Abstract
The cytotoxic T-lymphocyte (CTL) response against the murine cytomegalovirus (MCMV) immediate-early gene 1 (IE1) 89-kDa phosphoprotein pp89 plays a major role in protecting BALB/c mice against the lethal effects of the viral infection. CTL populations specific to MCMV early-phase and structural antigens are also generated during infection, but the identities of these antigens and their relative contributions to overall immunity against MCMV are not known. We previously demonstrated that DNA vaccination with a pp89-expressing plasmid effectively generated a CTL response and conferred protection against infection (J. C. Gonzalez Armas, C. S. Morello, L. D. Cranmer, and D. H. Spector, J. Virol. 70:7921–7928, 1996). In this report, we have sought (i) to identify other viral antigens that contribute to immunity against MCMV and (ii) to determine whether the protective response is haplotype specific. DNA immunization was used to test the protective efficacies of plasmids encoding MCMV homologs of human cytomegalovirus (HCMV) tegument (M32, M48, M56, M82, M83, M69, and M99), capsid (M85 and M86), and nonstructural antigens (IE1-pp89 and M84). BALB/c (H-2d) and C3H/HeN (H-2k) mice were immunized by intradermal injection of either single plasmids or cocktails of up to four expression plasmids and then challenged with sublethal doses of virulent MCMV administered intraperitoneally. In this way, we identified a new viral gene product, M84, that conferred protection against viral replication in the spleens of BALB/c mice. M84 is expressed early in the infection and encodes a nonstructural protein that shares significant amino acid homology with the HCMV UL83-pp65 tegument protein, a major target of protective CTLs in humans. Specificity of the immune response to the M84 protein was confirmed by showing that immunization with pp89 DNA, but not M84 DNA, protected mice against subsequent infection with an MCMV deletion mutant lacking the M84 gene. The other MCMV genes tested did not generate a protective response even when mice were immunized with vaccinia viruses expressing the viral proteins. However, the M84 plasmid was protective when injected in combination with nonprotective plasmids, and coimmunization of BALB/c mice with pp89 and M84 provided a synergistic level of protection in the spleen. Viral titers in the salivary glands were also reduced, but not to the same extent as observed in the spleen, and the decrease was seen only when the BALB/c mice were immunized with pp89 plus M84 or with pp89 alone. The experiments with the C3H/HeN mice showed that the immunity conferred by DNA vaccination was haplotype dependent. In this strain of mice, only pp89 elicited a protective response as measured by a reduction in spleen titer. These results suggest that DNA immunization with the appropriate combination of CMV genes may provide a strategy for improving vaccine efficacy.
Human cytomegalovirus (HCMV) is a serious opportunistic pathogen for both newborn and immunocompromised individuals. It is the leading viral cause of birth defects, affecting 6,000 to 7,000 infants yearly in the United States alone (16). In addition, nearly 10% of deaths of AIDS patients in 1992 were attributed to HCMV disease (45), while in bone marrow and solid organ transplant recipients, HCMV infection continues to lead to a high incidence of morbidity and even mortality (4). The severe medical problems associated with HCMV infection in these particularly vulnerable populations underline the necessity of developing a safe and effective vaccine. Studies of HCMV immunity and the testing of candidate vaccines have highlighted the importance of cell-mediated immunity for control of the infection and prevention of disease.
Because of the strict species specificity of the betaherpesviruses, animal models of CMV immunity have used murine cytomegalovirus (MCMV), which is similar to HCMV with regard to virion structure, genome organization, gene expression, tissue tropism, and latency. As with HCMV, the cellular immune response to MCMV has been shown to be critical for control of the acute infection (28). In the susceptible BALB/c strain, a single nonstructural protein—immediate-early protein 1–pp89 (IE1-pp89)— has been shown to be immunodominant for the generation of protective cytotoxic T lymphocytes (CTLs), and a single, nonpeptide epitope was determined to be targeted during natural infection (28). Delivery of this single epitope to BALB/c mice by vaccinia virus (14) or synthetic peptide (42) resulted in protective immunity. Similarly, immunization of BALB/c mice with a pp89-expressing plasmid generated similar levels of pp89-nonapeptide-specific CTL activity as did immunization with tissue culture-derived MCMV (17). However, this plasmid-mediated protection against MCMV was incomplete (17) compared to that generated by the attenuated virus. This suggests that, to be fully protective, the MCMV-specific CTL response may need to be directed against multiple viral antigens.
The number of MCMV antigens that contribute to the generation of protective CTL immunity is likely limited to those viral antigens that not only contain the appropriate epitopes for efficient binding to major histocompatibility complex (MHC) class I glycoprotein complexes and subsequent recognition by α/β T-cell receptors (TCRs) but are also capable of escaping the MCMV-mediated shutdown of antigen presentation. Similar to HCMV, immune evasion mechanisms affecting the MHC class I presentation pathway have evolved in MCMV to downregulate the presentation of MCMV-derived peptides on the cell surface (19). Seminal experiments characterizing MCMV immunity showed that, beginning in the early phase of viral replication, CTLs specific for an IE antigen could no longer recognize and lyse MCMV-infected cells (13). Levels of surface expression of MHC class I molecules did not appear to be affected at this time, and CTLs specific for an early antigen were able to lyse infected target cells, indicating the specific inhibition of pp89 presentation by early genes. However, later in the early phase, surface expression of MHC class I molecules is severely reduced as a result of MCMV early gene products blocking transport of peptide-loaded MHC class I complexes into the medial-Golgi compartment (12). This block appears to be mediated, at least in part, by the m152 early gene product (46, 50). This retention of class I complexes and the resultant prevention of antigen presentation were found to be abolished in infected cells treated with gamma interferon (IFN-γ) in vitro (20). Another MCMV gene, m06, was found to encode the glycoprotein gp48, which strongly associates with MHC class I molecules in the endoplasmic reticulum and reroutes them into the endosomal-lysosomal compartment for rapid proteolysis, thus preventing class I surface expression by an independent mechanism. Therefore, although CTLs directed against several viral epitopes may be generated during infection, there may exist only a limited number of epitopes that are sufficiently presented by the infected cell to trigger lysis of the infected cell at times early enough to be protective for the host.
Several lines of evidence point to the existence of other viral targets of CTLs during infection, although their identities have remained elusive. Reddehase and colleagues showed that target cells incubated with increasing levels of UV-inactivated virus became more susceptible to CTL-mediated cytolysis by splenocytes from MCMV-immune mice (37). Thus, target cells presenting virion proteins were specifically lysed by T cells generated during the natural infection. MHC-restricted T-cell clones have also been isolated and characterized as being specific for virion (36) and early-phase (13) antigens. Particularly noteworthy is a recent report in which pulmonary infiltrate cells in BALB/c bone marrow recipients were examined for relative CTL activities specific for the well-characterized IE1 nonapeptide versus the total CD8+ cytolytic activity. The total activity was measured by CD3ε-redirected lysis in order to bypass differences in TCR affinities for MHC-viral peptide complexes (21). A critical finding from these studies was that pulmonary CTLs recognized infected cells in all phases of the viral replicative cycle and that IE1-specific lysis accounted for only a fraction of the total CTL-mediated lysis in BALB/c mice. In addition, it was shown that target cells expressing early antigens were the most susceptible to lysis by pulmonary CTLs harvested during the peak of lytic activity. In view of the apparent shutdown of antigen presentation beginning during the early phase of viral gene expression, a requisite property of protective MCMV antigens may be that they either are expressed very early in the infection or are virion proteins introduced into the cytoplasm upon cell membrane-viral envelope fusion. However, the lack of knowledge regarding the specific identities of CTL targets of MCMV other than pp89 prevents this question from being rigorously addressed.
The goals of the work presented here were (i) to identify other MCMV antigens that generate protective cellular immunity in BALB/c mice and (ii) to determine whether this protection is limited to the H-2d haplotype. We previously showed that intradermal (i.d.) DNA immunization with a pp89-expressing plasmid elicits a protective CTL response in BALB/c mice. In this study, we extended these findings by constructing plasmid DNA vaccines encoding candidate MCMV antigens and testing their protective efficacies against subsequent MCMV challenge. Because MCMV-specific CTLs against early antigens as well as virion proteins are likely generated during the infection of BALB/c mice, we focused this study on an early antigen, M84, and the MCMV homologs of HCMV virion proteins. The M84 gene was chosen based on our previous work showing that the protein encoded by this gene was an early, nonstructural protein with strong homology to the HCMV CTL target pp65 (34). Other genes included in this study were the homologs of seven tegument and putative tegument antigens and two putative capsid antigens. Two inbred mouse strains of different H-2 haplotypes were immunized by injecting these plasmids either singly or in cocktails of multiple plasmids. We found that in BALB/c mice only plasmids expressing either the nonstructural pp89 or M84 protein could limit viral replication in the spleen following subsequent challenge with a wide range of sublethal MCMV doses. None of the other plasmids elicited consistent protection. Moreover, these genes were not protective even when delivered by immunizing the mice with recombinant vaccinia viruses expressing the same MCMV open reading frames (ORFs). The results also showed that coimmunization of BALB/c mice with the protective pp89 and M84 plasmids provided synergistic protection against viral replication in the spleen. The response to M84 to was found to be specific for M84 epitopes, as mice immunized with the M84-expressing plasmid were not protected against an MCMV deletion mutant lacking M84. The reductions in spleen titers were accompanied by smaller reductions in salivary gland titers, indicating that spread of virus to secondary organs of replication was only partially limited by these vaccines. Lastly, we found that the protective responses generated by our vaccines were haplotype dependent, as pp89 was the only antigen that provided detectable protection in C3H/HeN mice.
MATERIALS AND METHODS
Mice.
Only female, specific-pathogen-free inbred mice were used in these experiments. Mice were at least 6 weeks old upon arrival and were not injected with DNA or MCMV until at least 1 week later. BALB/c (H-2d) mice were obtained from Harlan-Sprague-Dawley, Inc., or Simonsen Laboratories, Inc., at 5 to 6 weeks of age. C3H/HeN (H-2k) mice were obtained from Simonsen Laboratories at 5 to 6 weeks of age. Mice were housed in microisolator-covered cages in a vivarium (University of California, San Diego) and given food and water ad libitum.
Cells and virus.
NIH 3T3 cells (ATCC CRL 1658), CV-1 cells (ATCC CCL 70), and HeLa S3 cells (ATCC CCL 2.2) were maintained in Dulbecco's modified Eagle's medium containing 10% (vol/vol) heat-inactivated bovine calf serum and 200 U of penicillin, 0.2 mg of streptomycin, 0.05 mg of gentamicin, 1.5 μg of amphotericin B, and 0.29 mg of l-glutamine per ml. MCMV strain K181 and the M84 deletion mutant ΔM84 (34) were prepared as salivary gland homogenates following intraperitoneal (i.p.) injection of female BALB/c mice as previously described (15). The titers of these viruses were determined on NIH 3T3 cells as described below.
Plasmid constructs.
