Genetic factors explain, at least in part, why some people resist infection more successfully than others. Rare gene disruptions cause fatal vulnerability to specific microbes, but more subtle differences are common and arise from minor variation in many genes. Recent advances in the human genome project and in high throughput genotyping technology will make it feasible within the next decade to screen the whole genome for genetic factors that determine susceptibility to HIV and AIDS, malaria, and tuberculosis. This will help to identify critical pathways of host defence and generate novel strategies for disease prevention. Understanding the evolutionary impact of infectious disease on the human genome may shed light on the origins of other common diseases, particularly those with an atopic or autoimmune component.
Why study genetic susceptibility to infection?
Historical accounts of the plague tell of individuals who survived unscathed in households where almost everyone else died. Each winter, British hospital wards are full of infants requiring oxygen therapy for bronchiolitis, but most infants infected with the same virus have little more than a runny nose. Over a million African children die each year of malaria, but many more remain in relatively good health despite being continually infected with the parasite.
To what extent does our genetic make up determine the different ways that we respond to the same infectious agent? This is difficult to answer because of the many other contributory factors involved, such as previous health, acquired immunity, and variability in the pathogen. Epidemiological analysis of the genetic component is confounded by environmental factors that cause familial clustering and is further complicated by the many different genes that are likely to be involved. Nevertheless, there is compelling evidence for a genetic component, including twin studies of tuberculosis,1 leprosy,2 malaria,3 and Helicobacter pylori infection4 and a large survey that found that individuals adopted in childhood had a markedly increased risk of death from infection if a biological parent had died prematurely of infection.5
Predicted developments
Description of full sequence of all 70 000-100 000 human genes and progress towards identification of all common polymorphisms (predicted to be in the order of 10 million)
Establishment of large sample collections for genetic epidemiology of infectious diseases and implementation of novel technologies for high throughput genotyping and bioinformatics
Identification of many common genetic variants that influence susceptibility to infection
Use of this information to discover genes encoding novel molecules that fight infection and to pinpoint critical pathways in immune regulation, leading to new therapeutic strategies for infectious and inflammatory disease
Discovery of additional genetic causes of severe immunodeficiency and further advances in gene therapy for such disorders
Unravelling the genetic and environmental determinants of infectious disease will soon be feasible. The human genome sequence provides the starting point for a systematic analysis of human genetic diversity (www.wellcome.ac.uk/en/genome). The most common form of DNA variation is a direct swap of one nucleotide for another, such as adenine for guanine, known as a single nucleotide polymorphism or SNP (pronounced “snip”). Polymorphisms that are present in at least 1-2% of normal individuals are found on average once in every 300-600 nucleotides, suggesting that some 10 million may be present across the whole genome.6 Although only a small proportion of these polymorphisms may be of functional relevance—by causing disruption or structural alteration of the protein encoded by a gene or by altering neighbouring regions of DNA that control gene regulation—all are of potential value as genetic markers for mapping regions of DNA that determine disease susceptibility. Much work is going into the development of DNA chips and other novel technologies for high throughput typing of single nucleotide polymorphisms that will make it feasible to screen many thousands of these markers in large study populations, with the ultimate goal of mapping disease associations across the whole genome. Efforts are being made to assemble the large DNA collections that will be required for this complex exercise.
What is the practical purpose of understanding the molecular genetic basis of susceptibility to infection? Efforts to develop vaccines and improved treatments for major diseases such as tuberculosis, HIV infection and AIDS, and malaria are hindered by our poor understanding of the molecular and cellular mechanisms that determine clinical outcome. Genetic epidemiology may identify hitherto unknown molecular mechanisms and improve understanding of critical events in the evolution of disease. For example, if an infectious disease is associated with high levels of a factor X in the blood, it is often difficult to know whether this is of pathogenic importance or simply an epiphenomenon of the disease process. But if the production of X is known to be determined by a genetic polymorphism, and if this polymorphism is shown to predispose to the disease in question, then there is a much stronger case for X playing a causal role.
