Abstract
The opportunistic fungal pathogen Candida parapsilosis is a major causative agent of candidiasis leading to death in immunocompromised individuals. Azoles are the first line of defense in treatment by inhibiting ERG11, involved in the synthesis of ergosterol, the main sterol fungal sterol. Resistance to azoles is on the increase worldwide including in Lebanon. The purpose of this study is to characterize nine hospital isolates labeled as C. parapsilosis: four resistant and five sensitive to fluconazole. Phenotypic characterization was achieved through a battery of tests that target pathogenicity attributes such as virulence, biofilm formation, stress resistance, and ergosterol content. Genotypic analysis was done through whole genome sequencing to mutations in key virulence and resistance genes. Phylogenetic comparison was performed to determine strain relatedness and clonality. Genomic data and phylogenetic analysis revealed that three of the nine C. parapsilosis isolates were misidentified; two as C. orthopsilosis and C. metapsilosis belonging to the C. parapsilosis complex, while the third was C. albicans. Moreover, several known and novel mutations in key drug resistance and virulence genes were identified such as ERG11, ERG3, ERG6, CDR1, and FAS2. Phylogenetic analysis revealed a high degree of relatedness and clonality within our C. parapsilosis isolates. Our results showed that resistant isolates had no increased ergosterol content, no statistically significant difference in virulence, but exhibited an increase in biofilm content compared to the sensitive isolates. In conclusion, our study, the first of its kind in Lebanon, suggests several mechanisms of antifungal drug resistance in C. parapsilosis hospital isolates.
Keywords: Candida parapsilosis, Fluconazole, Pathogenicity, Resistance, Clonality
Introduction
Infections brought on by invasive opportunistic fungal pathogens belonging to the Candida species have become much more common throughout the last 30 years. Up to 50% of all fatalities from fungal diseases can be attributed to invasive fungal infections, which are often linked to high rates of severe sickness [1]. C. parapsilosis presents a particular risk to newborns, recipients of organ transplants, and people on parenteral nutrition, and it belongs to the Candida clade that is characterized by the unique translation of the CUG codons as serine instead of leucine. Although C. parapsilosis infections have lower mortality and morbidity rates than C. albicans, several clinical isolates of C. parapsilosis have been reported to be less susceptible to echinocandins and resistant to azole treatment in some regions [2]. The C. parapsilosis complex is composed of three members: C. parapsilosis, C. orthopsilosis and C. metapsilosis. Before recent advances in genomics, they were indistinguishable and considered as one species, C. parapsilosis, up until 2005 [3]. Even nowadays, distinguishing between C. parapsilosis, C. orthopsilosis and C. metapsilosis is not possible through commercial systems, but only through DNA sequencing. Very few studies have investigated the virulence of C. parapsilosis, and even fewer that of C. orthopsilosis and C. metapsilosis. To date, only a few studies have shown that C. orthopsilosis and C. parapsilosis exhibit similar virulence and adhesion properties, while C. metapsilosis is generally considered the least virulent of the three species [4]. C. metapsilosis is also the least commonly isolated, causing between 0.6% and 6.9% of invasive candidiasis cases [5]. C. parapsilosis exhibits an oval, cylindrical round shape; it appears as white and creamy on potato dextrose agar with a shiny smooth or wrinkled colony phenotype. Unlike C. albicans and C. tropicalis, C. parapsilosis exists in the yeast and pseudohyphal form forming crepe-like and concentric patterns with no true hyphal extensions. Although all members of the C. parapsilosis species complex have diploid genomes, C. parapsilosis isolates are highly homozygous, while C. orthopsilosis and C. metapsilosis isolates exhibit extreme heterozygosity. These heterozygous characteristics likely stem from multiple hybridization events between closely related parents. A significant proportion of C. metapsilosis isolates have heterozygosity in the mating type-like locus, with introgression occurring at the MTLa locus. Mutations in the ERG11 gene or its overexpression are linked to azole resistance in Candida species [6, 7,8,9,10]. Resistance to drugs has also been attributed to an increase in cell wall thickness, particularly an upregulation of cell wall chitin biosynthesis, rendering the cell impermeable to the drug. C. parapsilosis complex was divided into III groups, group 1 refers to the parapsilosis species, while group II and group II later became known as orthopsilosis and metapsilosis; the DNA sequence of ITS1 sequences showed differences between the three groups.
The purpose of this study is to characterize nine C. parapsilosis Lebanese hospital isolates phenotypically and genotypically. Phenotypic characterization will be achieved through a battery of tests that target pathogenicity attributes such as virulence in a mouse model of disseminated candidosis, organ burden load, cell surface disruption, adhesion potential, ergosterol and biofilm formation. Genotypic analysis will be done through whole genome sequencing to identify documented and novel SNPs and mutations in key virulence and resistance genes. Phylogenetic comparison of isolates will also be done to analyze strain relatedness and clonality. The purpose is to attempt a correlation between the observed phenotypes and their specific genotypes.
Materials and Methods
C. parapsilosis strains
Nine C. parapsilosis hospital isolates were obtained from the Lebanese American University Medical Center, Rizk Hospital, Beirut in Lebanon. Species identification and broth microdilution antifungal susceptibility testing (Table 1) was performed by Husni et al. [11]. Four out of nine isolates were resistant to fluconazole, designated as isolates R1, R2, R3 and R4 and the five others designated as isolates S1, Ca, Co, S2 and Cm were sensitive to fluconazole. C. parapsilosis CDC317 MYA-4646 was used as a reference strain (GenBank accession no. GCF_000182765.1).
Table 1. Broth Microdilution Results.
Isolate R1, R2, R3 and R4 are resistant to fluconazole, whereas isolate S1, Ca, Co, S2 and Cm were sensitive.
| Isolates | Fluconazole (MIC) \varvecμ\varvecg/\varvecm\varvecL | |
|---|---|---|
| S1 | 0.5 | Sensitive |
| Ca | 1 | Sensitive |
| Co | 1 | Sensitive |
| R1 | 32 | Resistant |
| S2 | 2 | Sensitive |
| Cm | 0.5 | Sensitive |
| R2 | 8 | Resistant |
| R3 | 8 | Resistant |
| R4 | 16 | Resistant |
C. parapsilosis culture
C. parapsilosis isolates were inoculated in 5mL potato dextrose broth (PDB) (Conda Laboratories) then incubated at 30°C overnight with shaking 90 rpm overnight. Finally, isolates were streaked onto potato dextrose agar (PDA) and incubated at 30°C for 48h. One colony was inoculated in fresh PDB for biofilm quantification, DNA extraction, cell surface disruption, organ burden load, and virulence.
DNA extraction and whole genome sequencing
DNA was extracted using the “Quick-DNA Fungal/Bacterial Miniprep” kit (Zymo Research), following the manufacturer’s instructions. Elution was carried out using the BE buffer from the “NucleoSpin® DNA Yeast” kit by Macherey-Nagel. Quantity and quality of DNA were assessed using a NanoDrop spectrophotometer. Library preparation, Illumina sequencing at a depth of 90X coverage, adapter trimming, and genome assembly were conducted by MicrobesNG in Birmingham, UK. Using the ATCC CDC317 MYA 4646 as a reference strain, variant calling was performed by MicrobesNG to predict variants relative to their reference. Sequences were deposited in NCBI through BioProject ID number PRJNA1091179.