Restriction endonucleases, T4 DNA ligase, T4 DNA polymerase, Klenow fragment of DNA polymerase, and competent Escherichia coli cells (DH5α MAX) were obtained from Bethesda Research Laboratories, Inc., unless otherwise noted. DNA restriction fragments contained in agarose gels were purified using the Geneclean Kit (Bio 101) or by centrifugation of gel slices in 0.45-μm-pore-size Ultrafree-MC filter columns (Millipore) according to the manufacturer's recommendations. Unless otherwise specified, phosphorylated linker DNAs were obtained from Stratagene, Inc., and linker ligations and digestions were performed as described previously (40). When appropriate, orientations of MCMV sequences in recombinant clones were confirmed by restriction analysis. Construction and characterization of pACYC184-based plasmids containing the HindIII fragments of the MCMV genome (i.e., H3B, H3D, H3H, etc.) were described previously (33). To allow easier cloning of a number of ORFs into the vaccinia virus vector plasmid pSC11 (6), this plasmid was modified to place either a NotI, NheI, or KpnI restriction site at a position allowing expression of the cloned gene in recombinant vaccinia viruses under the control of the vaccinia virus p7.5 promoter. The pSC11 vector was digested with SmaI, then phosphorylated NotI (Promega), NheI, or KpnI linkers were ligated to the cut plasmid, and the plasmid was then digested with an excess of NotI, NheI, or KpnI, respectively. The linker-ligated plasmid was then isolated and recircularized, yielding pSC11(Not), pSC11(Nhe), or pSC11(Kpn), respectively. Restriction analysis in all three cases demonstrated placement of the introduced site at a position corresponding to the SmaI site of the parent plasmid.
For i.d. DNA immunization experiments, full ORFs encoding pp89 (IE1), M99 (pp28), M32, M82, M83, M84, M85, M86, M48, M56, and M69 were each subcloned into the eukaryotic expression vector pcDNA3 (Invitrogen). Construction of pcDNA3-pp89, pcDNA3-M82, pcDNA3-M83, and pcDNA3-M84 has been described previously (11). To subclone the M82 ORF into the vaccinia virus vector pSC11(Nhe), pcDNA3-M82 was cleaved with EcoRI, blunt ended, and ligated to a phosphorylated XbaI linker. The DNA was then cut with an excess of XbaI, and a 2.2-kbp fragment was isolated and ligated to NheI-cut pSC11(Nhe), yielding pSC11-M82. To clone the M84 ORF into a vaccinia virus vector plasmid, pcDNA3-M84 was cut with KpnI, and a 2-kbp fragment, containing the entire M84 ORF, was isolated and ligated to KpnI-cut pSC11(Kpn), yielding pSC11-M84. To clone the M99 ORF into pcDNA3, pGS-M99 (formerly pGS-UL99 [10]) was digested with BamHI and the 2.6-kbp fragment containing M99 was isolated and ligated to BamHI-cut pcDNA3.
The M32 ORF was subcloned from the HindIII B region of the MCMV genome into pcDNA3 as follows. pACYC184-H3B was digested with HindIII and SstII to release a 4.2-kbp SstII fragment containing the entire M32 ORF. This fragment was isolated and cloned into pBluescript II KS(+) (Stratagene) to yield pBS-4.2 H3B, a clone containing the 5′ end of the ORF proximal to the EcoRV site of the vector. To remove flanking sequences 5′ of the M32 ORF, pBS-4.2 H3B was cleaved with EcoRV and a 5.8-kbp fragment containing the M32 ORF, some upstream and downstream sequences, and the vector backbone was isolated and recircularized to yield pBS-M32. pBS-M32 was cut with EcoRV, and NotI linkers (Promega) were ligated to the ends. After NotI digestion, the plasmid was recircularized to yield pBS-5′Not M32. This plasmid was cut with SstII and DraI, blunt ended with T4 DNA polymerase, and then cut with HindIII to yield the entire M32 ORF on a 2.7-kbp fragment with a 5′ HindIII end and a blunt 3′ end. This 2.7-kbp fragment was isolated and ligated to HindIII- and EcoRV-cut pcDNA3 to yield pcDNA3-M32. To construct the M32-expressing vaccinia virus, pcDNA3-M32 was cut with NotI, and a 2.7-kbp fragment, containing the entire M32 ORF, was isolated and ligated to NotI-cleaved pSC11(Not), yielding pSC11-M32.
In order to construct pcDNA3-M85, the M85 ORF-containing plasmid H3C8.2-GEM (11) was digested with AatII and a 1.0-kbp fragment was isolated. This fragment was ligated to AatII-cut pGEM-7Zf(+), and a clone which contained the insert with the 5′ end of the M85 ORF proximal to the Sp6 promoter site of the vector was selected and designated pGEM-M85. pGEM-M85 was cut with AatII, and an adapter containing a NotI site and an AatII overhang, 5′-CAG CGG CCG CTG ACG T-3′ (Integrated DNA Technologies, Inc.), was ligated to the cut DNA. The end-adapted DNA was cut with NotI, and a 1.0-kbp fragment was isolated and ligated to NotI-cut pcDNA3, yielding pcDNA3-M85, or NotI-cut pSC11(Not), yielding pSC11-M85.
To construct pcDNA3-M86, the plasmid H3C14.2-GEM (11) was digested with KpnI and the 8.2-kbp fragment containing M86 was isolated and ligated to KpnI-cut pGEM-4Z. A clone with the 5′ end of the ORF proximal to the EcoRI site of the vector was isolated and designated H3C8.2-GEM(rev). To eliminate 3.1 kbp of 3′ flanking sequence from the M86 ORF, H3C8.2-GEM(rev) was cleaved with SphI, and the 7.8-kbp M86- and pGEM-4Z-containing fragment was isolated and recircularized to yield pGEM-iM86. This construct was digested with Eco47III (New England Biolabs) and SphI and then blunt ended. Phosphorylated XbaI linkers were then ligated to the blunt ends, and following digestion with XbaI, the 4.5-kbp fragment containing M86 was subcloned into XbaI-cleaved pcDNA3 to yield pcDNA3-M86 or NheI-cut pSC11(Nhe) to yield pSC11-M86.
To subclone the M69 ORF into pcDNA3, pACYC184-H3D was cleaved with SphI and the 3.8-kbp fragment was isolated. This M69-containing fragment was ligated to SphI-digested pGEM-4Z, and a clone containing the insert with the 3′ end of the M69 ORF proximal to the Sp6 promoter site of the vector was selected and designated pGEM-M69. This plasmid was cleaved with SnaBI (New England Biolabs), and HindIII linkers were ligated to the blunt ends. The HindIII-linkered DNA was then digested with HindIII, EcoRI, and DraI, and a 3.0-kbp 5′-HindIII to 3′-EcoRI fragment containing M69 was isolated and ligated to HindIII- and EcoRI-cleaved pcDNA3 to yield pcDNA3-M69.
To subclone the M48 ORF, pACYC184-H3H was digested with MscI and EcoRV, and HindIII linkers were ligated to the blunt ends. After digestion with HindIII, the 9.3-kbp HindIII fragment containing the M48 ORF was isolated and ligated to HindIII-cleaved pGEM-4Z to yield pGEM-M48. pGEM-M48 was cut with NheI, and the overhangs were filled in with Klenow fragment. NotI linkers were ligated to the blunt ends, and following digestion with NotI and HindIII, a 6.8-kbp HindIII-to-NotI fragment was isolated and ligated to HindIII- and NotI-cleaved pcDNA3 to yield pcDNA3-M48.
The M56 ORF was subcloned by first digesting pACYC184-H3D with HindIII and EcoRI. The 5.2-kbp EcoRI-to-EcoRI fragment containing M56 was isolated and subcloned into the EcoRI site of pGEM-4Z to yield pGEM-M56. pGEM-M56 was digested with BssHII, and the ends were filled in with Klenow fragment. After digestion with StuI, NotI linkers were ligated to the blunt ends. Following digestion with NotI, the 2.55-kbp fragment containing M56 was isolated and ligated into the NotI site of pcDNA3 to yield pcDNA3-M56.
To clone the MCMV early transcription unit e1 (5) into a vaccinia virus vector, pACYC184-EcoH, containing the 11-kbp EcoRI H fragment of MCMV, was cleaved with BamHI and EcoRI, and a 1.5-kbp fragment, containing the e1 gene, was isolated and ligated to BamHI- and EcoRI-cleaved pGEM1 (Promega), yielding e1-GEM. This plasmid was digested with AgeI (New England Biolabs) and EcoRI, and a 1.4-kbp fragment was isolated, blunt ended, and ligated to the SmaI-cleaved vaccinia virus vector pGS20, yielding pGS-e1.
Expression of MCMV ORFs.
Expression of full ORFs from the pcDNA3-based plasmids was assessed prior to immunization. Antigens were expressed and [35S]methionine-labeled by TnT (Promega) coupled in vitro transcription-translation using the T7 promoter in the vector and following the manufacturer's recommendations. Labeled translation products were added to Laemmli sodium dodecyl sulfate (SDS) sample buffer, heated to 100°C (or 42°C for M83 and M84) for 2 min, separated by SDS-polyacrylamide gel electrophoresis on gels with various acrylamide concentrations ranging from 5 to 15%, and visualized by fluorography or autoradiography.
Vaccinia virus construction.
Recombinant vaccinia viruses expressing the MCMV M32, M82, M84, M85, M86, and e1 ORFs or β-galactosidase were constructed using the vaccinia virus vector plasmids pSC11-M32, pSC11-M82, pSC11-M84, pSC11-M85, pSC11-M86, pGS-e1, and pSC11, respectively, and the WR strain of vaccinia virus as the parent virus (ATCC VR-119) using methods previously described (6, 23, 31, 47). The resulting recombinant viruses were designated M32-vacc, M82-vacc, M84-vacc, M85-vacc, M86-vacc, e1-vacc, and pSC11-vacc, respectively. Construction of the vaccinia virus recombinants expressing M83 (M83-vacc), M99 (M99-vacc, formerly UL99-vacc), and pp89 (pp89-vacc) was described previously (10, 11, 47). Recombinant viruses were grown and plaque purified on the 143B cell line (ATCC CRL 8303) under selection with 50 μg of 5-bromo-2′-deoxyuridine per ml. Southern blot analysis using probes to either the thymidine kinase region of the vaccinia virus genome or the MCMV gene to be inserted showed the expected hybridization pattern consistent with the appropriate insert DNA and also confirmed the homogeneity of the plaque-purified stock (data not shown). Large-scale viral stocks were prepared by growth of the plaque-purified virus on monolayers of HeLa S3 cells. Virus stocks were prepared by freeze-thawing infected cell pellets, prior to storage in aliquots at −70°C. Viral stock titers were determined by plaque assay on monolayers of CV-1 cells after treatment of the viral stock for 30 min at 37°C with 0.25% (vol/vol) trypsin. Production of immunoreactive products of the expected sizes from M99-vacc, M83-vacc, M84-vacc, M32-vacc, pp89-vacc, and e1-vacc was confirmed by Western blot analysis of infected cell extracts (data not shown).
Immunizations.
