Abstract
The current human immunodeficiency virus type 1 (HIV-1) shows an increasing number of distinct viral subtypes, as well as viruses that are recombinants of at least two subtypes. Although no biological differences have been described so far for viruses that belong to different subtypes, there is considerable sequence variation between the different HIV-1 subtypes. The HIV-1 long terminal repeat (LTR) encodes the transcriptional promoter, and the LTR of subtypes A through G was cloned and analyzed to test if there are subtype-specific differences in gene expression. Sequence analysis demonstrated a unique LTR enhancer-promoter configuration for each subtype. Transcription assays with luciferase reporter constructs showed that all subtype LTRs are functional promoters with a low basal transcriptional activity and a high activity in the presence of the viral Tat transcriptional activator protein. All subtype LTRs responded equally well to the Tat trans activator protein of subtype B. This result suggests that there are no major differences in the mechanism of Tat-mediated trans activation among the subtypes. Nevertheless, subtype-specific differences in the activity of the basal LTR promoter were measured in different cell types. Furthermore, we measured a differential response to tumor necrosis factor alpha treatment, and the induction level correlated with the number of NF-κB sites in the respective LTRs, which varies from one (subtype E) to three (subtype C). In general, subtype E was found to encode the most potent LTR, and we therefore inserted the core promoter elements of subtype E in the infectious molecular clone of the LAI isolate (subtype B). This recombinant LAI-E virus exhibited a profound replication advantage compared with the original LAI virus in the SupT1 T-cell line, indicating that subtle differences in LTR promoter activity can have a significant impact on viral replication kinetics. These results suggest that there may be considerable biological differences among the HIV-1 subtypes.
There are two viruses that cause AIDS in humans, namely, human immunodeficiency virus type 1 (HIV-1) and HIV-2. Both viruses have isogenic counterparts in chimpanzee and sooty mangabey simian immunodeficiency viruses (SIVcpz and SIVsm, respectively), and probably at least two cross-species transmissions of different retroviruses occurred from monkeys to humans (reviewed in reference 17). Most HIV-1 isolates identified to date in the pandemic belong to a group designated M for major. This group has spread worldwide within the last two decades (40). There are at least two additional HIV-1 groups that are confined to a more restricted geographical area in Africa. Several AIDS patients from west-central Africa have viruses from a distinct group designated O (outlier group). More recently, one member of a third group designated N (new group) was isolated from an AIDS patient in Cameroon (54). It is suspected that each group originated from a different SIVcpz transmission from monkeys to humans (18). There is no evidence to suggest that the O- and N-group viruses are less virulent or defective in transmission, and the worldwide spread of group M viruses may just result from a stochastic or chance process (63).
The group M viruses that comprise the current global pandemic have diversified during their worldwide spread. These isolates have been grouped according to their genomic sequences and can be divided into at least 10 distinct subtypes or clades termed A through J (40). Isolates from different subtypes may differ by 30 to 40% in the amino acid sequence of the Env protein, whereas variation ranges from 5 to 20% within a subtype. Subtypes are not stable entities because recombinants and even intergroup recombinants (57) with mosaic genomes are known to occur at an appreciable frequency (9, 19, 30, 48). The different subtypes are not distributed evenly throughout the world. For example, subtype B predominates in North America and Europe, and subtype E predominates in northern Thailand (17). There is at present no evidence for subtype-specific variation in virulence or transmission, and their diverse geographical distribution is likely to result from stochastic founder effects. Nevertheless, the possibility that the subtypes differ in their biological properties cannot be excluded, and this may affect their pathogenic potential. For instance, it has been suggested that subtype E viruses are particularly virulent and that they replicate more efficiently than other subtypes in Langerhans cells, which are potential target cells in heterosexual transmission (56), although follow-up studies could not confirm these results (15, 46). The relationship between virus subtype, biological properties, and pathogenicity is unknown, in part because virus replication studies have been performed almost exclusively with subtype B viruses.
Full-length genomic sequences of several subtypes of the HIV-1 group M have been reported (9, 19, 20, 30). Remarkable variation was observed in the nucleotide sequence of the long terminal repeat (LTR) region, which constitutes the transcriptional promoter (36, 37, 62). Despite accumulating sequence data on the HIV-1 subtypes, to data no subtype-specific differences in virus biology have been described. We therefore initiated an analysis of LTR sequence variation in the different HIV-1 subtypes and its functional consequences for viral transcription, replication, cell tropism, and pathogenicity. In this study, we present the LTR sequence of viral subtypes A through G and report functional differences of these transcriptional promoters as measured in transient transfection assays. Furthermore, we measured increased replication of the subtype B LAI isolate upon introduction of the LTR core promoter elements of subtype E.
MATERIALS AND METHODS
Patient samples, amplification, and sequencing of the HIV-1 LTR.
Human serum samples from patients suspected of having a non-subtype B HIV-1 infection were selected from the outpatient clinic of the Academic Medical Center of the University of Amsterdam, Amsterdam, The Netherlands, and the LTR-gag region of the viral genome was amplified by reverse transcription (RT)-PCR as will be described (13). We used this PCR material for a nested PCR with primer 5′T7-U3-M (5′ TAA TAC GAC TCA CTA TAG GGT TTT TAA AAG AAA AGG GGG GAC 3′), which contains the T7 promoter sequence (in italics) and primer 3′Sp6-R-M (5′ ATT TAG GTG ACA CTA TAG ATT GAG GCT TAA GCA GTG GG 3′), which contains an AflII-site (underlined) and an Sp6 promoter sequence (in italics). The PCR product of 12 serum samples was cloned in plasmid pCRII-TOPO according to the manufacturer's protocol (Invitrogen). Three positive clones of each serum sample were sequenced with the ET(−21M13fwd) primer and the DYEnamic™ direct cycle-sequencing kit (Amersham, Cleveland, Ohio) on an automatic sequencer (Applied Biosystems DNA sequencer 373A).
LTR-luciferase constructs.
One representative clone for each subtype was selected for subcloning (except for two samples for the C subtype, designated C1 and C2). The BseAI-AflII fragment (position −147 to +63) of the LTR was exchanged in an LTR-luciferase plasmid that is based on the sequence of the LAI isolate (subtype B). The pBlue3′LTR-luc plasmid is a pBluescript KS(+) derivative which is composed of a 1,426-bp BglI-XhoI fragment from pBluescript KS(+) containing the ColE1 ori, a 719-bp XhoI-HindIII LAI 3′ LTR fragment, a 1,951-bp HindIII-BamHI pGL3 luc gene, and a 1,625-bp BamHI-BglI fragment derived from pSV2CAT (27), which encompasses a simian virus 40 polyadenylation site and a pBluescript KS(+) fragment. The BseAI site is present in all clones except in two of the three subtype E clones, and we therefore used the third E sample for subcloning. We do not know whether the AflII site is present in the subtype LTR sequences because this region is in fact encoded by the downstream PCR primer (see Fig. 2). One additional construct was made in which the upstream TATAA box in subtype E was changed into TACAA. This was done with a mutagenic primer (5′ GCA TCC GGA GTA CTA CAA AGA CTG 3′) in a PCR with the standard 3′ primer. This product was subcloned as a BseAI-AflII fragment, and the sequence was verified. The pcDNA3-Tat vector was described previously (61).
FIG. 2.
Partial LTR sequence of subtypes A through G. The LTR region spanning position −177 to +67 of prototype virus LAI (subtype B) is shown at the top, with the position of several motifs and/or signals marked. Sequences were aligned with the sequence navigator program and optimized manually. Dashes indicate nucleotides that are identical to this prototype. Gaps are indicated by dots. Motifs present in the LAI sequence are underlined, whereas elements which are absent in LAI are boxed (e.g., AP-1). The nef stop codon in subtypes B and F is marked in boldface type (position −124 in LAI-B), and restriction sites used in subcloning are shown in italics. Structural details of the TAR element (position +1 to 56) are presented in Fig. 4. In subtype F, a 12-nucleotide duplication (15 nucleotides when one mismatch is allowed) explains the presence of two adjacent AP-1 sites. The BseA1-AflII fragment was used for subcloning in the LTR-luciferase plasmid. The sequences downstream of position +56 were encoded by the PCR primer and are therefore not shown for the subtypes.
Infectious HIV-1 molecular clones.
Molecular clones containing the basal promoter of subtype E in a subtype B background were made by exchanging the 1.7-kb BglI-XhoI fragment of pLAI (44) with the corresponding fragment of subtype E (or the Emut mutant), which was obtained by digestion of the respective pBlue3′LTR-luc plasmids. The clones are termed pLAI-E and pLAI-Emut.
