Abstract
Human endogenous retroviruses (HERVs) were first identified almost 20 years ago, and since then numerous families have been described. It has, however, been difficult to obtain a good estimate of both the total number of independently derived families and their relationship to each other as well as to other members of the family Retroviridae. In this study, I used sequence data derived from over 150 novel HERVs, obtained from the Human Genome Mapping Project database, and a variety of recently identified nonhuman retroviruses to classify the HERVs into 22 independently acquired families. Of these, 17 families were loosely assigned to the class I HERVs, 3 to the class II HERVs and 2 to the class III HERVs. Many of these families have been identified previously, but six are described here for the first time and another four, for which only partial sequence information was previously available, were further characterized. Members of each of the 10 families are defective, and calculation of their integration dates suggested that most of them are likely to have been present within the human lineage since it diverged from the Old World monkeys more than 25 million years ago.
Like those of other vertebrates, the human genome contains evidence for past infection by many different kinds of retroviruses (10, 27, 33, 61). Retroviral integration usually occurs within somatic cells, but occasionally such events take place within germ line cells, and when this occurs the retroviral sequences are passed vertically from parent to offspring (57). Such endogenous retroviruses generally remain replication competent until inactivated either by recombinational deletion between two repeat regions (termed long terminal repeats [LTRs]) situated at the 5′ and 3′ ends of the virus or by random mutation which occurs while the host genome is undergoing DNA replication (10, 53). During the period between insertion and inactivation, the viral copy number may increase via retrotransposition to different locations within the genome (53). The vertical transmission of these elements can occur over long periods of time; several replication-competent porcine retroviruses probably first infected their hosts more than 5 million years ago, and a number of defective endogenous human retroviruses are thought to have been present in the primate lineage for tens of millions of years (2, 8, 31, 43, 50).
Many types of human endogenous retroviruses (HERVs) have been characterized previously, and they have been classified into different groups, or families, partly on the basis of their sequence identity and partly according to the similarity of their primer binding sites (PBSs) to host tRNAs. Thus, members of the HERV.H family contain a PBS with a sequence similar to a region of tRNAHis, whereas the HERV.E family is primed by tRNAGlu. Despite the large amount of data available, the classification of the many different HERV families within an overall phylogenetic framework has been hampered for several reasons: (i) some highly divergent retroviruses are primed by the same type of tRNA; (ii) many HERV families have not been fully characterized, and the sequence information that has been reported is often derived from different genomic regions, making interfamily comparisons problematic; and (iii) the relative lack of sequence information on other host taxa has made it difficult to distinguish between genuinely monophyletic HERV families and polyphyletic families that appear monophyletic only because similar viruses in other hosts have not yet been described.
Recently these problems have been lessened both by the systematic isolation of endogenous retroviruses from many different vertebrate taxa and by the generation of large amounts of sequence data by the Human Genome Mapping Project (HGMP) (7, 16, 32). As of December 1998, sequence information was available for over 10,000 BACs, or cosmids, representing approximately 235,000,000 bp, or 7% of the human genome (5).
In this study, I investigated the relationships of the known HERV families to each other and to other nonhuman retroviruses, described and characterized six novel HERV families, and further characterized an additional four families for which only partial sequence information was previously available.
MATERIALS AND METHODS
Identification of HERVs within sequence data banks.
HERV sequences were obtained from the EMBL/GenBank/DDBJ database at the beginning of December 1998. Initially the computer data banks were screened by BLAST search (1) with part of the reverse transcriptase (RT) proteins (domains 1 to 7 described by Xiong and Eickbush [62]) from a number of distinct retroviral groups. These included several endogenous human retroviruses, such as HERV.L (12) (accession no. X89211), HERV.I (M92067) (29), HC-2 (Z70664) (19), HERV.H (K01891) (30), and HERV.K (M14123) (42), as well as other highly divergent retroviruses from nonhuman hosts, such as the walleye dermal sarcoma virus (WDSV) (L41838) and the dart poison frog Dendrobates ventimaculatus (Dev I; X95795) (18, 56). This search strategy was expected to result in the identification of most of the endogenous human retroviral sequences within the data bank which still encoded the appropriate region (in the sense that there had been no large postintegration deletion event) of the RT protein.
However, to confirm that this was indeed the case, a second method of screening was also performed. The endogenous human sequences identified during the first screening procedure were aligned, and a phylogeny was then constructed by the neighbor-joining approach with the program PAUP4d64 (written by D. L. Swofford). A representative from each of the monophyletic groups of HERVs present in the resultant phylogeny was then used in a BLAST search. The endogenous human viruses recovered by these searches were then examined (and added to the data set if they had not been identified previously) in descending order of similarity until representatives of the phylogenetic sister group (according to the neighbor-joining tree) of HERVs were encountered. An additional nine sequences were identified during this second screening.
The last group of HERVs included in the data set were the prototype members of each of the many previously identified HERV families (including those reviewed by Wilkinson et al. [61] and by Boeke and Stoye [10]). When this was not possible (either because the known members of the families were defective in the region of pol used or because the sequence of this region was not available), BLAST searches were performed, with an alternative region of the incomplete HERV being used as the probe; the most closely related cosmid sequences were then identified and recorded. Since these best- or closest-match cosmid clones also encoded the appropriate region of polymerase (Pol), it was possible to estimate the probable phylogenetic location of each incomplete HERV family in subsequent analyses.
Occasionally, cosmids with different accession numbers contained identical HERV sequences; however, in all cases, further investigation (using the chromosome and map information attached to each data bank submission) showed that the cosmids were originally derived from the same location within the human genome, and thus one sequence was excluded from subsequent analyses.
Alignment and phylogenetic reconstruction.
The HERV-derived RT sequences (162 in total, consisting of 152 sequences obtained from the HGMP and 10 prototypic HERV family sequences) were then aligned with a representative sample of nonhuman endogenous virus sequences (66 in total) as described by Xiong and Eickbush (62). The total number of sequences included in this alignment was therefore 228. Neighbor-joining trees were generated with PAUP4d64, using the protpars matrix (14) and all 228 elements in the data set. Using the same matrix, bootstrap values were obtained from 1,000 replicates.
Because of the long computation time periods required, the construction of maximum-parsimony trees by using the standard simple or random addition option was not practical. Instead, an alternative search strategy was employed. The data set was first reduced, or pruned, by using the output from the bootstrapped neighbor-joining trees, which indicated that in the phylogeny there were several well-supported terminal clusters of HERVs containing up to 42 members. When these clusters were supported by at least 95% of the bootstrapped neighbor-joining replicates, they were pruned by removing all but three of the taxa. This pruning resulted in the reduction of the number of taxa in the data set from 228 to 134. All of the maximum-parsimony-derived phylogenies were constructed using this reduced number of taxa and a search strategy described by Quicke et al. (D. Quicke, J. Taylor, and A. Purvis, submitted for publication) as follows. The data set was first subjected to 7,500 random additions by using an unordered matrix with tree bisection and reconnection, holding one tree in memory during each replicate. The shortest tree was then used in a heuristic search, with all optimal trees being saved; this resulted in the identification of a further 1,200 trees of the same length. This pool of minimum trees was then employed to reweight the data matrix, using the rescaled consistency index. Searches for minimum trees then continued with the reweighted data matrix. The minimum reweighted tree was identified and used as an input tree for another search in which the characters were again weighted to unity. Several rounds of reweighting followed by weighting to unity were performed; there was no further reduction in tree length.
Maximum-parsimony bootstrap replicates (100 in total) were subjected to 100 random additions by tree bisection and reconnection, with one tree being held in memory during each replicate. Although shorter trees would have been obtained if each bootstrap replicate had been subjected to a larger number of random-addition replicates (instead of 100 random additions per bootstrap replicate used), the computation time would have been excessive. However, it was probable that each of these bootstrapped trees was within a few steps of the minimum possible (unpublished results).
Calculation of integration dates.
The percentages of divergence between pairs of LTR sequences were calculated using their entire length, excluding regions containing deletions. These divergence figures were then corrected to account for the presence of multiple mutations at the same site, back mutations, and convergent substitutions, using the two-parameter model (21). Two estimates of the rate of change of the host genome were calculated, 2.1 × 10−9 and 1.3 × 10−9 per synonymous site per year, and hence two estimates of the integration date are provided. The first rate was based on a comparison of the percentage of divergence (11.6%) of synonymous sites in six genes in Old World monkeys and humans (25), whereas the second (7.3% divergence) used η-globin pseudogene and total single-copy DNA cross-hybridization data (25, 37). Both assumed a divergence date of humans and Old World monkeys of 27.5 million years ago (47).
RESULTS
HERV identification and data set construction.
A total of 152 novel HERV sequences were identified by screening the EMBL/GenBank/DDBJ nonredundant data banks. Two rounds of screening were performed; the first was based on similarity searches using part of the RT proteins derived from previously identified retroviruses, and the second was done with several of the newly identified HERVs themselves as the probes. The HERVs were named according to the cosmid or BAC in which they were identified, and multiple HERVs situated within the same vector were given an additional letter-based designation.
An alignment, based on part of the retroviral RT protein, of the novel HERV sequences and a representative sample (66 in total) of previously identified nonhuman endogenous retroviruses was performed. The prototypic members of several HERV families were also added to the data set, namely HERV.L (12), ERV-9 (23), HERV.I (29), HERV.H (30), HERV.HML6 (36), HERV.K (42), HERV.W (46), and HERV.E (48). Unfortunately, there is a lack of sequence information on some regions of the genomes of members of other putative HERV families (Table 1). It was therefore not possible to include most of them (with the exceptions of HERV.ADP [28] and HERV.FRD [49], which contain the appropriate region of pol) directly in the analyses presented here. Instead, the available sequence information from each of these partially characterized HERV families (ERV.1 [11], RRHERV.I [20], HERV.P [22], Hs5 [24], HERV.HML1-5 [35], HERV.R [40], HERV.FTD [49], NP-2 [51], HERV.S71 [59], and HERV.XA [60]) was used to identify the novel HERV-containing cosmid with the highest level of sequence similarity included in the data set, as shown in Table 2.
TABLE 1.
