Abstract
Prodigiosin (PG) is a red tripyrrole pigment from the prodiginine family that has attracted widespread attention due to its excellent biological activities, including anticancer, antibacterial and anti-algal activities. The synthesis and production of PG is of particular significance, as it has the potential to be utilized in a number of applications, including those pertaining to clinical drug development, food safety, and environmental management. This paper provides a systematic review of recent research on PG, covering aspects like chemical structure, bioactivity, biosynthesis, gene composition and regulation, and optimization of production conditions, with a particular focus on the biosynthesis and regulation of PG in Serratia marcescens. This provides a solid theoretical basis for the drug development and production of PG, and is expected to promote the further development of PG in medicine and other applications.
Keywords: Prodigiosin, Serratia marcescens, bioactivity, high-level synthesis, biosynthesis
1. Introduction
Prodigiosin (PG), a red pigment with a core structure of tripyrrole rings, represents a secondary metabolite derived from certain sources, including Serratia marcescens, actinomycetes, and marine bacteria. This metabolite exhibits a wide range of biological activities, including anticancer, antimicrobial, antibiofilm, antiparasitic, and antiviral properties. In oceanography, PG has demonstrated effectiveness as an algaecide against harmful algal blooms (HABs) (Zhang et al., 2020). Moreover, in the realm of food production, PG finds utility as antioxidants, additives, colorants, and more (Arivizhivendhan et al., 2018; Sajjad et al., 2018). Significantly, extensive research has been dedicated to exploring the anticancer potential of PG and its derivatives in treating various human cancers, such as small cell lung cancer (Langer et al., 2014) and breast cancer (Wang et al., 2016). With its favorable properties, PG has garnered significant attention across diverse fields.
Within the extensive prodiginine family, PG holds the distinction of being the first discovered and most widely recognized member. Other prodiginine family members, including cycloprodigosin, undecylprodigiosin (UP), metacycloprodigiosin, and streptorubin B, exhibit distinct molecular weights or structures compared to PG but share bioactive functions suitable for medical and clinical applications (Li et al., 2022).
This review provides a comprehensive description of the properties and synthesis of PG, offering insights into future perspectives and establishing a theoretical foundation for synthetic production. The primary emphasis of this review centers on elucidating the regulatory mechanisms of the biosynthetic gene cluster and optimizing production conditions for PG in S. marcescens, distinguishing it from other reviews on the subject.
2. Structures and properties of prodigiosin
2.1. Structure of prodigiosin
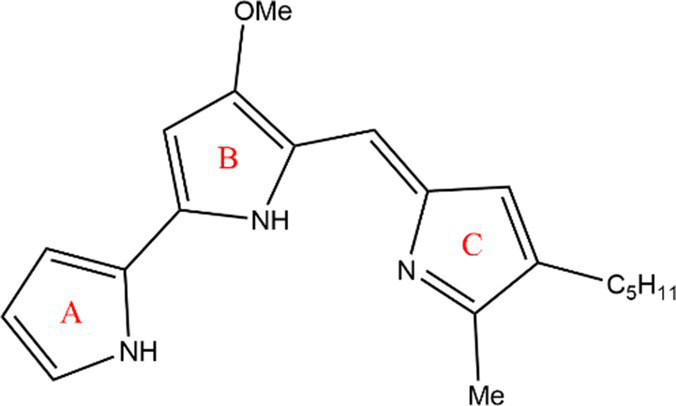
PG is a molecule with a red linear tripyrrole structure, which concludes A, B, and C rings. The three rings are pyrrole, 3-methoxypyrrole, and 2-methyl-3-pentylpyrrole, respectively. Two of the rings, A and B, are linked together to form a bipyrrole structure, and the C ring is connected to the B ring via a methylene group. The structures of some members of prodiginine family are shown in Table 1.
Table 1.
Structures, sources, and bioactivities of some major PGs.
| Name | Structure | Bacterial species | Bioactivity | Reference |
|---|---|---|---|---|
| Prodigiosin |
|
Serratia marcescens Serratia rubidaea Serratia 39,006 Streptomyces coelicolor Streptomyces griseoviridis Vibrio spartinae Janthinobacterium lividum Hahella chejuensis Pseudoalteromonas rubra Zooshikella rubidus Massilia frigida |
Anticancer Antibacterial Antifungal Antialgal Antimalarial Antibiofilm Antioxidant Antiaging Immunosuppressive |
Williamson et al. (2006), Kim et al. (2007), Kawasaki et al. (2009), Schloss et al. (2010), Setiyono et al. (2020), Vitale et al. (2020), Sun et al. (2021), Shaffer et al. (2023), Ma et al. (2024) |
| Undecylprodigiosin |
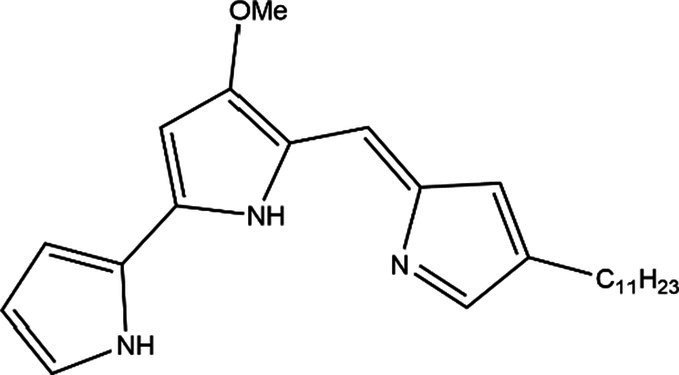
|
Serratia marcescens
Streptomyces coelicolor Streptomyces longispororuber Hahella |
Anticancer Antibacterial Antifungal Antimalarial Antialgal Immunosuppressive |
Williamson et al. (2005), Kim et al. (2007), Stankovic et al. (2014) |
| Decylprodigiosin |
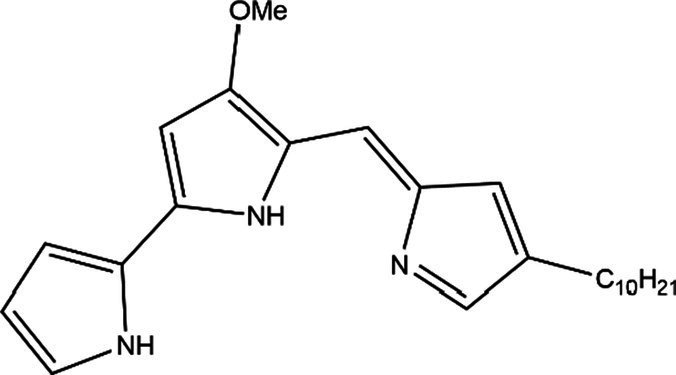
|
Streptomyces violaceoruber | Anticancer Antimicrobial |
Girão et al. (2024) |
| Cycloprodigiosin |
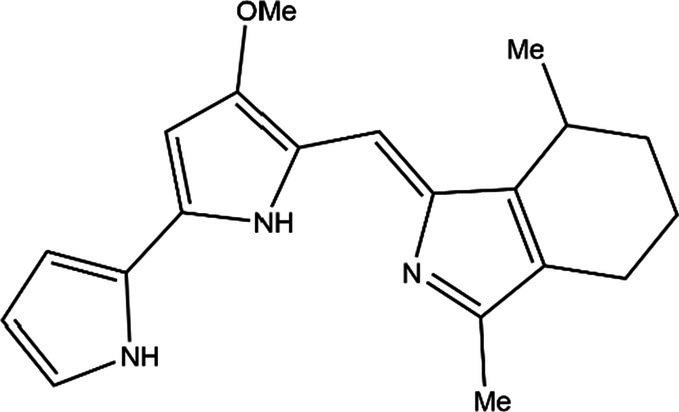
|
Pseudoalteromonas rubra
Zooshikella rubidus Massilia frigida Vibrio spartinae Vibrio gazogenes |
Anticancer Antibacterial Antifungal |
Setiyono et al. (2020), Shaffer et al. (2023) |
| Metacycloprodigiosin |
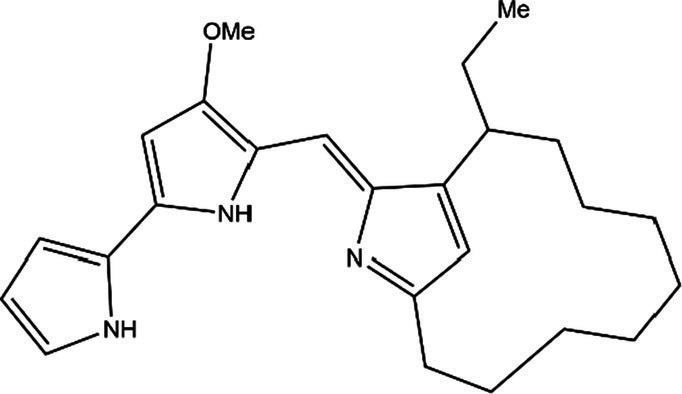
|
Streptomyces longispororuber | Anticancer Antimalarial Immunosuppressive |
Wasserman et al. (1969), Ikeda et al. (2016) |
| Streptorubin B |
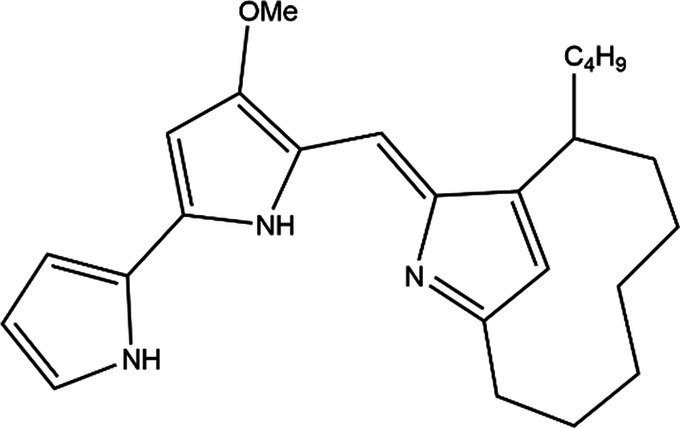
|
Streptomyces coelicolor | Antibacterial Antimalarial |
Yi et al. (2022) |
| Prodigiosin R1 |
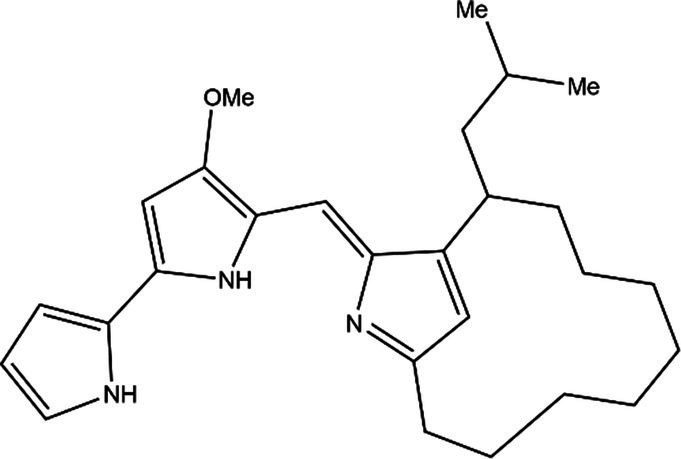
|
Streptomyces griseoviridis | Pending Research | Kawasaki et al. (2008) |
| Prodigiosin R2 |
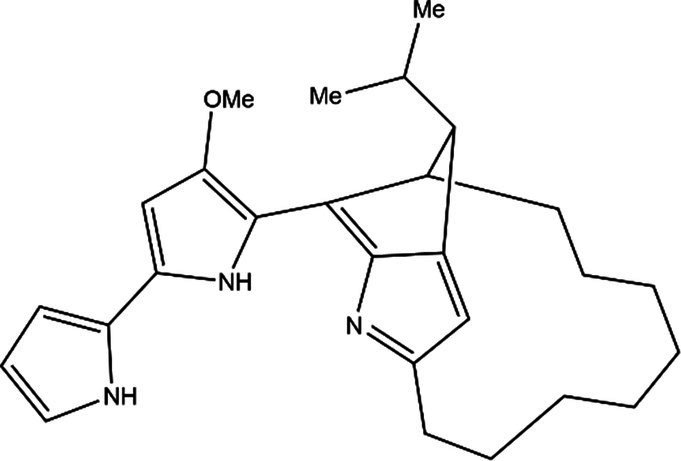
|
Streptomyces griseoviridis | Anticancer | Kimata et al. (2018) |
| Prodigiosin R3 |
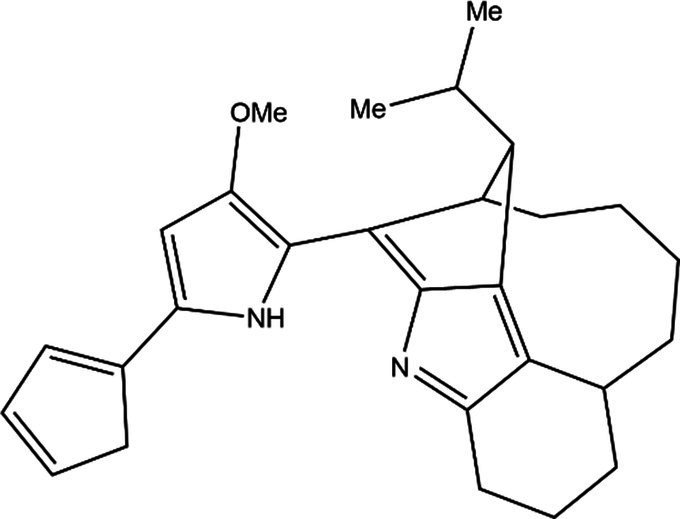
|
Streptomyces coelicolor | Anticancer | Kimata et al. (2022) |
In general, prodiginine can be divided into two main parts: linear and cyclic prodiginine. The main difference in linear prodiginine is the length of the carbon chain of the substituent group in the C-ring. The cyclic prodiginine can be divided into two subgroups, one of which forms a ring on the C-ring, such as cycloprodigiosin and Streptorubin B; Another subgroup is connecting the A ring with the C ring to form a ring, such as cyclononylprodigiosin and methylcyclooctylprodigiosin (Hu et al., 2016).