Cells and transfection assays.
The following adherent cell lines were used: the African green monkey kidney cell line COS, the cervix carcinoma cell line C33A (ATCC HTB31) (1), the human glioblastoma cell line U373 MG, the human astrocyte glioblastoma cell line U87, and HeLa cells. The cell lines were grown as a monolayer in Dulbecco's modified Eagle's medium supplemented with 10% (vol/vol) fetal calf serum, 20 mM glucose, and minimal essential medium nonessential amino acids at 37°C and in 5% CO2. These cell types were transfected by the calcium phosphate method as described previously (12). Various amounts of the specific plasmids were used in the transfection as indicated in the experiments, but the total amount of DNA was kept constant at 6 μg of plasmid DNA by addition of pcDNA3 (Invitrogen), in a final volume of 2 ml of 25 mM HEPES (pH 7.1), 125 mM NaCl, 0.75 mM Na2HPO4, and 0.12 M CaCl2.
Basal transcription of the different LTR-luciferase constructs was determined with 20 and 100 ng of plasmid DNA in at least two different transfections. To compare the different LTR activities, basal LTR activity was calculated relative to that of the LAI construct, which was arbitrarily set at a value of 1. There were no significant differences in the relative LTR activities measured with 20 or 100 ng of LTR-luciferase plasmid DNA, demonstrating that transcription was limited by the amount of plasmid DNA. Furthermore, all measurements were performed in the linear range of the luciferase assay. The Tat-activated levels of transcription were determined in at least two transfections with 30 and 100 ng pcDNA3-Tat in combination with 20 ng of LTR-luciferase construct. Tat-activated LTR activity was also calculated relative to the LAI construct. Relative Tat-activated LTR activity with 30 or 100 ng pcDNA3-Tat was similar, which shows that the measurements were in the linear range of Tat trans activation. The relative Tat responsiveness was calculated by dividing the relative Tat-induced LTR activity of each subtype by its own relative basal activity. All transfections used in the calculations were done with the same set of plasmids that were isolated simultaneously. The experiments in C33A cells were repeated with a different set of DNA preparations, producing similar results.
The human lymphocyte T-cell line SupT1 (55) was cultured in RPMI 1640 (Gibco BRL) supplemented with 10% (vol/vol) fetal calf serum. Transfections were carried out as previously described (35) using a Bio-Rad Gene Pulser. For the luciferase constructs, 5 μg of the pBlue3′LTR-luc construct with or without 500 μg of pcDNA3-Tat was used. For the molecular clones, 1 μg of plasmid DNA was used.
Luciferase assay.
Two days after transfection, the culture medium was removed and the cells were washed once with phosphate-buffered saline. The cells were lysed by the addition of 200 μl of reporter lysis buffer (Promega), and the sample was mixed for 45 min at room temperature. The lysate was collected in a tube, and the cell debris was removed by centrifugation for 15 min at 15,000 rpm in an Eppendorf centrifuge. The luciferase activity (in relative light units) was determined by a Berthold luminometer, model LB9501. A 30-μl sample was diluted with 270 μl of reaction buffer (3.3 mM ATP, 25 mM glycylglycine (pH 7.8), 15 mM MgSO4, 100 μg of bovine serum albumin (per ml) and 100 μl of 1 mM luciferin (Boehringer Mannheim). The luminometer was set for a 10-s measurement.
LTR nucleotide sequence analysis.
The LTR nucleotide sequences were aligned using the program sequence navigator (ABI) and adjusted manually. For phylogenetic analysis, we used the neighbor-joining method, and the distance matrix was generated by Kimura's two-parameter estimation as implemented in the TREECON program (59). The TFSEARCH program for the identification of transcription factor binding sites is constructed by Yutaka Akiyama and is accessible at the TRC Laboratory website, http://www.rwcp.or.jp/papia/. This program is based on the databases TRANSFAC, TRRD, and COMPEL, which store information about transcription factors and their binding sites (TRANSFAC), the regulatory hierarchy of whole genes (TRRD), and the structural and functional properties of composite elements (COMPEL). These databases are described in reference 24 and are accessible at http://www.transfac.gbf.de/TRANSFAC or http://www.bionet.nsc.ru/TRRD.
HIV-1 infections and CA-p24 measurements.
SupT1 cells were transfected with 1 μg of the molecular clones, and culture supernatants were harvested at the peak of infection and stored in aliquots at −70°C. An aliquot was used to determine the CA-p24 concentration by a twin-site enzyme-linked immunosorbent assay with D7320 (Biochrom, Berlin, Germany) as the capture antibody, alkaline phosphatase-conjugated anti-p24 monoclonal antibody (EH12-AP), and the AMPAK amplification system (Dako Diagnostics Ltd., ITK Diagnostics BV) as described previously (34, 38). Recombinant CA-p24 expressed in a baculovirus system was used as the reference standard. Viral infections were initiated with 5 ng of CA-p24 in a 5-ml SupT1 culture containing 106 cells. The viral infections were monitored by measuring CA-p24 levels.
Primer extension analysis.
Viral RNA was isolated from SupT1 cells at the peak of infection. A 1-ml sample was taken from the culture and centrifuged at 2,750 × g for 5 min to collect the cells. The cells were resuspended in 100 μl of extraction buffer (10 mM Tris-Cl [pH 7.5], 1 mM EDTA, 150 mM NaCl, and 500 μg of proteinase K per ml) and incubated at 56°C for 30 min. The volume of the mixture was increased by addition of 400 μl of 0.3 M sodium acetate (pH 5.2) and extracted twice with an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1). The purified RNA was precipitated by adding 3 volumes of 100% ethanol and subsequently collected by centrifugation in an Eppendorf centrifuge (15,000 rpm for 20 min at 4°C). The RNA pellet was washed once with 70% ethanol and, after drying, dissolved in 10 μl of H2O. Primer extension reactions were carried out in a final volume of 24 μl as follows. The viral RNA (3 μl) was mixed with excess lys3 DNA primer (2.9 pmol) in 12 μl of annealing buffer (83 mM Tris-Cl [pH 7.5], 125 mM KCl), heated for 2 min at 85°C and allowed to cool slowly to room temperature. The lys3 primer (5′ CAA GTC CCT GTT CGG GCG CCA 3′) anneals to the primer binding site and the three nucleotides directly downstream of it (positions +182 to +202 of the viral genome). Reverse transcription was initiated by the addition of 12 μl of 2× concentrated RT buffer (6 mM MgCl2, 20 mM dithiothreitol; 0.2 μl of avian myeloblastosis virus RT (Stratagene); 20 μM (each) of dGTP, dATP and dTTP; 10 μM dCTP; and 0.3 μl of [α-32P]dCTP [10 mCi/ml]). The final reaction mixture was incubated for 1 h at 37°C. Reverse transcription was terminated by the addition of 1 μl of 0.5 M EDTA [pH 8.0], and the cDNA products were ethanol precipitated and redissolved in formamide loading buffer (31). The samples were analyzed by polyacrylamide gel electrophoresis on a 6% sequencing gel (31). A 35S-labelled sequence reaction with the same lys3 primer and the pBlue3′LTR-luc plasmid was performed with the T7 Sequenase kit 2.0 according to the supplier's instructions (Amersham) and run alongside to determine the size of the cDNA products.
Nucleotide sequence accession numbers.
LTR nucleotide sequences from representative subtype clones have been deposited in the GenBank database. The accession numbers are AF1275566 (subtype A), AF1275567 (subtype C1), AF1275568 (subtype C2), AF1275569 (subtype D), AF1275570 (subtype E), AF1275571 (subtype F), AF1275572 (subtype G), and AF1275573 (subtype G").
RESULTS
Non-subtype B LTR sequences.