General properties of previously characterized HERV families investigated in this study
| Family | Primer | Size (kb) of sequenced region | Copy no.a | Available sequence datab |
|---|---|---|---|---|
| HERV.E | tRNAGlu | 8.8 | 85 | LTR-gag-pol-env-LTR |
| HERV.I | tRNAIle | 9.0 | 85 | LTR-gag-pol-env-LTR |
| HERV.W | tRNATrp | 7.6c | 115 | LTR-gag-pol-env-LTR |
| HERV.H | tRNAHis | 8.7d | 660 | LTR-gag-pol-env-LTR |
| HERV.L | tRNALeu | 6.5 | 575 | LTR-gag-pol-DUT-LTR |
| HERV.K | tRNALys | 9.1 | 170 | LTR-gag-DUT-pol-env-LTR |
| ERV-9 | tRNAArg | 1.8 + 3.8 | 70 | LTR + part gag-pol-Δenv-LTR |
| HERV.HML6 | tRNALys | 7.5e | 45 | LTR-gag-DUT-pol-env-LTR |
| RRHERV.I | tRNAIle | 3.3 | 15 | LTR-Δpol-env-LTR |
| HERV.R | tRNAArg | 5.0f | 15 | LTR + part gag + part pol + env-LTR |
| HERV.P | tRNAPro | 2× 0.6 + 0.4 | 70 | LTR + part pol + LTR |
| HERV.S71 | ? | 5.5 | 15 | gag-Δpol-LTR |
| HERV.XA | ? | 4.7 | 15 | Part pol-Δenv |
| HERV.ADP | ? | 1.5 | 60 | Part pol |
| HERV.FRD | ? | 2.8 | 15 | Part pol |
| HERV.HML5 | ? | 0.25 | 45 | Part pol |
Estimated copy number as determined by BLAST search and phylogenetic analysis (i.e., elements which contain domains 1 to 5 of RT, assuming a random distribution of elements within the human genome).
part, only part of the gene or region has been sequenced; Δ, entire gene or region has been sequenced but contains obvious deletions.
Combined total for nine cDNA clones.
env-containing elements.
Combined total for three clones.
Combined total for five subclones.
TABLE 2.
Previously described HERV families investigated in this study
| Family | Alternative name | Accession no. | Closest match
|
Reference | ||
|---|---|---|---|---|---|---|
| Cosmida | % Identity | Region of identity | ||||
| HERV.E | M10976 | 48 | ||||
| HERV.I | RTLV-I | M92067 | 29 | |||
| HERV.W | MSRV | AF009668 | 46,9 | |||
| HERV.H | RTLV-H | K01891, D11078 | 30,17 | |||
| HERV.L | X89211 | 12 | ||||
| HERV.K | M14123 | 42 | ||||
| ERV-9 | X57147 | 23 | ||||
| HERV.HML6 | HSU60268–HSU60270 | 36 | ||||
| HERV.ADP | ADP-pol | L14752 | 28 | |||
| HERV.FRD | U27240 | 49 | ||||
| HERV.R | ERV-3 | M12140 | AC004609 | 89 | 150 bp of pol | 40 |
| ERV-1 | K02916/7 | AC004609 | 82 | 2 100-bp regions of gag | 11 | |
| NP-2 | M15971 | AL023280 | 84 | 300 bp of env | 51 | |
| Hs5 | D10450 | AC004054 | 85 | 1,000 bp of env | 24 | |
| HERV.S71 | M32788 | AC002992a | 88 | 300 bp of RNase H gene | 59 | |
| RRHERV.I | M64936 | AC002992b | 90 | 500 bp of env | 20 | |
| HERV.P | HuRRS-P | —b | AC002069 | 99 | 130 amino acids of Pol | 22 |
| HERV.XA | U29659 | AC000378 | 88 | 500 bp of pol | 60 | |
| HERV.HML1 | U35102 | Z70280 | 95 | 230 bp of pol | 35 | |
| HERV.HML2 | U35104 | Z70280 | 80 | 230 bp of pol | 35 | |
| HERV.HML3 | U35153 | Z83745a | 94 | 230 bp of pol | 35 | |
| HERV.HML4 | U35160 | HERV.K10 | 86 | 50 bp of pol | 35 | |
| HERV.HML5 | U35161 | AC004536 | 88 | 230 bp of pol | 35 | |
| HERV-FTD | U27241 | AC004682 | 82 | 500 bp of pol | 49 | |
| HRES-1 | X16660 | No match | 45 | |||
Accession number of closest cosmid match if pol gene sequence from prototype isolate was not available.
—, see reference for sequence.
Sequence identity between the best-match HGDB sequence and the incomplete HERV sequences ranged from 82% (for cosmid AC004609 and ERV-1) to 99% (for cosmid AC002069 and HERV.P). In two cases, more than one HERV family had a best match with the same cosmid-derived HERV sequence: ERV.1 and HERV.R with the same region of AC004069, and HERV.HML1 and -2 with the same region of cosmid Z70820 (Table 2).
Phylogenetic analysis and nomenclature.
The alignment was subjected to phylogenetic analysis by both the neighbor-joining and maximum-parsimony approaches. Neighbor-joining trees were constructed using a data set consisting of 228 taxa (162 of which were of human origin [Table 3]). Due to the very long computation time periods necessitated, maximum-parsimony analyses utilized a smaller data set, numbering 134 taxa, of which 68 were of human origin (Table 3 shows which subset of HERV sequences were excluded from the maximum-parsimony analyses).
TABLE 3.
Location and classification of the HGMP-derived and prototype HERVs used in the present phylogenetic analyses
| Family and accession no.a | Chromosome locationb | Positionc (orientationd) |
|---|---|---|
| HERV.L family | ||
| HERV.L | (X89211) | |
| AL008713 | X | 22260 (+) |
| AL031119 | 6q24 | 95570 (−) |
| *AC004614 | 7p21-p22 | 51860 (−) |
| *AL021706 | Xq21.1-21.3 | 61100 (−) |
| *AC005261 | 19q13.4 | 26030 (−) |
| *AB000880 | 6p21.3 | 5580 (−) |
| *AC004544 | 4 | 60000 (+) |
| *Z98043 | 1q24 | 23600 (−) |
| *AC002449 | Xq23 | 117070 (−) |
| *AC002467 | 7q31 | 51010 (+) |
| *AC004225 | 5p | 32060 (−) |
| *AC004075 | X | 96290 (+) |
| *AF003528 | Xq13 | 60780 (+) |
| *AC003973 | 19 | 31200 (+) |
| *D86999 | 22q11.2 | 29120 (−) |
| *D86998 | 22q11.2 | 32700 (+) |
| *AC004045 (a) | 4q25 | 68940 (−) |
| *Z95124 | Xq21 | 43550 (+) |
| *Z70039 | X | 4831 (+) |
| *AC003975 | 7q31.3 | 15580 (−) |
| *AC002487 | 7q31 | 44600 (+) |
| *Z95331 | 22q11.2 | 6520 (−) |
| *AC004389 | X | 30390 (+) |
| *Z72519 | X | 31010 (−) |
| *Z69923 | 4p16.3 | 14690 (+) |
| *Z80771 | X | 75100 (+) |
| *AF070718 | 8q21 | 46200 (−) |
| *AF003529 | Xq26 | 122070 (+) |
| *AC004397 | 7q31.3 | 11500 (+) |
| *AC004541 | 7q11.2-21.1 | 43280 (+) |
| *AC003003 | 16p11.2-12 | 93430 (−) |
| *Z83841 | X | 57284 (+) |
| *AC004006 | 7q21-q22 | 100000 (+) |
| *AC002302 | 16p12.2-12 | 242050 (+) |
| *AJ006997 (b) | 21 | 27700 (+) |
| *Z84720 | X | 89900 (−) |
| *AL031054 | Xq27.2 | 137160 (+) |
| *AL023279 | Xq27.2 | 88420 (−) |
| *AC005530 | 6p21 | 43200 (−) |
| *AJ006997 (a) | 21 | 82360 (−) |
| *Z97054 | Xp11.2 | 83980 (−) |
| HERV.S family | ||
| AC004385 | X | 57520 (+) |
| AL009047 | X | 124667 (−) |
| AC002523 | Xq28 | 71140 (+) |
| Z68758 | 22q11.2-qter | 1146 (−) |
| AL009051 | 1q23-24 | 54501 (+) |
| HERV.E family | ||
| HERV.E | (M10976) | |
| AL023280 | Xq21.3-22.3 | 101960 (−) |
| AC000385 (a) | 11 | 153050 (−) |
| *AC000385 (b) | 11 | 132070 (−) |
| *AC002094 | 17 | 165330 (+) |
| *Z98257 | 1p35-36.2 | 11720 (+) |
| *AC004054 | 4q21 | 26830 (−) |
| RRHERV.I family, | ||
| AC002992 (b) | Y | 12550 (+) |
| HERV.R family, | ||
| AC004609 | 19p13.1 | 33950 (−) |
| HERV.S71 family, | ||
| AC002992 (a) | Y | 53730 (−) |
| HERV.ADP family | ||
| HERV.ADP | (L14752) | |
| AC004595 | 7p15.3-p21 | 96174 (−) |
| AL023579 | 6q26-27 | 42430 (−) |
| AC005741 | 5 | 67158 (−) |
| HERV.H family | ||
| HERV.H | (K01891) | |
| AC002384 | 7q22 | 51020 (+) |
| U95626 | 3 | 78690 (−) |
| *AC004456 (a) | 7q31 | 114150 (−) |
