Abstract
BACKGROUND AND AIMS:
In vivo induction of alcoholic chronic pancreatitis (ACP) causes significant acinar damage, increased fibroinflammatory response, and heightened activation of cyclic response element binding protein 1 (CREB) when compared with alcohol (A) or chronic pancreatitis (CP) mediated pancreatic damage. However, the study elucidating the cooperative interaction between CREB and the oncogenic KrasG12D/+ (Kras*) in promoting pancreatic cancer progression with ACP remains unexplored.
METHODS:
Experimental ACP induction was established in multiple mouse models, followed by euthanization of the animals at various time intervals during the recovery periods. Tumor latency was determined in these mice cohorts. Here, we established CREB deletion (Crebfl/fl) in Ptf1aCreERTM/+;LSL-KrasG12D+/−(KC) genetic mouse models (KCC−/−). Western blot, phosphokinase array, and qPCR were used to analyze the pancreata of Ptf1aCreERTM+/−, KC and KCC−/− mice. The pancreata of ACP-induced KC mice were subjected to single-cell RNA sequencing (scRNAseq). Further studies involved conducting lineage tracing and acinar cell explant cultures.
RESULTS:
ACP induction in KC mice had detrimental effects on the pancreatic damage repair mechanism. The persistent existence of acinar cell-derived ductal lesions demonstrated a prolonged state of hyperactivated CREB. Persistent CREB activation leads to acinar cell reprogramming and increased pro-fibrotic inflammation in KC mice. Acinar-specific Creb ablation reduced advanced PanINs lesions, hindered tumor progression, and restored acinar cell function in ACP-induced mouse models.
CONCLUSIONS:
Our findings demonstrate that CREB cooperates with Kras* to perpetuate an irreversible ADM and PanIN formation. Moreover, CREB sustains oncogenic activity to promote the progression of premalignant lesions toward cancer in the presence of ACP.
Keywords: cAMP response element binding protein (CREB), pancreatic intraepithelial neoplasia (PanIN), Pancreatic cancer, acinar-to-ductal metaplasia (ADM), alcoholic chronic pancreatitis (ACP)
Introduction
Alcoholic pancreatitis, whether acute or chronic, is a severe disease that negatively impacts the exocrine pancreas and acknowledged as of one the most significant risk factors for pancreatic cancer 1, 2. Notably, alcohol abuse is a prominent etiological factor responsible for chronic pancreatitis (CP), accounting for 60–90% of cases, which typically arises from recurrent episodes of acute pancreatitis 3, 4. The hallmark feature of alcoholic chronic pancreatitis (ACP) is its classification as a persistent inflammatory condition that causes a gradual decline in the exocrine function of the pancreas, eventually resulting in its atrophy and development of fibrosis 5.
The causal relationship between chronic inflammation and pancreatic cancers has been well-established in clinical studies 6, 7. Typically, the induction of chronic inflammation in the pancreas usually leads to tissue injury, prompting a transition in its cell fate, which is characterized by the loss of acinar cell differentiation and the acquisition of a ductal phenotype. This phenomenon of acinar to ductal metaplasia (ADM) serves as a physiological adaptive response to inflammation, aimed at limiting tissue damage 8. Nevertheless, an increasing amount of scientific evidence, supported by various experimental models, consistently demonstrates that the activation of inflammation in pancreatic tissue that expresses the oncogene KrasG12D/+ (Kras*) impedes its ability to regenerate and expedite the advancement of tumors 9–12. This chronic insult accelerates the development of neoplastic precursor lesions, such as ADM and pancreatic intraepithelial neoplasia (PanIN), which rapidly advance towards invasive carcinoma. However, the cooperative mechanisms of Kras* and chronic inflammation in the setting of ACP are not well understood.
Our recent research has uncovered the involvement of pancreas specific activation of PI3K/AKT/mTOR signaling node associated with the severity of ACP 13. Additionally, we have also seen substantial activation of cyclic AMP responsive element binding protein 1 (CREB) in an inflamed milieu of the pancreas, highlighting its potential involvement in the development of ACP pathogenesis 13. CREB undergoes phosphorylation at Ser133 by key upstream regulators, such as protein kinase A (PKA), Akt/protein kinase B (PKB), MAPK, and p90 ribosomal S6 kinase (p90RSK) 14. Once activated, CREB (pCREB) interacts with its coactivator and recruits additional transcriptional machinery to promote the development of cancer 15. In our previous study, we confirmed the involvement of CREB in propagating the aggressiveness of pancreatic cancer and showed its direct activation as a result of AKT signaling 16. In this context, CREB could be a valuable target as a crucial molecule in this signaling pathway, playing a vital role in cultivating ACP-induced pancreatic cancer progression.
This study conducts a comparative analysis of severe pancreatic damage induced by ACP in comparison to damage caused by alcohol or CP induction alone. Additionally, it explores the role of CREB in the development of pancreatic cancer driven by Kras* in the context of ACP. Here, we demonstrate persistent activation of CREB in acinar cells as they undergo a transition towards ductal phenotype in Kras* mice with ACP induction. When KC mice are induced with ACP, they show a greater number of high-grade PanIN lesions and an increased presence of fibro inflammation, which consequently leads to a reduced tumor latency period and accelerated pancreatic cancer progression. Deleting Creb in acinar cells of KC mice with ACP reduces the reprogramming of ADM, which in turn hinders tumor growth and prolongs their tumor latency period. Together, these findings provide a detailed understanding of how CREB contributes to the advancement of pancreatic cancer in the presence of chronic inflammation with Kras*.
Results
Establishing Alcoholic Chronic Pancreatitis (ACP) in Ptf1aCreERTM/+ Mice
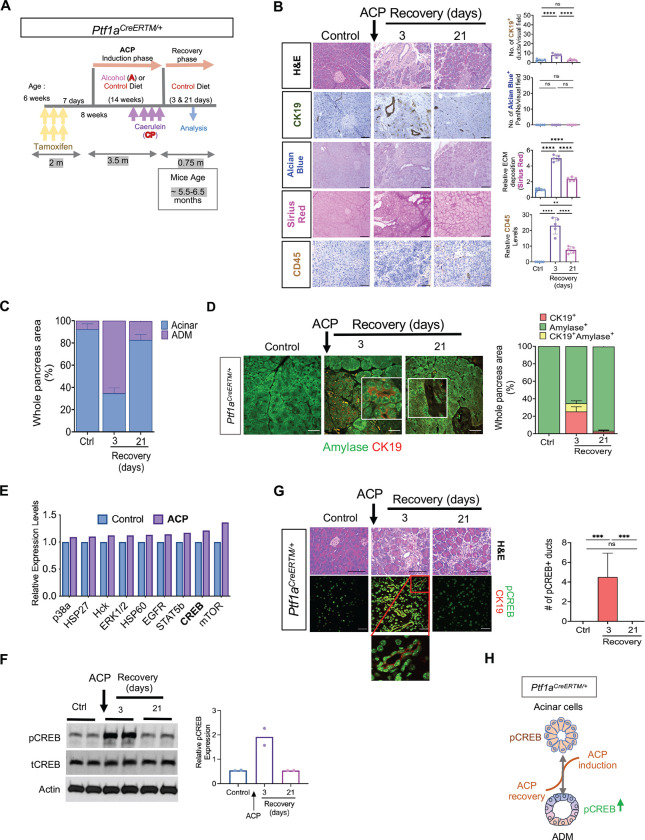
The experimental mouse model of ACP induction was established in Ptf1aCreERTM/+ mice as described previously 13. Mice were sacrificed after 3 or 21 days, also referred to as ACP recovery period (Figure 1A). Mice across all experimental cohorts exhibited weight gain with the maximum increase observed in vehicle and alcohol (A) alone cohorts. Cessation of exogenous ACP induction led to recovery of mice body weights (identical to control cohort) 21 days after ACP induction (Supplementary Figure S1A). Serum based evaluation of alcohol revealed increased concentration of blood alcohol levels in the mice cohort subjected to A or ACP induction as compared to control mice (Ctrl) (Supplementary Figure S1B). Ptf1aCreERTM/+ mice with ACP induction demonstrated significant reduction in the pancreas weight (implying pancreatic injury) on day 3 of the recovery period, when compared with other groups (Supplementary Figure S1C). In contrast, withdrawal of ACP exposure resulted in the complete recovery of pancreas weight to its normal level after 21 days (Supplementary Figure S1D). By measuring the serum amylase level, we observed its elevated levels in mice exposed to alcohol. Specifically, mice with ACP-induction exhibited the most substantial decline in serum amylase levels compared to control, A, and CP mice cohorts (Supplementary Figure S1E), suggestive of maximum acinar injury in this cohort. These findings were further substantiated using histological assessment of mice pancreas harvested after ACP induction (at day 3 of recovery period). The analysis revealed a profound injury to the pancreatic tissue architecture, including extensive acinar cell loss, presence of duct-like structures (CK-19+ positivity), activation of pancreatic stellate cells (PSCs) displaying aSMA positivity, increased collagen deposition (Sirius Red), and leukocyte infiltration (CD45+ immune cells) when compared with the pancreata of control, Aor CP exposed mice pancreata (Figure 1B and Supplementary Figure S1F). Similarly to previous findings 13, it was observed that the histological alterations during ACP induction in Ptf1aCreERTM/+ mice were transient in nature. This was confirmed by the complete reversal of pancreatic injury after discontinuing the exogenous ACP stimulus, (21-day recovery period) (Figure 1B). Notably, no histologically detectable PanINs were observed (Figure 1B and Supplementary Figure S1F) across all experimental cohorts. In addition, qPCR based transcriptional profiling of the ACP induced mice pancreata demonstrated significant downregulation of genes implicated in acinar cell regulation and function when compared to control mice pancreata (Supplementary Figure S2A).
Figure 1.