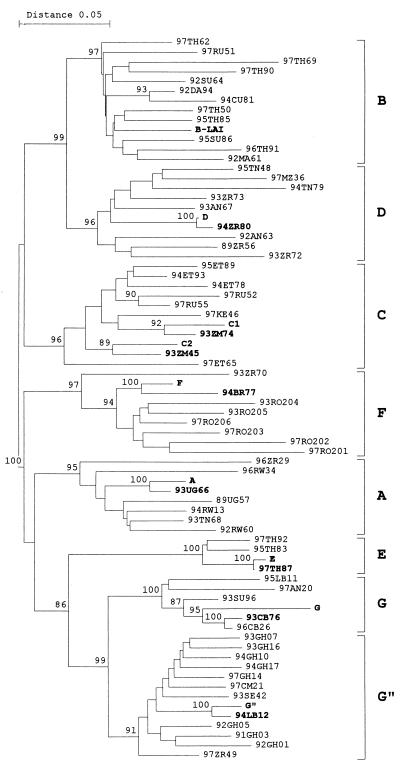
The LTR-gag region of the HIV-1 RNA genome was amplified by RT-PCR on serum samples from HIV-infected patients with a non-subtype B virus. Direct sequencing was performed on the PCR samples, which provides the most abundant or population sequence of the viral quasispecies in reference (13). The subtype was determined by comparison with the sequence of other subtype isolates (40). A detailed comparison of these primary isolates with the subtype reference sequences is provided previously (13). For this study, we selected serum samples representing subtypes A through G for cloning of the LTR promoter. The subtype A sample is actually an AC recombinant, but the LTR element is derived from subtype A. The subtype G" is not a distinct subtype but a cluster of AG recombinants (CRF-IbNG) with an LTR that is closely related to that of subtype G (11). To examine whether these clonal sequences correspond to the viral quasispecies present in the infected patient, we performed a phylogenetic analysis with both clonal and population-based sequences (Fig. 1). The LTR sequence of the prototype virus LAI of subtype B and other strains of subtypes A to G were included in this phylogenetic tree. Marked in boldface type are the patient isolates that were used for the cloning of individual LTRs (e.g., clone D originates from patient sample 94ZR80). We observed two distinct branches within the subtype C group and therefore included one isolate of each branch (samples C1 and C2). This phylogenetic tree indicates that the cloned LTRs are representative for the virus population in these patients and for their subtype. One interesting feature was observed for the subtype G sample, which is closely related to the population sequence of two patients, donor 93CB76 and patient 96CB26. It turned out that these two persons are heterosexual partners, suggesting that this virus spread by transmission between these persons.
FIG. 1.
Phylogenetic analysis of the HIV-1 subtype LTR clones. The analysis was performed with the population-based sequence of subtype PCR samples from several patients and the cloned LTR samples that were tested in detail in this study. Sequences were analyzed by the neighbor-joining method, and the distance matrix was generated by Kimura's two-parameter estimation as implemented in the TREECON program (59). Bootstrap values above 85 are indicated at nodes. The cloned LTRs and the corresponding patient sequences always cluster together, and both entries are marked in boldface type. The G" clone is derived from donor 93CB76 but also clusters closely to patient 96CB26. As these two patients are partners, the similarity in virus sequence is likely to reflect virus transmission from one person to the other.
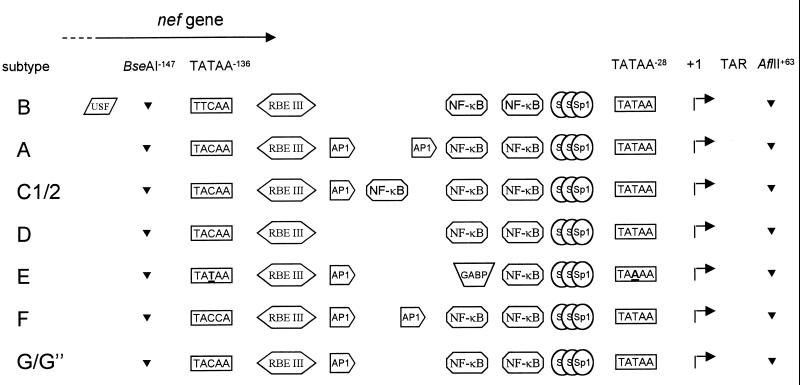
Numerous differences were observed in the subtype LTRs, and the nucleotide sequence of part of the LTR is shown in Fig. 2. We marked the position of important sequence elements in the LAI isolate of subtype B. This prototype LTR of isolate LAI contains a core promoter with a TATAA box, and three upstream binding sites for the transcription factor Sp1 are usually included in the core element. Several sequence changes in the Sp1 region were observed in the subtypes, but the putative effect on Sp1 binding remains unclear, in part because the HIV-1 LTR contains solely nonconsensus Sp1 sites (e.g., they are not found with the TFSEARCH program; see below). It remains possible that other members of the Sp1 family of transcription factors, e.g., the constitutively expressed Sp3 factor, bind some of the subtype LTRs. Located upstream of the core promoter are important enhancer elements, including binding sites for NF-κB, RBE III, and USF. We used the TFSEARCH program to analyze the subtype LTR sequences for the presence of transcription factor binding sites. Several notable differences are summarized in Fig. 3.
FIG. 3.
LTR promoter organization of HIV-1 subtypes A through G. Most experimental evidence for protein binding sites has been provided for the LTR of HIV-1 subtype B (reviewed in reference 21). Furthermore, recent evidence supports the conversion of the upstream NF-κB site of subtype E into a GABP binding site (62). The BseAI and AflII sites were used for subcloning in the LTR-luciferase reporter construct.
The number of NF-κB sites differs among the subtypes. Whereas prototype subtype B has two adjacent NF-κB sites, three NF-κB sites are present in both subtype C clones (see also reference 37). However, it is unlikely that all three sites will bind this transcription factor with equal affinity. In particular, the downstream site contains a subtype C-specific mutation that is predicted to negatively affect NF-κB binding. With the presence of two bona fide upstream NF-κB sites, it is tempting to speculate that this third site has evolved into a binding site for another transcription factor. The TFSEARCH program predicts reduced NF-κB binding potential, as the score is reduced from 97.5 to 85.4, but this program does not suggest that a new transcription factor-binding site is generated.
The TFSEARCH program does predict such an enhancer switch for the upstream NF-κB site of subtype E, which contains a typical deletion of a single T nucleotide (Fig. 2). The NF-κB binding score is reduced from 97.5 for the regular NF-κB site to below the threshold value of 85 for the upstream site of subtype E, with a concomitant rise of the score for the GABP transcription factor from undetectable to 87.7. Indeed, it was previously demonstrated that this minor sequence change interferes with NF-κB binding (37, 62). More interestingly, this mutant NF-κB site was shown to facilitate binding of GABP, a constitutively expressed transcription factor of the Ets family (62). This result testifies to the value of this computer-mediated search for transcription factor-binding sites.
Some of the transcription factor-binding sites upstream of the NF-κB region show subtype-specific variation, whereas other sites are well conserved. The RBE III site is absolutely conserved in all subtypes, which is consistent with previous reports (16). This cis-acting element is a binding site for RBF2 and is involved in the response to the protein-tyrosine kinase/Ras/Raf-signaling pathway (2). This site is often duplicated in patient isolates (16, 28), but the insert in subtype F does not represent a complete RBE III site. The USF binding site, which overlaps the nef gene, contributes to the LTR function of subtype B viruses (22, 53). Interestingly, we found this site exclusively present in the subtype B sequence (Fig. 2 and 3). In contrast, AP-1 binding sites have not been described for this region of the subtype B LTR, but such sites are predicted for most LTRs, except for subtypes B and D. The subtype B LTR has been suggested to encode AP-1 sites in the downstream U5 region of the LTR at position +154 (60), and we found several putative AP-1 sites in the upstream U3 region of the LTR promoter in several subtypes (results not shown). The presence of AP-1 motifs just upstream of the NF-κB sites and thus near the core promoter seems a significant difference between the subtypes. A single AP-1 site was predicted for subtypes C, E, G, and G". Two adjacent AP-1 sites are predicted for subtypes A and F. In the latter case, the tandem AP-1 site is most likely generated by duplication of a 12-nucleotide segment (15 nucleotides when one mismatch is allowed; see Fig. 2). These two AP-1 sites constitute the subtype F-specific insert immediately upstream of the NF-κBII site. This LTR region also encodes the C terminus of the Nef protein, and the other insert at position −130 in the subtype F sequence would theoretically extend the Nef open reading frame by two amino acids. However, the subtype F-specific insert encodes a new stop codon (at the equivalent of position −124 in subtype B), resulting in a Nef protein that is one amino acid shorter. Although speculative, this may represent a mechanism for subtype F to preclude the expression of a C-terminally extended Nef protein.