| *AC004456 (b) | 7q31 | 5660 (−) |
| *AF026251 | NDe | ND |
| *AL009031 | 6p22.3-24.1 | 138520 (+) |
| *AC002326 | 6 | 148750 (+) |
| *AF011889 | Xq28 | 85570 (+) |
| *AC004025 | Xp22 | 104670 (−) |
| *AC002066 | 7q31 | 36130 (−) |
| *AC004072 | X | 117925 (+) |
| *AF070717 | 8q21 | 16160 (−) |
| *AP000014 | 21q22.2 | 38340 (−) |
| *Z92543 | 6q22 | 30575 (+) |
| *AC003009 | 16p13.2-3 | 108950 (+) |
| *AC002530 | 7p15-p21 | 3475 (+) |
| *AC005276 | 7q31.3 | 3590 (+) |
| *AC000114 | Xq23 | 34280 (+) |
| *AC003091 | 7p21 | 116510 (+) |
| *Z71183 | 22q11.2 | 17985 (+) |
| *AL021327 | 6q21 | 28840 (+) |
| *Z76735 | X | 83560 (−) |
| *AC003078 | 7q21-q22 | 6200 (+) |
| *Z82210 | X | 19480 (−) |
| *AC004706 | 17 | 109705 (−) |
| *AC002526 | Xq23 | 70080 (−) |
| *U80460 | Xq13 | 65800 (+) |
| *AL021940 | 1q24 | 14875 (+) |
| *AC000064 (a) | 7q21-22 | 24110 (+) |
| *AC005145 | Xp22-166-169 | 75610 (−) |
| *AC005410 | 17 | 16905 (+) |
| *AC005549 | 17 | 130120 (−) |
| *AL031256 | 20q12 | 40955 (+) |
| *AC005392 | 19q13.2 | 174160 (−) |
| *AL023877 | Xp23-24 | 97410 (−) |
| *AF068862 | 8q21 | 75430 (−) |
| *Z99495 | 6p24 | 70115 (−) |
| *AC004514 | 16q21-22 | 41510 (−) |
| *AC005576 | 5q | 11040 (−) |
| *AC005386 | 10q25 | 175240 (+) |
| *AC002984 | 19q13.1 | 23490 (−) |
| HERV.F family, Z94277 | Xp11.3-Xp11.4 | 101180 (+) |
| HERV.F (type b) family | ||
| Z83745 (b) | X | 60210 (+) |
| AC002416 | X | 111780 (+) |
| HERV.XA family, AC000378 | 11 | 92770 (+) |
| HERV.I family | ||
| HERV.I | (M92067) | |
| AC004682 | 16q22.2 | 167493 (−) |
| AC004210 | 6p21 | 11996 (+) |
| *AL021878 | 22q13.1-13.2 | 29650 (−) |
| *AC004074 | X | 90318 (+) |
| *AC003100 (a) | 4q25 | 75436 (+) |
| *AC000394 | 9q34 | 19247 (−) |
| HERV.Z69907 family | ||
| Z69907 | 22q11.2 | 27132 (+) |
| AC004474 | Y | 106160 (+) |
| HERV.R (type b) family, | ||
| AC004045 (b) | 4q25 | 79040 (+) |
| ERV-9 family | ||
| ERV-9 | (X57147) | |
| AC004534 | 7q21-22 | 76280 (−) |
| Z84475 | 6q21 | 31440 (−) |
| *AC003087 | 7p15 | 43870 (+) |
| *AC004617 | Y | 168320 (−) |
| *Z99496 (a) | 6q22.1 | 2400 (−) |
| HERV.W family | ||
| HERV.W | (AF009668) | |
| AF045450 | 21q22.3 | 30400 (−) |
| AC000064 (b) | 7q21-22 | 33230 (+) |
| *U85196 | ND | 11680 (+) |
| *AC005187 | 4q25 | 108400 (+) |
| *AL023581 | 6 | 82630 (−) |
| *Z83850 | Xq22 | 92240 (+) |
| *AL022067 | 6 | 138140 (+) |
| HERV.P family | ||
| Z98255 | Xq11.2-13.3 | 148180 (+) |
| AC002069 | 7q21 | 42530 (+) |
| AL008987 | Xq21 | 47420 (−) |
| AC003969 | 11p14.3 | 116980 (−) |
| Z99496 (b) | 6q22.1 | 39850 (−) |
| HERV.HS49C23 family | ||
| AC002319 | 9q34 | 6388 (+) |
| AL023753 | 1p36.11 | 16270 (−) |
| HS49C23 | X | 62990 (+) |
| Z78021 | X | 28920 (−) |
| AL008706 | Xq27-28 | 10120 (−) |
| HERV-FRD family | ||
| HERV.FRD | (U27240) | |
| AC004022 | 7q21-22 | 91820 (+) |
| HERV.K family | ||
| HERV.K | (M14123) | |
| U91321 | 16p13.1 | 112740 (−) |
| Z70280 | X | 36610 (+) |
| AL022154 | Xq21.1-2 | 44890 (+) |
| Z83745 (a) | X | 44960 (−) |
| AC002464 | ND | 53850 (−) |
| AC003686 | 12q13.1 | 31140 (−) |
| Z99129 | 6q22 | 107600 (−) |
| AC003100 (b) | 4q25 | 23420 (+) |
| AC005318 | 15q26.1 | 16400 (+) |
| AL031601 | 10 | 264070 (+) |
| AL031393 | Xp11.3 | 9370 (−) |
| HERV-HML5 family | ||
| Z95437 | 6p21.3 | 8930 (−) |
| AC004536 | 7q31 | 36540 (+) |
| U69569 | Xq28 | 6380 (−) |
| HERV-HML6 family | ||
| HERV.HML6 | (HSU60269) | |
| Z84814 | 6p21.3 | 59370 (+) |
| AC005946 | 19q13.4 | 17800 (−) |
| AC003029 | 12q24 | 30850 (−) |
Asterisks indicate sequences that were excluded from maximum-parsimony analyses to reduce computation times (i.e., the sequence was only used in the neighbor-joining analyses).
Accession numbers (rather than locations) are given for prototype HERVs (in parentheses).
Approximate position of LPQG motif (or equivalent) in domain 4 of the RT protein.
Orientation with respect to the cosmid sequence.
ND, no data.
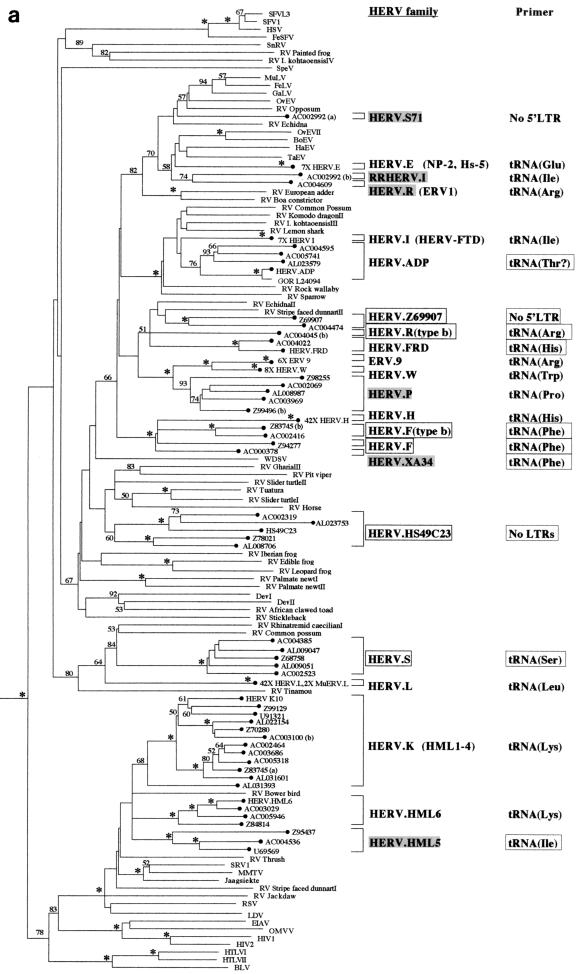
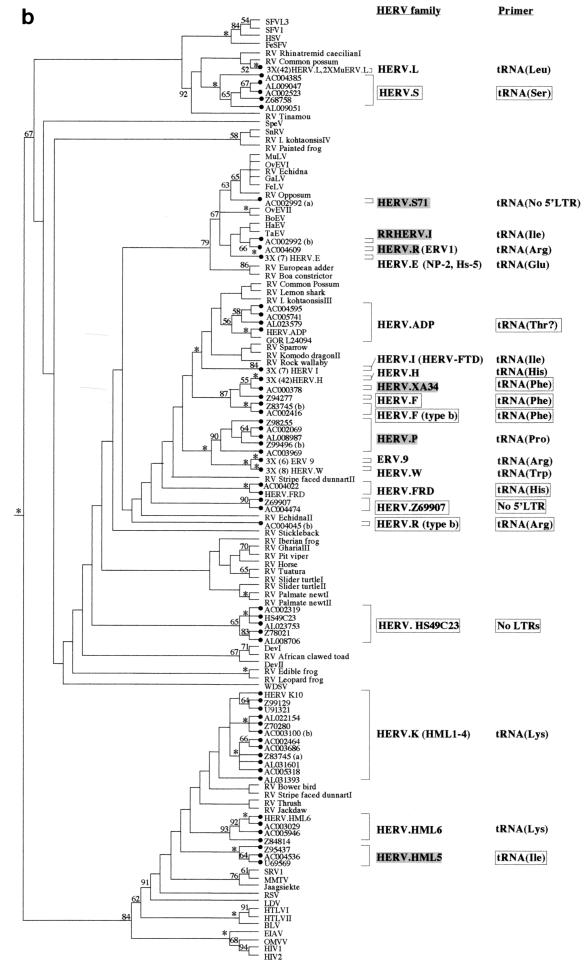
HERV families arise via a single horizontal transmission event followed by germ line integration and fixation within the host population. The copy number can then increase by retrotransposition or reinfection, and it is probable that most members of a particular family are separated by only a few rounds of viral replication (although they may well be separated by many rounds of host DNA replication) (10). Phylogenetic reconstruction of retroviral phylogenies containing HERV-derived sequences would therefore be expected to show well-supported clusters of such elements, with members of each HERV cluster being derived from the same family. Trees generated by both the neighbor-joining and maximum-parsimony approaches were broadly consistent with this hypothesis, showing numerous well-supported HERV lineages scattered across the phylogeny (Fig. 1).
FIG. 1.
Phylogenetic analyses of a 159-residue region of retroviral RT proteins. The trees were rooted on several gypsy LTR-retrotransposon sequences. To save space, multiple taxon names in some well-supported terminal clusters are not indicated. Instead, the number of taxa that actually clustered in that position are indicated on the taxon label. (a) Neighbor-joining tree with branch lengths proportional to the degree of divergence between the sequences. Figures on each branch represent percentage bootstrap support from 1,000 replicates; asterisks show support of at least 95%. HERVs are indicated by black circles. Novel HERV families are boxed. Previously described HERV families are shaded gray if the sequence of the pol gene was not available, and they were classified according to their closest cosmid matches (see Table 1). Elements in parentheses are likely to cluster with the adjacent family. Primer sites indicate the tRNA to which the viral PBS is most similar; these are boxed when this similarity is first reported in this study. (b) Strict consensus of 1,200 maximum-parsimony trees. The figure is labeled as in panel a except that branch lengths are not proportional to the divergence between the taxa. Also note that in contrast to panel a, maximum-parsimony data sets were pruned prior to analysis to reduce computation times. Thus, the first figure presented on some of the taxon labels represents the actual number of elements included in the analysis, whereas the number in parentheses represents the total estimated number that would cluster with the particular family if the additional elements had also been included in the data set (based on at least 95% bootstrap support by the neighbor-joining method; see Materials and Methods).