CREB activation is a hallmark of acinar-to-ductal metaplasia (ADM) in response to alcoholic chronic pancreatitis (ACP) in Ptf1aCreERTM/+ mice. (A) Schematic showing ACP induction in Ptf1aCreERTM/+ mice exposed to alcohol(ethanol) enriched liquid diet (A) and repetitive caerulein administration (CP). Mice were euthanized after 3 and 21 days of recovery period. (B) Representative images of mouse pancreas with H&E, images depicting ducts (CK19+), PanINs (Alcian Blue), collagen (Sirius red) and immune cells (CD45+) (left) and their quantification (right) (n=5 mice per group). (C) H&E based quantification of Ptf1aCreERTM/+ mice pancreata highlighting the presence of acinar and ADM regions, (n=3 mice per group). (D) Representative images of pancreas depicting amylase (green)/ CK19 (red) co-immunofluorescent labeling (left) and the corresponding quantification of CK19+ amylase+ cells (right), (n=3 mice per group). (E) Mouse kinase array analysis in control and ACP induced pancreata. (F) Western blot image (left) and quantification (right) showing increased phosphorylated levels of CREB in Ptf1aCreERTM/+ mice pancreatic tissue lysates of control and at 3-and 21- days of ACP recovery period (n = 2 mice per group). (G) Representative images depicting H&E and pCREB (green)/CK19 (red) co-immunofluorescent labeling (left) with the corresponding quantification (right) of pCREB+ ducts in pancreas harvested from ctrl and ACP-induced Ptf1aCreERTM/+ mice with 3 and 21 days of recovery period (n=6 mice per group). (H) Schematic demonstrating that ACP induces pancreatic tissue injury, resulting in a transient ADM with CREB activation, which subsequently returns to its baseline during the 21-day ACP recovery period. Scale bar, 50μm. ns nonsignificant; **p< 0.01; ***, p< 0.001; ****p<0.0001 by ANOVA.
Activation of CREB is a Hallmark of Acinar-To-Ductal Metaplasia (ADM) in Response to ACP
Pancreatic acinar cells display highest plasticity and dedifferentiate into ADM expressing ductal markers in response to inflammation induced damage 17. To further corroborate these observations, we conducted histological examinations in the pancreata of ACP induced mice. Using H&E (Figure 1C) and co immunofluorescence (I.F.) (Figure 1D) staining of CK19 and amylase in ACP-induced Ptf1aCreERTM/+ mice, we demonstrated the emergence of transient duct-like cells with 3 days of ACP induction. Interestingly, by the end of the 21-day recovery period, these cells underwent redifferentiation, repopulated the acinar compartment, providing evidence for the transient and reversible process nature of ADMs.
Intricate network of multiple signaling nodes regulate the dynamics of pancreatic tissue injury and repair 11. Therefore, to delve deeper into the molecular pathogenesis of ACP, we conducted phosphokinase array profiling on the pancreata of Ptf1aCreERTM/+ mice following ACP induction. Our analysis revealed significant upregulation of mTOR and CREB, which are two of the most prominently differentially upregulated effector kinases, compared to the control group of Ptf1aCreERTM/+ mice pancreas lysates (Figure 1E and Supplementary Figure S2B). Our earlier study showed that the induction of ACP promotes activation of the PI3K/mTOR pathway and orchestrates the fibroinflammatory program of ACP pathogenesis. Additionally, we discovered that inhibiting this signaling node effectively attenuates the severity of ACP in C57Bl/6 mice 13. Furthermore, our laboratory’s investigation into the aggressiveness of pancreatic cancer has identified CREB as a key modulator of oncogenic mutant KRAS signaling. The present study investigates the involvement of CREB in the pancreatic acinar compartment, which results in reprogramming of ADM towards ductal plasticity.
Further validation was conducted by performing immunoblotting and co-I.F. staining for pCREB/CK19. This analysis uncovered a significantly elevated expression of pCREB in duct-like structures after 3 days of ACP recovery, compared with control pancreata. However, by 21 days of ACP recovery, the pCREB expression returned to baseline levels, similar to the control group (Figure 1F and 1G.). Collectively, our findings suggest that the established ACP instigates pancreatic injury and fosters a fibroinflammatory milieu. Additionally, it enhances the activation of CREB in this experimental mouse model of ACP. Notably during recovery period, these alterations are reversed, resulting in attenuated CREB activation back to its baseline (Figure 1H).
ACP Induction Accelerates ADM Reprogramming to PanIN and Pancreatic Cancer in Oncogenic KrasG12D mutant mice
The persistent inflammatory stimulus, together with the presence of oncogenic driver mutation such as Kras in the acinar cells, expedite the formation of neoplastic lesions through the process of ADM and PanINs 18, 19. To further strengthen the evidence of ACP mediated reprogramming of acinar cells towards ductal phenotype, we utilized KC GEMM harboring mutant Kras*.
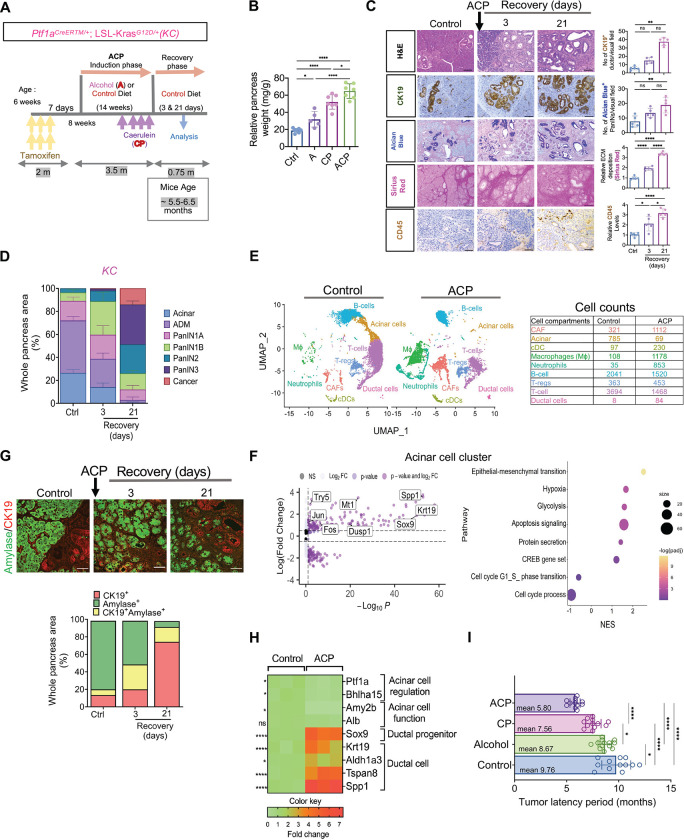
ACP induction was established by utilizing 6-week-old KC mice (Figure 2A). Both pancreas and blood samples were obtained from KC GEMM, aged between 5.5 – 6.5 months (both at 3 and 21-days recovery periods). Mice receiving an alcohol-based liquid diet showed a higher intake of alcohol, compared to control mice fed with regular diet (Supplementary Figure S3A). Inducing ACP in KC mice resulted in a pronounced increase in the relative pancreas weight (Figure 2B). Examination of the pancreas in the control KC mice aged 5.5–6.5 months showed mainly well-structured acinar compartments and low-grade PanIN lesions (Figure 2C and D). H&E stained pancreata from ACP induced KC mice revealed substantially higher presence of mucinous ductal lesions (low grade PanINs) with fewer acinar cells (after a 3-day recovery period) as compared with alcohol (A) or CP-induced KC mice pancreata (Figure 2D and Supplementary Figure S3C). Following ACP induction in KC GEMM, after a 21-day recovery period, histological analysis of pancreas tissue revealed progression of these mucinous PanINs towards high-grade lesions (Figure 2C and 2D), as well as significant increase in the CK-19+ ductal positivity, mucin content (alcian blue), activated PSCs (aSMA positivity), increased collagen deposition (Sirius Red), and infiltration of leukocyte (CD45+ immune cells) (Figure 2C). This increase was observed to be highest in ACP experimental cohort of KC mice pancreata as compared with alcohol (A) or CP-induction (Supplementary Figure S3B). The progression was also accompanied by a substantial increase in the deposition of extracellular matrix (ECM) and a heightened infiltration of immune cells (Figure 2C and Supplementary Figure S3B).
Figure 2.
ACP induction accelerates ADM reprogramming towards PanIN and pancreatic cancer in KrasG12D/+ mutant mice. (A) Schematic for ACP induction Ptf1aCreERTM/+; LSL-KrasG12D/+ (KC) mice exposed to alcohol enriched liquid diet (A) and repetitive caerulein administration (CP). Mice were euthanized after 3 and 21 days of recovery period. (B) Relative pancreas weight measurements in ctrl, A, CP, and ACP induced KC mice with n=5–7 mice per group. (C) Representative pancreatic images depicting H&E, of CK19+ ducts, PanINs (Alcian Blue), collagen (Sirius red) and immune cells (CD45+) in pancreata of control and ACP-induced KC mice with 3 and 21 days of recovery period (left) and their quantification (right) (n=5 mice per group). (D) Histopathological assessment using H&E staining was employed to quantify areas within the pancreas corresponding to acinar, ADMs, PanINs and cancer regions with n=3 mice per group. (E) UMAP projection displaying cell clusters (left) and cell count table (right) in the scRNA sequencing analysis of pancreata from control and ACP induction in KC mice. (F) Volcano plot (left) illustrating differentially expressed genes (DEG) and bubble plot (right) showing gene set enrichment analysis (GSEA) in the acinar cell subcluster of ACP vs. control in KC mice. (G) Representative pancreas images depicting amylase (green)/ CK19 (red) co-immunofluorescent labeling (top) and the corresponding quantification of CK19+ amylase+ cells (bottom) in ctrl and ACP-induced KC mice with 3 and 21 days of recovery period (n=3 mice per group). (H) Heat map illustrating the differential expression of genes associated with acinar or ductal phenotypes in the pancreas of control and ACP-induced KC mice with n=3 mice per group. (I) Measurement of tumor latency period in control, alcohol, CP, and ACP-induced KC mice (n=12–13 mice per group). Scale bar, 50μm. ns nonsignificant; *p<0.05; **p< 0.01; ****p<0.0001 by ANOVA or unpaired t test.
