Abstract
We have previously reported the construction and characterization of vindH1, an inducible recombinant in which expression of the vaccinia virus H1 phosphatase is regulated experimentally by IPTG (isopropyl-β-d-thiogalactopyranoside) (35). In the absence of H1 expression, the transcriptional competence and infectivity of nascent virions are severely compromised. We have sought to identify H1 substrates by characterizing proteins that are hyperphosphorylated in H1-deficient virions. Here, we demonstrate that the A14 protein, a component of the virion membrane, is indeed an H1 phosphatase substrate in vivo and in vitro. A14 is hyperphosphorylated on serine residues in the absence of H1 expression. To enable a genetic analysis of A14's function during the viral life cycle, we have adopted the regulatory components of the tetracycline (TET) operon and created new reagents for the construction of TET-inducible vaccinia virus recombinants. In the context of a virus expressing the TET repressor (tetR), insertion of the TET operator between the transcriptional and translational start sites of a late viral gene enables its expression to be tightly regulated by TET. We constructed a TET-inducible recombinant for the A14 gene, vindA14. In the absence of TET, vindA14 fails to form plaques and the 24-h yield of infectious progeny is reduced by 3 orders of magnitude. The infection arrests early during viral morphogenesis, with the accumulation of large numbers of vesicles and the appearance of “empty” crescents that appear to adhere only loosely to virosomes. This phenotype corresponds closely to that observed for an IPTG-inducible A14 recombinant whose construction and characterization were reported while our work was ongoing (47). The consistency in the phenotypes seen for the IPTG- and TET-inducible recombinants confirms the efficacy of the TET-inducible system and reinforces the value of having a second, independent system available for generating inducible recombinants.
Vaccinia virus is a complex DNA virus characterized by a high degree of genetic and physical autonomy from the host cell. Among the functions of the 200 gene products encoded by the 192-kb genome are those required for the expression of three temporal classes of genes, the replication of viral DNA, and the morphogenesis of nascent virions (27). The last is a complex process which remains poorly understood but is the subject of intense study. Electron microscopy, in conjunction with pharmacological intervention and the study of conditionally lethal mutants, has been useful in generating models of virion assembly (7, 9, 12, 20, 24, 25, 28, 31, 41, 44, 47, 49, 53, 55, 58, 59, 62, 63). Within the cytoplasm, morphogenesis is concentrated around areas of increased electron density which have been shown to contain viral DNA and the proteins to be encapsidated. Membrane crescents, which appear to elongate and curve until they enclose the electron-dense material within oval immature virions (IV), are the first hallmark of morphogenesis. The origin of these membranes is still a matter of controversy. De novo biogenesis of these membranes to form an IV delimited by a single lipid bilayer was an early and provocative model (10). Recent measurements of the membrane surrounding the IV are also consistent with the single-bilayer model (22), although this work did not address the origin of the membrane in question. Other investigators have presented data suggesting that the virion membrane derives from the intermediate compartment (49), a transitional zone of tubulovesicular components that reflects anterograde and retrograde transport between the endoplasmic reticulum and the Golgi apparatus. Membrane crescents, derived from tubulovesicular components of the intermediate compartment, would therefore be comprised of two membranes that have become so tightly apposed that the lumenal space is essentially absent. Clarification of which features of these distinct models are correct must be a central goal for researchers in the field.
IV formation is followed by a maturation process that involves morphological changes and a series of proteolytic processing events which generate infectious virions known as intracellular mature virons (IMV). A number of viral proteins have been shown to play a role in IV and IMV morphogenesis. Of great interest to our laboratory is the demonstration that nonpermissive infections performed with temperature-sensitive mutants carrying lesions within the F10 protein kinase arrest at the earliest stage of morphogenesis (53, 55). No crescents or immature or mature particles are seen during these infections, and no electron-dense virosomes appear. These data strongly suggest that protein phosphorylation plays an essential role in the membrane remodeling that initiates viral morphogenesis. Recent experiments in our laboratory have provided evidence supporting the characterization of F10 as a dual-specificity kinase that can phosphorylate serine, threonine, and tyrosine residues (13).
The importance of protein phosphorylation as a regulator of the viral life cycle is underscored by the fact that vaccinia virus encodes another protein kinase (B1) (2, 34, 42, 52) as well as a dual-specificity protein phosphatase, the product of the H1 gene. Using an inducible viral recombinant in which expression of the H1 phosphatase can be regulated experimentally (vindH1), we were able to demonstrate that H1 synthesis is not required for virion morphogenesis but is necessary to ensure the infectivity and transcriptional competence of nascent virions (35). Making the assumption that it was the phosphatase activity of H1 that was essential, we concentrated on identifying which virion proteins were H1 substrates. To do so, we examined 32P-labeled wild-type (wt) and H1-deficient (H1−) virions in order to determine which proteins were hyperphosphorylated in the absence of H1. Three proteins with apparent molecular weights of 25,000 (25K), 16K, and 11K were identified. We have previously shown that the 11K protein is the DNA-binding protein encoded by the F18 (F17 in the Copenhagen strain) gene and that the 25K protein is the product of the A17 gene (13, 35). The identification of the 16K phosphoprotein as the product of the A14 gene and its subsequent characterization are the subjects of this paper. As part of our analysis of the structure and function of the A14 protein, we also developed an alternative inducible system in which expression of viral genes can be regulated by the inclusion of tetracycline (TET) in the culture medium.
MATERIALS AND METHODS
Materials.
Restriction endonucleases, polynucleotide kinase, T4 DNA ligase, calf intestinal phosphatase, pancreatic RNase, S1 endonuclease, Taq polymerase, and DNA molecular weight standards were purchased from New England Biolabs, Inc. (Beverly, Mass.), or Boehringer Mannheim Biochemicals (Indianapolis, Ind.) and were used according to the instructions provided by the manufacturers. 32P-labeled nucleoside triphosphates, [32P]orthophosphate, and 35S-labeled methionine were acquired from Dupont/New England Nuclear Corp. (Boston, Mass.). Thin-layer cellulose F chromatography plates were purchased from EM Separations, Inc. (Gibbstown, N.J.). Constant boiling HCl was purchased from Pierce (Rockford, Ill.).
Cells and viruses.
Monolayer cultures of BSC40 primate cells, mouse L cells, or human thymidine kinase-deficient (TK−) cells were maintained in Dulbecco modified Eagle medium (DMEM; GIBCO-BRL, Gaithersburg, Md.) containing 5% fetal calf serum (FCS). wt vaccinia virus (WR strain), vindH1 (35), virus expressing the TET repressor (vtetR), and vindA14 (this report) were amplified in monolayer cultures of BSC40 cells or suspension cultures of L cells. Viral stocks were prepared from cytoplasmic lysates of infected cells by ultracentrifugation through 36% sucrose; titrations were performed on BSC40 cells. In some cases, virions were further purified by banding on 25 to 40% sucrose gradients (35). For vindA14, permissive infections were performed in the presence of TET (1 μg/ml). Where indicated, cytosine arabinoside (araC) was added to infected cultures a final concentration of 20 μg/ml.
Construction of vtetR.
The coding sequences of tetR were amplified from an appropriate plasmid using PCR and the following primers: TR1, 5′ GGGGATCCATGTCTAGATTAGATAA 3′, and TR2, 5′ GGGGATCCTTAAGACCCACTTTCACATT 3′. Primer TR1 introduced a BamHI site (bold) at the initiation codon (underlined), and primer TR2 introduced a BamHI site (bold) at the termination codon (underlined). The BamHI-cleaved PCR product was inserted into pGS53 vector DNA (37) which had been previously digested with BamHI and treated with calf intestinal alkaline phosphatase. The resultant plasmid places the tetR open reading frame (ORF) under the regulation of a constitutive vaccinia virus promoter; the flanking sequences allow insertion of this cassette into the nonessential TK gene of the viral genome via homologous recombination. BSC40 cells were infected with wt virus (multiplicity of infection [MOI], 0.03) and transfected with the pGS53/tetR plasmid. After 2 days, cells were harvested and TK− virus was purified by two rounds of plaque purification on human TK− cells in the presence of bromodeoxyuridine (25 μg/ml). The presence of the tetR gene within these viruses was confirmed by dot blot hybridization and PCR.
Construction of vindA14.
Overlap PCR was used to generate a 770-bp DNA fragment which included the 270-bp A14 ORF as well as 102 bp of downstream (towards A13) and 384 bp of upstream (towards A15) sequence. This construct also inserted the 19-bp operator sequence (O2) from the TET operon (5′ TCCCTATCAGTGATAGAGA 3′) (16, 17, 19) between the transcriptional and translational start sites of the A14 gene. The primers used to generate this fragment were 1, 5′ GGGGATCCATTGATCAAGTATGCAAT; B, 5′ CCTATCAGTGATAGAGAATGGACATGATGCTTA 3′; C, 5′ TCTATCACTGATAGGGATATTTAACTAATAAAAAT 3′; and 4, 5′ GGGGATCCTTATGCCAGATAATAGAT 3′. The template was viral genomic DNA. Primers 1 and 4 insert BamHI sites (bold) at the termini of the final PCR product; primers B and C insert the operator sequences (italicized) between the transcriptional and translational start sites (underlined) of the A14 ORF. In the first round of PCR, primers 1 plus B and C plus 4 were used to generate fragments of 385 and 400 bp that overlapped by 15 bp. The products were purified on glass beads (54); aliquots of each were taken and together served as the template for a second round of amplification that was performed using primers 1 and 4. The final 770-bp product was digested with BamHI and ligated to pUC/NEO DNA (31, 35) which had been digested with BamHI and treated with calf intestinal alkaline phosphatase.
Insertion of the TET operator-regulated A14 gene into the endogenous A14 locus was accomplished by transient dominant selection (14, 31, 35). Briefly, cells were infected with vtetR virus (MOI, 0.03) and transfected at 3 h postinfection (hpi) with pUC/NEO-opA14 DNA. From this point on, TET (1 μg/ml) was included in the culture medium at all times. At approximately 20 hpi, the medium was replaced with fresh medium containing G418 (2 to 3 mg/ml) and the infection was allowed to proceed for 2 to 3 days. Cells were then harvested, and G418-resistant virus was isolated by performing two rounds of plaque purification in the presence of G418. These viruses were predicted to have a partial duplication of the A14 locus and to contain endogenous and exogenous (operator-containing) copies of the gene flanking the inserted pUC/NEO sequences. Confirmation of the presence of both A14 alleles within the plaques was obtained by performing PCR with primers that flanked the transcriptional start site of the A14 gene. Products of 188 and 213 bp were amplified from the endogenous and exogenous alleles, respectively. These viruses were then subjected to two rounds of plaque purification in the absence of G418 in order to allow the duplicated region to resolve via homologous recombination. The same PCR was performed to distinguish those viruses that had resolved to the wt genotype from those that had resolved to contain only the operator-regulated allele. A plaque of the latter type was chosen for expansion and further study; this recombinant virus was named vindA14.
Construction of a TET-inducible β-Gal plasmid.