2.2. Physical properties of prodigiosin
The physical properties of PG play a crucial role in its application and identification during production. Despite being insoluble in water, PG exhibits moderate solubility in ether and alcohol, and can be dissolved in chloroform, methanol, acetonitrile, and dimethyl sulfoxide. The sensitivity of PG to light poses challenges in commercial dye applications (Someya et al., 2004). Spectrophotometric analysis under different pH conditions reveals that PG can manifest as either red (with a sharp peak at 535 nm) in acidic media or orange-yellow (with a broad curve centered on 470 nm) in alkaline environments (Jehlička et al., 2021). However, since PG analogs also have similar absorbance, this information is not sufficient for PG identification, so more sophisticated assays are required, such as the GC–MS, LC–MS or characteristic bands using RAMAN spectroscopy. Mass spectrometry analysis revealed that the mass-to-charge ratio(m/z) of PG was 324.2([M + H+]), and the molecular weight of PG is 324 Da. Rf values ranging from 0.9 to 0.95 are a distinctive feature of PG (Song et al., 2006; Bhagwat and Padalia, 2020).
2.3. Bioactivity of prodigiosin
2.3.1. Anticancer activity
Cancer, a non-communicable disease, is a major global cause of death, claiming nearly 10 million lives in 2020, making up about one-sixth of all deaths. Predominant cancers include breast, lung, colon, rectum, and prostate, with lung, colon, rectal, liver, stomach, and breast cancers showing the highest mortality rates (Sung et al., 2021). Effective cancer therapies are crucial for human health, with natural compounds being closely studied for medicinal potential.
PG has shown strong anti-cancer effects, with proven efficacy in treating melanoma, breast cancer, lung cancer, lymphocytic leukemia, and glioblastoma (Pan et al., 2012; Brown et al., 2015; Wang et al., 2016; Cheng et al., 2018; Chiu et al., 2018; Espona-Fiedler et al., 2022). It triggers apoptosis in cancer cells, including multidrug-resistant ones, with minimal toxicity to normal cells (Elahian et al., 2013). Obatoclax, a prodigiosin derivative, displays potent anti-cancer and pro-apoptotic properties. Clinical trials have demonstrated its effectiveness in treating relapsed chronic lymphocytic leukemia and extensive-stage small cell lung cancer, either alone or in combination with other drugs, resulting in positive therapeutic outcomes (Langer et al., 2014; Brown et al., 2015).
PG’s anti-cancer effects involve diverse mechanisms, with a key role played by promoting apoptosis in tumor cells. Apoptosis can be initiated through three pathways: the extrinsic (death receptor) pathway, the intrinsic (mitochondrial) pathway, and the endoplasmic reticulum (ER) pathway. PG primarily influences cellular apoptosis via the intrinsic (mitochondrial) pathway, which is regulated by the Bcl-2 protein family comprising both pro-apoptotic (Bax, Bak, Bid) and anti-apoptotic (Bcl-2, Bcl-W, Mcl-1) proteins. The balance between these proteins determines the initiation of cellular apoptosis (Wong, 2011). PG and Obatoclax bind to the BH3 domain of anti-apoptotic proteins, shifting the balance toward cell death by affecting the pro- and anti-apoptotic proteins. Experimental evidence in melanoma cells shows that PG disrupts the Mcl-1/Bak complex, leading to Bak release and activating the intrinsic apoptotic pathway (Hosseini et al., 2013). PG also induces apoptosis by altering pH levels, DNA cleavage, cell cycle arrest, and other mechanisms (Darshan and Manonmani, 2015).
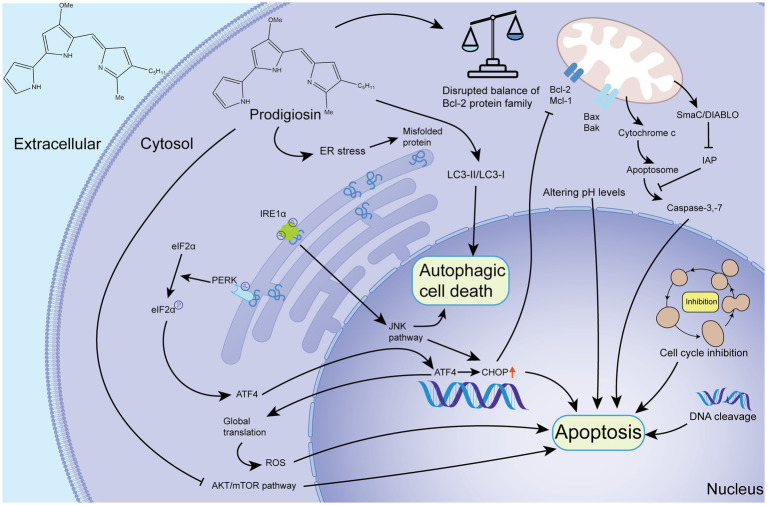
Research on the ER pathway of PG has revealed its role in inducing ER stress, which can trigger cell apoptosis through the IRE1α-JNK and PERK-eIF2α-ATF4-CHOP axes (Wang and Kaufman, 2016). PG induces ER stress by downregulating Bcl-2 and activating the PERK-eIF2α-ATF4-CHOP pathway or the IRE1α-JNK pathway in the ER (Pan et al., 2012). Furthermore, the ER pathway also contributes to autophagic cell death, as demonstrated in a study on human glioblastoma cells where PG activated the JNK pathway and inhibited the AKT/mTOR pathway, leading to autophagic cell death through ER stress (Cheng et al., 2018). A schematic of the mechanisms involved in apoptosis and autophagic cell death is shown in Figure 1.
Figure 1.
Schematic diagram of the mechanism of PG-mediated apoptosis and autophagic cell death. Upon entering the cell, PG can induce apoptosis by disrupting the balance of Bcl-2 family proteins and initiating the mitochondrial pathway. It can also induce apoptosis through the alteration of pH, cell cycle inhibition, and DNA cleavage. Additionally, PG can cause ER stress and autophagic cell death and apoptosis through the IRE1α-JNK and PERK-eIF2α-ATF4-CHOP pathway. The ATF4-CHOP pathway is responsible for autophagic cell death and apoptosis. In addition to the aforementioned pathways, PG can inhibit the AKT/mTOR pathway to cause apoptosis. Furthermore, PG can affect LC3 protein, which indicates that PG can cause autophagic cell death. IAP, inhibitor of apoptosis protein; ROS, reactive oxygen species; Smac, second mitochondrial-derived activator of caspases; DIABLO, director inhibitor of apoptosis-binding protein with Low pI (O’Brien and Kirby, 2008; Wong, 2011).
Inhibiting tumor cell migration and motility is essential for achieving anti-cancer effects. Research by Margarita Espona-Fiedler and her team found that PGs effectively reduce the adhesive capability of melanoma cells, impeding their ability to colonize distant organs and preventing metastasis (Espona-Fiedler et al., 2022). PG treatment also decreases the adhesive capacity of metastatic lung cancer cells and reduces filopodia formation in melanoma cells, which are crucial for cancer cell migration and proliferation. Furthermore, PG treatment downregulates genes associated with migration and invasion, such as Twist-1, MMP-1, MMP-2, and RhoA (Zhang et al., 2005).
2.3.2. Antimicrobial activity
Bacterial hazards have far-reaching impacts on human health, agriculture, the environment, and food safety, leading to infectious diseases, agricultural losses, environmental pollution, and antibiotic resistance. For instance, Staphylococcus aureus can result in food poisoning; Candida albicans can cause oral mucosal disease. It is essential to control the spread of antibiotic resistance to protect health, the environment, and food safety. Misuse of antibiotics has made treating infectious diseases difficult, emphasizing the need for new antibiotic discovery and development.