To assess the cellular heterogeneity and transcriptional changes associated with ACP induced accelerated progression of pancreatic cancer with Kras* in a GEMM, we employed single cell RNA sequencing (scRNAseq). Live single cell suspensions of KC mice pancreata in either control or with ACP were analyzed (at 5.5–6.5 months of age) using the 10x Genomics Chromium platform. In total, a total of 7903 cells for ctrl and 7797 cells for ACP group respectively were considered for analysis. The UMAP cell clusters were annotated by examining both classic cell type markers and previously described single cell gene signatures in pancreas tissue 20, 21. We successfully distinguished 9 distinct clusters representing different cell types (Figure 2E and Supplementary Figure S3D). Notably, there was increased abundance of cell types including ductal cells, macrophages, neutrophils, regulatory T cells (Tregs), and fibroblasts in ACP-induced KC mice when compared to control (Figure 2E). Furthermore, examination of transcriptional changes within the acinar cell compartment of the pancreas of KC mice showcased upregulation of mRNA transcripts related to inflammation induced stress, growth and proliferation with ACP induction compared to control (Figure 2F). Hallmark gene set enrichment pathway analysis (GSEA) identified an increase enrichment of several prominent cellular processes affiliated with ACP mediated acinar damage and ADM formation including cell apoptosis, protein secretion, epithelial mesenchymal transition (EMT) and CREB gene set (Figure 2F). To further corroborate these transcriptional observations of severe pancreatic injury with ACP, we conducted co-I.F. analysis for acinar (Amylase) and ductal cell type (CK19) markers in control KC and ACP-induced KC mice pancreata (Figure 2G). The uninjured ctrl KC pancreata showed higher presence of amylase+ acinar cells with CK19+ expression associated within the ductal structures only. However, in ACP-induced KC mice, the acinar compartment underwent a progressive transformation, gradually being replaced by substantial proportion of ductal structures from acinar origin (dual CK19+amylase+cells) at 3-day recovery period. Notably, analysis of pancreatic tissue at the end of 21-day recovery period, displayed ductal phenotype with highest proportion of CK19+ cells (Figure 2G). Subsequently, through qPCR analysis, validation of transcriptional alterations within the pancreata of ACP induced KC mice were further confirmed. This highlighted the reprogramming of acinar cells towards ductal metaplasia (Figure 2H).
The existence of advanced grade histological lesions progressing to pathologically evident cancer in the pancreata of KC with ACP induction correlated with significant shortened tumor latency period (Figure 2I, mean=5.80 months) as well, when compared to control (mean=9.76 months), A (mean= 8.67 months) or CP (mean=7.56 months) exposed KC mice, thereby confirming a heightened tumor burden induced by ACP in comparison to damage caused by alcohol or CP induction alone. Taken together, our findings demonstrate that ACP induction in KC GEMM not only impairs the pancreatic injury repair and acinar regeneration process, but also irreversibly reprogram these cells towards ductal phenotype to accelerate cancer progression.
CREB is Persistently Activated During PanIN Progression in KrasG12D mutant mice
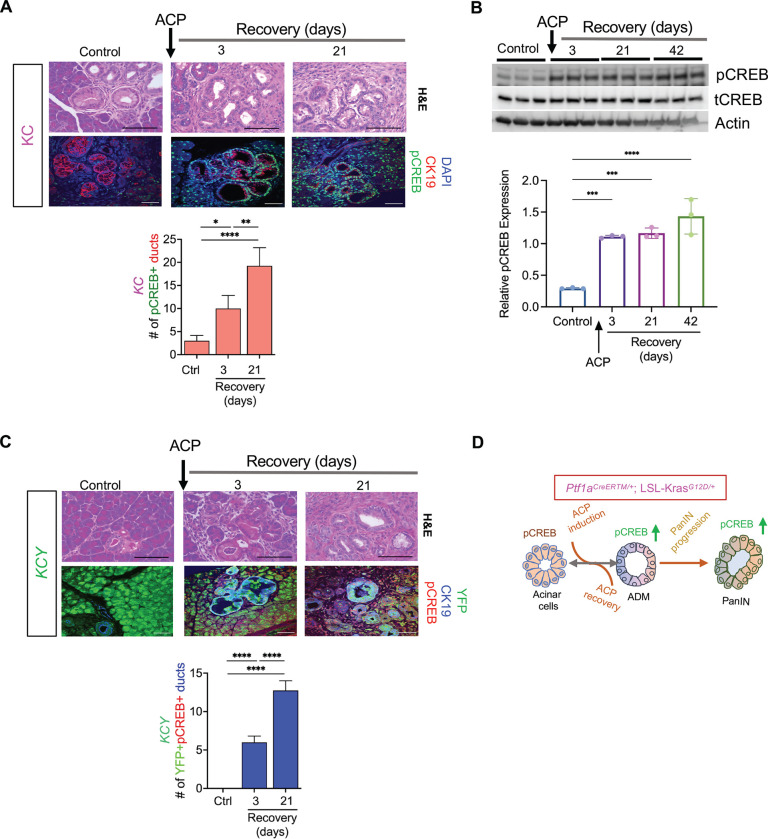
Next, we investigated the role of CREB in the reprogramming of acinar cells into a ductal phenotype in KC GEMM, in conjunction with ACP, during both the initiation and restoration stages. Histological assessment using co-I.F. analysis of pancreas tissues harvested after ACP induction in KC mice demonstrated a pronounced and substantial rise in the abundance of pCREB-positive staining within CK19+ ductal lesions during ACP recovery periods (3- and 21-days) in comparison to the pancreata of control KC mice (Figure 3A). Additional validation of CREB activation in ACP-induced KC mice was performed using Western blot analysis in the lysates of pancreas up to 42-days of recovery period (Figure 3B). The analysis confirmed that there was a heightened and sustained activation of CREB after 3, 21, and even up to 42 days of ACP recovery.
Figure 3.
Sustained hyperactivation of CREB in ACP KrasG12D /+ driven ADM and PanIN progression. (A) Histological assessment by H&E and co I.F. labeling of pCREB (green) and CK-19 (red) expression levels (top) along with corresponding quantification of pCREB+ ducts within the pancreata of control (ctrl) and KC mice at 3- and 21-day ACP recovery period (bottom) with n=4 mice per group. (B) Western blot image (top) and quantification (bottom) of pCREB in pancreatic tissue lysates of control and KC mice at 3-, 21- and 42-days of ACP recovery period (n=3 mice per group). (C) Representative H&E images of the pancreas along with co I.F. labeling of pCREB (red)/ CK19 (blue)/ YFP (green) (top) and corresponding of quantification (bottom) of YFP+ pCREB+ ducts in pancreatic tissues of ctrl and Ptf1aCreERTM/+; LSL-KrasG12D/+; R26R-EYFP (KCY) mice at 3- and 21-day of ACP recovery period (n=4 mice per group). (D) Schematic depicting sustained hyperactivation of CREB driving neoplastic reprogramming of acinar cells in KC mice upon ACP induction. Scale bar, 50μm. *p<0.05; **p< 0.01; ***, P < 0.001; ****p<0.0001 by ANOVA.
Next, we combined acinar targeted KC GEMM with a lineage tracer R26R-EYFP to generate Ptf1a-CreERTM/+;LSL-KrasG12D/+;R26R-EYFP(KCY) mouse model The KCY mice were subjected to ACP induction as detailed for KC GEMM mice in Figure 2A. The administration of Tamoxifen to KCY mice resulted in a recombination efficiency of around 95% in the acinar cells during a 24-hour period 22. In control KCY mice, YFP expression was mainly restricted within the acinar cells and excluded from CK19+ ducts. ACP-induced pancreatic injury in KCY mice led to an accelerated progression of PanIN lesions (like KC-ACP) with many of these ductal structures originating from the acinar cells (YFP+CK19+ducts) (Figure 3C). Notably, we observed sustained CREB activation within acinar-derived YFP/CK-19 dual-positive ductal lesions at both the 3- and 21-days of recovery period, post ACP induction. These results imply that CREB plays a role in the reprogramming of acinar cells into preneoplastic precursors following ACP induction (Figure 3D).