Plasmid DNAs were prepared from standard Luria-Bertani cultures using Qiagen Maxi and Mega columns according to the manufacturer's instructions. Plasmids were resuspended in Tris-EDTA (pH 8.0) made with endotoxin-free ddH2O (Life Technologies, Inc.). The endotoxin content of the DNA was reduced to 0.5 to 5 ng per mg of DNA, as measured by the Limulus amoebocyte lysate assay (Associates of Cape Cod, Inc., Woods Hole, Mass.), by four extractions with Triton X-114 essentially as described previously (35). Following ethanol precipitation, DNA pellets were washed with 70% ethanol (in endotoxin-free ddH2O) and resuspended in endotoxin-free 10 mM Tris-HCl (pH 8.0) to a concentration of approximately 2 mg per ml. DNA concentrations were measured by A260, and the plasmid DNA was verified by agarose gel electrophoresis to be >95% supercoiled, undegraded, and free of chromosomal DNA or RNA contamination. For injection, DNA was diluted in 10 mM Tris-buffered, endotoxin-free saline (pH 8.0).
For DNA immunizations, mice were i.d. injected with 30 to 50 μg of DNA (see Results) using a 0.5-ml insulin syringe equipped with a 28-gauge needle (U-100; Becton Dickinson & Co.). Mice were immunized three times in 10 to 14 days, with each immunization consisting of three injections of DNA into separate sites of the shaved skin of the lower back within 1.5 cm of the base of the tail.
For immunization with recombinant vaccinia viruses, mice were injected i.p. with 0.5 ml of phosphate-buffered saline (PBS) containing 107 PFU of recombinant vaccinia virus grown on HeLa S3 cells. In one experiment (see Results), mice were boosted 31 days following the initial vaccination. Mice were challenged i.p. with virulent MCMV K181 12 or 21 days following the last vaccination (see Results).
Virus challenge, tissue harvest, and plaque assay.
Two to four weeks after the last immunization, mice were challenged by i.p. injection of virulent MCMV strain K181 in 0.5 ml of sterile PBS. At various times postchallenge, mice were sacrificed by CO2 asphyxiation, spleens or salivary glands were aseptically removed, and 10% (wt/vol) homogenates were made in Dulbecco's modified Eagle's medium–10% bovine calf serum–10% dimethyl sulfoxide using sterile Pyrex homogenizers. Homogenates were stored at −70°C. All organ homogenates from a single experiment were diluted, and the titers were determined simultaneously by plaque assay on NIH 3T3 cells as previously described (17). Plaques were counted on day 4 or 5 postinfection (p.i.). Unless otherwise indicated, organ titers presented are the means of the log10 PFU per organ for four mice and error bars representing either the standard deviation or, for better visibility of data points in most line graphs, the standard error of the mean.
Statistical analysis.
MCMV organ titers were compared by one-factor analysis of variance (ANOVA) and Fisher's protected least significant difference post hoc test for pairwise comparisons between groups.
RESULTS
Expression of MCMV antigens from plasmid DNA vaccine vectors.
The genome of MCMV contains approximately 170 ORFs, and of all of the virally encoded antigens, only one IE antigen—IE1-pp89—has been identified as eliciting a protective CTL response in infected BALB/c mice. Based upon work by Koszinowski and Reddehase documenting the presence of other specific CTL populations, it appears that other CTL epitopes may be contained within viral early-phase and structural proteins (13, 36). In order to begin the identification of antigens that generate the protective CTL response against MCMV, we first isolated the coding sequences for several candidate MCMV antigens for subsequent testing in a plasmid DNA immunization-based protection assay. The virion proteins that we tested were the MCMV homologs of known HCMV tegument and capsid proteins (see Table 1 for list). The homology between the published DNA sequences of HCMV strain AD169 (7) and MCMV strain Smith (35) facilitated our subcloning of these MCMV homologs from the K181 genome into the mammalian expression vector pcDNA3 for strong HCMV major IE promoter-enhancer-driven expression of the ORFs in vivo. The tegument and putative tegument antigens of HCMV include UL32-pp150, UL48-p212 and UL56-p130 (3), pUL69 (49), UL82-pp71, UL83-pp65, and UL99-pp28, and their respective FASTA amino acid homologies are shown in Table 1. The major and minor capsid antigens of HCMV encoded by UL86 and UL85, respectively, share approximately 50% FASTA amino acid homology with their MCMV counterparts, M86 and M85, and both of these MCMV genes were subcloned for immunization trials.
TABLE 1.
HCMV antigens and their MCMV homologs used in DNA immunization experimentsa
| HCMV ORF | HCMV Ag | MCMV ORF | Identity (%)b |
|---|---|---|---|
| UL32 | pp150 | M32 | 25 |
| UL48 | p212 | M48 | 22 |
| UL56 | p130 | M56 | 42 |
| UL69 | pUL69 | M69 | 25 |
| UL82 | pp71 | M82 | 19 |
| UL83 | pp65 | M83 | 17 |
| M84 | 21 | ||
| UL85 | mCP | M85 | 55 |
| UL86 | MCP | M86 | 53 |
| UL99 | pp28 | M99 | 35 |
| UL123 | pp72 | m123 (pp89) | —c |
Abbreviations: Ag, antigen; mCP, minor capsid protein; MCP, major capsid protein.
Percent amino acid identity over the overlap identified by FASTA (35). Also see reference 11 for UL82-UL84 homologies with M82-M84.
No significant amino acid sequence homology, though position and function of pp72 and pp89 are conserved (35).
In our previous studies, we noted that the M84 ORF of MCMV possesses greater homology to UL83 than does the positional homolog M83 (11). Further analysis of the products of these two genes revealed that the M83 gene product was similar to HCMV pp65 by virtue of its late expression, phosphorylation in vivo, and virion association and that the M84-encoded protein is an early, nonstructural protein (11, 34). Based upon these latter findings, we included the M84-expressing plasmid in our immunization studies. Since we previously demonstrated that a pp89-expressing plasmid vaccine provides protection against MCMV infection in BALB/c mice (17), the pp89 cDNA was subcloned into pcDNA3 as a control for these immunization experiments.
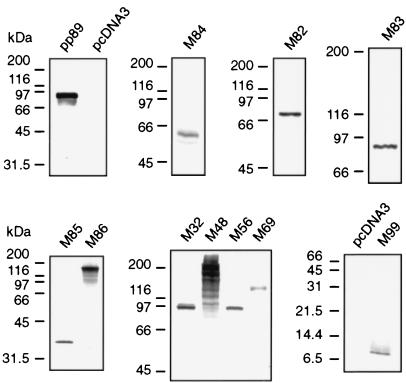
To confirm expression of antigens from the pcDNA3-based vaccine plasmids, all recombinant proteins were expressed by coupled in vitro transcription-translation reactions driven from the T7 promoter in the vector. Molecular masses of the [35S]methionine-labeled polypeptides (Fig. 1) correlated well with those predicted from the amino acid sequences (35), except that the M69 polypeptide migrated to a molecular mass approximately 25 kDa higher than the expected 93 kDa. This anomalous migration of M69 is likely due to large stretches of charged amino acids and/or a large percentage of serines predicted by its coding sequence.
FIG. 1.
Coupled in vitro transcription-translation of pcDNA3-based vaccine plasmids. All antigens encoded by the plasmids used for immunization of mice were expressed in vitro from the T7 promoter of the plasmid vector using the TnT (Promega) expression system. [35S]methionine-labeled translation products were denatured in reducing SDS-polyacrylamide gel electrophoresis buffer, electrophoresed on gels of various acrylamide concentrations ranging from 5 to 15%, and visualized by fluorography or autoradiography. Each panel represents a different expression experiment and gel with the migration of the molecular mass standards (in kilodaltons) in each gel shown at the left.
The relative protective effect elicited by pcDNA3-pp89 immunization increases with increased MCMV challenge dose.
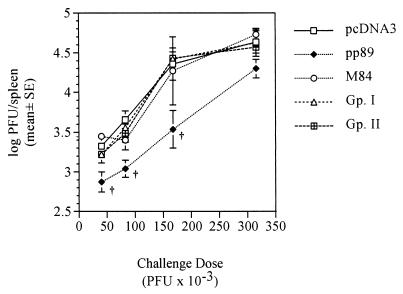
After demonstrating that immunization with pp89-expressing plasmid pcDNA-89 can protect BALB/c mice against both lethal and sublethal MCMV challenges (17), we proceeded to optimize the experimental conditions to increase our probability of identifying other potentially protective antigens. Although pcDNA-89 provides protection against lethal challenge with virulent virus, we found that this protection could be overwhelmed by increases of the lethal challenge dose occurring from normal experimental variation in virus administration. We therefore sought to develop a protection assay which used a sublethal challenge in order to prevent the small variations in challenge dose from acutely affecting the measure of protection. To this end, we titrated the level of challenge dose to find a sublethal dose that would not overwhelm the protective response from plasmid DNA immunization. We measured the replication of MCMV in the spleens of vaccinated mice on day 6 postchallenge, as these replication levels correlate well with mortality of BALB/c mice from MCMV infection. In addition, control of the acute MCMV infection in the spleens of BALB/c mice has been found to be CD8+ T cell mediated (24), and we previously demonstrated that a specific CTL response can be elicited by plasmid DNA immunization with a pp89-expressing plasmid (17).
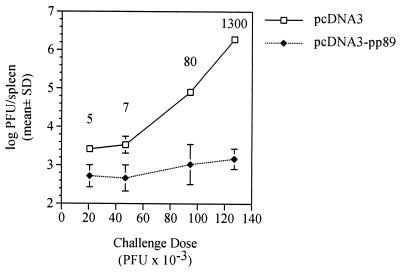
Four BALB/c mice per group were immunized by i.d. injections of 50 μg of pcDNA3 or pcDNA3-pp89 in endotoxin-free saline. Mice were injected three times over 10 days, and then, 2 weeks after the last immunization, the mice were i.p. challenged with either 21 × 103, 47 × 103, 95 × 103, or 127 × 103 PFU (approximately 0.13, 0.25, 0.5, or 0.75 × 50% lethal dose [LD50], respectively) of salivary gland-derived MCMV K181 (Fig. 2). Mice were sacrificed on day 6 postchallenge, and their spleens were aseptically removed, Dounce homogenized, and stored until plaque assay on NIH 3T3 cells. As seen in Fig. 2, the levels of MCMV replication in the spleens of the pcDNA3-immunized mice increased approximately 1,000-fold over the range of increasing challenge doses. In contrast, over the same range of MCMV challenge doses, the resulting spleen titers in the pcDNA3-pp89-immunized mice increased only 2.5-fold. At the greatest sublethal dose tested, corresponding to approximately 0.75 × LD50, MCMV titers in the pcDNA3-pp89-immunized mice were 1,300-fold lower than those in the pcDNA3-alone-immunized mice. Thus, for the pp89-expressing plasmid, the fold protection level relative to controls increased with increasing challenge dose, with the greatest protective effect observed after challenge with 0.75 × LD50. Since other antigens may elicit a response different in either nature or magnitude from that elicited against pp89, the protection of mice immunized with the other MCMV antigens was measured across this entire range of challenge doses. In a separate experiment, we also determined that similar protection was afforded by immunization with 10 or 50 μg per injection (data not shown), and therefore the doses of plasmid DNA did not appear to be limiting in this immunization assay.
FIG. 2.