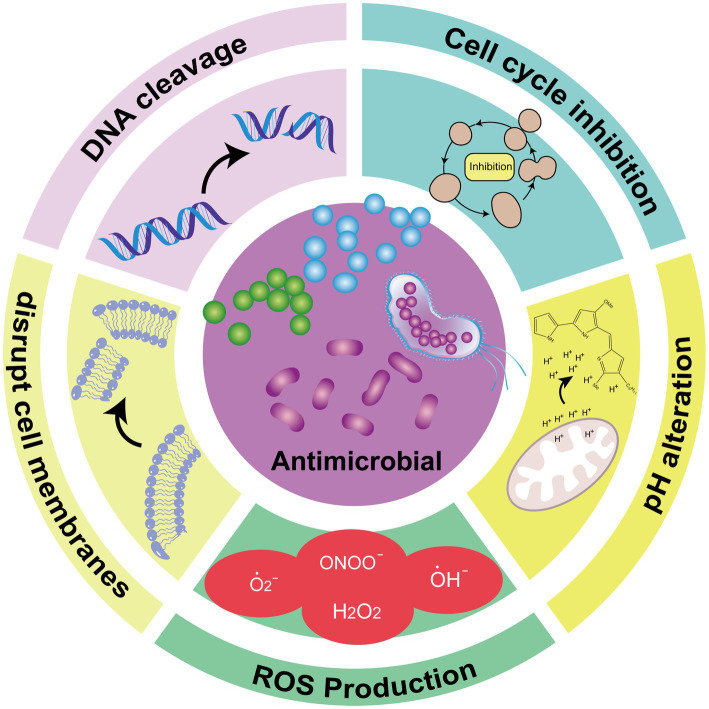
PGs exhibit broad-spectrum antimicrobial activity against a wide range of bacteria and fungi, including Enterococcus faecalis, Staphylococcus aureus, Streptococcus pyogenes, Salmonella typhimurium, Escherichia coli, Bacillus subtilis, and Candida albicans (Lapenda et al., 2015). However, compared to Gram-positive bacteria, PG’s inhibitory effects on Gram-negative bacteria are less pronounced. Research shows that cyclic PGs offer superior antibacterial effects compared to linear PGs (Lee et al., 2011). The lipophilic nature of PGs allows them to disrupt cell membranes through various mechanisms, leading to bacterial death (Ravindran et al., 2020). Additional antimicrobial properties of PGs include pH alteration, DNA cleavage, cell cycle inhibition, and induction of reactive oxygen species (ROS) production (Yip et al., 2019; Choi et al., 2021) (Figure 2). Due to this antimicrobial property, PG has been designed for use in food packaging. Amorim et al. added PG to the outer and inner layers of food packaging materials with good results, although it should be noted that PG is sensitive to light, and the stability of the outer layer under light conditions deserves to be studied (Amorim et al., 2022).
Figure 2.
Schematic diagram of the PG antimicrobial mechanism (Francisco et al., 2007).
2.3.3. Anti algae bloom
HABs pose a threat to aquatic ecosystems, marine life, and human health due to the toxins they produce. These blooms, fueled by nutrient overload, can result in oxygen depletion, fish kills, and disruptions in marine food chains. The economic impact includes losses in fisheries and tourism from beach closures and water quality problems. Effective monitoring and management strategies are crucial to mitigate these adverse effects. PG demonstrates effectiveness in controlling various algae species, including Prorocentrum donghaiense (Chen et al., 2021), red tide dinoflagellates (Jeong et al., 2005), Heterosigma akashiwo (Zhang et al., 2020), Phaeocystis globosa (Zhang et al., 2016), and Microcystis aeruginosa (Zhang et al., 2022), without harming non-target organisms such as fish. Its mechanism of action involves inducing ROS production and lipid peroxidation while inhibiting the cell cycle of harmful algae. This targeted approach makes PG a practical and effective solution for HAB mitigation (Zhang et al., 2016; Chen et al., 2021).
3. Synthesis of prodigiosin
PG can be produced through biosynthesis or chemical synthesis. Biosynthesis occurs inside specific bacteria like Serratia and Streptomyces, utilizing enzymes and genes for a natural and sustainable process suitable for large-scale production. In contrast, chemical synthesis involves a more complex multi-step organic process, offering precise control over the PG structure but potentially higher costs and operational complexity. When choosing a synthesis method, factors to consider include production scale, cost-effectiveness, and efficiency. This article provides an in-depth examination of PG biosynthesis for practical applications (Hu et al., 2016).
3.1. Biosynthesis of prodigiosin
The biosynthesis of PG was first articulated in 1966 and PG was synthesized by enzymatic condensation reaction of 2-methyl-3-pentylpyrrole (MAP) and 4-methoxy-2,2′-bipyrrole-5-carbaldehyde (MBC) (Morrison, 1966). Williams also revealed that PG’s precursors are derived from a number of amino acids and acetates. Among them, Wasserman’s study confirmed that MAP is derived from 2-octenal and pyruvate while MBC biosynthesis starts from L-proline and malonyl coenzyme A (Wasserman et al., 1973; Williams, 1973).
In the biosynthesis of PG, certain peptide products are produced via nonribosomal peptide synthesis (NRPSs) rather than ribosomes. NRPSs function by sequentially adding substrates to the peptide chain through a multienzyme complex utilizing the thiotemplate mechanism. These NRPSs comprise three key structural domains: adenylation domains for substrate selection and activation to amide-AMP, peptidyl carrier protein (PCP) domains for immobilizing activated substrates as thioesters to 4′-phosphopantetheine (Ppant), and condensation domains catalyzing the formation of peptide bonds between donor polypeptides and acceptor amino acids (Süssmuth and Mainz, 2017).
3.1.1. Genes responsible for PG synthesis
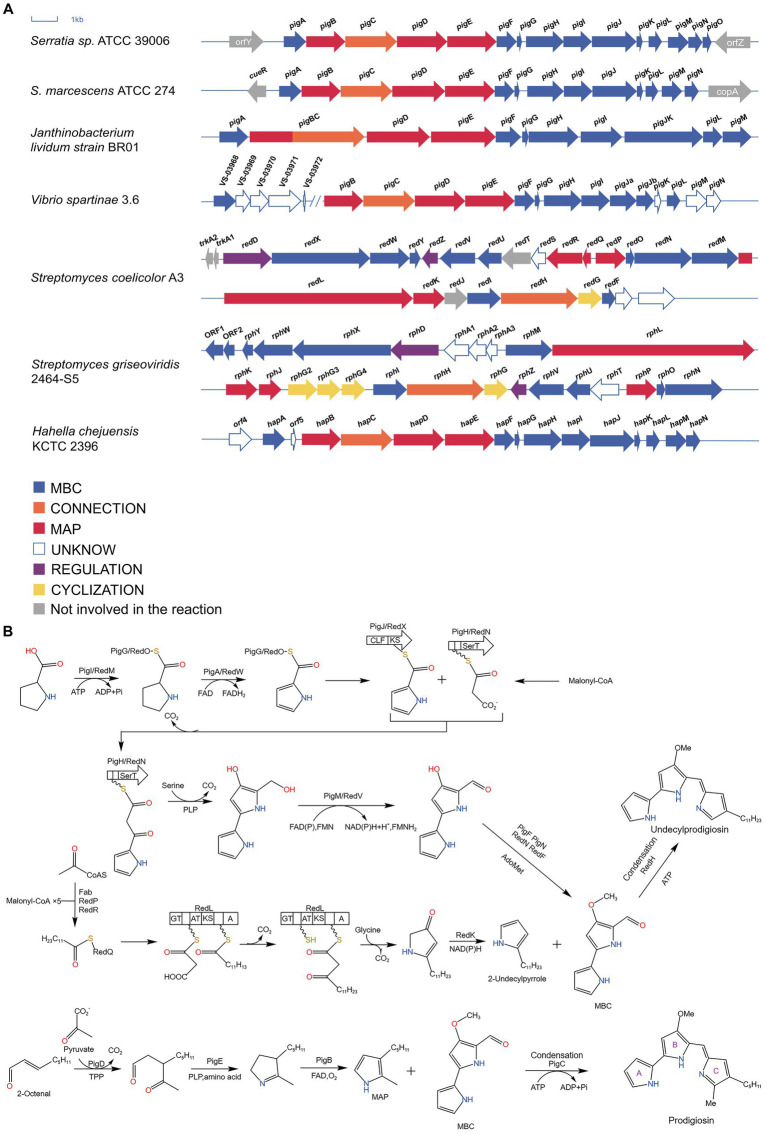
In 2004, Harris and his team cloned, sequenced, and expressed microorganisms that produce PG in heterologous hosts, including S. marcescens ATCC 274 (Sma274) and Serratia sp. ATCC 39006 (S39006). (Harris et al., 2004). Subsequently, Hahella chejuensis KCTC 2396 and Janthinobacterium lividum strain BR01 (JliBR01) were sequenced in 2006 and 2010, respectively, (Kim et al., 2006; Schloss et al., 2010). In 2020, through genomics and metabolomics, a new branch of PG production was discovered in marine Vibrio spartinae 3.6 (Vitale et al., 2020). The various microorganisms described above all contain 13–15 genes related to PG, while two Streptomyces species, Streptomyces coelicolor A3 and Streptomyces griseoviridis 2464-S5, have more complicated homologous gene clusters (Cerdeno et al., 2001; Kawasaki et al., 2009). The relevant gene clusters are shown in Figure 3.
Figure 3.
Biosynthesis of prodigiosin. (A) Clusters of genes related to the synthesis of PG in several microorganisms. Blue and red arrows indicate genes that synthesize MBC and MAP, respectively; orange arrows indicate genes that link MBC to MAP; yellow arrows indicate genes involved in ring formation; purple arrows indicate genes involved in regulation; and white and gray arrows indicate genes not involved in the synthesis reaction and unknown genes, respectively (Cerdeno et al., 2001; Harris et al., 2004; Kim et al., 2006; Kawasaki et al., 2009; Schloss et al., 2010; Vitale et al., 2020). (B) The biosynthetic pathway of PG (Williamson et al., 2006; Kim et al., 2007).
3.1.2. Biosynthesis of MAP
2-octenal, derived from the production of fatty acid biosynthesis enzymes or the autoxidation of unsaturated fatty acids, is a precursor for MAP synthesis. It reacts with pyruvate to form 3-acetyloctal under the action of pigD and thiamine pyrophosphate (TPP). pigD is homologous to acetolactate synthase and can catalyze the decarboxylation of pyruvate, binding the decarboxylated two-carbon fragment to 2-Octenal; then, pigE, which has transamination effect, acts on 3-acetyloctal to produce aminoketones, which self-cyclize to form H2MAP. Finally, H2MAP undergoes oxidative dehydrogenation to generate MAP (Williamson et al., 2005).