The goal was to construct a plasmid in which the β-galactosidase (β-Gal) gene was under the regulation of a late poxvirus promoter and the TET operator/repressor. A 1.3-kb fragment of plasmid p1246 (kindly provided by David Pickup, Duke University) containing the cowpox virus ATI promoter (40) was excised with EcoRI and cloned into pUC19. Clones in which the promoter was oriented towards the ClaI and BamHI sites in the pUC polylinker were chosen for further manipulation. The plasmid was digested with ClaI and BamHI and ligated to an oligonucleotide duplex which contained the sequence of the TET operator followed by an initiator methionine codon (underlined). This duplex was obtained by annealing the following two oligonucleotides: 5′ CGATCCCTATCAGTGATAGAGAATG 3′ (top) and 5′ GATCCATTCTCTATCACTGATAGGGAT 3′ (bottom). The annealed duplex contained the TET operator (bold) and a translational start site (underlined) flanked by 5′ single-strand extensions complementary to the extensions on the ClaI- and BamHI-cleaved plasmid DNA (italicized). Transformants containing the pUC/promoter/operator plasmid were identified, and the plasmid DNA was purified, cleaved with BamHI, and treated with calf intestinal alkaline phosphatase. The β-Gal gene was excised from plasmid pSC11 (8) as a 3.1-kb BamHI fragment and ligated into the BamHI-cleaved pUC/promoter/operator plasmid. The final plasmid was designated pLate/op/β-gal.
Construction of pInt/β-gal.
This plasmid was originally constructed in order to place the β-Gal ORF under the regulation of an intermediate vaccinia virus promoter. A 150-bp fragment containing the promoter of the vaccinia virus G8 gene was amplified by PCR using the HindIII G fragment of the viral genome as the template and the following primers: 1, 5′ GGTCTAGACAATTGTTTAGCTTTGG 3′, and 2, 5′ GGGGATCCATTTTAAATTTTTGTA 3′. These primers placed an XbaI site at the 5′ end of the promoter fragment (bold) and a BamHI site at its 3′ end (bold). The fragment was cleaved with XbaI and BamHI and inserted into pUC 19 DNA that had been similarly cleaved; Escherichia coli transformants carrying the pUC/pG8 plasmid were isolated. pUC/pG8 DNA was then purified, treated with BamHI and calf intestinal alkaline phosphatase, and ligated to a 3.1-kb BamHI fragment encoding the β-Gal gene that had been excised from plasmid pSC11 (8). Plasmids in which the β-Gal gene was in the proper orientation for expression from the G8 promoter were selected for further use and called pInt/β-gal.
Assay of β-Gal activity in infected cell lysates.
Thirty-five-millimeter-diameter dishes of BSC40 cells were infected with vtetR (MOI, 15) and transfected with 4 μg of the indicated plasmid. At 24 hpi, cells were harvested, washed once with phosphate-buffered saline (PBS), and resuspended in 100 μl of 10 mM Tris (7.8)–1 mM EDTA. Cell suspensions were subjected to three cycles of freezing and thawing and then clarified by centrifugation at 12,000 × g for 5 min at 4°C. One or 5 μl of these extracts was mixed with 400 μl of Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4, 40 mM β-mercaptoethanol) and incubated for 5 min, at which time 100 μl of Z buffer containing 4 mg of ONPG (o-nitrophenyl-β-d-galactopyranoside) per ml was added (1). The reaction mixtures were mixed, and the incubation was allowed to proceed for 30 min at room temperature before being stopped by the addition of 500 μl of 1 M sodium carbonate. The reaction mixtures were then analyzed by measuring their optical densities at 420 nm in a Beckman spectrophotometer.
Preparation of anti-A14 and anti-A17 antisera. (i) Construction of pATH:A14.
The A14 ORF was amplified by PCR using genomic DNA as the template. The upstream primer (5′ GGGAATTCCATATGGACATGATGCTTAT 3′) introduced EcoRI (italicized) and NdeI (bold) sites upstream of and overlapping the initiating ATG (underlined), respectively; the downstream primer (5′ GGGGATCCTTAGTTCATGGAAATAT 3′) introduced a BamHI (bold) site downstream of the termination codon (underlined). The 292-bp PCR product was digested with EcoRI and BamHI and inserted into pATH11 DNA (32) that had been previously digested with EcoRI and BamHI. The resultant plasmid fused the A14 ORF downstream of, and in frame with, the E. coli trpE gene.
(ii) Construction of pATH:A17.
The A17 ORF was amplified by PCR using genomic DNA as the template. The upstream primer (5′ CCCGCGTCTAGAATGAGTTATTTAAGATA 3′) introduced an XbaI site (italicized) upstream of the initiating ATG (underlined); the downstream primer (5′ GGGGGATCCTTAATAATCGTCAGTAT 3′) introduced a BamHI (italicized) site downstream of the termination codon (underlined). The 630-bp PCR product was digested with XbaI and BamHI and inserted into pATH23 DNA (32) that had been previously digested with XbaI and BamHI. The resultant plasmid fused the A17 ORF downstream of, and in frame with, the E. coli trpE gene.
(iii) Expression of trpE:A14 and trpE:A17 antigens.
E. coli transformants containing pATH:A14 directed the inducible synthesis of a 43-kDa fusion protein containing 34 kDa of E. coli trpE and 10 kDa of A14; pATH:A17 transformants expressed a 61-kDa fusion protein containing the E. coli trpE protein and the entire 25-kDa A17 protein. These fusion proteins were excised from sodium dodecyl sulfate (SDS)-polyacrylamide gels and used to immunize rabbits. The specificities of the resultant polyclonal antisera were confirmed by immunoblotting and immunoprecipitation assays.
Immunodetection analyses.
Ed Niles (State University of New York, Buffalo) kindly provided rabbit serum directed against the D8 protein. Rabbit anti-I3 (43), anti-L4 and anti-F18 (35), and anti-A14 and anti-A17 (this report) sera were developed in our laboratory. For immunoblot analysis, phosphatase inhibitors (1 mM sodium orthovanadate, 1 mM sodium fluoride, and 40 mM β-glycerol phosphate) were often included during the preparation of cell and virion extracts that were then subjected to fractionation by SDS-polyacrylamide gel electrophoresis (PAGE) and electrophoretic transfer to nitrocellulose (Schleicher and Schuell, Keene, N.H.) or Immobilon P (Millipore Corp., Bedford, Mass.) membranes as described previously (13). Primary sera are described above. Secondary antibodies (horseradish peroxidase-conjugated goat anti-rabbit antibodies) were obtained from Bio-Rad (Richmond, Calif.) and used according the manufacturer's instructions. Blots were developed colorimetrically or by enhanced chemiluminescence (DuPont NEN, Boston, Mass., or Pierce). Immunoprecipitation analyses were performed as described previously (13).
Preparation of protein sample for amino acid microsequencing.
Proteins were resolved on SDS-polyacrylamide gels and transferred to polyvinylidene difluoride (PVDF) membrane in CAPS transfer buffer (10 mM CAPS {3-[(cyclohexylamino)-1-propane-sulfonic acid} in 10% methanol, pH 11.3). Filters were stained with 0.1% amido black to reveal the relevant band, which was excised and submitted to the Protein Sequencing Facility of the Rockefeller University for microsequencing.
Metabolic labeling. (i) Metabolic labeling with 32PPi.
At the times indicated in the figures, infected monolayers were incubated with phosphate-free DMEM (ICN-Flow, Costa Mesa, Calif.) supplemented with l-glutamine, 5% phosphate-free FCS (prepared by dialysis against Tris-buffered saline [25 mM Tris-HCl, pH 7.4, 136 mM NaCl, 2.7 mM KCl]), and 50 μCi of [32P]orthophosphate per ml. Labeling was usually performed from 3 to 17 hpi. Cells were harvested and rinsed once with PBS; lysates were then prepared and analyzed by immunoprecipitation.
(ii) Metabolic labeling with [35S]methionine.
At the times postinfection indicated in the figures, cultures were rinsed with methionine-free DMEM (ICN-Flow) supplemented with l-glutamine and then fed with methionine-free DMEM supplemented with l-glutamine and 100 μCi of [35S]methionine (EXPRESS label; NEN-Dupont) per ml. The durations of the labeling reactions are indicated in the text or figure legends.
Preparation of 32P-labeled wt and H1− virions.
Confluent monolayers of BSC40 cells were infected with either wt virus or vindH1 at an MOI of 1. After adsorption of the inoculum for 1 h, cultures were maintained in the absence of IPTG in phosphate-free DMEM supplemented with l-glutamine and 32PPi (50 μCi/ml). Cells were harvested at 24 hpi, and 32P-labeled virions were purified by ultracentrifugation through sucrose cushions and sucrose gradients (35).
One-dimensional phosphoamino acid analysis.
The one-dimensional phosphoamino acid analysis procedure followed was essentially that described by the manufacturers of the Hunter Thin Layer Electrophoresis System, model HTLE-7000. 32P-labeled A14 was immunoprecipitated from metabolically labeled extracts. The immunoprecipitates were fractionated by SDS-PAGE and transferred to Immobilon P. The filter was exposed for autoradiography, and the film and filter were aligned to permit excision of the radiolabeled band from the filter. The protein was hydrolyzed in situ: the filter was rinsed in distilled water and then incubated at 110°C in 6 N constant boil HCI for 1 h. The liberated hydrolysate was collected by trichloroacetic acid precipitation. Samples were mixed with phosphoamino acid markers (20 nmol each) and spotted onto thin-layer cellulose F chromatography plates. Electrophoresis was performed in pyridine-glacial acetic acid-water (1:10:189) at 1,800 V for 25 min. The plates were dried, sprayed with ninhydrin, developed at 65°C, and exposed for autoradiography.
S1 nuclease mapping.
A 720-bp fragment containing the A14 ORF flanked by the TET operator, as well as upstream and downstream sequences, was amplified by PCR as described above under “Construction of vindA14.” The fragment was cleaved with AciI, treated with calf intestinal alkaline phosphatase, and terminally labeled with [γ-32P]ATP and polynucleotide kinase. The fragment was then cleaved with BamHI, and the resultant 613-bp fragment containing a single 5′ radiolabel was purified on glass beads and used as the probe for S1 nuclease analysis. Total RNA was prepared from BSC40 cells (RNeasy midi kit; Qiagen, Valencia, Calif.) 8 h after they were infected with vtetR or vindA14 (MOI, 10) in the presence or absence of TET (1 μg/ml). Purified RNA (10 μg) was mixed with 40 ng of probe (22,000 cpm) in a 30-μl reaction mixture containing 40 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 400 mM NaCl, and 1 mM EDTA. Samples were denatured by incubation at 65°C for 10 min and then allowed to anneal for 3 h at 37°C. Samples were then diluted 10-fold with a cocktail containing 280 mM NaCl, 30 mM NaAc, 4.5 mM ZnSO4, 5% glycerol, and 400 U of S1 nuclease per ml. The digestion was allowed to proceed for 30 min at 37°C. Samples were subjected to organic extraction and ethanol precipitation and were then analyzed by electrophoresis on a 6% denaturing acrylamide gel followed by autoradiography.
