 days after infection. FSC, forward scatter; FL, fluorescence.
Taken together, these data indicate that our CS-MLV-E vector represents a lentivirus-oncoretrovirus hybrid vector that has combined features of a lentivirus vector and the transcriptional unit of an oncoretrovirus vector, SRαLEGFP. Analysis of the transcriptional unit (MLV LTR plus the partial gag sequence of MoMLV) in the SIN vector showed that the MLV LTR promoter has a higher gene expression than the CMV promoter in T-cell lines, consistent with the high EGFP expression of the SRαLEGFP vector in these target cells.
We have also replaced the internal CMV promoter of CSCG with the MSV LTR and the partial gag sequence (Δgag803–1612) of the SRαLEGFP vector and found that such a vector (CS-MSV-E) shows no increase in EGFP expression compared to that of CSCG in T cells (Table 1), consistent with the different origin of the MSV LTR (28). This further confirms the fact that the strong EGFP expression of the SRαLEGFP vector in SUPT1 cells is, in part, associated with the MLV LTR promoter in the provirus.
Rh-MLV LTR, a novel MoMLV-derived LTR, has stronger promoter activity than MLV LTR.
A novel retrovirus (AMP-MCF) involving sequences derived from endogenous murine MCF-type viruses was isolated from the serum of one rhesus monkey that developed rapidly progressive T-cell lymphomas (29). It arises by recombination involving the genome of the vector, packaging-encoding sequences, and endogenous murine retroviral genomes in the producer clone, followed by further mutation in the rhesus monkey. We designated the 5′ LTR of this AMP-MCF virus the Rh-MLV LTR. The U3 segment of the Rh-MLV LTR has a 23-bp insertion, unlike the sequence of MLV LTR. The inserted segment is a direct copy of the 23 bp 3′ to the site of insertion and is inserted within the 5′ member of the two 75-bp direct repeats which compose the enhancer region of U3. We have replaced the internal CMV promoter of CSCG with this Rh-MLV LTR and the partial gag sequence (Δgag871–1612) of MoMLV (CS-Rh-MLV-E). Virus supernatant of CS-Rh-MLV-E was used to infect HeLa, 293T, SUPT1, and CEMX174 cell lines, and EGFP expression in the infected cells was analyzed.
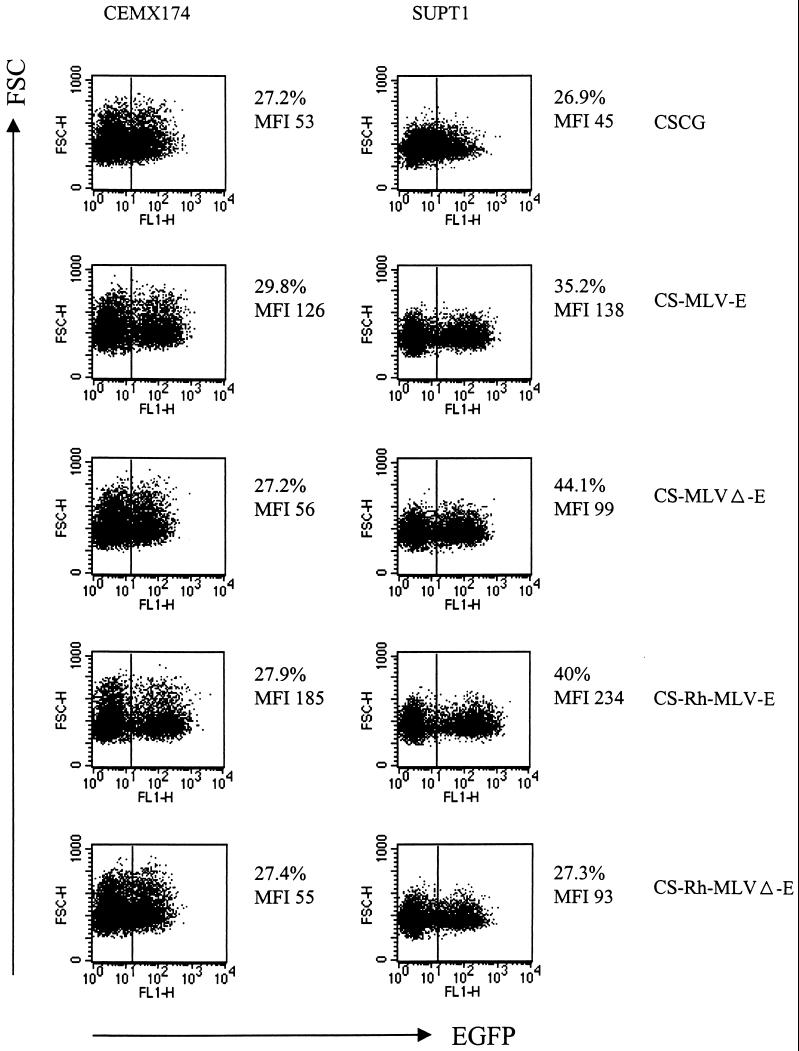
CS-MLV-E has two- to threefold-higher EGFP expression in T-cell lines than CSCG (Fig. 4 and Table 1). We compared CS-Rh-MLV-E with CS-MLV-E and CSCG at low concentrations of virus. CS-Rh-MLV-E and CS-MLV-E have comparable MFIs of EGFP in infected HeLa and 293T cells (Table 1). Interestingly, CS-Rh-MLV-E has an approximately twofold-higher MFI of EGFP expression in SUPT1 cells than CS-MLV-E, and it is fivefold higher than that of CSCG (Table 1 and Fig. 5). Based upon these results, we concluded that Rh-MLV is a stronger promoter than MLV in T cells and that the CS-Rh-MLV-E vector allows a higher level of gene expression in lymphoid-cell lines.