Single gene defects that cause severe immunodeficiency
Almost 100 major genetic defects of the immune system have been identified, most due to a rare mutation of a single gene. When a distinctive clinical phenotype is caused by a single mutation, it is generally possible for the gene to be tracked down by classic genetic detective work—that is, by linkage analysis in families with several affected members. Table 1 gives some examples of genetic defects that have been identified within each of the major categories of immunodeficiency disorder. Apart from the importance of this information for affected families, it has provided valuable insights into the molecular and cellular basis of host immunity against different microbial species.
Table 1.
Examples of severe immunodeficiency disorders*
| System involved | Typical clinical syndrome | Example of genetic defect |
|---|---|---|
| B lymphocyte | Recurrent bacterial infection due to defective antibody production | B cell cytoplasmic tyrosine kinase |
| CD40 ligand | ||
| T lymphocyte | Severe bacterial, viral, and fungal infection due to defective humoral and cellular immunity | Interleukin 2 receptor γ chain |
| Adenosine deaminase | ||
| Neutrophil | Severe bacterial infection due to defective phagocytosis | Cytochrome b |
| β2 integrin | ||
| Macrophage | Extreme susceptibility to infection with environmental mycobacteria | Interferon γ receptor |
| Complement | Recurrent Neisseria infection | Terminal complement components |
Almost 100 severe deficiency disorders have been identified. Each is caused by a rare mutation of a single gene. Different mutations in the same gene may cause subtle variations in clinical phenotype. Mutations of different genes may lead to similar clinical syndromes if they disrupt a common immune pathway.
This is illustrated by a recently described group of disorders that result in extreme susceptibility to intracellular bacteria. The story began when several children from the same village in Malta were investigated for atypical mycobacterial infections that were ultimately fatal. These did not match any known immunodeficiency syndrome, but dogged clinical investigation eventually revealed that the affected children were homozygous for a disruptive mutation of the interferon γ receptor 1 gene.7 Different mutations in this and related immune genes were subsequently found to predispose to infection with other mycobacteria and salmonella species. Some but not all of these mutations are fatal, and the clinical phenotype may be either dominant or recessive depending on the precise nature of the molecular defect.8,9 These findings show how genetic analysis may reveal critical host defence mechanisms against specific pathogens, which may ultimately lead to new approaches to disease prevention.
Complex genetic determinants of susceptibility to infection
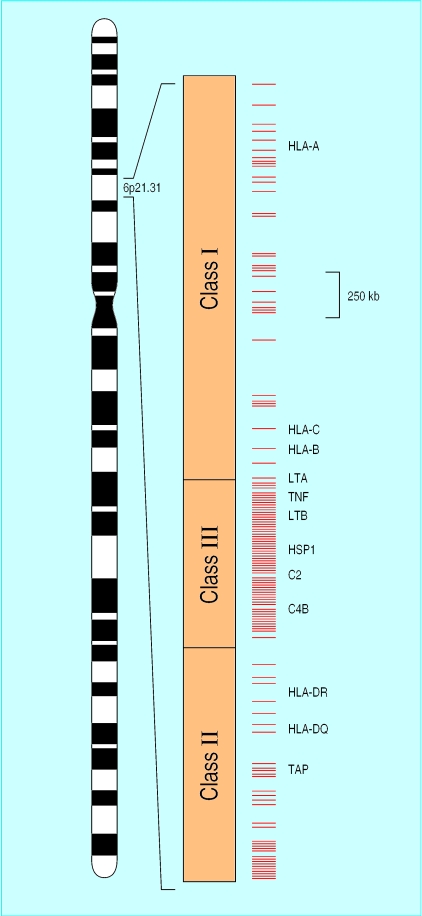
The classic example of genetic variation in the immune system is the major histocompatibility complex (MHC) on chromosome 6. This includes the highly polymorphic human leucocyte antigen (HLA) genes, best known to clinicians in the context of organ transplantation and autoimmune disease but increasingly recognised as a correlate of susceptibility to various infections including malaria,10 tuberculosis,11 HIV infection,12 and hepatitis B.13 The functional role of HLA is to present antigens to the immune system, and the extraordinary genetic diversity of HLA is postulated to have arisen as a host strategy to counter antigenic diversity in infectious organisms. HLA genes are located alongside a remarkably large number of other genes that are either known or predicted to have immunological functions (fig 1).14 These include genes for tumour necrosis factor, a key mediator of fever and the inflammatory response to infection, and for various complement and heat shock proteins.
