Abstract
Short hydrophobic regions referred to as fusion peptide domains (FPDs) at or near the amino terminus of the membrane-anchoring subunit of viral glycoproteins are believed to insert into the host membrane during the initial stage of enveloped viral entry. Avian sarcoma and leukosis viruses (ASLV) are unusual among retroviruses in that the region in the envelope glycoprotein (EnvA) proposed to be the FPD is internal and contains a centrally located proline residue. To begin analyzing the function of this region of EnvA, 20 substitution mutations were introduced into the putative FPD. The mutant envelope glycoproteins were evaluated for effects on virion incorporation, receptor binding, and infection. Interestingly, most of the single-substitution mutations had little effect on any of these processes. In contrast, a bulky hydrophobic substitution for the central proline reduced viral titers 15-fold without affecting virion incorporation or receptor binding, whereas substitution of glycine for the proline had only a nominal effect on EnvA function. Similar to other viral FPDs, the putative ASLV FPD has been modeled as an amphipathic helix where most of the bulky hydrophobic residues form a patch on one face of the helix. A series of alanine insertion mutations designed to interrupt the hydrophobic patch on the helix had differential effects on infectivity, and the results of that analysis together with the results observed with the substitution mutations suggest no correlation between maintenance of the hydrophobic patch and glycoprotein function.
The glycoproteins of enveloped viruses facilitate entry into the target cell by binding to a cell surface receptor and mediating fusion of the viral and target cell membranes. The envelope glycoprotein of the subgroup A avian sarcoma and leukosis viruses (ASLV), EnvA, is synthesized as a precursor protein (Pr95) which oligomerizes into a trimer (14). In a process similar to that observed for other retroviruses, Pr95 is proteolytically processed into two subunits, SU (gp85) and TM (gp37), and this processing is required for infection (10). As for other retroviral glycoproteins, the SU subunit contains the determinants for receptor specificity and binding (5, 6, 11, 43), while the TM subunit anchors the glycoprotein to the viral membrane and is postulated to be responsible for membrane fusion.
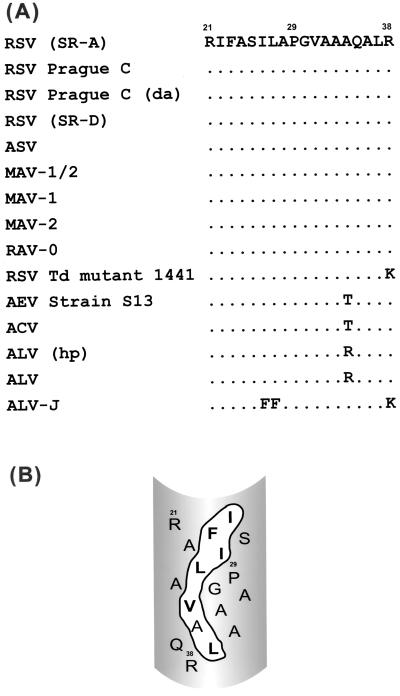
An early event in viral entry is, upon viral envelope activation, the insertion of a hydrophobic region of the viral envelope protein, the fusion peptide domain (FPD), into the cellular membrane (12, 23, 45). The FPD is found in the membrane-anchoring subunit and consists of 16 to 30 primarily hydrophobic amino acids displaying a hydrophobic index between those of a protein signal sequence and a transmembrane domain (49). FPDs are generally highly conserved within a viral family but not between families (24). Hydrophobic photoaffinity labeling of the influenza virus hemagglutinin (HA) FPD suggests that this domain forms an amphipathic helix during the fusion process, with the bulky hydrophobic residues exposed to the interior of the lipid bilayer (23). Infrared spectroscopy, circular dichroism, and electron spin resonance studies of peptides corresponding to the FPDs of human immunodeficiency virus type 1 (22), simian immunodeficiency virus (28, 35), Newcastle disease virus (7), and influenza virus (15, 30, 31, 33, 34, 40, 47) glycoproteins interacting with lipids indicate that these peptides conform to an amphipathic helix. Based on these and other studies, the ASLV FPD has been modeled as an amphipathic helix (Fig. 1B) (49). However, the requirement of an amphipathic helical conformation for FPDs remains controversial (19a), and there have been few studies directly testing this model in intact viral glycoproteins.
FIG. 1.
The putative FPD of ASLV. (A) Alignment of the putative FPD of ASLV. The viruses used for sequence alignment and their GenBank accession numbers (from top to bottom) are as follows: Rous sarcoma virus (RSV) Schmidt-Ruppin strain subgroup A [(SR-A)], P03397; RSV Prague strain subgroup C, P03396; RSV Prague strain subgroup C, duck adapted [(da)], S26419; RSV Schmidt-Ruppin strain subgroup D [(SR-D)], D10652; avian sarcoma virus, V01169; myeloblastosis-associated virus type 1/2 (MAV-1/2), L10923; MAV-1, L10922; MAV-2, L10924; Rous-associated virus type 0 (RAV-0), X07818; RSV Td mutant 1441, K00928; avian erythroblastosis virus (AEV) strain S13, A33902; avian carcinoma virus (ACV), M25158; avian leukosis virus (ALV) hypothetical protein [(hp)], S35437; ALV, S35427; ALV subgroup J (ALV-J), Z46390. (B) Model of the ASLV putative fusion peptide domain. The ASLV FPD has been modeled as an amphipathic helix with all of the bulky hydrophobic residues forming a patch on one face of the helix. The bulky hydrophobic residues are in boldface type, and the patch is outlined in black.
Although most retroviral FPDs are located at the amino terminus of the TM subunit, ASLV is unusual in that the putative FPD is located 20 residues internally to the amino terminus of the TM subunit. A feature common to a number of internal viral FPDs is a central proline which is believed to promote bending of the inserted FPD. Analysis of mutations to the central proline of the Semliki Forest virus E1 glycoprotein or the vesicular stomatitis virus G glycoprotein indicates a critical role for this residue in envelope glycoprotein function (18, 32, 51).
Here, we examine the effect of mutations in the putative internal FPD of ASLV EnvA. Twenty-six mutations within the ASLV EnvA FPD were generated and evaluated for virion incorporation into murine leukemia virus (MLV) pseudotypes, receptor binding, and infectivity. Our results indicate that the function of EnvA is refractory to single point mutations in the putative FPD and that the FPD centrally located proline is not required for infection. Moreover, the results of analysis of four insertion mutants designed to disrupt the register of the proposed FPD α-helix are not consistent with the amphipathic helix model for the ASLV putative FPD.
MATERIALS AND METHODS
Cell lines and plasmids.
293T cells and NIH 3T3 cells stably expressing the subgroup A ASLV receptor, Tva (3T3950) (43), were maintained in Dulbecco's modified Eagle's medium supplemented with 10% iron-supplemented calf serum, penicillin (100 U/ml), streptomycin (100 μg/ml), and 2 mM l-glutamine.
pjEnvA was constructed by ligating a 2.2-kb Schmidt-Ruppin EnvA BamHI fragment (20) into the expression vector pCB6 (8) that had been digested with BglII and BamHI. This construct eliminates the 5′ BamHI site while preserving the 3′ BamHI site. To construct pMyc-EnvA, oligonucleotides OS226 (5′-TCGGAGCAAAAGCTTATCTCCGAGGAAGATC-3′) and OS227 (5′-TCGAGATCTTCCTCGGAGATAAGCTTTTGC-3′), encoding the Myc epitope, were inserted at the unique XhoI site near the 5′ end of the EnvA coding region. The FPD mutations were generated by overlapping extension PCR (27), verified by DNA sequencing analysis, and inserted into pMyc-EnvA as a PpuMI-to-BamHI fragment. Other plasmids used include pCB6 EnvA Cleavage(−) [CL(−)] (21); pHit60, carrying MLV gag-pol; and pHit111, which expresses a retroviral genome that encodes β-galactosidase (β-gal) (44).