Sequence analysis and variant calling
Our sequence analysis approach consisted of scanning the genome for mutations in genes potentially involved in the development of azole resistance, biofilm formation and virulence. In total, we analyzed 96 genes (Supplementary Table 1). The genes of interest were translated using ExPASy translate tool (Swiss Institute of Bioinformatics) and the protein sequences were aligned using the Clustal Omega Multiple Sequence Alignment Tool (European Molecular Biology Laboratories, European Bioinformatics Institute) in order to determine amino acid substitutions in hospital isolates compared to the control strain. Mutations were verified by variant calling results performed by MicrobesNG. A literature search was performed in order to identify amino acid substitutions that have been previously documented.
Single Nucleotide Polymorphism (SNP) Detection
We conducted a SNP analysis within the 8 genomes of the C. parapsilosis complex, comparing them to the SNPs present in the initial references’ strains. We utilized the snippy multicommand (snippy-base application v4.5.0) developed by Seemann [12] to generate a multiple alignment of the core genome against a common reference, which was CDC317 MYA-4646 for the C. parapsilosis isolates and Co 90–125 for C. orthopsilosis and BP57 for C. metapsilosis. This reference genome was chosen as the basis for comparison. The pipeline was employed to identify and document the variants, generating individual files for each isolate listing the specific variations.
Annotation
Annotation of the reference genomes was achieved by using the following pipeline. First, gene prediction is done by using the combination of GeneMark-ES v4.71 and AUGUSTUS v3.4.0 by running BRAKER2 v2.1.6 with the fungus this step will identify the locus, CDS, and mRNA of the corresponding genes [13, 14,15,16,17,18,19,20]. Functional annotation was added by running the gff3 annotation file of the identified genes using Interproscan 5.50–84.0 [21] on the COG database, resulting in an XML file which in turn will be incorporated into the functional annotation pipeline Funannotate v1.8.7 [22,23,24]. The first step of this functional annotation is scanning the PFAM database using HMMscan default parameters (HMMer v3.3) (hmmer.org) and based on eggnog orthology data results using emapper v2.1.2 [25,26]. Furthermore, Diamond Blastp [27] used the UniProt DB v2023_02 and MEROPS v12.0 databases to generate separate functional annotation files that were later combined using Gene2Product v1.69. And finally, using Signalp v5.0 [28], secreted proteins were predicted.
Phylogenetic analysis and ITS sequencing
To determine the phylogenetic relationship between the isolates, the core genome sequence, recombination data, and single nucleotide polymorphisms (SNPs) in conjunction with parsnp v1.2, a tool available in the Harvest suite was utilized. The corresponding reference genome was used for this analysis. SNPs identified in local collinear blocks were then employed to construct an approximate maximum-likelihood tree using FastTree, incorporating the general time reversible (GTR) model of nucleotide substitution. We applied the Shimodaira–Hasegawa test, implemented in FastTree2, to evaluate the support for significant clustering observed in the phylogenetic tree. For graphical representation and annotation, we utilized the interactive tree of life (iTOL) [29,30]. The sequence of the Internal Transcribed Spacer region ITS1 near the 5.8 rRNA gene of isolate “Cm” was Blasted onto the NCBI database and compared with various C. genome ITS sequences to determine phylogeny.
Biofilm Assay
The protocol of biofilm biomass quantification was adopted from [6,31] with slight modifications. The culture medium adopted was PDB and OD measurements were taken at 590 nm. This experiment was performed in biological and technical triplicates and ATCC CDC317 MYA 4646 was used as a control reference.
Ergosterol
To extract plasma membrane ergosterol we used the protocol described by Arthington-Skaggs et al. [32]. Optical densities (ODs) were measured following the protocol detailed in Fattouh et al. [7] and these OD values were used to calculate ergosterol content by relying on the formulas in [7,32]. This experiment was performed in biological triplicates and CDC317 MYA 4646 and the control strain.
Sodium Dodecyl Sulfate (SDS) Susceptibility Surface Disruption Assay
Susceptibility to SDS was determined by microdilution; exponentially grown cultures were diluted to 106 cells/mL, and ten-fold serial dilutions were done. 5 μL of each dilution was spotted on PDA plates and incubated at 30°C on 0.025% SDS plates, and plates lacking SDS (control). All isolates were spotted in triplicates and were photographed at 24h 48h and 72h [6].
Virulence Assay and Organ Fungal Burden Load
To induce a disseminated systemic infection, the Fattouh et al. [7] protocol was adopted. 6 weeks old female BALB/c mice were injected via their tail vein with 5×108 cells in 200 μL 1X Phosphate Buffered Saline solution (PBS) to generate a systemic infection. 6 mice were injected per isolate, and a group of 6 mice were injected with 1X PBS as a control [6,7,33,34]. Mice manipulation followed all ethical standards of the Lebanese American University Institutional Care and Use Committee which approved the execution of this experiment in June 2022 under approval code LAU.ACUC.SAS.RK5.12/June/2022. For the organ burden load experiment, 6 weeks old female BALB/c mice were injected with 5×107 cells in 200 μL PBS via the tail vein to induce a systemic infection. Two mice were injected per isolate, and 2 mice were injected with 1X PBS as a control [34]. On day 4, mice were euthanized using carbon dioxide inhalation and designated organs (liver and kidneys) were taken and preserved in 5mL 1X PBS. Organs were homogenized using a Rotor Stator Homogenizer, 1 mL were taken from each homogenized organ and spread on a PDA plate. Plates were incubated at 30°C and colonies were counted after 48h [35].
Statistical analysis
Tables were generated using Microsoft Excel. Figures as well as all statistical analyses were executed using GraphPad Prism versions 7.00 or 10.1.2. For the virulence assay, the Mantel-Cox test was performed. Furthermore, for the quantification of fungal load, biofilm, and ergosterol, Kruskal Wallis test and Dunn’s multiple comparisons test were performed. Any p-value below or equal to 0.05 was considered significant.
Results
Species Identification and Phylogenetic analysis
Based on the MicrobesNG whole genome sequencing data, strains Co and Cm were deemed to be C. parapsilosis and Ca C. albicans. As a non parapsilosis complex species, The Ca strain was subsequently removed from all further phenotypic and genotypic analysis. A SNP based phylogenetic tree was generated for the sequenced samples and the reference genome (CDC317 MYA-4646). The tree showed that isolates Ca, Co and Cm were distinct from the rest with significant sequence divergence, whilst the other 6 isolates and the reference strain clustered together with high sequence similarity (Fig. 1 a). However, for two isolates Cm and Co, the variant calling did not match the C. parapsilosis CDC317-MYA4646 reference as the sequence exhibited many mutations and unmatched ORFs, so we suspected that they might belong to a different species. The Cm and Co sequences were blasted on NCBI, and Co exhibited very high sequence similarity to C. orthopsilosis. The remaining isolate, Cm did not match with the C. orthopsilosis (Co_90–125) reference sequence from NCBI exhibiting many mutations, and indels upon variant calling. To support this, a phylogenetic tree was generated for Cm alongside six C. orthopsilosis reference genomes (Fig. 1b). The phylogenetics tree showed that Cm is significantly divergent from all C. orthopsilosis reference strains, further justifying our suspicions that it is a different species. Finally, the ITS1 region of Cm was blasted against Candida ITS1 regions. The highest sequence similarity was for C. metapsilosis ITS1. As such the Cm isolate is C. metapsilosis.