Acinar Specific Ablation of CREB Attenuates Spontaneous KrasG12D mediated PanIN Progression
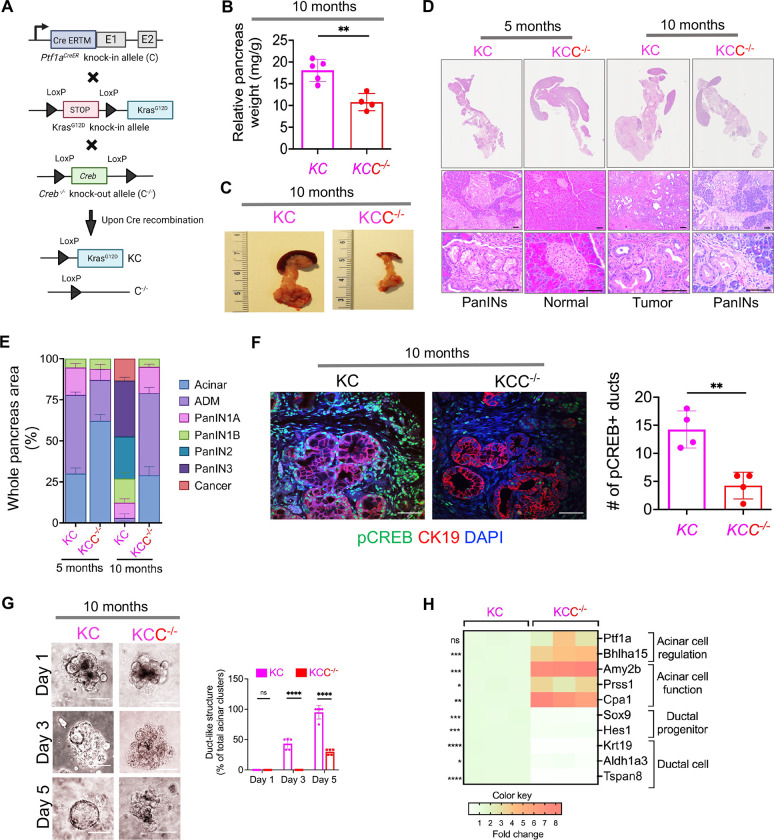
To further explore the impact of CREB on Kras* -driven ADM and PanIN formation, we generated acinar specific conditional Creb knockout mice in the presence of Kras* (Figure 4A) referred as Ptf1aCreERTM/+; LSL-KrasG12D/+; Crebfl/fl or KCC−/− mice whereas KC mice with intact or wild type Creb referred as KC. Pancreas were collected from these two mice cohorts at two different time intervals; 5 months and 10 months. Intriguingly, a marked reduction in relative pancreas weight was observed in the KC mice with Creb deletion (KCC−/−) as compared to wild type KC (Figure 4B and C). Next, on histological examination, the pancreata from 5-month-old KC mice displayed a higher prevalence of ADM, and low-grade PanIN lesions within the pancreas (Figure 4D and E). Conversely, pancreata of 5-month-old KCC−/− mice were devoid of precursor lesions and demonstrated normal pancreatic architecture. Additionally, on assessing the pancreata harvested from 10 months old KCC−/− mice displayed infrequent instances of ADM or PanIN1 lesions. On the contrary, in the pancreata of 10 months old KC mice, pancreas were marked by high-grade PanIN lesions and cancer (Figure 4D and E.)
Figure 4.
Acinar specific ablation of Creb attenuates spontaneous KrasG12D/+ mediated PanIN progression. (A) Breeding strategy for generation of acinar specific Creb deficient Kras* mutant mice (Ptf1aCreERTM/+; LSL-KrasG12D/+; Crebfl/fl or KCC−/−). (B) Comparative measurement of relative pancreas weight in 10-month-old Creb wild type (KC) and KCC−/− mice (n=4–5 mice per group). (C) Representative photomicrographs of whole pancreas depicting significantly less tumor burden in KCC−/− as compared to KC mice at 10 months of age. (D) Representative H&E images of the pancreas harvested from 5- and 10-month-old KC and KCC−/− mice. Scale bar, 10μm and 50μm. (E) Comparative assessment using H&E-based histology to examine the entire pancreas, illustrating the presence of acinar cells, ADMs, PanINs and cancerous regions in 5- and 10-month-old KC and KCC−/− mice (n=3 mice per group). (F) Co I.F. -image of pCREB (green), CK-19 (red) and DAPI (blue) (left) along with quantification (right) in pancreatic tissue sections harvested from 10-month-old KC and KCC−/− mice with n=4 mice per group. Scale bar, 50μm. (G) Representative bright-field photomicrographs of primary 3D acinar cell cultures (left) and quantification of duct like structures (right) established from 10-month-old KC and KCC−/− mice (scale bar, 20 μm) (n=5 mice per group). (H) qPCR of pancreatic tissue harvested from KC and KCC−/− mice for acinar and ductal genes (n=3 mice per group). ns nonsignificant; *p<0.05; **p< 0.01; ***p<0.001; ****p<0.0001 one way ANOVA or unpaired t test.
Subsequently, co-I.F. analysis was performed in the pancreas tissues harvested from KC and KCC−/− mice (at 10 months of age) to ascertain the activation of CREB (pCREB-Ser133) in ductal cell types using CK-19 marker (Figure 4F). This analysis confirmed a significant reduction of pCREB positivity in the ductal regions of the pancreas in KCC−/− mice, as compared to wildtype KC mice. Our results confirm that acinar cell specific CREB inactivation selectively attenuates the progression of pancreatic cancer. To further understand the effect of CREB on the ability of acinar cells to dedifferentiate into ductal cells, pancreatic acinar cells were isolated from both KC and KCC−/− mice pancreata for explant culture. Acinar-specific Creb ablation significantly blocked the formation of ductal-like structures compared with acinar cells with an intact CREB (Figure 4G). Furthermore, conducting qPCR analysis on pancreata obtained from 10-month-old KCC−/− mice unveiled a notable reduction in genes linked to ductal cell phenotype with a concomitant upregulation in genes associated with the acinar cell regulation, function, proliferation, and differentiation, when compared with corresponding age-matched wild type KC mice (Figure 4H). These findings collectively indicate that acinar-specific CREB ablation reduces the capability of acinar cells to undergo ADM reprogramming.
Acinar Specific Ablation of Creb attenuates Kras* induced PanINs to Pancreatic Cancer Progression with ACP
To further establish the association of acinar-specific CREB in advancing the propensity of ductal reprogramming with ACP induction, we first generated Ptf1aCreERTM/+ mice with Creb deletion (CC−/−) in the absence of Kras* (Supplementary Figure S4A). I.F. based histological assessment, confirmed loss of CREB expression within the pancreata of Ptf1aCreERTM/+;Crebfl/fl (CC−/−) as compared to wild type Ptf1aCreERTM/+ (C) (Supplementary Figure S4B).
Creb deleted mice exhibited significant enhancement in the regenerative potential of acinar cells after ACP induction when compared to Ptf1aCreERTM/+ mice with wild type Creb at 3-day recovery period (Supplementary Figure S4C). Additionally, this improved recovery in CC−/− mice was further associated with a significant reduction in CK19 + ducts and attenuation in the fibroinflammatory milieu in these mice when compared to Ptf1aCreERTM/+ mice, thereby implicating CREB as a pivotal factor in acinar regeneration even in the absence of oncogenic KrasG12D.
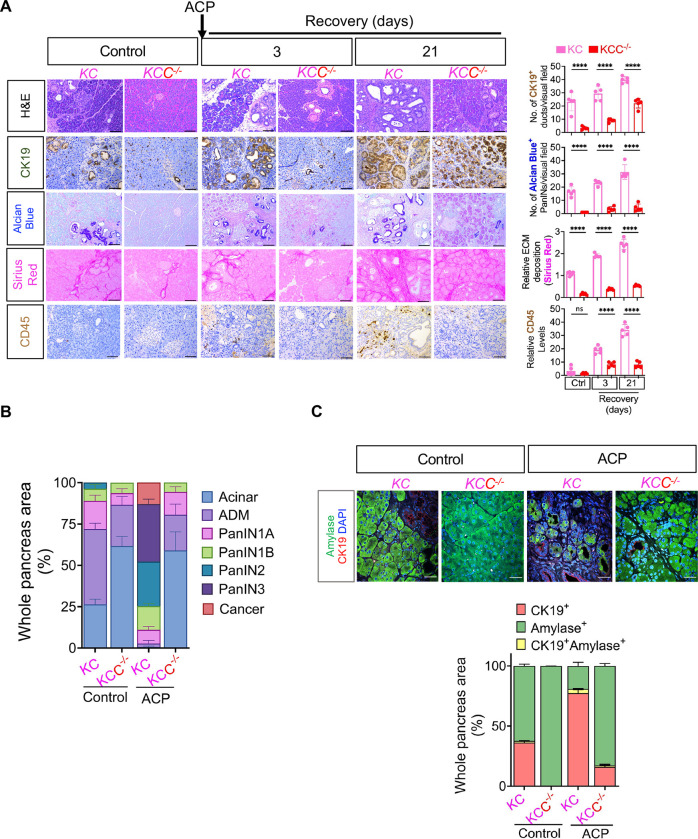
To comprehensively assess the capacity of acinar-specific Creb ablation to counteract Kras* driven ADM/PanIN originating from acini, we conducted both gross and histological comparison of KC and KCC−/− mice pancreata (both at 3- and 21-days of recovery period) post ACP induction as described in experimental schema of Supplementary Figure S5A. KC and KCC−/− mice maintained on a standard control liquid diet were employed as control animals to facilitate a comparative analysis of the distinct histological variations that emerge due to presence or absence of ACP induction arising due to CREB deletion (Figure 5A).
Figure 5.
Acinar specific ablation of Creb attenuates Kras* induced progression of ADM/PanINs towards pancreatic cancer with ACP. (A) Comparative histological evaluation of mouse pancreas, accompanied by representative photomicrographs, showcases H&E, CK-19, Alcian Blue, Sirius Red, and CD45+ staining within the pancreata of KC and KCC −/− mice in control and KC mice at 3- and 21-day ACP recovery period (left) and their quantification (right) (n=5 mice per group). (B) H&E-based histological quantification illustrating the presence of acinar cells, ADMs, PanINs and cancerous regions in control or within the pancreata of KC and KCC −/− mice harvested at 21 days of ACP recovery period (n=3 mice per group). (C) Representative pancreas images depicting amylase (green)/CK19 (red) and DAPI (blue) immunofluorescent labeling (top) and the corresponding quantification of CK19+ amylase+ cells (bottom) in control (KC and KCC −/−) or with ACP induction (n=3 mice per group). Scale bar, 50μm. ns nonsignificant; ****p<0.0001 by ANOVA.
ACP-induction in KC mice led to a pronounced increase in the relative pancreas weight (21 days of recovery period) confirming an increased tumor burden when compared to control KC mice (Supplementary Figure S5B). Notably, this marked increase in pancreas weight was less prominent in KCC−/− mice with ACP induction, where CREB had been deleted (Supplementary Figure S5B and C), altogether suggesting an association of attenuated tumor burden with loss of CREB expression.