Production of EnvA pseudotype MLV and infections.
Myc-EnvA pseudotype virus was generated by an adaptation of the previously described transient three-plasmid retroviral expression system (44). Briefly, 15 μg each of pMyc-EnvA, pHit60, and pHit111 was transfected into 6 × 106 293T cells in a 100-mm plate overnight by CaPO4. The medium was changed the following morning, and 36 h posttransfection the virus-containing media was clarified by two-step centrifugation at 430 × g and then 2,300 × g. The resulting viral supernatants were divided into aliquots and stored at −80°C.
Infections were performed by incubating 3T3950 cells overnight with 1 ml of either FPD mutant or wild-type Myc-EnvA pseudotype MLV. The following morning, 2 ml of media was added. The infection was allowed to proceed for 36 h, after which the cells were fixed with 2% paraformaldehyde and the titers were determined by X-Gal (5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid) staining infected cells for β-gal activity (50). The data presented are results from a minimum of three independent experiments using at least two different viral stocks.
Analysis of FPD mutant expression.
Cellular expression of the Myc-EnvA FPD mutants was analyzed by the following procedure. 293T cell monolayers from the three-plasmid transfection described above were washed twice with 1× phosphate-buffered saline (PBS; 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.7 mM KH2PO4; adjusted to a pH of 7.4) and then lysed with Triton lysis buffer (150 mM NaCl, 1% Triton X-100, 50 mM Tris [pH 8.0], 5 mM EDTA) on ice. Cellular debris was pelleted, the supernatants were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (10% acrylamide) and transferred to nitrocellulose, and then Myc-EnvA was detected by Western blotting with anti-Myc ascites (9E10).
To evaluate cell surface expression of the FPD mutants, 293T cells were transiently transfected overnight by CaPO4 with 15 μg of the appropriate FPD mutant envelope plasmids in parallel with Myc-EnvA and CL(−) plasmids. Thirty-six hours posttransfection the 293T monolayers were washed twice with 1× PBS, and the cells were then released from the dish with 10 ml of PBS containing 1 mM EDTA and 1 mM EGTA. The cells were pelleted at 430 × g for 10 min and resuspended in 3 ml of biotin-labeling buffer (130 mM NaCl, 20 mM HEPES [pH 7.9], 0.5 mM MgCl2) containing 1 mg of Sulfo-NHS-LC-biotin (Pierce, Rockford, Ill.) per ml. The biotinylation reaction was allowed to proceed for 45 min on ice, and the reaction was quenched with bovine serum albumin (BSA; fraction V; Boehringer Mannheim Corp.) (final concentration, 0.5 mg/ml) in the presence of 100 mM glycine. The cells were pelleted at 430 × g, washed twice with biotin buffer containing 20 mM glycine, and then lysed with Triton lysis buffer. Protein concentrations in the cell lysate were determined by the Bradford assay, and equivalent amounts of total protein were streptavidin-agarose-precipitated as described previously (42). Precipitates were resolved by SDS-PAGE (10% acrylamide), transferred to nitrocellulose, and then detected by Western blotting with 9E10 ascites.
Analysis of EnvA incorporation into virions.
Seven milliliters of viral supernatant from the three-way transfection described above was centrifuged in an SW41 rotor at 41,000 rpm for 22 min. Pelleted virus was resuspended in 3 ml of PBS, layered onto 20% sucrose, and ultracentrifuged in an SW55 rotor at 55,000 rpm for 20 min. The resulting virus pellet was lysed in 250 μl of RIPA buffer (140 mM NaCl, 10 mM Tris [pH 8.0], 5 mM EDTA, 1% Na-deoxycholate, 1% Triton X-100, 0.1% SDS). Viral lysates were resolved by SDS-PAGE (10% acrylamide), transferred to nitrocellulose, and then probed for Myc-EnvA by Western blot analysis using anti-avian myoblastosis virus polyclonal rabbit sera provided by Tom Matthews (Duke University) or for MLV p30 with an anti-MLV Gag goat polyclonal sera. The primary antibody reactivates were detected with 125I-protein A (NEN Dupont) and quantitated using a PhosphorImager and ImageQuant software (Molecular Dynamics, Sunnyvale, Calif.). Env/Gag ratios were determined and normalized to the wild-type Myc-EnvA Env/Gag ratio.
Analysis of receptor binding.
Binding analysis of the FPD mutant proteins was performed by enzyme-linked immunosorbent assay (ELISA) as described previously (2). Briefly, 96-well plates were coated overnight at 4°C with an anti-EnvA TM polyclonal antibody that recognizes the cytoplasmic domain of the ASLV Env (20). Myc-EnvA or the FPD mutant proteins from cell lysates were captured for 1 h at 4°C. The volume of lysate used was titrated to be sufficient to saturate the antibody on the plate. After envelope binding, the plates were washed and biotinylated sTva was added, after which the plates were incubated for 1 h at 4°C. Unbound sTva was washed away, and the bound sTva was detected using streptavidin-horseradish peroxidase and 2,2′-azinobis(3-ethylbenzthiazoline sulfonic acid) (ABTS; Pierce). After incubation for 30 min at room temperature, plates were read by using an ELISA reader at 405 nm (Molecular Dynamics). All samples were assayed in triplicate. Data were converted to percentages (percent wild-type Myc-EnvA binding) using the following equation: optical density at 405 nm of mutant Myc-EnvA/optical density at 405 nm of wild-type Myc-EnvA × 100.
RESULTS
Generation of epitope-tagged EnvA.
To facilitate analysis of expression and virion incorporation of EnvA, sequences encoding a Myc-epitope tag (17) were inserted at the amino terminus of the EnvA SU subunit (Fig. 2A). We have demonstrated previously that appending sequences to the 5′ end of the envA coding region has no discernible effect on EnvA glycoprotein expression and function (43; P. Bates, unpublished observation). To assess the function of Myc-EnvA, plasmids encoding wild-type EnvA and Myc-EnvA, respectively, were transiently transfected into 293T cells along with plasmids carrying MLV gag-pol and an MLV genome encoding β-gal (44). Incorporation of Myc-EnvA into MLV particles was evaluated by Western blot analysis of viruses released into the media using antibodies specific for MLV Gag and ASLV Env. The function of Myc-EnvA was compared to that of wild-type EnvA by analyzing the infectivity of MLV pseudotypes on NIH 3T3 cells stably expressing the subgroup A ASLV receptor, Tva (3T3950). No difference in cellular expression, processing, virion incorporation, or infectivity was seen between Myc-EnvA and wild-type EnvA (data not shown).
FIG. 2.