3.1.3. Biosynthesis of 2-undecylpyrrole
RedP condenses acetyl coenzyme A and malonyl redQ (ACP) to produce an acetoacetyl thioester with redQ attached; the carbonyl group is then reduced and dehydrated by the ketoreductase, dehydratase, and enoyl reductase of S. coelicolor fatty acid synthase (FAS) to produce butyryl ACP; the remaining four malonyl-redQ are catalyzed the subsequent extension step by redR, which transfers the resulting dodecyl group to the ACP structural domain of redL, accompanied by reduction and dehydration by FAS reductase; the other ACP attached to redL is then decarboxylated in the presence of ketosynthase, generating a β-ketomyristoyl thioester. The decarboxylation of β-ketomyristoyl thioester via pyridoxal phosphate (PLP) mediated condensation of glycine with an ACP-bound thioester releases CO2, and this intermediate is cyclized to produce 4-keto-2-undecylpyrroline. The final step is the dehydrogenation and reduction of 4-keto-2-undecylpyrroline by redK to yield 2-undecylpyrrole.
3.1.4. Biosynthesis of MBC
pigI/redM activates L-proline with ATP, and then transfers it to the adjacent PCP (pigG/redO) domain to form L-prolinyl-S-PCP; L-proline-S-PCP was then oxidized and dehydrogenated to obtain pyrrole-2-carboxy-S-PCP under the action of pigA/redW. Pyrrole-2-carboxy-S-PCP was transferred from pigG/redO to cysteine with pigJ/redX active centers to obtain pyrrole-2-carboxy-S-pigJ/redX; at the same time, the malonyl group is transferred from malonyl CoA to the 4′-phosphophenylene of an ACP in pigH/redN, and then the complex decarboxylates and attacks the thioester bond of pyrrole-2-carboxy-S-pigJ/redX, generating a pyrrole-β-ketothioester attached to pigH/redN. In the presence of pigH/redN pyrrole-β-ketothioester reacts with serine to form 4-hydroxy-2,2′-bipyrrole-5-methanol (HBM). The hydroxyl group of HBM was oxidized to form an aldehyde group in the presence of pigM/redV to give 4-hydroxy-2,2′-bipyrrole-5-carbaldehyde (HBC). Finally, HBC is methylated in the presence of pigF and pigN or redN and redF to generate MBC.
3.1.5. Condensation
pigC and can catalyze the condensation of MAP and MBC, and redH, a homolog of pigC, catalyzes the condensation of 2-undecylpyrrole and MBC to produce PG and UP, respectively. The understanding of the process of PG biosynthesis as well as the genes facilitates our use of microorganisms to synthesize PG in large quantities and to conduct research on PG.
3.2. Regulation of synthetic gene cluster
The transcription process of synthesizing PG is jointly regulated by multiple factors and has a relatively.
complex regulatory mechanism, including quorum sensing (QS) system, two component regulatory systems (TCSs) and various regulatory factors. The bacterial regulatory system QS, which responds to changes in bacterial density, is gaining attention. It is a communication process between bacterial cells involving the production, release, accumulation, and response to autoinducers (AI), extracellular signaling molecules (Mukherjee and Bassler, 2019). Gram-negative bacteria, like Serratia, utilize QS to regulate gene expression using N-acyl homoserine lactones (acyl-HSLs) as AI (AI-1) (Wei and Lai, 2006). In Serratia sp. ATCC 39006, PG production is regulated by QS through the LuxIR homologs SmaI/SmaR and SpnI/SpnR. SmaR inhibits the synthesis of PG by inhibiting the transcription of PG gene clusters, while SmaI can synthesize C4-HSL and bind to the transcription inhibitor SmaR. However, at low cell density, the synthesis of C4-HSL is insufficient to relieve the inhibitory effect of SmaR on transcription, and PG production is inhibited; When the cell density is high, the concentration of C4-HSL reaches the threshold and binds to SmaR, reducing the inhibitory effect and increasing PG production. A similar system is observed with SpnI/SpnR. Besides AI-1’s role in regulating PG synthesis, LuxS/AI-2 can also regulate PG synthesis. The LuxS enzyme catalyzes bacterial methyl metabolism, producing AI-2 as a metabolic byproduct. In S. marcescens 274, researchers have demonstrated that the PG yield significantly decreases in luxS mutants compared to the wild-type S. marcescens 274. Subsequently, through transversely expressing LuxS, it was proven that the direct cause of reduced PG production is the absence of LuxS (Coulthurst et al., 2004). Hence, AI-1 and AI-2 can be ingeniously employed in the process of PG synthesis to enhance PG yield. TCSs are composed of two proteins, namely transmembrane sensor histidine kinases (HK) and response regulatory (RR) factors, which are widely distributed in the bacterial community and have important roles in regulating intercellular communication and secondary metabolism. The basic mechanism of this system is that when HK recognizes a specific signal, its conserved histidine residue undergoes self-phosphorylation, and then the phosphate group is transferred to the conserved aspartic acid residue of RR. RR phosphorylation can promote specific gene transcription (Zschiedrich et al., 2016). The sensing kinase GacS and response regulatory factor GacA are typical TCS that are widely present in Gram negative bacteria. They regulate bacterial QS, secondary metabolism, biofilm formation, and movement through Gac/Rsm signaling pathways, including PG synthesis (Song et al., 2023). The PigQ/PigW mentioned earlier is a homolog of GacA/GacS and participates in PG synthesis. In addition, there are also EepR/EepS (Stella et al., 2015), PhoB/PhoR (Gristwood et al., 2009), RssB/RssA (Horng et al., 2010), RcsB/RcsC (Pan et al., 2021), BarA/UvrY (Liu et al., 2023), EnvZ/OmpR (Jia et al., 2022), and CpxA/CpxR (Sun et al., 2020) involved in the synthesis of PG. Among them, RssB/RssA, RcsB/RcsC and BarA/UvrY are negatively regulated and can inhibit PG synthesis; the rest are positively regulated.
In addition to these two systems, various regulatory factors can control the transcription of gene clusters. The σ factor is an important subunit in the RNA polymerase holoenzyme, and the core enzyme lacking the σ factor has almost no RNA synthesis activity, as the role of this factor is to recognize the transcription start point. RpoS is a typical σ factor in Gram-negative bacteria, and its influence on PG synthesis varies across different bacterial model experiments. In the study of Serratia sp. ATCC 39006, it was found that RpoS can negatively regulate the synthesis of PG; while in the study of S. marcescens 1912768R, RpoS can positively regulate the synthesis of PG. The specific mechanisms for these two cases are still unclear, but it can be demonstrated that the regulation of PG by RpoS is diverse. The reasons for this diversity may be attributed to differences in gene clusters or their association with QS (Wilf and Salmond, 2012; Qin et al., 2020). In addition to the σ factor, researchers have also investigated the ω factor, which also belongs to RNA polymerase. The rpoZ gene encodes the synthesis of the ω factor, playing a crucial role in UP synthesis and in the absence of the rpoZ gene, the production of UP decreases considerably, which can be partially restored by the supplementation of the rpoZ gene (Santos-Beneit et al., 2011).
Fumarate and nitrate reduction regulatory protein (Fnr) is a global regulatory factor of the Crp/Fnr family that controls gene transcription. At the transcriptional level, Fnr has a negative impact on the production of PG in Serratia sp. ATCC 39006 under aerobic conditions. There is a Fnr binding site between the −10 and − 35 regions of pigA, which inhibits the synthesis of PG by disrupting the binding of the promoter and RNA polymerase (Sun et al., 2021).
Stringent starvation protein A (SspA) is an RNA polymerase-associated regulatory protein. While studying Pseudoalteromonas sp. strain R3, the researchers found that strains lacking SspA are unable to produce prodiginine. The reason for this is related to the iron carrier-dependent iron uptake pathway: SspA positively regulates the expression of the siderophore biosynthetic gene (pvd) cluster. Its absence hinders the uptake of iron through the iron carrier-dependent pathway, leading to iron deficiency and subsequently affecting prodiginine synthesis (Yin et al., 2021).
The PtrA protein is transcribed from the ptrA gene, which promotes transcription of the pig gene in S. marcescens FZSF02. PsrA is a transcriptional regulator belonging to the LysR family. It binds directly to the promoter region of the pig operon at a regulatory binding site (RBS) and an activating binding site (ABS) in the promoter region of the pig manipulator. This positive regulation leads to an increase in PG transcription in S. marcescens. Both were detected by real-time quantitative PCR (RT-qPCR) and there was an increase in gene expression in pigA-pigN (Pan et al., 2022; Lin et al., 2023).
3.3. Factors affecting synthesis
PG is a secondary metabolite that emerges in the advanced growth stages, with its production susceptibility to an array of factors such as light, temperature, pH level, microbial characteristics, and culture medium composition. Efforts are continuously focused on investigating the most favorable production conditions and refining production techniques to enhance PG yields.
It is well known that PG is light-sensitive and demonstrates instability in illuminated environments (Someya et al., 2004). Consequently, the yield fluctuates with light intensity, resulting in a lower yield under light conditions compared to dark conditions (Ryazantseva et al., 2012). Wang et al. elucidated that PG absorbs light and causes phototoxicity on bacterial cell membranes, leading to PG leakage (Wang et al., 2013).
Temperature affects PG synthesis in two main ways. Firstly, the activity of the condensing enzyme produced by pigC. Lower temperatures below 22°C reduce enzyme activity, leading to decreased PG production, while higher temperatures above 30°C render the enzyme inactive, halting PG synthesis. Secondly, the cpx (TCS) in Gram-negative bacteria is vital in temperature-dependent PG regulation. At elevated temperatures, the sensor HK protein CpxA is activated, triggering a phosphorylation cascade that ultimately inhibits transcription of pig gene clusters and reduces PG production (Sun et al., 2020). Additionally, pH levels also impact pigment deposition, with pH 7–9 being optimal, while extremes of below 3.0 or above 10.0 are unfavorable (Sumathi et al., 2014).
The level of dissolved oxygen in the culture medium has a profound effect on PG production and agitation and aeration are important for dissolved oxygen levels (Heinemann, 1970). Research by C. Sumathi et al. demonstrated that the most optimal yield of PG was achieved at a stirring rate of 200 r.p.m. and aeration of 3 vvm. Both too high and too low rates of stirring and aeration led to decreased PG production (Sumathi et al., 2014). While previous studies suggested a high oxygen transfer rate (OTR) was beneficial for PG production, recent findings during UP synthesis indicated that lower OTRs actually increased yield. This is attributed to oxygen limitation prompting a shift from primary to secondary metabolism, ultimately enhancing production (Gamboa-Suasnavart et al., 2018). Therefore, identifying the correct stirring and aeration rates to achieve an optimal OTR is essential for maximizing PG yield. Negative feedback regulation impacts PG production in batch cultures, prompting the development of various bioreactors to improve yields. Extraction columns using HP-20 adsorbent resin demonstrated a 31% increase in pigment production compared to traditional batch fermentation methods (Kim et al., 2000; Song et al., 2006). The aforementioned optimizations for production conditions are based on Serraria and may not be applicable to the remaining strains that produce PG.