Figure 1.
a: SNP based phylogenetic tree of hospital isolates. C. parapsilosis isolates R1, R2, R3, R4, S1 and S2 cluster together and are phylogenetically close as opposed to Co and Cm that cluster together separately. Isolate Ca is phylogenetically divergent from all the other isolates.
b: SNP based phylogenetic tree of Cm with 6 C. orthopsilosis genomes. Cm is significantly divergent from all C. orthopsilosis reference sequences, suggesting it might belong to a different species.
Mutational analysis
Whole genome sequencing was performed on all 8 isolates, followed by variant calling. For the C. parapsilosis isolates, mutational analysis on the genes in Table S1 was performed. For the orthopsilosis and metapsilosis isolates, the mutational analysis is shown in Tables S2 and S3. Note the high clonality amongst C. parapsilosis isolates whereby similar mutations are observed in different strains.
Biofilm Assay
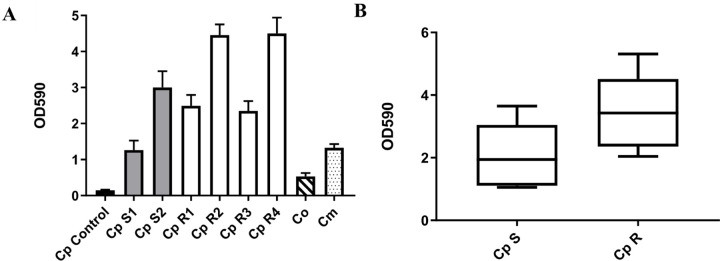
All isolates exhibited an increase in biofilm formation compared to the control strain (Fig. 2A). However resistant isolates exhibited more biofilm formation on average compared to sensitive strains (Fig. 2B). Fluconazole resistant isolates (R1, R2, R3, R4) revealed an increase in biofilm formation compared to sensitive strains (S1, S2) with (x ~1.75) fold increase. Fluconazole sensitive isolates showed (x ~ 2.13) folds increase in the potential of biofilm formation when compared to the reference CDC317 MYA-4646, while resistant isolates exhibited a (x ~ 4.5) increase in biofilm formation compared to the control. Interestingly, both Co and Cm showed less biofilm formation than sensitive and resistant C. parapsilosis isolates.
Figure 2. Biofilm formation.
(A)Biofilm formation is represented as measurements of optical density at 590 nm (OD590) for all C. parapsilosiscomplex isolates. The black bar graph represents the C. parapsilosisreference CDC317 MYA-4646, control, while the gray bar represents the fluconazole sensitive C. parapsilosis isolates, and the white bar graphs represent the fluconazole resistant C. parapsilosis isolates. The hatched bar is for C. orthopsilosis, and the dotted one is for C. metapsilosis. For all isolates and the control, quantification of biofilm amounts was performed in biological triplicates. Mann-Whitney test: Cp-S vs. Cp-R: p = 0.0055 is significant, Cp-S vs. Co: p = 0.0001 is significant, Cp-S vs. Cm: p = 0.2908 is not significant, Cp-R vs. Co: p < 0.0001 is significant, Cp-R vs. Cm: p < 0.0001 is significant, and Co vs. Cm: p = 0.0022 is significant. (B) Kruskal Wallis test. Boxplot graph was generated to compare the potential of biofilm formation in fluconazole-resistant clinical C. parapsilosis isolate groups (R1, R2, R3, R4) with that of the reference CDC317 MYA-4646 and the fluconazole-susceptible isolates group (S1, S2). Resistant isolates exhibited more potential of biofilm formation than the sensitive strains. For all isolates, the experiment was performed in biological triplicates. p < 0.0001 for Cp sensitive isolates compared to control is significant, p < 0.0001 for Cp resistant isolates compared to control is significant.
Ergosterol Assay
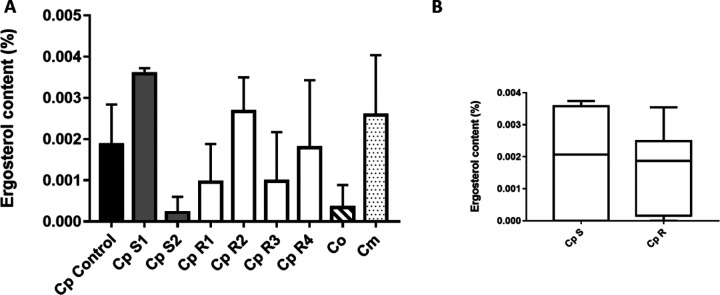
One of the main mechanisms involved in drug resistance in Candida species is the upregulation of ergosterol. When ergosterol deposition increases, the plasma membrane becomes thicker blocking fluconazole from entering the cell, resulting in resistance. We thus measured ergosterol content in both our susceptible and resistant isolates. Although we observed an increase in ergosterol content in some of the sensitive and resistant isolates, no correlation was found between drug resistance and ergosterol content. Isolate Cp S1 showed 92% increase in ergosterol content compared to Cp S2 that exhibited a decrease in approximately 102% of its ergosterol content compared to the control. On the other hand, Cp R1, Cp R3 and Cp R4 showed a decrease in ergosterol content compared to the control, while Cp R2 showed an ergosterol increase of 43%. Our data suggests that ergosterol content might not be the only mechanism to achieve resistance in our isolates. Based on the Mann-Whitney test, observed differences were not statistically significant even though a slight increase in ergosterol content in sensitive parapsilosis isolates was found.
Virulence Assay
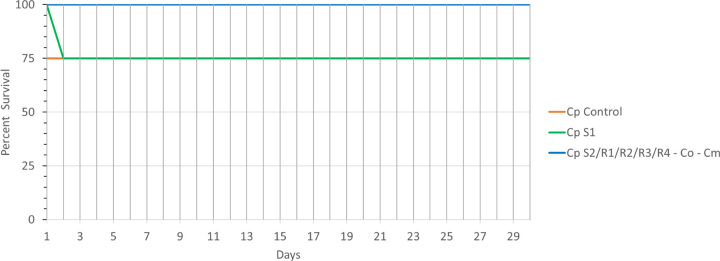
Each isolate was injected into the tail vein of 4 mice in a disseminated model of infection. One out of 4 mice injected with the sensitive isolate S1 was moribund by day 2, whereas no death was observed in isolates S2, Co, Cm, R1, R2, R3 and R4 (Fig. 3). Overall, our data analysis shows that fluconazole resistance is associated with a slight attenuation of virulence in the parapsilosis complex as one mouse injected with S1 and one mouse injected with the sensitive parapsilosis ATCC control strain died, but none of the mice injected with resistant strains were moribund.
Figure 3. Ergosterol content.
(A) In the Mann-Whitney test, the black bar graph represents the C. parapsilosis reference CDC317 MYA-4646, control, while the gray bar represents the fluconazole sensitive C. parapsilosis isolates, and the white bar graphs represent the fluconazole resistant C. parapsilosis isolates. The hatched bar is for C. orthopsilosis, and the dotted one is for C. metapsilosis. For all isolates and the control, quantification of ergosterol amounts were performed in biological triplicates. Cp-S vs. Cp-R: p = 0.6786 is not significant; Cp-S vs. Co: p = 0.5238 is not significant; Cp-S vs. Cm: p = 0.5 is not significant; Cp-R vs. Co: p = 0.1824 is not significant; Cp-R vs. Cm: p = 0.2901 is not significant; Co vs. Cm: p = 0.1 is not significant. (B) In the Kruskal Wallis test & Dunn’s multiple comparisons test, the boxplot graph was generated to represent the ergosterol content in fluconazole sensitive and fluconazole resistant clinical C. parapsilosis isolate groups and upon comparison with the control, CDC317 MYA-4646. p < 0.0001 for Cp sensitive isolates (S1 and S2) compared to control (significant); p < 0.0001 for Cp resistant isolates (R1, R2, R3, R4) compared to control is significant too. For all isolates, the experiment was performed in biological triplicates.