Electron microscopy. (i) Transmission electron microscopy.
Cells were prepared for electron microscopy as previously described (31, 53). Briefly, 60-mm-diameter dishes of BSC40 cells were infected with vindA14 in the absence or presence of TET (1 μg/ml) as indicated in the figures. At 17 hpi, the medium was removed and the cells were rinsed with PBS and then fixed in situ with 2% glutaraldehyde. Cells were then scraped from the dish, collected by sedimentation, and subjected to fixation with osmium tetroxide, dehydration, and embedding as previously described (53).
(ii) Immunoelectron microscopy.
Cells were infected as described above and then fixed in situ with 4% paraformaldehyde–0.1% glutaraldehyde in PBS (see Fig. 4) or 0.1 M sodium cacodylate (pH 7.4) (see Fig. 9). Cells were then scraped from the dish, collected by sedimentation, overlaid with additional fixative, and subjected to graded dehydration and embedding in LR White resin, which was cured at 50 to 55°C. For Fig. 9, pellets were embedded in 4% agarose and then washed in 5% sucrose–0.1 M sodium cacodylate (pH 7.4) prior to dehydration. Sections were collected on nickel grids. The labeling procedure was as follows: sections were blocked with 1× TBS (137 mM NaCl, 2.7 mM KCl, 25 mM Tris [pH 7.4]) containing 5% FCS (see Fig. 4) or 5% bovine serum albumin (see Fig. 9) for 15 to 30 min prior to being incubated for 40 to 60 min with the clarified primary antiserum in 1× TBS supplemented with 1% of the blocking agent. Grids were then washed in 1× TBS four times for 5 min each time and then incubated for 40 to 60 min with 10-nm-diameter gold bead-conjugated goat anti-rabbit immunoglobulin G (Goldmark Biologicals, Phillipsburg, N.J. [for the results shown in Fig. 4], or Amersham Scientific [for the results shown in Fig. 9]) in 1× TBS supplemented with 1% of the blocking agent. Grids were then washed six times for 5 min each time in 1× TBS, fixed for 5 min in 1% glutaraldehyde, rinsed with 1× TBS [see Fig. 4], rinsed with distilled water, and then poststained with 2% uranyl acetate (aqueous) for 5 to 10 min. The grids were examined on a model JEOL 100XC-II or Hitachi H-600 microscope.
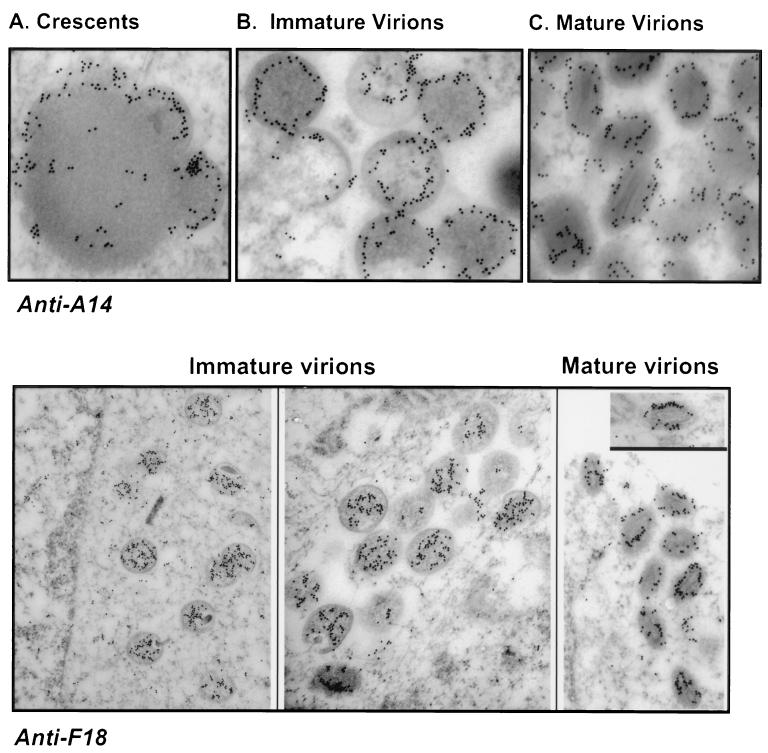
FIG. 4.
Immunoelectron microscopic localization of the A14 and F18 proteins within wt vaccinia virions. Cells were infected with wt virus (MOI, 2); at 16 hpi, cells were fixed in situ and processed as described in Materials and Methods. Sections were labeled with anti-A14 or anti-F18 serum and a secondary serum conjugated to 10-nm-diameter gold particles. (A to C) A14 is immunolocalized within the membranes of emerging crescents, immature virions, and mature virions. Magnifications, ×59,000 (A), ×56,000 (B), and ×60,000 (C). (Lower panels) F18 is immunolocalized within the interiors of immature virions. Within mature virions, it appears to localize in a ring that outlines the periphery of the lucent core. Magnifications, ×23,000 (left), ×35,000 (middle), and ×34,000 (right).
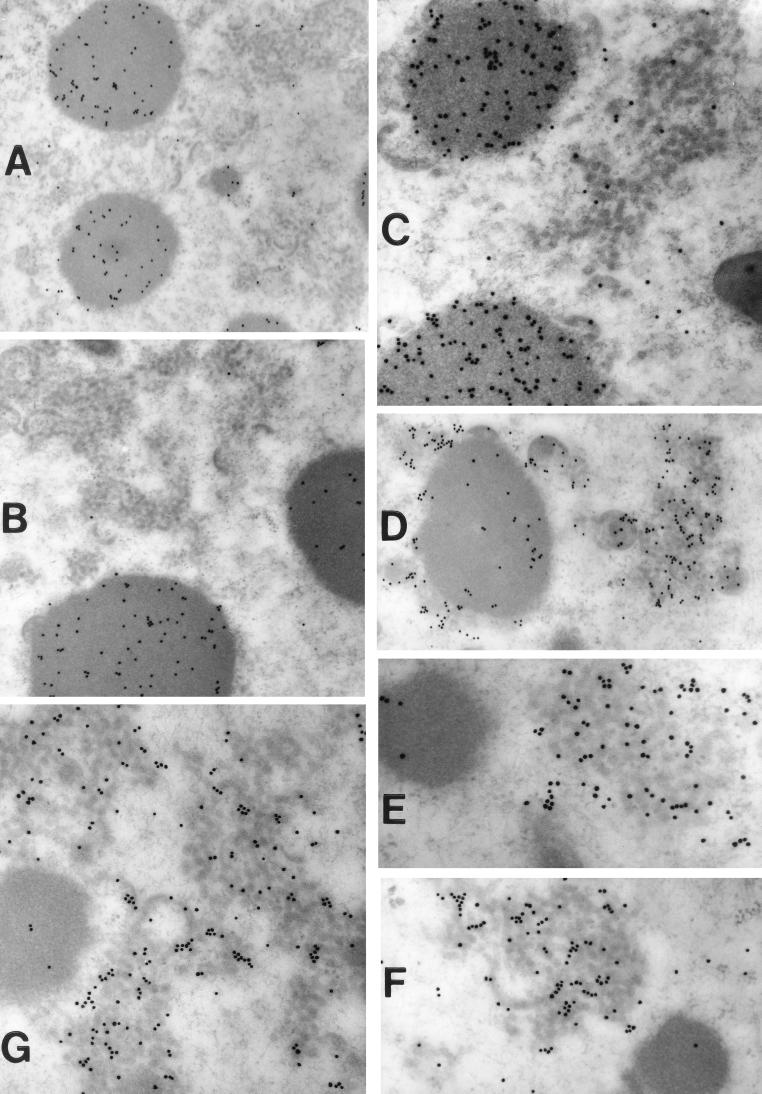
FIG. 9.
Immunoelectron localization of the L4 and F18 DNA-binding proteins and the D8 and A17 membrane proteins in cells infected nonpermissively with vindA14. Cells infected with vindA14 under nonpermissive conditions were processed for postembedding immunolectron microscopy as described in Materials and Methods. Cells were labeled with the following primary antisera: anti-L4 serum (A and B), anti-F18 serum (C), anti-A17 serum (D and E), and anti-D8 serum (F and G). In all cases, the secondary antiserum was conjugated to 10-nm-diameter gold particles. Prominent labeling of the virosomes is seen in panels A to C; in contrast, panels D to G show prominent labeling of the vesicle clusters. Magnifications: ×28,000 (A), ×34,000 (B and D), ×57,000 (C, F, and G), and ×68,000 (E).
Preparation of the figures.
Original data were scanned on a SAPHIR scanner (Linotype-Hell Co., Hauppauge, N.Y.) using Adobe Photoshop software (Adobe Systems, Inc., San Jose, Calif.). Labeling of the figures was performed with Canvas software (Deneba Systems, Inc., Miami, Fla.); graphs were prepared using SigmaPlot software (SPSS, Chicago, Ill.). Figures 8 and 9 were prepared photographically rather than digitally.
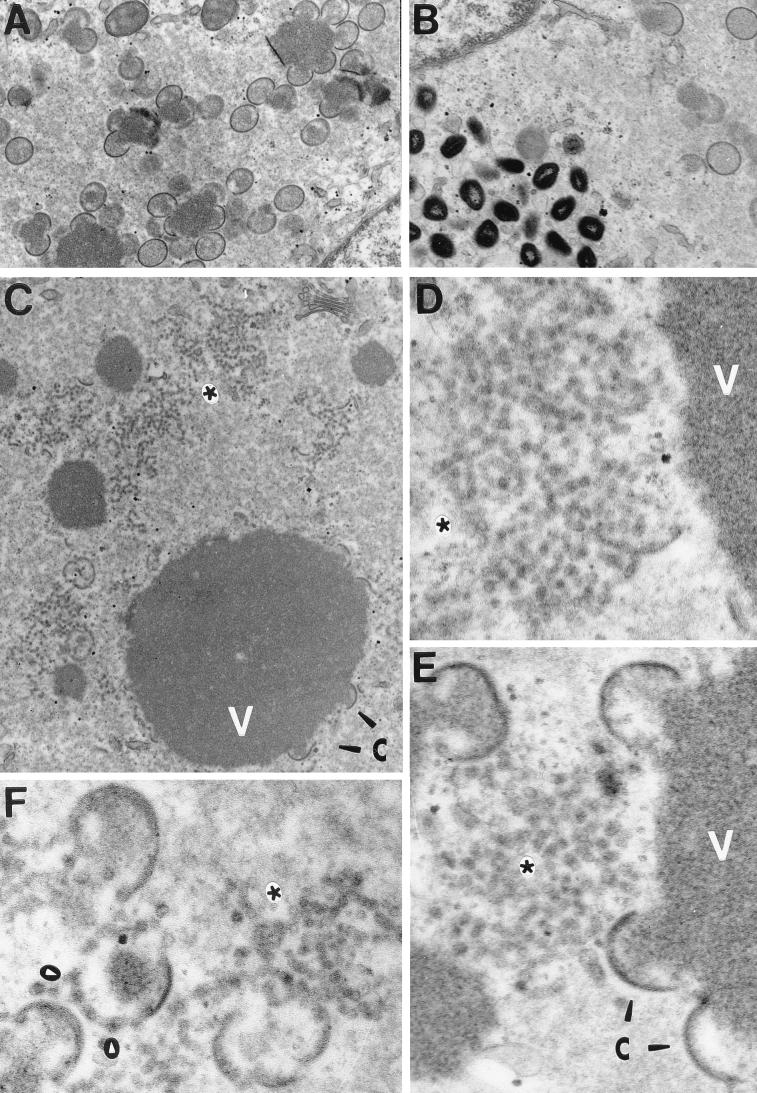
FIG. 8.