Figure 1.
Structure of the human major histocompatibility complex (MHC) on chromosome 6, which has been fully sequenced.14 Within 3.6 megabases there are an estimated 128 expressed genes, of which 40% are predicted to have immune function. Red bars mark the locations of putative immune genes, some of which are labelled (TNF=tumour necrosis factor, LT=lymphotoxin, HSP=heat shock protein, C2 and C4=complement genes, TAP=antigen peptide transporter)
A high degree of linkage disequilibrium is observed across the MHC, meaning that allelic variation in one gene tends to be strongly correlated with that in a neighbouring gene. So, although different genes within the MHC may independently determine susceptibility to infection (and in malaria there is strong evidence for this15), it is also clear that a disease association observed with one gene may simply reflect what is going on in a neighbouring gene. Detailed mapping studies are required to distinguish polymorphisms that cause disease from those of linked genetic markers.
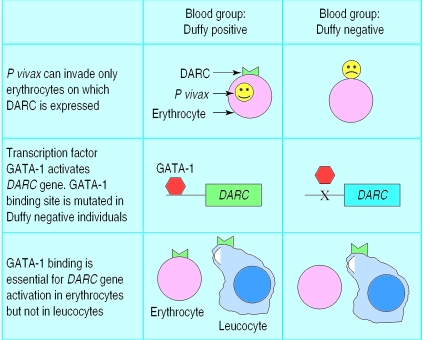
Congregations of immune genes outside the MHC include a dense cluster of cytokine genes in a segment of chromosome 5 that shows genetic linkage with susceptibility to schistosomiasis and asthma.16,17 The candidate gene approach, in which investigators first pick on a gene of interest and then search for polymorphisms to screen in their study population, has uncovered a growing number of intriguing associations (table 2) but in most cases further work is required to ascertain their functional importance. Knowledge of the functional basis of a genetic association may be highly revealing about basic disease mechanisms: a beautiful example is the relation between the Duffy blood group antigen and susceptibility to Plasmodium vivax malaria (fig 2).
Table 2.
Examples of candidate gene associations with common infectious diseases*
| Infection | Candidate gene region | Main function of protein encoded | Selected references |
|---|---|---|---|
| HIV and AIDS | CCR5, CCR2 | Chemokine receptor | Carrington et al,12 Martin et al18 |
| HLA class I | Antigen presentation | ||
| Hepatitis B | HLA class II | Antigen presentation | Thursz et al13 |
| Meningococcal disease | C5 to C9 | Terminal complement pathway | Hibberd et al19 |
| MBL | Opsonisation | ||
| Tuberculosis | HLA-DR | Antigen presentation | Singh et al,11 Bellamy et al20 |
| NRAMP1 | Divalent cation transporter | ||
| VDR | Vitamin D receptor | ||
| Leprosy | HLA class II | Antigen presentation | Todd et al,21 Roy et al22 |
| TNF | Pro-inflammatory cytokine | ||
| Malaria | DARC | Chemokine receptor | Hill et al,10 Knight et al,15 Miller et al,23 Tournamille et al,24 Flint et al,25 Ruwende et al26 |
| α and β globin | Haemoglobin subunits | ||
| G6PD | Carbohydrate metabolism | ||
| HLA class I and II | Antigen presentation | ||
| TNF | Pro-inflammatory cytokine | ||
| Leishmaniasis | TNF | Pro-inflammatory cytokine | Cabrera et al27 |
Gene regions in which allelic variants have been associated with increased or decreased susceptibility to infectious disease. Variant allele is often common in the population (such as above 10%), but alteration in disease risk is relatively modest (twofold to fourfold). These examples probably represent a small minority of the total number of genetic factors involved.
Figure 2.
