Abstract
The genus Abies is widely distributed across the world and is of high importance for forestry. Since chloroplasts are usually uniparentally inherited, they are an important tool for specific scientific issues like gene flow, parentage, migration and, in general, evolutionary analysis. Established genetic markers for organelles in conifers are rather limited to RFLP markers, which are more labour and time intensive, compared with SSR markers. Using QUIAGEN CLC Workbench 23.03, we aligned two chloroplast genomes from different Abies species (NCBI accessions: NC_039581, NC_042778, NC_039582, NC_042410, NC_035067, NC_062889, NC_042775, NC_057314, NC_041464, MH706706, MH047653 and MH510244) to identify potential SSR candidates. Further selection and development of forward and reverse primers was performed using the NCBI Primer Blast Server application. In this article, we introduce a remarkably polymorphic SSR marker set for various Abies species, which can be useful for other conifer genera, such as Cedrus, Pinus, Pseudotsuga or Picea. In total, 17 cpSSRs showed reliable amplification and polymorphisms in A. grandis with a total of 68 haplotypes detected. All 17 cpSSRs amplified in the tested Abies spp. In the other tested species, except for Taxus baccata, at least one primer was polymorphic.
Keywords: Abies, chloroplast, conifer, grand fir, microsatellite, SSR
We introduce a remarkably polymorphic SSR marker set for various Abies species, which can be useful for other conifer genera, such as Cedrus, Pinus, Pseudotsuga or Picea. In total, 17 cpSSRs showed reliable amplification and polymorphisms in Abies grandis with a total of 68 haplotypes detected. All 17 cpSSRs amplified in the tested Abies spp. In the other tested species, except for Taxus baccata, at least one primer was polymorphic.
1. INTRODUCTION
Organelle genomes are commonly known to be inherited in various pathways in plants. Depending on the species, both, chloroplasts and mitochondria, are either inherited maternally, paternally or even by both parents together. Even within conifers, reports about different inheritance pathways are published (Adams, 2019), which is why comprehensive knowledge about the target species is obligatory for genetic studies in conifers. For Abies spp. and Pinaceae, in general, inheritance of chloroplasts usually follows the paternal line and mitochondria the maternal linen (Isoda et al., 2000; Vendramin & Ziegenhagen, 1997; Ziegenhagen et al., 1995). However, all conifer organelle genomes have in common, that their genomes´ silent mutation rates are much lower compared to nuclear sequences (Drouin et al., 2008; Wolfe et al., 1987). Hence, they tend to be more conservative compared with nuclear markers. This combined with the inheritance pathway makes genetic markers in organelles well suitable for gene flow, parentage, migration and, in general, evolutionary analysis. Established conifer chloroplast markers often use restriction enzymes to detect different alleles. However, such RFLP markers tend to be more labour intensive and less polymorphic compared to SSR and SNP markers (Powell et al., 1996). In this publication, we aligned 12 chloroplast genomes of nine different Abies species to detect regions of variation within the genus Abies. Subsequently, we designed and tested chloroplast SSR primer pairs for promising loci aiming for a polymorphic cpSSR set in Abies grandis. Primer candidates were tested in an A. grandis test sample set, which covers the complete native range of the species, as well as additional conifer species. In the present article, we introduce a highly polymorphic, easy–to‐use cpSSR marker set for A. grandis and additional conifer species.
2. MATERIALS AND METHODS
2.1. SSR identification and primer design
We aligned 12 chloroplast genomes of different Abies species (Table 1) using the QIAGEN CLC Workbench 23.03 (Qiagen, Hilden, Germany) to detect chloroplast DNA variation in the Abies genus. All sequences originate from the NCBI database (Sayers et al., 2021) in FASTA format. Manual browsing for regions of variation in short repetitive motives in the alignment revealed potential SSR candidates, of which we chose only those, flanked by conservative nucleotide sequences. The NCBI Primer blast server application (Ye et al., 2012) was used to develop forward and reverse primers in highly conservative strand regions around the SSR candidates. Position in the chloroplast genome (Table A1) of the selected (Table 2) polymorphic and the nonpolymorphic rejected (Table A2) markers was determined by blasting against the NCBI database.
TABLE 1.
NCBI accession code and according to publication for aligned chloroplast genome sequences from different Abies species.
| Species | Origin | Accession | Publication |
|---|---|---|---|
| A. concolor | North America | NC_039581 | Gernandt et al. (2018) |
| A. balsamea | North America | NC_042778 | Wu et al. (2019) |
| A. religiosa | Central America | NC_039582 | Gernandt et al. (2018) |
| A. alba | Europe | NC_042410 | Li et al. (2019) |
| A. sibirica | Europe | NC_035067 | Wu et al. (unbublished) |
| A. ferreana | Asia | NC_062889 | Wang et al. (2022) |
| A. fargesii | Asia | NC_042775 | Guo and Xu (2019) |
| A. fabri | Asia | NC_057314 | Shao et al. (2020) |
| A. chensiensis | Asia | NC_041464 | Zhao et al. (2019) |
| A. chensiensis | Asia | MH706706 | Su et al. (2019) |
| A. chensiensis | Asia | MH047653 | Liu et al. (2018) |
| A. chensiensis | Asia | MH510244 | Li et al. (2018) |
TABLE 2.
