Abstract
The placenta plays a crucial role in successful mammalian reproduction. Ruminant animals possess a semi-invasive placenta characterized by a highly vascularized structure formed by maternal endometrial caruncles and fetal placental cotyledons, essential for full-term fetal development. The cow placenta harbors at least two trophoblast cell populations: uninucleate (UNC) and binucleate (BNC) cells. However, the limited capacity to elucidate the transcriptomic dynamics of the placental natural environment has resulted in a poor understanding of both the molecular and cellular interactions between trophoblast cells and niches, and the molecular mechanisms governing trophoblast differentiation and functionalization. To fill this knowledge gap, we employed Stereo-seq to map spatial gene expression patterns at near single-cell resolution in the cow placenta at 90 and 130 days of gestation, attaining high-resolution, spatially resolved gene expression profiles. Based on clustering and cell marker gene expression analyses, key transcription factors, including YBX1 and NPAS2, were shown to regulate the heterogeneity of trophoblast cell subpopulations. Cell communication and trajectory analysis provided a framework for understanding cell-cell interactions and the differentiation of trophoblasts into BNCs in the placental microenvironment. Differential analysis of cell trajectories identified a set of genes involved in regulation of trophoblast differentiation. Additionally, spatial modules and co-variant genes that help shape specific tissue structures were identified. Together, these findings provide foundational insights into important biological pathways critical to the placental development and function in cows.
Keywords: Spatial transcriptomics, Dairy cow, Placenta, Gestation
INTRODUCTION
Efficient reproduction is crucial to the profitability and sustainability of the beef and dairy cattle industries (Sánchez et al., 2018; Wiltbank et al., 2016). Pregnancy loss is significant in both beef and dairy cattle, even in well-managed herds, playing a major role in productivity (Diskin et al., 2016). Multiple factors affect the establishment and maintenance of cow (Bos taurus) pregnancies, including fertilization, embryo cleavage, and functional placental development (Moraes et al., 2018; Spencer & Hansen, 2015). A functional placenta is crucial for the absorption and transportation of nutrients necessary for fetal growth, especially in the second and third trimesters, affecting offspring survival and adult health (Davenport et al., 2023a; Sartori et al., 2010). The placenta also produces unique hormones that influence fetal growth and maternal adaptation to pregnancy.
With its unique “polycotyledonary” structure, the cow placenta appeals to reproductive biology researchers. Complex interactions between clusters of fetal villi (“cotyledon”) and maternal crypts (“caruncle”) create a “placentome” (Johnson et al., 2018; Schmidt et al., 2006). The presence of a synepitheliochorial interhemal barrier in the placentome plays a vital role in anchoring the placenta to the uterus and in facilitating the exchange of substances between the mother and fetus (Awad et al., 2013; Dai et al., 2020). Trophoblast cells, originating from the outer layer of the blastomere, are essential in placental formation, and are classified into two distinct types in cows and ruminants: uninucleate trophoblast cells (UNCs) and trophoblast binucleated cells (BNCs) (Wooding, 2022). BNCs differentiate from UNCs either through nuclear division without cell division or through endoreduplication, a process of genome replication without cell division. These cells invade the endometrial epithelium and embed in the endometrial tissue, becoming part of the placenta (Green et al., 2021). UNCs secrete interferon tau (IFNT), a maternal recognition molecule in pregnancy, while BNCs secrete specific molecules critical for successful pregnancy, including chorionic somatomammotropin hormone 1 (CSH1), prolactin-related proteins (PRP), and pregnancy-associated glycoproteins (PAGs) (Wooding & Wathes, 1980). Despite detailed studies on the morphology and histology of cow fertilization and the placenta, our understanding of placental regulation and pregnancy maintenance, as well as the cellular and molecular biology of trophoblast cell differentiation, remains incomplete. Single-cell RNA sequencing (scRNA-seq), a high-throughput molecular analysis technique, offers a valuable approach for investigating placental development by capturing the heterogeneity of gene expression profiles in placental cells at each stage of pregnancy (Li et al., 2020; Liu et al., 2022; Vento-Tormo et al., 2018). Nonetheless, due to cell dissociation, scRNA-seq fails to accurately map the developmental atlas of placental cells within the complex native environment of the placenta. In addition, the characteristic large diameters of BNCs in the ruminant placenta often exceed the maximum pore size of conventional scRNA-seq techniques (40 µm), further increasing the difficulty in using this approach to study placental development. Furthermore, scRNA-seq typically fails to capture the spatial interactions between placental cell lineages, limiting our understanding of placental development (Chen et al., 2022).