Organ fungal burden Load
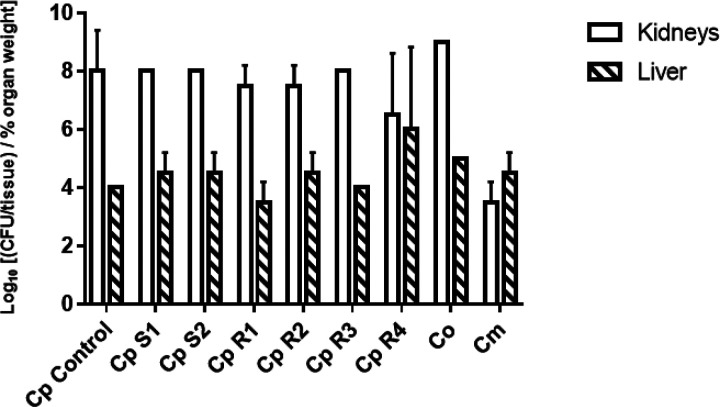
Since no significant differences were observed in our virulence assay, we decided to assess fungal burden load for each of the isolates to determine whether discrepancies amongst isolates are observed. However, no significant difference was observed between sensitive and resistant isolates as far as organ burden load. All isolates exhibited an increased burden load in the kidneys compared to the liver (Fig. 4) with a significant p value of 0.0024. On the other hand, fungal burden load in kidneys and liver for sensitive versus resistant strains compared with the control was not significant with a p value > 0.9. We did we observe any significant differences in organ burden load within the three species.
Figure 4. Figure 3: Murine Model Assay. Virulence potential in a murine disseminated model of infection.
BALB/c mice were injected with 108 cells of CDC317 MYA-4646 and fluconazole-sensitive and resistant isolates and monitored for survival over 30 days. The survival rate in most fluconazole-susceptible and all resistant isolates was 100%. Mantel-Cox test (log-rank test): p = 0.5366 (not significant). Logrank test for trend: P = 0.0552 (not significant).
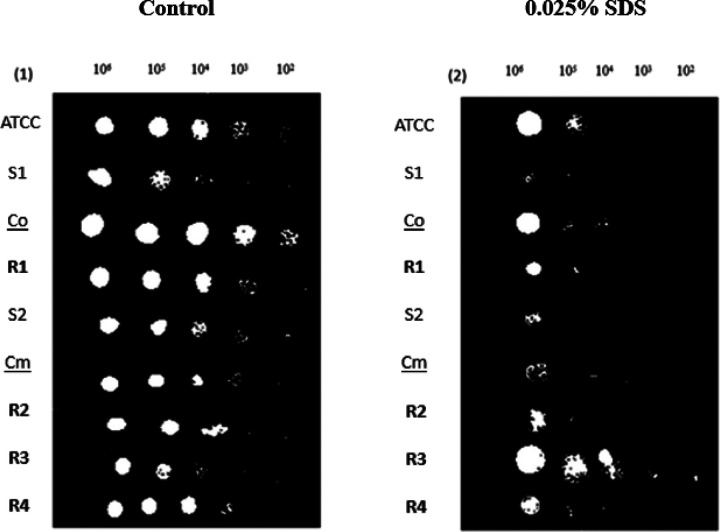
Cell Wall Disruption
The cell wall is a crucial component of fungal cells that plays a critical role in maintaining cell shape, protection from osmotic stress, and the host. Sodium Dodecyl Sulfate (SDS) is a detergent that can disrupt the integrity of the fungal cell wall by solubilizing its components, such as glucans, chitin, and proteins. This disruption can cause fungal cells to become more susceptible to lysis, stress, and other environmental insults. Strains were 10x serially diluted and grown in various concentrations of SDS. No growth was observed at concentrations above 0.025% SDS. Fluconazole resistant isolate R3 was significantly more resistant to SDS than the control strain, while fluconazole sensitive strains S1, S2, and Cm were less resistant to SDS% (Fig. 5). Decreased growth in the sensitive strains implies a relatively weak and permeable cell wall, which can be easily disrupted by SDS. In contrast, resistant fungal strains may have a thicker, more complex, or more impermeable cell wall that is less susceptible.
Figure 5. Figure 4: Fungal Burden Load.
BALB/c mice were intravenously injected in the tail vein with 108 cells for each of the indicated strains (S1, S2, R1, R2, R3, R4, Co, Cm). Data are expressed as the log10 colony-forming units (CFUs)/tissue of % organ weigh from kidneys and liver homogenates at day 4 after the challenge. The log10 CFUs from both kidneys were combined and averaged. Log10 counts were compared for statistical significance by Mann-Whitney test with a p = 0.0024 (significant). A p-value <0.05 was considered to be statistically significant. Moreover, C. parapsilosis sensitive vs. C. parapsilosis resistant kidneys fungal load has a p = 0.4182 (not significant), for the liver fungal load p = 0. 5899 (not significant). C. parapsilosis sensitive vs. C. orthopsilosis kidneys fungal load have a p = 0.0667 (not significant), for the liver fungal load p > 0.9999 (not significant). C. parapsilosis sensitive vs. C. metapsilosis kidneys fungal load has a p = 0.0667 (not significant), for the liver fungal load p = 0.4667 (not significant). C. parapsilosis resistant vs. C. orthopsilosis kidneys fungal load has a p= 0.0222 (significant), for the liver p = 0.9333 (not significant). C. parapsilosis resistant vs. C. metapsilosis kidneys fungal load has a p = 0.222 (significant), for the Liver p = 0.2444 (not significant). C. orthopsilosis vs. C. metapsilosis for the kidneys fungal load is p = 0.3333 (not significant), and for the liver p > 0.9999 (not significant).
Discussion
In Lebanon, as well as on a global level, there has been limited research conducted on the infections of the fungal pathogen C. parapsilosis and its emerging azole resistance in hospital isolates. Most studies focus on C. albicans as it is the most commonly isolated Candida pathogen [8]. Furthermore, there has been no research as to the relationship between resistance patterns and pathogenic attributes of this yeast species. As such, the primary objective of this research was to characterize our hospital isolates, both from a phenotypic and genotypic perspective. Phenotypic characterization entailed a series of tests targeting pathogenesis traits, such as virulence in a mouse model of disseminated candidiasis, organ burden load, biofilm formation, ergosterol content, and cell wall disruption. Genotypic analysis was conducted through whole genome sequencing and SNPs identification to identify known and previously uncharacterized mutations in critical virulence and resistance genes. SNP analysis was done through 3 different methods MicrobesNG, manually through EXPASY and Clustal Omega Multiple Sequence Alignment Tool, and through snippy multicommand, to confirm our results. Additionally, we performed a phylogenetic analysis of the isolates to examine their relatedness and clonality. The overarching goal is to establish a relationship between the observed phenotypes, resistance profiles, and specific genotypes.