Researchers have harnessed genetic engineering techniques or optimized culture conditions to maximize PG production. In the realm of genetic engineering, researchers possess the ability to selectively target and modify the genes of the host cell to obtain the desired genotype.
Firstly, we can knock out or modify genes that are unfavorable for PG production or enhance the expression of genes favorable for PG production to achieve higher yields. Serratia sp. ATCC 39006 has a gene called ser39006_013370 can encode a regulatory protein Fnr. Di Sun et al. inserted mini-Tn5 transposon into ser39006_ In 013370, the production of PG per cell unit was higher than that of the wild-type. It is worth noting whether changing the gene structure will have an impact on the normal life activities of bacteria. In addition to playing a negative regulatory role in PG synthesis, Fnr regulators also regulate carbapenem biosynthesis and flagellar genes. For the former, Fnr negatively regulates carbapenem biosynthesis through CarR, a gene that can encode a pathway specific activator of bacteriostatic carbapenems; For the latter, Fnr can enhance the expression of flagellar genes, which is conducive to bacterial motility. Therefore, it can be seen that knocking out or modifying a gene has both advantages and disadvantages for the bacteria, and the appropriate method should be selected according to the actual situation (Sun et al., 2021). In one study, researchers introduced clusters of biosynthetic genes encoding PGs into Pseudomonas putida and used genome editing tools to make auxiliary proteins required for polyketide synthases (PKSs) and NRPSs more available, leading to increased PG production (Cook et al., 2021).
Secondly, we can introduce genes related to PG production into suitable host cells for mass production. The ligase encoded by pigC is the key enzyme that links MAP and MBC, and the synthesis rate and amount of this enzyme are important for PG production. Researchers introduced the pigC gene into E. coli, which greatly increased the expression of E. coli pigC through self-induction and a good induction strategy, as well as increased PG production due to pigC’s function of linking MAP and MBC (You et al., 2018).
In terms of optimizing cultivation conditions, a large number of researchers have conducted research from aspects such as medium composition, production conditions, and microbial species. Culture conditions that result in a relatively high increase in PG yield are summarized in Table 2. Before large-scale synthesis, optimal production conditions are typically determined experimentally in the laboratory, encompassing the identification of the optimal temperature, pH, carbon and nitrogen sources, and their suitable ratio (Kurbanoglu et al., 2015). Researchers have conducted extensive studies on the media for PG production, starting with commercial media such as nutrient broth, Luria-Bertani medium, and peptone-glycerol broth. Adding raw materials for the synthesis of MBC and MAP to these media can enhance PG synthesis, such as proline and fatty acids. Later on, researchers found that there is a great potential for the production of medium composed of inexpensive agricultural and fishery by-products, such as peanut seed, peanut oil (Giri et al., 2004), potato starch, cassava (de Araujo et al., 2010), brown sugar (Aruldass et al., 2014), crab shells, squid pens (Nguyen et al., 2019), and shrimp shells (Nguyen et al., 2021). These high-yield, low-cost, and environmentally significant substrates are considered the focus of large-scale PG synthesis (Bhagwat and Padalia, 2020; Wang et al., 2020). For example, in the production of PG using Tannery Fleshing, the yield was around 47 mg/mL under optimum production conditions (Sumathi et al., 2014); the highest yield of 27.65 mg/mL was obtained when using soybean oil as a carbon source and peptone as a nitrogen source with a C/N of 100/10 (Liu et al., 2021).
Table 2.
Summary of the optimization of production conditions.
| Strains | Medium composition | Temperature (°C) | PH | Yield (mg/L/h) | References |
|---|---|---|---|---|---|
| S. marcescens UCP 1549 | Solid medium; 6% cassava wastewater supplemented with 2% mannitol | 28 | 7 | 1031.25 PG | de Araujo et al. (2010) |
| S. marcescens UCP 1549 | SSF; WB impregnating salts solution containing 5% WSO | 28 | ND | 499.17 PG | Dos Santos et al. (2021) |
| S. marcescens NPLR1 | Fermentor; Salt solution containing 3% Tannery Fleshing | 30 | 8 | 1175.00 PG | Sumathi et al. (2014) |
| S. marcescens WT | Erlenmeyer flask; 2% powdered peanut seed in distilled water | 28 | 7 | 1076.38 PG | Giri et al. (2004) |
| S. marcescens WT | Erlenmeyer flask; 1 g/L potato starch powder, 0.6 g/L casein and 1 g/L yeast extract | 30 | 7 | 100.00 PG | Suryawanshi et al. (2014) |
| H. chejuensis (KCTC 2396) | Bioreactor; marine broth | 25 | 6.89 | 452.38 PG | Jeong et al. (2021) |
| S. marcescens TKU011 | Erlenmeyer flask; 1% α-chitin, 0.6% casein, 0.05% K2HPO4, and 0.1% CaSO4 | 25 | 5.65–6.15 | 96.25 PG | Nguyen et al. (2019) |
| S. marcescens FZSF02 | Erlenmeyer flask; glucose, peanut powder, beef extract, and olive oil | 26 | 7 | 214.20 PG | Lin et al. (2019) |
| S. marcescens C3 | Erlenmeyer flask; starch/peptone = 16 g/L/10.67 g/L, trace elements and alginate beads. | 30 | 7 | 216.67 PG | Chen et al. (2013) |
|
S. marcescens TNU01 |
Bioreactor; 1.12% de-SSP/0.48% casein and 0.02% K2SO4 and 0.05% K2HPO4 | 27 | 7 | 775.00 PG | Nguyen et al. (2021) |
|
S. marcescens TNU02 |
Bioreactor; 1.12% de-CSP/0.48% casein and 0.02% (NH4)2SO4, 0.1% K2HPO4 | 27 | 6.15 | 637.50 PG | Nguyen et al. (2020) |
| S. marcescens SS-1 | Erlenmeyer flask; YE medium and 10 g/L Proline | 30 | 8 | 104.17 UP | Wei et al. (2005) |
| S. marcescens WT | Erlenmeyer flask; powdered peanut seed broth |
28 | 7 | 783.33 PG | Shahitha (2012) |
| S. marcescens UTM1 | Bioreactor; 10% brown sugar | 25 | 7 | 337.88 PG | Aruldass et al. (2014) |
|
S. marcescens BWL1001 |
Erlenmeyer flask; Soybean oil 100 g/L, peptone 10 g/L | 28 | 5 | 768.06 PG | Liu et al. (2021) |
SSF, solid-state fermentation; WB, wheat bran; WSO, waste soybean oil; ND, data not available; WT, wild type; de-SSP, demineralized shrimp shell powders; YE, yeast extract; UP, undecylprodigiosin.
4. Discussion
The discovery of prodigiosin (PG) has sparked interdisciplinary research interest in its bioactivities, such as its chemical makeup, bioactivity mechanisms, synthetic pathways, regulation, and production optimization within the prodiginine family. The identification of new PG members has highlighted their widespread presence in nature, inspiring further investigation into their bioactivity and potential for large-scale production. The mechanism of PG bioactivity is a focus of research. The mechanisms responsible for the anti-cancer effects of PG are multifaceted and complex, involving various aspects such as cell proliferation, apoptosis, angiogenesis, signaling pathways, and antioxidation. The advantage of PG as a natural product lies in its relatively low-toxicity, making it a highly anticipated potential candidate for pharmaceutical development. Despite the potential therapeutic effects observed in cellular and animal models, further in-depth research and experiments are still required before clinical application. PG demonstrates broad-spectrum antimicrobial activity, effectively targeting various microorganisms including bacteria and fungi. This characteristic makes it a valuable tool in combating complex infectious pathogens. While some antibacterial mechanisms have been identified, further research is necessary to gain a detailed molecular-level understanding of PG’s mechanisms.
It is important to note that nanocomposite technology utilizing PG for disease treatment has become a popular area of research due to PG’s strong biological activity. Nanomaterials can address PG’s hydrophobicity and low bioavailability, overcome its limitations in disease treatment, and improve its ability to transport and release in the body, resulting in better therapeutic outcomes (Araujo et al., 2022; Majumdar et al., 2022). This area of nanocarrier delivery is relatively less explored compared to other research directions, and some novel nanoparticles such as dendrimer and PG can be investigated next to explore better therapeutic effects.
Since researchers have studied the gene clusters involved in direct PG synthesis and obtained different gene clusters for different strains, there has also been a great deal of research on transcriptional regulatory systems or factors that play important roles in PG synthesis, such as QS, TCSs, and other regulatory factors. However, we have a poor understanding of some of the regulatory mechanisms behind the effects of synthesis, for example, the exact mechanism of the effect of LuxS/AI-2 on PG synthesis is still unknown.
Regarding large-scale production, the expanding application range and increasing market demand for PG have led to research on optimized cultivation techniques, high-yielding microbial strains, and extraction and purification techniques. This research lays the foundation for the industrial production of PG. Regarding culture medium, the focus of research is on using inexpensive and high-yield agricultural by-products to produce PG in large quantities, which also has environmental significance. As for microbial strains, S. marcescens remains the most commonly used. It is worth mentioning that S. marcescens is an opportunistic pathogen and exposure to this bacterium in immunocompromised individuals can cause various diseases such as wound infections, eye infections, meningitis, septicemia, and others. In case of infection, antibiotics like ciprofloxacin and gentamicin can be taken (Zivkovic Zaric et al., 2023). There have been experiments to transform wild-type strains into stable and excellent strains, which can then be combined with high-quality culture medium for mass production.
5. Conclusion
In our comprehensive review, we delve into recent advancements in understanding the chemical structure, physicochemical properties, gene regulation, and optimized production techniques of PG, shedding light on the underlying mechanisms at play. We underscore the extraordinary biological activity of PG and the factors that impact its production efficiency. Despite significant progress, there are still numerous areas within the mechanistic studies that warrant further investigation. To deepen our comprehension of PG’s interactions with diverse pathogens, it is imperative to conduct thorough examinations into its toxicity profile, pharmacokinetics, drug compatibility, and other pertinent characteristics. Future research endeavors should prioritize exploring the synergistic potential of PG in combination with other therapeutic modalities and delineating their tailored applications across various cancer types and infections. With regards to large-scale production, optimizing culture conditions in alignment with elucidated regulatory mechanisms, selecting and enhancing high-yielding strains through exploring natural microorganisms or implementing mutagenesis and genetic engineering techniques, and ultimately integrating superior culture conditions with high-performing strains are pivotal steps toward achieving elevated production yields. These strategies are pivotal in laying a robust foundation for advancing PGs as potent anticancer and antimicrobial agents.
Author contributions
YL: Visualization, Writing – original draft. DL: Visualization, Writing – original draft. RJ: Visualization, Writing – original draft. ZL: Writing – review & editing. XG: Conceptualization, Writing – review & editing, Supervision.