Spatial enhanced resolution omics sequencing (Stereo-seq), a spatial transcriptomics technique that enables adjustable resolution, high sensitivity, and a wide field of view, has recently been developed (Chen et al., 2022). In the current study, we applied Stereo-seq to analyze cow placental samples and construct a spatial transcriptomic atlas of placental development, thereby localizing cell types in cotyledons and intercotyledonary regions with high precision and revealing variations in composition, regulatory factors, and their spatial interactions. Placental samples from cows at 90- and 130-day post-gestation, marking the transition into the second trimester, were collected to capture a diverse population of trophoblast cells for transcriptomic characterization and modeling of their formation (Hayashi et al., 2013). Overall, this study provides a comprehensive framework and resource for advancing our understanding of placental development, function, and molecular regulation in ruminants.
MATERIALS AND METHODS
Ethics statement
All applicable international, national, and/or institutional guidelines for the care and use of animals were strictly followed. All relevant procedures involving animal experiments presented in this study were compliant with the ethical regulations regarding animal research and conducted under the approval of the Animal Care and Use Committee of Northwest A&F University (Approval No.: NWAFAC1019).
Dairy cows
A group of healthy Holstein cows were selected for artificial insemination under the same feeding environment and subsequently assessed for pregnancy status after 30 days.
Tissue collection and cryopreservation
Placentas were dissected from two female Holstein cows at different gestational periods (days 90 and 130), one sample per period. The placental cotyledons and surrounding tissues, containing maternal and fetal tissues, were isolated from the uterus, and washed with Dulbecco’s Phosphate-Buffered Saline (DPBS) to remove residual blood. Each sample included both caruncular/placentomal and adjacent intercaruncular/interplacentomal areas, subsequently placed in OCT-containing cassettes. During the cleaning and embedding processes, great care was taken to preserve the adhesion between fetal and maternal tissues, with sample collection completed within 5 min each time. Cryosections were cut to 10 μm thickness using a Leica CM1950 cryostat (Germany).
Stereo-seq library preparation and sequencing
Preparation of the Stereo-Seq chip and library were performed as described previously (Chen et al., 2022; Liu et al., 2022). Tissue sections were adhered to a Stereo-seq chip, fixed in methanol at −20°C for 30 min, then subjected to nucleic acid staining (Thermo Fisher, Q10212, USA) and imaging (Ti-7 Nikon Eclipse microscope, Japan). The tissue sections were then permeated at 37°C for 5 min. The cDNA was purified using VAHTS DNA CleanBeads (Vazyme, N411, China). The indexed scRNA-seq library was generated according to the manufacturer’s protocols. The sequencing library was quantified using the Qubit ssDNA assay. DNA nanospheres (DNBs) were loaded into a patterned nanoarray and sequenced on a MGI DNBSEQ-Tx sequencer (50 bp for read 1 and 100 bp for read 2).
Stereo-seq raw data processing
A Fastq file was generated using a MGIDNBSEQ-Tx sequencer (BGI, China). The CID and MID were contained in read 1 (CID: 1–25 bp, MID: 26–35 bp), while read 2 was composed of cDNA sequences. The CID sequence on the first read was mapped to the in situ-obtained chip design coordinates taken from the first round of sequencing, allowing a one-base mismatch to correct the sequence and polymerase chain reaction (PCR) errors. Readings of MIDs containing N bases or two or more bases and with a quality score of less than 10 were excluded. The CID and MID associated with each read were added to the header of each read. Reads retained using STAR were then matched against the reference genome (ARS-UCD1.3) (Dobin et al., 2013), and mapped reads with MAPQ>10 were counted and annotated with the corresponding gene. Unique Molecular Identifiers (UMIs) sharing the same CID and locus were collapsed, allowing a mismatch to correct sequence and PCR errors. Finally, this information was used to generate an expression profile matrix that contained the CID. The entire methodology was integrated into the SAW pipeline, available at https://github.com/BGIResearch/SAW.
Binning data of spatial Stereo-seq data
Raw data were processed with the same procedure described in Chen et al. (2022). Transcripts collected from 100×100 DNBs were merged into one Bin 100, with Bin 100 treated as a unit of fundamental analysis. Bin IDs were synthesized on the capture chip by their spatial adjustment (spatial_x and spatial_y). In particular, the DNB at the bottom left of Bin 100 was selected to represent the location of Bin 100. The sample was manually outlined to exclude containers not derived from the tissue sample. For comparison with other resolutions, DNBs on day 90 were also binned at other sizes, i.e., Bin 20, 50, 80, 100, 150, and 200, followed by unsupervised clustering.