The relative protective effect elicited by pcDNA3-pp89 immunization, as measured by replication in the spleen, increases with increasing viral challenge dose. Naive BALB/c mice were immunized three times in 10 days by i.d. injections of 50 μg of either pcDNA3 or pcDNA3-pp89 as described in Materials and Methods. Two weeks after the last immunization, mice were i.p. challenged with one of four serial dilutions of salivary gland-derived MCMV corresponding to 21 × 103, 47 × 103, 95 × 103, and 127 × 103 PFU in 0.5 ml of PBS per mouse. Six days postchallenge, spleens were removed and the viral titers were determined. Spleen titer values presented are the means of the log10 PFU per spleen for four mice with the standard deviation (SD) indicated by error bars. Values above each challenge dose indicate the fold reductions of the nonlogarithmic mean titers (i.e., PFU per organ) of pcDNA3-pp89-immunized mice relative to the corresponding pcDNA3-immunized controls.
Plasmid DNA immunization with M84, either alone or in combination with other expression plasmids, protects BALB/c mice against challenge with increasing sublethal doses of MCMV.
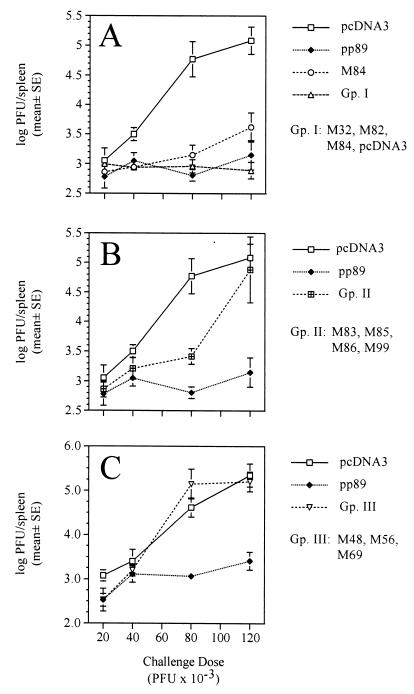
Since we were able to elicit consistent protection against a wide range of sublethal challenge doses following plasmid immunization with pp89, we used this challenge strategy as a measure of the protective abilities of the other plasmids encoding MCMV antigens. In order to efficiently screen a dozen additional antigens in mice of various MHC backgrounds, we chose to immunize groups of mice with mixtures of the different plasmids rather than immunize with each plasmid alone. This method is roughly analogous to that used by Barry and coworkers (1), who screened pools of expression clones from sublibraries made from the genome of Mycoplasma pulmonis, although their immunization pools contained large numbers of uncharacterized plasmids. If we found a particular plasmid pool to be protective in the initial screen, then its constituent plasmids would be used singly in a later immunization experiment to determine which antigen(s) provided the protective effect. Because the results of a pilot immunization experiment using a single sublethal challenge dose suggested that pcDNA3-M84 may provide protection (data not shown), the M84-expressing plasmid was singled out to be used alone as well as in combination with other expression plasmids. The pcDNA3-based vaccine plasmids were divided into three groups of plasmid cocktails. pcDNA3-M84 was included in group I (Gp. I) with M32 and M82—two antigens which did not confer protection in pilot immunization or recombinant vaccinia virus immunization experiments (see below). Gp. I consisted of equal masses of pcDNA3-M32, -M82, and -M84, as well as pcDNA3 vector alone added to normalize the injected DNA mass with that of Gp. II. Gp. II contained pcDNA3-M83, -M85, -M86, and -M99, and Gp. III contained pcDNA3-M48, -M56, and -M69.
In the first screen of the vaccine plasmids, four BALB/c mice per group were immunized i.d. three times over 2 weeks with pcDNA3, pcDNA3-pp89, pcDNA3-M84, Gp. I, or Gp. II. Each injection consisted of 40 μg of total DNA, with 10 μg of each antigen-expressing plasmid and the balance made up with pcDNA3. Two weeks after the last immunization, mice were challenged with one of four challenge doses of virulent virus as described above, and the spleen titers on day 6 postchallenge were determined. For display purposes, the data for Gp. I and II are presented in two separate panels of Fig. 3 with the results for pcDNA3 and pp89 included in both panels for comparison. It should be noted that when the titer of an organ was below the limits of assay sensitivity, generally 102 PFU/organ, the titer for that organ was arbitrarily set to the limit of sensitivity for calculation of the mean and standard deviation. Thus, values corresponding to the organ titer reduction for that group of mice relative to the pcDNA3-alone-immunized controls were underestimates. Consistent with our previous findings, we found that the MCMV titers in the spleens of pcDNA3-immunized mice increased from 103 to greater than 105 PFU per spleen over the range of challenge doses tested, while the titers in the pp89-alone group remained at a relatively constant 102.9 PFU per spleen (Fig. 3A and B). Compared to the pcDNA3 group, spleen titers in the M84-immunized group were also consistently reduced at all of the challenge doses, although the suppression of viral replication afforded by the M84 plasmid was less than that for pp89. The average spleen titers in the M84 group increased from 102.8 at the lowest challenge dose to only 103.6 at the highest dose, an increase of approximately sixfold. When pcDNA3-M84 was included in Gp. I with M32 and M82, the resulting protective effect of M84 was still observed and even slightly increased compared to that for M84 administered alone (Fig. 3A). Immunization with the Gp. II DNAs (Fig. 3B) resulted in some protection at the lower challenge doses, with an approximately 20- to 25-fold reduction in MCMV titers compared to that for the pcDNA3-immunized mice subjected to the same challenge dose of 80 × 103 PFU per mouse. However, unlike the protection observed in the pp89-, M84-, and Gp. I-immunized groups, protection elicited by the Gp. II plasmids was overwhelmed at the highest challenge dose. Furthermore, the antigens represented in Gp. II were not found to generate consistent protection against a variety of challenge doses when used singly and delivered by recombinant vaccinia viruses (see below and data not shown). Moreover, when the M82, M83, and M32 plasmids were tested singly for protective efficacy after we increased the plasmid dose from 10 to 50 μg per injection, we found results similar to those above (data not shown).
FIG. 3.
Plasmid DNA immunization of BALB/c mice with pcDNA3-based vectors used alone or in combination and the reductions in spleen titers following increasing sublethal challenge doses of virulent MCMV. BALB/c mice were i.d. immunized three times over 2 weeks with 40 μg of DNA consisting of either pcDNA3 or a cocktail of 10 μg of each antigen-expressing plasmid to be tested and the balance of 40 μg consisting of pcDNA3. Two weeks after the last immunization, mice were i.p. challenged with either 20 × 103, 40 × 103, 80 × 103, or 120 × 103 PFU, and spleens were harvested 6 days postchallenge for MCMV titer determination. Spleen titer values presented are the means of the log10 PFU per spleen for four mice with the standard error (SE) of the mean indicated by error bars. (A) Spleen titers of mice immunized with either pcDNA3, pcDNA3-pp89, pcDNA3-M84, or Gp. I. (B) Spleen titers from the same pcDNA3- or pcDNA3-pp89-immunized mice as in panel A as well as the Gp. II-immunized mice. (C) Spleen titers from an independent experiment in which mice were immunized with 30 μg of pcDNA3, 10 μg of pcDNA3-pp89 plus 20 μg of pcDNA3 to normalize masses), or 30 μg of Gp. III (10 μg of each plasmid shown).
In a separate experiment, the protective abilities of the MCMV homologs of the HCMV gene products UL48, UL56, and UL69 were tested using the approach described above. Mice were immunized with either pcDNA3, pcDNA3-pp89, or Gp. III and then challenged 2 weeks after the last immunization with i.p. inoculation of the same virus and doses used previously. In this experiment, while pp89 provided protection at all of the challenge doses, most notably at the higher doses, the viral titers in the spleens of the Gp. III-immunized mice were comparable to those of the pcDNA3-immunized mice (Fig. 3C). We conclude from this result that M48, M56, and M69 do not elicit a protective response in BALB/c mice.
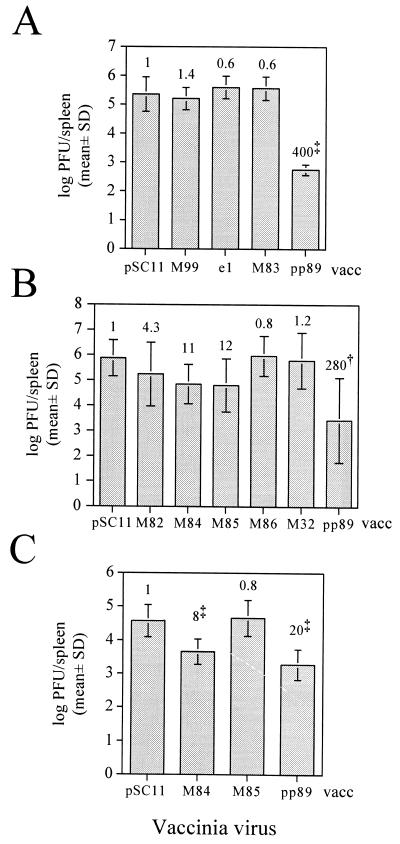
In addition to using plasmid DNA vaccine-based immunization of mice to identify protective MCMV antigens, we also immunized mice with recombinant vaccinia viruses as an alternative delivery and expression system for some of the MCMV antigens tested above. Vaccinia virus has been used as a model system for intracellular expression of antigens for generation of CTL responses, and this model was used extensively to characterize pp89-mediated immunity to MCMV. We constructed vaccinia virus recombinants of strain WR that express the same M32, M82, M83, M84, M85, M86, M99, and pp89 coding sequences used for plasmid vaccine construction such that expression was driven by the vaccinia virus p7.5 promoter. These MCMV antigen-encoding vaccinia viruses were designated M32-vacc, M82-vacc, M83-vacc, M84-vacc, M85-vacc, M86-vacc, M99-vacc, and pp89-vacc, respectively. In addition, we constructed a recombinant vaccinia virus, e1-vacc, expressing the early transcription unit e1 (5). Finally, a β-galactosidase-expressing vaccinia virus recombinant, pSC11-vacc, was constructed as a negative control for immunization experiments.
In the first immunization trial, three BALB/c mice per group were i.p. immunized with 107 PFU of the vaccinia virus recombinant pSC11-vacc, M99-vacc, e1-vacc, M83-vacc, or pp89-vacc and then i.p. boosted 31 days after the first vaccination. Twelve days following the second vaccination, mice were i.p. challenged with 40 × 103 PFU of virulent MCMV, and on day 6 postchallenge, spleens were harvested as described above for MCMV titer determination. Similar to the plasmid DNA immunization results above, we found that only the pp89-expressing vaccinia virus provided significant protection in the spleen, with a 400-fold reduction in MCMV titers in the spleens of pp89-vacc-immunized mice relative to the pSC11-vacc-immunized controls (Fig. 4A). In a separate trial, four mice per group were immunized as described above with a single dose of 107 PFU of the other vaccinia viruses expressing putative structural antigens, M82-vacc, M85-vacc, M86-vacc, and M32-vacc, as well the nonstructural antigen-expressing M84-vacc and pp89-vacc. On day 21 postvaccination, mice were challenged with 30 × 103 PFU of MCMV, and on day 6 postchallenge, MCMV titers in the spleen were determined. As in the first vaccinia virus trial, we found that immunization with pp89-vacc provided significant protection (P = 0.005) with a 280-fold reduction in spleen titer relative to that for pSC11-vacc-immunized mice (Fig. 4B). We also found 11- and 12-fold titer reductions in the mice immunized with M84-vacc and M85-vacc, respectively (Fig. 4B). However, these reductions were not statistically significant (P > 0.15). A third trial was performed using 12 mice per group in order to more accurately assess the protective efficacies of M84-vacc and M85-vacc. Following a single immunization as described above, mice were i.p. challenged with 80 × 103 PFU of MCMV. Although MCMV replication in the spleens of the pSC11-vacc-immunized mice was approximately 10-fold lower than that in the other two trials, mice receiving either M84-vacc or pp89-vacc showed statistically significant (P < 0.001) reductions in day 6 spleen titers (Fig. 4C). In this trial, M85-vacc-immunized mice showed MCMV titers similar to those of the pSC11-vacc controls, indicating that M85-vacc-mediated protection may not be consistently protective. Of note, plasmid DNA Gp. II contains the M85 plasmid, and immunization with this plasmid cocktail did not provide protection at the highest challenge dose. In summary, immunization trials using vaccinia virus recombinants expressing the MCMV ORFs yielded results similar to those from the plasmid DNA immunization experiments.