Funding Statement
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Amorim L. F. A., Mouro C., Riool M., Gouveia I. C. (2022). Antimicrobial food packaging based on Prodigiosin-incorporated double-layered bacterial cellulose and chitosan composites. Polymers (Basel) 14:315. doi: 10.3390/polym14020315, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araujo R. G., Zavala N. R., Castillo-Zacarías C., Barocio M. E., Hidalgo-Vázquez E., Parra-Arroyo L., et al. (2022). Recent advances in Prodigiosin as a bioactive compound in nanocomposite applications. Molecules 27:4982. doi: 10.3390/molecules27154982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arivizhivendhan K. V., Mahesh M., Boopathy R., Swarnalatha S., Regina Mary R., Sekaran G. (2018). Antioxidant and antimicrobial activity of bioactive prodigiosin produces from Serratia marcescens using agricultural waste as a substrate. J. Food Sci. Technol. 55, 2661–2670. doi: 10.1007/s13197-018-3188-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aruldass C. A., Venil C. K., Zakaria Z. A., Ahmad W. A. (2014). Brown sugar as a low-cost medium for the production of prodigiosin by locally isolated Serratia marcescens UTM1. Int. Biodeterior. Biodegradation 95, 19–24. doi: 10.1016/j.ibiod.2014.04.006 [DOI] [Google Scholar]
- Bhagwat A., Padalia U. (2020). Optimization of prodigiosin biosynthesis by Serratia marcescens using unconventional bioresources. J. Genet. Eng. Biotechnol. 18:26. doi: 10.1186/s43141-020-00045-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. R., Tesar B., Yu L., Werner L., Takebe N., Mikler E., et al. (2015). Obatoclax in combination with fludarabine and rituximab is well-tolerated and shows promising clinical activity in relapsed chronic lymphocytic leukemia. Leuk. Lymphoma 56, 3336–3342. doi: 10.3109/10428194.2015.1048441, PMID: [DOI] [PubMed] [Google Scholar]
- Cerdeno A. M., Bibb M. J., Challis G. L. (2001). Analysis of the prodiginine biosynthesis gene cluster of Streptomyces coelicolor A3(2): new mechanisms for chain initiation and termination in modular multienzymes. Chem. Biol. 8, 817–829. doi: 10.1016/S1074-5521(01)00054-0, PMID: [DOI] [PubMed] [Google Scholar]
- Chen Y., Luo G., Chen S., Zhang D., Xie W., Wang Z., et al. (2021). The potential of prodigiosin for control of Prorocentrum donghaiense blooms: Algicidal properties and acute toxicity to other marine organisms at various trophic levels. Ecotoxicol. Environ. Saf. 228:112913. doi: 10.1016/j.ecoenv.2021.112913, PMID: [DOI] [PubMed] [Google Scholar]
- Chen W.-C., Yu W. J., Chang C. C., Chang J. S., Huang S. H., Chang C. H., et al. (2013). Enhancing production of prodigiosin from Serratia marcescens C3 by statistical experimental design and porous carrier addition strategy. Biochem. Eng. J. 78, 93–100. doi: 10.1016/j.bej.2013.02.001 [DOI] [Google Scholar]
- Cheng S. Y., Chen N. F., Kuo H. M., Yang S. N., Sung C. S., Sung P. J., et al. (2018). Prodigiosin stimulates endoplasmic reticulum stress and induces autophagic cell death in glioblastoma cells. Apoptosis 23, 314–328. doi: 10.1007/s10495-018-1456-9, PMID: [DOI] [PubMed] [Google Scholar]
- Chiu W. J., Lin S. R., Chen Y. H., Tsai M. J., Leong M., Weng C. F. (2018). Prodigiosin-emerged PI3K/Beclin-1-independent pathway elicits Autophagic cell death in doxorubicin-sensitive and -resistant lung Cancer. J. Clin. Med. 7:321. doi: 10.3390/jcm7100321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi S. Y., Lim S., Yoon K. H., Lee J. I., Mitchell R. J. (2021). Biotechnological activities and applications of bacterial pigments Violacein and Prodigiosin. J. Biol. Eng. 15:10. doi: 10.1186/s13036-021-00262-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook T. B., Jacobson T. B., Venkataraman M. V., Hofstetter H., Amador-Noguez D., Thomas M. G., et al. (2021). Stepwise genetic engineering of Pseudomonas putida enables robust heterologous production of prodigiosin and glidobactin a. Metab. Eng. 67, 112–124. doi: 10.1016/j.ymben.2021.06.004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthurst S. J., Kurz C. L., Salmond G. P. C. (2004). luxS mutants of Serratia defective in autoinducer-2-dependent ‘quorum sensing’ show strain-dependent impacts on virulence and production of carbapenem and prodigiosin. Microbiology (Reading) 150, 1901–1910. doi: 10.1099/mic.0.26946-0, PMID: [DOI] [PubMed] [Google Scholar]
- Darshan N., Manonmani H. K. (2015). Prodigiosin and its potential applications. J. Food Sci. Technol. 52, 5393–5407. doi: 10.1007/s13197-015-1740-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Araujo H. W., Fukushima K., Takaki G. M. (2010). Prodigiosin production by Serratia marcescens UCP 1549 using renewable-resources as a low cost substrate. Molecules 15, 6931–6940. doi: 10.3390/molecules15106931, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos R. A., Rodríguez D. M., da Silva L. A. R., de Almeida S. M., de Campos-Takaki G. M., de Lima M. A. B. (2021). Enhanced production of prodigiosin by Serratia marcescens UCP 1549 using agrosubstrates in solid-state fermentation. Arch. Microbiol. 203, 4091–4100. doi: 10.1007/s00203-021-02399-z, PMID: [DOI] [PubMed] [Google Scholar]
- Elahian F., Moghimi B., Dinmohammadi F., Ghamghami M., Hamidi M., Mirzaei S. A. (2013). The anticancer agent prodigiosin is not a multidrug resistance protein substrate. DNA Cell Biol. 32, 90–97. doi: 10.1089/dna.2012.1902, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espona-Fiedler M., Manuel-Manresa P., Benítez-García C., Fontova P., Quesada R., Soto-Cerrato V., et al. (2022). Antimetastatic properties of Prodigiosin and the BH3-mimetic Obatoclax (GX15-070) in melanoma. Pharmaceutics 15:97. doi: 10.3390/pharmaceutics15010097, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco R., Pérez-Tomás R., Gimènez-Bonafé P., Soto-Cerrato V., Giménez-Xavier P., Ambrosio S. (2007). Mechanisms of prodigiosin cytotoxicity in human neuroblastoma cell lines. Eur. J. Pharmacol. 572, 111–119. doi: 10.1016/j.ejphar.2007.06.054, PMID: [DOI] [PubMed] [Google Scholar]
- Gamboa-Suasnavart R. A., Valdez-Cruz N. A., Gaytan-Ortega G., Reynoso-Cereceda G. I., Cabrera-Santos D., López-Griego L., et al. (2018). The metabolic switch can be activated in a recombinant strain of Streptomyces lividans by a low oxygen transfer rate in shake flasks. Microb. Cell Factories 17:189. doi: 10.1186/s12934-018-1035-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girão M., Freitas S., Martins T. P., Urbatzka R., Carvalho M. F., Leão P. N. (2024). Decylprodigiosin: a new member of the prodigiosin family isolated from a seaweed-associated Streptomyces. Front. Pharmacol. 15:485. doi: 10.3389/fphar.2024.1347485, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri A. V., Anandkumar N., Muthukumaran G., Pennathur G. (2004). A novel medium for the enhanced cell growth and production of prodigiosin from Serratia marcescens isolated from soil. BMC Microbiol. 4:11. doi: 10.1186/1471-2180-4-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gristwood T., Fineran P. C., Everson L., Williamson N. R., Salmond G. P. (2009). The PhoBR two-component system regulates antibiotic biosynthesis in Serratia in response to phosphate. BMC Microbiol. 9:112. doi: 10.1186/1471-2180-9-112, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A. K. P., Williamson N. R., Slater H., Cox A., Abbasi S., Foulds I., et al. (2004). The Serratia gene cluster encoding biosynthesis of the red antibiotic, prodigiosin, shows species- and strain-dependent genome context variation. Microbiology (Reading) 150, 3547–3560. doi: 10.1099/mic.0.27222-0, PMID: [DOI] [PubMed] [Google Scholar]
- Heinemann B. (1970). Influence of dissolved oxygen levels on production oflasparaginase and prodigiosin by Serratia marcescens. Appl. Environ. Microbiol. 19, 800–804. doi: 10.1128/am.19.5.800-804.1970, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horng Y.-T., Chang K. C., Liu Y. N., Lai H. C., Soo P. C. (2010). The RssB/RssA two-component system regulates biosynthesis of the tripyrrole antibiotic, prodigiosin, in Serratia marcescens. Int. J. Med. Microbiol. 300, 304–312. doi: 10.1016/j.ijmm.2010.01.003, PMID: [DOI] [PubMed] [Google Scholar]
- Hosseini A., Espona-Fiedler M., Soto-Cerrato V., Quesada R., Pérez-Tomás R., Guallar V. (2013). Molecular interactions of prodiginines with the BH3 domain of anti-apoptotic Bcl-2 family members. PLoS One 8:e57562. doi: 10.1371/journal.pone.0057562, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D. X., Withall D. M., Challis G. L., Thomson R. J. (2016). Structure, chemical synthesis, and biosynthesis of Prodiginine natural products. Chem. Rev. 116, 7818–7853. doi: 10.1021/acs.chemrev.6b00024, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda H., Shikata Y., Watanapokasin R., Tashiro E., Imoto M. (2016). Metacycloprodigiosin induced cell death selectively in β-catenin-mutated tumor cells. J. Antibiot. 70, 109–112. doi: 10.1038/ja.2016.75, PMID: [DOI] [PubMed] [Google Scholar]
- Jehlička J., Edwards H. G. M., Oren A.Analysis of brown, violet and blue pigments of microorganisms by Raman spectroscopy. (2021).