Under ideal circumstances, with the complete sampling of retroviral diversity across all hosts in which they occur, the total number of HERV families could be calculated simply by counting the number of HERV lineages intermingled with the nonhuman retroviruses in the phylogeny (in effect, this counts the number of retroviral host switches into the human lineage). However, this is not the case; there is in fact much better sampling of endogenous retroviruses from humans than from other vertebrate taxa. These differences in sampling efficiency mean that some HERVs may appear to belong to the same family (i.e., they cluster together after phylogenetic reconstruction) simply because closely related viruses in nonhuman hosts have yet to be identified. Thus, in order to obtain a reasonable estimate of the actual number of HERV families, several assumptions about their evolution had to be made.
First, it was assumed that HERVs with alternative PBS homologies (even if they clustered together in the phylogenies) were derived from separate cross-species transmission events and were therefore independent families. For example, in Fig. 1, HERV.P, HERV.W, and ERV-9 (primed by tRNAArg) all cluster together with robust bootstrap support, but because they all have different PBS homologies, they are here regarded as three separate families. This is consistent with previous reports (46, 61).
A second assumption was that HERVs encoding the same PBS which were polyphyletic with respect to viruses in nonhuman hosts also represented separate families. Thus, in Fig. 1a, HERV.K and HERV.HML6 are classified as separate families because they form a polytomy with RV-Bower bird (i.e., the presence of only a few nonhuman viruses in this region of the phylogeny was enough to split up the HERV.K and HERV.HML6 lineages in some trees, and the addition of further nonhuman viruses would be likely to split them in all trees). The fourth member of the polytomy, HERV.HML5, has an alternative PBS and is therefore already regarded as an independent family.
Last, it was assumed that HERV families which were paraphyletic with respect to nonhuman viruses or to HERVs with alternative PBS homologies were also independently derived. For example, in Fig. 1b, HERV.XA, HERV.F, and HERV.F (type b), which are all primed by tRNAPhe, are paraphyletic with respect to HERV.H and thus represent separate families. In this case, the presence of multiple families was also confirmed by investigating sequence similarity between the LTR and gag regions of the four families as described below.
The above criteria suggested the presence of 22 endogenous retroviral families within the 7% of the human genome that has been sequenced to date (both neighbor-joining and maximum-parsimony analyses gave the same figure). Of these, 12 contained prototypic members which have been well characterized in previous reports (although the pol genes of 4 of these 12, HERV.S71, RRHERV.I, HERV.R, and HERV.P, were defective or incomplete, and so these families are represented by closest cosmid matches [Table 2]), 4 contained prototypic members which had generally been less well characterized (of these, HERV.HML5 and HERV.XA are based on closest cosmid matches), and 6 represented novel families. A variety of other previously characterized human endogenous elements are also likely to be members of one of the 22 families (they are shown in parentheses in Fig. 1). In contrast, BLAST searches with the element HRES-1, which has been reported to be related to human T-cell leukemia virus type 1 HTLV-1 (45), failed to show a match with any HERV-containing cosmid (or indeed with any HGMP sequence), and therefore this element is not represented. The cosmids containing members of each of the families identified after phylogenetic reconstruction are shown in Table 3.
Because HERV families have often been classified according to the similarity of their PBSs to specific types of host tRNA, I attempted to identify these regions in all six of the novel HERV families, the four partially characterized families, and HERV.S71 (i.e., those families for which PBS data were not previously available). This analysis was performed by first identifying the LTR sequences upstream and downstream of the pol sequences employed in the phylogenies (using the program BLAST to BLAST [54]) and then comparing the sequence immediately 3′ to the 5′ LTR against a tRNA sequence database (52). From this approach it was apparent that the partially characterized HERV families are primed by a variety of tRNAs, as follows: HERV.XA by tRNAPhe, HERV.ADP (probably) by tRNAThr, HERV.FRD by tRNAHis, and HERV.HML5 by tRNAIle (Table 4). I was unable to find any 5′ LTRs in members of the HERV.S71 family (the only other previously characterized family for which PBS data were not available). Despite the new data on the PBS homologies of these families, their nomenclature has been left unchanged in this report.
TABLE 4.
General properties of partially characterizeda and novel HERV families
| Family | Cosmid | Primer | Size (kb) | Copy no.b | Genomic organization |
|---|---|---|---|---|---|
| Partially characterized | |||||
| HERV.ADP | AC005741 | tRNAThr(?) | 8.4 | 60 | LTR-gag-pol-env-LTR |
| HERV.FRD | AC004022 | tRNAHis | 10.8 | 15 | LTR-ORF?-gag-pol-env-LTR |
| HERV.XA | AC000378 | tRNAPhe | 6.2 | 15 | LTR-gag-Δpol-Δenv-LTR |
| HERV.HML5 | AC004536 | tRNAIle | 7.8 | 45 | LTR-Δgag-DUT-pol-env-LTR |
| Novel | |||||
| HERV.Z69907 | Z69907 | No 5′ LTR | ∼6.6 | 30 | gag-pol-Δ-env-LTR |
| HERV.R (type b) | AC004045(b) | tRNAArg | 8.7 | 15 | LTR-gag-pol-env-LTR |
| HERV.F | Z94277 | tRNAPhe | 8.7 | 15 | LTR-gag-pol-env-LTR |
| HERV.F (type b) | AC002416 | tRNAPhe | 6.8 | 30 | LTR-gag-Δpol-Δenv-LTR |
| HERV.S | AC004385 | tRNASer | 6.7 | 70 | LTR-gag-pol-env-LTR |
| HERV.H49C23 | H49C23 | No LTRs | ∼6.0 | 70 | Δgag-pol-Δenv |
Properties of closely related HERV-containing cosmid clones.
Number of elements containing domains 1 to 5 of RT, estimated by BLAST search followed by phylogenetic reconstruction.
Of the six novel HERV families, two were primed by tRNAPhe and two others were primed by tRNASer and tRNAArg and they have therefore been termed HERV.F, HERV.F (type b), HERV.S, and HERV.R (type b); an independently derived HERV.R family has been described previously by O'Connell and Cohen (41). There were no obvious 5′ LTR sequences in the two remaining families, and thus they have been designated HERV.Z69907 and HERV.HS49C23 after the cosmids in which the prototypic members are located.
HERV families have often been broadly divided into two classes, with class I HERVs being related to the mammalian type C retrovires, exemplified by feline leukemia virus (FeLV) and gibbon ape leukemia virus (GaLV), and class II HERVs being most similar to the mammalian type B and D retroviruses or avian leukosis viruses, such as mouse mammary tumor virus (MMTV), simian retrovirus type 1 (SRV-1), or Rous sarcoma virus (RSV) (10, 61). Recently, the presence of a third HERV class (class III) has been proposed (3, 26) based on the similarity of HERV.L to spumaviruses such as the human spumavirus (HSV) (12). Although the topologies of the trees shown in Fig. 1 differed according to the method of reconstruction (largely due to the previously identified weak support across the backbone of the retroviral phylogeny [16]), it is clear from this analysis that very few of the HERV families are actually closely related to these retroviral genera. In particular, both the HERV.L and HERV.S families appear to be only distantly related to the spumaviruses, and the HERV.HS49C23 family clusters with several groups of viruses isolated from nonmammalian vertebrates rather than with the type C retroviruses. Furthermore, no HERV family appears to be most closely related to either the previously identified mammalian type B and D retroviruses or the avian leukosis viruses. Instead, the HERVs in this region of the tree tended to cluster with endogenous retroviral fragments derived from birds and nonplacental mammals (although the exact relationships revealed differed depending to the method of phylogenetic reconstruction).
Characterization of novel HERV families.
Prototypic members of each of the novel HERV families were investigated further in order to provide some background information on the general properties of these elements. Although it was difficult to identify the exact 5′ and 3′ ends of the gag, pol, and env genes (due to small insertions or deletions and in-frame stop codons), their presence or absence could still be established by the identification of certain motifs conserved among different retroviruses (58). Furthermore, by analyzing the distances between these motifs, it was also possible to determine whether a large deletion had occurred in the particular gene under study.
(i) HERV.S.
The prototype member of the HERV.S family is located on cosmid AC004385, which is derived from the X chromosome (Table 3). It is approximately 6.7 kb in length and has a typical retroviral structure which appears relatively intact, with no appreciable deletions in gag, pol, or env (Table 4). There are, however, multiple in-frame stop codons and small deletions, indicating that the element is unlikely to be able to express any major gene products. The five members of the HERV.S family described in this report were found via BLAST searches of 7% of the human genome, suggesting (assuming that these elements are more or less randomly distributed) that the copy number of this family is at least 70 per haploid genome.
The HERV.S provirus contains typical, though relatively short, proviral LTR structures (a 5′ LTR of 317 bp and a 3′ LTR of 318 bp) bounded by the inverted terminal repeats TG and CA (see Fig. 3a). Potential promoter (TATAAA) and polyadenylation (AATAAA) sequences were also apparent. The PBS and polypurine tract (the primer sites for minus- and plus-strand DNA synthesis) immediately follow the 5′ LTR and precede the 3′ LTR, respectively, with the PBS showing 16 of 18 matches to the 3′ end of the human serine tRNA; the PPT is 11 bp in length. The observed percentage of divergence between the two LTRs was 12.4%, corresponding to a corrected divergence (taking into account back mutations and multiple substitutions at the same site) of 13.6%. Because the LTRs were presumably identical when the element first integrated into the genome (55), the approximate length of time the element has been vertically passaged can be estimated by using this figure and the rate of change within the primate lineage, which I calculated as being 0.13 or 0.21% per million years. This gives an integration date for the HERV.S element within cosmid AC004385 of between 32 and 52 million years ago. These figures have a large range since there is some uncertainty over the level of divergence between Old World monkeys and humans (25, 37). Furthermore, I have assumed rate constancy within the primate lineage, but there is evidence suggesting that this is not the case; it is probable that there has been some degree of slowdown during hominid evolution (4, 25).
FIG. 3.
LTR sequences of five novel HERV families (a) and four partially characterized families (b). When both the 5′ and 3′ LTRs could be identified, they were aligned against each other, with dashes representing missing residues. The PPT (before the start of the 3′ LTR) and PBSs (after the end of the 5′ LTR) are underlined, as are the direct repeats flanking each end of the element and the inverted repeats flanking each LTR. The promoter and polyadenylation signals are boxed. In some cases, not all these features could be identified for each element (or they differ slightly from the expected sequence). This is probably due to postintegration mutation. Similarly, this is also likely to account for the observed differences between the PBS and closest-match tRNA sequence shown below each alignment. The estimated integration dates of each HERV are also shown.