FIG. 5.
Partial gag sequence of MoMLV in CS-MLV-E and CS-Rh-MLV-E vectors is involved in enhanced EGFP expression in CEMX174 and SUPT1. CEMX174 and SUPT1 cells were infected by virus supernatant of CSCG (virus titer, 0.3 × 106 IU/ml; MOI, 0.075), CS-MLV-E (virus titer, 1.1 × 106 IU/ml; MOI, 0.055), CS-MLVΔ-E (virus titer, 1.3 × 106 IU/ml; MOI, 0.065), CS-Rh-MLV-E (virus titer, 1.9 × 106 IU/ml; MOI, 0.095), and CS-Rh-MLVΔ-E (virus titer, 1.2 × 106 IU/ml; MOI, 0.06). The percentage of EGFP+ cells and the MFI of EGFP expression of the infected cells were analyzed by flow cytometry 3 days after infection. FSC, forward scatter; FL, fluorescence.
It has been shown that SUPT1 cells are transduced efficiently with a SIN vector containing the internal housekeeping PGK gene promoter (RRL-PGK-EGFP-SIN18) (31). We therefore compared our CS-Rh-MLV-E with RRL-PGK-EGFP-SIN18 in SUPT1 cells. As described previously, CS-Rh-MLV-E consistently shows a fivefold increase in EGFP expression compared to that of CSCG (Tables 1 and 2). In contrast, RRL-PGK-EGFP-SIN18 showed only <2-fold enhancement of EGFP expression compared to that of CSCG (Table 2).
TABLE 2.
Summary of level of EGFP expression by various lentivirus-retrovirus hybrid vectors in SUPT1 cells
| Construct | Parent SIN vector construct | Internal promoter | Presence of partial gag sequencea | Relative EGFP expressionb (± SD) |
|---|---|---|---|---|
| CSCG | CS-G | CMV | − | 1 |
| CS-CMV-ΔgagE | CS-G | CMV | + | 0.74 ± 0.09 |
| CS-MSV-E | CS-G | MSV | + | 0.98 ± 0.09 |
| CS-MLV-E | CS-G | MLV | + | 2.9 ± 0.5 |
| CS-MLVΔ-E | CS-G | MLV | − | 2.3 ± 0.6 |
| CS-Rh-MLV-E | CS-G | Rh-MLV | + | 4.9 ± 1.0 |
| CS-Rh-MLVΔ-E | CS-G | Rh-MLV | − | 2.5 ± 0.5 |
| RRL-PGK-EGFP-SIN18 | SIN-18 | PGK | − | 1.6 ± 0.1 |
| SIN-18-PGK-ΔgagE | SIN-18 | PGK | + | 0.63 ± 0.16 |
| SIN-18-Rh-MLV-E | SIN-18 | Rh-MLV | + | 9.9 ± 1.9 |
+, present; −, absent.
Relative to the EGFP mean fluorescence level of the CSCG construct.
The partial gag sequence of MoMLV in CS-MLV-E and CS-Rh-MLV-E is essential for enhanced EGFP expression in CEMX174 cells.
We investigated whether the partial, untranslated gag sequence of MoMLV present in both the CS-MLV-E and CS-Rh-MLV-E vectors affects EGFP expression in infected cells. The partial gag sequence has previously been shown to encode packaging signal for MoMLV retrovirus that markedly increases the titer of in vitro-packaged retroviral vectors (5, 17). We deleted this partial gag sequence of MoMLV (Δgag871–1612) in CS-MLV-E (CS-MLVΔ-E) and CS-Rh-MLV-E (CS-Rh-MLVΔ-E) and compared their EGFP expressions in HeLa, 293T, SUPT1, and CEMX174 cell lines with those of CS-MLV-E and CS-Rh-MLV-E, respectively. This sequence should have no effect upon packaging of lentivirus vectors, and as indicated, we observed no change in these lentivirus vector titers. Deletion of the partial gag sequence in CS-MLVΔ-E and CS-Rh-MLVΔ-E did not significantly decrease the MFI of EGFP in HeLa or 293T cells, possibly due to the relatively low EGFP expression under the control of MLV or Rh-MLV LTRs in these cells (Table 1). However, an approximately twofold decrease in EGFP expression was observed in the infected SUPT1 and CEMX174 cells when the partial gag sequence was deleted (Fig. 5). These results demonstrated that (i) the partial gag sequence of MoMLV in these vectors is essential for the enhanced EGFP expression in these two T-cell lines; (ii) MLV or Rh-MLV LTR alone gave rise to a twofold increase in EGFP expression in SUPT1 cells compared to that of the CMV promoter in CSCG; (iii) the two- to threefold increase in EGFP expression of the CS-MLV-E- and CS-Rh-MLV-E-infected CEMX174 cells, in comparison to that of CSCG, could be largely attributed to the presence of this partial gag sequence.
We further tested the effect of the partial gag sequence of MoMLV (Δgag871–1612) on EGFP expression in T cells in two other SIN vector settings. We cloned Δgag871–1612 downstream of the CMV and PGK promoters in CSCG (CS-CMV-ΔgagE) and RRL-PGK-EGFP-SIN18 (SIN-18-PGK-ΔgagE), respectively, thus comparing its effect on the level of EGFP expression under the control of an internal CMV promoter and a housekeeping PGK promoter. Interestingly, we found that inclusion of this partial gag sequence in both constructs resulted in an approximately two- to threefold decrease in the EGFP expression in SUPT1 cells (Table 2) or primary T-cell blasts (data not shown) compared to those in their parental constructs (CSCG or RRL-PGK-EGFP-SIN18). Thus, the effect of the gag sequence in enhancing gene expression is dependent on being in the context of an MLV LTR.