Primer sequences, repeat motifs, approximate fragment lengths, melting temperatures, GC content, number of observed alleles and polymorphic information content (PIC) in Abies grandis for 17 introduced polymorphic primer pairs.
| Primer | Primer sequence (5′‐3′) | Repeat motif | Fragment length | Tm (°C) | GC content (%) | Observed alleles | PIC |
|---|---|---|---|---|---|---|---|
| AGcp01‐F | TTCCAACCCCAAGTCTGGTC | C(12) | 202 | 59.52 | 55 | 4 | 0.200 |
| AGcp01‐R | GGATCGGAAATTGCATAAGCCTC | 60.06 | 48 | ||||
| AGcp02‐F | AAGTAGCTCCTGGATGGGGA | A(18) | 309 | 59.66 | 55 | 4 | 0.439 |
| AGcp02‐R | TTTCAACAAGTCGCACACCC | 59.26 | 50 | ||||
| AGcp03‐F | TCACACTGCTTTTCGGAGGG | T(17) | 124 | 60.25 | 55 | 2 | 0.699 |
| AGcp03‐F | TCGTGAAGCGAGAAAGGTGT | 59.61 | 50 | ||||
| AGcp04‐F | TCATTGGGTTCYTTGGRCCTT | G(12) | 214 | 60.13 | 48 | 3 | 0.207 |
| AGcp04‐R | TTGCCTCTCCTGATGGTTGG | 59.67 | 55 | ||||
| AGcp05‐F | TCGATCCATTTCCACCGGTAT | T(15) | 154 | 58.96 | 48 | 5 | 0.419 |
| AGcp05‐R | TCCGATCTGAATTACGGAAACCT | 59.55 | 43 | ||||
| AGcp06‐F | TGGACCATGAAAATGAAAGAATGGA | A(16) | 205 | 59.22 | 36 | 5 | 0.634 |
| AGcp06‐R | TGGGTAAGTCTTAGGACCCG | 58.14 | 55 | ||||
| AGcp09‐F | ACCTCAGCTATGTCCCTCGT | A(11) | 201 | 60.03 | 55 | 3 | 0.338 |
| AGcp09‐R | CGATCGGTCGCCAGGATAAA | 59.97 | 55 | ||||
| AGcp11‐F | GGGAAGAAAGAACATTTGGAAAACA | T(16) | 205 | 58.65 | 36 | 6 | 0.674 |
| AGcp11‐R | ACGTAATCTCCGGGATCCTTATT | 58.84 | 43 | ||||
| AGcp15‐F | ACCATTCAACCATACCCGCA | AT(7) | 173 | 59.67 | 50 | 3 | 0.223 |
| AGcp15‐R | AATGAAGGTGCTCAAGGGAGG | 59.99 | 52 | ||||
| AGcp18‐F | TTGGTACGGCACTTGAGAGA | T(17) | 169 | 58.67 | 50 | 7 | 0.510 |
| AGcp18‐R | AGTGACATCAATAACTGGTCCAA | 57.76 | 39 | ||||
| AGcp19‐F | CATGCCAACCACTCAACTCAC | A(13)T(5) | 230 | 59.73 | 52 | 3 | 0.201 |
| AGcp19‐R | TGACGTGGTGGAAGTCATCAG | 60 | 52 | ||||
| AGcp20‐F | GTGTTCCTCTATCCGTGGAGT | T(20) | 207 | 58.9 | 52 | 8 | 0.569 |
| AGcp20‐R | GAGGCGTACATCTCTTCTGGT | 59.25 | 52 | ||||
| AGcp21‐F | GCTTACCCTACATGGTCGAGA | A(19) | 123 | 58.97 | 52 | 8 | 0.803 |
| AGcp21‐R | TTCCTGGTATTGTCCAAGAATAGT | 57.36 | 38 | ||||
| AGcp22‐F | CTGCTGGATGCAGAGGAACT | T(14) | 161 | 59.75 | 55 | 3 | 0.493 |
| AGcp22‐R | TCCGATGGATTGTTACTGTGTATTG | 59.18 | 40 | ||||
| AGcp23‐F | TGGATTCGGTCCATTGATTGC | T(9)‐A‐C(9)T(7) | 178 | 58.98 | 48 | 8 | 0.780 |
| AGcp23‐R | CCATATTAGTTGACACRAKMTTTCA | 57.23 | 36 | ||||
| AGcp24‐F | GACCGATCATTGCGGGTACA | T(16) | 334 | 60.18 | 55 | 6 | 0.510 |
| AGcp24‐R | ATCCTCATGGAGGTGGGGAA | 59.95 | 55 | ||||
| AGcp25‐F | ACCCTTTTCCGAGGGGTAGT | T(23) | 351 | 60.18 | 55 | 10 | 0.773 |
| AGcp25‐R | ACCTCATACGGCTCCTCCTT | 60.03 | 55 |
2.2. Material
Sample material from A. grandis originates from a sample collection in the German IUFRO provenance trials in Lower Saxony, Hesse and North Rhine Westphalian. In total, 96 A. grandis individuals from all seed provenance regions (Rau et al., 2008) in the natural distribution range were included (Table A3). Sample material for other conifer species stem eighter from a sample collection in the Forest Botanical Garden of the University of Göttingen and the Arboretum in Hørsholm of the University of Copenhagen or were stored DNA samples of projects by the Department of Forest Genetics and tree breeding of the University of Göttingen or by ISOGEN GmbH & Co. KG (for details see Table A4). Also, 96 samples including 12 Abies spp., 2 Cederus spp., Larix decidua, Picea abies, Pinus sylvestris, Pseudotsuga menziesii and Taxus baccata were used in the transferability tests.
2.3. DNA extraction
From all fresh sample, genomic DNA was extracted from mature needle tissue. We used approximately 50 mg fresh or 25 mg dried materials cut into small pieces, frozen in liquid nitrogen and ground in an MM300 ball mill (Retsch, Haan, Germany) for 2 min at 30 Hz. For the extraction, we used the DNeasyTM 96 Plant Kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Stored DNA samples were previously extracted in the same way. For P. abies, 5 mL of 26% polyvinylpyrrolidone solution was added to the 90.5 mL lysis buffer during the extraction.