Stereo-seq data processing, integration, and visualization
Seurat objects were generated from the previously filtered UMI count tables for each sample independently, followed by data normalization and identification of the top 3 000 variable genes. Subsequently, Harmony was used to integrate all datasets from each gestation period into a single Seurat object to rectify batch effects. For each gestation-specific integrated Seurat object, Stereo-seq data from 71 899 barcoded spatial spots were SC-transformed using Seurat (v.4.0.5, https://github.com/satijalab/seurat), then used for batch correction. The top 3 000 variable genes were then selected for spatial RNA-seq clustering using SCTransform (https://satijalab.org/seurat). Principal component analysis (PCA) was then performed on a matrix of spot and gene expression (UMI) counts, reducing data dimensionality to the top 20 principal components (PCs). Uniform Manifold Approximation and Projection (UMAP) was initialized in this PCA space to visualize the data on reduced UMAP dimensions. The spots were clustered on PCA space using the Shared Nearest Neighbor (SNN) algorithm implemented as FindNeighbors and FindClusters in Seurat v.4.0.5 with the parameters k=30 and resolution=0.5. The resulting spot clusters, indicative of tissue anatomical regions, were then visualized on UMAP space using the SpatialDimPlot command. Significant differences between early (EG) and mid-gestation (MG) cluster proportions were calculated using the Fisher’s Exact Test, followed by Benjamini-Hochberg P-value adjustment. Significance was defined as clusters with a false discovery rate (FDR)≤0.05 and an odds ratio≥1.5 or ≤−1.5. An odds ratio of ±infinity was plotted as ±2. The top 10 positive markers per cluster were identified using FindAllMarkers with default parameters. The slide was set as a latent variable and genes detected in at least 10% of one population were considered expressed.
Differential expression analysis within and between cell types
Spots within each cell-type cluster were stratified by EG and MG. Differentially expressed genes (DEGs) between the two pregnancy groups were calculated using FindMarkers, with the parameters: ident.1–“day130”, group.by=“group”, subset.ident=“subset”, and logfc.threshold=0 to output all expressed genes. Statistical significance for differential expression was defined by adjusted P≤0.05, log2 fold-change≥1 or ≤−1, and ≥60% spots in one group (pct.1 or pct.2) expressing the gene.
Identification of maternal and fetal single cells
The Souporcell python tool was applied to determine the genetic origin of each barcoded cell without prior knowledge of individual genotypes (Heaton et al., 2020). Souporcell inferred the identities of Bin1 by analyzing each SAW-aligned BAM file from the cow placental samples against the ARS-UCD1.3 genome and SAW-filtered barcodes. Following Souporcell analysis, each Bin was classified by status (singlet, doublet, or unassigned) and assignment (maternal, fetal, or ambiguous origin), with the proportion of cell sources (Bin 100) quantified based on the resolution adopted for the study, employing a 90% threshold. These identities were then linked to the transcriptome-based cell dimensionality reduction data to confirm the maternal or fetal identity of each annotated cell type.
Cell-type mapping
To validate the cell-type identity characterized in the cow placental sections, published annotated scRNA-seq profiles of cow placenta were downloaded for comparison (Davenport et al., 2023b). In short, cells from Stereo-seq and scRNA-seq underwent SCTransform, FindIntegrationAnchors, and IntegrateData, using mutual dominant components from Seurat, and were further co-embedded in low-dimensional space through UMAP. After co-embedding, a K-nearest neighbor (KNN)-based method was used to connect the cell clusters between Stereo seq and scRNA-seq.
Gene Ontology (GO) analysis of DEGs
Significant genes were selected based on their P-values from the differential expression test. The ClusterProfiler R package was used for enrichment analysis of GO terms representing various biological processes (BP).
Identification of spatially auto-correlated gene modules
The Hotspot tool (v.0.9.1) was used to identify modules of spatially correlated genes (Detomaso & Yosef, 2021). Analysis considered gene expression in all placental sections across two developmental stages, setting a minimum UMI of 50 for both days 90 and 130. Input data were then normalized against the total number of UMIs for these genes in each Bin. Subsequently, a KNN graph for these genes was created using the create_knn_graph function with the parameter n_neighbors=6/10(day90/day130). A gene exhibiting spatial autocorrelation was identified, employing an FDR<0.05 for further analyses. Modules were identified using the “create_modules” function with the parameters: min_gene_threshold (40 genes for day 90 and 30 genes for day 130) and fdr_threshold=0.05. For cell-type enrichment analysis of each module, cell-type composition of Bins with high module scores (>3) was calculated, as described in detail in Hotspot.
Protein-protein interaction (PPI) network analysis of spatial modules
STRING (https://string-db.org) was used to identify potential interactions between genes in one/multiple modules. The PPI network was visualized using Cytoscape v.3.8.2 (https://cytoscape.org/), in which nodes corresponded to proteins and node size was proportional to the relative connectivity in each network.
Ligand-receptor interaction and transcription factor network construction
The CellChat package was applied for ligand-receptor interaction analysis (Jin et al., 2021), with the “secretory signaling”, “extracellular matrix (ECM) receptor”, and “cell-cell contact” paired datasets selected to analyze cellular communication. For transcription factor regulation network analysis, a total of 1 220 cow transcription factors in AnimalTFDB and a cell-gene matrix were taken as input using the GENIE3 package. In the output regulator-target table, only pairs with weights greater than 0.1 were retained and the first 2–6 transcription factors were further displayed.