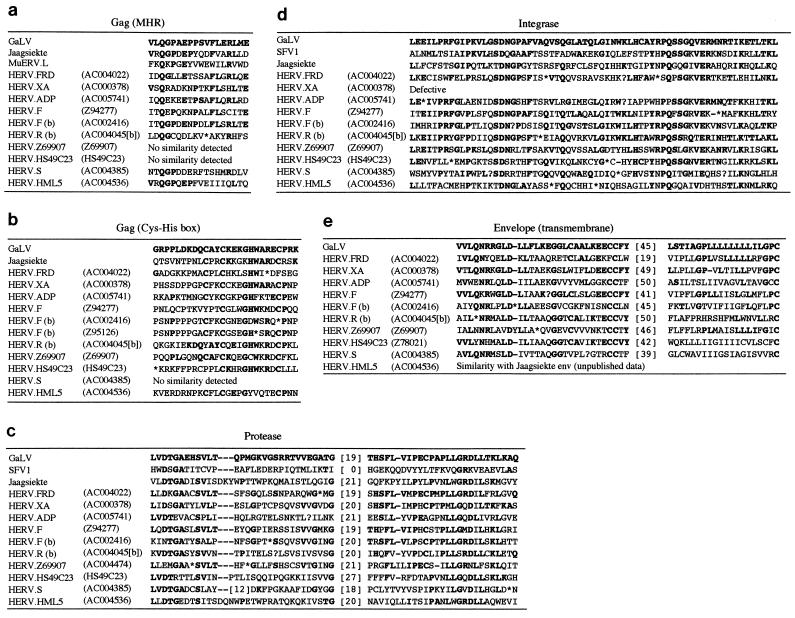
Most retroviral gag genes encode a short conserved region, termed the major homology region (MHR), in the capsid protein and a Cys-His motif in the nucleocapsid of the form CX2CX4HX4C, which is thought to be involved in binding to nucleic acids (58). Alignments of these regions are shown for several of the HERV families investigated in this study (see Fig. 4a and b). A putative MHR was identified in the gag gene of HERV.S, but the Cys-His motif appears to be absent (several different members of the family were investigated for its presence). Both HERV.L and murine endogenous retrovirus type L (MuERV.L) also contain an MHR but lack the Cys-His motif, whereas the spumaviruses do not appear to encode either region (6, 12, 58). BLAST searches using the 3′ end of the Gag protein from AC004385 as the probe (i.e., the region in which the Cys-His motif is situated within other retroviruses) demonstrated a low level of similarity to HERV.L and MuERV.L, but no matches were obtained with other retroviral isolates. The HERV.S family pol gene was found to have a typical retroviral organization, encoding motifs associated with the protease (Pro), RT and integrase (Int) proteins but no other gene products (Fig. 2) (see also 4c and d). BLAST searches with the region between the end of 3′ Pol and the 3′ LTR generally showed few matches, with the exception of two short amino acid motifs related to part of the transmembrane proteins of other retroviruses (Fig. 4e).
FIG. 4.
Alignment of conserved amino acid motifs present within the HERV families identified in this report. The figures in parentheses indicate the cosmids from which the sequences were derived. GaLV, Jaagsiekte, HSV, and MuERV.L are provided for comparative purposes. (a) Gag alignment spanning nucleotide positions 1660 to 1713 within the GaLV sequence (13); (b) Gag alignment spanning positions 2074 to 2157; (c) Pro alignment spanning positions 2266 to 2490; (d) Int alignment spanning positions 4969 to 5166; (e) Env alignment spanning positions 7220 to 7504.
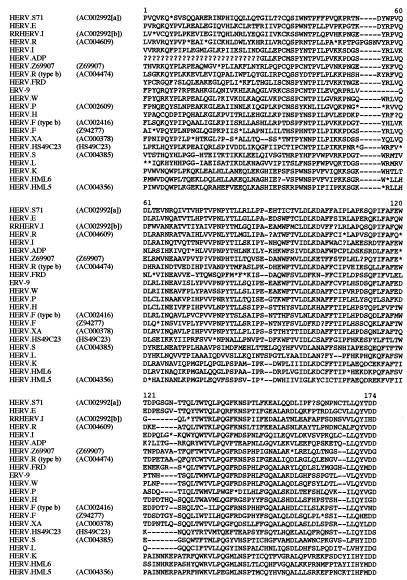
FIG. 2.
Partial RT sequence alignment of representative members of each of the 22 independent HERV families discussed in this report. The sequence of this region is not available for the prototype members of some of the HERV families. In these cases, the cosmid from which the sequence was derived is also shown. The similarity of the cosmid sequence to the prototypic family member and its location within the cosmid are indicated in Tables 2 and 3, respectively.
It is interesting that two of the viruses most closely related to the HERV.S family, namely HERV.L and MuERV.L (6, 12), show significant differences in genomic organization. For example, HERV.L and MuERV.L both lack an env gene, which appears to be present in HERV.S, whereas HERV.L and MuERV.L encode a dUTPase between pol and their 3′ LTRs (6, 12), and this gene was absent from all members of the HERV.S family (unpublished results).
(ii) HERV.R (type b).
Only one member of the HERV.R (type b) family was identified during this study, in cosmid AC004045(b), which is located at q25 on chromosome 4 (Table 3). This element is 8.7 kb in length, has the structure LTR-gag-pol-env-LTR (Table 4), and, like the prototypic member of the HERV.S family, does not contain any large deletions, but it is probably incapable of replication due to the presence of in-frame stop codons and frameshifts. Because only one member of this family was identified, any estimate of copy number is subject to considerable uncertainty, but it will probably be low. The HERV.R (type b) 5′ LTR is 643 bp in length, and its 3′ LTR is 692 bp, with the promoter and polyadenylation sequences situated toward the 3′ end (Fig. 3a). The two LTRs have an observed divergence of 11.4% (12.4% corrected), corresponding to an estimated integration date of 30 to 47 million years ago. The element contains a 17-bp PPT and shows 17 of 18 matches to the 3′ end of the mouse arginine tRNA. Two other previously identified HERV families, ERV-9 and HERV.R, also use an arginine tRNA primer (23, 41). The gag, pol, and env genes all contain the expected conserved motifs (Fig. 2 and 4).
The observed relationship of the HERV.R (type b) family to other retroviruses differed somewhat depending on the method used for phylogenetic reconstruction. In neighbor-joining trees, the family clustered with the HERV.FRD and HERV.Z69907 families (see below) as well as two partially characterized endogenous retroviruses derived from marsupials and monotremes (Fig. 1a). In contrast, maximum-parsimony analyses placed the family basal to a large clade of viruses that included 11 other HERV families (Fig. 1b). This discrepancy is probably due to the poor resolution of this region of the retroviral tree; the backbone of the phylogeny from which many of these sequences emerge is not at all well supported, as discussed above (16).
(iii) HERV.F.
Cosmid Z94277 contains an HERV sequence with a PBS exhibiting 16 of 18 matches with the human phenylalanine tRNA, and thus this family has been termed HERV.F (Fig. 3a). This HERV, located at position p11.3-11.4 on the X chromosome (Table 3), was the only member of the family to be identified by BLAST searches and phylogenetic reconstruction, which indicated that these elements are likely to be present at low copy numbers within the human genome. Both neighbor-joining and maximum-parsimony analyses suggested a close relationship with the HERV.H family (Fig. 1), which, in contrast with HERV.F, is present at a very high copy number, with at least 600 pol-containing members (30). The HERV.F genome is approximately 8.7 kb in length and encodes motifs indicative of the presence of gag, pol, and env (Fig. 2 and 4); there was no evidence of other gene products. Although all three genes appeared similar in size to those of other retroviruses, suggesting that there have been no large deletions since integration of the provirus, only small open reading frames ORFs were present within them (Table 4). The two LTRs (a 470-bp 5′ LTR and a 515-bp 3′ LTR with a 45-bp duplication adjacent to the PPT) were found exhibit 13.0% divergence (14.6% corrected), suggesting that the element first integrated into the primate lineage 35 to 56 million years ago.
(iv) HERV.F (type b).
A second HERV.F family was also apparent from my phylogenetic analyses (17 of 18 matches with human phenylalanine tRNA [Fig. 3a]). This family, termed HERV.F (type b), has also been independently identified by Lindeskog (26). The two families are closely related, both to each other and to the previously characterized family HERV.XA (60), which is also primed by phenylalanine (see below). Despite the similarity of their PBSs and their close phylogenetic relationship, I think that they should be regarded as separate families for two reasons. First, elements from the three families were not placed into a single, well-supported monophyletic clade in my analyses (and were paraphyletic with respect to the HERV.H family), as is the case for many other families, such as HERV.E, HERV.H, and HERV.I (Fig. 1). Second, although the RT-based amino acid alignment demonstrated a high level of similarity, this was not the case when other regions of the viral genomes were compared. For example, there was no obvious nucleotide sequence similarity between the LTRs of the different families, and only low levels were observed in comparisons of gag and env (unpublished results).
Only two members of the HERV.F (type b) family were identified during this study, indicating a copy number of approximately 30. Both elements are situated on the X chromosome. The prototypic member (in cosmid AC002416) is 6.8 kb in length and contains a pol gene with a deletion 5′ to the Int motif shown in Fig. 4d) and an env gene with a large deletion upstream of the transmembrane region (Table 4 and unpublished results). Although the gag gene in AC002416 is full length and contains an MHR (Fig. 4a), some of the conserved residues in the Cys-His motif have probably been altered by postinsertion mutation, and thus the equivalent region from the second member of the family (in cosmid Z95126) is also shown in Fig. 4b. The 5′ and 3′ LTRs (387 and 388 bp, respectively) have diverged by 7.5% (7.9% corrected), with an estimated integration date for the element of 19 to 30 million years ago.
(v) HERV.Z69907.
The prototypic member of the HERV.Z69907 family is located at q11.2 on chromosome 22. The only other member recovered by BLAST searches is present in cosmid AC004474, and this suggests a copy number of approximately 30. Both elements were highly defective; neither appeared to possess a 5′ LTR, and in only one (Z69907) was there a suggestion of a PPT and, thus, a 3′ LTR. The putative LTR did not appear to contain obvious promoter or polyadenylation signals. The Z69907 element encodes motifs that suggest the presence of the gag, pol, and env genes (Fig. 2 and 4 and Table 4). However, it contains a deletion in Pro (the alternative element in this family is therefore shown in the alignment in Fig. 4c), and the distance between the Int and transmembrane motifs was shorter than that observed in other viruses; thus, env is also likely to contain a large deletion (unpublished data). Furthermore, no obvious MHR could be detected in either virus. Like HERV.R (type b), the phylogenetic position of this family was dependent on the method used in tree reconstruction, being placed as a sister group to a marsupial-derived element in neighbor-joining analysis and toward the base of a large, poorly supported clade (which includes numerous HERV families) with maximum parsimony.