The ASLV putative FPD mutants. (A) Schematic of ASLV Env. The SU and TM subunits are covalently linked by a disulfide bond. The different fill patterns indicate functional domains within each subunit, as follows: checkerboard, Myc-epitope tag; stippling, subgroup-determining region; wavy lines, FPD; diagonal lines, heptad repeat; windowpane, CX6CC motif; all black, membrane-spanning domain. The sequence of the putative FPD is shown below its region within TM. The first residue of TM is number 1. (B) ASLV FPD point mutations. (C) ASLV FPD proline mutations. (D) ASLV FPD insertion and deletion mutations. Dots denote homology and dashes indicate deletions.
Effect of point mutations within the ASLV FPD on infectivity, receptor binding, and virion incorporation.
An alignment of the ASLV TM sequences encoding the putative FPD shows a high degree of homology between all the ASLV envelopes. Variation is seen in only 4 of 17 residues comprising the putative FPD (Fig. 1A). One FPD variant substitutes Lys for Arg38, suggesting that conserving a basic residue is preferred at this position of the ASLV Env protein. Interestingly, Ala34, which is predicted by the amphipathic helix model to lie on the less hydrophobic face of the helix (Fig. 1B), tolerates polar and charged amino acid substitutions, indicating that there is not a strict hydrophobic requirement at this position. Similarly, the only other variant (avian leukosis virus subgroup J) substitutes bulky hydrophobic residues, but this does not affect the nature of the amphipathic helix.
To begin analyzing the sequence requirements of the ASLV FPD, we generated 16 site-directed mutations within the putative ASLV FPD (Fig. 2A). In general, these alterations were designed to conserve the overall hydrophobicity of this important functional domain. To directly address the amphipathic helix model for the ASLV putative FPD, we attempted to disrupt the proposed hydrophobic patch on the helical face (Fig. 1B) by replacement of the bulky hydrophobic residues Phe23, Ile26, Leu27, and Leu37 with either alanine or glycine. To test whether flexibility at the middle of the putative FPD was necessary, a centrally located glycine residue (Gly30) was replaced with the β-branched amino acid valine. Substitutions at position 35 of the ASLV TM were designed to test the significance of a polar residue in a primarily hydrophobic domain. Finally, the conserved carboxy-terminal basic residue was replaced with glycine to address the functional significance of a basic residue at this position (Fig. 2B).
The FPD point mutants were transiently expressed in 293T cells and subsequently labeled with a membrane-impermeable biotin compound which only labels cell surface proteins. SDS-PAGE and Western blotting of cell lysates revealed that the FPD mutants were all expressed and processed to levels similar to those of wild-type Myc-EnvA (Table 1). Similar analysis of streptavidin-agarose-precipitated proteins from cell lysates indicated that all the FPD point mutants are present on the cell surface at levels similar to those of wild-type Myc-EnvA (data not shown).
TABLE 1.
Summary of ASLV FPD mutants
| Envelope glycoproteina | Titer (105 IU/ml)b | Expressionc | Env/Gag ratiod | % Receptor bindingf |
|---|---|---|---|---|
| Controls | ||||
| Myc-EnvA | 4.3 ± 0.8 (100) | +++ | 1.00 | 100 |
| CL(−) | <10−5 (0) | +++ | 1.50 | 110 |
| No Env | <10−5 (0) | − | 0.03 | 0 |
| Point mutations | ||||
| F23A | 0.30 ± 0.06 (7) | +++ | 0.12 | 160 |
| F23W | 0.90 ± 0.61 (21) | +++ | 0.46 | 98 |
| I26A | 4.9 ± 1.4 (110) | +++ | 0.83 | ND |
| L27A | 2.2 ± 1.0 (51) | +++ | 0.61 | ND |
| L27T | 3.1 ± 1.0 (72) | +++ | 0.56 | ND |
| L27W | 5.7 ± 3.1 (130) | +++ | 0.46 | ND |
| I26W/L27W | 1.4 ± 0.3 (33) | +++ | 0.20 | 132 |
| G30P | 1.5 ± 0.1 (35) | +++ | 0.69 | ND |
| G30V | 3.6 ± 1.1 (85) | +++ | 0.64 | ND |
| Q35G | 2.0 ± 0.9 (47) | +++ | 0.64 | ND |
| Q35N | 2.8 ± 1.0 (65) | +++ | 1.05 | 120 |
| Q35R | 4.2 ± 2.3 (98) | +++ | 0.72 | 97 |
| Q35W | 2.5 ± 1.8 (58) | +++ | 0.84 | ND |
| A36G | 2.2 ± 0.7 (51) | +++ | 0.64 | ND |
| L37G | 5.5 ± 3.5 (130) | +++ | 0.77 | 190 |
| R38G | 4.4 ± 1.8 (100) | +++ | 0.80 | ND |
| Proline mutations | ||||
| P29G | 1.0 ± 0.0 (23) | +++ | 0.81 | 130 |
| P29V | 0.30 ± 0.06 (7) | +++ | 0.38 | 150 |
| P29G/G30P | 4.5 ± 0.6 (100) | +++ | 0.83 | ND |
| Insertion and deletion mutations | ||||
| Δ24-26 | <10−5 (0) | +++ | 0.06e | 61 |
| L27T, ΔI26/ΔP29 | −10−5 (0) | +++ | 0.09e | 89 |
| A24[A]S25 | 0.60 ± 0.08 (15) | +++ | 0.75 | 170 |
| P29[A]G30 | 1.9 ± 0.5 (44) | +++ | 1.10 | 110 |
| A34[A]Q35 | 0.03 ± 0.08 (0.6) | +++ | 1.10 | 94 |
| L37[A]R38 | 0.44 ± 0.19 (10) | +++ | 0.96 | 130 |
Envelope glycoproteins in bold are defective for infectivity.
Values in parentheses are percentages [percent wild-type, determined by the following equation: (mutant titer/Myc-EnvA titer) × 100].
Mutant Env expression compared to Myc-EnvA expression in cell lysates, as determined by Western blotting. +++, ≥50% of wild-type levels; −, no Env detected.
Ratios determined by phosphorimage analysis of Western blots of virions for Env and Gag and normalized to Myc-EnvA.
Env-Gag ratios determined for processed envelope glycoprotein.
Determined by ELISA (2). Data normalized to Myc-EnvA binding to sTva. ND, not done.
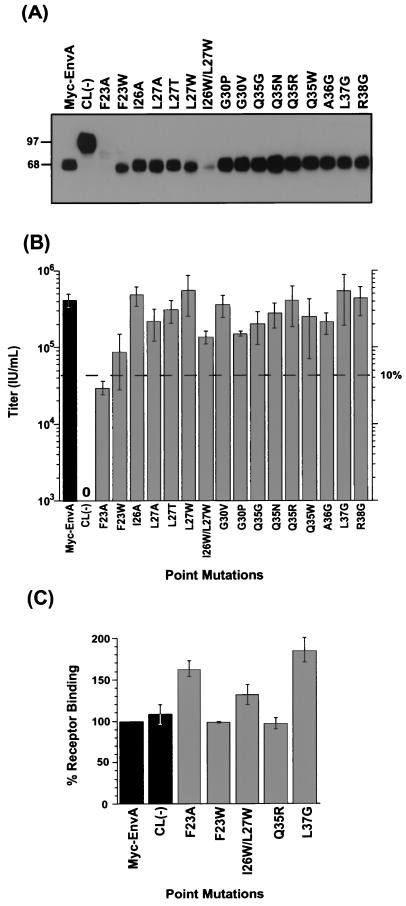
The Myc-EnvA mutants were used to produce MLV pseudotypes carrying a β-gal marker gene by transient transfection of 293T cells. To determine if the FPD mutants were efficiently incorporated into MLV virions, pseudotyped viral preparations were purified through sucrose and analyzed by SDS-PAGE and Western blotting, and Env/Gag ratios were determined by phosphorimage analysis. As shown in Fig. 3A and Table 1, most of the FPD point mutants are efficiently incorporated into virions. Two notable exceptions are F23A and I26W/L27W, a double mutation. These two mutants are, respectively, incorporated 10- and 5-fold less efficiently than wild-type Myc-EnvA. Moreover, both F23A and, to a lesser extent, F23W are incompletely processed.