Lanosterol 14-demethylase, part of the cytochrome P450 enzyme family, plays a crucial role in ergosterol synthesis. The ERG11 (CPAR2_303740) gene encodes this enzyme, and it serves as a target for azole medications. Azoles, with fluconazole being a prominent example, are frequently employed in the management of candidiasis. ERG11 mutations are known to be associated with azole resistance, and within azole-resistant strains, the Y132F substitution in ERG11 is the sole substitution observed in the literature [36]. Moreover, in a study on azole resistance mechanisms in C. parapsilosis by Grossman et al. [37], the sequences of ERG11 were examined and the Y132F substitution was found in 56.7% of their fluconazole-resistant isolates. They concluded that this mutation likely contributes significantly to fluconazole resistance in C. parapsilosis. Berkow et al. [38] also found this mutation, in addition to another substitution, R398I. The R398I substitution was present as the sole mutation in susceptible isolates, or alongside the Y132F substitution. Our findings are compatible with their data as R398I was detected in sensitive isolates S1 and S2, and the F132Y detected in resistant isolates R1, R2, R3, and R4. Furthermore, previous research showed that mutations in ERG3 and ERG6 are also associated with azole resistance, [39]. Stefanek et al. [40] observed two mutations S208G and S304G in the ERG6 gene involved in azole resistance; these two mutations are present in all our C. parapsilosis resistant isolates, but even in our sensitive isolates suggesting that other mechanisms might be responsible for resistance. One other mechanism of drug resistance is the upregulation of efflux pumps; these efflux pumps pump the drug out of the intracellular membrane. CDR1 (CPAR2_405290) is one of the major efflux pumps involved in fluconazole resistance; CDR1 serves as a versatile ATP-Binding cassette (ABC) efflux transporter, responsible for pumping out and expelling azoles from the cell [10]. This action decreases the intracellular drug accumulation, ultimately leading to the development of resistance. In our isolates, CDR1 was mutated in all the resistant isolates R1 (I1287V, I969T), R2 (N1132D), R3 (I1287V) and R4 (I1287V) possibly leading to drug resistance. Interestingly we did not observe a upregulation of ergosterol content in our isolates, which lends further credibility to the fact that pumping out the drug might be a more plausible mechanism of resistance in our isolates.
The formation of biofilm in microorganisms is regarded as both a characteristic of pathogenicity and a protective response to unfavorable conditions, such as fluctuations in pH, temperature, nutrient availability, or exposure to antifungal drugs. Candida accomplishes this by generating a dense network of extracellular polymeric substances, which acts as a protective barrier against external stresses and provides an environment conducive for growth and survival. This increased concentration of the matrix, compared to planktonic cells, offers greater protection against drugs and results in enhanced resistance. FAS2 (CPAR2_807400) was found to be essential for the proper development of biofilms with absence of FAS2 led to reduced virulence in a systemic mouse infection model [41]. CFEM (Common in Fungal Extracellular Membranes) proteins were associated with abnormal biofilm formation and decrease in virulence. Interestingly S1, S2, R1, R2, R3 and R4 all have the same T99A mutation in FAS2 and all have attenuated virulence. In addition, strains S1 and S2 have a mutation in CPAR2_407410 (MP65) which is known to be involved in biofilm formation; mutated forms of the gene are known to produce less biofilm [42,43]. Taff et al. [44] showed that azole susceptible strains produce less biofilm; S1 and S2 in our study produce less biofilm in accordance with the literature. We also examined EFG1, which is a gene associated with biofilm formation, and it turned out to be mutated, which might cause abnormal biofilm formation [45].
Lockhart et al. [46], reported that the combined prevalence of C. orthopsilosis and C. metapsilosis in C. parapsilosis complex infections is generally less than 10%. In our case, they account for 25% of our samples. However, it should be noted that one reason for this could be the low sample size in our study. Lockhart et al. [46] also noted that C. orthopsilosis and C. metapsilosis isolates tend to remain susceptible to fluconazole. In our study, we found that C. metapsilosis (Cm) and C, orthopsilosis (Co) exhibited susceptibility to fluconazole, with MIC values of 0.5 and 1, respectively, in accordance with their study. In addition to the susceptible C. metapsilosis and C. orthopsilosis isolates, C. parapsilosis resistant and sensitive isolates did not exhibit strong virulence in murine disseminated infections which is correlated to previous studies that show no correlation between drug resistance and virulence [9].
As far as orthopsilosis, we analyzed ERG11 and found multiple mutations such as Q211K, A421V, and V485I in isolates that retained susceptibility to fluconazole. This aligns with the findings of Xiang et al. [47]. Additionally, we observed mutations in other ERGs, but these mutations were not previously documented in the literature. In our study, we discovered mutations in the MDR1 and MRR1 efflux pumps in C. orthopsilosis (Co). However, we did not find any prior literature that specifically mentioned these mutations (supplementary table). Mutations in the CHS3 gene affect chitin deposition and virulence [48]. These mutations may be responsible for the observed reduction in virulence in Co strains with the following mutations.
Significant heterozygosity and diversity are prominent features of the genetic landscape of C. orthopsilosis and C. metapsilosis, which could have been influenced by hybridization events. The shortage of ATCC references, resulting in a lack of well-characterized strains for comparison, further complicates mutational analysis. However, our study confirms the high rate of diversity found in these isolates which is a hallmark of orthopsilosis and metapsilosis as they are the result of hybridization events which can be seen from the vast number of mutations documented in the literature[3,49], and observed in tables S2 and S3 in our isolates. It is widely thought that orthopsilosis is an ancestor to parapsilosis, justifying why there is more sequence diversity in orthopsilosis versus parapsilosis, that is a “newer” species and that has not yet had enough time to diverge significantly [3].
One interesting but unexpected aspect of our study is the rate of isolate-misidentification 50. All our isolates were labeled by our tertiary care center as C. parapsilosis [11]. However, based on our whole genome sequencing data a significant percentage of our isolates- 3 out of 9- were misidentified. For two of the isolates this misidentification might not have significant clinical implications since C. orthopsilosis and C. metapsilosis are part of the same family as parapsilosis and treatment is very similar for all three [48]. However, misidentification as C. albicans is more serious and can have adverse effects on treatment, and patient wellbeing. This stresses the need of using DNA sequencing data for proper microorganism identification.
Our study is based on 9 isolates, a relatively low sample size as far as statistical analysis is concerned. The reason for this is that the rate of C. parapsilosis hospital infection is low to begin with compared to C. albicans or bacterial pathogens for example. The fact that a third of them turned out to be non parapsilosis Candida affected our ability to determine statistically significant correlations between resistant and sensitive isolates.
An interesting result from our study is the high clonality and sequence similarity amongst our C. parapsilosis isolates. This can be seen in the phylogenetic tree analysis whereby all C. parapsilosis isolates cluster tightly together with a very low rate of substitutions amongst them as can be seen through the very short branch lengths that typically represent nucleotide substitutions per site. As opposed to the high rate of substitutions within orthopsilosis and metapsilosis isolates we observed a relatively low mutation rate and diversity within our parapsilosis isolates, similar to previous findings. The genes FAS2, MDR1, ALS6, ALS11, CPAR2_700020, SAP7, HWP1, UPA and UME6 had mutations which were exclusively found in resistant isolates and mutations in CPAR2_302400 and CPAR2_110220 found in the sensitive ones. This high rate of clonality is supported by our mutation analysis whereby similar mutations appear in multiple isolates. Utilizing mutation analysis to unveil the genetic basis of resistance represents a critical initial step in enhancing our understanding of Candida pathophysiology and in devising more potent strategies for combating drug resistance. Isolates with high sequence similarity and similar mutations isolated from the same ward at the same time, which is the case for our isolates, is an indication of a hospital outbreak. However, the lack of hospital data and patient history prevents us from further analysis and conclusions.