Immunohistochemistry
Fresh placental tissues were fixed in 4% paraformaldehyde for 24 h, then removed for tissue trimming, dehydration, and embedding in paraffin. The trimmed paraffin blocks were cooled on a −20°C freezing table, then placed on a paraffin slicer and sliced at a thickness of 4 μm. Subsequently, the sections were deparaffined and rehydrated at room temperature, followed by heat-induced antigen retrieval, and blocking with goat serum for 1 h. The sections were then incubated with primary antibodies (YB-1/YBX1 Rabbit mAb (A3534, 1:100) or NPAS2 Rabbit pAb (A16930,1:100)) at 4°C overnight. After washing the following day, the sections were incubated with secondary antibody (horseradish peroxidase (HRP)-anti-rabbit IgG) for 1 h at room temperature. The sections were then washed and stained with 3,3'-diaminobenzidine (DAB) and counterstained with hematoxylin and eosin (H&E).
RESULTS
Spatial clustering and molecular characterization of spatiotemporal transcriptomic atlas of cow placenta
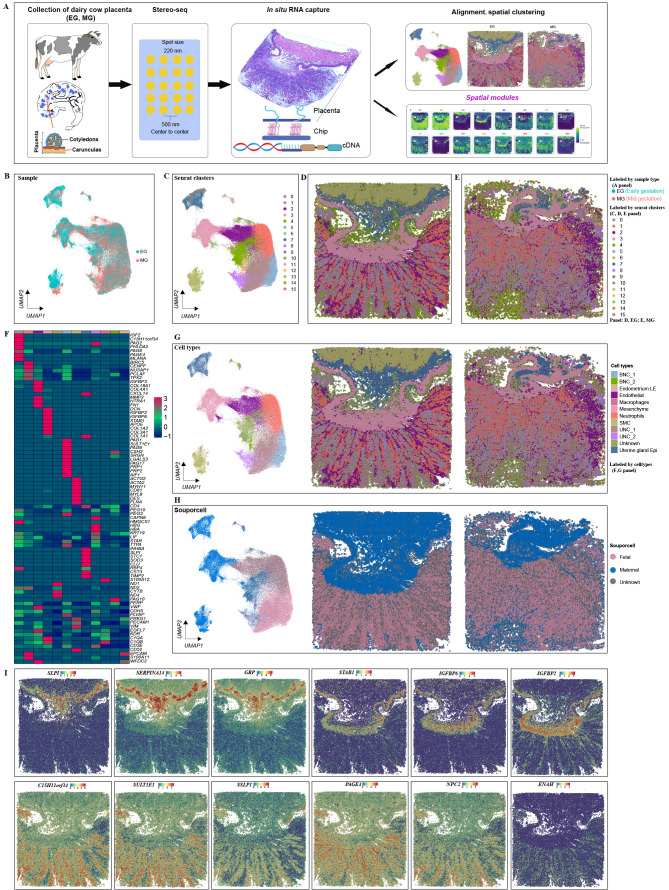
To study the interplay between differentiation and morphogenesis in placental development, we collected longitudinal sections of mushroom-shaped structures from cows at different stages (90- and 130-day post-gestation) for spatial transcriptomic analysis. High-resolution Stereo-seq chips (spot size: 220 nm; center-to-center size: 500 nm; chip size: 1 cm2) were used for the in situ capture of RNA transcripts (Chen et al., 2022; Liu et al., 2022) (Supplementary Figure S1A; Figure 1A), based on the trade-off between capturing fine-scale spatial information and adequate gene expression counts. Comparing the spatial range from 100 μm (Bin 200, 200×200 DNB) to 10 μm (Bin 20, 20×20 DNB, equivalent to approximately one cell diameter), we found that 50 μm (Bin 100, 100×100 DNB) provided a satisfactory number of genes and counts for further analyses, with high resolution (Supplementary Figure S1). Transcripts captured by 100×100 DNBs were then merged as one Bin 100, treated as the fundamental analysis unit. A total of 71 899 Bins were retrieved at this resolution, with an average number of captured genes per Bin ranging from 832 on day 130 to 1 263 on day 90 and UMIs ranging from 1 793 on day 130 to 3 189 on day 90 (Supplementary Figure S1B, C). These variations indicate differences in gene and UMI distribution between sections from different time points and spatial motifs.
Figure 1.
Spatial transcriptomic atlas of dairy cow placenta
A: Spatial transcriptome profiling workflow depicting collection, tissue handling, library preparation of two sections per capture area, sequencing, and analysis (modified from Abdisa (2018), Chen et al. (2022), and Liu et al. (2022)). B–E: Unbiased identification of cell-type heterogeneity and biological variance in day 90 (EG) versus day 130 (MG) cow placenta. B: UMAP clustering is colored by sample group (EG or MG). C: Identified cell clusters. D, E: Spatial mapping of defined clusters. Spatial distribution of main clusters defined for EG (D panel) and MG (E panel) tissue slices. Mapping of clusters on tissue demonstrates appropriate alignment with morphology. F: Gene expression of cell type-specific markers. G: UMAP plot of new clusters defined by gene expression similarity and subsequently labeled according to their congruence with tissue morphology. H: UMAP plot and spatial map showing fetal or maternal origins of filtered single cells based on single nucleotide polymorphism (SNP) analysis. I: Spatially resolved expression of marker genes in partial clustering.