2.4. Primer pair amplification and scoring
Primer testing was done performing PCR in a Biometra TProfessional thermocycler (Analytik Jena, Jena, Germany) with a touchdown protocol, consisting of 15 minutes initial denaturation (95°C), followed by 10 cycles of 60 s of denaturation (94°C), 60 s annealing (60°C; Δ‐1°C each cycle) and 60 s extension (72°C). Subsequently, another 25 cycles were performed, using the same temperatures and times, but maintaining 50°C in the annealing step. The protocol was finalized by 20 minutes of extension (72°C). Each volume (15 μL) contained about 2 ng of sample DNA, 1.05 pmol Tris–HCl and 0.263 pmol (NH4)2SO4, 37.5 pmol MgCl2, 2.5 pmol of each dNTP and 1U HOT FIREPol® Taq polymerase (Solis BioDyne, Tartu, Estonia). Polymorphic primer pairs were multiplexed with up to four primer pairs. Each sample volume contained 2.5 pmol M13 Primer (fluorescent dye labelled), 0.5 pmol forward and 1.25 pmol reverse primer of each tested primer pair in the respective multiplex. Each forward primer had an M13‐tail addition to the designed sequence.
The fragment sizes of the PCR products were determined using an ABI Genetic Analyzer 3130xl (Applied Biosystems, Foster City, CA, United States) and the accompanying Genemapper software v3.7 (Applied Biosystems, Foster City, CA, United States) (Figure A2).
Initial tests for successful amplification, fragment polymorphisms and good delimitation between fragment sizes were assessed using 16 samples of four distinct seed provenances. Subsequently, the polymorphic and well‐distinguishable primer pairs were tested in an enlarged data set of 96 A. grandis samples across the distribution range to estimate the degree of polymorphism (Table A3). Seed provenance regions follow the classification according to Rau et al. (2008). To test the reproducibility and verify the scoring of the data, an independent PCR, according to the protocol described above, was performed on these 96 samples. Additionally, we tested all polymorphic primer pairs in a sample set composed of 12 different related Abies and seven other conifer species (Table 3/Table A4).
TABLE 3.
Number of alleles for each of the polymorphic AGcp primer pairs in various conifer species.
| N | AGcp‐No. | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01 | 02 | 03 | 04 | 05 | 06 | 09 | 11 | 15 | 18 | 19 | 20 | 21 | 22 | 23 | 24 | 25 | ||
| Abies alba | 8 | 2 | 4 | 4 | 2 | 2 | 3 | 2 | 2 | 1 | 5 | 4 | 2 | 3 | 2 | 5 | 2 | 5 |
| Abies balsamea | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| Abies borisii regis | 8 | 1 | 3 | 5 | 2 | 3 | 3 | 3 | 2 | 1 | 5 | 4 | 3 | 4 | 4 | 3 | 4 | 4 |
| Abies cephalonica | 4 | 1 | 2 | 3 | 2 | 2 | 2 | 3 | 1 | 1 | 3 | 2 | 2 | 2 | 2 | 2 | 3 | 3 |
| Abies concolor | 3 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 | 1 | 1 | 2 | 2 | 2 | 2 | 2 | 1 | 2 |
| Abies fargesii | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 1 | 3 | 2 | 3 | 1 | 3 | 3 | 3 | 2 | 3 |
| Abies holophylla | 5 | 3 | 2 | 3 | 3 | 2 | 3 | 1 | 1 | 1 | 2 | 2 | 2 | 4 | 3 | 2 | 1 | 2 |
| Abies homolepis | 5 | 1 | 3 | 3 | 2 | 3 | 2 | 1 | 1 | 1 | 3 | 3 | 1 | 3 | 2 | 3 | 3 | 4 |
| Abies koreana | 3 | 2 | 1 | 2 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 2 | 1 | 1 | 1 | 2 | 1 | 3 |
| Abies magnifica | 2 | 1 | 1 | 2 | 1 | 2 | 2 | 1 | 2 | 1 | 2 | 2 | 2 | 2 | 2 | 2 | 1 | 2 |
| Abies nordmanniana | 5 | 1 | 2 | 4 | 2 | 2 | 2 | 3 | 1 | 1 | 4 | 3 | 2 | 4 | 2 | 3 | 3 | 3 |
| Abies pinsapo | 4 | 1 | 1 | 2 | 1 | 2 | 3 | 1 | 1 | 1 | 1 | 3 | 2 | 2 | 2 | 3 | 1 | 3 |
| Cedrus atlantica | 4 | 1 | 1 | 2 | 2 | 0 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 |
| Cedrus libani | 4 | 2 | 1 | 1 | 1 | 0 | 2 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 3 |
| Larix decidua | 8 | 3 | 1 | 3 | 3 | 1 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 |
| Picea abies | 8 | 2 | 1 | 1 | 2 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | 2 |
| Pinus sylvestris | 8 | 1 | 1 | 1 | 1 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 2 |
| Pseudotsuga menziesii | 8 | 3 | 1 | 1 | 4 | 1 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 1 | 0 | 2 | 3 |
| Taxus baccata | 4 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 |
Note: Color code highlights whether loci are polymorphic (green), did successfully amplify (yellow) or did not sufficiently amplify (no color) in the tested species. The number of individuals used per species (N) is indicated in italic.
2.5. Haplotype network construction and PCI calculation
A network of the detected haplotype in the 96 samples of A. grandis was constructed with NETWORK 10.2.0.0 (fluxus‐engineering.com) using the median‐joining algorithm (Bandelt et al., 1999). Visualization was performed in Gephi 0.10 (Bastian et al., 2009) using Yifan Hu's proportional layout algorithm (Hu, 2005) followed by manual adjusting for readability, avoiding overlaps and accounting for mutation steps, final post‐processing of the figure was done in Inkscape 1.1 (Inkscape Project, 2021) including inserting pie charts of occurrence frequency in different provenances generated in R 4.2.2 (R Core Team, 2022). Polymorphic information content (PCI) was calculated using the PopGenUtils 0.1.8 R package (Tourvas, 2021).
3. RESULTS
We developed 17 highly polymorphic primer pairs, which exhibited up to 10 different alleles in a comparatively small data set of 96 individuals (Table 2). All developed primer pairs amplify fragment sizes between 70 and 450 bp, contain between 36 and 63% Guanine and Adenine (mean ~ 50.1%) and are characterized by low chances of forming hair‐pin structures (Table A5). Melting temperatures ranged between 57°C and 61°C for all primers, with less than 2°C difference between primers of the same pair. The majority of the promising SSR candidates (repetitive motifs, polymorphic between different genomes) amplify single base pair repetitive motifs. We selected two double base pair motives, as well. However, only one of those (AGcp15) occur to be polymorphic in the primer tests. Despite the mostly single base pair repeats reproducibility was >99% over all primer in the 96 A. grandis samples in the comparison of the two independently performed PCRs and scorings.