In conclusion, this is the first study of its kind to address C. parapsilosis complex isolates from both a phenotypic and genomic perspective. Our data has revealed variations in both the physical characteristics and genetic makeup of azole-resistant and azole-sensitive isolates. While our parapsilosis isolates displayed largely similar physical traits, the key distinction was observed in their ability to form biofilms, with the resistant isolates showing a higher propensity for enhanced biofilm formation. Our study also revealed hospital misidentification rates. Finally, it is well known that in addition to genomic mutations, drug resistance can be caused by upregulation of expression. Future work entailing a larger set of isolates with patient history, coupled with a transcriptomic approach would be of interest.
Figure 6. Figure 5: Cell Surface Disruption Assay.
Strains were 10X serially diluted and plated on media containing 0.025% SDS. Colony formation was photographed after 24h. (1) Shows the control plates with no SDS added. (2) Shows the growth of sensitive and resistant strains with the presence of SDS. R3 showed more resistance to SDS and more survival potential than the other resistant isolates, followed by R4 and R2; for the sensitive isolates, the S1 and S2 were susceptible to SDS, and their cell wall was being disrupted.
Table 2.
List of C. parapsilosis mutations
| Genes | Roles | isolates | Protein Mutations |
|---|---|---|---|
| FAS2; CPAR2_807400 | Virulence | S1, S2 | A328V; I330T; S1349N |
| Biofilm Formation | R1, R3, R4 | I330T; S1349N | |
| CPAR2_110220 | Resistance to SRS | S1 | P441T; R478K |
| Virulence | |||
| Resistance to caspofungin | |||
| Resistance to fluconazole | |||
| ERG11, CPAR2_303740 | Resistance to azole | S1, S2 | F132Y; R398I |
| Ergosterol | R1, R2, R3, R4 | F132Y | |
| FKS1, CPAR2_106400 | Resistance to caspofungin | S2 | A1422G; M1499I |
| Biofilm Formation | |||
| UPC2, CPAR2_207280 | Resistance to ketoconazole | R1 | N455R |
| CFEM4, CPAR2_402890 | Biofilm formation | S1, S2, R1, R3, R4 | T99A |
| Virulence | |||
| ATP6; CPAR2_203290 | S1, S2 | A120T | |
| R1 | T191A | ||
| CPAR2_207540 | Resistance to fluconazole | R3, R4 | I489V |
| Virulence | |||
| CDR1; CRAR2_405290 | Resistance to fluconazole | R1 | I1287V, I969T |
| R2 | N1132D | ||
| R3, R4 | I1287V | ||
| ERG4; CPAR2_502980 | Virulence | R1, R3 | P63S |
| Ergosterol | R3 | P63S | |
| ERG6; CPAR2_405010 | Biofilm formation | S1, S2, R1, R2, R3, R4 | S208G; S304G |
| Virulence | |||
| Ergosterol | |||
| UPC2; CPAR2_207280 | Resistance to ketoconazole | R1 | N455D |
| Ergosterol | |||
| STP4; CPAR2_211740 | Resistance to caspofungin | S1, S2, R1, R3, R4 | L195P |
| Resistance to SDS | |||
| CHS1; CPAR2_805640 | Virulence | R1 | P52L |
| MP65; CPAR2_407410 | Adhesion | S1, S2 | F369L |
| Biofilm formation | |||
| Virulence | |||
| SAPP3; CPAR2_102420 | Virulence increased | R1, R3, R4 | T359I |
| GCN4; CPAR2_806570 | Biofilm formation: decreased | S2 | G120D |
| NTC1; CPAR2_803760 | Resistance to caspofungin | S1, S2, R1, R3, R4 | L671Q |
| CHS3; CPAR2_801800 | Virulence | S1, S2, R1, R3, R4 | L38R |
| ALS6; CPAR2_404790 | Adhesion | R1 | S2190T |
| R3, R4 | I1430T, S2190T | ||
| ALS7; CPAR2_404800 | Virulence | R2, R3, R4 | E765D |
| ALS11; CPAR2_404780 | Virulence | S1, S2 | G94E, L350V |
| R1, R3, R4 | L350V, G1000R | ||
| CFEM5; CPAR2_300110 | Biofilm Abnormal | R1, R3 | S224T |
| R4 | G807S, P805L, A803T, G797S, L735P, T733A, G717S, P715L, S224T, P795L, A793T, A783T, S737G | ||
| CPAR2_700020 | S1, S2 | V171I | |
| CDRV CPAR2_700030 | Resistance to fluconazole | R1 | L591I |
| R3, R4 | D127Y | ||
| CPAR2_700100 | Virulence | S1, S2, R1,R3, R4 | F46I |
| CPAR2_700140 | Resistance to caspofungin | R1, R3, R4 | N140S |
| EFG1; CPAR2_701620 | Resistance to SDS | S1, S2, R1, R3, R4 | E452G |
| Resistance to caspofungin | |||
| Virulence | |||
| Biofilm Formation | |||
| SAP7; CPAR2_105640 | Virulence decreased | R1, R3, R4 | Q66H |
| SAP9; CPAR2_102610 | Virulence decreased | R1, R3, R4 | S439G |
| CPAR2_302400 | R3, R4 | I114M | |
| ERG25; CPAR2_801410 | Resistance to fluconazole | R3 | D9N |
| Ergosterol | |||
| HWP1; CPAR2_403520 | Biofilm formation | S1, S2 | L461S |
| Virulence | R1, R4 | G1129E, Q1128P, G1127R, E1126G, S1114G, L1113R P975S, S974G, L973P, L461S, P1115S | |
| R3 | G1129E, Q1128P G1127R, E1126G, L461S | ||
| EPA; FUN31; CPAR2_808450 | Resistance to caspofungin | R1, R3, R4 | D396G |
| EAP1; CPAR2_805000 | Resistance to SDS | R3 | T663S |
| UME6; CPAR2_803820 | Virulence | S1, S2 | C547G, T46A |
| R1, R3, R4 | C547G | ||
| FLO8; CPAR2_601080 | Biofilm formation | S1, S2, R1, R3, R4 | G1052S |
| Virulence |
Acknowledgements
The authors are thankful to Mr. Elias Abi Ramia and Mr. Charbel Yazbeck from the Animal Facility at the Lebanese American University, Byblos for valuable assistance. We are also grateful to the Department of Natural Sciences and LAU’s PIRF grant for funding.
Funding
This work was financially supported by the Department of Natural Sciences at the Lebanese American University, and by the PIRF intramural grant.
Funding Statement
This work was financially supported by the Department of Natural Sciences at the Lebanese American University, and by the PIRF intramural grant.
Footnotes
Competing Interests The authors do not have any pertinent financial or non-financial interests to declare.
Ethics approval
Ethical approval for sample collection from patients was obtained on April 26, 2016, by the Institutional Review Board of the Lebanese American University. All ethical standards of the Lebanese American University Institutional Care and Use Committee which approved the execution of this experiment in under approval code IRB#: LAU.SOM.RH1.26/Apr/2016. Written informed consent for participation was not required for this study in accordance with the national legislation and the institutional requirements.
Ethical approval for mice manipulation was obtained in June 2022 by the Institutional Review Board of the Lebanese American University under approval code: LAU.ACUC.SAS.RK5.12/June/2022
Supplementary Files
Contributor Information
Reine El Hady, Lebanese American University - Byblos Campus.
Nour Fattouh, Saint George University of Beirut.
Marc Finianos, Charles University: Univerzita Karlova.
Ibrahim Bitar, Charles University: Univerzita Karlova.