The transcription start site (position +1) is located 24 nucleotides downstream of the TATAA box. Perhaps most intriguing is the sequence change in the TATAA box of subtype E at position −28 into TAAAA (37). The LTR sequence of a total of 18 subtype E isolates has been determined, and all isolates contain this typical mutation (36, 37, 62; unpublished results from our laboratory). Another striking feature of the subtype E LTR is the presence of an upstream TATAA box at position −136, which differs from the sequence of other subtypes at one or two positions. Interestingly, this TATAA−136 box has been suggested to functionally replace the mutated TAAAA−28 box (37). To test this idea, we used PCR mutagenesis to change the upstream TATAA−136 box into TACAA−136, which is the most common sequence in the other subtypes, and this Emut LTR was included in the subsequent promoter assays and virus replication studies.
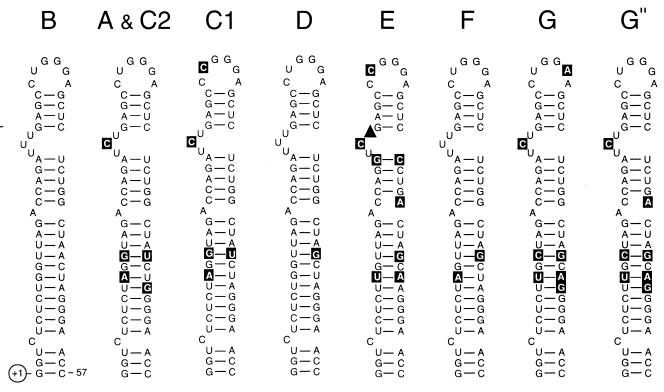
The TAR motif is encoded in the transcribed region and acts as an RNA enhancer through binding of the viral Tat trans activator protein and the cellular cyclin T factor (7, 14, 64). Because the RNA secondary structure of this motif is critical for function, we analyzed the typical hairpin structure for the different subtype sequences (Fig. 4). The secondary structure of the TAR hairpin is based on mutational (5) and phylogenetic (3) evidence. It is clear that the subtypes have distinctive mutations in the TAR hairpin, which are mostly located in the lower stem region. Most sequence changes represent either basepair variations (e.g., A-U to G-U) or basepair covariations (e.g., A-U to G-C) that do not disturb the secondary structure.
FIG. 4.
Comparison of the TAR RNA secondary structure in different HIV-1 subtypes. The hairpin structure of subtype B isolate LAI was used as prototype. Nucleotide changes occurring in the other subtypes are in reverse contrast. Nucleotide deletion is indicated by ▴. A detailed TAR phylogenetic analysis of different HIV-1 subtype B sequences and SIVs has been reported previously (3; 4).
Differential activity of the HIV-1 subtype LTR promoters.
The sequencing results indicate that the subtype LTRs are likely to differ in their ability to bind cellular transcription factors, and this may obviously affect the LTR promoter activity. To test this, we inserted the subtype promoters upstream of the luciferase gene in a reporter construct. This cloning strategy was designed to allow insertion of non-subtype B LTR sequences in the LAI molecular clone (subtype B) for replication studies. This necessitates the conservation of the nef gene, which overlaps part of the regulatory LTR DNA motifs (nef UGA stop codon at position −124) (Fig. 2). We therefore inserted the BseAI-AflII fragment (position −147 to +63) (Fig. 2 and 3) of the subtype LTR into the standard LTR-luciferase construct, which contains the LTR of subtype B virus LAI. Thus, the recombinant LTRs maintain the subtype B-specific USF site but have exchanged all transcription motifs that are located further downstream, including the TAR element.
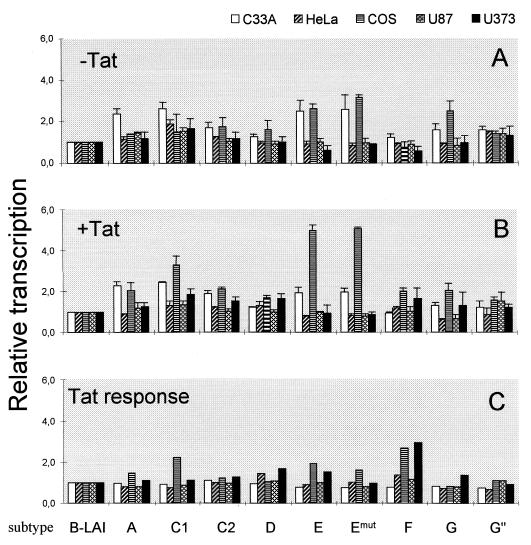
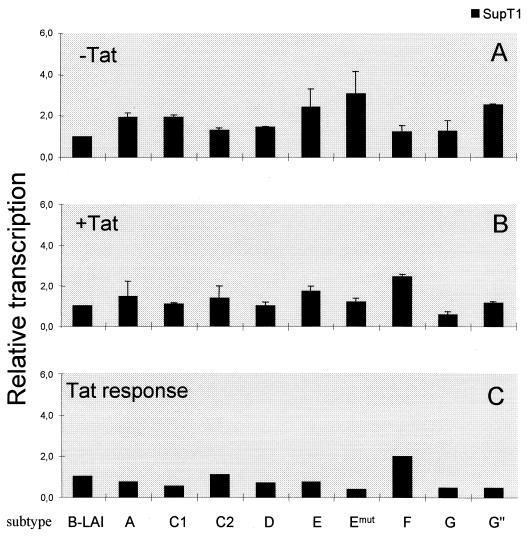
The LTR-luciferase constructs of subtype A through G, including the two subtype C samples and the modified LTR of subtype E with a mutation in the upstream TATAA−136 box (Emut), were subsequently tested for promoter activity in different cell lines. We compared LTR activities in three cell lines that are regularly used for transient transfection studies: the human cervix carcinoma cell lines C33A and HeLa, of which the latter is transformed by human papilloma virus type 18, and the African green monkey kidney cell line COS, which is transformed by simian virus 40. We also compared the LTR activity in two human astrocyte glioblastoma cell lines (U87 and U373). The cells were transiently transfected in the absence and presence of a second plasmid encoding the Tat trans activator protein of subtype B. Basal and Tat-activated LTR activities were measured and used to calculate relative transcriptional activity and standard deviation. These values are plotted in Fig. 5A and B, respectively, with the basal and Tat-activated activities of the subtype B LTR promoter each arbitrarily set at a value of 1. These two activities were used to calculate the Tat responsiveness of each LTR by dividing the Tat-activated level by the basal activity for each subtype. This Tat responsiveness was also related to that of subtype B (Fig. 5C).
FIG. 5.
Differential activity of the HIV-1 subtype LTR promoters. The subtype LTRs were tested for basal activity without Tat (A) and for induced transcription in the presence of Tat (B), and these two values were used to calculate the Tat response (C). These three transcriptional parameters were related to that of the prototype LAI LTR (subtype B), of which the values were each arbitrarily set at a value of 1. Each LTR activity is the average of at least four independent measurements, and the standard deviation is given.
All subtype LTR promoters demonstrated low basal and high Tat-induced transcription levels, a pattern of gene expression very similar to that documented previously for subtype B (reviewed in reference 21). This result may not be surprising because the LTRs were derived from actively replicating viruses that should have a functional and Tat-responsive LTR promoter. Nevertheless, differences in promoter activity of the different subtype LTRs were observed, in particular without Tat protein. In fact, the basal LTR activity of most non-B subtypes was significantly higher than that of the subtype B LTR (Fig. 5A). The LTRs of subtype A, E, and G and the two subtype C samples were approximately two- to threefold more active than the subtype B LTR in C33A and COS cells. Less variation in promoter activity was measured in HeLa, which also produced a distinct activity pattern for the subtypes. A small but significant increase was measured for subtype G" and the two subtype C samples in HeLa cells. The basal activity in the two astrocyte cell lines was similar for all subtypes, with only a small increase for subtype C.
We next compared the subtype LTR promoters in the presence of Tat trans activator protein of isolate LAI. Pronounced induction levels were measured in all cell lines. For instance, with the prototype LTR of the LAI virus and 30 ng of Tat plasmid, we obtained an approximately 6-fold induction in C33A cells, 35-fold induction in HeLa cells, and 20-fold induction in COS cells. With 100 ng of Tat plasmid, which is also within the linear range of Tat-mediated activation, we measured an 11-fold induction in C33A cells, a 64-fold induction in HeLa cells, and a 40-fold induction in COS cells. Note that these values are both in the linear range of Tat transactivation and are not the maximum level of Tat transactivation. Although the different LTRs encode distinct TAR hairpin motifs (Fig. 4), we measured no significant difference in Tat response of the subtype LTRs. In other words, the subtype LTR activity pattern observed in the presence of Tat largely mimics the pattern obtained without an activator, with only minor variation between cell types. This demonstrates that the LAI-Tat protein recognizes all subtype TAR sequences. To address the Tat response of the subtype LTRs more accurately, we calculated the actual fold induction and plotted the relative Tat response (Fig. 5C). No significant differences were measured in C33A cells. Subtypes D and F demonstrated an improved Tat response of approximately 40% in HeLa cells. The most significant changes in Tat response were observed in COS cells, with a 50% increase for subtype A and up to a twofold increase for subtype C1, E, and (in particular) F. The other subtype C sample (C2) did not show this pattern, indicating that differences in promoter activity do also exist among different isolates of a single subtype. There were no significant differences in the Tat responsiveness in U87 and U373 cells.