Electron microscopic examination reveals a profound morphogenesis defect in cells infected nonpermissively with vindA14. Cells were infected with vindA14 in the presence (A and B) or absence (C to F) of TET for 17 h, at which time they were fixed in situ and processed for conventional transmission electron microscopy. During permissive infections (A and B), all the normal intermediates of viral morphogenesis are seen, including virosomes with adjacent crescents, immature virions, and mature virions. During nonpermissive infections, the profile is strikingly different. Virosomes (V) are abundant, and abnormal crescents are seen at their periphery (arrowheads labeled C in panels C and E). These crescents appear empty and detached from the virosomal contents. The most striking feature of these nonpermissively infected cells is the accumulation of large groups of vesicles (asterisks). In some cases, discontinuous crescents that appear to consist of closely abutted vesicles are also seen (white arrowheads in panel F). Magnifications: ×21,000 (A), ×26,000 (B), ×25,000 (C), ×83,000 (D and E), and ×72,000 (F).
RESULTS
The 16-kDa protein hyperphosphorylated in H1-deficient virions is the product of the A14 gene.
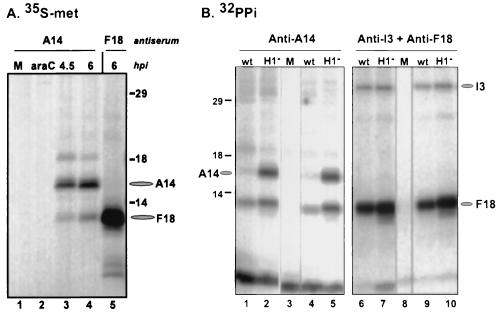
In order to identify substrates of the H1 phosphatase among the proteins encapsidated in vaccinia virions, we prepared 32P-labeled virions from cells infected with wt virus or with vindH1 in the absence of IPTG. As shown in Fig. 1A, three proteins of 11, 16, and 25 kDa were hyperphosphorylated in H1-deficient virions. When these virions were treated with NP-40 plus dithiothreitol (DTT) in order to prepare membrane and core fractions (13, 35), the 16-kDa phosphoprotein was released into the solubilized membrane fraction (not shown). Visualization of the membrane fraction by SDS-PAGE and silver staining indicated that the 16-kDa protein appeared as a doublet in H1-deficient virions and that this doublet was resolved to a single band upon treatment of permeabilized virions with exogenous, recombinant H1 phosphatase (Fig. 1B) but not upon treatment with a catalytically inert mutant of H1 (C110S; not shown) (35). These data confirmed that the 16-kDa phosphoprotein was indeed an H1 substrate.
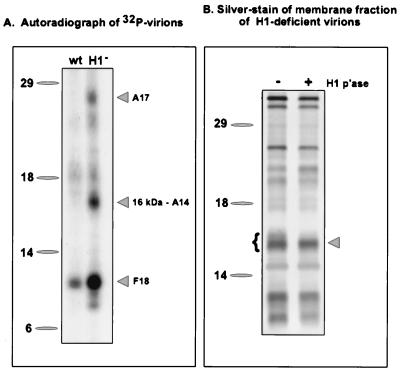
FIG. 1.
Identification of the A14 protein as a 16-kDa component of the virion membrane that is hyperphosphorylated in H1-deficient virions. (A) Sucrose-banded 32P-labeled virions were prepared from cells infected with wt virus or vindH1 in the absence of IPTG. Equivalent quantities of virions were analyzed by electrophoresis on an SDS–17% polyacrylamide gel and visualized by autoradiography. Three hyperphosphorylated proteins of 25, 16, and 11 kDa were seen in H1-deficient virions. The identification of the 16-kDa protein as the product of the A14 gene is reported in this study. For both panels, the electrophoretic migrations of protein standards are shown at the left, with their molecular masses indicated in kilodaltons. (B) H1-deficient virions (5 μg) were permeabilized with NP-40 plus DTT, and the membrane fraction was prepared. Half of the material was analyzed directly (lane −), and half was treated with recombinant H1 phosphatase (p'ase) (35) for 3 h at 37°C (lane +). The samples were then fractionated on an SDS–17% polyacrylamide gel and visualized by silver staining. The brace at the left indicates the multiple species of the 16-kDa protein seen in H1-deficient virions; these bands are converted to the most rapidly migrating form upon treatment with the H1 phosphatase. This dephosphorylated species was subjected to microsequencing analysis and found to be the product of the A14 gene.
H1-deficient virions were permeabilized and treated with recombinant H1, and the solubilized membrane fraction was resolved on an SDS–17% polyacrylamide gel which was then transferred to a PVDF membrane. Proteins were visualized by amido black staining; a band representing the 16-kDa species was excised from the filter and submitted for N-terminal amino acid sequence analysis. The peptide sequence obtained (MDMMLMIGNYFSGVLPA) was compared to sequences in the available databases, and 16 of 17 residues matched perfectly with the A14L ORF of vaccinia virus (Copenhagen strain) (18). This 270-bp ORF is preceded by the consensus sequence for late transcription, TAAATG. It is predicted to encode a 90-amino-acid polypeptide with a calculated molecular weight of 9,994 and a pI of 9.3. The predicted sequence is highly hydrophobic; overall, 54.4% of the residues are nonpolar. The N terminus of the protein is unusually rich in methionine (4 of the first 10 residues), and residues 13 to 31 and 45 to 64 are predicted to form membrane-spanning helices. The protein has two cysteine residues that might participate in disulfide bond formation; there are 10 serine residues (see below) that are possible sites of phosphorylation. The C-terminal region of the protein is less hydrophobic and is unusually rich in histidine residues. It is not clear which of these sequence features account(s) for the anomalous electrophoretic migration of the A14 protein (16 rather than 10 kDa).
A14 is hyperphosphorylated on serine residues when H1 phosphatase expression is repressed.
Having identified the A14 protein as a hyperphosphorylated component of H1-deficient virions, we were interested in investigating its synthesis and posttranslational modification further. Cells were infected with wt virus or vindH1 and metabolically labeled with [35S]Met for 1.5 h at various points after infection; cell lysates were prepared and subjected to immunoprecipitation analyses. In Fig. 2A, the temporal profile of A14 synthesis is shown. A14 is not expressed in the presence of an inhibitor of DNA replication (araC) (lane 2), confirming that it is a late protein; synthesis is evident, however, at 4.5 and 6 hpi (lanes 3 and 4) and continues thereafter (not shown). Two additional species of 18 and 11 kDa are routinely seen in these immunoprecipitations. The 11-kDa species comigrates with the F18 protein, as is evident when lanes 4 and 5 are compared; we have not determined the identity of the 18-kDa species. When similar immunoprecipitations were performed on extracts prepared from cells labeled for longer time periods or subjected to a 2.5-h chase period after a 45-min pulse, the 21-kDa, mature form of the A17 protein (5) was routinely coimmunoprecipitated (not shown).
FIG. 2.
Immunoprecipitation of A14 from [35S]methionine and 32PPi-labeled protein extracts. (A) Cells were left uninfected (lane M) or infected with wt virus (in the presence or absence of 20 μg of araC per ml) at an MOI of 15 and radiolabeled with [35S]Met for 1.5 h prior to being harvested at the times indicated. The araC sample was harvested at 4 hpi. Samples were subjected to immunoprecipitation with anti-A14 serum (lanes 1 to 4) or anti-F18 serum (lane 5), fractionated by SDS-PAGE, and visualized by fluorography. The radiolabeled species corresponding the A14 and F18 proteins are indicated at the right with ovals; the lines at the right indicate the electrophoretic migration pattern of the protein standards, with their molecular masses indicated in kilodaltons. (B) Cells were left uninfected (lanes M) or infected with wt virus or vindH1 (in the absence of IPTG [H1−]) at an MOI of 2 and radiolabeled with 32PPi from 3 to 17 hpi. Samples were then harvested and subjected to immunoprecipitation analysis with anti-A14 serum (lanes 1 to 5) or a mixture of anti-I3 and anti-F18 sera (lanes 6 to 10). The immunoprecipitates were then fractionated on an SDS–17% polyacrylamide gel and visualized by autoradiography. The positions of the A14, I3, and F18 proteins are indicated with ovals. All infections were performed in duplicate. Protein standards are indicated at the left, with their molecular masses shown in kilodaltons.
The results of immunoprecipitations performed with extracts prepared from 32PPi-labeled cells are shown in Fig. 2B. Whereas phosphorylated A14 was difficult to detect in wt-infected cells, a strong signal was seen when H1 phosphatase expression was repressed (compare lanes 1 and 4 with lanes 2 and 5). Interestingly, we always detected an additional phosphoprotein that comigrated with F18 in these immunoprecipitations. This 11-kDa species was not recovered when the immunoprecipitations were performed with the anti-I3 serum alone (not shown), suggesting that there may indeed be a bona fide interaction between the A14 and F18 proteins.
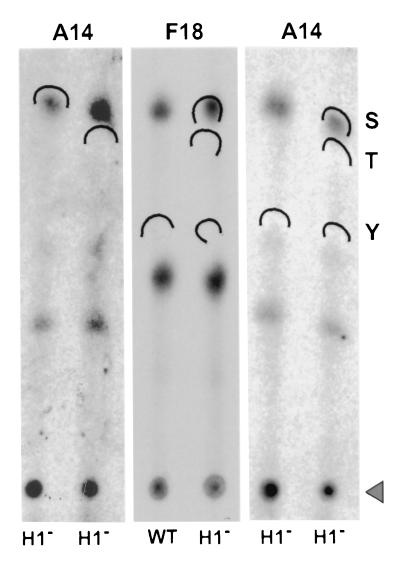
The hyperphosphorylation of A14 seen during nonpermissive vindH1 infections is specific, since the phosphorylation of the I3 protein is not altered (compare lanes 6 and 9 with lanes 7 and 10). F18 is hyperphosphorylated when the H1 phosphatase is repressed (compare lanes 6 and 9 with lanes 7 and 10), as we have reported previously (35). Immunoprecipitates comparable to those shown in lanes 2 and 5 of panel B were fractionated by SDS-PAGE and transferred to PVDF membranes, and the portion of the filter containing the hyperphosphorylated A14 was subjected to acid hydrolysis and one-dimensional phosphoamino acid analysis. The phosphorylated F18 protein was studied in parallel. As shown in Fig. 3, both F18 and A14 appeared to be phosphorylated only on serine residues. We know that our experimental protocol would have revealed phosphothreonine or phosphotyrosine had they been present (13).