The genetic depletion of Creb in the pancreata of KCC−/− mice resulted in a remarkable protective effect against ACP-triggered ADM and PanIN progression, as evident through histological analyses involving H&E, CK19, and alcian blue staining (Figure 5A and B). Detailed examination of KC mice pancreata after ACP induction displayed higher prevalence of high grade PanIN lesions along with the presence of pancreatic cancer, on the contrary the pancreatic tissue architecture of KCC−/− mice despite ACP induction displayed mostly normal organization of acinar cells with fewer ADMs and low grade PanIN lesions (Figure 5A and B). Additionally, a notable reduction in the fibroinflammatory milieu (Sirius red and CD45+ cells) was also observed (3 and 21 days of ACP recovery period) as compared to KC GEMM with intact CREB expression. Similar beneficial histological trends were also noted in the control KCC−/− mice group when compared against the control KC mice (Figure 5A and B).
Additionally, co-I.F. staining for amylase/CK19+ within the pancreatic tissues of KC vs. KCC−/− mice, showcased a notably heightened proportion of amylase-positive acinar cells in the Creb-depleted KCC−/− mice, suggesting a profound protective effect against ACP-induced injury during the recovery periods (Figure 5C). On the contrary, the pancreata of KC mice exhibited substantial damage resulting from ACP induction, as observed through high abundance of CK-19+ cells with concomitant loss of amylase+ acini.
Acinar Specific Ablation of CREB Attenuates ACP Induced Acinar to Ductal Reprogramming and Increases Tumor Latency Period
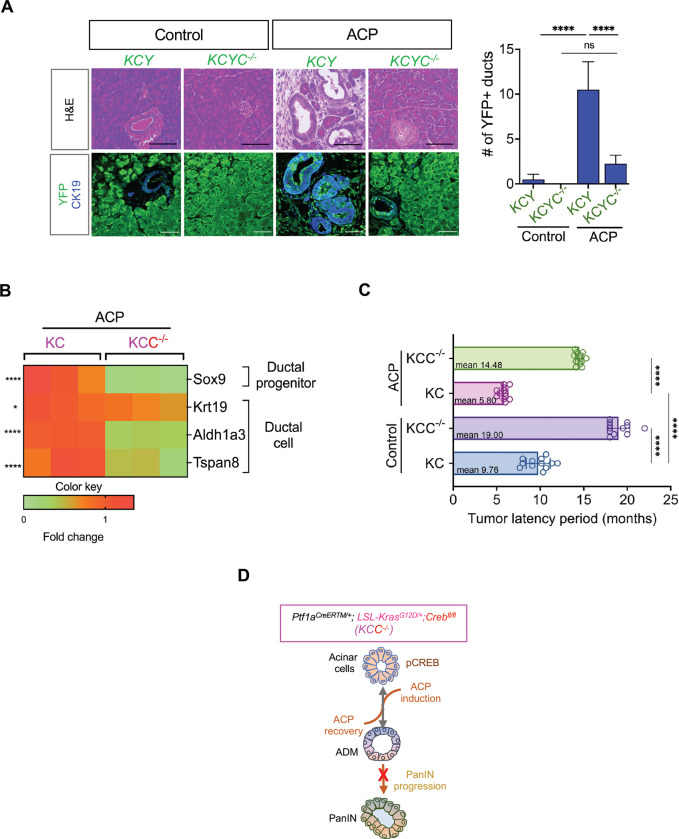
We next studied ACP-induced pancreatic injury in a lineage tracing mice model of KC (KCY; Ptf1a-CreERTM/+;LSL-KrasG12D/+;R26R-EYFP) and KCYC−/− (Creb deletion in KCY) mice (. Co-I.F. staining displayed an increased number of YFP+/CK19+ ductal lesions concurrent with the loss of acinar cell compartment upon ACP induction in KCY mice as compared to controls. We also found that a substantial reduction in the presence of ductal structures characterized by a minimal number of cells exhibiting dual positivity of YFP+/CK19+ during ACP induced KCYC−/− when compared to KCY mice. We verified these results that CREB promotes ductal phenotype using qPCR-based transcriptional profiling (Figure 6B). Our qPCR analysis confirmed notable decrease in the mRNA expression levels of genes associated with the ductal cell identity and function (Sox9, Krt19, Aldh1a3, and Tspan8) in the pancreas of ACP induced KCC−/− than in KC mice.
Figure 6.
Acinar specific ablation of Creb attenuates ADM reprogramming and increases tumor latency period with ACP induction in KC mice. (A) Representative H&E images of the pancreas along with co-immunofluorescent labeling (left) and quantification (right) of YFP (green)/CK19 (blue) in Ptf1a-CreERTM/+;LSL-KrasG12D/+;R26R-EYFP (KCY) and Crebfl/fl (KCYC−/−) as control or with ACP induction (n=4 mice per group). (B) qPCR-based analysis of pancreatic tissue harvested from KC and KCC−/− mice with ACP induction for acinar and ductal genes (n=3 mice per group). (C) Quantitative assessment of tumor latency period in ctrl or with ACP induction in KC and KCC−/− mice (n=12 mice per group). (D) Schematic representation illustrating that acinar-specific Creb ablation diminishes the progression of ADM/PanINs toward pancreatic cancer in the presence of ACP induction in KC mice. Scale bar, 50μm. nsnonsignificant; *p<0.05; ****p<0.0001 by ANOVA or unpaired t test.
Despite persistent inflammatory activation of ACP, Creb-deficient GEMMs (KCC−/− and KCYC−/−) displayed low-grade PanIN lesions and diminished pancreatic ductal phenotype, which further reflected in tumor latency (Figure 6C). ACP-induced KCC−/− mice pancreata had a mean pathologically apparent malignancy duration of 14.48 months, compared to 5.80 months for KC-ACP mice, on the contrary control KC harboring wild type Creb and KCC−/− exhibited greater tumor latency (9.76 vs. 19 months) (Figure 6C). Overall, these results suggest that acinar-specific Creb ablation reduces ACP-induced acinar-to-ductal reprogramming and Kras*-induced neoplastic progression, delaying tumor burden (Figure 6D).
Discussion
Chronic alcohol consumption results in the development of persistent inflammation, leading to detrimental effects on the pancreas, and elevate the likelihood of cancer development. However, the precise mechanisms underlying this phenomenon are not fully understood. The present study entails developing alcoholic chronic pancreatitis (ACP) in multiple mice models by administering alcohol conjunction with supraphysiological doses of caerulein. This model of chronic pancreatic injury involving ACP induces more substantial damage to the pancreas compared to the effects observed with alcohol or CP alone. ACP influences various subcellular compartments within the pancreas, exhibiting characteristics previously identified in human disease, as substantiated by prior studies13, 23–28. The chronic inflammatory stimulus of ACP causes transient injury to pancreatic tissue even without the presence of oncogenic Kras*. This injury is resolved upon removal of the external insult. Nevertheless, when Kras* is present, ACP exhibits synergistic cooperation, sustaining pancreatic tissue injury and promoting high-grade PanIN lesions along with heightened fibroinflammatory milieu which leads to the development of pancreatic cancer. Significantly, we have seen persistent activation of CREB in acinar cells that are undergoing dedifferentiation towards ductal structures. This persistent CREB activation is identified as a critical molecular factor driving inflammation-associated carcinogenesis. The CREB activation synergizes with ACP induction and Kras*. Moreover, by employing lineage tracing, histological analysis, and molecular profiling, studies conducted on acinar-specific Creb deletion in KC mice has established the activation of regenerative mechanisms in the pancreas and mitigates PanIN progression, regardless of the presence or absence of ACP induction. The findings provide a comprehensive molecular framework focused on CREB, which enhances our understanding of how genetic determinants and inflammation interact to promote the development of pancreatic cancer originating from acinar cells.
The adult pancreas contains pancreatic acinar cells that exhibit significant plasticity and can transform into a progenitor-like cell type with ductal characteristics. This process is known as acinar to ductal metaplasia (ADM) and plays a crucial role in the repopulation of the pancreas during prolonged inflammatory damage 25, 29–31. Consistent with this finding, our study shows that in Ptf1aCreERTM+/− mice without Kras*, there is rapid and reversible adaptability of ADM and reduction of fibroinflammatory milieu which appears to contribute to the histological resolution of injured pancreas tissue upon ACP induction.
Approximately 90–95% of patients with pancreatic cancer have activating, oncogenic KRAS mutations 32, 33. Prior studies using animal models have shown that the combination of Kras* and chronic inflammation causes pancreatic acinar cells to undergo irreversible transdifferentiation towards ADMs, leading to the rapid progression of advanced PanINs and pancreatic cancer 19, 29, 30, 34. In accordance with these findings, our analysis of the histological and molecular profiling reveals that confounding ACP induction in KC mice greatly impairs the regeneration of pancreatic parenchyma. Furthermore, it leads to irreversible reprogramming of acinar cells into ADM and high-grade PanIN lesions, accompanied by a notable accumulation of fibrotic stroma. This, in turn, confirmed that the presence of Kras* hinders tissue repair following ACP induction.
Chronic pancreatic injury leads to local inflammation, which not only potentiate intrinsic alterations in acinar cell fate but also orchestrate a fibroinflammatory response 13, 25. This work showcases a significant synergistic interaction between Kras* and ACP, resulting in a substantial accumulation of collagen deposition. This is accompanied by an augmented presence of activated pancreatic stellate cells (PSCs) in the pancreas. This suggests a potential correlation between increased fibrosis and the heightened occurrence of activated PSCs or fibroblasts. However, additional studies are needed to investigate the precise mechanisms and potential role of CREB regulated secreted factors for activation of PSCs in relation to ACP.