FIG. 4.
Immunization of BALB/c mice with recombinant vaccinia viruses expressing the MCMV ORFs. BALB/c mice were i.p. immunized with the recombinant vaccinia viruses and challenged with virulent MCMV, and spleens were harvested 6 days postchallenge for MCMV titer determination. Spleen titer values presented are the means of the log10 PFU per spleen for each group with the standard deviation (SD) indicated by error bars. Values above each titer bar indicate the fold reductions of the nonlogarithmic mean titers (i.e., PFU per spleen) of that immunized mouse group relative to the corresponding pSC11-vacc-immunized controls, with statistically significant titer reductions denoted as superscripts (∗, P < 0.05; †, P < 0.01; ‡, P < 0.001; one-factor ANOVA and Fisher's protected least significant difference test). (A) Three mice per group were i.p. immunized with 107 PFU of one of the recombinant vaccinia viruses expressing the MCMV antigens shown or the β-galactosidase-expressing pSC11-vacc. A booster injection was given 31 days postimmunization, and 12 days following the boost, mice were i.p. challenged with 40 × 103 PFU of MCMV. (B) Four mice per group were given a single i.p. immunization with 107 PFU of the vaccinia virus recombinants shown and, 21 days postimmunization, were challenged with 30 × 103 PFU of MCMV. (C) Twelve mice per group were immunized as described for panel B and challenged with 80 × 103 PFU of MCMV.
Taken together, DNA immunization of BALB/c mice with plasmids encoding either the immunodominant pp89 protein or the pp65 homolog M84 can protect BALB/c mice from splenic viral replication over a wide range of sublethal challenge doses. Relative to controls, the fold suppression of MCMV replication elicited by either of these plasmids increased with challenge dose, with the protection by pp89 being consistently greater than that afforded by M84. While titers in both of these groups remained low relative to those in vector-alone-immunized controls, the titers were above the limits of detection, indicating that protection was still incomplete. The rationale for screening multiple plasmids by coinjecting them in cocktails was justified by the observation that the protective ability of pcDNA3-M84 was not negatively affected by its coimmunization with nonprotective expression plasmids. However, it is not known whether the immune responses to other protective MCMV antigens could be negatively affected by coimmunization in a plasmid cocktail. Other than pp89 and M84, none of the other antigens tested by plasmid DNA or vaccinia virus immunization were capable of generating significant protection against viral replication in the spleen.
Coimmunization of BALB/c mice with plasmids expressing pp89 and M84 results in synergistic reduction in spleen replication following challenge with a high sublethal dose.
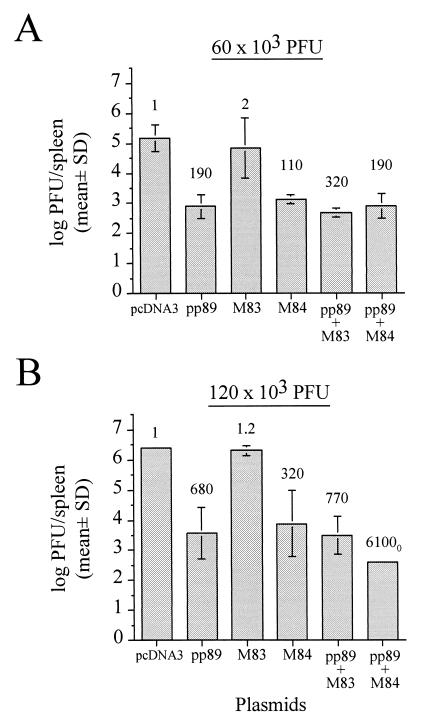
As described above, screening of 11 antigen-expressing plasmids in two inbred mouse strains revealed only two antigens which could protect BALB/c mice from challenge, the major IE1 protein pp89 and the pp65 homolog M84. Protection elicited by either plasmid against MCMV replication was incomplete as evidenced by the presence of detectable virus in the spleen. We therefore sought to determine whether a combination of these two MCMV genes could augment the antiviral response. BALB/c mice were i.d. immunized three times over 2 weeks with either 30 μg of pcDNA3; 15 μg of pp89, M83, or M84 (each combined with 15 μg of pcDNA3 to normalize DNA masses); or a cocktail of 15 μg each of pp89 and M83 or pp89 and M84. Three weeks after the last immunization, vaccinated mice were i.p. challenged with 60 × 103 or 120 × 103 PFU of virulent MCMV. On day 6 postchallenge, spleens were homogenized for MCMV titer determination.
Consistent with the above data, immunization with pp89 or M84 alone resulted in reductions in titer following infection with either challenge dose (Fig. 5). Relative to the spleen titers of pcDNA3-immunized mice, titer reductions of 190-fold for pp89 and 110-fold for M84 were observed after the 60 × 103 PFU challenge (Fig. 5A), and 680- and 320-fold reductions were measured for pp89 and M84, respectively, following the 120 × 103 PFU challenge (Fig. 5B). In contrast, titers in M83-immunized mice were nearly indistinguishable from those in vector-alone controls at either challenge dose. Moreover, mice coinjected with pp89 and the nonprotective M83 plasmids and then challenged with either viral dose showed no significant reduction in spleen titer below that elicited by pp89 immunization alone (Fig. 5). After challenge of the pp89-M84-coimmunized group with the lower challenge dose, the resulting titer reduction level was comparable to that observed with pp89 alone (Fig. 5A). However, when pp89 and M84 plasmids were coinjected into the group of mice and mice were then challenged with 120 × 103 PFU, the resulting spleen titers were reduced to over 6,100-fold below those in the pcDNA3 group, a level approximately 10-fold lower than that for the pp89-alone group and 20-fold lower than that for the M84-alone group (Fig. 5B). In addition, since MCMV was detectable in the spleen of only one of the four mice, the actual reduction in spleen titer might even have been greater, as titers of MCMV-negative mice were arbitrarily set to the detection limit of 102 PFU/spleen (denoted by the subscript 0 in Fig. 5B). Note that, in this coimmunized group, the standard deviation of spleen titers was not sufficiently large to be depicted with an error bar. Taken together, the above results suggest that the immune responses against pp89 and M84 are able to complement each other and interact synergistically. However, the high dose of challenge virus needed to produce this effect suggests that the immune cells primed by the plasmids require a large input viral dose to be rapidly restimulated or to cause them to migrate from the immunization site to the tissues where they are able to alter the course of the systemic infection.
FIG. 5.
Plasmid DNA immunization of BALB/c mice with protective and nonprotective plasmids, either alone or in combination, and the resulting viral spleen titers following sublethal MCMV challenge. BALB/c mice were i.d. immunized three times with either 30 μg of pcDNA3 or a cocktail of 15 μg of each antigen-expressing pcDNA3-based plasmid to be tested and the remainder of 30 μg consisting of pcDNA3. Two weeks after the last immunization, mice were i.p. challenged with either 60 × 103 (A) or 120 × 103 (B) PFU of MCMV. On day 6 postchallenge, spleens were harvested, and the resultant spleen titers are shown as the means of the log10 PFU per spleen for four mice with the standard deviation (SD) indicated by error bars. Values above each titer bar indicate the fold reductions of the nonlogarithmic mean titers (i.e., PFU per organ) of that group relative to the corresponding pcDNA3-immunized controls. The subscript 0 indicates that three of the four mice in this group had MCMV titers below the detection limit of 102 PFU/spleen and that the titer values of each of these three mice were arbitrarily set to the detection limit for mean titer calculation. Note also that the standard deviation of this group was too low to depict with an error bar.
The immune response against pcDNA3-M84 does not protect against challenge with an MCMV mutant lacking M84 expression.
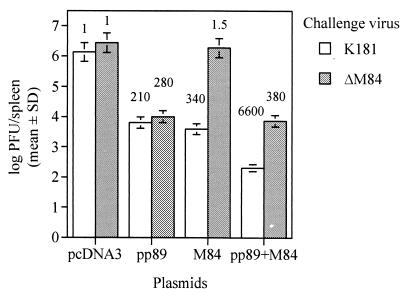
The above results showed that prior immunity against M84 is protective against subsequent MCMV challenge and that coimmunization with both M84 and pp89 results in a synergistic level of protection against a high challenge dose. We previously demonstrated that plasmid DNA immunization with a pp89-expressing plasmid induces CTL responses against the dominant pp89 nonapeptide epitope as strong as those generated during infection with tissue culture-derived MCMV (17). Because M84 is also a nonstructural protein (34), we would expect that the protection generated by the M84-expressing plasmid is also a cell-mediated response. From our experiments above, however, we could not exclude the possibility that protection mediated by pcDNA3-M84 (or M84-vacc) was due to a cytokine response that could result in a more rapid nonspecific priming of naive CTLs or activation of natural killer (NK) cells or, in the case of pp89-M84-coimmunized mice, a more rapid induction of pp89 memory CTLs into cytolytic effectors. We therefore tested whether the protection mediated by pcDNA3-M84 required that the infected cells express the M84 protein. We previously found that M84 was dispensable for viral replication in tissue culture and that a K181 deletion mutant, ΔM84, was able to replicate with slightly attenuated growth levels in the target organs of BALB/c and other strains of mice (34). We therefore tested whether this M84 deletion mutant could escape pcDNA3-M84-based immunity in plasmid DNA-immunized mice.
Four BALB/c mice per group were immunized with pcDNA3, pp89, M84, or the pp89-M84 cocktail as described for Fig. 5. Two weeks after the last immunization, mice were i.p. challenged with 120 × 103 PFU of salivary gland-derived stocks of either wild-type K181 or ΔM84. We found that on day 6 postchallenge, MCMV titers in the spleens of pcDNA3-alone-immunized mice were comparable whether they were inoculated with the wild-type or ΔM84 virus (Fig. 6). Mice immunized with pcDNA3-pp89 alone were found to control replication of either wild-type or ΔM84 viruses to equal levels. Since we previously showed that the levels and kinetics of pp89 protein expression in ΔM84-infected NIH 3T3 cells were identical to those in the wild-type-virus-infected cells (34), the similar viral titers in the spleens of the pp89-immunized mice suggest that the presentation of pp89 in infected cells in vivo is not significantly affected in the absence of M84 expression. Most importantly, Fig. 6 shows that when M84-immunized mice are challenged with wild-type or ΔM84 viruses, only the M84-expressing wild-type virus is reduced in titer with a similar magnitude as that afforded by immunization with pp89. ΔM84, in contrast, replicated in M84-immunized mice to a level similar to that in the pcDNA3-alone-immunized controls. Thus, pcDNA3-M84-mediated responses require that infected cells express M84 in order to be protective. When mice were coimmunized with pp89 and M84 plasmids and challenged with wild-type MCMV, we observed a synergistic reduction level of 6,600-fold in spleen titer relative to that in pcDNA3-immunized–K181-challenged mice. This reduction level is consistent with the pp89-M84 coimmunization-mediated protection shown in Fig. 5. However, when pp89-M84-coimmunized mice were challenged with ΔM84, the synergistic protection was abrogated and a titer reduction level of 380-fold was obtained. This level of protection closely matched that provided against ΔM84 following immunization with only the pp89 plasmid. Therefore, the protective response elicited by pp89 immunization appears to be independent of the M84 response, as the inclusion of the M84 plasmid with the pp89 plasmid did not augment the pp89-specific response against ΔM84.