CS-MLV-E and CS-Rh-MLV-E have higher EGFP expression in infected, primary PBMC culture.
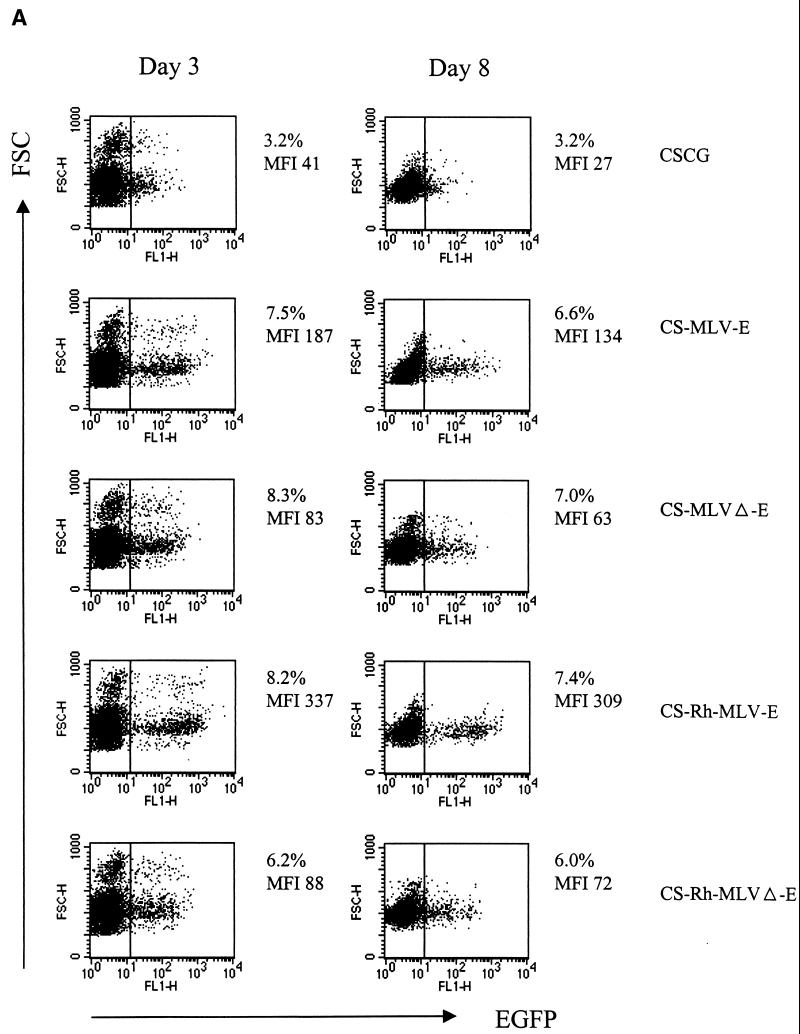
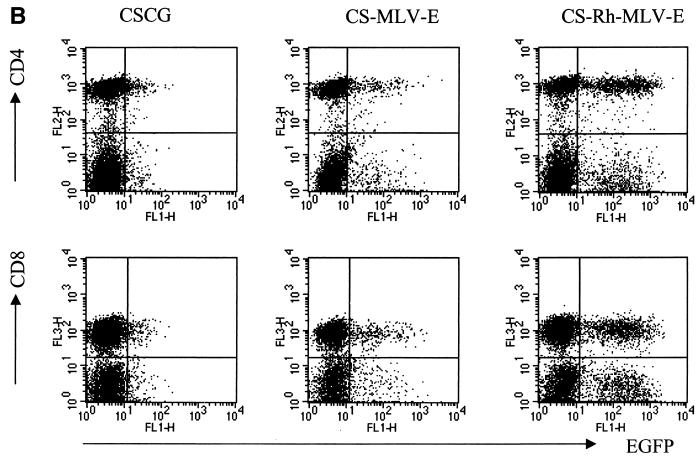
The above-mentioned results show that, in infected SUPT1 cells, the MLV LTR is a stronger promoter than CMV (comparing CS-MLVΔ-E with CSCG), the Rh-MLV LTR is a stronger promoter than the MLV LTR (comparing CS-Rh-MLV-E with CS-MLV-E), and the partial untranslated gag sequence of MoMLV (Δgag871–1612) is required for further enhancement of gene expression (comparing CS-MLV-E with CS-MLVΔ-E and CS-Rh-MLV-E with CS-Rh-MLVΔ-E). We tested our vectors in a primary PBMC culture that was activated by CD3 and CD28 antibodies. Activated T-cell blasts were then infected by virus supernatants of CSCG, CS-MLV-E, and CS-Rh-MLV-E and analyzed for EGFP expression 3 days postinfection. As shown in Fig. 6A, we found that CS-MLV-E and CS-Rh-MLV-E had four- and eightfold-higher EGFP expression, respectively, than CSCG, which was fully consistent with the results obtained in SUPT1 and CEMX174 cells. The expression of EGFP remained high and stable when we analyzed the same infected culture 8 days postinfection. These results confirm that the CS-MLV-E and CS-Rh-MLV-E vectors have higher gene expression levels in T lymphoid cells than CSCG and that Rh-MLV is a stronger promoter than MLV in T cells. In addition, we have shown that control of EGFP expression under these vectors is similar in both CD4+ and CD8+ T cell subsets (Fig. 6B).
FIG. 6.