(vi) HERV.HS49C23.
The members of the HERV.HS49C23 family are unusual in that they tend to cluster with viruses derived from nonmammalian vertebrates, in contrast to many of the other HERV families shown in Fig. 1. The prototypic member is located on chromosome X, and its phylogenetic relationship has been briefly described in a previous report (16). It now appears that members of this family are present at approximately 70 copies in the human genome and that they are probably all highly defective. No LTR sequences could be identified for any member of the group, and BLAST searches suggested that only three (those on cosmids HS49C23, AC002319, and Z78021) contained anything more than small fragments with similarity to other retroviruses; the first two elements could be aligned across a 6-kb region of their genomes (unpublished results). Despite the presence of multiple in-frame stop codons and frameshifts, it was possible to derive some information on these elements because HS49C23 itself appears to contain a complete pol gene (i.e., there are no large deletions evident) and the Cys-His motif in Gag. There was no evidence of a transmembrane motif in env, but one was present in Z78021 (Fig. 4e). The lack of LTRs precluded attempts at estimating the integration dates of members of the family, but their highly defective nature suggests that they may be among the oldest of the HERVs.
Additional characterization of previously identified families. (i) HERV.HML5.
HERV.HML5 was first identified from a 250-bp PCR-amplified pol gene fragment by Medstrand and Blomberg (35). Like the other HERV.HML family constituents, it a member of the class II HERV superfamily, whose previously characterized members have been found to be primed by lysine tRNA. The HERV.HML5 pol gene fragment appears closely related to elements within three cosmid clones (Table 3), and one of them, in AC004536 (with which there was 88% nucleotide identity across a 250-bp region), was used to investigate the genomic organization of the family. The element in cosmid AC004536, which was found to be 7.8 kb in length, inserted into a 6-bp target site. The two 5′ and 3′ LTRs were 482 and 488 bp, respectively, and contained the expected inverted repeats and control sequences (Fig. 3b). The estimated integration date was 33 to 53 million years ago (based on a corrected LTR divergence figure of 13.7%). Unlike all other previously identified class II HERVs, members of the HERV.HML5 family are primed by an isoleucine tRNA rather than by tRNALys. The element within AC004536 showed 14 of 18 matches to the human Ile tRNA, and a second member of the family (within cosmid Z95437) showed 17 of 18 (Fig. 3b). The structure of the HERV.HML5 family is similar to those of the other well-characterized viruses to which it is most closely related, such as HERV.K10, MMTV, and jaagsiekte retrovirus (38, 42, 63). The Pro, RT, and Int motifs encoded within the pol gene and the MHR and Cys-His motifs encoded by gag were all apparent, and as with HERV.K10 and MMTV, these elements also appear to contain a dUTPase between the protease and RT genes. An alignment of the HERV.HML5 dUTPase and a human dUTPase (34) sequence is shown in Fig. 5. Although there was no similarity to the transmembrane motif within the envelope (Env) protein shown in Fig. 4e, a BLAST-derived match was obtained with the Env proteins of MMTV, HERV.K10, and jaagsiekte retrovirus (unpublished results).
FIG. 5.
Alignment of the dUTPase motif within the HERV.HML5 family (derived from the element within cosmid AC004536) with a human dUTPase (34).
(ii) HERV.ADP.
HERV.ADP was originally described by Lyn et al. (28). This element is defective, consisting of a solitary pol gene of approximately 1.5 kb in length. Southern hybridization analysis indicated that the members of this family are present at high copy numbers, and sequence analysis suggested that they are most closely related to HERV.I (28). Analyses presented here show that members of this family are likely to have copy numbers of around 60 and confirm their close relationship with HERV.I. The neighbor-joining tree in Fig. 1a placed HERV.I and HERV.ADP in a monophyletic group, although this was not the case for the maximum-parsimony tree, which instead placed the HERV.ADP family next to viruses derived from several different vertebrate classes. Analysis of the PBS sequences of HGMP cosmid clones closely related to HERV.ADP (AC005741 shows 83% nucleotide identity to HERV.ADP across 800 bp of pol, for example) indicated that these elements are probably not primed by isoleucine tRNA (the tRNA primer for the HERV.I family); instead, the element within AC005741 showed 14 of 18 matches to the human threonine tRNA, as shown in Fig. 3b. It should be noted, however, that the same PBS sequence also showed 13 of 18 matches to the murine leukemia virus proline tRNA PBS, and thus the binding affinity of this family cannot yet be considered definitive (unpublished results).
For the above-stated reasons, it is likely that HERV.ADP and HERV.I represent separate families. Investigation of the AC005741 HERV sequence indicated that it has a length of 8.4 kb and that full-length (but defective) gag, pol, and env genes are present (Fig. 2 and 4 and Table 4). The estimated integration date of this element, based on a corrected LTR divergence figure of 12.4%, was 30 to 48 million years ago. This figure is consistent with the reported insertion of the ADP-ribosyltransferase pseudogene into HERV.ADP, which is thought to have occurred at least 27 million years ago (28).
(iii) HERV.FRD.
HERV.FRD was isolated by reverse transcription-PCR from retrovirus-like particles released from the human breast cancer cell line T47-D (49). Characterization of a 2.8-kb region of the pol gene indicated that this element was most closely related to class I HERVs such as ERV-9 (49). I identified a full-length member of this family within cosmid AC004022 at q21-22 on chromosome 7 (Table 3). The two elements exhibited 99% nucleotide identity across the 2.8 kb of pol and clustered with robust bootstrap support in my analyses. Further investigation of AC004022 revealed that the element is longer, at 10.8 kb, than other endogenous retroviruses (Table 4). The LTRs measured 715 bp (5′) and 703 bp (3′) in length and differed by over 19% (Fig. 3b). The corrected divergence of 22.6% suggests an integration date of 53 to 87 million years ago, implying that this HERV family may be one of the oldest and that it is probably present in most, if not all, extant primate species. The PBS sequences indicate that these viruses, like the HERV.H family members (30), are probably primed by tRNAHis; there were 14 matches of 18 to the human histidine tRNA (Fig. 3b).
The AC004022 provirus has remained reasonably intact and contains all of the amino acid motifs associated with gag, pol, and env (Fig. 2 and 4), although only short ORFS were apparent in all three genes. The size of each appeared to be roughly equivalent to those of other retroviruses, and the unexpectedly large size of the AC004022 element was due to the presence of a 2-kb region inserted between the 5′ LTR and the gag gene (the 5′ end of gag in AC004022 was identified by comparison with the GaLV gag gene [unpublished results]). There are two possible explanations for the presence of this region: (i) it represents a later insertion into a preexisting HERV sequence and was not part of the original provirus, or (ii) members of this family encode an additional gene upstream of gag. If the second scenario were the case, then the FRD family would be unique among HERVs in encoding a gene in addition to gag, dUTPase, pol, and env. It is difficult to estimate the probabilities of the alternative scenarios since only one member of the family has been characterized in this region. Furthermore, BLAST searches failed to reveal any obvious similarities to repetitive sequences (which would support scenario i) or to other proteins (which would support scenario ii) (unpublished results). Determination of the presence or absence of this putative gene in other members of the HERV.FRD family will therefore ultimately resolve this question.
The relationship of the HERV.FRD family to other viruses (in common with several of the other HERV families described in this report) was difficult to resolve due to inconsistencies between trees constructed by different methods, with the HERV.FRD members being placed in a location similar to that of the HERV.R (type b) and HERV.Z69907 families (Fig. 1).
(iv) HERV.XA.
Members of the HERV.XA family are present at low copy numbers (16 per haploid genome) in humans and are related to HERV.H. Similar viruses have been identified in the Old and New World monkeys, indicating that this family is at least 40 to 45 million years old (60). To date, five members of this family have been partially characterized, with the most complete characterization (HERV.XA38) extending from the central region of the pol gene to the end of a somewhat truncated env gene (60). Part of the HERV.XA pol gene was found to be 88% identical to a 500-bp portion of cosmid AC000378 (Table 2), and further investigation suggested that this cosmid contains a 6.2-kb element with a full-length gag gene and a large deletion spanning the 3′ end of the pol gene and the 5′ region of env (Table 4). Unlike previously characterized members of this family, AC000378 contained intact LTRs (other members have one or more Alu repeats at their 3′ ends [60]). The 5′ and 3′ LTRs were 433 and 438 bp in length and exhibited 9.2% divergence (10.0% corrected), implying that this element first integrated into the primate lineage 24 to 38 million years ago. Like members of the two HERV.F families, AC000378 is primed by a phenylalanine tRNA, with 17 of 18 matches conserved between its PBS and the 3′ end of the human tRNAPhe.
The identification of only one member of this family by BLAST search and the phylogenetic position of this member are in accord with the previously observed low copy number and relatively close relationship to HERV.H. The lack of PBS homology with HERV.H and the low levels of sequence similarity to HERV.F and HERV.F (type b) (to which the element within AC000378 is most closely related) outside of RT indicate that HERV.XA probably constitutes a separate family.
DISCUSSION
This report describes one of the first attempts to systematically identify and characterize endogenous retroviruses identified by the HGMP and to examine their relationship to other vertebrate retroviruses. Recently, Lindeskog (26) used a somewhat different approach, based largely on pol gene sequence similarity, to classify the HERVs into 13 groups, several of which contained more than one family. My analyses, which were based on phylogenetic criteria rather than sequence divergence, led to the identification of 22 independent HERV families. Several of the families described here have not been identified previously, but there are also other differences between our analyses. For example, Lindeskog (as in this report) identified two families of HERV.I-related elements. However, in his case, they were HERV.I itself and HERV.FTD, whereas my analyses suggest that HERV.FTD and HERV.I are very similar and that both are closely related to the HERV.ADP family (which was not included in the other data set [26]). However, in contrast, Lindeskog identified five ERV-9/HERV.W/HERV.P-related families, whereas my analyses indicated that there are only three.