All polymorphic primers amplified in all of the 12 tested Abies species (Table 3). Despite the small sample sizes (2 ≤ n ≤ 8) for each species apart from A. grandis, we observed polymorphisms in the majority of successfully amplified primer pairs across all Abies species. For conifer genera apart from Abies, amplification was often not successful for single primer pairs. However, we observed polymorphic loci in some genera, such as Cedrus, Larix, Picea, Pinus and Pseudotsuga. Only within the Taxus genus none of the primer pairs occurred to be polymorphic.
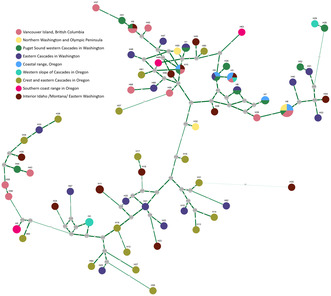
The haplotype network (Figure A1) shows a group of more closely related haplotypes, including those haplotypes that occur multiple times and in different regions of origin. Within the network, no clear separation of regions based on haplotypes can be observed.
4. DISCUSSION
The introduced cpSSR set for A. grandis is remarkably polymorphic also in other conifers especially Abies spp. Using 17 novel cpSSRs, we were able to define 68 different haplotypes from a sample set of only 96 A. grandis individuals. Previous studies already provided evidence for high diversity in Abies chloroplast haplotypes (e.g. Parducci et al., 2001; Vendramin et al., 1999), even using a limited amount of cpSSR markers. However, the introduced marker set of 17 cpSSR markers provides a valuable addition to the available marker set. Using additional genetic markers enables higher resolution of the genetic diversity, especially in geographically widespread studies. All of those novel cpSSR markers are also well suited for multiplex approaches and might thus contribute to a time and labour effective workflow. Finally, the transferability of some markers to additional species provides a functional tool for chloroplast haplotype definition in species of other genera, such as Pseudotsuga menziesii, or Picea abies. Certainly, the broad coverage of the distribution range contributed to this high diversity. Moreover, with up to 10 different alleles per primer pair, it is very likely to find even more haplotypes in larger datasets. Together with the option to use the marker set in several different Abies species, its highly polymorphic character makes it a well‐suited toolkit for future studies across a wide range of Abies species. The low number of polymorphic cpSSRs in A. balsamea might be due to the fact that only two seeds from the same seed source (Canda, Quebec) were analysed. The primer pairs AGcp01, AGcp02, AGcp03 and AGcp24 amplified in all tested conifer species and additional primer pairs amplified also in some of the non‐Abies species from different conifer genera. However, for very few primer pair‐species combinations, the PCR protocol might be not ideal and could be optimized for usage. The majority of the peaks were very clearly recognizable and easy to delimit. Despite their small sample sizes (n ≤ 8), some of the primer pairs, such as AGcp01, AGcp04, AGcp24 and AGcp25, even had various alleles in some tested non‐Abies species. Thus, it is likely, that some of the loci exhibit additional alleles in the tested species since our test plate contained only a small sample count of each species. Therefore, primer pairs, which were successfully amplified in the tested species, are promising candidates for those genera.
Despite the predominantly single base pair repeat motifs, the introduced cpSSR set exhibited reliable peaks, which could easily be kept apart and showed >99% reproducibility between repetitions. We chose highly conservative regions to design both, the forward and reverse primers on the upstream and downstream strands. No primers were developed for polymorphic regions in the alignment. SSR primer pairs from such regions would probably not result in reliable results since every possible primer pair would harbour the risk of several SNPs within the binding region. Such polymorphisms could compromise the annealing of the primers, possibly leading to non‐amplification of the targeted region in some samples. To examine such high polymorphic gene regions, SNP screening would probably be more promising due to the high abundance of SNPs in this region. However, some of the introduced primer pairs are positioned within gene sequences (Table S1) and, therefore, might be correlated to adaptive traits. Primer pairs excluded after the first test with 16 A. grandis samples could still be of interest in other Abies spp. Comprehensive information about all remaining primer pairs is reported in Table S2.
5. CONCLUSION
We have developed a reliable and user‐friendly cpSSR marker set for the relatively sparsely investigated species A. grandis. In a relatively small sample set, the marker set has already shown remarkable diversity. Further studies building on this marker set will contribute to the understanding of genetic patterns within the natural distribution range of A. grandis. Additionally, we demonstrated the usability of the set in other Abies spp. and conifer genera, improving upon the mostly RFLP‐based marker systems available for the chloroplast genome in conifers.
AUTHOR CONTRIBUTIONS
Jeremias Götz: Data curation (equal); formal analysis (equal); investigation (lead); writing – original draft (lead); writing – review and editing (equal). Ludger Leinemann: Funding acquisition (equal); resources (equal); supervision (equal); writing – review and editing (equal). Oliver Gailing: Funding acquisition (equal); resources (equal); supervision (equal); writing – review and editing (equal). André Hardtke: Resources (equal); writing – review and editing (equal). Oliver Caré: Conceptualization (lead); data curation (equal); formal analysis (equal); investigation (supporting); methodology (lead); resources (equal); visualization (lead); writing – review and editing (equal).
FUNDING INFORMATION
This research was funded by the German Federal Ministry of Food and Agriculture (BMEL) represented by the Fachagentur Nachwachsende Rohstoffe e. V. (FNR) grant number FKZ 2220NR313A. We acknowledge support by the Open Access Publication Funds of the Göttingen University.
CONFLICT OF INTEREST STATEMENT
The authors declare no competing interests that are relevant to the content of this article.