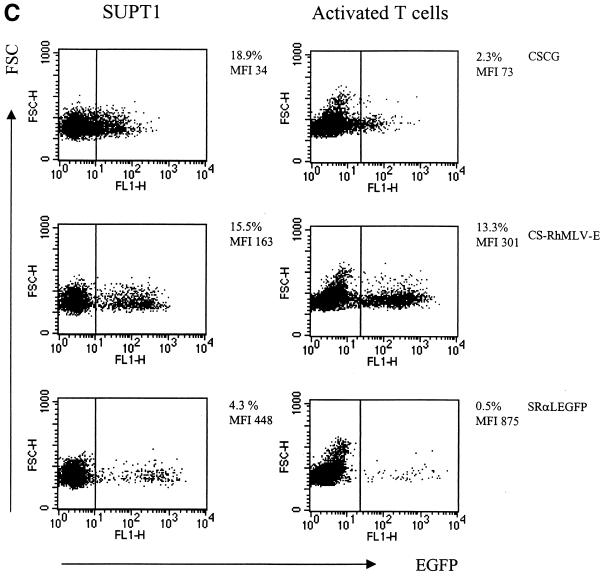
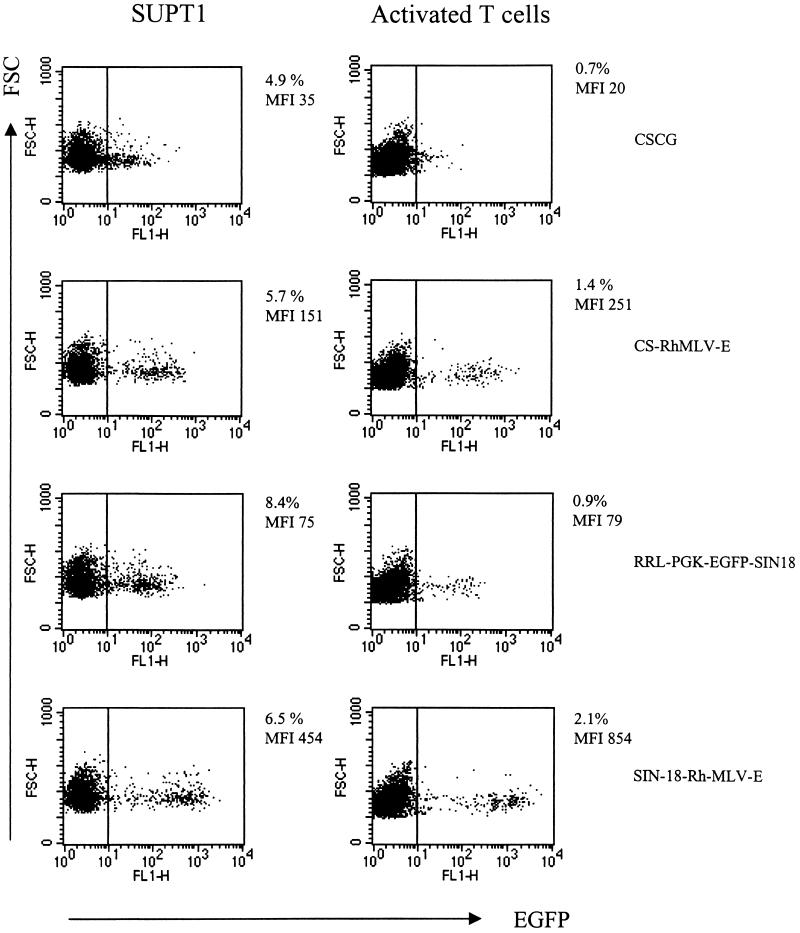
CS-Rh-MLV-E vector allows a high level of EGFP expression in primary activated T cells. (A) Flow cytometry analysis of CSCG-, CS-MLV-E-, and CS-Rh-MLV-E-infected T cells on day 3 or day 8 postinfection. (B) EGFP expressions in the infected CD4+ and CD8+ T-cell subsets were similar. Human PBMC were activated by plate-bound anti-CD3 and anti-CD28 MAbs for 60 h. Activated T-cell blasts were infected by virus supernatant of CSCG (virus titer, 0.3 × 106 IU/ml; MOI, 0.75), CS-MLV-E (virus titer, 1.1 × 106 IU/ml; MOI, 0.55), CS-MLVΔ-E (virus titer, 1.3 × 106 IU/ml; MOI, 0.65), CS-Rh-MLV-E (virus titer, 1.9 × 106 IU/ml; MOI, 0.48), and CS-Rh-MLVΔ-E (virus titer, 1.2 × 106 IU/ml; MOI, 0.3). The percentage of EGFP+ cells and the MFI of EGFP expression of the infected cells were analyzed by flow cytometry on days 3 and 8 postinfection. On day 8, aliquots of the infected cells were also stained for surface expression of CD4 and CD8 molecules and analyzed by fluorescence-activated cell sorter. (C) Flow cytometry analysis of the CSCG-, CS-RhMLV-E-, and SRαLEGFP-infected SUPT1 and activated primary T cells. SUPT1 cells were infected by virus supernatant of CSCG (virus titer, 1 × 106 IU/ml; MOI, 2.5), CS-RhMLV-E (virus titer, 1.4 × 106 IU/ml; MOI, 0.07), and SRαLEGFP (virus titer, 7 × 104 IU/ml; MOI, 0.18) at the following dilutions: undiluted (1×) for CSCG, 1/50× dilution for CS-Rh-MLV-E, and 1× for SRαLEGFP. Activated T-cell blasts were infected by undiluted virus supernatant of CSCG (virus titer, 1 × 106 IU/ml; MOI, 2.5) and CS-RhMLV-E (virus titer, 1.4 × 106 IU/ml; MOI, 3.5), except for SRαLEGFP, where 140×-concentrated virus supernatant (virus titer, 9.8 × 106 IU/ml; MOI, 25) was used in the infection. The percentage of EGFP+ cells and the MFI of EGFP expression of the infected cells were analyzed by flow cytometry on day 3 postinfection. FSC, forward scatter; FL, fluorescence.