The LTR of subtype E is a TATAA-less promoter.
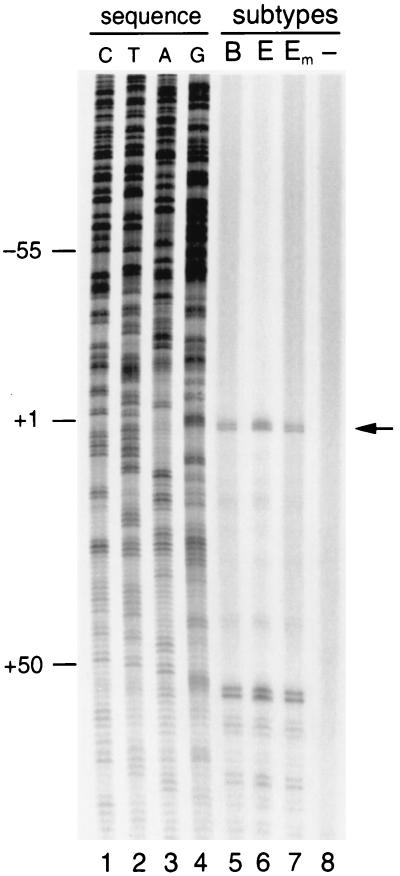
We next addressed whether the upstream TATAA−136 box in subtype E is used to compensate for mutation of the regular TATAA box (TAAAA−28 in subtype E). We constructed the Emut promoter, in which the upstream TATAA−136 box was changed into TACAA. The activity of this Emut LTR was indistinguishable from that of the wild-type E promoter, in both the absence and presence of Tat (Fig. 5). This result indicates that the subtype E-specific TATAA−136 box does not contribute to promoter activity. Thus, the subtype E LTR is an efficient promoter despite mutation of the regular TATAA−28 box, suggesting that the subtype E LTR belongs to the class of TATAA-less promoters. Because the TATAA-box plays a role in positioning of the transcription initiation complex, we analyzed the RNA start site usage of subtype E by primer extension analysis. The same start site was found for viral transcripts initiated from the subtype B or E promoter (Fig. 6, compare lanes 5 and 6). Furthermore, we confirmed that mutation of the upstream TATAA−136 box does not affect the activity of the subtype E LTR or its start site usage (Fig. 6, lane 7).
FIG. 6.
Primer extension to map the RNA start site of HIV-1 subtypes B and E. Total cellular RNA was isolated from infected SupT1 cells for primer extension analysis. Lane 5, LAI; lane 6, LAI-E; lane 7, LAI-Emut; lane 8, uninfected SupT1 cells. A sequence reaction was performed on the pBlue3′LTR-luc plasmid with the same primer as used in the primer extension reaction (lanes 1 to 4). The signals around position +55 represent RT pauses due to the secondary structure of the TAR hairpin in the HIV-1 template, as was described previously (26).
Subtype LTR activity in T cells and the effect of tumor necrosis factor alpha (TNF-α) stimulation.
The promoter activity of the LTR-luciferase constructs was further analyzed in a lymphocyte T-cell line that represents a natural host cell type for HIV-1 infection. SupT1 cells were transfected with the LTR-luciferase constructs in the absence or presence of the Tat-expressing plasmid, yielding a 37-fold induction for the reference LAI construct representing subtype B. The relative basal and activated LTR activities and the relative Tat responsiveness were calculated, and these data are plotted in Fig. 7. The subtype E basal activity is nearly three times higher than that of subtype B, and the basal activity of subtypes A, C1 and G" is also significantly increased (Fig. 7A). Promoter activity in the presence of Tat (Fig. 7B and C) is similar for all subtypes except for a strong Tat response in subtype F, which was also observed in COS cells (Fig. 5).
FIG. 7.
Differential activity of the HIV-1 subtype LTR promoters in SupT1 cells. The subtype LTRs were tested for basal activity without Tat (A) and for induced transcription in the presence of Tat (B), and these two values were used to calculate the Tat response (C). These three transcriptional parameters were related to that of the prototype LAI LTR (subtype B), of which the values were arbitrarily set at a value of 1. The average LTR activity and the standard deviation are given.
TNF-α stimulates the HIV-1 LTR through activation of NF-κB (43). Since the number of NF-κB sites varies from one to three for the subtypes (Fig. 3), we measured TNF-α responsiveness of the different LTRs. SupT1 cells were transfected with 5 μg of LTR-luciferase construct, and the cells were split after 24 h and cultured for an additional 24 h with or without 30 ng of TNF-α per ml. Luciferase activity was determined, and the TNF-α stimulation was calculated by dividing the luciferase activity from cells cultured in the presence of TNF-α by the corresponding cells cultured without TNF-α. The results (Fig. 8) indicate correlation between the number of NF-κB sites and the level of TNF-α stimulation. Subtype E, with one NF-κB site, is induced 1.5-fold by TNF-α; the subtypes with two NF-κB sites show a 2.5- to 3-fold stimulation; and subtype C, with three NF-κB sites, is activated 3.4-fold.
FIG. 8.
TNF-α responsiveness of the HIV-1 subtype LTRs. SupT1 cells were transfected by electroporation with 5 μg of LTR-luciferase. The cells were split after 24 h and cultured for another 24 h without or with TNF-α (30 ng/ml). Luciferase activity was determined, and the TNF-α response was calculated as the ratio of these activities.
Viruses with a subtype E LTR replicate faster than the LAI reference strain.
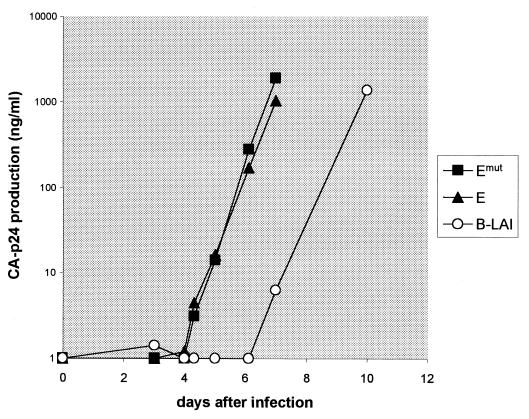
The LTR promoter architecture of subtype E is rather distinct, and this LTR represents the most active basal promoter in SupT1 cells. We therefore selected this subtype for further studies. The subtype B molecular clone LAI was used to insert the core promoter elements of subtype E (and the Emut mutant). The region spanning position −147 to +82 of the 3′ LTR was exchanged. However, only the U3 region of the 3′ LTR is inherited by the viral progeny, and the recombinant progeny will thus contain the −147 to −1 region of subtype E, including the unique TAAAA box and NF-κB and Sp1 sites. The recombinant viruses inherit the R region of the 5′ LTR (results not shown), which encodes the TAR element of subtype B isolate LAI. Viral stocks were used for infection of SupT1 cells, and replication was followed by measuring CA-p24 production in the culture supernatant. Both LAI-E recombinants reached a peak infection about 3 days before the LAI virus (Fig. 9). These results indicate that the upstream TATAA−136 box in subtype E is not important for virus replication, which is consistent with the results of the LTR-luciferase assays. Most importantly, these results indicate that the subtype E LTR profoundly increases the replication capacity of the LAI virus in SupT1 cells. This result was confirmed in three independent infection experiments.
FIG. 9.
Replication of subtype B virus LAI with the core LTR of subtype E. The molecular clone pLAI and two derivatives with the LTR fragment (position −147 to −1) of subtype E (LAI-E and LAI-Emut) were used to generate viral stocks in SupT1 cells. Infections were started with equal amounts of virus (5 ng of CA-p24). Virus replication was followed by measuring CA-p24 production in the culture supernatant. B-LAI (○) is the wild-type LAI virus; LAI-E (■) and LAI-Emut (▴) are described in the text. Similar results were obtained in three independent infections.