Of the 22 families shown in Fig. 1, 6 had not been characterized previously (although HERV.F [type b] has been independently identified [26]), and the affinities of a further 6 families which had been previously described were based on closest matches to HGMP-derived cosmid sequences. However, it is likely that even the figure of 22 is a conservative one, for two reasons. First, some families are probably harbored at sufficiently low copy number that they are not present in the 7% of the human genome investigated here. Indeed, available sequence data for several families is currently limited to a single element. Second, it is likely that a number of the families which appear monophyletic in this study are (due to relatively poor sampling of nonhuman viruses in some regions of the retroviral phylogeny) actually polyphyletic, and these will eventually be split. For example, the phylogenies shown in Fig. 1 suggest that there are three families of HERV.K-like viruses within the human genome, namely HERV.K itself, HERV.HML5, and HERV.HML6. However, previous reports have suggested (on the basis of sequence divergence) the presence of up to 10 HERV.K-like families (3, 15, 35), and it is possible that the isolation of additional vertebrate retroviral sequences will cause some of the class II HERVs to be broken up into additional families. The relatively low support for the monophyly of many of the HERVs in this region of the tree supports this notion; individual HERV families would typically be expected to cluster with robust bootstrap support. For the same reasons, it is possible that the HERV.HS49C23- and HERV.L-related elements are actually derived from more than one family. The monophyly of two HERV.HS49C23 family lineages have less than 66% bootstrap support in the two trees, and HERV.L could not be phylogenetically distinguished from the rodent virus MuERV.L (6). In contrast, several other previously reported elements are unlikely to represent independent families since they appear to be very closely related to other HERVs. Thus, Np2 and HS-5 are probably members of the HERV.E family, ERV.1 is probably a member of the HERV.R family, and HERV.FTD probably belongs to the HERV.I family. HRES-1 did not have any obvious similarity to any retroviral sequence, and its inclusion with the other HERVs must therefore be considered doubtful.
This report underscores the problems presently associated with HERV nomenclature. The most widely used system classifies families according to the tRNA used to prime DNA synthesis (10, 61). However, this information has often not been available, and thus other HERV families have been named according to a variety of criteria, such as a nearby gene (e.g., HERV.ADP [28]), a clone number (e.g., HERV.S71 [59]), or even an amino acid motif present within the sequence (e.g., HERV.FRD [49]). Furthermore, the term HERV family is itself problematic since the Retroviridae as a whole has also been given family designation (39). The next hierarchical level places related families into classes (originally two but recently three), with class I elements being related to the mammalian type C retroviruses (such as FeLV and GaLV), class II to being related to the mammalian type B and D retroviruses (MMTV and SRV-1) and avian leukosis viruses (RSV), and class III being related to the spumaviruses (HSV) (3, 12, 26, 61).
The greatest drawback with the former system is that it is based on a single character (the tRNA complementary to the viral PBS) which does not correspond closely to the viral phylogeny, the most obvious example being the three HERV families primed by tRNAIle. All three are phylogenetically more closely related to other HERV families which use alternative tRNA primers, and, furthermore, HERV.HML5 clusters with the class II HERVs whereas both of the others are class I. The class level designations work better because they are based on viral relatedness. According to the phylogenies presented in Fig. 1, there are currently 17 class I, 3 class II, and 2 class III families (the 2 class III families being HERV.L [12] and HERV.S). However, it should be noted that some HERV families within the same class are only very distantly related to each other. For example, HERV.R and HERV.HS49C23 exhibit only 33% amino acid identity across the most highly conserved region of RT, as shown in Fig. 2 (unpublished data). Furthermore, in some retroviral phylogenies (such as that shown in Fig. 1a), the spumaviruses appear more closely related to the class I HERVs than to the class III HERVs. Despite these problems, it is my opinion that the current systems should remain in place, at least until the human genome sequence has been completed and the full complement of HERV families has been determined.
All of the families identified in this study encode in-frame stop codons or frameshift mutations, and several also contain large deletions. This is consistent with the characterization of previously identified HERV families, virtually all of which are highly defective (10, 61). The most-intact element identified so far is HERV.K10, which has a single stop codon within both gag and env and possibly a small deletion, also within env (42). Another unusual feature of HERV.K10 is the low level of divergence between its LTRs (of about 0.2%), suggesting that it probably integrated into the primate lineage in the recent past (42). This is in contrast to other HERV families, many of which are thought to have been passaged vertically since the divergence of the common ancestor of humans and Old World monkeys some 25 to 30 million years ago (2, 31, 44, 47, 50). LTR divergence data suggest that many of the families described here have also been present since the human and Old World monkey lineages diverged and that one family, HERV.FRD, may be exceptionally old: the two integration dates estimated for this element were 53 and 87 million years. Although these figures should be treated with some caution, since they assume a molecular clock within the primate lineage (4, 25), they still suggest that the HERV.FRD family may be of similar age to the HERV.H family (which is thought to have entered the primate lineage more than 50 million years ago [2, 31, 47]). Another potentially ancient family is HERV.HS49C23. No LTRs could be identified in any member of the family, but none contain ORFs of more than 200 amino acids (unpublished data), and they have an unusual phylogenetic position, being only distantly related to other mammalian viruses.
HERV genomic organization as described here is also consistent with reports of previously identified endogenous elements in that gag, pol, and env but are generally encoded there are few, if any, other gene products. There were two exceptions to this: the HERV.HML5 family (like with the other class II HERVs) encodes a dUTPase between the Pro and RT genes, and the HERV.FRD family may encode an additional (∼2-kb) gene product between its 5′ LTR and gag gene.
Finally, the HGMP sequence library provides an excellent opportunity to study the long-term evolutionary biology and retrotransposition dynamics of endogenous retroviral families, and we are currently using these data to track the evolution of these elements within the primate lineage.
ACKNOWLEDGMENTS
This work was supported by the Royal Society.
I thank J. Martin for the unpublished caecilian (I. kohtaoensisIII and -IV) and RV Common possumII sequences and P. Kabat for the unpublished RV EchidnaII, RV Jackdaw, and RV Thrush sequences. Thanks also to J. Taylor and D. Quicke for permission to use their tree searching strategy prior to publication and to J. Martin, C. Gosling, and R. Gifford for useful discussions.
ADDENDUM IN PROOF
The classification of endogenous human retroviruses is currently being reviewed by the ICTV Retroviridae committee. Comments or suggestions regarding HERV nomenclature can be sent via e.mail to Roswitha Löwer at: loero@pei.de (Paul Ehrlich Institut, Langen, Germany).
REFERENCES
- 1.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D L. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderssen S, Sjottem E, Svineng G, Johansen T. Comparative analyses of LTRs of the ERV-H family of primate-specific retrovirus-like elements isolated from marmoset, African green monkey, and man. Virology. 1997;234:14–30. doi: 10.1006/viro.1997.8590. [DOI] [PubMed] [Google Scholar]
- 3.Andersson M L, Lindeskog M, Medstrand P, Westley B, May F, Blomberg J. Diversity of human endogenous retrovirus class II-like sequences. J Gen Virol. 1999;80:255–260. doi: 10.1099/0022-1317-80-1-255. [DOI] [PubMed] [Google Scholar]
- 4.Bailey W J, Fitch D H A, Tagle D A, Czelusniak J, Slightom J L, Goodman M. Molecular evolution of the ψη-globin gene locus: gibbon phylogeny and the hominid slowdown. Mol Bol Evol. 1991;8:155–184. doi: 10.1093/oxfordjournals.molbev.a040641. [DOI] [PubMed] [Google Scholar]
- 5.Beck S, Sterk P. Genome scale DNA sequencing: where are we? Curr Opin Biotechnol. 1998;9:116–120. doi: 10.1016/s0958-1669(98)80094-3. [DOI] [PubMed] [Google Scholar]
- 6.Bénit L, De Parseval N, Casella J-F, Callebaut I, Cordonnier A, Heidmann T. Cloning of a new murine endogenous retrovirus, MuERV-L, with strong similarity to the human HERV-L element and with a gag coding sequence closely related to the Fv1 restriction gene. J Virol. 1997;71:5652–5657. doi: 10.1128/jvi.71.7.5652-5657.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bénit L, Lallemand J-B, Casella J-F, Philippe H, Heidmann T. ERV-L elements: a family of endogenous retrovirus-like elements active through the evolution of mammals. J Virol. 1999;73:3301–3308. doi: 10.1128/jvi.73.4.3301-3308.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benveniste R, Todaro G. Evolution of type C viral genes: preservation of ancestral murine type C viral sequences in pig cellular DNA. Proc Natl Acad Sci USA. 1975;72:4090–4094. doi: 10.1073/pnas.72.10.4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blond J-L, Besème F, Duret L, Bouton O, Bedin F, Perron H, Mandrand B, Mallet F. Molecular characterization and placental expression of HERW-W, a new human endogenous retrovirus family. J Virol. 1999;73:1175–1185. doi: 10.1128/jvi.73.2.1175-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boeke J D, Stoye J P. Retrotransposons, endogenous retroviruses, and the evolution of retroelements. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 343–435. [PubMed] [Google Scholar]
- 11.Bonner T I, O'Connell C, Cohen M. Cloned endogenous retroviral sequences from human DNA. Proc Natl Acad Sci USA. 1982;79:4709–4713. doi: 10.1073/pnas.79.15.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cordonnier A, Casella J-F, Heidmann T. Isolation of novel human endogenous retroviral-like elements with foamy virus-related pol sequence. J Virol. 1995;69:5890–5897. doi: 10.1128/jvi.69.9.5890-5897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delassus S, Sonigo P, Wain-Hobson S. Genetic organisation of gibbon ape leukaemia virus. Virology. 1989;173:205–213. doi: 10.1016/0042-6822(89)90236-5. [DOI] [PubMed] [Google Scholar]
- 14.Felsenstein J. PHYLIP (phylogeny inference package), version 3.5c. Seattle: Department of Genetics, University of Washington; 1993. [Google Scholar]