We directly compared the efficiency of T-lymphoid-cell transduction by CS-Rh-MLV-E and SRαLEGFP. Unconcentrated virus supernatant of SRαLEGFP and lentivirus vectors (CSCG or CS-Rh-MLV-E) generally yielded titers of 104 and 106 IU/ml, respectively. When unconcentrated virus supernatant of SRαLEGFP was used to infect SUPT1 cells at an MOI of 0.175, we found that 4.3% of infected cells were EGFP+ and that EGFP expression (MFI, 448) was high (Fig. 6C). However, no EGFP+ cells could be detected when unconcentrated virus supernatant of SRαLEGFP was used to infect activated T-cell blasts at the same MOI (data not shown). We also used concentrated virus supernatant of SRαLEGFP to infect activated T cells in the same experiment. We found that 0.5% of EGFP+ cells (with the highest EGFP expression [MFI, 875]) were detected in the culture when an MOI of 25 was used (Fig. 6C). In contrast, 13.3% of CS-Rh-MLV-E-infected primary activated T-cell blasts were EGFP+ (MFI, 301) at an MOI of 3.5 (Fig. 6C). In fact, we could detect 4.8% EGFP+ activated T-cell blasts when CS-Rh-MLV-E was used at an MOI of 0.35 (data not shown). These results indicate that CS-Rh-MLV-E is more efficient than SRαLEGFP in T-lymphoid-cell transduction.
Further enhancement of EGFP expression by the Rh-MLV LTR and the partial gag sequence (Δgag871–1612) in a SIN vector that has further deletions in the U3 region.
In the CS-G SIN vector setting, 133 nucleotides in the U3 region of the 3′ LTR, including the TATA box and binding sites for transcription factors Sp1 and NF-κB, were deleted (19). A more extensive deletion in the U3 region of the 3′ LTR (eliminating all of the transcriptional elements located upstream of the SP1 binding sites) was used in the SIN vector (SIN-18) used in the construction of RRL-PGK-EGFP-SIN18 (31).
We tested whether further elimination of any cis-acting effects of the HIV-1 LTR would affect the control of EGFP expression under the internal promoter. The Rh-MLV promoter and the partial gag sequence (Δgag871–1612) were cloned into the SIN-18 vector (SIN-18-Rh-MLV-E). Virus supernatant of SIN-18-Rh-MLV-E was used to infect T lymphoid cells (SUPT1 and activated T-cell blasts) and was compared to that of CSCG, CS-Rh-MLV-E, and RRL-PGK-EGFP-SIN18. SIN-18-RH-MLV-E has ∼10-fold-higher EGFP expression in T cells than CSCG (Fig. 7 and Table 2). We compared SIN-18-RH-MLV-E and CS-Rh-MLV-E and found that SIN-18-RH-MLV-E has a two- to threefold-higher MFI of EGFP expression in T cells than CS-Rh-MLV-E (Fig. 7 and Table 2). Furthermore, in an identical SIN vector setting (SIN-18), SIN-18-Rh-MLV-E shows a 6- to 10-fold-higher MFI of EGFP expression than RRL-PGK-EGFP-SIN18 (Fig. 7 and Table 2), demonstrating that the combination of Rh-MLV LTR and Δgag871–1612 performs better than the housekeeping PGK gene promoter.
FIG. 7.
Further enhancement of EGFP expression by the Rh-MLV LTR and the partial gag sequence in a SIN vector that has further deletions in the U3 region. SUPT1 cells were infected by virus supernatant of CSCG (virus titer, 0.5 × 106 IU/ml; MOI, 0.025), CS-RhMLV-E (virus titer, 0.5 × 106 IU/ml; MOI, 0.025), RRL-PGK-EGFP-SIN18 (virus titer, 0.3 × 106 IU/ml; MOI, 0.075), and SIN18-Rh-MLV-E (virus titer, 0.6 × 106 IU/ml; MOI, 0.03) at the following dilutions: 1/50× for CSCG, CS-RhMLV-E, and SIN18-Rh-MLV-E and 1/10× for RRL-PGK-EGFP-SIN18. For T-cell blast infection experiments, 1× undiluted virus supernatants of CSCG (MOI, 1.25), CS-Rh-MLV-E (MOI, 1.25), RRL-PGK-EGFP-SIN18 (MOI, 0.75), and SIN-18-Rh-MLV-E (MOI, 1.5) were used. The percentage of EGFP+ cells and the MFI of EGFP expression of the infected cells were analyzed by flow cytometry 3 days after infection. FSC, forward scatter; FL, fluorescence.