FIG. 3.
A14 and F18 are hyperphosphorylated on serine residues when H1 phosphatase expression is repressed. Immunoprecipitates of A14 or F18 (indicated above the lanes) prepared from 32P-labeled cells infected with wt virus or vindH1 in the absence of IPTG (H1−) (indicated below the lanes) were resolved by SDS-PAGE and transferred to a PVDF membrane. The areas of filter corresponding the A14 or F18 protein were excised, hydrolyzed in situ, and subjected to thin-layer electrophoresis in the presence of phosphoserine (S), phosphothreonine (T), and phosphotyrosine (Y) standards. The standards were visualized by ninhydrin staining, and the positions to which they migrated are indicated by the wickets; the origin is indicated by the triangle. The radiolabeled phosphoamino acids released from the A14 and F18 proteins were visualized by autoradiography. The A14 sample was analyzed in quadruplicate.
A14 localizes to the virion membranes of crescents, IV, and IMV but is not exposed on the external surface of IMV.
In our initial identification of the A14 phosphoprotein, we found that it localized to the membrane fraction of virions that had been subjected to NP-40 plus DTT fractionation. Partitioning of A14 to the virion membrane was confirmed by using Triton X-114 to prepare detergent-soluble and aqueous fractions (data not shown). We also tested the accessibility of encapsidated A14 to trypsin, chymotrypsin, or proteinase K treatment; whereas the viral D8 protein was cleaved under these conditions, A14 was not (not shown). The conclusion that A14 is localized to an inner face of the virion membrane is in agreement with observations published by other investigators using different anti-A14 antisera (46, 48). As a final comparison to their studies, we performed immunoelectron microscopy with our antiserum to visualize the association of the A14 protein with the membranes of nascent virions. As shown in the upper panels of Fig. 4, the membranes of crescents (A), IV (B), and IMV (C) show distinct labeling with anti-A14 serum. A14 is clearly associated with virion membranes throughout their maturation. Although it is difficult to be precise about the exact localization of the encapsidated A14 molecules, the images obtained suggest that A14 is removed from the outermost edge and occupies a more internal position within the virion membrane.
We also utilized our anti-F18 antiserum to localize the F18 protein within nascent and mature virions. The coimmunoprecipitation data noted above suggested that the A14 and F18 proteins might interact in vivo. F18 is known to be the major proteinaceous component of the virion core and has a high affinity for DNA (15, 29, 30). Our data indicated that F18 was localized throughout the interiors of immature virions but, surprisingly, was not found within condensed nucleoids (Fig. 4, lower panels). Whether F18 is really excluded from nucleoids or whether the epitopes recognized by our serum are masked within this subviral structure is not known. Genetic analyses have indicated that the F18 protein is indeed required for the formation of normal nucleoids and for the transition of virions from the immature to the mature form (62). We also examined the localization of F18 within mature virions, as shown in the rightmost images of the lower panels. F18 was localized in a ring that appeared to occupy a position midway between the outer membrane and the lucent core of a virion. This localization, unexpectedly, was not markedly different from that observed for the A14 protein.
Construction of a new system for inducible gene expression in vaccinia virus and utilization of the TET operator and repressor.
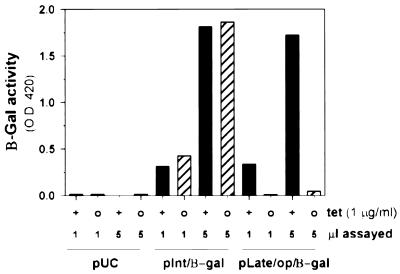
Temperature-sensitive mutants, drug-resistant mutants, and IPTG-inducible recombinants have been invaluable tools in unraveling the functions of a growing number of vaccinia virus gene products. No temperature-sensitive mutants carrying lesions in A14 have been isolated, and the IPTG-inducible recombinant discussed by Rodriguez et al. (47) had not yet been published at this point in our study. As we initiated the construction of an inducible A14 recombinant, we envisioned the utility of being able to independently regulate the expression of more than one viral gene product in a given recombinant. Therefore, we were interested in developing a second system for inducible gene expression. We chose the TET operator and repressor system (16, 17, 19) and generated a virus expressing tetR under the regulation of the constitutive 7.5K promoter (vtetR). As an initial test of the efficacy of the system, we constructed a plasmid in which β-Gal was placed under the regulation of the cowpox ATI promoter and the TET operator (pLate/op/β-gal). As a control, we used a second plasmid in which β-Gal was under the regulation of the G8 intermediate promoter (pInt/β-gal). Cells were infected with vtetR and then transfected with pUC19, pInt/β-gal, or pLate/op/β-gal. Duplicate infections were performed in the absence or presence of TET (1 μg/ml). At 24 hpi, cells were harvested and lysates were assayed for β-Gal activity. As shown in Fig. 5, the expression of β-Gal from pLate/op/β-gal was indeed dependent on the inclusion of TET in the culture medium; comparison of the values obtained when 5-μl samples of the extracts were assayed indicated that omission of TET reduced the level of β-Gal activity to 2.3% of that seen when TET was included. As expected, expression from pInt/β-gal was independent of TET.
FIG. 5.
Tetracycline-regulated gene expression in the context of vaccinia virus infection. Cells were infected with the vtetR recombinant that expresses the TET repressor protein from a constitutive viral promoter. Cells were then transfected with one of three plasmids, pUC19, pInt/β-gal, and pLate/op/β-gal, and maintained in the absence (○) or presence (+) of 1 μg of TET per ml. At 24 hpi, cells were harvested and lysates were assayed for β-Gal activity using a colorimetric assay in which enzyme activity generates an increase in optical density at 420 nm (O.D. 420). One and 5 μl of each lysate were assayed. Values obtained from cells infected in the presence of TET are shown with black bars; those obtained from cells infected in the absence of TET are shown in hatched bars.
Construction of vindA14, a viral recombinant in which expression of A14 is dependent upon the inclusion of TET in the culture medium.
Having confirmed the efficacy of the system, we generated the vindA14 virus, in which the TET operator has been placed between the transcriptional and translational start sites of the endogenous A14 gene. The construct was generated using overlap PCR, and the recombinant virus was isolated using transient dominant selection, as described in Materials and Methods and used previously in our laboratory (31, 35).
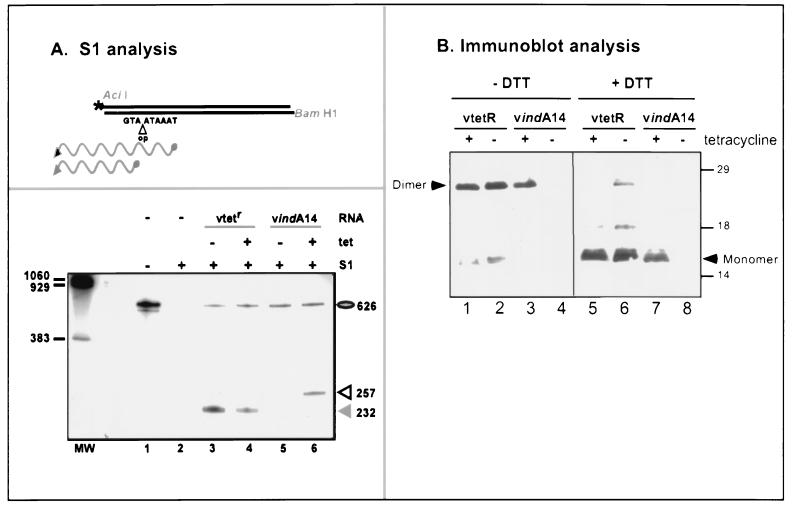
The presence of the tetR gene (within the TK locus) and the TET operator (within the A14 gene) in the vindA14 genome was confirmed by PCR. To determine whether transcription of the A14 gene was in fact dependent upon TET, cultures were infected with vtetR or vindA14 in the absence or presence of TET; at 8 hpi, cells were harvested and total RNA was prepared. S1 nuclease protection was used to monitor the levels and start sites of A14 transcripts, as shown in Fig. 6. The drawing at the top of Fig. 6A illustrates the position of the terminally radiolabeled probe relative to those of the predicted A14 mRNA transcripts. The probe was derived from the operator-containing construct. Therefore, the complementarity between the probe and the wt transcript extends to the ATG of the translational start site, whereas the homology with the vindA14 transcript extends through the transcribed operator sequences to the transcriptional start site. The experimental results are shown in the lower box of panel A. The probe is shown in lane 1; its sensitivity to S1 nuclease digestion in the absence of RNA is evident in lane 2. Lanes 3 and 4 confirm that vtetR-infected cultures express a wt A14 transcript whether or not TET is included in the culture medium. In contrast, lanes 5 and 6 indicate that no A14 transcript is detectable when cultures are infected with vindA14 in the absence of TET; in the presence of TET, a transcript of the appropriate size (i.e., larger) is expressed. Quantitation of the protected fragments using a phosphorimager and ImageQuant software confirmed that the levels of mRNA transcripts that accumulate during infections performed in the absence of TET are <1 to 2% of those seen in induced infections. The level of A14 gene expression achieved during induced vindA14 infections appears comparable to that seen in vtetR-infected cells.
FIG. 6.
TET regulates the accumulation of both A14 mRNA transcripts and A14 protein during infections with vindA14. (A) S1 nuclease analysis of A14 mRNA transcripts. The upper box depicts the radiolabeled probe used in the S1 nuclease analysis. The position of the 5′ radiolabel at the AciI site is marked with an asterisk. The insertion of the TET operator (op) between the transcriptional (TAAATA) and translational (ATG) start sites of the inducible A14 allele within the vindA14 recombinant is also shown. The upper wavy line depicts the protected fragment that would be observed upon hybridization of the probe with the vindA14-derived A14 transcript; the lower wavy line depicts the protected fragment that would be observed upon hybridization with a wt A14 transcript. The lower box represents the results of the S1 nuclease protection assay. Cells were infected with either vtetR or vindA14 (MOI, 10) in the presence (+) or absence (−) of TET (1 μg/ml), and RNA was isolated during the late phase of infection. Reactions were analyzed by denaturing PAGE and visualized by autoradiography. Control samples included those in which the probe was incubated in the absence of RNA, without or with subsequent S1 nuclease digestion (lanes 1 and 2). The expected protected fragment of 232 nucleotides was seen when the probe was hybridized with RNA isolated from vtetR infections performed in either the absence or the presence of TET (lane 3 or 4, respectively). No protected fragment was seen when the probe was hybridized with RNA isolated from cells infected with vindA14 in the absence of TET; a protected fragment of 257 nucleotides was seen after hybridization of the probe with RNA isolated from cells infected with vindA14 in the presence of TET. (B) Immunoblot analysis. Cells were infected with vtetR or vindA14 (MOI, 2) in the presence or absence of TET (1 μg/ml) and harvested at 16 hpi. Half of each sample was resuspended in sample buffer lacking DTT, and half was resuspended in sample buffer containing DTT. The samples were then boiled, fractionated on an SDS–17% polyacrylamide gel, and transferred to nitrocellulose for immunoblot analysis with anti-A14 serum. The blot was developed by chemiluminescence. The position of the dimeric (25-kDa) and monomeric (16-kDa) forms of A14 are indicated. No immunodetectable A14 was seen in samples prepared from vindA14 infections performed in the absence of TET. The dashes at the right indicate the electrophoretic profile of protein standards, with their molecular masses indicated in kilodaltons.