FIG. 6.
Protection of plasmid DNA-immunized mice against either wild-type K181 or the M84 deletion mutant ΔM84. Four BALB/c mice per group were i.d. immunized as described for Fig. 5, and 2 weeks following the last immunization, mice were i.p. challenged with 120 × 103 PFU of MCMV from salivary gland-derived stocks of either wild-type K181 or ΔM84. Titer values and error bars are as described for Fig. 5. Values above each titer bar indicate the fold reductions of the nonlogarithmic mean titers (i.e., PFU per organ) of that group relative to the corresponding pcDNA3-immunized controls challenged with the same virus. SD, standard deviation.
Immunization with pcDNA3-pp89 alone or in combination with pcDNA3-M84, but not immunization with pcDNA3-M84 alone, results in consistent reductions in salivary gland titer.
Having observed augmented protection from viral replication in the spleens of BALB/c mice coimmunized with the plasmids expressing pp89 and M84, we sought to determine if the limited replication in the spleen (and possibly other abdominal organs) could substantially reduce viral spread to the salivary glands. We found previously that immunization with a pp89 plasmid vaccine resulted in reduced salivary gland titers relative to those observed in mice immunized with vector backbone alone (17), but titers were still approximately 106 PFU/organ. In an independent coimmunization experiment, BALB/c mice were i.d. immunized with the same DNAs as described above. The range of challenge doses chosen (2 × 103, 10 × 103, 30 × 103, and 60 × 103 PFU) was lower than that used when measuring spleen titer reductions because salivary gland titers are maximal following injection of lower input viral doses. Day 10 postchallenge salivary gland titers for the mice immunized with pcDNA3, -pp89, or -84 or pp89-M84 can be seen in Fig. 7. Salivary gland titers of pcDNA3-alone-immunized mice increased from 106.5 to 107.2 PFU/organ over the range of challenge doses and remained maximal at approximately 107 PFU/organ. In mice immunized with pp89 and challenged with the three lowest viral doses, there were approximately twofold (P < 0.05), sixfold (P < 0.001), and eightfold (P < 0.01) reductions in salivary gland titers relative to those for pcDNA3-immunized controls, respectively, while at the highest dose the reduction was threefold (P < 0.05). Thus, the protective effect of pcDNA3-pp89 in the salivary gland increased with increasing challenge dose, but unlike the protection found in the spleen, the titer reductions approached only a single order of magnitude. In contrast to pp89, salivary gland titers in the M84 group were not reduced relative to those for pcDNA3-immunized mice over the range of challenge doses given. When pp89 and M84 were used for coimmunization, salivary gland titer reductions of 3-fold (P < 0.05), 9-fold (P < 0.001), and 11-fold (P < 0.01) were observed following the lower challenge doses and a reduction of 5-fold (P < 0.01) was observed with the highest challenge dose: reductions at least as great as or just measurably greater than those with pp89 immunization alone. However, coimmunization with pp89 and M84 still allowed the virus to replicate to titers of 106 PFU in salivary glands. Thus, while protection in the salivary glands mediated by immunization with pp89 alone or in combination with M84 was statistically significant following the range of the challenge doses tested, the magnitude was less than that observed in the spleen.
FIG. 7.
Plasmid DNA immunization of BALB/c mice with protective plasmids, used alone or in combination, and the resulting salivary gland viral titers following sublethal MCMV challenge. BALB/c mice were i.d. immunized as described for Fig. 5 and 2 weeks after the last immunization were i.p. challenged with either 2 × 103, 10 × 103, 30 × 103, or 60 × 103 PFU of MCMV. Day 10 postchallenge salivary gland titers are shown as the means of the log10 PFU per salivary gland for four mice with the standard error (SE) of the mean indicated by error bars. Statistically significant titer reductions relative to the pcDNA3-immunized mice are indicated as follows: ∗, P < 0.05; †, P < 0.01; and ‡, P < 0.001 (one-factor ANOVA and Fisher's protected least significant difference test).
Plasmid DNA immunization with pcDNA3-pp89, but not pcDNA3-M84, provides detectable protection in C3H/HeN mice.
An optimal vaccine should include antigen(s) that is presented by MHC molecules derived from wide immunogenetic backgrounds. In the MCMV model of immunity, the BALB/c inbred strain has been the primary animal used for determining which antigens were immunodominant for CTL recognition and protection (28). However, it has since been found that the BALB/c strain may rely upon CD8+ T lymphocytes for the control of MCMV to a greater extent than do other inbred mouse strains (29). Other inbred strains possess higher resistance to MCMV due to both H-2 haplotype and non-H-2-linked genes that are responsible for activating NK cells early in the infection before CTLs are fully active. Because the C3H/HeN inbred strain is highly resistant to MCMV due to both its H-2k haplotype (18) and its rapid activation of NK cells by secretion of high levels of IFN-γ, we chose to test the efficacy of the plasmid DNA immunization cocktails in these mice.
Four female C3H/HeN mice per group were i.d. immunized three times over 14 days with pcDNA3, pp89, M84, Gp. I, or Gp. II. Two weeks after the last immunization, mice were i.p. challenged with approximately 40 × 103, 80 × 103, 160 × 103, or 320 × 103 PFU per mouse (see Fig. 8 for actual doses administered). Previous i.p. challenge experiments of mock-immunized or unimmunized C3H/HeN mice had provided us with a challenge dose range which would result in MCMV titers in the spleen which were measurably above the detection limits by plaque assay. Because of the rapid and efficient ability of the C3H strain to control MCMV infection in spleen tissue, spleen titers at day 3, rather than day 6, postchallenge were measured.
FIG. 8.
Plasmid DNA immunization of C3H/HeN mice with pcDNA3-based vectors used either alone or in combination and the reductions in spleen titers following increasing sublethal challenge doses of virulent MCMV. C3H/HeN mice were immunized as described for Fig. 3 and then i.p. challenged 2 weeks following the last immunization with either approximately 40 × 103, 80 × 103, 160 × 103, or 320 × 103 PFU of MCMV. Day 3 postchallenge spleen titer values presented are the means of the log10 PFU per spleen for four mice with the standard error (SE) of the mean indicated by error bars. Statistically significant titer reductions relative to the pcDNA3-immunized mice are indicated as in Fig. 7. Spleen titers are those of mice immunized with either pcDNA3, pcDNA3-pp89, pcDNA3-M84, Gp. I, or Gp. II.
As shown in Fig. 8, only pp89 plasmid immunization was able to generate protection in the C3H/HeN mice. MCMV titers in the pp89-immunized mice rose exponentially as the challenge dose was increased, and the slope was nearly the same as that in the pcDNA3-immunized mice, with the titers in the pp89 groups remaining approximately two- to sevenfold below those for the controls (Fig. 8). While the titer reductions in the pp89-immunized group relative to those for the pcDNA3-immunized controls were statistically significant (P < 0.01) at the three lowest challenge doses, the overall reduction trend contrasts with that observed for BALB/c mice. For BALB/c mice, increasing doses of virus resulted in an increasing relative protection level afforded by the protective plasmid(s). Similar results were also consistently observed in two other independent experiments measuring pp89-mediated protection in the C3H/HeN strain. We also found that, in contrast to the observed response in BALB/c mice, i.d. immunization of C3H/HeN mice with M84 alone or in combination with other MCMV genes (Gp. I) was unable to elicit protection in the spleen against any of the viral challenge doses (Fig. 8). Moreover, Gp. II was not protective in the C3H/HeN strain at any of the challenge doses.
DISCUSSION
The goal of this study was to identify the other potential targets of protective CTLs in BALB/c mice and to determine whether similar protective responses would be generated in a strain with a different H-2 haplotype. Our approach was to use plasmid DNA immunization, an effective and straightforward method for generating CTL responses, to elicit cell-mediated immunity specific for an MCMV early antigen and several MCMV homologs of HCMV virion-associated proteins. These antigens were chosen by virtue of their ability to be presented on the cell surface in association with MHC class I complexes before the early-phase MHC class I shutdown. CTLs directed against these antigens may theoretically lyse the infected cell prior to release and spread of progeny virus. Although CTLs are likely generated to an array of MCMV antigens during the course of infection, our assay was designed specifically to identify those antigens that not only generate CTL responses in vivo but also are protective against challenge with virulent virus.
The results of the experiments presented here showed that of the two MCMV homologs of HCMV UL83-pp65, the M84 protein was protective while the positional homolog M83 was not. Protection was observed in BALB/c, but not C3H/HeN, mice. We previously reported the homologies and possible evolutionary relationships of the M83 and M84 proteins with their HCMV homologs UL83 and UL84 (11). The positional homologs M83 and UL83 are both late proteins that are virion associated. However, UL84 is an early, nonstructural protein, and the M84 protein is also expressed early and is not detectable in the virion (34). In addition, both M83 and UL83 proteins are targets of humoral responses during infection of their respective hosts. Whereas pp65 has been documented to be targeted by the HCMV-specific CTLs across various HLA types in healthy seropositive individuals (2, 26, 32, 48), only the M84 homolog conferred protection to mice following DNA immunization. Because the M84 protein is nonstructural, the protective immunity elicited by the M84 plasmid is almost certainly cell mediated. This is further supported by the demonstration that the M84-mediated protection from challenge required M84 expression in the infected cell. Therefore, the M84 protein likely contains one or more epitopes that may be presented on class I molecules of the H-2d BALB/c strain. The consistently weaker protection levels afforded by M84 relative to pp89 could be due to several factors including in vivo protein expression levels from the vaccine plasmids, relative efficiencies of peptide processing and loading, TCR-MHC affinities, or the relative availability of M84 protein during infection for proteolysis and presentation. Interestingly, recent experiments reevaluating the relative frequencies of CTL responses against HCMV IE1-pp72 and UL83-pp65 in healthy HCMV-seropositive blood donors showed that, in 12 donors tested, all 12 had CTLs specific for either pp72 or pp65, but only 4 had CTLs for both antigens (26).