DISCUSSION
The LTR promoter region of HIV-1 subtypes A through G was sequenced and tested for transcriptional activity. Several notable differences were observed in the core promoter-enhancer region (Fig. 2 and 3). Some of these differences in subtypes C and E have been reported previously (37), but we now report a complete LTR analysis of subtypes A through G. We observed differences in the number of particular motifs (three instead of the regular two NF-κB sites in subtype C). Furthermore, we observed a switch to a new binding specificity (NF-κB to GABP in subtype E), and the subtype-specific loss or gain of motifs (e.g., USF is subtype B specific, and the number of AP-1 sites varies from zero to two for the different subtypes). These genetic changes appear to be characteristic for the respective subtypes. For instance, the NF-κB–to–GABP switch is present in all 18 subtype E sequences reported to date (62).
All subtype LTRs were found to be functional promoters with a low basal activity and a high Tat-induced activity. In fact, all subtype LTRs responded equally well to the Tat trans activator protein of subtype B. This result suggests that there are no major differences in the mechanism of Tat-mediated trans activation among the subtypes. Nevertheless, distinct cell type-specific differences in basal promoter activity were measured for the subtype LTRs. Cell type-specific differences in the concentration and/or activity of nuclear transcription factors interacting with the LTR are likely to form the basis for these differences. Although the differences in promoter activity reported in this study may not seem very dramatic, a twofold difference in LTR activity may be very important in terms of viral fitness. The replication experiment with the subtype B LAI virus with the core LTR elements of subtype E demonstrates that a relative small difference in promoter activity can have a significant impact on virus replication. It is likely that the gain of LTR function in subtype E versus B is due, at least in part, to the NF-κB–to–GABP enhancer switch (62). Further studies are required to evaluate the contribution of the subtype LTRs to regulated viral transcription and (cell type-specific) replication. In particular, these regulatory sequences could serve to specify the proficiency at which the virus can integrate cellular activation signals (29) or to define the optimal cellular environment for viral gene expression. For instance, we measured significant differences in the TNF-α response, which correlated with the number of NF-κB sites in the LTR.
A striking feature of the fully active subtype E LTR is the mutation within the TATAA box to TAAAA. Three theoretical possibilities can be suggested for the promoter function of this TATAA-less LTR. First, there may be another TATAA element in this LTR promoter. Second, the subtype E LTR may encode an initiator element. Third, the TAAAA motif may be functional as an alternative TATAA box. Transcriptional promoters usually contain either a TATAA box 25 to 30 nucleotides upstream of the transcription initiation site or an initiator element overlapping this start site. However, promoters can have both or neither of these motifs (47). The TATAA box is recognized by the general transcription factor TFIID, which consists of the TATAA-binding protein and TATAA-binding protein-associated factors. Subsequently, a preinitiation complex is assembled through binding of other general transcription factors (47, 49). The initiator functions similarly to the TATAA box in directing accurate transcription by RNA polymerase II (37) and can function independently or synergistically with the TATAA box.
The first possibility is that another TATAA element takes over the TATAA function. This idea was raised previously because the subtype E LTR is unique in having another TATAA sequence at position −136 (37) (Fig. 2 and 3). This possibility was tested in this study by mutation of this upstream motif (TATAA−136 to TACAA). However, this mutant promoter was fully active in LTR-luciferase assays and did support virus replication, thereby ruling out a functional role of the upstream TATAA box. This possibility is unlikely for other reasons. The same TATAA−28-to-TAAAA mutation is present in subtype I (19) and some AG recombinant viruses (11, 41), apparently without the compensatory generation of an upstream TATAA box. Furthermore, usage of the upstream TATAA box at position −136 will move the transcriptional start site to around position −110, and this relocation of the U3-R border will have profound consequences for viral replication. For instance, the TAR hairpin signal will move to an internal position in the viral transcript, which interferes with the TAR function in Tat-mediated transcriptional activation (5, 52). Finally, we determined experimentally that subtype E uses the regular transcriptional initiation site. These combined results demonstrate that the upstream TATAA−136 box in the LTR promoter of subtype E is not functional. Experiments are underway to test whether this LTR uses an initiator element or the alternative TAAAA box to interact with the transcription machinery.
The subtype E promoter is inactivated by substitution of the TAAAA box for the regular TATAA sequence (36). Because this regular TATAA box is present in all other subtypes, and found to be important in the subtype B LTR (6, 42), the subtype E promoter may have compensatory changes elsewhere in the LTR promoter to facilitate the function of the TAAAA−28 motif (36, 37). It remains to be tested whether there is such cross talk between the alternative TAAAA box and subtype E-specific promoter motifs. Our replication studies show that the subtype E core promoter functions efficiently in the subtype B context, which includes the TAR and Tat elements. Thus, the proposed cross talk (36) between TAAAA and the TAR motif in subtype E is unlikely. In addition, a recent paper did not find any support for these combined mutations (41). Another candidate motif is the flanking Sp1 region. For instance, the TATAA-less promoter of the mouse aprt gene was found to rely exclusively on multiple Sp1 sites, including some nonconsensus sites, to trigger transcription (39). For the HIV-1 subtype B LTR, it has been found that the location of the Sp1 sites, relative to the TATAA box, is an important determinant for achieving maximal transcriptional activity (25). There is also some genetic evidence for a functional TATAA-Sp1 interaction. HIV-1 mutants lacking the Sp1 region are replication impaired, but revertant viruses can be selected that have typical changes that extend the CATATAA box to TATATAA (50). Perhaps more striking, some of these revertants also acquired the same mutation as observed in subtype E viruses (TAAAA). Further experimentation is underway to test these putative functional interactions in the subtype LTRs.
We report considerable variation in the LTR promoter-enhancer motifs of viruses that belong to different subtypes of HIV-1 group M. This finding is not without precedent in the field of retrovirology. For instance, we recently described duplication of the complete Sp1 region through prolonged culturing of an attenuated HIV-1 subtype B virus, yielding a stronger LTR promoter with six Sp1 sites and a fitter virus (8). There is also evidence for variation in the number of Sp1 binding sites in the LTR promoter of natural HIV-1 isolates. Several HIV-infected persons were found to contain isolates with four Sp1 sites (28), and one natural isolate with five Sp1 sites was recently identified (51). These examples represent relatively blatant LTR rearrangements, but minor sequence alterations can also have a dramatic effect on LTR function and virus replication. For animal retroviruses, there is ample evidence for changes in host cell tropism or modulation of the viral oncogenic or pathogenic properties by minor sequence variation in the LTR (reviewed in reference 58). For instance, a point mutation in the Moloney murine leukemia virus LTR was shown to increase transcription and enable replication in embryonal cells because of the generation of an Sp1 binding site (23). The equine infectious anemia virus of the Lentivirus genus provides another interesting example where the presence of an Ets-1 binding site in the LTR is essential for productive replication in macrophages (10, 33). Similarly, the nucleotide sequence and functional variations between the LTRs of different avian leukosis viruses have important biological consequences, and a direct correlation between pathogenicity-oncogenicity and LTR transcriptional activity was found (58).
Subtype E, which shows the most distinct promoter architecture, was examined in more detail by performing replication studies with the subtype B isolate LAI with the core promoter elements of subtype E. Both variants driven by the subtype E promoter (LAI-E and LAI-Emut) replicated significantly faster than the LAI virus. These initial results indicate that there may be notable differences in the replication of HIV-1 subtypes due to genetic variation in the LTR promoter. Furthermore, we have observed significant replication differences for the other subtype LTR recombinant viruses in various cell types (results not shown). Obviously, such differences may have a direct impact on the pathogenicity of these viruses. Although there is no published evidence for significant differences in pathogenicity of the HIV-1 subtypes, such biological variation does exist among different immunodeficiency viruses (17). Both HIV-1 and HIV-2 cause AIDS in humans, but epidemiological studies suggest that HIV-2 is not as easily transmittable as HIV-1, and the incubation period for the development of disease is longer for HIV-2 (32, 45). Furthermore, disease progression is not an inevitable outcome of infection by an immunodeficiency virus, since African green monkeys and sooty mangabey monkeys can be persistently infected with SIV without development of disease. Because it is likely that viral genetic factors determine at least in part the course of disease progression in vivo, it is important to study in more detail the biological differences between the HIV-1 subtypes that constitute the current pandemic.
ACKNOWLEDGMENTS
This study was supported by grants from the Dutch AIDS Fund (AIDS Fonds, Amsterdam, The Netherlands). M.A.-U. is a Socrates exchange student from the University of Barcelona.