FIG. 3.
Effects of ASLV fusion peptide domain point mutations on virion incorporation, infectivity, and receptor binding. (A) Analysis of mutant ASLV envelope glycoprotein incorporation into MLV virions. MLV pseudotyped with ASLV FPD point mutant envelope glycoproteins was pelleted through 20% sucrose and lysed in RIPA buffer. Viral lysates were separated by SDS-PAGE and analyzed by Western blot analysis with 9E10 anti-Myc antibody (17). SU migrates at 68 kDa, and uncleaved Env migrates at 97 kDa. (B) Effects of FPD point mutations on viral infectivity. MLV pseudotyped viral stocks incorporating the ASLV FPD point mutant envelope glycoproteins and containing a retroviral genome encoding β-gal were used to infect NIH 3T3 cells stably expressing Tva (3T3950). Forty-eight hours postinfection, the cells were fixed and stained for β-gal expression. Viral titers were determined by enumeration of β-gal-positive cells. The results shown are the averages of at least three independent experiments, with standard deviations (error bars). Dashed line indicates the titer that is 10% of the wild-type titer. Glycoproteins displaying activity below this level are defined as significantly impaired. Myc-EnvA and CL(−) are positive and negative controls, respectively. (C) Receptor binding activity of FPD point mutant Env proteins. Mutant and wild-type envelope glycoproteins were captured by an anti-TM antibody onto a 96-well plate, and then receptor binding was determined by ELISA (2). For panels B and C, bar shading indicates whether results are for control Env glycoproteins (black bars) or FPD mutant proteins (gray bars).
The effect of the FPD mutations on glycoprotein function was assessed using a single-cycle infectivity assay (44). Viral titers were determined by enumeration of cells expressing β-gal. The results shown in Fig. 3B are the averages of at least three independent experiments using at least two different viral stocks. The uncleaved ASLV envelope [CL(−)] serves as a negative control, since it has previously been demonstrated to yield noninfectious viruses (10).
As shown in Fig. 3B, 15 of the 16 point mutations in the ASLV FPD had little or no effect on glycoprotein function, as measured by viral infectivity. Given the strong conservation seen in the ASLV FPD, this result is somewhat surprising. One exception is F23A (Fig. 3B and Table 1), where viral infection was impaired nearly 15-fold compared to that of wild-type Myc-EnvA. A more conservative substitution at this position, F23W, modestly reduced titers by approximately fivefold compared to wild-type Myc-EnvA titers.
We next determined if changes in the FPD affect the ability of Myc-EnvA to bind to Tva, the receptor for subgroup A ASLV, using an ELISA-based binding assay (3). Analysis of receptor binding by three FPD mutants that display wild-type infection demonstrates that these envelope proteins retain full receptor binding activity (Fig. 3C). The uncleaved EnvA [CL(−)], which is defective for infection, binds receptor as well as Myc-EnvA, demonstrating that cleavage activation is not required for receptor binding. The two FPD point mutants with decreased infectivity, F23A and F23W, also displayed full receptor binding activity. Thus, these alterations to the FPD in the TM subunit had no effect on the receptor binding domain in the SU subunit, which suggests that these two mutant envelope glycoproteins are not grossly misfolded.
The F23A mutation had a significant (10-fold or greater) effect on viral infectivity; however, this mutation also substantially reduced incorporation into MLV, virions complicating interpretation of this mutant. Comparison to the I26W/L27W mutant, which also displays reduced incorporation but maintains wild-type infectivity, supports the hypothesis that alteration of residue 23 impairs envelope function. Additionally, the F23W mutant was similar to wild-type EnvA for virion incorporation, perhaps suggesting that the modest reduction in infectivity observed with this mutant results from the conserved substitution of Trp for Phe at residue 23 affecting membrane fusion.
Effect of proline mutations on virion incorporation, infectivity, and receptor binding.
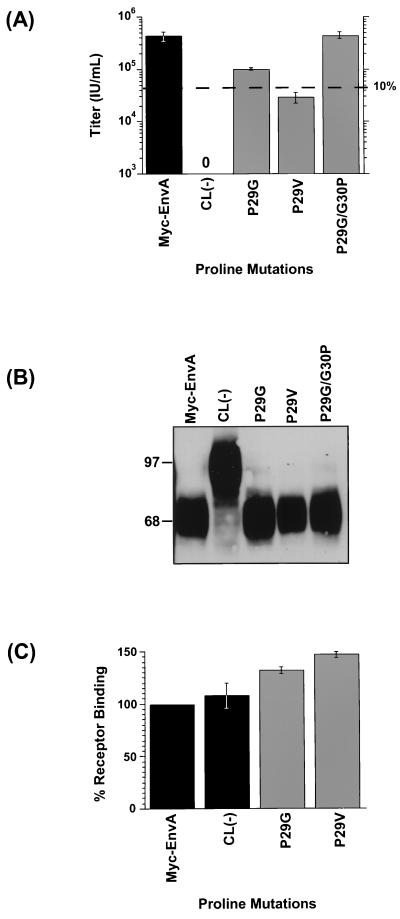
Since previous data in other viral systems has shown the functional importance of a proline near the middle of an internal FPD, we investigated the role of the centrally located proline in the putative ASLV FPD. Proline causes a bend, or kink, in the polypeptide backbone; therefore, we introduced two mutations to test if flexibility in the middle of the putative FPD is required for envelope function (Fig. 2C). A Pro-to-Gly mutation at position 29 (P29G) that should still allow nonrestricted bending of the domain at this position was introduced as was a valine substitution (P29V) that would be predicted to constrain this region from bending because of the bulky β-branched side chain of this residue. A Pro29 and Gly30 swap (P29G/G30P) was also generated to evaluate if the exact placement of the proline is critical for ASLV envelope function.
Transient transfection of 293T cells was used to produce MLV pseudotypes with each of the Myc-EnvA proline mutants, and glycoprotein function was evaluated using the single-cycle infection assay. Substitution for proline by valine (P29V) had a significant effect on ASLV envelope function, reducing titers by approximately 15-fold in replicate experiments, compared to the wild-type Myc-EnvA titer (Fig. 4A and Table 1), whereas a Gly substitution at this position (P29G) had a modest effect on infectivity, reducing titers approximately fourfold compared to the wild-type titer. Changing the relative position of the central proline (P29G/G30P) did not affect infection, suggesting that there is not a stringent spatial requirement that a Pro be at position 29.