To minimize technical batch effects, the expression profiles for each capture area (Bin 100) were combined for further analyses. Initial UMAP clustering was performed on tissue-covered spots from both EG and MG tissue sections, yielding 16 unique clusters (Figure 1B, C; Supplementary Figures S2, S3). The cluster points were mapped back to the original coordinates in the tissue slices (Figure 1D, E). The genetic variants within the Stereo-seq reads were used to distinguish between fetal and maternal Bins (Heaton et al., 2020) (Figure 1H). The top 10 DEGs per cluster (Figure 1F; Supplementary Table S1) were then analyzed to categorize the clusters into 10 major and biologically relevant cell types (Figure 1G), including: two different types of UNCs (marked by PAG2, C15H11orf34, PAG8, and MLANA) (Green et al., 2000; Hashizume et al., 2007; Xie et al., 1995), neutrophils (Nes, marked by BIRC5, CENPF, NUSAP1, PCLAF, and TPX2) (Davenport et al., 2023b), endothelial (Endo, marked by CDH5, EGFL7, and KDR) and endometrium luminal epithelial cells (Endometrium LE, marked by EPCAM, SERPINE1, and WFDC2), mesenchymal cells (Mes, marked by COL1A1, COL3A1, DCN and IGFBP2) (Olaya-C et al., 2015; Yang et al., 2021), uterine gland epithelial cells (U_Epi, marked byCXCL14, GRP, SLPI, and SERPINA14) (Budipitojo et al., 2003; Kimura et al., 2006; King, 1993; Ulbrich et al., 2009), two populations of BNCs (marked by PAG1, PAG4, CSH2, PRP1, PAG5, PAG6, and PAG10) (Green et al., 2000), macrophages (marked by S100A12, C1QA, and C1QB) (Averill et al., 2012; Ryckman et al., 2003; Wang et al., 2018), and smooth muscle cells (SMCs, marked by ACTG2, ACTA2, MYH11, and DES) (Wang et al., 2022). One unknown cell population was also present, displaying marker genes related to mitochondrial function, which was not identified and categorized. Notably, all clusters contained EG and MG cells (Supplementary Figure S2). Analysis also identified that the expression levels of special AT-rich sequence binding protein 1 (SATB1), gastrin releasing peptide (GRP), serpin peptidase inhibitor (SERPINA14), and secreted seminal-vesicle Ly-6 protein 1 (SSLP1) were spatially restricted to the developing endometrial stromal and gland layers (Figure 1I). Spatial maps of marker genes are shown in Supplementary Figure S4.
Stereo-seq can dissect tissues at cellular resolution (Chen et al., 2022). To validate cell-type identification by placental Stereo-seq, deconvolution analysis of the Stereo-seq data was performed using the KNN method in Seurat (Supplementary Figure S5). To understand the spatial organization of cell types in broad anatomical regions, spots were labeled by cell type and visualized on H&E-stained sections from respective stages (Figure 1G). As expected, spatial profiling of the defined clusters on original tissue sections aligned strongly with morphology. To determine how cell-type composition changed with placental development, the proportions of each cell type were compared between the EG and MG groups, showing that UNCs and BNCs were the main cell types in placentas (Supplementary Figure S6A). General proportions of cluster abundance were significantly imbalanced between EG and MG placentas in seven out of 10 clusters (Supplementary Figure S6C). For instance, the UNC_2, BNC_2, and macrophage clusters were significantly more abundant in the MG (day 130) placentas.
Specific expression patterns and spatial distribution of PAGs and PRPs in cow placenta during pregnancy
PAGs are abundantly expressed in the placentas of species within the Cetartiodactyla order, especially in Bovidae, performing a range of functions in both the epitheliochorial and synepitheliochorial placental forms (Szafranska et al., 2001). Within B. taurus, the PAG family consists of 21 members, categorized into 13 modern and eight ancient phylogenetic groups based on the evolutionary timeline of their emergence (Hughes et al., 2000; Touzard et al., 2013). To detect the spatial and temporal expression of PAGs, gene expression analysis was performed between cell clusters at different stages. Results showed that two types of PAGs were primarily expressed in the cotyledon and to a lesser extent in the caruncle. PAG4, PAG14, PAG18, PAG19, and PAG20 were mostly restricted to the placental cotyledon. PAG4, PAG10, PAG11, PAG15, PAG18, PAG19, PAG20, and PAG21 exhibited lower expression levels in maternal-fetal interface cells at both EG and MG (Supplementary Figure S7A). One exception in the ancient group was PAG2 on day 130, which showed higher expression in the caruncle. In the cotyledon, PAG1, PAG17, and PAG21 from the modern group showed similar patterns of spatial expression to PAG2.