Supporting information
Tables S1–S2
ACKNOWLEDGEMENTS
The authors are grateful for the support in sample collection in the IUFRO provenance trials by Bernhard Hosius (ISOGEN GmbH & Co. KG) and the forest rangers of the Hessian, North Rhine Westphalian and Lower Saxonian state forest. We would also like to show our gratitude to Volker Meng (Forest Botanical Garden and Arboretum of the University of Göttingen), Ole Kim Hansen and Erik Dahl Kjær (University of Copenhagen, Department of Geosciences and Natural Resource Management, Section for Forest, Nature and Biomass) for information and permission for the sample collection of different Abies spp. in the Forest Botanical Garden of the University of Göttingen and the Arboretum in Hørsholm of the University of Copenhagen, respectively. Open Access funding enabled and organized by Projekt DEAL.
TABLE A1.
Position of 17 polymorphic AGcp SSR amplicons, relative to Abies chloroplast genes.
| Primerpair | Closest genes/proteins | Amplicon position, relative to genes |
|---|---|---|
| AGcp01 | psbK | Overlapping amplicon, adjacent to 3′ end of gene sequence, overlapping |
| AGcp02 | psbH | Overlapping amplicon, adjacent to 5′ end of gene sequence, overlapping |
| AGcp03 | L16, S12 | Inside gene sequence |
| AGcp04 | rps12, psbZ | Inside rps12 sequence, adjacent to 3′ end of psbZ sequence, overlapping |
| AGcp05 | rps12 | Inside gene sequence |
| AGcp06 | rps12, rsp15 | Inside rps12 gene sequence, adjacent to 3′ end of rsp15 sequence, overlapping |
| AGcp07 | rps12 | Inside gene sequence |
| AGcp08 | trnL‐CAA | Adjacent to 3′ end of gene, overlapping amplicon |
| AGcp09 | trnK‐UUU | Inside gene sequence |
| AGcp10 | trnK‐UUU | Adjacent to 5′ end of gene, non‐overlapping amplicon |
| AGcp11 | psbI | Adjacent to 3′ end of gene, non‐overlapping amplicon |
| AGcp12 | psbT, psbN | Amplicon between gene sequences, overlapping both sequences |
| AGcp13 | rps12, petD | Inside both gene sequences |
| AGcp14 | rps12, rlp14, rlp16 | Inside rsp 12gene sequence, overlapping 5´end of rlp14 gene sequence and 3´end of rlp 16 gene sequence. |
| AGcp15 | rps12, trnG‐UCC | Inside rsp 12gene sequence, adjacent to 5′ end of trnG‐UCC gene sequence, overlapping amplicon |
| AGcp16 | rps12 | Inside gene sequence |
| AGcp17 | rps12 | Inside gene sequence |
| AGcp18 | rps12 | Inside gene sequence |
| AGcp19 | rps12, trnV‐GAC | Inside rsp12 gene sequence, amplicon adjacent to 5′ end of trnV‐GAC gene sequence, overlapping |
| AGcp20 | ‐ | No genes nearby |
| AGcp21 | ‐ | No genes nearby |
| AGcp22 | trnI‐CAU | Adjacent to 5′ end of gene, non‐overlapping amplicon |
| AGcp23 | ycf2 | Adjacent to 5′ end of gene, non‐overlapping amplicon |
| AGcp24 | rps12, rsp19, rpl2 | inside rsp 12 gene sequence, amplicon adjacent to 5′ end of rsp19, and 3# end of rpl2 gene sequence, overlapping both sequences |
| AGcp25 | rps12, ycf3 | Inside both gene sequences |
TABLE A2.
Information about the initially non‐polymorphic chloroplast SSR markers, developed for Abies grandis.
| Primer | Primer sequence (5′‐3′) | Pepeat motif | Fragment length | Tm (°C) | GC content (%) |
|---|---|---|---|---|---|
| AGcp07‐F | TACCATCCCCATCAGAACGA | T(13) | 259 | 57.83 | 50 |
| AGcp07‐R | GACGATCATAGGTCGGGAGT | 58.39 | 55 | ||
| AGcp08‐F | ATCGATATCAATACTCCAATACGCT | TA(8) | 149 | 57.9 | 36 |
| AGcp08‐R | TTTGAGTCTCGCGTGTCTACC | 60.07 | 52 | ||
| AGcp10‐F | GTYGCACGTTGCTTTCTACC | T(5) | 70 | 59.84 | 55 |
| AGcp10‐R | TGATTCGAGATCAGSAGGGAG | 59.31 | 52 | ||
| AGcp12‐F | TTCTTCCGAGAACCACCCAA | C(4) | 177 | 58.87 | 50 |
| AGcp12‐R | TGGGCAACCCTCTGAACAAC | 60.47 | 55 | ||
| AGcp13‐F | ATGGTCCTCGGATTTCTCCT | A(5) | 241 | 57.82 | 50 |
| AGcp13‐R | TGACTGATGCTTTTCCACCCA | 59.86 | 48 | ||
| AGcp14‐F | CGGGCTCCACTGTTATCCG | A(14) | 436 | 60.23 | 63 |
| AGcp14‐R | GCAGATAGAAGCCGGACGAA | 59.9 | 55 | ||
| AGcp16‐F | TGAAGCGAATGGAGACAAGTTA | C(8) | 124 | 57.73 | 41 |
| AGcp16‐R | TCCCTACCCTCTCGTGTCAA | 59.59 | 55 | ||
| AGcp17‐F | TGGAGACTGGGAATTRACGG | T(16) | 232 | 59.39 | 55 |
| AGcp17‐R | GCACGGATAGAAGTGGGTGA | 59.46 | 55 |
TABLE A3.
Origin of the genetic material of the 96 Abies grandis samples from the different German IUFRO trials.
| Origin | Trial | Sample size |
|---|---|---|
| Vancouver Island, Brit. Kolumbien | IUFRO Lauterberg, Germany | 16 |
| Olympic Peninsula, Washington | IUFRO Lauterberg, Germany | 4 |
| Puget Sound, Washington | IUFRO Lauterberg, Germany | 12 |
| Central Washington | IUFRO Hochstift, Germany | 12 |
| Coastal mountain range, Oregon | IUFRO Lauterberg, Germany | 12 |
| Western Oregon, Main land | IUFRO Lauterberg, Germany | 4 |
| Southern coast, Oregon | IUFRO Lauterberg, Germany | 4 |
| Central Oregon | IUFRO Lauterberg, Germany | 16 |
| Idaho | IUFRO Langen, Germany | 16 |
TABLE A4.