Having confirmed that the transcription of the endogenous A14 locus was being controlled by the TET repressor and operator, we analyzed the levels of A14 protein that accumulated during induced and repressed infections. Infected cell lysates were subjected to immunoblot analysis using a polyclonal anti-A14 antiserum, as shown in Fig. 6B. Because A14 had previously been shown to have a tendency to form disulfide-linked dimers (26, 46, 47, 51), we analyzed the samples in the presence (lanes 5 to 8) and absence (lanes 1 to 4) of reducing agent (DTT). vtetR-infected cells accumulated equivalent levels of A14 whether or not TET was included in the culture medium (compare lanes 1 and 2 with lanes 5 and 6); A14 dimers (25 kDa) were the dominant species in the absence of DTT (lanes 1 and 2) and monomers (16 kDa) were predominant in the presence of DTT (lanes 5 and 6). In contrast, A14 protein was virtually undetectable when vindA14 infections were maintained in the absence of TET (lanes 4 and 8); in the presence of TET, the pattern of dimer and monomer forms (lanes 3 and 7) was equivalent to that seen with the parental vtetR virus. Thus, both the RNA and protein analyses indicated that the A14 locus was efficiently repressed when vindA14 infections were performed in the absence of TET.
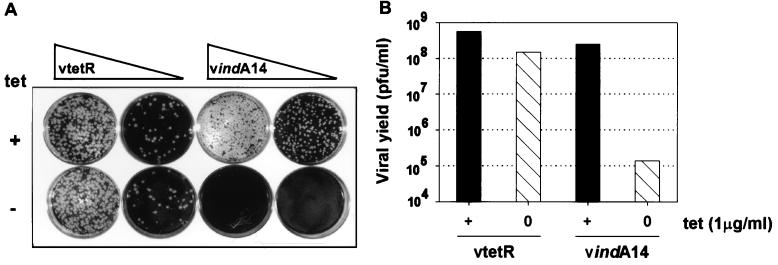
Repression of A14 expression abrogates plaque formation and reduces the yield of infectious IMV by 3 orders of magnitude.
As a first test of whether the repression of A14 expression compromised viral infection, plaque assays were performed with vtetR and vindA14 in the presence or absence of TET. As shown in Fig. 7A, plaque formation by vtetR was unaffected by TET. In contrast, plaque formation by vindA14 was completely abrogated in the absence of TET, whereas wt-sized plaques developed in the presence of TET. We next infected cultures with vtetR or vindA14 at an MOI of 2 and harvested the infected cells at 24 hpi. The yield of virus was then titrated in the presence of TET. This experiment was designed to provide a more accurate quantification of the impact of A14 repression on virus production. Moreover, this experiment allowed us to determine whether A14 expression was required for the production of infectious IMV or only for the further maturation of a subset of IMV into the cell-associated and extracellular enveloped that lead to plaque formation (6). We found that the production of IMV by vindA14-infected cultures was reduced by 3 orders of magnitude when TET was omitted from the culture medium (Fig. 7B); in contrast, IMV production by vtetR-infected cells was not affected by the omission of TET.
FIG. 7.
vindA14 is a conditionally lethal mutant, and plaque formation and the production of infectious viral progeny are dependent upon TET. (A) vtetR and vindA14 were titrated on monolayers of BSC40 cells in the presence (+) or absence (−) of TET. (B) Cells were infected with vtetR or vindA14 for 24 h in either the presence (+) or the absence (○) of TET; cell lysates were prepared and virus was titrated on BSC40 cells in the presence of TET.
To determine where the infectious cycle arrested in the absence of A14 expression, we monitored the temporal profile of protein synthesis in metabolically labeled cells infected with vtetR or vindA14 in the presence or absence of TET. No perturbation in the pattern of gene expression was seen when the expression of A14 was prevented (not shown). However, pulse-chase protocols revealed that the proteolytic processing of several virion proteins that normally accompanies IMV maturation (p4a to 4a, p4b to 4b, pre-L4 to L4) failed to occur when A14 synthesis was repressed (not shown). These data suggested that the primary role of the A14 protein was likely to be in virion morphogenesis.
Repression of A14 synthesis leads to aberrant and abortive virion morphogenesis.
Cultures infected with vindA14 in the presence or absence of TET were examined by electron microscopy. In the presence of TET, we observed all of the normal intermediates in virion assembly: virosomes, crescents, and immature and mature virions (Fig. 8A and B). In the absence of TET, morphogenesis was clearly aberrant and incomplete (Fig. 8C to F). There was a conspicuous absence of any immature or mature virions. Dense virosomes were apparent and numerous. Crescents (Fig. 8C and E) were seen surrounding some of the virosomes, although their appearance was abnormal because of their apparent emptiness. During wt infections, crescents appeared to associate with and encircle the dense virosomal material. In the uninduced vindA14-infected cultures, however, the crescents appeared to be detached from the virosomal material. In addition, some of the crescents appeared discontinuous (Fig. 8F), as if they represented an accumulation of abutting but discrete vesicles. The most striking feature of these infected cells, however, was the presence of a vast number of what appeared to be membrane vesicles (Fig. 8C to F). These vesicles were clustered in large groups that were often distant and removed from the virosomes.
Immunolocalization of DNA-binding proteins and viral membrane components in vindA14-infected cells.
To provide further insight into the composition of the vesicle clusters and electron-dense structures seen in cells infected nonpermissively with vindA14, we performed immunoelectron microscopy. Cells were labeled with antisera directed against the L4 (3, 4, 56, 57, 60, 61) or F18 (29, 30, 62) DNA-binding proteins or the D8 (23, 29) or A17 (5, 33, 44, 59) membrane proteins. Representative images are shown in Fig. 9. As seen in panels A and B, the electron-dense virosomes labeled robustly with the anti-L4 serum whereas the nearby clusters of vesicles remained unlabeled. Similarly, labeling with the anti-F18 serum was essentially restricted to the virosomes (panel C), which labeled prominently with this serum. The inverse of this pattern can be seen in panels D and E, which represent sections labeled with anti-A17 serum, and F and G, which represent sections labeled with anti-D8 serum. The clusters of vesicles labeled heavily with both sera, whereas virtually no labeling of the virosomes was seen. In panel D, labeling of the vesicular and crescent membranes surrounding the virosome by the anti-A17 serum can be seen. Together, these data provide compelling evidence that the abundant and unusual clusters of vesicles found in cells infected nonpermissively with vindA14 are indeed membranous in nature and contain proteins that are well-characterized components of the viral membrane. Furthermore, size comparison of the 10-nm-diameter gold particles with the vesicles suggests that the vesicles have an average diameter of approximately 25 nm.
DISCUSSION
We show in this report that the encapsidated A14 protein is one of the substrates of the H1 phosphatase. Hyperphosphorylation of A14 when H1 synthesis is repressed can clearly be seen by a dramatic elevation in the level of 32PPi that can be incorporated into A14. Moreover, the electrophoretic migration of the protein changes from a single band with an apparent molecular weight of 16K to a doublet created by the appearance of a more slowly migrating species. A14, A17, and F18 have now been shown to represent virion proteins which are hyperphosphorylated in H1-deficient virions (13, 35). Although hyperphosphorylation of A14, a component of the virion membrane, is unlikely to be responsible for the transcriptional incompetence and lack of infectivity of the H1-deficient virions, it might well contribute to the structural instability that we have previously reported (35). We noted that treatment of H1-deficient virions with NP-40 plus DTT released significant levels of proteins that normally remain tightly associated with the core, such as the DNA-binding proteins F18 and L4. Since, as mentioned above, we often find that F18 is coimmunoprecipitated with A14 during analyses of extracts prepared at late times of infection, the protein-protein interactions that stabilize virion integrity may be compromised by hyperphosphorylation. Although the interaction of the membrane protein A14 and the DNA-binding protein F18 was initially surprising, several lines of data suggest that they might well interact in vivo. First, our ultrastructural localization of the F18 protein within mature virions places it in a ring that surrounds the central, lucent core; in fact, the pattern of F18 localization within mature virions is quite similar to that seen for the A14 protein. Second, electron spectroscopic imaging of encapsidated DNA has also suggested that the genome may be localized in a ring that surrounds the central, lucent core of the virion (21). Taken together, these data suggest that the nucleoprotein complex may associate with proteins located on the inner face the viral membrane. These protein-protein interactions might provide an explanation for the puzzling observation that, in A32-deficient virions, the F18 protein can still be efficiently encapsidated even though the viral DNA is not (7).
Little was known about the A14 gene product when we first identified it as an encapsidated phosphoprotein and an H1 substrate. It had been identified as one of the structural components of the vaccinia virion and characterized as readily forming disulfide-linked dimers (26, 51). Indeed, we observed that the electrophoretic mobility of A14 was consistent with an apparent molecular weight of 25K in the absence of DTT but 16K in the presence of DTT. Since then, several investigators have published reports regarding the structure and function of the A14 protein (5, 46–48). There is general agreement that A14 is a component of membranes, that it tends to form disulfide-linked dimers, and that it is phosphorylated. Both our study and that of Salmons et al. (48) support the localization of A14 within the viral membrane in such a way that it is not exposed to proteolytic digestion. Further support for the conclusion that A14 is not exposed on the surfaces of virions was obtained by our demonstration that incubation of anti-A14 serum with IMV failed to neutralize viral infectivity (not shown). An association of A14 with the A17 protein was first demonstrated by Rodriguez et al. (46) and is supported by the work of Betakova et al. (5) and by our experiments (not shown).
Rodriguez et al. (46) proposed that A14 was likely to be phosphorylated on a particular threonine residue, but both our study and that of Betakova et al. (5) have shown that phosphorylation is exclusively on serine residues and appears to depend upon the F10 kinase. Although Betakova et al. (5) could easily detect the phosphorylation of A14 during wt infections, we found that 32P-labeling of A14 is difficult to detect under these conditions with our reagents and experimental protocols. Only when H1 synthesis is repressed can we see robust evidence of A14 phosphorylation; this difference may reflect a difference in the antisera used to detect A14 or other experimental variables. Our data do suggest, however, that the phosphorylation state of A14 is exquisitely sensitive to the levels of H1 phosphatase, which implies that A14 is either a poor substrate or an inaccessible one for cellular phosphatases.