The placenta generates different peptides and steroid hormones that regulate fetal growth and placental function. PRPs are placenta-produced peptides that belong to the growth hormone/prolactin family and show structural similarities to prolactin and placental prolactin. Although several PRP genes have been detected in the cow placenta, their spatiotemporal expression profiles are not clear. Based on the Stereo-seq dataset, our results showed that PRP1, PRP2, PRP3, PRP4, PRP6, PRP8, PRP9, and PRP14 were predominantly expressed in the cotyledon, exhibiting similar spatial expression patterns as PAGs, with PRP1 showing high expression and PRP4, PRP9, and PRP14 showing low expression in the cotyledon (Supplementary Figure S7B).
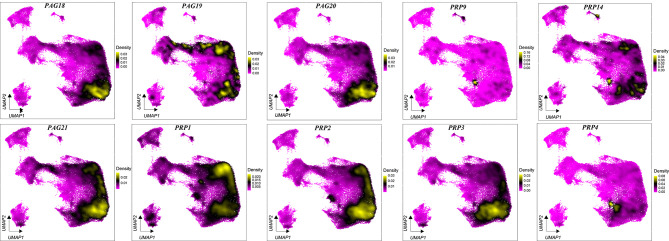
Density plots showed the distribution of PAG and PRP gene expression in the different clusters (Figure 2). PAGs and PRPs were enriched in UNC and BNC clusters (Figure 2) (Polei et al., 2020a). In addition, PAG12, PAG9, and PAG14 were expressed in only a small fraction of cells in the UNC and BNC clusters, suggesting that these clusters are heterogeneous. Ancient PAG8 was mainly expressed in UNCs but not in BNCs, consistent with previous research (Wiedemann et al., 2018). Several PAGs and PRPs, such as PRP1, PAG1, PAG3, PAG5, and PAG7, exhibited high density distributions in neutrophils. These results highlight the specific expression and cellular localization of PAGs and PRPs.
Figure 2.
Density distribution of PAGs and PRPs in different cell clusters in cow placentas during pregnancy
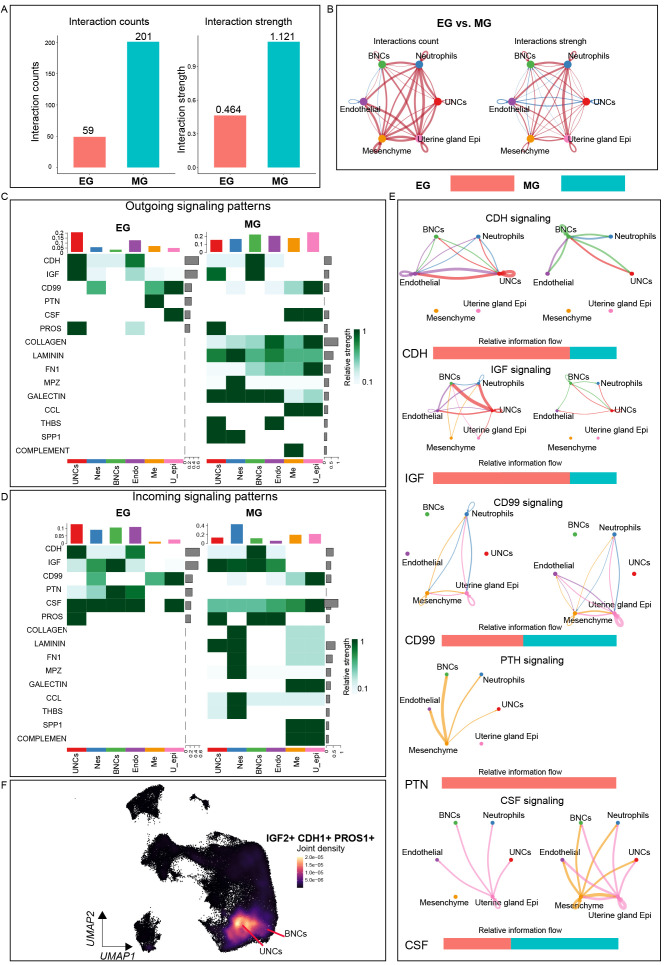
Cell-cell interaction analysis of placentas in different gestation periods
Given the successful creation of spatial transcriptome profiles of the cow placenta and characterization of the molecular landscape in different regions, unbiased “ligand-receptor”, “ECM-receptor”, and “cell-cell contact” interaction analyses were next performed using CellChat to investigate the complex signaling networks in the placental microenvironment (Ge et al., 2020; Ramilowski et al., 2015). In total, 59 and 201 significant interactions were identified in the EG and MG placentas, respectively (Figure 3A, B). Cellular communication strength over placental space is shown in Supplementary Figure S8A. Among the six clusters, UNCs and BNCs exhibited the strongest outgoing signaling pattern, indicating that these cells are essential in microenvironment homeostasis regulation during cow pregnancy (Figure 3C). Unexpectedly, several conserved (CDH, IGF, CD99, CSF, and PROS) and specific patterns (COLLAGEN, LAMNIN, FN1, and MPZ) of signal input and output were observed at both EG and MG (spatial expression patterns are displayed in Supplementary Figure S8B). Among these signaling interactions, many classical cell adhesion, cell migration, and epithelial cell proliferation, differentiation, and apoptosis signaling pathways were identified with strong signaling output patterns in UNCs and BNCs, including IGF, CDH, and PROS (Figure 3C, E), with the expression density of genes mainly in UNCs and BNCs. Certain signals with different patterns were observed between EG and MG, including PTH, CSF, and CCL (Figure 3F). These signaling network results may help further our understanding of the physiological processes occurring in the cow placental microenvironment.