Origin of the genetic material of the 96 samples of other Abies spp. and other conifers.
| Species | Sample origin | Provenance | Sample size | |
|---|---|---|---|---|
| Abies alba | Arboretum Copenhagen | Lapos, Romania | 1 | 8 |
| ISOGEN GmbH& Co.KG | Lower Saxony, Germany | 7 | ||
| Abies balsamea | Arboretum Copenhagen | Quebec, Canada | 2 | 2 |
| Abies borisii regis | Forest Botanical Garden Göttingen | x | 2 | 8 |
| ISOGEN GmbH & Co.KG | Slavjanka Mountanis, Greece/Bulgaria | 6 | ||
| Abies cephalonica | Forest Botanical Garden Göttingen | x | 4 | 4 |
| Abies concolor | Arboretum Copenhagen | New Mexico, Santa Fee, Blue Bird Mesa, USA | 1 | 3 |
| Forest Botanical Garden Göttingen | x | 2 | ||
| Abies fargesii | Arboretum Copenhagen | Gansu (2) and Sichuan (1), China | 3 | 3 |
| Abies holophylla | Arboretum Copenhagen | Gangwon, South Korea (3) and Forest in Denmark (2) | 5 | 5 |
| Abies homolepis | Arboretum Copenhagen | Shizouka, Japan | 2 | 5 |
| Forest Botanical Garden Göttingen | x | 3 | ||
| Abies koreana | Arboretum Copenhagen | Jeju Iland, South Korea | 3 | 3 |
| Abies magnifica | Forest Botanical Garden Göttingen | x | 2 | 2 |
| Abies nordmanniana | Arboretum Copenhagen | Sups. equi‐trojani, Denmark | 1 | 5 |
| ISOGEN GmbH & Co.KG | Sups. bornmuelleriana Kökez, Bolu, Turkey (2), sups. ambrolauri Caucasus, Georgia (2) | 4 | ||
| Abies pinsapo | Forest Botanical Garden Göttingen | x | 1 | 4 |
| Arboretum Copenhagen | Cadiz, Spain (1) and Rif Mts., Marocco (2) | 3 | ||
| Cedrus atlantica | ISOGEN GmbH & Co.KG | North Rhine‐Westphalia, Germany | 4 | 4 |
| Cedrus libani | ISOGEN GmbH & Co.KG | North Rhine‐Westphalia, Germany | 4 | 4 |
| Larix decidua | ISOGEN GmbH & Co.KG | Kusel, Rhineland‐Palatinate, Germany | 8 | 8 |
| Picea abies | Department of Forest Genetics and Forest Tree breeding | Low‐elevation phenotype, Harz Mts.; Germany | 4 | 8 |
| High elevation phenotype, Thuringia, Germany | 4 | |||
| Pinus sylvestris | ISOGEN GmbH & Co.KG | Thuringia, Germany | 8 | 8 |
| Pseudotsuga menziesii | ISOGEN GmbH & Co.KG | var. menziesii, North Rhine‐Westphalia, Germany | 4 | 8 |
| Department of Forest Genetics and Forest Tree breeding | var. glauca, IUFRO trial Germany | 4 | 8 | |
| Taxus baccata | Department of Forest Genetics and Forest Tree breeding | Bavaria, Germany | 4 | |
Note: Sample origin refers to the place of sampling or the institution where samples were collected/stored and provenance for known geographic origin of the material.
TABLE A5.
Self complementarity scores of primers according to NCBI Primer blast server application (Ye et al., 2012).
| Primer | Self complementarity | Self 3′ complementarity |
|---|---|---|
| AGcp01‐F | 3 | 1 |
| AGcp01‐R | 4 | 1 |
| AGcp02‐F | 6 | 0 |
| AGcp02‐R | 3 | 0 |
| AGcp03‐F | 3 | 0 |
| AGcp03‐F | 3 | 0 |
| AGcp04‐F | 4 | 0 |
| AGcp04‐R | 2 | 0 |
| AGcp05‐F | 6 | 2 |
| AGcp05‐R | 6 | 0 |
| AGcp06‐F | 5 | 0 |
| AGcp06‐R | 7 | 2 |
| AGcp09‐F | 4 | 0 |
| AGcp09‐R | 6 | 0 |
| AGcp11‐F | 4 | 0 |
| AGcp11‐R | 6 | 1 |
| AGcp15‐F | 3 | 0 |
| AGcp15‐R | 3 | 0 |
| AGcp18‐F | 4 | 0 |
| AGcp18‐R | 5 | 3 |
| AGcp19‐F | 4 | 0 |
| AGcp19‐R | 4 | 1 |
| AGcp20‐F | 4 | 2 |
| AGcp20‐R | 6 | 0 |
| AGcp21‐F | 4 | 2 |
| AGcp21‐R | 5 | 2 |
| AGcp22‐F | 6 | 1 |
| AGcp22‐R | 4 | 1 |
| AGcp23‐F | 4 | 2 |
| AGcp23‐R | 4 | 2 |
| AGcp24‐F | 4 | 2 |
| AGcp24‐R | 6 | 0 |
| AGcp25‐F | 4 | 1 |
| AGcp25‐R | 2 | 0 |
FIGURE A1.