A complex interplay of transcriptional gene networks, acinar cell homeostasis, exocrine function, and cellular signaling during pancreatic regeneration has been reported previously35. This coordinated transition from a mesenchymal, progenitor-like state to a proliferative acinar cell appears to be dysregulated in Kras* mice35. In line with our findings, we have discovered grave damage to the acinar cells in the context of ACP. Our analysis of scRNA transcriptomics within the acinar subcluster unveiled notable enrichments in pathways associated with both apoptosis and cell proliferation. ScRNAseq analysis demonstrates the emergence of ductal progenitors (ADM) in KC mice with ACP induction, suggesting increased propensity of epithelial-mesenchymal transition (EMT). This highlights the transcriptional reprograming of acinar cells towards a ductal phenotype.
The various signaling nodes, downstream of the Kras* pathway, activate different transcription factors that have been linked to the irreversible development of ADM/PanINs 11. We present new evidence that demonstrates hyper activation of transcription factor CREB in acinar cells undergoing acinar to ductal reprogramming, which occurs during the induction of ACP. In this study, we showed that the expression of CREB was essential for the formation of ADM in an ex-vivo explant culture studies. Furthermore, genetic ablation of acinar specific CREB significantly attenuated the development of both spontaneous and ACP-induced ADM/PanIN formation, while simultaneously promoting the regeneration of acinar cells in KC mice. These data suggest that CREB plays a crucial role in driving the growth of pancreatic cancer and facilitates the synergism between Kras* and ACP. Previous studies, including our own, have demonstrated the crucial function of CREB as an oncogenic transcription factor associated with severity of this disease16, 36. Upon phosphorylation at Ser-133, CREB becomes active and interacts with coactivator, the CREB-binding protein (CBP), enabling the recruitment of other essential transcriptional machinery elements necessary to drive oncogenic molecular processes 15. Within this context, Kim et al., have recently demonstrated that the activation of CREB, downstream of Kras* signaling, interacts with mutant p53, subsequently this leads to the activation of transcriptional programs to promote metastasis of pancreatic cancer 36. In addition, the use of small molecule inhibitors to target this oncogenic factor has been shown to attenuate the severity of the disease in both GEMM and orthotopic models of pancreatic cancer 16, 36, 37. Although some aspects of CREB’s role in promoting pancreatic tumorigenesis have been clarified by these studies, particularly its interaction and regulation with cancer cell intrinsic mediators, Therefore, further research on the KCC−/− epithelium is essential to fully comprehend how the loss of acinar cell intrinsic Creb mitigates ADM/ PanIN progression in the presence of Kras*, with or without ACP induction.
We conducted a comprehensive analysis of the KCC −/− mice model with Creb deletion to determine its involvement in the advancement of pancreatic cancer. While activation of CREB with Kras* promotes transdifferentiation of acinar cells towards ductal phenotype in wild type KC mice, genomic loss of Creb in KCC −/− mice pancreata results in regression of ADM/PanIN lesions and promotes re-differentiation of transformed ductal cells to acinar cells. Our current study has shown that deleting Creb in acinar cells leads to profound remodeling of surrounding fibroinflammatory reaction, suggesting that Creb plays a role in affecting the surrounding microenvironment. Previously, our research has determined that CREB promotes transcriptional upregulation and secretion of pro-inflammatory chemokines/cytokines 38. CREB mediated control in the secretion of immune modulatory factors helps facilitate the communication between epithelial and myeloid cells in pancreatic cancer. Myeloid cells, which are part of innate immune system, have an instructive role in regulating epithelial cell identity and plasticity in the presence of Kras* 17. Hence, it is possible that the increased presence of ductal lesions and the accelerated onset of tumor formation in ACP-induced KC mice, could be ascribed to the augmented oncogenic activity of CREB, which propels the immune regulatory mechanisms. Ongoing investigations in our laboratory are dedicated in deciphering the molecular mechanism of CREB mediated control of the innate immune regulation and its interaction with the epithelial-myeloid cross talk.
In summary, our study demonstrates that the induction of ACP causes acinar cells to transform into aggressive neoplasms through irreversible ADM reprogramming them towards a ductal phenotype in the presence of Kras*. Notably, the continuous presence of ductal lesions originating from acinar cells exhibits sustained hyperactivation of CREB associated with this phenotypic shift when compared to the pancreas of mice in the control group. Acinar-specific Creb ablation attenuated the formation of advanced PanIN lesions, impeding tumor progression in KC mice induced with ACP. Our research establishes a compelling foundation for further investigations into the involvement of CREB as a critical molecular determinant in the advancement of pancreatic cancer driven by ACP, in conjunction with Kras*. These findings provide a foundation for the potential development of rational therapeutic strategies focused on identifying, preventing, and intercepting cancers before they progress to an intractable stage.
Methods
Creb Deletion in Genetically Engineered Ptf1aCreERTM/+;LSL-KrasG12D+/− (KC) Mice Model
Ptf1aCreERTM/+ knock-in allele mice were obtained from Jackson Laboratory (Bar Harbor, ME; stock number: 019378) 22 and crossed with LSL-KrasG12D/+ to generate acinar-specific KrasG12D/+ mutant mice, referred to as Ptf1aCreERTM/+;LSL-KrasG12D+/− (KC) mice. To generate acinar-specific Creb1 (Creb) knockout mice, Ptf1aCreERTM/+; Crebfl/fl mice were crossed with the LSL-KrasG12D/+; Crebfl/fl to generate Ptf1aCreERTM/+;LSL-KrasG12D/+; Crebfl/fl (KCC−/−) mice. The Crebfl/fl mice were obtained from Professor Eric Nestler (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY 11724)39. More precisely, Creb was selectively excised from acinar cells in LSL-KrasG12D/+ mice by utilizing inducible Cre recombinase, which was consistently expressed by the Ptf1aCreERTM/+ promoter. Pancreas-specific Cre recombinase activity was induced in 6-week-old mice through the administration of tamoxifen (Sigma-Aldrich, cat. # T5648) daily for 6 consecutive days at a dose of 0.15 mg/g body weight followed by rest for a week before onset of ACP induction phase.
Lineage Tracing in R26REYFP Reporter Mice
R26REYFP mice (obtained from The Jackson Laboratory; stock number: 006148) 40 were procured for lineage tracing investigations. To precisely trace the lineage of recombined acinar cells, Ptf1aCreERTM/+ mice, containing the R26REYFP reporter, were crossed with LSL-KrasG12D/+ mice to generate Ptf1aCreERTM/+;LSL-KrasG12D/+; R26REYFP. These mice were then bred with Crebfl/fl mice to produce Ptf1aCreERTM/+;LSL-KrasG12D/+; R26REYFP; Crebfl/fl (hereafter referred to as KCYC−/−) mice.
Mouse Genotyping
Genotyping was conducted by an automated genotyping service provider Transnetyx (Cordoba, TN, USA). The sequence of probes used for genotyping analysis has been listed in Supplementary Table S1.
Establishing experimental induction of Alcoholic Chronic pancreatitis (ACP) in-vivo
The experimental mice were pair-fed with alcohol for 14 weeks using Lieber-DeCarli alcohol-based liquid diet (A) (Bioserv Inc. cat. #F1259SP), containing 5% vol/vol ethanol, while control mice received a standard control-liquid diet (C) (Bioserv Inc. cat. #F1259SP.) with 28% carbohydrates instead of ethanol. During the final 4 weeks of alcohol exposure, chronic pancreatitis (CP) was induced by administering caerulein, solubilized in PBS to achieve a concentration of 10 mg/mL (ACP induction period). This feeding regimen has previously been reported to mimic pancreatic damage due to chronic alcohol use in humans 13, 41, 42. Caerulein was delivered intraperitoneally at a dose of 50 μg/kg through hourly injections (6 times daily, for 3 days per week) over a 4-week period. This combined impact of alcohol and caerulein treatment characterized the ACP group. Animals were euthanized in a humane manner to harvest blood and pancreas after ACP induction on days 3 or 21 (ACP recovery period). A comprehensive overview of the treatment groups, spanning both induction and recovery phases, is provided in Supplementary Table S2.
Tumor Latency Period Estimation
In the context of GEMMs, tumor latency period refers to the duration between tumor initiation or formation to the point at which these pancreatic tumors became grossly discernible during biweekly abdominal palpation examinations. Mice exhibiting noticeable signs of illness attributed to an increased tumor burden were euthanized. Subsequently, their pancreatic tissues were collected and subjected to H&E staining for pathological analysis.
Animal Studies
Mice of both sexes weighing 20–25 grams used in this study were housed in pathogen-free conditions under a 12-hour light dark diurnal cycle with a controlled temperature of (21°C–23°C) and maintained on standard rodent chow diet (Harlan Laboratories) before the onset of the experimental induction of ACP protocol. Mice were euthanized upon manifestation of signs of compromised health, including weight loss, accelerated respiration, hunched posture, piloerection, and reduced activity. All animal experiments were approved and performed in compliance with the regulations and ethical guidelines for experimental and animal studies of the Institutional Animal Care (IACUC) and the University of Miami (Miami, FL; Protocol No. 15–057, 15–099, 18–081 and 21–093).
Serum Alcohol Analysis
The alcohol concentration in the serum was estimated using an ethanol assay kit (Abcam, cat. # ab65343) as per the manufacturer’s protocol and has been detailed previously 13.
Serum Amylase Analysis
The serum amylase levels in blood samples collected from various study groups of mice were quantified through a colorimetric assay (Abcam, cat. # ab102523). This analysis was conducted in accordance with the manufacturer’s provided protocol and described in detail previously 13.