DISCUSSION
We studied gene expression under the control of an internal CMV immediate-early promoter and MSV, MLV, and Rh-MLV LTRs in the SIN lentiviral vector. Transduction of SUPT1 cells by the vector that contains the internal MLV LTR resulted in a twofold increase in EGFP expression over the CMV promoter. The EGFP expression can be further increased by twofold when a gag sequence (Δgag803–1612) derived from MoMLV is included in the CS-MLV-E construct. Interestingly, replacement of MLV by Rh-MLV LTR (CS-Rh-MLV-E) allowed a twofold-higher EGFP expression than CS-MLV-E and a five- to eightfold-higher EGFP expression than the CMV promoter. There was a further 2- to 3-fold increase in EGFP expression (thus, a 10-fold-higher gene expression than CMV) when the Rh-MLV promoter and Δgag871–1612 were used in a SIN vector setting (SIN-18) that has a further deletion in the U3 region of the HIV-1 LTR. Thus, the combination of the Rh-MLV LTR and the gag sequence (Δgag871–1612) of MoMLV in SIN-18-Rh-MLV-E in the context of the SIN-18 vector represented the strongest promoter activity in the T-cell lines and activated primary T cells tested.
The CS-MLV-E vector differs from CSCG only in the internal promoter (MLV LTR instead of CMV) and the inclusion of a partial gag sequence of MoMLV. Thus, this lentivirus-based vector contains a transcriptional promoter-enhancer unit closely resembling that of the murine oncoretrovirus SRαLEGFP provirus. Perhaps not surprisingly, we found that the control of EGFP expression under the CS-MLV-E vector in HeLa, 293T, SUPT1, and CEMX174 cells resembles that under the SRαLEGFP vector. Notably, CS-MLV-E maintained two- to threefold-higher EGFP expression than CSCG independent of the MOI. However, we found that EGFP expression of CS-MLV-E, despite having a higher percentage of infectivity (EGFP-positive cells), is still three- to fivefold lower than that of SRαLEGFP. It is not clear why SRαLEGFP expresses EGFP at a higher level than CS-MLV-E. There are three mutually nonexclusive possibilities. First, the size of the SRαLEGFP proviral genome is smaller than that of CS-MLV-E, thus allowing a more efficient transcription and translation. Second, there exists a DNA sequence downstream of the 3′ MLV LTR promoter in the SRαLEGFP construct that we did not clone into CS-MLV-E, and the context of the MLV LTR may influence expression. Third, the transcriptional elements located upstream of the SP1 binding sites in the U3 region of the 3′ LTR of the CS-G SIN vector may interfere with the activity of the internal MLV promoter. In this regard, it is of interest to compare the levels of gene expression of CS-Rh-MLV-E and SIN-18-Rh-MLV-E because the vectors contain identical Rh-MLV internal promoters and the Δgag871–1612 sequence. One major difference between the two vectors is the SIN setting—CS-Rh-MLV-E vector contains a 133-nucleotide deletion in the U3 region of the 3′ LTR that includes the TATA box and binding sites for transcription factors Sp1 and NF-κB, whereas SIN-18-Rh-MLV-E has a more extensive 400-nucleotide deletion in the U3 region of the 3′ LTR that has eliminated all of the transcriptional elements located upstream of the SP1 binding sites. The observation that SIN-18-Rh-MLV-E has two- to threefold-higher EGFP expression than CS-Rh-MLV-E supports the hypothesis of residual promoter interference in the CS-Rh-MLV-E vector.
We compared the efficiency of T-lymphoid-cell transduction by SRαLEGFP and our lentivirus-oncoretrovirus hybrid vector. Despite the high level of gene expression in T cells by SRαLEGFP, we reasoned that this retrovirus vector is undesirable for gene therapy of T cells for two reasons. First, SRαLEGFP virus titer is consistently 10- to 100-fold lower than that of the lentivirus (or lentivirus-oncoretrovirus hybrid) vector, making it more difficult to produce a large quantity for gene therapy purposes. Second, although SRαLEGFP is expressed efficiently in T cells, it requires a much higher MOI to achieve a low level of gene transduction in primary T-cell blasts compared to that required by CS-Rh-MLV-E (Fig. 6C). SIN-18-Rh-MLV-E expressed EGFP at a level comparable to that of SRαLEGFP in T lymphoid cells with relatively high transduction efficiency (Fig. 7). Thus, SIN-18-Rh-MLV-E may prove useful in T-lymphoid-cell gene therapy because of its relatively high virus titer, ability to infect T cells efficiently, and high level of gene expression in T cells.
The partial gag sequence has previously been shown to encode a packaging signal for MoMLV retrovirus that markedly increases the titer of in vitro-packaged retrovirus vectors. However, the MoMLV-packaging sequence should not affect HIV-1 packaging. In fact, we show that inclusion of this sequence in CS-MSV-E, CS-MLV-E, or CS-Rh-MLV-E does not significantly affect the virus titer when compared to the parental CSCG vector. Interestingly, we found that deletion of this extended packaging sequence of MoMLV resulted in an approximately twofold decrease in EGFP expression observed in the infected SUPT1 and CEMX174 cells. These results demonstrated a novel, unappreciated feature of this extended packaging sequence in gene expression. There are two possible explanations for the enhanced gene expression in these vectors. First, the extended packaging sequence stabilizes the mRNA transcript and therefore allows higher gene expression. Second, it may allow higher gene expression through a more efficient mRNA nuclear export. Recently, King et al. reported that incorporation of this extended packaging sequence into Rex-dependent expression vectors based on the human T-cell leukemia virus allows Rex-independent gene expression (14). Using fluorescent in situ hybridization combined with confocal microscopy, they showed that the 312-nucleotide element can replace Rex-mediated nuclear export and expression of transcripts containing HTLV-1 cis-acting repressive elements. Furthermore, it is interesting to note that introduction of this partial gag sequence (Δgag871–1612) downstream of the CMV promoter (CS-CMV-ΔgagE) or PGK promoter (SIN-18-PGK-ΔgagE) diminished gene expression when compared with the CMV (CSCG) and PGK (RRL-PGK-EGFP-SIN18) promoters alone, suggesting that the activity is dependent on being in the context of an MLV LTR.