The malaria parasite Plasmodium vivax invades human erythrocytes by binding to Duffy antigen/chemokine receptor (DARC) expressed on the erythrocyte surface. Many west Africans have a single nucleotide polymorphism in the DARC promoter region that prevents binding of the erythroid transcription factor GATA-1, thus suppressing DARC expression in erythrocytes but not other cell types. This confers complete protection against infection with P vivax but not against other species of malaria parasite, which invade erythrocytes through different receptors23,24
Large epidemiological surveys are needed to examine the complex interactions that may be involved in susceptibility to common infections—for example, to what extent do genetic factors influence the likelihood that a smoker will develop chronic bronchitis? In addition microbes themselves possess a huge amount of genetic diversity. Some argue that the most successful microbes are those that have co-evolved with their host to enable asymptomatic chronic infection and that virulent pathogens which cause severe pathology are often an evolutionary accident. The availability of data on genomic sequences for HIV, Mycobacterium tuberculosis, Neisseria meningitides, Plasmodium falciparum, and other important human pathogens will make it much easier to address these complex issues (www.ncbi.nlm.nih.gov/).
Genetic epidemiological approaches
The classic tool of molecular genetics is linkage analysis, in which highly polymorphic markers distributed throughout the genome are used to identify chromosomal regions that segregate with disease susceptibility within families. This is the preferred method for identifying genes that exert a major effect on disease susceptibility, but it is less likely to be successful if disease susceptibility is determined by several genetic determinants with small individual effects. With most infectious diseases, a more practicable approach is to analyse genetic association by comparing allele frequencies in diseased individuals with those of controls recruited from exactly the same ethnic group. Because of the difficulty of ensuring exact ethnic matching, an alternative is to study intrafamilial association, in which the distribution of genotypes among index cases is compared with that predicted from the parental genotypes.28 As the number of polymorphisms grows, sample size requirements also escalate, since a P value of 0.05 may be of little significance if this is for only one of 100 polymorphisms that are being assessed in a study. With a long term prospect of screening diseases for association with polymorphic markers in every known human gene, sample sizes in the order of 2000 will be required to validate genetic associations that give a doubling or a halving of relative risk.29
Discovering a genetic association is not the end of the story. Every polymorphism shows a greater or lesser degree of association with neighbouring polymorphisms (linkage disequilibrium). So when we find a disease association with a polymorphism in gene X, the next step is to search for other polymorphisms in the region of X and then to compare the strength of disease association with different combinations of linked polymorphisms (known as haplotypes) in order to dissect the causative polymorphism from linked markers that are functionally irrelevant. If gene X is just one of several environmental and genetic factors that determine disease susceptibility then the process of fine mapping may be extremely complex (thus, it is much more difficult for diabetes than for cystic fibrosis), and this is the area where the most questions remain about the feasibility of the new genetic approach to the analysis of common human diseases.
What will be the clinical dividends?
The human genome project has generated huge expectations, but it will take decades for the full clinical implications to be revealed. With this caveat in mind, what are the potential benefits for the management and prevention of infectious disease?
For severe immunodeficiency syndromes, increasingly detailed understanding of underlying genetic causes will improve diagnostic screening and genetic counselling and will spawn new treatment strategies including gene therapy. Information about more subtle genetic risk factors for common diseases may be valuable in prescribing appropriate antimicrobial drugs or deciding which patients are most likely to benefit from a vaccine, and life threatening infections such as septic shock may be managed more effectively if genetic risk factors and prognostic predictors can be identified on admission. But the most important advances in the long term will be at the level of the population rather than at that of the individual patient. Genetic epidemiology provides a potentially powerful way of identifying the critical molecular events required for an infectious agent to invade a human host, and for the host to eradicate or succumb to the infection. This information is likely to revolutionise drug and vaccine development, a point that has already been recognised by many of the major pharmaceutical agencies (http://genetics.glaxowellcome.com/genetics.asp).
Humans evolved in a hostile microbial environment, and natural selection by infectious disease may be one of the major causes of human genetic diversity, particularly in the immune system. The high frequency of the sickle haemoglobin gene in west Africa, which has arisen because of its protective effect against malaria and despite the lethal nature of the homozygous state, gives some idea of the strength of genetic selection that may be involved. It is possible that some Western ailments such as atopy and autoimmune disease are a legacy of the evolutionary impact of infectious disease, with immune gene variants selected for protection against parasites and other infectious pathogens having deleterious effects in an increasingly hygienic environment. A deeper understanding of the genetic factors that determine susceptibility to infection may ultimately provide clues to the prevention of a much wider range of common diseases.