Immunohistochemistry (IHC) and Immunofluorescence (IF) Tissue Staining
Harvested pancreas tissues from mice necropsies were fixed in 10% neutral buffered formalin and embedded in paraffin to carry out histological staining procedures including Hematoxylin and Eosin (H&E), Sirius Red, Masson’s trichrome and Alcian Blue. For immunohistochemistry (IHC) based detection, antigen retrieval was performed using citrate (pH 6.0) or Tris-EDTA buffer as previously described prior to incubation with BlockAid Blocking Solution (Thermo Fisher). For IHC, endogenous peroxidase activity was blocked by incubating with 3% H2O2. Tissue sections were stained with primary antibodies at specified concentrations (Supplementary Table S3) overnight at 4°C. IHC slides were developed using 3,3’ diaminobenzidine (DAB) substrate (Vector) followed by counterstain using Meyer’s hematoxylin and imaged using DM750 microscope (Leica Microsystems). For IF based staining, primary antibody was detected using species-specific Alexa Fluor 594 and/or Alexa Fluor 488 (Thermo Fisher) secondary antibodies incubated on sections for 1 hour at room temperature. Nuclear staining was performed using Hoechst 33342 dye (Thermo Fisher). Imaging was performed using the Olympus Fluoview1000 confocal microscope. Stained tissues samples were quantified using ImageJ image analysis software (NIH, Bethesda, MD) as a percentage of positive staining.
H&E based Assessment of Pancreatic Lesions
Pancreatic tissue sections from age-matched mice in each group were stained with H&E and subsequently examined for ADMs, pancreatic lesions and carcinoma in situ. The percentage of acinar area and number of ducts that contained any grade of PanIN lesions were measured by examining 10 H&E-stained high-power fields (40× magnification) per slide 43. PanINs were graded according to established criteria44. In PanIN1 ducts, the normal cuboidal pancreatic epithelial cells transition to columnar architecture and can gain polyploid morphology. PanIN2 lesions are associated with a loss of polarity. PanIN3 lesions (or in-situ carcinoma) show cribriform morphology, the budding off cells and luminal necrosis with marked cytological abnormalities, without invasion beyond the basement membrane. The data were expressed as a percentage of total lesions in the whole pancreas.
Western blot Analysis
The freshly harvested pancreas tissue was promptly flash-frozen and preserved at −80°C for long-term storage. To prepare protein lysates for Western blot analysis, the frozen tissues were thawed and homogenized in RIPA buffer (0.1% SDS, 50 mM Tris·HCl, 150 mM NaCl, 1% NP-40, and 0.5% Na deoxycholate) with protease inhibitor cocktail (Sigma, St. Louis, MO) and PhosSTOP phosphatase inhibitor (Roche, Indianapolis, IN, USA). Lysates were sonicated and centrifuged at 10,000 g for 15 minutes at 4°C to collect supernatant. The protein concentration of the cell and tissue lysate was determined by Bio-Rad protein assay kit (Bio-Rad, Hercules, CA). 35 μg of whole-cell lysate or whole-tissue lysate was separated on NuPAGE Novex 4–12% Bis-Tris Gels and transferred on iBlot transfer stack using iBlot dry blotting transfer system (Life Technologies). For immune-detection, membranes were incubated with antibodies listed in Supplementary Table S2. The membranes were subsequently incubated with secondary anti-mouse or anti-rabbit secondary antibodies conjugated with horseradish peroxidase (Jackson ImmunoResearch). Finally, the immunoreactive bands were developed with Pierce ECL Western Blotting Substrate (Thermo Scientific) and recorded on blue basic autoradiography film (Bioexpress). Uncropped images of the blots are shown in Supplementary Figure S6.
Phosphokinase Array Analysis
The phosphorylation profiles of multiple effector kinases were screened using a phospho-kinase antibody array (R&D System, cat. #ARY003B). In brief, 200 mg of equal protein from pancreatic tissue lysates were loaded onto a nitrocellulose membrane with duplicate capture antibody spots. Phosphorylated protein levels were determined with phospho-specific antibodies and detected through chemiluminescent based reaction chemistry. Spot density on the membrane was quantified using HL++ image analysis software.
Pancreas Digestion for Single cell RNA Sequencing and Library Generation
Mouse pancreas tissue harvested from control KC and KC with ACP induction were mechanically dissociated to generate single cell suspensions as described previously 20. Pancreas fragments of size 1–2 mm were then immersed in 0.02% trypsin C-EDTA 0.05% (Biological Industries) for 10 minutes at 37 °C with agitation, followed by washing with 10% FCS/DMEM. Subsequently, for the next dissociation step, the cells underwent a wash with HBSS × 1 containing 1 mg/ml collagenase P, 0.2 mg/ml bovine serum albumin (BSA), and 0.1 mg/ml trypsin inhibitor. After incubation for 20–30 minutes at 37 °C with agitation, the samples were pipetted up and down and returned to 37 °C and passed through a 70 mm nylon mesh (Corning #431751), and washed twice with HBSS × 1 containing 4% BSA and 0.1 mg/ml DNase I. The samples were divided into two equal volumes, with one sample subjected to centrifugation and washed three times at 60g to isolate large cells containing acinar cells. Simultaneously, the second sample underwent centrifugation and three washes at 300g to collect all cells. In cases where an abundance of red blood cells was observed in the second sample, the cells were treated with red blood cell lysing buffer (Sigma Aldrich). Live cells were isolated using the MACS dead cells removal kit (Miltenyi Biotech #130–090-101). Finally, the two samples were combined at a cell ratio of 30% from the 60g samples and 70% from the 300g samples and processed for single cell RNA sequencing library preparation by Oncogenomics-Shared Resource Facility (OGSR, University of Miami). In brief, cells were counted, and 10,000 cells were loaded per lane on the 10x Chromium microfluidic chips. Single-cell capture, barcoding, and library preparation were performed using the Chromium 85 Controller, Chromium Next GEM Single Cell 3’ GEM, Library & Gel Bead Kit v3.1, and the Chromium Next GEM Chip G kit (10xGenomics) with a cell recovery target of 100,000 cells as per manufacturer’s guidelines. cDNA and libraries were then sequenced on an Illumina NovaSeq 6000. The Cell Ranger pipeline (version 7.0, 10xGenomics) was employed to transform Illumina base call files into FASTQ files, alignment of these FASTQ files to the GRCm38 reference genome, and generation of digital gene-cell counts matrix was carried out by the Biostatistics and Bioinformatics Shared Resource facility (BBSR, University of Miami). Subsequently, the count matrices were imported into R version 3.5.0 and subjected to analysis using the R package Seurat version 4.0.13–15. All scRNA seq datasets produced here will be deposited on the Gene Expression Omnibus platform.
Cluster Identification and Annotation of Single-cell RNA Sequencing Dataset
Gene-cell matrices were analyzed with Seurat package (v4.3.0 RStudio). Cells fewer than 200 transcripts and ≤ 7.5% mitochondrial counts were removed. Feature measurements were normalized using the Normalize Data function with a scale factor of 10,000 and the LogNormalize normalization method. Variable genes were identified using the FindVariableFeatures function. Data were scaled and centered using linear regression on the counts and the cell cycle score difference. Principal Component Analysis (PCA) was performed with the RunPCA function using the previously defined variable genes to identify necessary dimensions for >90% variance within the data. Violin plots were then used to filter the data according to user-defined criteria. Cell clusters were obtained via the FindNeighbors and FindClusters functions, using a resolution of 0.7 for all samples, and non-linear dimensional reduction was then performed using Uniform Manifold Approximation and Projection (UMAP) clustering. A FindAllMarkers table was created, and clusters were defined by user-defined criteria. Clusters which co-expressed known distinct marker genes were merged for subsequent analysis.
Differential Gene Expression Analysis of Single-cell RNA Sequencing Dataset
Prior to differential gene expression analysis, each cluster of interest was subjected to normalization, scaling, and PCA. Next, the function “FindMarkers” from Seurat v4.0 R package was utilized to finds the differentially expressed genes for identity classes. Genes were considered differentially expressed if detected in at least 25% of clusters, with default log fold change of 0.25. Wilcoxon Rank Sum test was used. Volcano plots were generated based on these output files using the EnhancedVolcano package. Gene set enrichment analysis (GSEA) was performed using the “fgsea” R package on differentially expressed genes (log(FC) >0.5 and adjusted p-value<0.05). “MSigDB” R package was utilized to access the following databases: C2 (KEGG, REACTOME, PID, BIOCARTA), C5 (GO:BP) and H (Hallmarks). Among different databases, the gene sets that met the statistical requirements were then curated, visualized via “ggplot2” R package, and ordered by Normalized Enrichment Score (NES). RNA Isolation and qPCR Analysis
RNA was isolated from flash frozen pancreas tissues using the RNeasy Kit (Qiagen) according to the manufacturer’s protocol. cDNA generated after performing reverse transcription of RNA product was subjected to quantitative PCR (qPCR) analysis using gene-specific predesigned primers (RT2 qPCR Primer Assay, Qiagen), listed in Supplementary Table S4. Gene expression was normalized to the housekeeping gene GAPDH using the comparative CT (ΔΔCT) method and reported as fold change relative to control.
Isolation of Primary Pancreatic Acinar cells and 3D Explant Culture
Isolation of primary pancreatic acinar cells and establishment of 3D explant cultures was performed as described previously 45. Briefly, the acinar cells were incorporated into a mixture of collagen and Waymouth medium. On days 1, 3, and 5, duct-like structures were counted and quantified in a blinded fashion. The area of the ducts was determined using ImageJ software. Quantification of ductal structures involved the manual counting of atleast four distinct fields under 10× magnification in triplicates.
Statistical Analysis
Descriptive statistics were calculated using Prism software (GraphPad Software Inc). Results are shown as values of means ± SD unless otherwise indicated. To assess multiple comparisons, one way ANOVA was applied followed by Tukey’s or Dunnett’s tests when deemed appropriate. A two-tailed Student’s t test was used for two group comparisons. Statistical significance was defined using a cutoff of 0.05, except when indicated in the figure legend otherwise.