Although Rodriguez et al. reported an essential role for the A14 protein in virion morphogenesis in 1997 (47), no such information was available to us at the time that we identified the A14 phosphoprotein as an H1 substrate. Motivated by the need to create an inducible recombinant in which A14 expression could be regulated, and by the desire to augment the genetic tools available for the study of vaccinia virus, we adapted the regulatory components of the TET operon for use in the vaccinia virus genome. The TET system has several advantages, including the nontoxicity of TET and the high affinity of the repressor for the operator. We created the vtetR virus, in which a p7.5-regulated TET repressor gene has been inserted within the nonessential TK locus. (We have also created the vtetRvlacI virus, in which both the TET and lactose repressors are inserted within the TK locus (B. Unger and P. Traktman, unpublished data). We first demonstrated the utility of this virus in transient-transfection assays by monitoring the expression of a β-Gal gene regulated by the TET operator sequence. Expression was minimal in the absence of TET and high in the presence of TET. Starting with the parental virus vtetR, we then created vindA14, in which the TET operator has been placed between the transcriptional and translational start sites of the A14 gene. (Alteration of the promoter sequence from TAAATG to TAAATA, in order to avoid translational initiation upstream of the operator sequence, is not predicted to exert a significant effect on the transcriptional efficiency of the gene [11]). Our data show that the accumulation of mRNA transcripts and the synthesis and accumulation of the A14 protein are dependent upon the inclusion of TET in the medium during vindA14 infections; omission of TET represses gene expression by 2 orders of magnitude. TET was not seen to have any effect on the parental virus, vtetR. These data confirm that the TET-inducible system provides an efficient means for regulating gene expression in the context of vaccinia virus infection and so represents a new approach to the generation of conditional mutants for functional studies.
Using vindA14, we demonstrated that the repression of A14 synthesis abrogated plaque formation and reduced the 24-h yield of infectious virus by 3 orders of magnitude. No impact on the profile of viral gene expression was seen, although the proteolytic processing of the core proteins that usually accompanies virion maturation was absent. Our electron microscopic analysis revealed an interesting phenotype which is consistent with that observed by Rodriguez et al. in their study of an IPTG-inducible A14 recombinant (47). We observed no mature or immature virions. Electron-dense virosomes were plentiful, and “empty” crescents were often found at their periphery. These structures had the general shape of crescents, but they appeared detached from the virosome and did not enclose any of the electron-dense material that normally “fills” the crescent membrane. Some of these crescents were discontinuous, appearing as a string of contiguous vesicles. The most striking feature of the cells, however, was the accumulation of enormous numbers of vesicles in large clusters that were often removed from the virosomes. We confirmed that these vesicles contain the A17 and D8 proteins, known components of the virion membrane.
Vesicles have not previously been considered a normal intermediate in the biogenesis of the viral particle. Although crescents have been portrayed as emanating from the membranes of the intermediate compartment (49), a vesicular intermediate has not been given emphasis. However, it is clear that the hallmark of A14 repression is the accumulation of massive numbers of detached vesicles. In contrast, repression of A17 synthesis leads to the appearance of vesicles which form a corona around the virosome (M. Sauter and P. Traktman, unpublished data; 45, 59). When nonpermissive infections are performed with a temperature-sensitive mutant carrying lesions in the F10 kinase, however, no evidence of such vesicle accumulation is seen (53, 55). We propose, therefore, that vesicle diversion from the intermediate compartment is likely to be a normal step in virion biogenesis that is dependent on the F10 kinase but that the vesicles are short-lived in the presence of the A14 and A17 proteins due to their rapid maturation into crescents. In the absence of A14, the recruitment of vesicles to the virosome, their adhesion to the virosomal contents, and their apparent fusion into crescents are grossly deficient. In the absence of A17, recruitment to the virosome appears normal but maturation into crescents is never seen. This model, in the context of the numerous genetic and immunologic reagents that are now available, opens the way for mechanistic investigations of how F10, A14, and A17 work together in virion morphogenesis.
The implications of the involvement of a protein kinase and two phosphoproteins at these earliest stages of virion morphogenesis are inescapable. Analysis of the role of protein phosphorylation in regulating morphogenesis has been made more intriguing by the recent observations that A17 is phosphorylated on tyrosine as well as serine/threonine residues in a manner that is genetically dependent upon the F10 kinase (5, 13). Moreover, we have recently reported that F10 is likely to be a dual-specificity kinase, since bacteria induced to express F10 coordinately acquire significant amounts of phosphotyrosine on a number of proteins (13). The participation of protein kinases and phosphatases in cellular vesicle trafficking is now well documented. For example, at the onset of mitosis, the Golgi apparatus disassembles and vesiculates. The docking of vesicles derived from the endoplasmic reticulum and destined for the Golgi apparatus is blocked, primarily due to the phosphorylation of the GM130 protein on the Golgi membranes. GM130 phosphorylation impairs its interaction with the p115 protein found on the incoming vesicles (36, 38) and hence prevents the “consumption” of these vesicles by the Golgi apparatus. Cell cycle-regulated phosphorylation events that control the localization of p115 on vesicle membranes have also been reported (50). We suggest that the F10 kinase might mediate analogous phosphorylation events that facilitate the diversion of intracellular vesicles away from their normal trafficking pathway, enabling the maturation of these vesicles into virion membranes. Analysis of the details of this process should enhance our understanding of vaccinia virus biogenesis and resolve some of the controversy over the origin and number of the lipid bilayers surrounding mature viral particles (10, 22, 49). Moreover, this analysis should serve as a genetically tractable model for organelle biogenesis.
ACKNOWLEDGMENTS
We gratefully acknowledge the contribution of M. Sauter to the initial preparation of the anti-A17 serum and thank Poi Lee and B. Topaz for technical assistance in generating the anti-A14 and anti-A17 sera, respectively. The electron microscopic analyses would not have been possible without the superb guidance and technical support of Leona Cohen-Gould and Gang Ning.
This work was supported by a grant to P.T. from the NIH (2 R01 GM 53601) and by a special group of donors from the Dorothy Rodbell Cohen Foundation.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1997. [Google Scholar]
- 2.Banham A H, Smith G L. Vaccinia virus gene B1R encodes a 34-kDa serine/threonine protein kinase that localizes in cytoplasmic factories and is packaged into virions. Virology. 1992;191:803–812. doi: 10.1016/0042-6822(92)90256-o. [DOI] [PubMed] [Google Scholar]
- 3.Bayliss C D, Smith G L. Vaccinia virion protein VP8, the 25 kDa product of the L4R gene, binds single-stranded DNA and RNA with similar affinity. Nucleic Acids Res. 1997;25:3984–3990. doi: 10.1093/nar/25.20.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bayliss C D, Wilcock D, Smith G L. Stimulation of vaccinia virion DNA helicase I8R, but not A18R, by a vaccinia core protein L4R, an ssDNA binding protein. J Gen Virol. 1996;77:2827–2831. doi: 10.1099/0022-1317-77-11-2827. [DOI] [PubMed] [Google Scholar]
- 5.Betakova T, Wolffe E J, Moss B. Regulation of vaccinia virus morphogenesis: phosphorylation of the A14L and A17L membrane proteins and C-terminal truncation of the A17L protein are dependent on the F10L kinase. J Virol. 1999;3:3534–3543. doi: 10.1128/jvi.73.5.3534-3543.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blasco R, Moss B. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J Virol. 1992;66:4170–4179. doi: 10.1128/jvi.66.7.4170-4179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cassetti M C, Merchlinsky M, Wolffe E J, Weisberg A S, Moss B. DNA packaging mutant: repression of the vaccinia virus A32 gene results in noninfectious, DNA-deficient, spherical, enveloped particles. J Virol. 1998;72:5769–5780. doi: 10.1128/jvi.72.7.5769-5780.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chakrabarti S, Brechling K, Moss B. Vaccinia virus expression vector: coexpression of galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol. 1985;5:3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dales S, Milanovanovitch V, Pogo B G T, Weintraub S B, Huima T, Wilton S, McFadden G. Biogenesis of vaccinia: isolation of conditional lethal mutants and electron microscopic characterization of their phenotypically expressed defects. Virology. 1978;844:403–428. doi: 10.1016/0042-6822(78)90258-1. [DOI] [PubMed] [Google Scholar]
- 10.Dales S, Mosbach E H. Vaccinia as a model for membrane biogenesis. Virology. 1968;35:564–583. doi: 10.1016/0042-6822(68)90286-9. [DOI] [PubMed] [Google Scholar]
- 11.Davison A J, Moss B. The structure of vaccinia virus late promoters. J Mol Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- 12.DeMasi J, Traktman P. Clustered charge-to-alanine mutagenesis of the vaccinia virus H5 gene: isolation of a dominant, temperature-sensitive mutant with a profound defect in morphogenesis. J Virol. 2000;74:2393–2405. doi: 10.1128/jvi.74.5.2393-2405.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Derrien M, Punjabi A, Khanna M, Grubisha O, Traktman P. Tyrosine phosphorylation of A17 during vaccinia virus infection: involvement of the H1 phosphatase and the F10 kinase. J Virol. 1999;73:7287–7296. doi: 10.1128/jvi.73.9.7287-7296.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Falkner F G, Moss B. Transient dominant selection of recombinant vaccinia viruses. J Virol. 1990;64:3108–3111. doi: 10.1128/jvi.64.6.3108-3111.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fogelsong P D, Bauer W R. Effects of ATP and inhibitory factors on the activity of vaccinia virus type I topoisomerase. J Virol. 1984;49:1–8. doi: 10.1128/jvi.49.1.1-8.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gatz C, Kaiser A, Wendenburg R. Regulation of a modified CaMV 35S promoter by the Tn10-encoded Tet repressor in transgenic tobacco. Mol Gen Genet. 1991;227:229–237. doi: 10.1007/BF00259675. [DOI] [PubMed] [Google Scholar]
- 17.Gatz C, Quail P H. Tn-10-encoded tet repressor can regulate an operator-containing plant promoter. Proc Natl Acad Sci USA. 1988;85:1394–1397. doi: 10.1073/pnas.85.5.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goebel S J, Johnson G P, Perkus M E, David S W, Winslow J P, Paoletti E. The complete DNA sequence of vaccinia virus. Virology. 1990;179:247–266. doi: 10.1016/0042-6822(90)90294-2. [DOI] [PubMed] [Google Scholar]