Figure 3.
Cell-cell communication network within placental microenvironment
A: Bar plot showing total interaction counts and strength within placental microenvironment in EG and MG placentas. B: Circle plots showing up- or down-regulation of cell-cell communication network between EG and MG placentas. Red and blue lines represent up- and down-regulated interactions, respectively. C, D: Heatmap of outgoing/incoming signals of each cluster between EG and MG placentas. Gradient of white to dark green indicates low to high expression weight values in heatmap. E: Circle plots showing differential strength (right) of specific signals in cell-cell communication network between EG and MG. Red or blue edges represent increased or decreased signals in EG state, respectively. F: Joint density map of selected signaling pathways expressed in placental cell clusters.
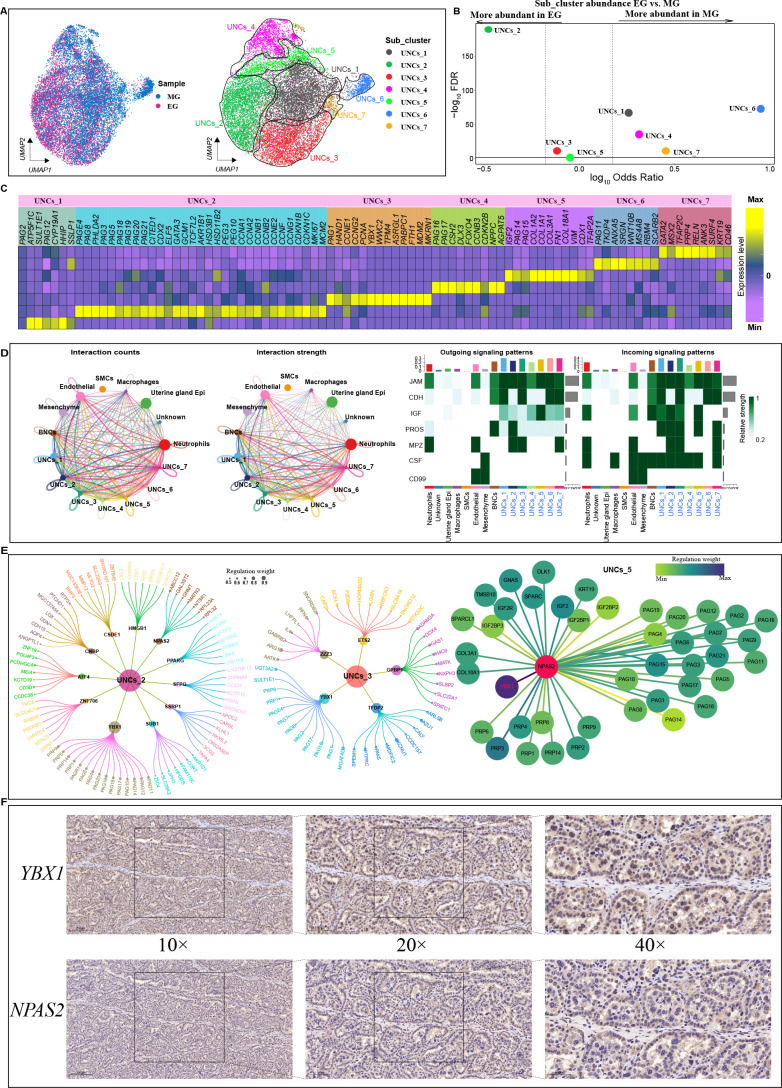
Heterogeneity and spatial localization of UNC cluster in EG and MG
Given that UNCs play a significant role in implantation, establishment, and maintenance of placentation during pregnancy in cows (Sağsöz et al., 2022), populations of UNCs at both gestation stages were collected for subpopulation analysis. Subcluster analysis of UNCs revealed seven transcriptionally unique subpopulations, including UNCs_1 (SULT1E1 and CYP19A1), UNCs_2 (PAG8, PEG3, PEG10, and PHLDA2), UNCs_3 (PAG1, HAND1, and PCNA), UNCs_4 (CSH2, PAG16, and PAG17), UNCs_5 (IGF2 and TFAP2A), UNCs_6 (PAG11 and TKDP4), and UNCs_7 (GATA2 and MSX2) (Figure 4A–C). The relative proportions of all seven populations were significantly different between EG and MG, with subclusters 2 and 3 decreasing and subclusters 1, 4, 6, and 7 increasing during placental development (Figure 4B; Supplementary Figure S9 for cell subpopulation spatial locations). These results indicate that different stages of gestation can broadly impact the relative abundance of UNC subpopulations.