- Jeong Y., Kim H. J., Kim S., Park S. Y., Kim H. R., Jeong S., et al. (2021). Enhanced large-scale production of Hahella chejuensis-derived Prodigiosin and evaluation of its bioactivity. J. Microbiol. Biotechnol. 31, 1624–1631. doi: 10.4014/jmb.2109.09039, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H., Yim J. H., Lee C., Choi S. H., Park Y. K., Yoon S. H., et al. (2005). Genomic blueprint of Hahella chejuensis, a marine microbe producing an algicidal agent. Nucleic Acids Res. 33, 7066–7073. doi: 10.1093/nar/gki1016, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X., Zhao K., Liu F., Lin J., Lin C., Chen J. (2022). Transcriptional factor OmpR positively regulates prodigiosin biosynthesis in Serratia marcescens FZSF02 by binding with the promoter of the prodigiosin cluster. Front. Microbiol. 13:1041146. doi: 10.3389/fmicb.2022.1041146, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki T., Sakurai F., Hayakawa Y. (2008). A prodigiosin from the roseophilin producer Streptomyces griseoviridis. J. Nat. Prod. 71, 1265–1267. doi: 10.1021/np7007494, PMID: [DOI] [PubMed] [Google Scholar]
- Kawasaki T., Sakurai F., Nagatsuka S. Y., Hayakawa Y. (2009). Prodigiosin biosynthesis gene cluster in the roseophilin producer Streptomyces griseoviridis. J. Antibiot. (Tokyo) 62, 271–276. doi: 10.1038/ja.2009.27, PMID: [DOI] [PubMed] [Google Scholar]
- Kim C. H., Kim S. W., Hong S. I. (2000). An integrated fermentation-separation process for the production of red pigment by Serratia sp. KH-95. Process Biochem. 35, 485–490. doi: 10.1016/S0032-9592(99)00091-6 [DOI] [Google Scholar]
- Kim D., Lee J. S., Park Y. K., Kim J. F., Jeong H., Oh T. K., et al. (2007). Biosynthesis of antibiotic prodiginines in the marine bacterium Hahella chejuensis KCTC 2396. J. Appl. Microbiol. 102, 937–944. doi: 10.1111/j.1365-2672.2006.03172.x, PMID: [DOI] [PubMed] [Google Scholar]
- Kim D., Park Y. K., Lee J. S., Kim J. F., Jeong H., Kim B. S., et al. (2006). Analysis of a prodigiosin biosynthetic gene cluster from the marine bacterium Hahella chejuensis KCTC 2396. J. Microbiol. Biotechnol. 16, 1912–1918. [Google Scholar]
- Kimata S., Matsuda T., Kawasaki T., Hayakawa Y. (2022). Prodigiosin R3, a new multicyclic prodigiosin formed by prodigiosin cyclization genes in the roseophilin producer. J. Antibiot. 76, 14–19. doi: 10.1038/s41429-022-00572-0, PMID: [DOI] [PubMed] [Google Scholar]
- Kimata S., Matsuda T., Suizu Y., Hayakawa Y. (2018). Prodigiosin R2, a new prodigiosin from the roseophilin producer Streptomyces griseoviridis 2464-S5. J. Antibiot. 71, 393–396. doi: 10.1038/s41429-017-0011-1, PMID: [DOI] [PubMed] [Google Scholar]
- Kurbanoglu E. B., Ozdal M., Ozdal O. G., Algur O. F. (2015). Enhanced production of prodigiosin by Serratia marcescens MO-1 using ram horn peptone. Braz. J. Microbiol. 46, 631–637. doi: 10.1590/S1517-838246246220131143, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer C. J., Albert I., Ross H. J., Kovacs P., Blakely L. J., Pajkos G., et al. (2014). Randomized phase II study of carboplatin and etoposide with or without obatoclax mesylate in extensive-stage small cell lung cancer. Lung Cancer 85, 420–428. doi: 10.1016/j.lungcan.2014.05.003 [DOI] [PubMed] [Google Scholar]
- Lapenda J. C., Silva P. A., Vicalvi M. C., Sena K. X. F. R., Nascimento S. C. (2015). Antimicrobial activity of prodigiosin isolated from Serratia marcescens UFPEDA 398. World J. Microbiol. Biotechnol. 31, 399–406. doi: 10.1007/s11274-014-1793-y, PMID: [DOI] [PubMed] [Google Scholar]
- Lee J. S., Kim Y. S., Park S., Kim J., Kang S. J., Lee M. H., et al. (2011). Exceptional production of both prodigiosin and cycloprodigiosin as major metabolic constituents by a novel marine bacterium, Zooshikella rubidus S1-1. Appl. Environ. Microbiol. 77, 4967–4973. doi: 10.1128/AEM.01986-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., He S., Zhang X., Gao Q., Liu Y., Liu L. (2022). Structures, biosynthesis, and bioactivities of prodiginine natural products. Appl. Microbiol. Biotechnol. 106, 7721–7735. doi: 10.1007/s00253-022-12245-x, PMID: [DOI] [PubMed] [Google Scholar]
- Lin C., Jia X., Fang Y., Chen L., Zhang H., Lin R., et al. (2019). Enhanced production of prodigiosin by Serratia marcescens FZSF02 in the form of pigment pellets. Electron. J. Biotechnol. 40, 58–64. doi: 10.1016/j.ejbt.2019.04.007 [DOI] [Google Scholar]
- Lin J., Yu Y., Zhao K., Zhao J., Rensing C., Chen J., et al. (2023). PtrA regulates prodigiosin synthesis and biological functions in Serratia marcescens FZSF02. Front. Microbiol. 14:1240102. doi: 10.3389/fmicb.2023.1240102, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Xu D., Wu D., Xu M., Wang Y., Wang W., et al. (2023). BarA/UvrY differentially regulates prodigiosin biosynthesis and swarming motility in Serratia marcescens FS14. Res. Microbiol. 174:104010. doi: 10.1016/j.resmic.2022.104010, PMID: [DOI] [PubMed] [Google Scholar]
- Liu W., Yang J., Tian Y., Zhou X., Wang S., Zhu J., et al. (2021). An in situ extractive fermentation strategy for enhancing prodigiosin production from Serratia marcescens BWL1001 and its application to inhibiting the growth of Microcystis aeruginosa. Biochem. Eng. J. 166:107836. doi: 10.1016/j.bej.2020.107836 [DOI] [Google Scholar]
- Ma Z., Xiao H., Li H., Lu X., Yan J., Nie H., et al. (2024). Prodigiosin as an Antibiofilm agent against the bacterial biofilm-associated infection of Pseudomonas aeruginosa. Pathogens 13:145. doi: 10.3390/pathogens13020145, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumdar S., Mandal T., Mandal D. D. (2022). Chitosan based micro and nano-particulate delivery systems for bacterial prodigiosin: optimization and toxicity in animal model system. Int. J. Biol. Macromol. 222, 2966–2976. doi: 10.1016/j.ijbiomac.2022.10.072, PMID: [DOI] [PubMed] [Google Scholar]
- Morrison D. A. (1966). Prodigiosin synthesis in mutants of Serratia marcesens. J. Bacteriol. 91, 1599–1604. doi: 10.1128/jb.91.4.1599-1604.1966, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Bassler B. L. (2019). Bacterial quorum sensing in complex and dynamically changing environments. Nat. Rev. Microbiol. 17, 371–382. doi: 10.1038/s41579-019-0186-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V. B., Chen S. P., Nguyen T. H., Nguyen M. T., Tran T. T. T., Doan C. T., et al. (2019). Novel efficient bioprocessing of marine chitins into active anticancer Prodigiosin. Mar. Drugs 18:15. doi: 10.3390/md18010015, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V. B., Nguyen D. N., Nguyen A. D., Ngo V. A., Ton T. Q., Doan C. T., et al. (2020). Utilization of crab waste for cost-effective bioproduction of Prodigiosin. Mar. Drugs 18:523. doi: 10.3390/md18110523, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T.-H., Wang S. L., Nguyen D. N., Nguyen A. D., Nguyen T. H., Doan M. D., et al. (2021). Bioprocessing of marine Chitinous wastes for the production of bioactive Prodigiosin. Molecules 26:3138. doi: 10.3390/molecules26113138, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien M. A., Kirby R. (2008). Apoptosis: a review of pro-apoptotic and anti-apoptotic pathways and dysregulation in disease. J. Vet. Emerg. Crit. Care 18, 572–585. doi: 10.1111/j.1476-4431.2008.00363.x [DOI] [Google Scholar]
- Pan M. Y., Shen Y. C., Lu C. H., Yang S. Y., Ho T. F., Peng Y. T., et al. (2012). Prodigiosin activates endoplasmic reticulum stress cell death pathway in human breast carcinoma cell lines. Toxicol. Appl. Pharmacol. 265, 325–334. doi: 10.1016/j.taap.2012.08.034, PMID: [DOI] [PubMed] [Google Scholar]
- Pan X., Tang M., You J., Liu F., Sun C., Osire T., et al. (2021). Regulator RcsB controls Prodigiosin synthesis and various cellular processes in Serratia marcescens JNB5-1. Appl. Environ. Microbiol. 87:e02052-20. doi: 10.1128/AEM.02052-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X., Tang M., You J., Osire T., Sun C., Fu W., et al. (2022). PsrA is a novel regulator contributes to antibiotic synthesis, bacterial virulence, cell motility and extracellular polysaccharides production in Serratia marcescens. Nucleic Acids Res. 50, 127–148. doi: 10.1093/nar/gkab1186, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin H., Liu Y., Cao X., Jiang J., Lian W., Qiao D., et al. (2020). RpoS is a pleiotropic regulator of motility, biofilm formation, exoenzymes, siderophore and prodigiosin production, and trade-off during prolonged stationary phase in Serratia marcescens. PLoS One 15:e0232549. doi: 10.1371/journal.pone.0232549, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran A., Anishetty S., Pennathur G. (2020). Molecular dynamics of the membrane interaction and localisation of prodigiosin. J. Mol. Graph. Model. 98:107614. doi: 10.1016/j.jmgm.2020.107614, PMID: [DOI] [PubMed] [Google Scholar]
- Ryazantseva I. N., Saakov V. S., Andreyeva I. N., Ogorodnikova T. I., Zuev Y. F. (2012). Response of pigmented Serratia marcescens to the illumination. J. Photochem. Photobiol. B 106, 18–23. doi: 10.1016/j.jphotobiol.2011.08.006, PMID: [DOI] [PubMed] [Google Scholar]
- Sajjad W., SajjadAziz I. A., SikanderHasan S., FarihaShah A. A. (2018). Antiproliferative, antioxidant and binding mechanism analysis of prodigiosin from newly isolated radio-resistant Streptomyces sp. strain WMA-LM31. Mol. Biol. Rep. 45, 1787–1798. doi: 10.1007/s11033-018-4324-3, PMID: [DOI] [PubMed] [Google Scholar]
- Santos-Beneit F., Barriuso-Iglesias M., Fernández-Martínez L. T., Martínez-Castro M., Sola-Landa A., Rodríguez-García A., et al. (2011). The RNA polymerase omega factor RpoZ is regulated by PhoP and has an important role in antibiotic biosynthesis and morphological differentiation in Streptomyces coelicolor. Appl. Environ. Microbiol. 77, 7586–7594. doi: 10.1128/AEM.00465-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P. D., Allen H. K., Klimowicz A. K., Mlot C., Gross J. A., Savengsuksa S., et al. (2010). Psychrotrophic strain of Janthinobacterium lividum from a cold Alaskan soil produces prodigiosin. DNA Cell Biol. 29, 533–541. doi: 10.1089/dna.2010.1020, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setiyono E., Adhiwibawa M. A. S., Indrawati R., Prihastyanti M. N. U., Shioi Y., Brotosudarmo T. H. P. (2020). An Indonesian marine bacterium, Pseudoalteromonas rubra produces antimicrobial prodiginine pigments. ACS Omega 5, 4626–4635. doi: 10.1021/acsomega.9b04322, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer J. M. C., Giddings L. A., Samples R. M., Mikucki J. A. (2023). Genomic and phenotypic characterization of a red-pigmented strain of Massilia frigida isolated from an Antarctic microbial mat. Front. Microbiol. 14:33. doi: 10.3389/fmicb.2023.1156033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahitha G. (2012). Enhanced production of Prodigiosin production in Serratia Marcescens. J. Appl. Pharm. Sci. 2, 138–140. doi: 10.7324/JAPS.2012.2823 [DOI] [Google Scholar]