Rola Husni, Lebanese American University School of Medicine.
Roy khalaf, Lebanese American University - Byblos Campus.
References
- 1.Brown G. D., Denning D. W., Gow N. A. R., Levitz S. M., Netea M. G., & White T. C. (2012). Hidden killers: Human fungal infections. Science Translational Medicine, 4(165), 165rv13. 10.1126/scitranslmed.3004404 [DOI] [PubMed] [Google Scholar]
- 2.PFALLER M. A., MOET G. J., MESSER S. A., JONES R. N., & CASTANHEIRA M. (2011). Candida bloodstream infections: Comparison of species distributions and antifungal resistance patterns in community-onset and nosocomial isolates in the SENTRY antimicrobial surveillance program, 2008–2009. Antimicrobial Agents and Chemotherapy, 55(2), 561–566. 10.1128/AAC.01079-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tavanti A., Davidson A. D., Gow N. A. R., Maiden M. C. J., & Odds F. C. (2005). Candida orthopsilosis and candida metapsilosis spp. nov. to replace candida parapsilosis groups II and III. Journal of Clinical Microbiology, 43(1), 284–292. 10.1128/JCM.43.1.284-292.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gácser A., Trofa D., Schäfer W., & Nosanchuk J. D. (2007). Targeted gene deletion in candida parapsilosis demonstrates the role of secreted lipase in virulence. The Journal of Clinical Investigation, 117(10), 3049–3058. 10.1172/JCI32294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gomez-Lopez A., Alastruey-Izquierdo A., Rodriguez D., Almirante B., Pahissa A., Rodriguez-Tudela J. L., Cuenca-Estrella M., & Barcelona Candidemia Project Study Group. (2008). Prevalence and susceptibility profile of candida metapsilosis and candida orthopsilosis: Results from population-based surveillance of candidemia in spain. Antimicrobial Agents and Chemotherapy, 52(4), 1506–1509. 10.1128/AAC.01595-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toutounji M., Tokajian S., & Khalaf R. A. (2019). Genotypic and phenotypic characterization of C. albicans Lebanese hospital isolates resistant and sensitive to caspofungin. Fungal Genetics and Biology, 127, 12–22. [DOI] [PubMed] [Google Scholar]
- 7.Fattouh N., Hdayed D., Geukgeuzian G., Tokajian S., & Khalaf R. A. (2021). Molecular mechanism of fluconazole resistance and pathogenicity attributes of Lebanese C. albicans hospital isolates. Fungal Genetics and Biology, 153, 103575. [DOI] [PubMed] [Google Scholar]
- 8.Younes S. S., & Khalaf R. A. (2013). The candida albicans Hwp2p can complement the lack of fomentation of a saccharomyces cerevisiae flo11 null strain. Microbiology (Society for General Microbiology), 159(Pt 6), 1160. 10.1099/mic.0.067249-0 [DOI] [PubMed] [Google Scholar]
- 9.Khalaf R. A., Fattouh N., Medvecky M., & Hrabak J. (2021). Whole genome sequencing of a clinical drug resistant candida albicans isolate reveals known and novel mutations in genes involved in resistance acquisition mechanisms. Journal of Medical Microbiology, 70(4) 10.1099/JMM.0.001351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fattouh N., Khalaf R. A., & Husni R. (2024). Candida glabrata hospital isolate from Lebanon reveals micafungin resistance associated with increased chitin and resistance to a cell surface disrupting agent. Journal of global antimicrobial resistance, S2213–7165(24)00039–0. Advance online publication. 10.1016/jjgar.2024.02.012 [DOI] [PubMed] [Google Scholar]
- 11.Husni R., Bou Zerdan M., Samaha N., Helou M., Mahfouz Y., Saniour R., Hourani S., Kolanjian H., Afif C., Azar E., El Jisr T., Mokhbat J., Abboud E., Feghali R., Abboud E., Matta H., Karayakouboglo G., Matar M., Moghnieh R., & Daoud Z. (2023). Characterization and susceptibility of non-albicans candida isolated from various clinical specimens in lebanese hospitals. Frontiers in Public Health, 11, 1115055–1115055. 10.3389/fpubh.2023.1115055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Seemann T. (2015). Snippy: Fast Bacterial Variant Calling From NGS Reads [Internet]. San Francisco, CA: github. [Google Scholar]
- 13.Altschul S. F., Gish W., Miller W., Myers E. W., & Lipman D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(2), 403–410. doi: 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 14.Stanke M., Diekhans M., Baertsch R., and Haussler D. (2008). Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics 24, 637–644. doi: 10.1093/bioinformatics/btn013 [DOI] [PubMed] [Google Scholar]
- 15.Stanke M., Schoffmann O., Morgenstern B., and Waack S. (2006). Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinform. 7:62. doi: 10.1186/1471-2105-7-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ter-Hovhannisyan V., Lomsadze A., Chernoff Y. O., and Borodovsky M. (2008). Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res. 18, 1979–1990. doi: 10.1101/gr.081612.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., et al. (2009). BLAST+: architecture and applications. BMC Bioinform. 10:421. doi: 10.1186/1471-2105-10-421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoff K. J., Lange S., Lomsadze A., Borodovsky M., and Stanke M. (2016). BRAKER1: unsupervised RNA-seq-based genome annotation with GeneMarkET and AUGUSTUS. Bioinformatics 32, 767–769. doi: 10.1093/bioinformatics/btv661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoff K. J., Lomsadze A., Borodovsky M., and Stanke M. (2019). Whole-genome annotation with BRAKER. Methods Mol. Biol. 1962, 65–95. doi: 10.1007/978-1-4939-9173-0_5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruna T., Hoff K. J., Lomsadze A., Stanke M., and Borodovsky M. (2021). BRAKER2: automatic eukaryotic genome annotation with GeneMark-EP+ and AUGUSTUS supported by a protein database. NAR Genom. Bioinform. 3: lqaa108. doi: 10.1093/nargab/lqaa108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones P., Binns D., Chang H. Y., Fraser M., Li W., McAnulla C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blachowicz A., Chiang A. J., Elsaesser A., Kalkum M., Ehrenfreund P., Stajich J. E., et al. (2019). Proteomic and metabolomic characteristics of extremophilic fungi under simulated mars conditions. Front. Microbiol. 10:1013. doi: 10.3389/fmicb.2019.01013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vasquez-Gross H., Kaur S., Epstein L., and Dubcovsky J. (2020). A haplotype phased genome of wheat stripe rust pathogen Puccinia striiformis f. sp. tritici, race PST-130 from the Western USA. PLoS One 15: e0238611. doi: 10.1371/journal.pone.0238611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith C. A. (2021). Macrosynteny analysis between lentinula edodes and lentinula novae-zelandiae reveals signals of domestication in lentinula edodes. Sci. Rep. 11:9845. doi: 10.1038/s41598-021-89146-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huerta-Cepas J., Forslund K., Coelho L. P., Szklarczyk D., Jensen L. J., von Mering C., et al. (2017). Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 34, 2115–2122. doi: 10.1093/molbev/msx148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huerta-Cepas J., Szklarczyk D., Heller D., Hernandez-Plaza A., Forslund S. K., Cook H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky 1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buchfink B., Xie C., and Huson D. H. (2015). Fast and sensitive protein alignment using diamond. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176 [DOI] [PubMed] [Google Scholar]