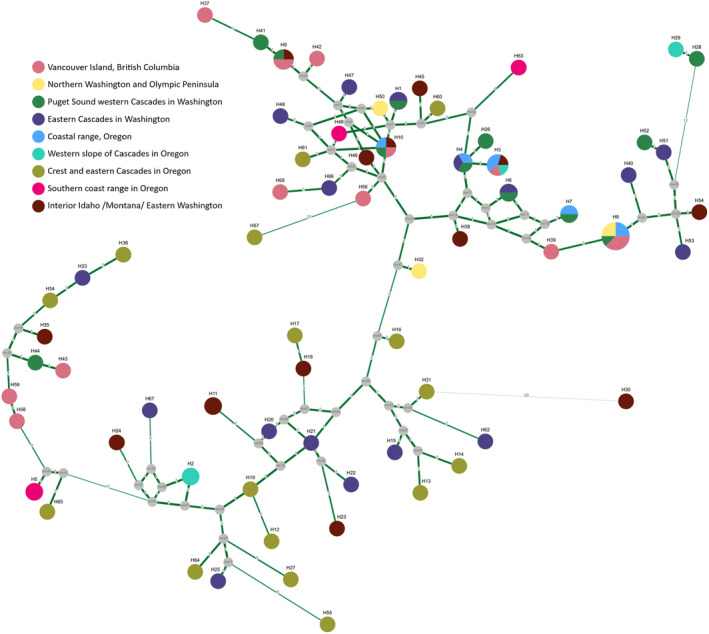
Median vector haplotype network of the detected 68 variants at 11 cpSSRs in 96 samples of Grand Fir (Abies grandis) across the distribution range. Haplotypes are named numerically with the prefix ‘H’ and coloured according to the origin of the sample/s containing respective haplotype. Median vectors are displayed as grey circles. Mutation steps are indicated as numbers at and thickness of the connecting edges. Construction of the network was performed with NETWORK 10.2.0.0 (fluxus‐engineering.com) using the median‐joining algorithm (Bandelt et al., 1999).
FIGURE A2.
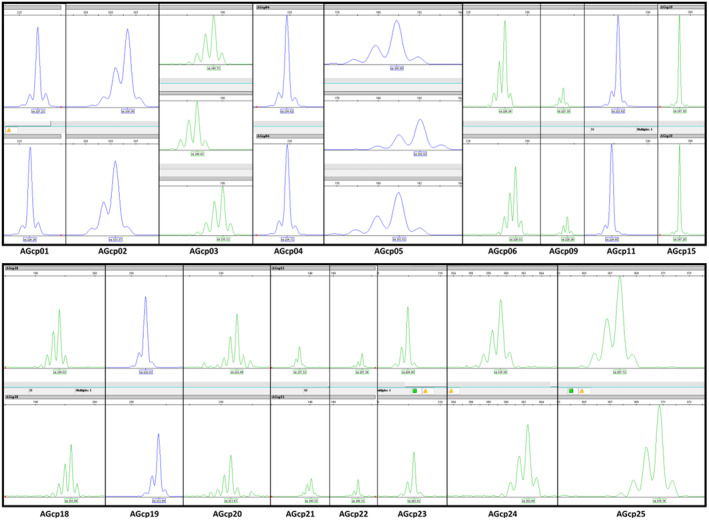
Representative peaks of different alleles for each of the introduced Abies grandis SSR markers as displayed in the ABI Genetic Analyzer 3130xl.
Götz, J. , Leinemann, L. , Gailing, O. , Hardtke, A. , & Caré, O. (2024). Development of a highly polymorphic chloroplast SSR set in Abies grandis with transferability to other conifer species—A promising toolkit for gene flow investigations. Ecology and Evolution, 14, e11593. 10.1002/ece3.11593
DATA AVAILABILITY STATEMENT
Genotype data are available as a Tables S1 and S2 with the online version of the current paper.
REFERENCES
- Adams, R. P. (2019). Inheritance of chloroplasts and mitochondria in conifers: A review of paternal, maternal, leakage and facultative inheritance. Phytologia, 101(2), 134–138. [Google Scholar]
- Bandelt, H. J. , Forster, P. , & Rohl, A. (1999). Median‐joining networks for inferring intraspecific phylogenies. Molecular Biology and Evolution, 16, 37–48. 10.1093/oxfordjournals.molbev.a026036 [DOI] [PubMed] [Google Scholar]
- Bastian, M. , Heymann, S. , & Jacomy, M. (2009). Gephi: An open source software for exploring and manipulating networks. Proceedings of the 3rd International AAAI Conference on Weblogs and Social Media, ICWSM 2009, 3, 361–362. 10.1609/icwsm.v3i1.13937 [DOI] [Google Scholar]
- Drouin, G. , Daoud, H. , & Xia, J. (2008). Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Molecular Phylogenetics and Evolution, 49, 827–831. 10.1016/j.ympev.2008.09.009 [DOI] [PubMed] [Google Scholar]
- Gernandt, D. S. , Reséndiz Arias, C. , Terrazas, T. , Aguirre Dugua, X. , & Willyard, A. (2018). Incorporating fossils into the Pinaceae tree of life. American Journal of Botany, 105, 1329–1344. 10.1002/ajb2.1139 [DOI] [PubMed] [Google Scholar]
- Guo, C. , & Xu, Y. (2019). Next‐generation sequencing yields the complete chloroplast genome of Abies fargesii . Mitochondrial DNA Part B: Resources, 4, 1273–1274. 10.1080/23802359.2019.1591230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. (2005). Efficient and high quality force‐directed graph drawing. Mathematica Journal, 10, 37–71. [Google Scholar]
- Inkscape Project . (2021). Incscape . Retrieved from https://inkscape.org
- Isoda, K. , Shiraishi, S. , Watanabe, S. , & Kitamura, K. (2000). Molecular evidence of natural hybridization between Abies veitchii and A. Homolepis (Pinaceae) revealed by chloroplast, mitochondrial and nuclear DNA markers. Molecular Ecology, 9, 1965–1974. 10.1046/j.1365-294x.2000.01088.x [DOI] [PubMed] [Google Scholar]
- Li, G. Y. , Wu, W. J. , Zhang, Y. X. , Mao, J. H. , Song, Y. J. , Han, X. Y. , Song, M. H. , Guo, Z. N. , Dong, Y. , & Liu, X. M. (2019). Next‐generation sequencing yields the complete chloroplast genome of Abies alba . Mitochondrial DNA Part B: Resources, 4, 575–576. 10.1080/23802359.2018.1535856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Chen, C. , Bai, G. , Li, B. , Chen, H. , Zhou, Y. , & Li, S. (2018). The complete chloroplast genome sequence of Abies chensiensis (Pinaceae: Abietoideae), an endangered species endemic to China. Mitochondrial DNA Part B: Resources, 3, 986–987. 10.1080/23802359.2018.1507636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M.‐L. , Bai, J.‐Q. , Dong, W.‐L. , Wang, R.‐N. , Dong, P.‐B. , Wang, N.‐ , Liu, H.‐Y. , & Fang, M.‐F . (2018). Characterization of the whole plastid genome sequence of Abies chensiensis (Pinaceae), an endangered endemic conifer in China. Mitochondrial DNA Part B, 3, 1141–1142. 10.1080/23802359.2018.1521312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parducci, L. , Szmidt, A. E. , Madaghiele, A. , Anzidei, M. , & Vendramin, G. G. (2001). Genetic variation at chloroplast microsatellites (cpSSRs) in Abies nebrodensis (Lojac.) Mattei and three neighboring Abies species. Theoretical and Applied Genetics, 102, 733–740. 10.1007/s001220051704 [DOI] [Google Scholar]