Footnotes
Competing interests: None declared.
References
- 1.Comstock GW. Tuberculosis in twins: a re-analysis of the prophit survey. Am Rev Respir Dis. 1978;117:621–624. doi: 10.1164/arrd.1978.117.4.621. [DOI] [PubMed] [Google Scholar]
- 2.Fine PE. Immunogenetics of susceptibility to leprosy, tuberculosis, and leishmaniasis. An epidemiological perspective. Int J Lepr Other Mycobact Dis. 1981;49:437–454. [PubMed] [Google Scholar]
- 3.Jepson AP, Banya WA, Sisay-Joof F, Hassan-King M, Bennett S, Whittle HC. Genetic regulation of fever in Plasmodium falciparum malaria in Gambian twin children. J Infect Dis. 1995;172:316–319. doi: 10.1093/infdis/172.1.316. [DOI] [PubMed] [Google Scholar]
- 4.Malaty HM, Engstrand L, Pedersen NL, Graham DY. Helicobacter pylori infection: genetic and environmental influences. A study of twins. Ann Intern Med. 1994;120:982–986. doi: 10.7326/0003-4819-120-12-199406150-00002. [DOI] [PubMed] [Google Scholar]
- 5.Sorensen TI, Nielsen GG, Andersen PK, Teasdale TW. Genetic and environmental influences on premature death in adult adoptees. N Engl J Med. 1988;318:727–732. doi: 10.1056/NEJM198803243181202. [DOI] [PubMed] [Google Scholar]
- 6.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 7.Newport MJ, Huxley CM, Huston S, Hawrylowicz CM, Oostra BA, Williamson R, et al. A mutation in the interferon-gamma-receptor gene and susceptibility to mycobacterial infection. N Engl J Med. 1996;335:1941–1949. doi: 10.1056/NEJM199612263352602. [DOI] [PubMed] [Google Scholar]
- 8.Altare F, Jouanguy E, Lamhamedi S, Doffinger R, Fischer A, Casanova JL. Mendelian susceptibility to mycobacterial infection in man. Curr Opin Immunol. 1998;10:413–417. doi: 10.1016/s0952-7915(98)80114-3. [DOI] [PubMed] [Google Scholar]
- 9.Jouanguy E, Lamhamedi-Cherradi S, Lammas D, Dorman SE, Fondaneche MC, Dupuis S, et al. A human IFNGR1 small deletion hotspot associated with dominant susceptibility to mycobacterial infection. Nat Genet. 1999;21:370–378. doi: 10.1038/7701. [DOI] [PubMed] [Google Scholar]
- 10.Hill AV, Allsopp CE, Kwiatkowski D, Anstey NM, Twumasi P, Rowe PA, et al. Common west African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- 11.Singh SP, Mehra NK, Dingley HB, Pande JN, Vaidya MC. Human leukocyte antigen (HLA)-linked control of susceptibility to pulmonary tuberculosis and association with HLA-DR types. J Infect Dis. 1983;148:676–681. doi: 10.1093/infdis/148.4.676. [DOI] [PubMed] [Google Scholar]
- 12.Carrington M, Nelson GW, Martin MP, Kissner T, Vlahov D, Goedert JJ, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 13.Thursz MR, Kwiatkowski D, Allsopp CE, Greenwood BM, Thomas HC, Hill AV. Association between an MHC class II allele and clearance of hepatitis B virus in the Gambia. N Engl J Med. 1995;332:1065–1069. doi: 10.1056/NEJM199504203321604. [DOI] [PubMed] [Google Scholar]
- 14.Complete sequence and gene map of a human major histocompatibility complex. The MHC sequencing consortium. Nature. 1999;401:921–923. doi: 10.1038/44853. [DOI] [PubMed] [Google Scholar]
- 15.Knight JC, Udalova I, Hill AV, Greenwood BM, Peshu N, Marsh K, et al. A polymorphism that affects OCT-1 binding to the TNF promoter region is associated with severe malaria. Nat Genet. 1999;22:145–150. doi: 10.1038/9649. [DOI] [PubMed] [Google Scholar]