- Someya N., Nakajima M., Hamamoto H., Yamaguchi I., Akutsu K. (2004). Effects of light conditions on prodigiosin stability in the biocontrol bacterium Serratia marcescens strain B2. J. Gen. Plant Pathol. 70, 367–370. doi: 10.1007/s10327-004-0134-7 [DOI] [Google Scholar]
- Song M. J., Bae J., Lee D. S., Kim C. H., Kim J. S., Kim S. W., et al. (2006). Purification and characterization of prodigiosin produced by integrated bioreactor from Serratia sp. KH-95. J. Biosci. Bioeng. 101, 157–161. doi: 10.1263/jbb.101.157, PMID: [DOI] [PubMed] [Google Scholar]
- Song H., Li Y., Wang Y. (2023). Two-component system GacS/GacA, a global response regulator of bacterial physiological behaviors. Engineering. Microbiology 3:100051. doi: 10.1016/j.engmic.2022.100051 [DOI] [Google Scholar]
- Stankovic N., Senerovic L., Ilic-Tomic T., Vasiljevic B., Nikodinovic-Runic J. (2014). Properties and applications of undecylprodigiosin and other bacterial prodigiosins. Appl. Microbiol. Biotechnol. 98, 3841–3858. doi: 10.1007/s00253-014-5590-1 [DOI] [PubMed] [Google Scholar]
- Stella N. A., Lahr R. M., Brothers K. M., Kalivoda E. J., Hunt K. M., Kwak D. H., et al. (2015). Serratia marcescens cyclic AMP receptor protein controls transcription of EepR, a novel regulator of antimicrobial secondary metabolites. J. Bacteriol. 197, 2468–2478. doi: 10.1128/JB.00136-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumathi C., MohanaPriya D., Swarnalatha S., Dinesh M. G., Sekaran G. (2014). Production of prodigiosin using tannery fleshing and evaluating its pharmacological effects. ScientificWorldJournal 2014:290327. doi: 10.1155/2014/290327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Wang L., Pan X., Osire T., Fang H., Zhang H., et al. (2020). Improved Prodigiosin production by relieving CpxR temperature-sensitive inhibition. Front. Bioeng. Biotechnol. 8:344. doi: 10.3389/fbioe.2020.00344, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D., Zhou X., Liu C., Zhu J., Ru Y., Liu W., et al. (2021). Fnr negatively regulates Prodigiosin synthesis in Serratia sp. ATCC 39006 during aerobic fermentation. Front. Microbiol. 12:734854. doi: 10.3389/fmicb.2021.734854, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R. L., Laversanne M., Soerjomataram I., Jemal A., et al. (2021). Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. doi: 10.3322/caac.21660, PMID: [DOI] [PubMed] [Google Scholar]
- Suryawanshi R. K., Patil C. D., Borase H. P., Salunke B. K., Patil S. V. (2014). Studies on production and biological potential of Prodigiosin by Serratia marcescens. Appl. Biochem. Biotechnol. 173, 1209–1221. doi: 10.1007/s12010-014-0921-3 [DOI] [PubMed] [Google Scholar]
- Süssmuth R. D., Mainz A. (2017). Nonribosomal peptide synthesis—principles and prospects. Angew. Chem. Int. Ed. 56, 3770–3821. doi: 10.1002/anie.201609079, PMID: [DOI] [PubMed] [Google Scholar]
- Vitale G. A., Sciarretta M., Palma Esposito F., January G. G., Giaccio M., Bunk B., et al. (2020). Genomics-metabolomics profiling disclosed marine Vibrio spartinae 3.6 as a producer of a new branched side chain Prodigiosin. J. Nat. Prod. 83, 1495–1504. doi: 10.1021/acs.jnatprod.9b01159, PMID: [DOI] [PubMed] [Google Scholar]
- Wang M., Kaufman R. J. (2016). Protein misfolding in the endoplasmic reticulum as a conduit to human disease. Nature 529, 326–335. doi: 10.1038/nature17041, PMID: [DOI] [PubMed] [Google Scholar]
- Wang Z., Li B., Zhou L., Yu S., Su Z., Song J., et al. (2016). Prodigiosin inhibits Wnt/beta-catenin signaling and exerts anticancer activity in breast cancer cells. Proc. Natl. Acad. Sci. USA 113, 13150–13155. doi: 10.1073/pnas.1616336113, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Luo H., Song G., Liu C., Wang J., Xu J., et al. (2013). Prodigiosin found in Serratia marcescens y2 initiates phototoxicity in the cytomembrane. Electron. J. Biotechnol. 16:4. doi: 10.2225/vol16-issue4-fulltext-7 [DOI] [Google Scholar]
- Wang S. L., Nguyen V. B., Doan C. T., Tran T. N., Nguyen M. T., Nguyen A. D. (2020). Production and potential applications of bioconversion of chitin and protein-containing fishery Byproducts into Prodigiosin: a review. Molecules 25:2744. doi: 10.3390/molecules25122744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman H. H., Rodgers G. C., Keith D. D. (1969). Metacycloprodigiosin, a tripyrrole pigment from Streptomyces longisporus ruber. J. Am. Chem. Soc. 91, 1263–1264. doi: 10.1021/ja01033a065, PMID: [DOI] [PubMed] [Google Scholar]
- Wasserman H. H., Sykes R. J., Peverada P., Shaw C. K., Cushley R. J., Lipsky S. R. (1973). Biosynthesis of prodigiosin. Incorporation patterns of C-labeled alanine, proline, glycine, and serine elucidated by fourier transform nuclear magnetic resonance. J. Am. Chem. Soc. 95, 6874–6875. doi: 10.1021/ja00801a080, PMID: [DOI] [PubMed] [Google Scholar]
- Wei J. R., Lai H. C. (2006). N-acylhomoserine lactone-dependent cell-to-cell communication and social behavior in the genus Serratia. Int. J. Med. Microbiol. 296, 117–124. doi: 10.1016/j.ijmm.2006.01.033, PMID: [DOI] [PubMed] [Google Scholar]
- Wei Y.-H., Yu W.-J., Chen W.-C. (2005). Enhanced undecylprodigiosin production from Serratia marcescens SS-1 by medium formulation and amino-acid supplementation. J. Biosci. Bioeng. 100, 466–471. doi: 10.1263/jbb.100.466, PMID: [DOI] [PubMed] [Google Scholar]
- Wilf N. M., Salmond G. P. C. (2012). The stationary phase sigma factor, RpoS, regulates the production of a carbapenem antibiotic, a bioactive prodigiosin and virulence in the enterobacterial pathogen Serratia sp. ATCC 39006. Microbiology (Reading) 158, 648–658. doi: 10.1099/mic.0.055780-0 [DOI] [PubMed] [Google Scholar]
- Williams R. P. (1973). Biosynthesis of Prodigiosin, a secondary metabolite of Serratia marcescens. Appl. Environ. Microbiol. 25, 396–402. doi: 10.1128/am.25.3.396-402.1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson N. R., Fineran P. C., Leeper F. J., Salmond G. P. C. (2006). The biosynthesis and regulation of bacterial prodiginines. Nat. Rev. Microbiol. 4, 887–899. doi: 10.1038/nrmicro1531, PMID: [DOI] [PubMed] [Google Scholar]
- Williamson N. R., Simonsen H. T., Ahmed R. A. A., Goldet G., Slater H., Woodley L., et al. (2005). Biosynthesis of the red antibiotic, prodigiosin, in Serratia: identification of a novel 2-methyl-3-n-amyl-pyrrole (MAP) assembly pathway, definition of the terminal condensing enzyme, and implications for undecylprodigiosin biosynthesis in Streptomyces. Mol. Microbiol. 56, 971–989. doi: 10.1111/j.1365-2958.2005.04602.x, PMID: [DOI] [PubMed] [Google Scholar]
- Wong R. S. (2011). Apoptosis in cancer: from pathogenesis to treatment. J. Exp. Clin. Cancer Res. 30:87. doi: 10.1186/1756-9966-30-87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi J. S., Kim J. M., Kang M. K., Kim J. H., Cho H. S., Ban Y. H., et al. (2022). Whole-genome sequencing and analysis of Streptomyces strains producing multiple antinematode drugs. BMC Genomics 23:610. doi: 10.1186/s12864-022-08847-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J., Ding M., Zha F., Zhang J., Meng Q., Yu Z. (2021). Stringent starvation protein regulates Prodiginine biosynthesis via affecting Siderophore production in Pseudoalteromonas sp. strain R3. Appl. Environ. Microbiol. 87:e02949-20. doi: 10.1128/AEM.02949-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip C. H., Yarkoni O., Ajioka J., Wan K. L., Nathan S. (2019). Recent advancements in high-level synthesis of the promising clinical drug, prodigiosin. Appl. Microbiol. Biotechnol. 103, 1667–1680. doi: 10.1007/s00253-018-09611-z, PMID: [DOI] [PubMed] [Google Scholar]
- You Z., Zhang S., Liu X., Wang Y. (2018). Enhancement of prodigiosin synthetase (PigC) production from recombinant Escherichia coli through optimization of induction strategy and media. Prep. Biochem. Biotechnol. 48, 226–233. doi: 10.1080/10826068.2017.1421965, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang J., Shen Y., Liu J., Wei D. (2005). Antimetastatic effect of prodigiosin through inhibition of tumor invasion. Biochem. Pharmacol. 69, 407–414. doi: 10.1016/j.bcp.2004.08.037, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang B., Yang Y., Xie W., He W., Xie J., Liu W. (2022). Identifying Algicides of Enterobacter hormaechei F2 for control of the harmful alga Microcystis aeruginosa. Int. J. Environ. Res. Public Health 19:7556. doi: 10.3390/ijerph19137556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Zheng W., Wang H. (2020). Physiological response and morphological changes of Heterosigma akashiwo to an algicidal compound prodigiosin. J. Hazard. Mater. 385:121530. doi: 10.1016/j.jhazmat.2019.121530 [DOI] [PubMed] [Google Scholar]
- Zhang H., Peng Y., Zhang S., Cai G., Li Y., Yang X., et al. (2016). Algicidal effects of Prodigiosin on the harmful algae Phaeocystis globosa. Front. Microbiol. 7:602. doi: 10.3389/fmicb.2016.00602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zivkovic Zaric R., Zaric M., Sekulic M., Zornic N., Nesic J., Rosic V., et al. (2023). Antimicrobial treatment of Serratia marcescens invasive infections: systematic review. Antibiotics 12:367. doi: 10.3390/antibiotics12020367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zschiedrich C. P., Keidel V., Szurmant H. (2016). Molecular mechanisms of two-component signal transduction. J. Mol. Biol. 428, 3752–3775. doi: 10.1016/j.jmb.2016.08.003, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]