In our present study, we found that a combination of the novel MoMLV-derived LTR (Rh-MLV) and the partial gag sequence of MoMLV allowed the strongest EGFP expression in the T-cell lines and activated primary T cells. This primate-derived murine retrovirus LTR, Rh-MLV LTR, is identified in the AMP-MCF retrovirus that was isolated from the serum of one rhesus monkey that developed rapidly progressive T-cell lymphomas (29). It is important to note that multiple copies of this AMP-MCF virus genome were only present in tumor DNA of one of the three animals that developed lymphoma and that tumor cells are of clonal origin with respect to the retroviral integration sites. These data are consistent with a pathogenic mechanism in which cell transformation and clonal tumor evolution arise from insertional mutagenesis of critical growth control genes by retrovirus integration. As with oncogenesis by weakly oncogenic retroviruses (those of birds, for example), cancer is induced by rare integration events resulting from a high level of chronic replication of the retroviruses. The use of such a primate-derived LTR, therefore, would not result in a new risk for the use of such vectors in clinical applications. Future experiments will be necessary to address other safety issues (such as the potential recombination between the endogenous retroviral sequences and the Δgag871–1612 sequence of MuLV).
Genetic modifications of hematopoietic progenitor cells (1, 3, 7, 20) could lead to expression of genes of interest in lymphoid cells, thus allowing potential treatment of dysfunctional T cells, which are often disrupted by an inherited or acquired immunodeficiency, such as HIV-1 disease (10). Therefore, it is desirable to obtain SIN HIV-1-based vectors that allow a high level of gene expression in T lymphocytes. CS-Rh-MLV-E and SIN-18-Rh-MLV-E show 5- and 10-fold-higher EGFP expression, respectively, than the CMV promoter (CSCG). In addition, a direct comparison of SIN-18-Rh-MLV-E with a PGK-containing vector, RRL-PGK-EGFP-SIN18, shows that there was a 6- to 10-fold-higher MFI of EGFP expression with SIN-18-Rh-MLV-E (Fig. 7 and Table 2), demonstrating that the combination of the Rh-MLV LTR and Δgag871–1612 sequence performs better than the housekeeping PGK gene promoter. In summary, we have generated lentivirus vectors that have combined useful features of HIV-1 and MoMLV. These should be useful in allowing a higher level of therapeutic gene expression in lymphoid cells in gene therapy.
ACKNOWLEDGMENTS
We thank I. M. Verma for providing CSCG and CS-G, D. Trono for RRL-PGK-EGFP-SIN18, and A. W. Nienhuis for 5-3BS plasmids. We thank Kouki Morizono for advice and technical assistance and Liz Duarte and Rosie Taweesup for manuscript preparation.
This work was supported by NIH grants AI 39975 and AI36555. S.K.P.K. is a Research Fellow of the National Cancer Institute of Canada supported with funds provided by the Terry Fox Run.
REFERENCES
- 1.Akkina R K, Rosenblatt J D, Campbell A G, Chen I S, Zack J A. Modeling human lymphoid precursor cell gene therapy in the SCID-hu mouse. Blood. 1994;84:1393–1398. [PubMed] [Google Scholar]
- 2.An D S, Wersto R P, Agricola B A, Metzger M E, Lu S, Amado R G, Chen I S Y, Donahue R E. Marking and gene expression by a lentiviral vector in transplanted human and non-human primate CD34+ cells. J Virol. 2000;74:1286–1295. doi: 10.1128/jvi.74.3.1286-1295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An D S, Koyanagi Y, Zhao J Q, Akkina R, Bristol G, Yamamoto N, Zack J A, Chen I S. High-efficiency transduction of human lymphoid progenitor cells and expression in differentiated T cells. J Virol. 1997;71:1397–1404. doi: 10.1128/jvi.71.2.1397-1404.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.An D S, Morizono K, Li Q X, Mao S H, Lu S, Chen I S. An inducible human immunodeficiency virus type 1 (HIV-1) vector which effectively suppresses HIV-1 replication. J Virol. 1999;73:7671–7677. doi: 10.1128/jvi.73.9.7671-7677.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender M A, Palmer T D, Gelinas R E, Miller A D. Evidence that the packaging signal of Moloney murine leukemia virus extends into the gag region. J Virol. 1987;61:1639–1646. doi: 10.1128/jvi.61.5.1639-1646.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blomer U, Naldini L, Kafri T, Trono D, Verma I M, Gage F H. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonyhadi M L, Moss K, Voytovich A, Auten J, Kalfoglou C, Plavec I, Forestell S, Su L, Bohnlein E, Kaneshima H. RevM10-expressing T cells derived in vivo from transduced human hematopoietic stem-progenitor cells inhibit human immunodeficiency virus replication. J Virol. 1997;71:4707–4716. doi: 10.1128/jvi.71.6.4707-4716.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Case S S, Price M A, Jordan C T, Yu X J, Wang L, Bauer G, Haas D L, Xu D, Stripecke R, Naldini L, Kohn D B, Crooks G M. Stable transduction of quiescent CD34(+)CD38(−) human hematopoietic cells by HIV-1-based lentiviral vectors. Proc Natl Acad Sci USA. 1999;96:2988–2993. doi: 10.1073/pnas.96.6.2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Donahue R E, Kessler S W, Bodine D, McDonagh K, Dunbar C, Goodman S, Agricola B, Byrne E, Raffeld M, Moen R. Helper virus induced T cell lymphoma in nonhuman primates after retroviral mediated gene transfer. J Exp Med. 1992;176:1125–1135. doi: 10.1084/jem.176.4.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Greenberg P D, Riddell S R. Deficient cellular immunity—finding and fixing the defects. Science. 1999;285:546–551. doi: 10.1126/science.285.5427.546. [DOI] [PubMed] [Google Scholar]