FIG. 4.
Effects of ASLV FPD proline mutations on infectivity, virion incorporation, and receptor binding. (A) Effects of FPD proline mutations on viral infectivity. Titers were determined as described in the legend to Fig. 3B. The results shown are the averages of at least three independent experiments, with standard deviations (error bars). Dashed line indicates the titer that is 10% of wild type. (B) Analysis of mutant ASLV envelope glycoprotein incorporation into MLV virions. MLV virions pseudotyped with ASLV FPD proline mutant envelope glycoproteins were analyzed as described in the legend to Fig. 3A. (C) Receptor binding activity of FPD proline mutant glycoproteins. An ELISA-based assay was performed as described in the legend to Fig. 3C. For panels A and C, bar shading indicates whether results are for control envelope glycoproteins (black bars) or FPD mutant proteins (gray bars).
Western blot analysis of MLV pseudotyped virions purified by centrifugation through 20% sucrose was used to examine the incorporation of the three proline mutants. Similar to wild-type Myc-EnvA, the proline mutants appear to be correctly processed and incorporated into virions (Fig. 4B and Table 1), suggesting that the phenotypes observed for P29V and P29G are not due to reduced EnvA incorporation into MLV [note some proteolytic degradation observed in the CL(−) sample].
To determine if the alterations to Pro29 had an effect on receptor binding, the ELISA-based receptor binding assay was employed. As illustrated in Fig. 4C and Table 1, mutations P29V and P29G had no effect on the ability of these envelope proteins to bind receptor compared to wild-type Myc-EnvA binding ability. This finding, coupled with the efficient incorporation of these mutant proteins into virions, indicates that the effect of the substitutions at Pro29 on viral entry is postreceptor binding and suggests that alteration of the central region of the ASLV FPD affects membrane fusion.
Effect of insertion and deletion mutations on virion incorporation, infectivity, and receptor binding.
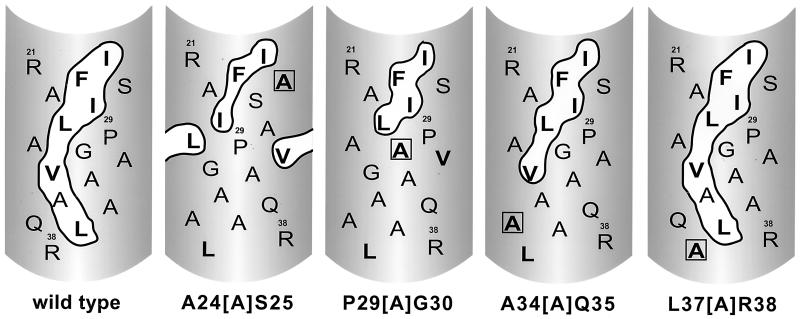
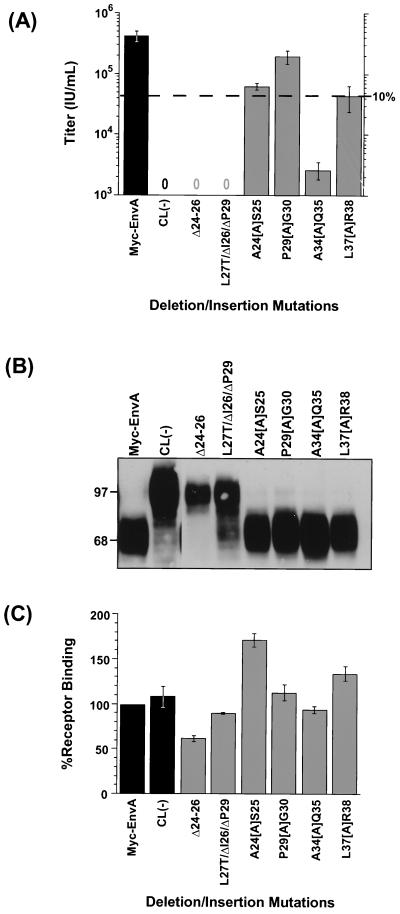
An amphipathic helix model has been proposed for the putative ASLV FPD. However, for the ASLV Env and many other viral glycoproteins, this model has not been rigorously tested. To test whether the amphipathic helix model appropriately describes the subgroup A ASLV FPD, we generated a series of insertion and deletion mutations. Two deletion mutants removing one or two bulky hydrophobic residues within the proposed hydrophobic patch were generated (Fig. 2D, Δ24-26 and L27T/ΔI26/ΔP29). Four alanine insertion mutants that should alter the register of the proposed amphipathic helix, thereby affecting the size of the hypothetical hydrophobic patch, were also created (Fig. 2D and see Fig. 6).
FIG. 6.
Potential effect of ASLV FPD alanine insertion mutations on the proposed bulky hydrophobic patch. Helical net representation of wild-type and ASLV FPD alanine insertion mutations. Proposed bulky hydrophobic patch is encircled. Bulky hydrophobic residues are in boldface type. Inserted alanine residue is boxed.
The mutant envelope glycoproteins were pseudotyped into MLV particles by transient transfection of 293T cells, and glycoprotein function was analyzed using the single-cycle infection assay. Neither of the deletion mutations yielded infectious virus. In contrast, the alanine insertion mutations had varying effects on infectivity, with insertions near the carboxy terminus showing a trend toward greater decreases in infectivity. The insertion mutants near the amino terminus were slightly less severe or had no effect on viral infection. A24[A]S25 yielded virus with titers reduced approximately sevenfold, while P29[A]G30 was comparable to the wild type for infection. The A34[A]Q35 mutation resulted in an approximately 170-fold reduction in the viral titers, while the L37[A]R38 mutation produced virions with 10-fold lower infectivity.
Analysis of envelope glycoproteins from lysates of transiently expressing cells demonstrated that all of the insertion and deletion Myc-EnvA FPD mutants were competent to bind receptor (Fig. 5C and Table 1). However, Western blot analysis of virions indicated that mostly unprocessed Δ24-26 and L27T/ΔI26/ΔP29 glycoproteins were incorporated into MLV virions [note some proteolytic degradation observed in the CL(−) and L27T/ΔI26/ΔP29 samples] (Fig. 5B and Table 1), suggesting that the nonfunctional phenotype displayed by these two envelope proteins is due to inefficient processing of the precursor envelope into SU and TM subunits prior to incorporation into viruses. In contrast, all the insertion mutant envelope proteins are efficiently processed and incorporated into MLV particles at levels similar to those of wild-type Myc-EnvA (Fig. 5B and Table 1). Taken together, these results demonstrate that the effect on glycoprotein function displayed by the insertion mutants A24[A]S25, A34[A]Q35, and L37[A]R38 occurs post-receptor binding and is consistent with these FPD mutants directly affecting membrane fusion.
FIG. 5.
Effects of ASLV FPD insertion and deletion mutations on infectivity, virion incorporation, and receptor binding. (A) Effects of FPD insertion and deletion mutations on viral infectivity. Titers were determined as described in the legend to Fig. 3B. The results shown are the averages of at least three independent experiments, with standard deviations (error bars). Dashed line indicates the titer that is 10% of wild type. (B) Analysis of mutant ASLV envelope glycoprotein incorporation into MLV virions. MLV virions pseudotyped with ASLV FPD insertion and deletion mutant proteins were analyzed as described in the legend to Fig. 3A. (C) Receptor binding activity of FPD insertion and deletion mutant glycoproteins. An ELISA-based assay was performed as described in the legend to Fig. 3C. For panels A and C, bar shading indicates whether results are for control envelope glycoproteins (black bars) or FPD mutant proteins (gray bars).