Although at least some virion proteins elicit specific CTLs during MCMV infection, none of the plasmids or recombinant vaccinia viruses encoding structural or putative structural MCMV antigens was protective in vivo. It could be reasoned that either none of these proteins contained epitopes that could be presented on cells of either of the H-2 haplotypes tested or the immunization methods failed to elicit CTL responses strong enough to alter the course of infection. It is noteworthy that only the two plasmids coding for nonstructural proteins, pp89 and M84, were protective in BALB/c mice. In contrast to the late expression patterns generally observed for the structural proteins, peak expression of pp89 and M84 is at IE and early times, respectively. DNA immunization may have been able to elicit responses too weak to detect the relatively small amounts of virion-associated protein entering the cytoplasm upon viral penetration but strong enough to detect the presentation of the de novo-synthesized antigens at the peak of their expression. Although CTLs are thought to be exquisitely sensitive to peptide-loaded class I molecules (9), adequate levels of these complexes may be required for the migration and activation of memory cells from the immunization site to the infected target organs. Even in measuring the activity of CTLs from MCMV-immune mice using target cells presenting only virion-associated antigens, high doses of UV-inactivated virus particles (200 PFU equivalents per cell) and an excess of in vitro restimulated effectors (5:1 to 25:1 effector/target ratios) are required to achieve levels of specific lysis similar to those using target cells undergoing de novo expression of viral antigens (36, 37). The additional need for the migration of DNA-primed effector cells may be reflected in the augmented protection reported from priming mice with plasmid DNA and then boosting with the appropriate antigen-expressing recombinant vaccinia virus (43, 44). The vaccinia virus infection may draw the DNA-primed CTLs into tissues where they can encounter their specific antigen, proliferate, and provide increased immunosurveillance in the organs targeted by the challenge virus. Experiments examining the protection levels following immunization and challenge of mice in proximal or distal sites may help determine if these spatial relationships apply to this system.
Our studies also showed that in the H-2k strain C3H/HeN, immunization with the pp89-expressing plasmid provided some protection against splenic viral replication. Although the reductions in spleen titer were within 10-fold compared with vector-alone-immunized controls, our results suggest that one or more pp89 epitopes are presented in association with the MHC H-2k heavy chain. C3H mice are among the most resistant to MCMV infection, partially due to their H-2k haplotype (18). Early activation of NK cells through IFN-γ production also helps to rapidly control viral replication in visceral organs. Results from experiments utilizing monoclonal antibody-mediated depletion of T-lymphocyte subsets in such resistant strains suggest that CD8+ T-cell populations may play less of a role in viral clearance during the acute infection than innate responses (29). Thus, even moderate CTL responses generated by the pp89 plasmid may have been overshadowed by the strong innate response. However, it has been shown for mutant and wild-type 129 strains that previous vaccination with an attenuated MCMV deletion mutant significantly limits the splenic replication upon subsequent virulent MCMV challenge regardless of the IFN-γ receptor-mediated mechanisms of viral control (30). The efficacy of the vaccine was compromised, however, in β2 microglobulin null 129 mice. Taken together, these data suggest that effective vaccination with an attenuated MCMV should provide T-cell-mediated protection in mice with intact IFN-γ responses. In addition, the efficient control of splenic viral replication in C3H forced us to measure viral replication at day 3 postchallenge, a time that was perhaps too early for CTLs primed by DNA immunization to become fully active effectors. Previous work by Del Val et. al demonstrated that CTLs generated in C57BL/6 (H-2b) mice following infection with MCMV or a pp89-expressing vaccinia virus specifically lyse syngeneic target cells expressing IE-phase genes (13). Thus, it appears as though at least one pp89-derived CTL epitope may be presented in association with MHC class I complexes from H-2d, H-2b, and H-2k strains. Levels of specific lysis in vitro of target cells expressing IE or IE plus early antigens by C57BL/6 haplotype-derived CTLs, however, were reduced approximately two- to threefold relative to those obtained from the BALB/c mice (13). Thus, the protective ability of pp89 in the C3H/HeN strain may be less pronounced than that in the BALB/c strain due to a lower relative affinity of pp89-derived epitopes for H-2k class I complexes and/or non-CD8+ T-lymphocyte-mediated effectors that dominate the antiviral response in this strain.
When BALB/c mice were coimmunized with pp89 and M84 DNAs, a synergistic reduction in spleen titers was observed relative to those after immunization with either plasmid alone. However, this synergistic effect was observed only when the mice were challenged with the highest doses of virus. This suggests that, upon more stringent challenge conditions, the observed interaction between the two CTL epitopes may have been synergistic. Since these two CTL epitopes are located on proteins from different phases of viral gene expression, it is possible that inclusion of MCMV genes encoding both IE and early proteins in a DNA vaccine accounts for such a substantial increase in protection.
In a recent study, the breadth of the CTL response against MCMV was analyzed using a bone marrow transplantation model in BALB/c mice (21). The authors found that, when pulmonary CTL activity peaked at 4 weeks p.i., CTLs displayed the highest CD3ε-redirected cytolytic activity against target cells presenting early (12 h p.i.) antigens, while target cells presenting only virion proteins were not significantly lysed. Pulmonary CTLs isolated at 3 weeks p.i., 1 week before activities were optimal, however, displayed the highest levels of activity against IE and late proteins. Thus, in order for complete CTL-mediated protection to occur in the lung, the CTL repertoire may need to encompass multiple epitopes in order to adapt to changing conditions in the infected tissue such as the relative availability of viral antigens for MHC class I presentation. Although in our experiments the antigens in Gp. II did not provide protection against the highest MCMV challenge dose, the inclusion of one or more of them in the pp89-M84 DNA vaccine may help diversify the resulting CTL responses and provide more complete protection against viral replication.
In contrast to the reduction of spleen titers to nearly undetectable levels in pp89-and-M84-coimmunized BALB/c mice, viral replication in the salivary glands was reduced only up to 10-fold below that of control mice. Previously, we reported salivary gland titer reductions of approximately 50-fold following pp89 DNA immunization (17). However, pp89 was expressed from pcDNA-I/Amp, and the use of the pcDNA3 vector in this report may account for differences in immunity. Through depletion studies of immune T lymphocytes, it was previously found that CD4+ cells are the effectors of viral clearance from this organ (24, 25). Although the protective immunity provided by CD8+ lymphocytes may suppress viral replication in the abdominal organs, virus that seeds the salivary glands replicates to high levels until CD4+ lymphocyte-mediated clearance can occur. Therefore, it appears likely that, although our plasmid DNA immunization-mediated protection may dramatically reduce MCMV replication in the spleen to nearly undetectable levels, the ability of the virus to disseminate to and amplify in the salivary gland was not significantly impeded in the immunized animals. This is not surprising, since plasmid DNA immunization favors the generation of CD8-mediated T-cell responses. Taken together, these data suggest that, in order for vaccination to significantly reduce salivary gland titers, adequate CD4+ antiviral activity in the salivary gland must be established prior to or coinciding with viral seeding of that organ.
DNA-mediated immunization has become a valuable research tool and holds great promise for future use in preventing infectious diseases. The field is moving rapidly, and with continued innovations in augmenting immune responses against plasmid-encoded antigens by coadministration of cytokines or cytokine-expressing plasmids (8, 22, 27), utilizing immunostimulatory DNA sequences (41), improving chemical or physical delivery systems, and delivering antigens as immune-response-enhancing ubiquitin fusion proteins (38, 39), it seems likely that it will be possible to vigorously prime both arms of the acquired cellular immune response to MCMV and provide complete protection against acute infection and the establishment of latency. The identification of MCMV antigens that elicit these types of protective responses will be crucial to directing this prophylactic immunity. The insights provided from animal models can then be used for the development of a safe and effective vaccine against HCMV.
ACKNOWLEDGMENTS
We thank Kimberly Koller for her help in subcloning the M48, M56, and M69 genes and Allison Hirsch for excellent general technical assistance. We also acknowledge the members of the laboratory for critical reading of the manuscript.
This work was supported by research grant numbers 6-FY98-0650 and 6-FY97-0409 from the March of Dimes Birth Defects Foundation and by NIH grant AI20954. L. D. Cranmer was supported in part by a grant from the Life and Health Insurance Medical Research Fund and by NIH-NIGMS predoctoral training grant GM07198.
REFERENCES
- 1.Barry M A, Lai W C, Johnston S A. Protection against mycoplasma infection using expression-library immunization. Nature. 1995;377:632–635. doi: 10.1038/377632a0. [DOI] [PubMed] [Google Scholar]
- 2.Boppana S, Britt W. Recognition of human cytomegalovirus gene products by HCMV-specific cytotoxic T cells. Virology. 1996;222:293–296. doi: 10.1006/viro.1996.0424. [DOI] [PubMed] [Google Scholar]
- 3.Bradshaw P A, Duran-Guarino M R, Perkins S, Rowe J I, Fernandez J, Fry K E, Reyes G R, Young L, Foung S K. Localization of antigenic sites on human cytomegalovirus virion structural proteins encoded by UL48 and UL56. Virology. 1994;205:321–328. doi: 10.1006/viro.1994.1648. [DOI] [PubMed] [Google Scholar]
- 4.Britt W, Alford W. Cytomegaloviruses. In: Fields B, Knipe D, Howley P, editors. Fields virology. 2nd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 2493–2523. [Google Scholar]
- 5.Buhler B, Keil G M, Weiland F, Koszinowski U H. Characterization of the murine cytomegalovirus early transcription unit e1 that is induced by immediate-early proteins. J Virol. 1990;64:1907–1919. doi: 10.1128/jvi.64.5.1907-1919.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chakrabarti S, Brechling K, Moss B. Vaccinia virus expression vector: coexpression of beta-galactosidase provides visual screening of recombinant virus plaques. Mol Cell Biol. 1985;5:3403–3409. doi: 10.1128/mcb.5.12.3403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chee M S, Bankier A T, Beck S, Bohni R, Brown C M, Cerny R, Horsnell T, Hutchison III C A, Kouzarides T, Martignetti J A, Preddie E, Satchwell S C, Tomlinson P, Weston K M, Barrell B G. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- 8.Chow Y H, Huang W L, Chi W K, Chu Y D, Tao M H. Improvement of hepatitis B virus DNA vaccines by plasmids coexpressing hepatitis B surface antigen and interleukin-2. J Virol. 1997;71:169–178. doi: 10.1128/jvi.71.1.169-178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christinck E R, Luscher M A, Barber B H, Williams D B. Peptide binding to class I MHC on living cells and quantitation of complexes required for CTL lysis. Nature. 1991;352:67–70. doi: 10.1038/352067a0. [DOI] [PubMed] [Google Scholar]