Supplementary Material
Acknowledgements
The authors thank Dr. Erin Dickey for her assistance in the editing process and Dr. Oliver McDonald for his help in assessing the histopathology sections. We gratefully acknowledge Analytical Imaging Shared Resource (AISR), Onco-Genomics Shared Resource (OGSR); research histopathological services provided by Cancer Modeling Shared Resource (CMSR), and Biostatistics and Bioinformatics Shared Resource (BBSR) core facilities provided by University of Miami, Sylvester Comprehensive Cancer Center (SCCC). BioRender was used for the creation of Figure 4A (License agreement SC265R8O4S) and Supplementary Figure S4A (License agreement RS265R8Z28).
Funding
The research described in this study received financial support from the National Institutes of Health (NIH) National Cancer Institute (NCI) through the R01 CA262526 grant. Additionally, funding was provided by the Florida Department of Health grant 22K06, specifically through the James Esther and King Biomedical Research Program, awarded to N.S. Nagathihalli. The Histopathology Core Service was conducted with the assistance of the Sylvester Comprehensive Cancer Center (SCCC) support grant, under the supervision of N. Nagathihalli. The research presented in this paper received financial support from the NCI of the NIH through Award Number P30 CA240139. The authors bear full responsibility for the content and the opinions expressed in this work, which may not necessarily reflect the official perspectives of the NIH.
Footnotes
Conflict of Interest: The authors declare no potential conflicts of interest.
References
- 1.Peery AF, Crockett SD, Murphy CC, et al. Burden and Cost of Gastrointestinal, Liver, and Pancreatic Diseases in the United States: Update 2021. Gastroenterology 2022;162:621–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nieto LM, Salazar M, Kinnucan J, et al. Incidence, Burden, and Predictors of Readmission for Acute Alcoholic Pancreatitis: A National Analysis over 11 Months. Dig Dis Sci 2023;68:423–433. [DOI] [PubMed] [Google Scholar]
- 3.Singer MV. Effect of ethanol and alcoholic beverages on the gastrointestinal tract in humans. Rom J Gastroenterol 2002;11:197–204. [PubMed] [Google Scholar]
- 4.Dufour MC, Adamson MD. The epidemiology of alcohol-induced pancreatitis. Pancreas 2003;27:286–90. [DOI] [PubMed] [Google Scholar]
- 5.Yadav D, Lowenfels AB. The epidemiology of pancreatitis and pancreatic cancer. Gastroenterology 2013;144:1252–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masamune A, Watanabe T, Kikuta K, et al. Roles of Pancreatic Stellate Cells in Pancreatic Inflammation and Fibrosis. Clinical Gastroenterology and Hepatology 2009;7:S48–S54. [DOI] [PubMed] [Google Scholar]
- 7.Bosetti C, Lucenteforte E, Silverman DT, et al. Cigarette smoking and pancreatic cancer: an analysis from the International Pancreatic Cancer Case-Control Consortium (Panc4). Annals of Oncology 2012;23:1880–1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strobel O, Dor Y, Alsina J, et al. In vivo lineage tracing defines the role of acinar-to-ductal transdifferentiation in inflammatory ductal metaplasia. Gastroenterology 2007;133:1999–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris JPt, DA Cano, Sekine S, et al. Beta-catenin blocks Kras-dependent reprogramming of acini into pancreatic cancer precursor lesions in mice. J Clin Invest 2010;120:508–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kopp JL, von Figura G, Mayes E, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell 2012;22:737–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat Rev Gastroenterol Hepatol 2017;14:296–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Storz P, Crawford HC. Carcinogenesis of Pancreatic Ductal Adenocarcinoma. Gastroenterology 2020;158:2072–2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mehra S, Srinivasan S, Singh S, et al. Urolithin A attenuates severity of chronic pancreatitis associated with continued alcohol intake by inhibiting PI3K/AKT/mTOR signaling. Am J Physiol Gastrointest Liver Physiol 2022;323:G375–g386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johannessen M, Delghandi MP, Moens U. What turns CREB on? Cell Signal 2004;16:1211–27. [DOI] [PubMed] [Google Scholar]
- 15.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem 1999;68:821–61. [DOI] [PubMed] [Google Scholar]
- 16.Srinivasan S, Totiger T, Shi C, et al. Tobacco Carcinogen-Induced Production of GM-CSF Activates CREB to Promote Pancreatic Cancer. Cancer Res 2018;78:6146–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y, Yan W, Mathew E, et al. Epithelial-Myeloid cell crosstalk regulates acinar cell plasticity and pancreatic remodeling in mice. Elife 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gidekel Friedlander SY, Chu GC, Snyder EL, et al. Context-dependent transformation of adult pancreatic cells by oncogenic K-Ras. Cancer Cell 2009;16:379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guerra C, Collado M, Navas C, et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell 2011;19:728–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schlesinger Y, Yosefov-Levi O, Kolodkin-Gal D, et al. Single-cell transcriptomes of pancreatic preinvasive lesions and cancer reveal acinar metaplastic cells’ heterogeneity. Nat Commun 2020;11:4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hosein AN, Huang H, Wang Z, et al. Cellular heterogeneity during mouse pancreatic ductal adenocarcinoma progression at single-cell resolution. JCI Insight 2019;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kopinke D, Brailsford M, Pan FC, et al. Ongoing Notch signaling maintains phenotypic fidelity in the adult exocrine pancreas. Developmental Biology 2012;362:57–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vonlaufen A, Phillips PA, Xu Z, et al. Withdrawal of alcohol promotes regression while continued alcohol intake promotes persistence of LPS-induced pancreatic injury in alcohol-fed rats. Gut 2011;60:238–46. [DOI] [PubMed] [Google Scholar]
- 24.Lugea A, Tischler D, Nguyen J, et al. Adaptive unfolded protein response attenuates alcohol-induced pancreatic damage. Gastroenterology 2011;140:987–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu S, Chheda C, Ouhaddi Y, et al. Characterization of Mouse Models of Early Pancreatic Lesions Induced by Alcohol and Chronic Pancreatitis. Pancreas 2015;44:882–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dawra R, Sah RP, Dudeja V, et al. Intra-acinar trypsinogen activation mediates early stages of pancreatic injury but not inflammation in mice with acute pancreatitis. Gastroenterology 2011;141:2210–2217.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sah RP, Dudeja V, Dawra RK, et al. Cerulein-induced chronic pancreatitis does not require intra-acinar activation of trypsinogen in mice. Gastroenterology 2013;144:1076–1085.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tao J, Cheema H, Kesh K, et al. Chronic pancreatitis in a caerulein-induced mouse model is associated with an altered gut microbiome. Pancreatology 2022;22:30–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Logsdon CD, Ji B. Ras Activity in Acinar Cells Links Chronic Pancreatitis and Pancreatic Cancer. Clinical Gastroenterology and Hepatology 2009;7:S40–S43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rooman I, Real FX. Pancreatic ductal adenocarcinoma and acinar cells: a matter of differentiation and development? Gut 2012;61:449–458. [DOI] [PubMed] [Google Scholar]
- 31.Liou G-Y, Döppler H, Necela B, et al. Macrophage-secreted cytokines drive pancreatic acinar-to-ductal metaplasia through NF-κB and MMPs. Journal of Cell Biology 2013;202:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.di Magliano MP, Logsdon CD. Roles for KRAS in pancreatic tumor development and progression. Gastroenterology 2013;144:1220–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey P, Chang DK, Nones K, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature 2016;531:47–52. [DOI] [PubMed] [Google Scholar]
- 34.Guerra C, Schuhmacher AJ, Cañamero M, et al. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 2007;11:291–302. [DOI] [PubMed] [Google Scholar]
- 35.Kong B, Bruns P, Behler NA, et al. Dynamic landscape of pancreatic carcinogenesis reveals early molecular networks of malignancy. Gut 2018;67:146–156. [DOI] [PubMed] [Google Scholar]
- 36.Kim MP, Li X, Deng J, et al. Oncogenic KRAS Recruits an Expansive Transcriptional Network through Mutant p53 to Drive Pancreatic Cancer Metastasis. Cancer Discov 2021;11:2094–2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arensman MD, Telesca D, Lay AR, et al. The CREB-binding protein inhibitor ICG-001 suppresses pancreatic cancer growth. Mol Cancer Ther 2014;13:2303–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bianchi A, De Castro Silva I, Deshpande NU, et al. Cell-Autonomous Cxcl1 Sustains Tolerogenic Circuitries and Stromal Inflammation via Neutrophil-Derived TNF in Pancreatic Cancer. Cancer Discovery 2023;13:1428–1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Covington HE 3rd, Maze I, Sun H, et al. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron 2011;71:656–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.DelGiorno KE, Hall JC, Takeuchi KK, et al. Identification and Manipulation of Biliary Metaplasia in Pancreatic Tumors. Gastroenterology 2014;146:233–244.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perides G, Tao X, West N, et al. A mouse model of ethanol dependent pancreatic fibrosis. Gut 2005;54:1461–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Souza El-Guindy NB, Kovacs EJ, De Witte P, et al. Laboratory models available to study alcohol-induced organ damage and immune variations: choosing the appropriate model. Alcohol Clin Exp Res 2010;34:1489–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aykut B, Pushalkar S, Chen R, et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 2019;574:264–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hruban RH, Adsay NV, Albores-Saavedra J, et al. Pancreatic intraepithelial neoplasia: a new nomenclature and classification system for pancreatic duct lesions. Am J Surg Pathol 2001;25:579–86. [DOI] [PubMed] [Google Scholar]
- 45.Liou GY, Döppler H, Braun UB, et al. Protein kinase D1 drives pancreatic acinar cell reprogramming and progression to intraepithelial neoplasia. Nat Commun 2015;6:6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.