- 11.Hershfield M S. Adenosine deaminase deficiency: clinical expression, molecular basis, and therapy. Semin Hematol. 1998;35:291–298. [PubMed] [Google Scholar]
- 12.Kafri T, Blomer U, Peterson D A, Gage F H, Verma I M. Sustained expression of genes delivered directly into liver and muscle by lentiviral vectors. Nat Genet. 1997;17:314–317. doi: 10.1038/ng1197-314. [DOI] [PubMed] [Google Scholar]
- 13.Kim V N, Mitrophanous K, Kingsman S M, Kingsman A J. Minimal requirement for a lentivirus vector based on human immunodeficiency virus type 1. J Virol. 1998;72:811–816. doi: 10.1128/jvi.72.1.811-816.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King J A, Bridger J M, Gounari F, Lichter P, Schulz T F, Schirrmacher V, Khazaie K. The extended packaging sequence of MoMLV contains a constitutive mRNA nuclear export function. FEBS Lett. 1998;434:367–371. doi: 10.1016/s0014-5793(98)00948-x. [DOI] [PubMed] [Google Scholar]
- 15.Landau N R, Littman D R. Packaging system for rapid production of murine leukemia virus vectors with variable tropism. J Virol. 1992;66:5110–5113. doi: 10.1128/jvi.66.8.5110-5113.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyaard L, Schuitemaker H, Miedema F. T-cell dysfunction in HIV infection: anergy due to defective antigen-presenting cell function? Immunol Today. 1993;14:161–164. doi: 10.1016/0167-5699(93)90279-T. [DOI] [PubMed] [Google Scholar]
- 17.Miller A D, Rosman G J. Improved retroviral vectors for gene transfer and expression. BioTechniques. 1989;7:980–986. , 989. [PMC free article] [PubMed] [Google Scholar]
- 18.Miyoshi H, Takahashi M, Gage F H, Verma I M. Stable and efficient gene transfer into the retina using an HIV-based lentiviral vector. Proc Natl Acad Sci USA. 1997;94:10319–10323. doi: 10.1073/pnas.94.19.10319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miyoshi H, Blomer U, Takahashi M, Gage F H, Verma I M. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyoshi H, Smith K A, Mosier D E, Verma I M, Torbett B E. Transduction of human CD34+ cells that mediate long-term engraftment of NOD/SCID mice by HIV vectors. Science. 1999;283:682–686. doi: 10.1126/science.283.5402.682. [DOI] [PubMed] [Google Scholar]
- 21.Mochizuki H, Schwartz J P, Tanaka K, Brady R O, Reiser J. High-titer human immunodeficiency virus type 1-based vector systems for gene delivery into nondividing cells. J Virol. 1998;72:8873–8883. doi: 10.1128/jvi.72.11.8873-8883.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naldini L, Blomer U, Gage F H, Trono D, Verma I M. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naldini L, Blomer U, Gallay P, Ory D, Mulligan R, Gage F H, Verma I M, Trono D. In vivo gene delivery and stable transduction of nondividing cells by a lentiviral vector. Science. 1996;272:263–267. doi: 10.1126/science.272.5259.263. [DOI] [PubMed] [Google Scholar]
- 24.Purcell D F J, Broscius C M, Vanin E F, Buckler C E, Nienhuis A W, Martin M A. An array of murine leukemia virus-related elements is transmitted and expressed in a primate recipient of retroviral gene transfer. J Virol. 1996;70:887–897. doi: 10.1128/jvi.70.2.887-897.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roe T, Reynolds T C, Yu G, Brown P O. Integration of murine leukemia virus DNA depends on mitosis. EMBO J. 1993;12:2099–2108. doi: 10.1002/j.1460-2075.1993.tb05858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell S M, Tayebi N, Nakajima H, Riedy M C, Roberts J L, Aman M J, Migone T S, Noguchi M, Markert M L, Buckley R H. Mutation of Jak3 in a patient with SCID: essential role of Jak3 in lymphoid development. Science. 1995;270:797–800. doi: 10.1126/science.270.5237.797. [DOI] [PubMed] [Google Scholar]
- 27.Sutton R E, Wu H T, Rigg R, Bohnlein E, Brown P O. Human immunodeficiency virus type 1 vectors efficiently transduce human hematopoietic stem cells. J Virol. 1998;72:5781–5788. doi: 10.1128/jvi.72.7.5781-5788.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tsichlis P N, Lazo P A. Virus-host interactions and the pathogenesis of murine and human oncogenic retroviruses. Curr Top Microbiol Immunol. 1991;171:95–171. doi: 10.1007/978-3-642-76524-7_5. [DOI] [PubMed] [Google Scholar]
- 29.Vanin E F, Kaloss M, Broscius C, Nienhuis A W. Characterization of replication-competent retroviruses from nonhuman primates with virus-induced T-cell lymphomas and observations regarding the mechanism of oncogenesis. J Virol. 1994;68:4241–4250. doi: 10.1128/jvi.68.7.4241-4250.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zufferey R, Nagy D, Mandel R J, Naldini L, Trono D. Multiply attenuated lentiviral vector achieves efficient gene delivery in vivo. Nat Biotechnol. 1997;15:871–875. doi: 10.1038/nbt0997-871. [DOI] [PubMed] [Google Scholar]
- 31.Zufferey R, Dull T, Mandel R J, Bukovsky A, Quiroz D, Naldini L, Trono D. Self-inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]