We acknowledge Wim van Est for preparation of the artwork.
REFERENCES
- 1.Auersperg N. Long-term cultivation of hypodiploid human tumor cells. J Natl Cancer Inst. 1964;32:135–163. [PubMed] [Google Scholar]
- 2.Bell B, Sadowski I. Ras-responsiveness of the HIV-1 LTR requires RBF-1 and RBF-2 binding sites. Oncogene. 1996;13:2687–2697. [PubMed] [Google Scholar]
- 3.Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Progr Nucleic Acid Res Mol Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 5.Berkhout B, Jeang K T. Detailed mutational analysis of TAR RNA: critical spacing between the bulge and loop recognition domains. Nucleic Acids Res. 1991;19:6169–6176. doi: 10.1093/nar/19.22.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berkhout B, Jeang K T. Functional roles for the TATA promoter and enhancers in basal and Tat-induced expression of the human immunodeficiency virus type 1 long terminal repeat. J Virol. 1992;66:139–149. doi: 10.1128/jvi.66.1.139-149.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berkhout B, Silverman R H, Jeang K T. Tat trans-activates the human immunodeficiency virus through a nascent RNA target. Cell. 1989;59:273–282. doi: 10.1016/0092-8674(89)90289-4. [DOI] [PubMed] [Google Scholar]
- 8.Berkhout B, Verhoef K, van Wamel J L B, Back N K T. Genetic instability of live, attenuated human immunodeficiency virus type 1 vaccine strains. J Virol. 1999;73:1138–1145. doi: 10.1128/jvi.73.2.1138-1145.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carr J K, Salminen M O, Albert J, Sanders-Buell E, Gotte D, Birx D L, McCutchan F E. Full genome sequences of human immunodeficiency virus type 1 subtypes G and A/G intersubtype recombinants. Virology. 1998;247:22–31. doi: 10.1006/viro.1998.9211. [DOI] [PubMed] [Google Scholar]
- 10.Carvalho M, Kirkland M, Derse D. Protein interactions with DNA elements in variant equine infectious anemia virus enhancers and their impact on transcriptional activity. J Virol. 1993;67:6586–6595. doi: 10.1128/jvi.67.11.6586-6595.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cornelissen M, van den Burg R, Zorgdrager F, Goudsmit J. Spread of distinct human immunodeficiency virus type 1 AG recombinant lineages in Africa. J Gen Virol. 2000;81:515–523. doi: 10.1099/0022-1317-81-2-515. [DOI] [PubMed] [Google Scholar]
- 12.Das A T, Klaver B, Berkhout B. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Baar M P, De Ronde A, Berkhout B, Cornelissen M, Van der Horn K H M, Van der Schoot A M, De Wolf F, Lukashov V V, Goudsmit J. Subtype-specific sequence variation of the HIV type 1 long terminal repeat and primer binding site. AIDS Res Hum Retrovir. 2000;16:499–504. doi: 10.1089/088922200309160. [DOI] [PubMed] [Google Scholar]
- 14.Dingwall C, Ernberg I, Gait M J, Green S M, Heaphy S, Karn J, Lowe A D, Singh M, Skinner M A, Valerio R. Human immunodeficiency Virus 1 tat protein binds trans-activating-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci USA. 1989;86:6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dittmar M T, Simmons G, Hibbitts S, O'Hare M, Louisrirotchanakul S, Beddows S, Weber J, Clapham P R, Weiss R A. Langerhans cell tropism of human immunodeficiency virus type 1 subtype A through F isolates derived from different transmission groups. J Virol. 1997;71:8008–8013. doi: 10.1128/jvi.71.10.8008-8013.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Estable M C, Bell B, Hirst M, Sadowski I. Naturally occurring human immunodeficiency virus type 1 long terminal repeats have a frequently observed duplication that binds RBF-2 and represses transcription. J Virol. 1998;72:6465–6474. doi: 10.1128/jvi.72.8.6465-6474.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fauci A S, Desrosiers R C. Pathogenesis of HIV and SIV. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 587–636. [PubMed] [Google Scholar]
- 18.Gao F, Bailes E, Robertson D L, Chen Y, Rodenburg C M, Michael S F, Cummins L B, Arthur L O, Peeters M, Shaw G M, Sharp P M, Hahn B H. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature. 1999;397:436–441. doi: 10.1038/17130. [DOI] [PubMed] [Google Scholar]
- 19.Gao F, Robertson D L, Carruthers C D, Li Y, Bailes E, Kostrikis L G, Salminen M O, Bibollet-Ruche F, Peeters M, Ho D D, Shaw G M, Sharp P M, Hahn B H. An isolate of human immunodeficiency virus type 1 originally classified as subtype I represents a complex mosaic comprising three different group M subtypes (A, G, and I) J Virol. 1998;72:10234–10241. doi: 10.1128/jvi.72.12.10234-10241.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao F, Robertson D L, Carruthers C D, Morrison S G, Jian B, Chen Y, Barre-Sinoussi F, Girard M, Srinivasan A, Abimiku A G, Shaw G M, Sharp P M, Hahn B H. A comprehensive panel of near-full-length clones and reference sequences for non-subtype B isolates of human immunodeficiency virus type 1. J Virol. 1998;72:5680–5698. doi: 10.1128/jvi.72.7.5680-5698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gaynor R B. Regulation of HIV-1 gene expression by the transactivator protein Tat. Curr Top Microbiol Immunol. 1995;193:51–77. doi: 10.1007/978-3-642-78929-8_3. [DOI] [PubMed] [Google Scholar]
- 22.Giacca M, Gutierrez M I, Menzo S, D'adda di Fabrizio F, Falaschi A. A human binding site for transcription factor USF/MLTF mimics the negative regulatory element of human immunodeficiency virus type 1. Virology. 1992;186:133–147. doi: 10.1016/0042-6822(92)90067-y. [DOI] [PubMed] [Google Scholar]
- 23.Grez M, Zörnig M, Nowock J, Ziegler M. A single point mutation activates the Moloney murine leukemia virus long terminal repeat in embryonal stem cells. J Virol. 1991;65:4691–4698. doi: 10.1128/jvi.65.9.4691-4698.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel A E, Kel O V, Ignatieva E V, Ananko E A, Podkolodnaya O A, Kolpakov F A, Podkolodny N L, Kolchanov N A. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang L M, Jeang K T. Increased spacing between Sp1 and TATAA renders human immunodeficiency virus type 1 replication defective: implication for Tat function. J Virol. 1993;67:6937–6944. doi: 10.1128/jvi.67.12.6937-6944.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klasens B I, Huthoff H T, Das A T, Jeeninga R E, Berkhout B. The effect of template RNA structure on elongation by HIV-1 reverse transcriptase. Biochim Biophys Acta. 1999;1444:355–370. doi: 10.1016/s0167-4781(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 27.Klaver B, Berkhout B. Comparison of 5′ and 3′ long terminal repeat promoter function in human immunodeficiency virus. J Virol. 1994;68:3830–3840. doi: 10.1128/jvi.68.6.3830-3840.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koken S E C, van Wamel J L, Goudsmit J, Berkhout B, Geelen J L. Natural variants of the HIV-1 long terminal repeat: analysis of promoters with duplicated DNA regulatory motifs. Virology. 1992;191:968–972. doi: 10.1016/0042-6822(92)90274-s. [DOI] [PubMed] [Google Scholar]
- 29.Leitman D C, Mackow E R, Williams T, Baxter J D, West B L. The core promoter region of the tumor necrosis factor alpha gene confers phorbol ester responsiveness to upstream transcriptional activators. Mol Cell Biol. 1992;12:1352–1356. doi: 10.1128/mcb.12.3.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lole K S, Bollinger R C, Paranjape R S, Gadkari D, Kulkarni S S, Novak N G, Ingersoll R, Sheppard H W, Ray S C. Full-length human immunodeficiency virus type 1 genomes from subtype C-infected seroconverters in India, with evidence of intersubtype recombination. J Virol. 1999;73:152–160. doi: 10.1128/jvi.73.1.152-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 32.Markovitz D M. Infection with human immunodeficiency virus type 2. Ann Intern Med. 1993;118:211–218. doi: 10.7326/0003-4819-118-3-199302010-00010. [DOI] [PubMed] [Google Scholar]