- 15.Franklin G C, Chretien S, Hanson I M, Rochefort H, May F E B, Westley B R. Expression of human sequences related to those of mouse mammary tumor virus. J Virol. 1988;62:1203–1210. doi: 10.1128/jvi.62.4.1203-1210.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herniou E, Martin J, Miller K, Cook J, Wilkinson M, Tristem M. Retroviral diversity and distribution in vertebrates. J Virol. 1998;72:5955–5966. doi: 10.1128/jvi.72.7.5955-5966.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hirose Y, Takamatsu M, Harada F. Presence of env genes in members of the RTLVH family of human endogenous retrovirus-like elements. Virology. 1993;192:52–61. doi: 10.1006/viro.1993.1007. [DOI] [PubMed] [Google Scholar]
- 18.Holzschu D L, Martineau D, Fodor S K, Vogt V M, Bowser P R, Casey J W. Nucleotide sequence and protein analysis of a complex piscine retrovirus, walleye dermal sarcoma virus. J Virol. 1995;69:5320–5331. doi: 10.1128/jvi.69.9.5320-5331.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kabat P, Tristem M, Opavsky R, Pastorek J. Human endogenous retrovirus HC2 is a new member of the S71 retroviral subgroup with a full-length pol gene. Virology. 1996;226:83–94. doi: 10.1006/viro.1996.0630. [DOI] [PubMed] [Google Scholar]
- 20.Kannan P, Buettner R, Pratt D, Tainsky M. Identification of a retinoic acid-inducible endogenous retroviral transcript in the human teratocarcinoma-derived cell line PA-1. J Virol. 1991;65:6343–6348. doi: 10.1128/jvi.65.11.6343-6348.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:116–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 22.Kröger B, Horak I. Isolation of novel human retrovirus-related sequences by hybridization to synthetic oligonucleotides complementary to the tRNAPro primer-binding site. J Virol. 1987;61:2071–2075. doi: 10.1128/jvi.61.7.2071-2075.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.La Mantia G, Maglione D, Pengue G, Di Cristofano A, Simeone A, Lanfrancone L, Lania L. Identification and characterization of novel human endogenous retroviral sequences preferentially expressed in undifferentiated embryonal carcinoma cells. Nucleic Acids Res. 1991;19:1513–1520. doi: 10.1093/nar/19.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levy L, Lobelle-Rich P, Elder J, Payne S, Montelaro R. An unusual retrovirus-like sequence identified in human DNA. J Gen Virol. 1990;71:1613–1618. doi: 10.1099/0022-1317-71-7-1613. [DOI] [PubMed] [Google Scholar]
- 25.Li W-H, Tanimura M. The molecular clock runs more slowly in man than in apes and monkeys. Nature. 1987;336:93–96. doi: 10.1038/326093a0. [DOI] [PubMed] [Google Scholar]
- 26.Lindeskog M. Transcription, splicing and genetic structure within the human endogenous retroviral HERV.H family. Ph.D. thesis. Lund, Sweden: Department of Infectious Diseases and Medical Microbiology, Lund University; 1999. [Google Scholar]
- 27.Lower R, Lower J, Kurth R. The viruses in all of us: characteristics and biological significance of human endogenous retrovirus sequences. Proc Natl Acad Sci USA. 1996;93:5177–5184. doi: 10.1073/pnas.93.11.5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lyn D, Deaven L L, Istock N, Smulson M. The polymorphic ADP-ribosyltransferase (NAD+) pseudogene 1 in humans interrupts an endogenous pol-like element on 13q34. Genomics. 1993;18:206–211. doi: 10.1006/geno.1993.1456. [DOI] [PubMed] [Google Scholar]
- 29.Maeda N, Kim H S. Three independent insertions of retrovirus-like sequences in the haptoglobin gene cluster of primates. Genomics. 1990;8:671–683. doi: 10.1016/0888-7543(90)90254-r. [DOI] [PubMed] [Google Scholar]
- 30.Mager D L, Henthorn P S. Identification of a retrovirus-like repetitive element in human DNA. Proc Natl Acad Sci USA. 1984;81:7510–7514. doi: 10.1073/pnas.81.23.7510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mager D L, Freeman J D. HERV.H endogenous retroviruses—presence in the New-World branch but amplification in the Old-World primate lineage. Virology. 1995;213:395–404. doi: 10.1006/viro.1995.0012. [DOI] [PubMed] [Google Scholar]
- 32.Martin J, Herniou E, Cook J, Waugh O'Neill R, Tristem M. Interclass transmission and phyletic host tracking in the murine leukemia-related retroviruses. J Virol. 1999;73:2442–2449. doi: 10.1128/jvi.73.3.2442-2449.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin M, Bryan T, Rasheed S, Khan A. Identification and cloning of endogenous retroviral sequences present in human DNA. Proc Natl Acad Sci USA. 1981;78:4892–4896. doi: 10.1073/pnas.78.8.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McIntosh E M, Ager D D, Gadsden M H, Haynes R H. Human dUTP pyrophosphatase: cDNA sequence and potential biological importance of the enzyme. Proc Natl Acad Sci USA. 1992;89:8020–8024. doi: 10.1073/pnas.89.17.8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medstrand P, Blomberg J. Characterization of novel reverse transcriptase encoding human endogenous retroviral sequences similar to type A and type B retroviruses: differential transcription in normal human tissues. J Virol. 1993;67:6778–6787. doi: 10.1128/jvi.67.11.6778-6787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medstrand P, Mager D L, Yin H, Dietrich U, Blomberg J. Structure and genomic organisation of a novel human endogenous retrovirus family: HERV.K (HML-6) J Gen Virol. 1997;78:1731–1744. doi: 10.1099/0022-1317-78-7-1731. [DOI] [PubMed] [Google Scholar]
- 37.Miyamoto M M, Goodman M. DNA systematics and evolution of primates. Annu Rev Ecol Syst. 1990;21:197–220. [Google Scholar]
- 38.Moore R, Dixon M, Smith R, Peters G, Dickson C. Complete nucleotide sequence of a milk-transmitted mouse mammary tumor virus: two frameshift suppression events are required for translation of gag and pol. J Virol. 1987;61:480–490. doi: 10.1128/jvi.61.2.480-490.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D. Virus taxonomy: the classification and nomenclature of viruses. The Sixth Report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer-Verlag; 1995. [Google Scholar]
- 40.O'Connell C, O'Brien S, Nash W, Cohen N. ERV-3, a full-length human endogenous provirus: chromosomal localization and evolutionary relationships. Virology. 1984;138:225–235. doi: 10.1016/0042-6822(84)90347-7. [DOI] [PubMed] [Google Scholar]
- 41.O'Connell C, Cohen M. The long terminal repeats of a novel human endogenous retrovirus. Science. 1984;226:1204–1206. doi: 10.1126/science.6505687. [DOI] [PubMed] [Google Scholar]
- 42.Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986;60:589–598. doi: 10.1128/jvi.60.2.589-598.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Patience C, Takeuchi Y, Weiss R A. Infection of human cells by an endogenous retrovirus of pigs. Nat Med. 1997;3:282–286. doi: 10.1038/nm0397-282. [DOI] [PubMed] [Google Scholar]
- 44.Patience C, Wilkinson D A, Weiss R A. Our retroviral heritage. Trends Genet. 1997;13:116–120. doi: 10.1016/s0168-9525(97)01057-3. [DOI] [PubMed] [Google Scholar]
- 45.Perl A, Rosenblatt J, Chen I, Di Vincenzo J, Bever R, Poiesz J, Abraham G. Detection and cloning of new HTLV-related endogenous sequences in man. Nucleic Acids Res. 1989;17:6841–6854. doi: 10.1093/nar/17.17.6841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perron H, Garson J A, Bedin F, Beseme F, Paranhos-Baccala G, Komurian-Pradel F, Mallet F, Tuke P W, Voisset C, Blond J L, Lalande B, Seigneurin J M, Mandrand B the Collaborative Research Group on Multiple Sclerosis. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. Proc Natl Acad Sci USA. 1997;94:7583–7588. doi: 10.1073/pnas.94.14.7583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Purvis A. A composite estimate of primate phylogeny. Philos Trans R Soc Lond Ser B. 1995;348:405–421. doi: 10.1098/rstb.1995.0078. [DOI] [PubMed] [Google Scholar]
- 48.Repaske R, Steele P E, O'Neill R R, Rabson A B, Martin M A. Nucleotide sequence of a full-length endogenous retroviral segment. J Virol. 1985;54:764–772. doi: 10.1128/jvi.54.3.764-772.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seifarth W, Skladny H, Krieg-Schneider F, Reichert A, Hehlmann R, Leib-Mosch C. Retrovirus-like particles released from the human breast cancer cell line T47-D display type B- and C-related endogenous retroviral sequences. J Virol. 1995;69:6408–6416. doi: 10.1128/jvi.69.10.6408-6416.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shih A, Coutavas E E, Rush M G. Evolutionary implications of primate endogenous retroviruses. Virology. 1991;182:495–502. doi: 10.1016/0042-6822(91)90590-8. [DOI] [PubMed] [Google Scholar]
- 51.Silver J, Rabson A, Bryan T, Willey R, Martin M. Human retroviral sequences on the Y chromosome. Mol Cell Biol. 1987;7:1559–1562. doi: 10.1128/mcb.7.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sprinzl M, Horn C, Brown M, Ioudovitch A, Steinberg S. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1998;26:148–153. doi: 10.1093/nar/26.1.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sverdlov E D. Perpetually mobile footprints of ancient infections in human genome. FEBS Lett. 1998;428:1–6. doi: 10.1016/s0014-5793(98)00478-5. [DOI] [PubMed] [Google Scholar]
- 54.Tatusova T A, Madden T L. Blast 2 sequences—a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–250. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 55.Telesnitsky A, Goff S P. Reverse transcription and the generation of viral DNA. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 121–160. [PubMed] [Google Scholar]
- 56.Tristem M, Herniou E, Summers K, Cook J. Three retroviral sequences in amphibians are distinct from those in mammals and birds. J Virol. 1996;70:4864–4870. doi: 10.1128/jvi.70.7.4864-4870.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vogt P K. Historical introduction to the general properties of retroviruses. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 1–25. [PubMed] [Google Scholar]
- 58.Vogt V M. Retroviral virions and genomes. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 27–71. [PubMed] [Google Scholar]
- 59.Werner T, Brack-Werner R, Leib-Mosch C, Backhaus H, Erfle V, Hehlmann R. S71 is a phylogenetically distinct human retroviral element with structural and sequence homology to simian sarcoma virus (SSV) Virology. 1990;174:225–238. doi: 10.1016/0042-6822(90)90071-x. [DOI] [PubMed] [Google Scholar]
- 60.Widegren B, Kjellman C, Aminoff S, Sahlford L G, Sjogren H-O. The structure and phylogeny of a new family of human endogenous retroviruses. J Gen Virol. 1996;77:1631–1641. doi: 10.1099/0022-1317-77-8-1631. [DOI] [PubMed] [Google Scholar]
- 61.Wilkinson D A, Mager D L, Leong J-A C. Endogenous human retroviruses. In: Levy J A, editor. The Retroviridae. III. New York, N.Y: Plenum Press; 1994. pp. 465–535. [Google Scholar]
- 62.Xiong Y, Eickbush T H. Origin and evolution of retroelements based on their reverse transcriptase sequences. EMBO J. 1990;9:3353–3362. doi: 10.1002/j.1460-2075.1990.tb07536.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.York D F, Vigne R, Verwoerd D W, Querat G. Nucleotide sequence of jaagsiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J Virol. 1992;66:4930–4939. doi: 10.1128/jvi.66.8.4930-4939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]