- 28.Almagro Armenteros J. J., Tsirigos K. D., Sønderby C. K., Petersen T. N., Winther O., Brunak S., von Heijne G., & Nielsen H. (2019). SignalP 5.0 improves signal peptide predictions using deep neural networks. Nature biotechnology, 37(4), 420–423. 10.1038/s41587-019-0036-z [DOI] [PubMed] [Google Scholar]
- 29.Kraftova L., Finianos M., Studentova V., Chudejova K., Jakubu V., Zemlickova H., Papagiannitsis C. C., Bitar I., & Hrabak J. (2021). Evidence of an epidemic spread of KPC-producing Enterobacterales in Czech hospitals. Scientific reports, 11(1), 15732. 10.1038/s41598-021-95285-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reslan L., Araj G. F., Finianos M., El Asmar R., Hrabak J., Dbaibo G., & Bitar I. (2022). Molecular Characterization of Candida auris Isolates at a Major Tertiary Care Center in Lebanon. Frontiers in microbiology, 12, 770635. 10.3389/fmicb.2021.770635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silva-Dias A., Miranda I. M., Branco J., Monteiro-Soares M., Pina-Vaz C., & Rodrigues A. G. (2015). Adhesion, biofilm formation, cell surface hydrophobicity, and antifungal planktonic susceptibility: Relationship among C. spp. Frontiers in Microbiology, 6(205). 10.3389/fmicb.2015.00205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Arthington-Skaggs BA, Jradi H, Desai T, Morrison CJ. Quantitation of ergosterol content: Novel method for determination of fluconazole susceptibility of Candida albicans. J Clin Microbiol 1999; 37:3332–7. 10.1128/JCM.37.10.3332-3337.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Conti H. R., Huppler A. R., Whibley N., & Gaffen S. L. (2014). Animal models for candidiasis. Current protocols in immunology, 105(1), 19–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang N., Strugnell R., Wijburg O., Brodnicki T. (2011). Measuring Bacterial Load and Immune Responses in Mice Infected with Listeria monocytogenes. Journal of Visualized Experiments (JoVE), 54, e3076. doi: 10.3791/3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Y., Min F., Pan J., Wang J., Yuan W., Zhang Y., Huang R., & Zhang L. (2015). Systemic C. parapsilosis Infection Model in Immunosuppressed ICR Mice and Assessing the Antifungal Efficiency of Fluconazole. Veterinary Medicine International, 2015, 370641. 10.1155/2015/370641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Magobo R. E., Lockhart S. R., Govender N. P., Wadula J., Rensburg v., van Rensburg C. J., Whitelaw A., Zietsman I., Miller N., Smith P., van Greune J., Brink A., Hoosen A., Perovic O., Nchabaleng M., Orth H., Coovadia Y., Badenhorst L., Moolman J., … Patel J. (2020). Fluconazole-resistant candida parapsilosis strains with a Y132F substitution in the ERG11 gene causing invasive infections in a neonatal unit, south africa. Mycoses, 63(5), 471–477. 10.1111/myc.13070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grossman N. T., Pham C. D., Cleveland A. A., & Lockhart S. R. (2015). Molecular mechanisms of fluconazole resistance in candida parapsilosis isolates from a U.S. surveillance system. Antimicrobial Agents and Chemotherapy, 59(2), 1030–1037. 10.1128/AAC.04613-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Berkow E. L., Manigaba K., Parker J. E., Barker K. S., Kelly S. L., & Rogers P. D. (2015). Multidrug Transporters and Alterations in Sterol Biosynthesis Contribute to Azole Antifungal Resistance in C. parapsilosis. Antimicrobial agents and chemotherapy, 59(10), 5942–5950. 10.1128/AAC.01358-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.VANDEPUTTE P., TRONCHIN G., BERGES T., HENNEQUIN C., CHABASSE D., & BOUCHARA J. (2007). Reduced susceptibility to polyenes associated with a missense mutation in the ERG6 gene in a clinical isolate of candida glabrata with pseudohyphal growth. Antimicrobial Agents and Chemotherapy, 51(3), 982–990. 10.1128/AAC.01510-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Štefánek M., Wenner S., Borges V., Pinto M., Gomes J. P., Rodrigues J., Faria I., Pessanha M. A., Martins F., Sabino R., Veríssimo C., Nogueira I. D., Carvalho P. A., Bujdakova H., & Jordao L. (2022). Antimicrobial resistance and biofilms underlying catheter-related bloodstream coinfection by enterobacter cloacae complex and candida parapsilosis. Antibiotics (Basel), 11(9), 1245. 10.3390/antibiotics11091245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen L. N., Trofa D., & Nosanchuk J. D. (2009). Fatty acid synthase impacts the pathobiology of candida parapsilosis in vitro and during mammalian infection. PloS One, 4(12), e8421–e8421. 10.1371/journal.pone.0008421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sandini S., Stringaro A., Arancia S., Colone M., Mondello F., Murtas S., Girolamo A., Mastrangelo N., & De Bernardis F. (2011). The MP65 gene is required for cell wall integrity, adherence to epithelial cells and biofilm formation in candida albicans. BMC Microbiology, 11(1), 106–106. 10.1186/1471-2180-11-106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu Z., Huang T., Du M., Soteyome T., Lan H., Hong W., Peng F., Fu X., Peng G., Liu J., & Kjellerup B. V. (2022). Regulatory network controls microbial biofilm development, with candida albicans as a representative: From adhesion to dispersal. Bioengineered, 13(1), 253–267. 10.1080/21655979.2021.1996747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taff H. T., Mitchell K. F., Edward J. A., & Andes D. R. (2013). Mechanisms of candida biofilm drug resistance. Future Microbiology, 8(10), 1325–1337. 10.2217/fmb.13.101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramage G., VandeWalle K., López-Ribot J. L., & Wickes B. L. (2002). The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development in Candida albicans. FEMS microbiology letters, 214(1), 95–100. https://doi.org/10.1111Zj.1574-6968.2002.tb11330.x [DOI] [PubMed] [Google Scholar]
- 46.Lockhart S. R., Messer S. A., Pfaller M. A., & Diekema D. J. (2008). Geographic distribution and antifungal susceptibility of the newly described species candida orthopsilosis and candida metapsilosis in comparison to the closely related species candida parapsilosis. Journal of Clinical Microbiology, 46(8), 2659–2664. 10.1128/JCM.00803-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiang M., Liu J., Ni P., Wang S., Shi C., Wei B., Ni Y., & Ge H. (2013). Erg11 mutations associated with azole resistance in clinical isolates of C andida albicans. FEMS Yeast Research, 13(4), 386–393. 10.1111/1567-1364.12042 [DOI] [PubMed] [Google Scholar]
- 48.Bulawa C. E., Miller D. W., Henry L. K., & Becker J. M. (1995). Attenuated virulence of chitin-deficient mutants of candida albicans. Proceedings of the National Academy of Sciences - PNAS, 92(23), 10570–10574. 10.1073/pnas.92.23.10570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.O’Brien C. E., Zhai B., Ola M., Bergin S. A., Ó Cinneide E., O’Connor L, Rolling T., Miranda E., Babady N. E., Hohl T. M., & Butler G. (2022). Identification of a novel Candida metapsilosis isolate reveals multiple hybridization events. G3 (Bethesda, Md.), 12(1), jkab367. 10.1093/g3journal/jkab367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barada G., Basma R., & Khalaf R. A. (2008). Microsatellite DNA identification and genotyping of Candida albicans from Lebanese clinical isolates. Mycopathologia (1975), 165(3), 115–125. 10.1007/s11046-008-9089-0 [DOI] [PubMed] [Google Scholar]