DISCUSSION
Residues 21 to 38 located internally in the TM subunit of the ASLV envelope glycoprotein are believed to encode the fusion peptide. Several lines of evidence suggest definition of this region as the putative FPD of the ASLV Env. First, hydropathy plot analysis of the TM subunit of ASLV implicates this region to be the FPD and this domain is highly conserved among ASLV, as is generally found for FPDs (Fig. 1). Importantly, the proposed FPD is juxtaposed to the heptad repeat region, as has been observed for a number of other viral FPDs (9, 19). In addition, this region shares characteristics with other putative internal FPDs (49). Finally, recent mutational analysis demonstrated that alteration of Val31 to Glu within the putative FPD of a soluble form of EnvA impairs receptor-induced liposome binding, believed to represent the initial step of glycoprotein-mediated membrane fusion (25).
Mutational analysis of envelope glycoproteins has been instrumental in the identification and functional characterization of a number of viral FPDs. To define the role of the putative ASLV FPD in viral infection, an extensive number of conservative point mutations and deletion and insertion mutations were introduced into the ASLV EnvA FPD, and the mutant FPDs assayed for infectivity, receptor binding activity, and incorporation into MLV virions. Of the 22 mutants that were processed and incorporated efficiently into MLV virions, only one point mutation and two insertion mutations significantly affected ASLV envelope function, decreasing titers by 10-fold or more. None of these three mutations decreased receptor binding, supporting the idea that they directly affect membrane fusion mediated by the ASLV envelope glycoprotein.
An interesting finding of this study is that most of the point mutations introduced into the ASLV FPD had little effect on viral entry. This is in sharp contrast to mutational analysis of other viral FPDs. Conservative alterations to the putative FPDs of vesicular stomatitis virus G (18, 51), Semliki Forest virus E1 (32), and influenza virus HA (46) glycoproteins resulted in a decrease in fusion activity. Conversely, individual Gly-to-Ala mutations in the simian virus 5 F protein putative FPD increased the fusogenic activity of this glycoprotein (29) by increasing the kinetics of fusion (1). One possibility for the lack of ASLV EnvA mutant phenotype may be that the single-cycle infection assay is not sensitive enough to detect subtle differences in infectivity between FPD mutant Myc-EnvA glycoproteins. However, using a similar assay to analyze the Moloney MLV putative FPD, numerous infection defective mutants, including glycoproteins containing conservative substitutions, were detected (52), indicating that a single-cycle infection assay is suitable for this type of analysis. In addition, Hernandez and White showed that nonconservative substitutions to the subtype A ASLV putative FPD significantly impaired glycoprotein function, using an infectivity assay nearly identical to the one employed here (26).
Our observation that numerous single point mutations within the putative FPD have little effect on ASLV EnvA function may suggest that no individual amino acid in this region is critical for EnvA membrane fusion activity. A centrally located proline in the FPD is a common feature of internal viral FPDs (49) and may allow a bend, or kink, in the proposed amphipathic helix. Previous mutational analysis showed that alterations to the centrally located proline of the Semliki Forest virus FPD in the E1 subunit resulted in retention of this subunit in the rough endoplasmic reticulum (32). Furthermore, when Pro127 in the vesicular stomatitis virus G protein putative FPD was change to Asp, Leu, or Gly, cell-cell fusion was dramatically affected (18, 51). These results may imply a requirement for this residue in internal FPDs. Similarly, our data demonstrate that altering the central proline in the ASLV FPD to Val (P29V) impaired viral infectivity. In contrast, a mutant in which the ASLV FPD Pro29 was changed to Gly (P29G) retained 25% of wild-type envelope function. One hypothesis to explain this observation is that the functional requirement in this region is not specific for proline but rather for an ability to bend or flex near the center of the internal FPD. Thus, the smaller glycine residue is tolerated in place of the proline residue while a bulky valine residue, which should impede bending, is not tolerated. This hypothesis is supported by the observation that three additional mutants in this region, P29G/G30P, P29[A]G30, and G30P, all of which should support bending in the middle of the FPD (Fig. 2), have infectious titers similar to that of wild-type EnvA.
Peptide studies employing the FPDs of several viral glycoproteins have yielded significant information on the structure of amino-terminal FPDs in a lipid environment. These studies demonstrated that for human immunodeficiency virus type 1, simian immunodeficiency virus, influenza virus, and Newcastle disease virus, peptides corresponding to the FPD adopt an amphipathic helix conformation (7, 22, 31) and insert into the lipid bilayer at an oblique orientation (7, 22, 28, 30, 33, 48). In many cases, these peptides cause liposome leakage (13, 15, 37, 39, 40) as well as liposome fusion (13, 16, 35, 36, 41), suggesting that they are biologically active. These results, in addition to photoaffinity labeling studies of the influenza virus HA FPD (23), have led to generalization of the amphipathic helix model to other viral fusion peptides including ASLV (49). In order to determine if this model aptly describes the structure of the putative ASLV FPD during membrane fusion, insertion mutants that would affect the size of the proposed hydrophobic patch were produced and analyzed. The results suggest that, for ASLV, this region does not conform to an amphipathic helix. Indeed, for the ASLV mutants analyzed, there is no correlation between the size of the hydrophobic patch and infectivity (Fig. 6; Table 1). Mutants A24[A]S25 and P29[A]G30, both of which disrupt most of the proposed hydrophobic patch, have a modest effect and no effect on viral titer, respectively. This is in sharp contrast to the severely impaired mutant, A34[A]Q35, which retains most of the hydrophobic patch. The observation that the insertion mutations near the carboxy terminus of the putative ASLV FPD have a more dramatic effect on viral infectivity may suggest that the orientation of the FPD with respect to the downstream heptad repeat domain must be preserved. Alternatively, during the fusion process, the FPD may need to interact with other regions of Env. By inserting an alanine residue at or near the carboxy terminus, the register of the FPD may be altered such that intrasubunit interactions required in steps of the membrane fusion process other than FPD insertion are impeded.
It seems likely that internal FPDs may have functional requirements distinct from those of the more thoroughly studied amino-terminal FPDs. Internal FPDs may not adopt an amphipathic helix conformation due to constraints asserted by the location of this domain within the membrane-anchoring subunit. While the internal FPD of PH-30, the sperm fusion protein, has been modeled as an amphipathic helix (4), Muga et al. have demonstrated that a peptide corresponding to this domain does not have α-helical properties when inserted into a membrane (38). The studies presented here suggest that the ASLV FPD may also not be an amphipathic α-helix.
The observation that the mutants F23W, P29V, A24[A]S25, A34[A]Q35, and L37[A]R38 diminish viral entry without affecting receptor binding is consistent with the hypothesis that these mutations perturb membrane fusion. However, additional experiments analyzing receptor-induced structural rearrangements, receptor-triggered liposome binding, and development of a quantitative cell-cell fusion assay will be required to more precisely define the mechanistic effects of these mutations on glycoprotein-mediated membrane fusion.