- 10.Cranmer L D, Clark C, Spector D H. Cloning, characterization, and expression of the murine cytomegalovirus homologue of the human cytomegalovirus 28-kDa matrix phosphoprotein (UL99) Virology. 1994;205:417–429. doi: 10.1006/viro.1994.1662. [DOI] [PubMed] [Google Scholar]
- 11.Cranmer L D, Clark C L, Morello C S, Farrell H E, Rawlinson W D, Spector D H. Identification, analysis, and evolutionary relationships of the putative murine cytomegalovirus homologs of the human cytomegalovirus UL82 (pp71) and UL83 (pp65) matrix phosphoproteins. J Virol. 1996;70:7929–7939. doi: 10.1128/jvi.70.11.7929-7939.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Val M, Hengel H, Häcker H, Hartlaub U, Ruppert T, Lucin P, Koszinowski U H. Cytomegalovirus prevents antigen presentation by blocking the transport of peptide-loaded major histocompatibility complex class I molecules into the medial-Golgi compartment. J Exp Med. 1992;176:729–738. doi: 10.1084/jem.176.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Val M, Münch K, Reddehase M J, Koszinowski U H. Presentation of CMV immediate-early antigen to cytolytic T lymphocytes is selectively prevented by viral genes expressed in the early phase. Cell. 1989;58:305–315. doi: 10.1016/0092-8674(89)90845-3. [DOI] [PubMed] [Google Scholar]
- 14.Del Val M, Schlicht H J, Volkmer H, Messerle M, Reddehase M J, Koszinowski U H. Protection against lethal cytomegalovirus infection by a recombinant vaccine containing a single nonameric T-cell epitope. J Virol. 1991;65:3641–3646. doi: 10.1128/jvi.65.7.3641-3646.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elliott R, Clark C, Jaquish D, Spector D H. Transcription analysis and sequence of the putative murine cytomegalovirus DNA polymerase gene. Virology. 1991;185:169–186. doi: 10.1016/0042-6822(91)90765-4. [DOI] [PubMed] [Google Scholar]
- 16.Fowler K B, Stagno S, Pass R F, Britt W J, Boll T J, Alford C A. The outcome of congenital cytomegalovirus infection in relation to maternal antibody status. N Engl J Med. 1992;326:663–667. doi: 10.1056/NEJM199203053261003. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez Armas J C, Morello C S, Cranmer L D, Spector D H. DNA immunization confers protection against murine cytomegalovirus infection. J Virol. 1996;70:7921–7928. doi: 10.1128/jvi.70.11.7921-7928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grundy J E, Mackenzie J S, Stanley N F. Influence of H-2 and non-H-2 genes on resistance to murine cytomegalovirus infection. Infect Immun. 1981;32:277–286. doi: 10.1128/iai.32.1.277-286.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hengel H, Koszinowski U H. Interference with antigen processing by viruses. Curr Opin Immunol. 1997;9:470–476. doi: 10.1016/s0952-7915(97)80097-0. [DOI] [PubMed] [Google Scholar]
- 20.Hengel H, Lucin P, Jonjic S, Ruppert T, Koszinowski U H. Restoration of cytomegalovirus antigen presentation by gamma interferon combats viral escape. J Virol. 1994;68:289–297. doi: 10.1128/jvi.68.1.289-297.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holtappels R, Podlech J, Geginat G, Steffens H P, Thomas D, Reddehase M J. Control of murine cytomegalovirus in the lungs: relative but not absolute immunodominance of the immediate-early 1 nonapeptide during the antiviral cytolytic T-lymphocyte response in pulmonary infiltrates. J Virol. 1998;72:7201–7212. doi: 10.1128/jvi.72.9.7201-7212.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iwasaki A, Stiernholm B J, Chan A K, Berinstein N L, Barber B H. Enhanced CTL responses mediated by plasmid DNA immunogens encoding costimulatory molecules and cytokines. J Immunol. 1997;158:4591–4601. [PubMed] [Google Scholar]
- 23.Jonjic S, del Val M, Keil G M, Reddehase M J, Koszinowski U H. A nonstructural viral protein expressed by a recombinant vaccinia virus protects against lethal cytomegalovirus infection. J Virol. 1988;62:1653–1658. doi: 10.1128/jvi.62.5.1653-1658.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jonjic S, Mutter W, Weiland F, Reddehase M J, Koszinowski U H. Site-restricted persistent cytomegalovirus infection after selective long-term depletion of CD4+ T lymphocytes. J Exp Med. 1989;169:1199–1212. doi: 10.1084/jem.169.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jonjic S, Pavic I, Lucin P, Rukavina D, Koszinowski U H. Efficacious control of cytomegalovirus infection after long-term depletion of CD8+ T lymphocytes. J Virol. 1990;64:5457–5464. doi: 10.1128/jvi.64.11.5457-5464.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kern F, Surel I P, Faulhaber N, Frommel C, Schneider-Mergener J, Schonemann C, Reinke P, Volk H. Target structures of the CD8+-T-cell response to human cytomegalovirus: the 72-kilodalton major immediate-early protein revisited. J Virol. 1999;73:8179–8184. doi: 10.1128/jvi.73.10.8179-8184.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim J J, Ayyavoo V, Bagarazzi M L, Chattergoon M A, Dang K, Wang B, Boyer J D, Weiner D B. In vivo engineering of a cellular immune response by coadministration of IL-12 expression vector with a DNA immunogen. J Immunol. 1997;158:816–826. [PubMed] [Google Scholar]
- 28.Koszinowski U H, Reddehase M J, Del Val M. Principles of cytomegalovirus antigen presentation in vitro and in vivo. Semin Immunol. 1992;4:71–79. [PubMed] [Google Scholar]
- 29.Lathbury L J, Allan J E, Shellam G R, Scalzo A A. Effect of host genotype in determining the relative roles of natural killer cells and T cells in mediating protection against murine cytomegalovirus infection. J Gen Virol. 1996;77:2605–2613. doi: 10.1099/0022-1317-77-10-2605. [DOI] [PubMed] [Google Scholar]
- 30.MacDonald M R, Li X Y, Stenberg R M, Campbell A E, Virgin H W. Mucosal and parenteral vaccination against acute and latent murine cytomegalovirus (MCMV) infection by using an attenuated MCMV mutant. J Virol. 1998;72:442–451. doi: 10.1128/jvi.72.1.442-451.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mackett M, Smith G L, Moss B. General method for production and selection of infectious vaccinia virus recombinants expressing foreign genes. J Virol. 1984;49:857–864. doi: 10.1128/jvi.49.3.857-864.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLaughlin-Taylor E, Pande H, Forman S J, Tanamachi B, Li C R, Zaia J A, Greenberg P D, Riddell S R. Identification of the major late human cytomegalovirus matrix protein pp65 as a target antigen for CD8+ virus-specific cytotoxic T lymphocytes. J Med Virol. 1994;43:103–110. doi: 10.1002/jmv.1890430119. [DOI] [PubMed] [Google Scholar]
- 33.Mercer J A, Marks J R, Spector D H. Molecular cloning and restriction endonuclease mapping of the murine cytomegalovirus genome (Smith strain) Virology. 1983;129:94–106. doi: 10.1016/0042-6822(83)90398-7. [DOI] [PubMed] [Google Scholar]
- 34.Morello C S, Cranmer L D, Spector D H. In vivo replication, latency, and immunogenicity of murine cytomegalovirus mutants with deletions in the M83 and M84 genes, the putative homologs of human cytomegalovirus pp65 (UL83) J Virol. 1999;73:7678–7693. doi: 10.1128/jvi.73.9.7678-7693.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rawlinson W D, Farrell H E, Barrell B G. Analysis of the complete DNA sequence of murine cytomegalovirus. J Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reddehase M J, Bühring H J, Koszinowski U H. Cloned long-term cytolytic T-lymphocyte line with specificity for an immediate-early membrane antigen of murine cytomegalovirus. J Virol. 1986;57:408–412. doi: 10.1128/jvi.57.1.408-412.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reddehase M J, Keil G M, Koszinowski U H. The cytolytic T lymphocyte response to the murine cytomegalovirus. II. Detection of virus replication stage-specific antigens by separate populations of in vivo active cytolytic T lymphocyte precursors. Eur J Immunol. 1984;14:56–61. doi: 10.1002/eji.1830140111. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez F, An L L, Harkins S, Zhang J, Yokoyama M, Widera G, Fuller J T, Kincaid C, Campbell I L, Whitton J L. DNA immunization with minigenes: low frequency of memory cytotoxic T lymphocytes and inefficient antiviral protection are rectified by ubiquitination. J Virol. 1998;72:5174–5181. doi: 10.1128/jvi.72.6.5174-5181.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rodriguez F, Zhang J, Whitton J L. DNA immunization: ubiquitination of a viral protein enhances cytotoxic T-lymphocyte induction and antiviral protection but abrogates antibody induction. J Virol. 1997;71:8497–8503. doi: 10.1128/jvi.71.11.8497-8503.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sato Y, Roman M, Tighe H, Lee D, Corr M, Nguyen M D, Silverman G J, Lotz M, Carson D A, Raz E. Immunostimulatory DNA sequences necessary for effective intradermal gene immunization. Science. 1996;273:352–354. doi: 10.1126/science.273.5273.352. [DOI] [PubMed] [Google Scholar]
- 42.Scalzo A A, Elliott S L, Cox J, Gardner J, Moss D J, Suhrbier A. Induction of protective cytotoxic T cells to murine cytomegalovirus by using a nonapeptide and a human-compatible adjuvant (Montanide ISA 720) J Virol. 1995;69:1306–1309. doi: 10.1128/jvi.69.2.1306-1309.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schneider J, Gilbert S C, Blanchard T J, Hanke T, Robson K J, Hannan C M, Becker M, Sinden R, Smith G L, Hill A V. Enhanced immunogenicity for CD8+ T cell induction and complete protective efficacy of malaria DNA vaccination by boosting with modified vaccinia virus Ankara. Nat Med. 1998;4:397–402. doi: 10.1038/nm0498-397. [DOI] [PubMed] [Google Scholar]
- 44.Sedegah M, Jones T R, Kaur M, Hedstrom R, Hobart P, Tine J A, Hoffman S L. Boosting with recombinant vaccinia increases immunogenicity and protective efficacy of malaria DNA vaccine. Proc Natl Acad Sci USA. 1998;95:7648–7653. doi: 10.1073/pnas.95.13.7648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Selik R M, Chu S Y, Ward J W. Trends in infectious diseases and cancers among persons dying of HIV infection in the United States from 1987 to 1992. Ann Intern Med. 1995;123:933–936. doi: 10.7326/0003-4819-123-12-199512150-00006. [DOI] [PubMed] [Google Scholar]
- 46.Thäle R, Szepan U, Hengel H, Geginat G, Lucin P, Koszinowski U H. Identification of the mouse cytomegalovirus genomic region affecting major histocompatibility complex class I molecule transport. J Virol. 1995;69:6098–6105. doi: 10.1128/jvi.69.10.6098-6105.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volkmer H, Bertholet C, Jonjic S, Wittek R, Koszinowski U H. Cytolytic T lymphocyte recognition of the murine cytomegalovirus nonstructural immediate-early protein pp89 expressed by recombinant vaccinia virus. J Exp Med. 1987;166:668–677. doi: 10.1084/jem.166.3.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wills M R, Carmichael A J, Mynard K, Jin X, Weekes M P, Plachter B, Sissons J G. The human cytotoxic T-lymphocyte (CTL) response to cytomegalovirus is dominated by structural protein pp65: frequency, specificity, and T-cell receptor usage of pp65-specific CTL. J Virol. 1996;70:7569–7579. doi: 10.1128/jvi.70.11.7569-7579.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winkler M, Stamminger T. A specific subform of the human cytomegalovirus transactivator protein pUL69 is contained within the tegument of virus particles. J Virol. 1996;70:8984–8987. doi: 10.1128/jvi.70.12.8984-8987.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ziegler H, Thale R, Lucin P, Muranyi W, Flohr T, Hengel H, Farrell H, Rawlinson W, Koszinowski U H. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity. 1997;6:57–66. doi: 10.1016/s1074-7613(00)80242-3. [DOI] [PubMed] [Google Scholar]