- 16.Marquet S, Abel L, Hillaire D, Dessein H, Kalil J, Feingold J, et al. Genetic localization of a locus controlling the intensity of infection by Schistosoma mansoni on chromosome 5q31-q33. Nat Genet. 1996;14:181–184. doi: 10.1038/ng1096-181. [DOI] [PubMed] [Google Scholar]
- 17.A genome-wide search for asthma susceptibility loci in ethnically diverse populations. The collaborative study on the genetics of asthma (CSGA) Nat Genet. 1997;15:389–392. doi: 10.1038/ng0497-389. [DOI] [PubMed] [Google Scholar]
- 18.Martin MP, Dean M, Smith MW, Winkler C, Gerrard B, Michael NL, et al. Genetic acceleration of AIDS progression by a promoter variant of CCR5. Science. 1998;282:1907–1911. doi: 10.1126/science.282.5395.1907. [DOI] [PubMed] [Google Scholar]
- 19.Hibberd ML, Sumiya M, Summerfield JA, Booy R, Levin M. Association of variants of the gene for mannose-binding lectin with susceptibility to meningococcal disease. Meningococcal Research Group. Lancet. 1999;353:1049–1053. doi: 10.1016/s0140-6736(98)08350-0. [DOI] [PubMed] [Google Scholar]
- 20.Bellamy R, Ruwende C, Corrah T, McAdam KP, Whittle HC, Hill AV. Variations in the NRAMP1 gene and susceptibility to tuberculosis in West Africans. N Engl J Med. 1998;338:640–644. doi: 10.1056/NEJM199803053381002. [DOI] [PubMed] [Google Scholar]
- 21.Todd JR, West BC, McDonald JC. Human leukocyte antigen and leprosy: study in northern Louisiana and review. Rev Infect Dis. 1990;12:63–74. doi: 10.1093/clinids/12.1.63. [DOI] [PubMed] [Google Scholar]
- 22.Roy S, McGuire W, Mascie-Taylor CG, Saha B, Hazra SK, Hill AV, et al. Tumor necrosis factor promoter polymorphism and susceptibility to lepromatous leprosy. J Infect Dis. 1997;176:530–532. doi: 10.1086/517282. [DOI] [PubMed] [Google Scholar]
- 23.Miller LH, Mason SJ, Clyde DF, McGinniss MH. The resistance factor to Plasmodium vivax in blacks. The Duffy-blood-group genotype, FyFy. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 24.Tournamille C, Colin Y, Cartron JP, Le Van Kim C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy-negative individuals. Nat Genet. 1995;10:224–228. doi: 10.1038/ng0695-224. [DOI] [PubMed] [Google Scholar]
- 25.Flint J, Hill AV, Bowden DK, Oppenheimer SJ, Sill PR, Serjeantson SW, et al. High frequencies of alpha-thalassaemia are the result of natural selection by malaria. Nature. 1986;321:744–750. doi: 10.1038/321744a0. [DOI] [PubMed] [Google Scholar]
- 26.Ruwende C, Khoo SC, Snow RW, Yates SN, Kwiatkowski D, Gupta S, et al. Natural selection of hemi- and heterozygotes for G6PD deficiency in Africa by resistance to severe malaria. Nature. 1995;376:246–249. doi: 10.1038/376246a0. [DOI] [PubMed] [Google Scholar]
- 27.Cabrera M, Shaw MA, Sharples C, Williams H, Castes M, Convit J, et al. Polymorphism in tumor necrosis factor genes associated with mucocutaneous leishmaniasis. J Exp Med. 1995;182:1259–1264. doi: 10.1084/jem.182.5.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lander ES, Schork NJ. Genetic dissection of complex traits. Science. 1994;265:2037–2048. doi: 10.1126/science.8091226. [DOI] [PubMed] [Google Scholar]
- 29.Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. doi: 10.1126/science.273.5281.1516. [DOI] [PubMed] [Google Scholar]