ACKNOWLEDGMENTS
We thank Yasamin Mir-Shekari and Carrie Balliet for review of the manuscript and Joseph Rucker, Rouven Wool-Lewis, Lijun Rong, and members of the Bates laboratory for useful discussions. We acknowledge the generosity of Tom Matthews, who provided rabbit polyclonal anti-AMV sera.
This work was supported by grants to P.B. from the National Institutes of Health (CA63531 and CA76256) and the American Heart Association (95015200). J.W.B. is a trainee of grant T32-AI-07325 from the National Institutes of Health.
REFERENCES
- 1.Bagai S, Lamb R A. A glycine to alanine substitution in the paramyxovirus SV5 fusion peptide increases the initial rate of fusion. Virology. 1997;283:283–290. doi: 10.1006/viro.1997.8858. [DOI] [PubMed] [Google Scholar]
- 2.Balliet J W, Berson J, D'Cruz C M, Huang J, Crane J, Gilbert J M, Bates P. Production and characterization of a soluble, active form of Tva, the subgroup A avian sarcoma and leukosis virus receptor. J Virol. 1999;73:3054–3061. doi: 10.1128/jvi.73.4.3054-3061.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bates P, Young J A, Varmus H E. A receptor for subgroup A Rous sarcoma virus is related to the low density lipoprotein receptor. Cell. 1993;74:1043–1051. doi: 10.1016/0092-8674(93)90726-7. [DOI] [PubMed] [Google Scholar]
- 4.Blobel C P, Wolfsberg T G, Turck C W, Myles D G, Primakoff P, White J M. A potential fusion peptide and an integrin ligand domain in a protein active in sperm-egg fusion. Nature. 1992;356:248–252. doi: 10.1038/356248a0. [DOI] [PubMed] [Google Scholar]
- 5.Bova C A, Manfredi J P, Swanstrom R. env genes of avian retroviruses: nucleotide sequence and molecular recombinants define host range determinants. Virology. 1986;152:343–354. doi: 10.1016/0042-6822(86)90137-6. [DOI] [PubMed] [Google Scholar]
- 6.Bova C A, Olsen J C, Swanstrom R. The avian retrovirus env gene family: molecular analysis of host range and antigenic variants. J Virol. 1988;62:75–83. doi: 10.1128/jvi.62.1.75-83.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brasseur R, Lorge P, Goormaghtigh E, Ruysschaert J M, Espion D, Burny A. The mode of insertion of the paramyxovirus F1 N-terminus into lipid matrix, an initial step in host cell/virus fusion. Virus Genes. 1988;1:325–332. doi: 10.1007/BF00257096. [DOI] [PubMed] [Google Scholar]
- 8.Brewer C B. Cytomegalovirus plasmid vectors for permanent lines of polarized epithelial cells. Methods Cell Biol. 1994;43:233–245. doi: 10.1016/s0091-679x(08)60606-8. [DOI] [PubMed] [Google Scholar]
- 9.Chambers P, Pringle C R, Easton A J. Heptad repeat sequences are located adjacent to hydrophobic regions in several types of virus fusion glycoproteins. J Gen Virol. 1990;71:3075–3080. doi: 10.1099/0022-1317-71-12-3075. [DOI] [PubMed] [Google Scholar]
- 10.Dong J Y, Dubay J W, Perez L G, Hunter E. Mutations within the proteolytic cleavage site of the Rous sarcoma virus glycoprotein define a requirement for dibasic residues for intracellular cleavage. J Virol. 1992;66:865–874. doi: 10.1128/jvi.66.2.865-874.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dorner A J, Coffin J M. Determinants for receptor interaction and cell killing on the avian retrovirus glycoprotein gp85. Cell. 1986;45:365–374. doi: 10.1016/0092-8674(86)90322-3. [DOI] [PubMed] [Google Scholar]
- 12.Durrer P, Galli C, Hoenke S, Corti C, Gluck R, Vorherr T, Brunner J. H+-induced membrane insertion of influenza virus hemagglutinin involves the HA2 amino-terminal fusion peptide but not the coiled coil region. J Biol Chem. 1996;271:13417–13421. doi: 10.1074/jbc.271.23.13417. [DOI] [PubMed] [Google Scholar]
- 13.Duzgunes N, Shavnin S A. Membrane destabilization by N-terminal peptides of viral envelope proteins. J Membr Biol. 1992;128:71–80. doi: 10.1007/BF00231872. [DOI] [PubMed] [Google Scholar]
- 14.Einfeld D, Hunter E. Oligomeric structure of a prototype retrovirus glycoprotein. Proc Natl Acad Sci USA. 1988;85:8688–8692. doi: 10.1073/pnas.85.22.8688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Epand R M, Cheetham J J, Epand R F, Yeagle P L, Richardson C D, Rockwell A, Degrado W F. Peptide models for the membrane destabilizing actions of viral fusion proteins. Biopolymers. 1992;32:309–314. doi: 10.1002/bip.360320403. [DOI] [PubMed] [Google Scholar]
- 16.Epand R M, Epand R F. Relationship between the infectivity of influenza virus and the ability of its fusion peptide to perturb bilayers. Biochem Biophys Res Commun. 1994;202:1420–1425. doi: 10.1006/bbrc.1994.2089. [DOI] [PubMed] [Google Scholar]
- 17.Evan G I, Lewis G K, Ramsay G, Bishop J M. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fredericksen B L, Whitt M A. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J Virol. 1995;69:1435–1443. doi: 10.1128/jvi.69.3.1435-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gallaher W R, Ball J M, Garry R F, Griffin M C, Montelaro R C. A general model for the transmembrane proteins of HIV and other retroviruses. AIDS Res Human Retrovir. 1989;5:431–440. doi: 10.1089/aid.1989.5.431. [DOI] [PubMed] [Google Scholar]
- 19a.Gallaher W R, Segrest J P, Hunter E. Are fusion peptides really “sided” insertional helices? Cell. 1992;70:531–532. doi: 10.1016/0092-8674(92)90423-a. [DOI] [PubMed] [Google Scholar]
- 20.Gilbert J M, Bates P, Varmus H E, White J M. The receptor for the subgroup A avian leukosis-sarcoma viruses binds to subgroup A but not to subgroup C envelope glycoprotein. J Virol. 1994;68:5623–5628. doi: 10.1128/jvi.68.9.5623-5628.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert J M, Hernandez L D, Balliet J W, Bates P, White J M. Receptor-induced conformational changes in the subgroup A avian leukosis and sarcoma virus envelope glycoprotein. J Virol. 1995;69:7410–7415. doi: 10.1128/jvi.69.12.7410-7415.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gordon L M, Curtain C C, Zhong Y C, Kirkpatrick A, Mobley P W, Waring A J. The amino-terminal peptide of HIV-1 glycoprotein 41 interacts with human erythrocyte membranes: peptide conformation, orientation and aggregation. Biochim Biophys Acta. 1992;1139:257–274. doi: 10.1016/0925-4439(92)90099-9. [DOI] [PubMed] [Google Scholar]
- 23.Harter C, James P, Bachi T, Semenza G, Brunner J. Hydrophobic binding of the ectodomain of influenza hemagglutinin to membranes occurs through the “fusion peptide.”. J Biol Chem. 1989;264:6459–6464. [PubMed] [Google Scholar]