- Powell, W. , Morgante, M. , Andre, C. , Hanafey, M. , Vogel, J. , Tingey, S. , & Rafalski, A. (1996). The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Molecular Breeding, 2, 225–238. 10.1007/BF00564200 [DOI] [Google Scholar]
- R Core Team . (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing. [Google Scholar]
- Rau, H. , König, A. , & Ruetz, W. (2008). Ergebnisse des westdeutschen IUFRO‐ Küstentannen‐Provenienzversuches im Alter 27. In Beiträge Aus Der Nordwestdeutschen Forstlichen Versuchsanstalt, Band 4″ in Den Universitätsdrucken Im Universitätsverlag Göttingen (p. 62). Nordwestdeutsche Forstliche Versuchsanstalt (NW‐FVA). 10.17875/gup2008-271 [DOI] [Google Scholar]
- Sayers, E. W. , Bolton, E. E. , Brister, J. R. , Canese, K. , Chan, J. , Comeau, D. C. , Connor, R. , Funk, K. , Kelly, C. , Kim, S. , Madej, T. , Marchler‐Bauer, A. , Lanczycki, C. , Lathrop, S. , Lu, Z. , Thibaud‐Nissen, F. , Murphy, T. , Phan, L. , Skripchenko, Y. , … Sherry, S. T. (2021). Database resources of the National Center for biotechnology information. Nucleic Acids Research, 49, D10–D17. 10.1093/nar/gkab1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao, Y. Z. , Chen, Y. , Zhang, X. C. , & Xiang, Q. P. (2020). Species delimitation and phylogeography of Abies delavayi complex: Inferred from morphological, molecular, and climatic data. Journal of Systematics and Evolution, 58, 234–246. 10.1111/jse.12500 [DOI] [Google Scholar]
- Su, L. , Zhao, P.‐F. , Lu, X.‐F. , & Shao, Y.‐Z. (2019). The complete chloroplast genome sequence of Abies chensiensis (Pinaceae). Mitochondrial DNA Part B Resources, 4, 3262–3263. 10.1080/23802359.2018.1542992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourvas, N. (2021). popGenUtils: A collection of useful functions to deal with genetic data in R . R package version 0.1, 6.
- Vendramin, G. G. , Degen, B. , Petit, R. J. , Anzidei, M. , Madaghiele, A. , & Ziegenhagen, B. (1999). High level of variation at Abies alba chloroplast microsatellite loci in Europe. Molecular Ecology, 8(7), 1117–1126. 10.1046/j.1365-294X.1999.00666.x [DOI] [PubMed] [Google Scholar]
- Vendramin, G. G. , & Ziegenhagen, B. (1997). Characterisation and inheritance of polymorphic plastid microsatellites in Abies . Genome, 40, 857–864. 10.1139/g97-811 [DOI] [PubMed] [Google Scholar]
- Wang, D. , Zeng, Q. Y. , & Han, X. M. (2022). The complete chloroplast genome of Chinese endemic species Abies ferreana (Pinaceae) and its phylogenetic analysis. Mitochondrial DNA Part B: Resources, 7, 1282–1284. 10.1080/23802359.2022.2097028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe, K. H. , Li, W. H. , & Sharp, P. M. (1987). Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proceedings of the National Academy of Sciences of the United States of America, 84, 9054–9058. 10.1073/pnas.84.24.9054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, S. , Lu, X. F. , Zhao, P. F. , & Shao, Y. Z. (2019). Next‐generation sequencing yields the complete chloroplast genome of Abies balsamea . Mitochondrial DNA Part B: Resources, 4, 1445–1446. 10.1080/23802359.2019.1598824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z. , Arvestad, L. & Thompson, S. L. (unbublished) Five gymnosperm plastomes reveal rampant rearrangements in Cupressophytes and the retention of ndh pseudogenes in Abies sibirica and Pinus sylvestris .
- Ye, J. , Coulouris, G. , Zaretskaya, I. , Cutcutache, I. , Rozen, S. , & Madden, T. L. (2012). Primer‐BLAST: A tool to design target‐specific primers for polymerase chain reaction. BMC Bioinformatics, 13, 134. 10.1186/1471-2105-13-134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Zhou, T. , Chen, X. , Zhang, X. , Bai, G. , & Zhao, G. (2019). Characterization of the complete chloroplast genome of Abies chensiensis (Pinaceae), an endemic to China. Mitochondrial DNA Part B: Resources, 4, 23–24. 10.1080/23802359.2018.1535851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegenhagen, B. , Kormutak, A. , Schauerte, M. , & Scholz, F. (1995). Restriction site polymorphism in chloroplast dna of silver fir. Forest Genetics, 2, 9. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S2
Data Availability Statement
Genotype data are available as a Tables S1 and S2 with the online version of the current paper.