- 19.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimley P M, Rosenblum E N, Mims S J, Moss B. Interruption by rifampin of an early stage in vaccinia virus morphogenesis: accumulation of membranes which are precursors of virus envelopes. J Virol. 1970;6:519–533. doi: 10.1128/jvi.6.4.519-533.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harauz G, Evans D H, Beniac D R, Arsenault A L, Rutherford B, Ottensmeyer F P. Electron spectroscopic imaging of encapsidated DNA in vaccinia virus. Can J Microbiol. 1995;41:889–894. doi: 10.1139/m95-122. [DOI] [PubMed] [Google Scholar]
- 22.Hollinshead M, Vanderplasschen A, Smith G L, Vaux D J. Vaccinia virus intracellular mature virions contain only one lipid membrane. J Virol. 1999;73:1503–1517. doi: 10.1128/jvi.73.2.1503-1517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiao J C, Chung C S, Chang W. Vaccinia virus envelope D8L protein binds to cell surface chondroitin sulfate and mediates the adsorption of intracellular mature virions to cells. J Virol. 1999;73:8750–8761. doi: 10.1128/jvi.73.10.8750-8761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X, Carroll L J, Wolffe E J, Moss B. De novo synthesis of the early transcription factor 70-kilodalton subunit is required for morphogenesis of vaccinia virions. J Virol. 1996;70:7669–7677. doi: 10.1128/jvi.70.11.7669-7677.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu X, Wolffe E J, Weisberg A S, Carroll L J, Moss B. Repression of the A8L gene, encoding the early transcription factor 82-kilodalton subunit, inhibits morphogenesis of vaccinia virions. J Virol. 1998;72:104–112. doi: 10.1128/jvi.72.1.104-112.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ichihashi Y, Oie M, Tsuruhara T. Location of DNA-binding proteins and disulfide-linked proteins in vaccinia virus structural elements. J Virol. 1984;50:929–938. doi: 10.1128/jvi.50.3.929-938.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson G P, Goebel S J, Paoletti E. An update on the vaccinia virus genome. Virology. 1993;196:381–401. doi: 10.1006/viro.1993.1494. [DOI] [PubMed] [Google Scholar]
- 28.Kane E M, Shuman S. Vaccinia virus morphogenesis is blocked by a temperature-sensitive mutation in the I7 gene that encodes a virion component. J Virol. 1993;67:2689–2698. doi: 10.1128/jvi.67.5.2689-2698.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao S, Ressner E, Kates J, Bauer W R. Purification and characterization of a superhelix binding protein from vaccinia virus. Virology. 1981;111:500–508. doi: 10.1016/0042-6822(81)90352-4. [DOI] [PubMed] [Google Scholar]
- 30.Kao S Y, Bauer W R. Biosynthesis and phosphorylation of vaccinia virus structural protein VP11. Virology. 1987;159:399–407. doi: 10.1016/0042-6822(87)90479-x. [DOI] [PubMed] [Google Scholar]
- 31.Klemperer N, Ward J, Evans E, Traktman P. The vaccinia virus I1 protein is essential for the assembly of mature virions. J Virol. 1997;71:9285–9294. doi: 10.1128/jvi.71.12.9285-9294.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koerner T J, Hill J E, Myers A M, Tzagaloff A. High-expression vectors with multiple cloning sites for construction of trpE fusion genes: pATH vectors. Methods Enzymol. 1991;194:477–490. doi: 10.1016/0076-6879(91)94036-c. [DOI] [PubMed] [Google Scholar]
- 33.Krijnse-Locker J, Schleich S, Rodriguez D, Goud B, Snijder E J, Griffiths G. The role of a 21-kDa viral membrane protein in the assembly of vaccinia virus from the intermediate compartment. J Biol Chem. 1996;271:14950–14958. doi: 10.1074/jbc.271.25.14950. [DOI] [PubMed] [Google Scholar]
- 34.Lin S, Chen W, Broyles S S. The vaccinia virus B1R gene product is a serine/threonine protein kinase. J Virol. 1992;66:2717–2723. doi: 10.1128/jvi.66.5.2717-2723.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu K, Lemon B, Traktman P. The dual-specificity phosphatase encoded by vaccinia virus, VH1, is essential for viral transcription in vivo and in vitro. J Virol. 1995;69:7823–7834. doi: 10.1128/jvi.69.12.7823-7834.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lowe M, Rabouille C, Nakamura N, Watson R, Jackman M, Jamsa E, Rahman D, Pappin D J C, Warren G. Cdc2 kinase directly phosphorylates the cis-Golgi matrix protein GM130 and is required for Golgi fragmentation in mitosis. Cell. 1998;94:783–793. doi: 10.1016/s0092-8674(00)81737-7. [DOI] [PubMed] [Google Scholar]
- 37.Mackett M, Smith G L, Moss B. General method for production and selection of infectious vaccinia virus recombinants expressing foreign genes. J Virol. 1984;49:857–864. doi: 10.1128/jvi.49.3.857-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nakamura N, Lowe M, Levine T P, Rabouille C, Warren G. The vesicle docking protein p115 binds GM130, a cis-Golgi matrix protein, in a mitotically regulated manner. Cell. 1997;89:445–455. doi: 10.1016/s0092-8674(00)80225-1. [DOI] [PubMed] [Google Scholar]
- 39.Niles E G, Seto J. Vaccinia virus gene D8 encodes a virion transmembrane protein. J Virol. 1988;62:3772–3778. doi: 10.1128/jvi.62.10.3772-3778.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patel D D, Ray C A, Drucker R P, Pickup D J. A poxvirus-derived vector that directs high levels of expression of cloned genes in mammalian cells. Proc Natl Acad Sci USA. 1988;85:9431–9435. doi: 10.1073/pnas.85.24.9431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ravanello M P, Hruby D E. Conditional lethal expression of the vaccinia virus L1R myristylated protein reveals a role in virion assembly. J Virol. 1994;68:6401–6410. doi: 10.1128/jvi.68.10.6401-6410.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rempel R E, Traktman P. Vaccinia virus B1 kinase: phenotypic analysis of temperature-sensitive mutants and enzymatic characterization of recombinant proteins. J Virol. 1992;66:4413–4426. doi: 10.1128/jvi.66.7.4413-4426.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rochester S C, Traktman Characterization of the single-stranded DNA binding protein encoded by the vaccinia virus I3 gene. J Virol. 1998;72:2917–2926. doi: 10.1128/jvi.72.4.2917-2926.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rodriguez D, Esteban M, Rodriguez J R. Vaccinia virus A17L gene product is essential for an early step in virion morphogenesis. J Virol. 1995;69:4640–4648. doi: 10.1128/jvi.69.8.4640-4648.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rodriguez D, Risco C, Rodriguez J R, Carrascosa J L, Esteban M. Inducible expression of the vaccinia virus A17L gene provides a synchronized system to monitor sorting of viral proteins during morphogenesis. J Virol. 1996;70:7641–7653. doi: 10.1128/jvi.70.11.7641-7653.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rodriguez J R, Risco C, Carrascosa J L, Esteban M, Rodriguez D. Characterization of early stages in vaccinia virus membrane biogenesis: implications of the 21-kilodalton protein and a newly identified 15-kilodalton envelope protein. J Virol. 1997;71:1821–1833. doi: 10.1128/jvi.71.3.1821-1833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodriguez J R, Risco C, Carrascosa J L, Esteban M, Rodriguez D. Vaccinia virus 15-kilodalton (A14L) protein is essential for assembly and attachment of viral crescents to virosomes. J Virol. 1998;72:1287–1296. doi: 10.1128/jvi.72.2.1287-1296.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salmons T, Kuhn A, Wylie F, Schleich S, Rodriguez J R, Rodriguez D, Esteban M, Griffiths G, Locker J K. Vaccinia virus membrane proteins p8 and p16 are cotranslationally inserted into the rough endoplasmic reticulum and retained in the intermediate compartment. J Virol. 1997;71:7404–7420. doi: 10.1128/jvi.71.10.7404-7420.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sodeik B, Doms R W, Ericsson M, Hiller G, Machamer C E, van't Hof W, van Meer G, Moss B, Griffiths G. Assembly of vaccinia virus: role of the intermediate compartment between the endoplasmic reticulum and the Golgi stacks. J Cell Biol. 1993;121:521–541. doi: 10.1083/jcb.121.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sohda M, Misumi Y, Yano A, Takami N, Ikehara Y. Phosphorylation of the vesicle docking protein p115 regulates its association with the Golgi membrane. J Biol Chem. 1998;273:5385–5388. doi: 10.1074/jbc.273.9.5385. [DOI] [PubMed] [Google Scholar]
- 51.Takahashi T, Oie M, Ichihashi Y. N-terminal amino acid sequences of vaccinia virus structural proteins. Virology. 1994;202:844–852. doi: 10.1006/viro.1994.1406. [DOI] [PubMed] [Google Scholar]
- 52.Traktman P, Anderson M K, Rempel R E. Vaccinia virus encodes an essential gene with strong homology to protein kinases. J Biol Chem. 1989;264:21458–21461. [PubMed] [Google Scholar]
- 53.Traktman P, Caligiuri A, Jesty S A, Liu K, Sankar U. Temperature-sensitive mutants with lesions in the vaccinia virus F10 kinase undergo arrest at the earliest stage of virion morphogenesis. J Virol. 1995;69:6581–6587. doi: 10.1128/jvi.69.10.6581-6587.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vogelstein B, Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci USA. 1979;76:615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang S, Shuman S. Vaccinia virus morphogenesis is blocked by temperature-sensitive mutations in the F10 gene, which encodes protein kinase-2. J Virol. 1995;69:6376–6388. doi: 10.1128/jvi.69.10.6376-6388.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wilcock D, Smith G. Vaccinia virions lacking core protein VP8 are deficient in early transcription. J Virol. 1996;67:934–943. doi: 10.1128/jvi.70.2.934-943.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wilcock D, Smith G L. Vaccinia virus core protein VP8 is required for virus infectivity, but not for core protein processing or for INV and EEV formation. Virology. 1994;202:294–304. doi: 10.1006/viro.1994.1346. [DOI] [PubMed] [Google Scholar]
- 58.Williams O, Wolffe E J, Weisberg A S, Merchlinsky M. Vaccinia virus WR gene A5L is required for morphogenesis of mature virions. J Virol. 1999;73:4590–4599. doi: 10.1128/jvi.73.6.4590-4599.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wolffe E J, Moore D M, Peters E J, Moss B. Vaccinia virus A17L open reading frame encodes an essential component of nascent viral membranes that is required to initiate morphogenesis. J Virol. 1996;70:2797–2808. doi: 10.1128/jvi.70.5.2797-2808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang W-P, Bauer W R. Biosynthesis and post-translational cleavage of vaccinia virus structural protein VP8. Virology. 1988;167:585–590. [PubMed] [Google Scholar]
- 61.Yang W-P, Bauer W R. Purification and characterization of vaccinia virus structural protein VP8. Virology. 1988;167:578–584. [PubMed] [Google Scholar]
- 62.Zhang Y, Moss B. Vaccinia virus morphogenesis is interrupted when expression of a gene encoding an 11-kilodalton phosphorylated protein is prevented by the Escherichia coli lac repressor. J Virol. 1991;65:6101–6110. doi: 10.1128/jvi.65.11.6101-6110.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang Y, Moss B. Immature viral envelope formation is interrupted at the same stage by lac operator mediated repression of the vaccinia virus D13L gene and by the drug rifampicin. Virology. 1992;187:643–653. doi: 10.1016/0042-6822(92)90467-4. [DOI] [PubMed] [Google Scholar]