- 33.Maury W. Regulation of equine infectious anemia virus expression. J Biomed Sci. 1998;5:11–23. doi: 10.1007/BF02253351. [DOI] [PubMed] [Google Scholar]
- 34.McKeating J A, McKnight A, Moore J P. Differential loss of envelope glycoprotein gp120 from virions of human immunodeficiency virus type 1 isolates: effects on infectivity and neutralization. J Virol. 1991;65:852–860. doi: 10.1128/jvi.65.2.852-860.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melkonyan H, Sorg C, Klempt M. Electroporation efficiency in mammalian cells is increased by dimethyl sulfoxide (DMSO) Nucleic Acids Res. 1996;24:4356–4357. doi: 10.1093/nar/24.21.4356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Montano M A, Nixon C P, Essex M. Dysregulation through the NF-κB enhancer and TATA box of the human immunodeficiency virus type 1 subtype E promoter. J Virol. 1998;72:8446–8452. doi: 10.1128/jvi.72.10.8446-8452.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Montano M A, Novitsky V A, Blackard J T, Cho N L, Katzenstein D A, Essex M. Divergent transcriptional regulation among expanding human immunodeficiency virus type 1 subtypes. J Virol. 1997;71:8657–8665. doi: 10.1128/jvi.71.11.8657-8665.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 39.Mummaneni P, Yates P, Simpson J, Rose J, Turker M S. The primary function of a redundant Sp1 binding site in the mouse aprt gene promoter is to block epigenetic gene inactivation. Nucleic Acids Res. 1998;26:5163–5169. doi: 10.1093/nar/26.22.5163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Myers G, Korber B, Hahn B H, Jeang K-T, Mellors J H, McCutchan F E, Henderson L E, Pavlakis G N. Human retroviruses and AIDS. A compilation and analysis of nucleic acid and amino acid sequences. Los Alamos, N.Mex: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory; 1995. [Google Scholar]
- 41.Naghavi M H, Schwartz S, Sonnerborg A, Vahlne A. Long terminal repeat promoter/enhancer activity of different subtypes of HIV type 1. AIDS Res Hum Retrovir. 1999;15:1293–1303. doi: 10.1089/088922299310197. [DOI] [PubMed] [Google Scholar]
- 42.Olsen H S, Rosen C A. Contribution of the TATA motif to Tat-mediated transcriptional activation of human immunodeficiency virus gene expression. J Virol. 1992;66:5594–5597. doi: 10.1128/jvi.66.9.5594-5597.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Osborn L, Kunkel S, Nabel G J. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc Natl Acad Sci USA. 1989;86:2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Peden K, Emerman M, Montagnier L. Changes in growth properties on passage in tissue culture of viruses derived from infectious molecular clones of HIV-1LAI, HIV-1MAL, and HIV-1ELI. Virology. 1991;185:661–672. doi: 10.1016/0042-6822(91)90537-l. [DOI] [PubMed] [Google Scholar]
- 45.Pepin J, Morgan G, Dunn D, Gevao S, Mendy M, Gaye I, Scollen N, Tedder R, Whittle H. HIV-2-induced immunosuppression among asymptomatic West African prostitutes: evidence that HIV-2 is pathogenic, but less so than HIV-1. AIDS. 1991;5:1165–1172. [PubMed] [Google Scholar]
- 46.Pope M, Frankel S S, Mascola J R, Trkola A, Isdell F, Birx D L, Burke D S, Ho D D, Moore J P. Human immunodeficiency virus type 1 strains of subtypes B and E replicate in cutaneous dendritic cell-T-cell mixtures without displaying subtype-specific tropism. J Virol. 1997;71:8001–8007. doi: 10.1128/jvi.71.10.8001-8007.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pugh B F. Mechanisms of transcription complex assembly. Curr Opin Cell Biol. 1996;8:303–311. doi: 10.1016/s0955-0674(96)80002-0. [DOI] [PubMed] [Google Scholar]
- 48.Robertson D L, Hahn B H, Sharp P M. Recombination in AIDS viruses. J Mol Evol. 1995;40:249–259. doi: 10.1007/BF00163230. [DOI] [PubMed] [Google Scholar]
- 49.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 50.Ross E K, Buckler-White A J, Rabson A B, Englund G, Martin M A. Contribution of NF-κB and Sp1 binding motifs to the replicative capacity of human immunodeficiency virus type 1: distinct patterns of viral growth are determined by T-cell types. J Virol. 1991;65:4350–4358. doi: 10.1128/jvi.65.8.4350-4358.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rousseau C, Abrams E, Lee M, Urbano R, King M-C. Long terminal repeat and nef gene variants of human immunodeficiency virus type 1 in perinatally infected long-term survivors and rapid progressors. AIDS Res Hum Retrovir. 1997;13:1611–1623. doi: 10.1089/aid.1997.13.1611. [DOI] [PubMed] [Google Scholar]
- 52.Selby M J, Bain E S, Luciw P A, Peterlin B M. Structure, sequence, and position of the stem-loop in tar determine transcriptional elongation by tat through the HIV-1 long terminal repeat. Genes Dev. 1989;3:547–558. doi: 10.1101/gad.3.4.547. [DOI] [PubMed] [Google Scholar]
- 53.Sieweke M H, Tekotte H, Jarosch U, Graf T. Cooperative interaction of Ets-1 with USF-1 required for HIV-1 enhancer activity in T cells. EMBO J. 1998;17:1728–1739. doi: 10.1093/emboj/17.6.1728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Muller-Trutwin M C, Saragosti S, Georges-Courbot M C, Barre-Sinoussi F, Brun-Vezinet F. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4:1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 55.Smith S D, Shatsky M, Cohen P S, Warnke R, Link M P, Glader B E. Monoclonal antibody and enzymatic profiles of human malignant T-lymphoid cells and derived cell lines. Cancer Res. 1984;44:5657–5662. [PubMed] [Google Scholar]
- 56.Soto-Ramirez L E, Renjifo B, McLane M F, Marlink R, O'Hara C, Sutthent R, Wasi C, Vithayasai P, Vithayasai V, Apichartpiyakul C, Auewarakul P, Pena Cruz V, Chui D S, Osathanondh R, Mayer K, Lee T H, Essex M. HIV-1 Langerhans cell tropism associated with heterosexual transmission of HIV. Science. 1996;271:1291–1293. doi: 10.1126/science.271.5253.1291. [DOI] [PubMed] [Google Scholar]
- 57.Takehisa J, Zekeng L, Ido E, Yamaguchi-Kabata Y, Mboudjeka I, Harada Y, Miura T, Kaptué L, Hayami M. Human immunodeficiency virus type 1 intergroup (M/O) recombination in Cameroon. J Virol. 1999;73:6810–6820. doi: 10.1128/jvi.73.8.6810-6820.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tsichlis P N, Lazo P A. Virus-host interactions and the pathogenesis of murine and human oncogenic retroviruses. In: Kung H J, Vogt P K, editors. Retroviral insertion and oncogene activation. Berlin, Germany: Springer-Verlag; 1991. pp. 95–171. [DOI] [PubMed] [Google Scholar]
- 59.Van de Peer Y, De Wachter R. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput Appl Biosci. 1997;13:227–230. doi: 10.1093/bioinformatics/13.3.227. [DOI] [PubMed] [Google Scholar]
- 60.Van Lint C, Amella C A, Emiliani S, John M, Jie T, Verdin E. Transcription factor binding sites downstream of the human immunodeficiency virus type 1 transcription start site are important for virus infectivity. J Virol. 1997;71:6113–6127. doi: 10.1128/jvi.71.8.6113-6127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Verhoef K, Koper M, Berkhout B. Determination of the minimal amount of Tat activity required for human immunodeficiency virus type 1 replication. Virology. 1997;237:228–236. doi: 10.1006/viro.1997.8786. [DOI] [PubMed] [Google Scholar]
- 62.Verhoef K, Sanders R W, Fontaine V, Kitajima S, Berkhout B. Evolution of the human immunodeficiency virus type 1 long terminal repeat promoter by conversion of an NF-κB enhancer element into a GABP binding site. J Virol. 1999;73:1331–1340. doi: 10.1128/jvi.73.2.1331-1340.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wain-Hobson S. More ado about HIV's origins. Nat Med. 1998;4:1001–1002. doi: 10.1038/1986. [DOI] [PubMed] [Google Scholar]
- 64.Wei P, Garber M E, Fang S-M, Fisher W H, Jones K A. A novel CDK9-associated C-type cyclin interacts directly with HIV-1 Tat and mediates its high-affinity, loop-specific binding to TAR RNA. Cell. 1998;92:451–462. doi: 10.1016/s0092-8674(00)80939-3. [DOI] [PubMed] [Google Scholar]