- 24.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 25.Hernandez L D, Peters R J, Delos S E, Young J A T, Agard D A, White J M. Activation of a retroviral membrane fusion protein: soluble receptor-induced liposome binding of the ASLV envelope glycoprotein. J Cell Biol. 1997;139:1–10. doi: 10.1083/jcb.139.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez L D, White J M. Mutational analysis of the candidate internal fusion peptide of the avian leukosis and sarcoma virus subgroup A envelope glycoprotein. J Virol. 1998;72:3259–3267. doi: 10.1128/jvi.72.4.3259-3267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ho S N, Hunt H D, Horton R M, Pullen J K, Pease L R. Site-directed mutagenesis by overlap extension using the polymerase chain reaction. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 28.Horth M, Lambrecht B, Khim M C, Bex F, Thiriart C, Ruysschaert J M, Burny A, Brasseur R. Theoretical and functional analysis of the SIV fusion peptide. EMBO J. 1991;10:2747–2755. doi: 10.1002/j.1460-2075.1991.tb07823.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath C M, Lamb R A. Studies on the fusion peptide of a paramyxovirus fusion glycoprotein: roles of conserved residues in cell fusion. J Virol. 1992;66:2443–2455. doi: 10.1128/jvi.66.4.2443-2455.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishiguro R, Kimura N, Takahashi S. Orientation of fusion-active synthetic peptides in phospholipid bilayers: determination by Fourier transform infrared spectroscopy. Biochemistry. 1993;32:9792–9797. doi: 10.1021/bi00088a034. [DOI] [PubMed] [Google Scholar]
- 31.Lear J D, DeGrado W F. Membrane binding and conformational properties of peptides representing the NH2 terminus of influenza HA-2. J Biol Chem. 1987;262:6500–6505. [PubMed] [Google Scholar]
- 32.Levy-Mintz P, Kielian M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J Virol. 1991;65:4292–4300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luneberg J, Martin I, Nussler F, Ruysschaert J M, Herrmann A. Structure and topology of the influenza virus fusion peptide in lipid bilayers. J Biol Chem. 1995;270:27606–27614. doi: 10.1074/jbc.270.46.27606. [DOI] [PubMed] [Google Scholar]
- 34.Macosko J C, Kim C H, Shin Y K. The membrane topology of the fusion peptide region of influenza hemagglutinin determined by spin-labeling EPR. J Mol Biol. 1997;267:1139–1148. doi: 10.1006/jmbi.1997.0931. [DOI] [PubMed] [Google Scholar]
- 35.Martin I, Defrise-Quertain F, Mandieau V, Nielsen N M, Saermark T, Burny A, Brasseur R, Ruysschaert J M, Vandenbranden M. Fusogenic activity of SIV (simian immunodeficiency virus) peptides located in the GP32 NH2 terminal domain. Biochem Biophys Res Commun. 1991;175:872–879. doi: 10.1016/0006-291x(91)91646-t. [DOI] [PubMed] [Google Scholar]
- 36.Martin I, Ruysschaert J M. Comparison of lipid vesicle fusion induced by the putative fusion peptide of fertilin (a protein active in sperm-egg fusion) and the NH2-terminal domain of the HIV2 gp41. FEBS Lett. 1997;405:351–355. doi: 10.1016/s0014-5793(97)00213-5. [DOI] [PubMed] [Google Scholar]
- 37.Martin I, Schaal H, Scheid A, Ruysschaert J M. Lipid membrane fusion induced by the human immunodeficiency virus type 1 gp41 N-terminal extremity is determined by its orientation in the lipid bilayer. J Virol. 1996;70:298–304. doi: 10.1128/jvi.70.1.298-304.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muga A, Neugebauer W, Hirama T, Surewicz W K. Membrane interaction and conformational properties of the putative fusion peptide of PH-30, a protein active in sperm-egg fusion. Biochemistry. 1994;33:4444–4448. doi: 10.1021/bi00181a002. [DOI] [PubMed] [Google Scholar]
- 39.Rafalski M, Lear J D, DeGrado W F. Phospholipid interactions of synthetic peptides representing the N-terminus of HIV gp41. Biochemistry. 1990;29:7917–7922. doi: 10.1021/bi00486a020. [DOI] [PubMed] [Google Scholar]
- 40.Rafalski M, Ortiz A, Rockwell A, van Ginkel L C, Lear J D, DeGrado W F, Wilschut J. Membrane fusion activity of the influenza virus hemagglutinin: interaction of HA2 N-terminal peptides with phospholipid vesicles. Biochemistry. 1991;30:10211–10220. doi: 10.1021/bi00106a020. [DOI] [PubMed] [Google Scholar]
- 41.Rapaport D, Shai Y. Interaction of fluorescently labeled analogues of the amino-terminal fusion peptide of Sendai virus with phospholipid membranes. J Biol Chem. 1994;269:15124–15131. [PubMed] [Google Scholar]
- 42.Rong L, Bates P. Analysis of the subgroup A avian sarcoma and leukosis virus receptor: the 40-residue, cysteine-rich, low-density lipoprotein receptor repeat motif of Tva is sufficient to mediate viral entry. J Virol. 1995;69:4847–4853. doi: 10.1128/jvi.69.8.4847-4853.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rong L, Edinger A, Bates P. Role of basic residues in the subgroup-determining region of the subgroup A avian sarcoma and leukosis virus envelope in receptor binding and infection. J Virol. 1997;71:3458–3465. doi: 10.1128/jvi.71.5.3458-3465.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Soneoka Y, Cannon P M, Ramsdale E E, Griffiths J C, Romano G, Kingsman S M, Kingsman A J. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 1995;23:628–633. doi: 10.1093/nar/23.4.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stegmann T, White J M, Helenius A. Intermediates in influenza induced membrane fusion. EMBO J. 1990;9:4231–4241. doi: 10.1002/j.1460-2075.1990.tb07871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Steinhauer D A, Wharton S A, Skehel J J, Wiley D C. Studies of the membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi S. Conformation of membrane fusion-active 20-residue peptides with or without lipid bilayers. Implication of alpha-helix formation for membrane fusion. Biochemistry. 1990;29:6257–6264. doi: 10.1021/bi00478a021. [DOI] [PubMed] [Google Scholar]
- 48.Tatulian S A, Hinterdorfer P, Baber G, Tamm L K. Influenza hemagglutinin assumes a tilted conformation during membrane fusion as determined by attenuated total reflection FTIR spectroscopy. EMBO J. 1995;14:5514–5523. doi: 10.1002/j.1460-2075.1995.tb00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.White J M. Viral and cellular membrane fusion proteins. Annu Rev Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- 50.Wool-Lewis R J, Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang L, Ghosh H P. Characterization of the putative fusogenic domain in vesicular stomatitis virus glycoprotein G. J Virol. 1994;68:2186–2193. doi: 10.1128/jvi.68.4.2186-2193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu N, Cannon P M, Chen D, Anderson W F. Mutational analysis of the fusion peptide of Moloney murine leukemia virus transmembrane protein p15E. J Virol. 1998;72:1632–1639. doi: 10.1128/jvi.72.2.1632-1639.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]