Figure 4.
Heterogeneity within UNC clusters
A: UMAP clustering of UNC spots based on two developmental stages (left) and gene expression (right). Cells are colored according to their developmental stage or types. B: Differences in cluster abundance plotted as log10 odds ratios against adjusted P-value –log10 FDR. Solid horizontal line represents significance threshold of 0.05 and vertical dotted lines represent a 1.5-fold change in either direction. C: Heatmap of DEGs in each UNC subcluster based on two developmental stage samples. D: Left: Number (left) and weight (right) of interactions between UNC subcluster and all clusters. Different colors represent different signal sources. Line width indicates number/weight of interactions. Right: Heatmap of CellChat signaling in each UNC subcluster. Left panel shows outgoing signaling patterns (expression weight value of signaling molecules) and right panel shows incoming signaling patterns (expression weight value of signaling receptors). E: Top candidate master regulators and their target genes for UNCs_2 (left), UNCs_3 (center) and UNCs_5 (right) subclusters. Bubble size indicates weight from high to low. F: Immunohistochemical results of NPAS2 and YBX1 in cow placenta. Scale bar: 200 μm (10×); 100 μm (20×); 50 μm (40×).
To further explore the UNC subpopulations and their biological functions, differential expression analysis was performed among the seven UNC subpopulations. Although the general PAG and PRP genes were expressed in all UNC subclusters, some PAG and PRP types showed different expression patterns in the seven UNC subclusters (Figure 4C). For example, PAG8, PAG3, PAG18, PAG19, PAG20, PAG21, and PAG5 were highly expressed in UNCs_2, while CSH2, PAG16, and PAG17 were highly expressed in UNCs_5. PRP4 showed high expression in UNCs_7, while CYP19A1 and ATP5F1C were highly expressed in UNCs_1. The CYP19A1 gene, which encodes aromatase, is involved in the biosynthesis of estrogens, required for successful establishment and maintenance of pregnancy and proper fetal development (Chatuphonprasert et al., 2018), whereas the ATP5F1C gene is involved in the assembly and function of adenosine triphosphate (ATP) synthase, playing a crucial role in cellular energy metabolism (Zeng et al., 2022). The UNCs_2 subpopulation, in addition to expressing typical trophoblast cell markers, also expressed transcription factors (CITED1, CDX2, GATA3, TCF7L2), proliferation markers (MKI67, MCM2), and cell cycle markers (CCNA1, CCNA2, CCNB1, CCNB2, CCNE2, CCNF, CCNG1, CCNG2, CDKN1B, CDKN1C), and was thus defined as proliferative UNCs. Interestingly, relatively higher expression levels of related ECM components, including type I collagen proteins (COL18A1, COL3A1, COL1A1, and COL1A2), were observed in UNCs_5 and UNCs_2. These findings are consistent with Polei et al. (2020b), revealing the expression of collagen proteins in some trophoblast cells. Subsequent GO and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses revealed that the molecular functions and biological processes regulated by DEGs in UNCs_5 and UNCs_6 were related to the ECM (Supplementary Figure S10E). KEGG pathway analysis indicated that UNCs_1 and UNCs_7 were involved in the vascular endothelial growth factor (VEGF) signaling pathway, while UNCS_2 and UNCs_3 were enriched with the HIF-1 signaling pathway. The VEGF and HIF-1 signaling pathways are implicated in modulating trophoblast cell invasion and angiogenesis during placental development (Anbo et al., 2019; Yu et al., 2019).
As the UNCs_2 subcluster showed high expression of pregnancy-specific proteins, with an increased proportion in early pregnancy, we hypothesized that this subcluster may be associated with pregnancy-specific peptide hormone secretion in EG. To determine which signaling pathway regulates phenotypic transformation between UNC subpopulations, a cell interaction network was constructed between UNCs and other placental cells using CellChat (Figure 4D). Results indicated that the UNCs_2 subcluster was primarily involved in IGF, JAM, PROS, MPZ, and CSF signaling, while the UNCs_1 and UNCs_3 subclusters showed the strongest signal output and reception capacity among the seven clusters (Figure 4E; Supplementary Figure S10C), underscoring their important regulatory functions in the placental microenvironment. UNCs_2 and UNCs_5 subpopulations exhibited strong IGF output and input signaling, suggesting involvement in trophoblast migration and differentiation under the regulation of paracrine IGF signaling. To further explore the key regulators of each UNC subcluster, we analyzed 1 220 cow transcription factors and their downstream gene sets using GENIE3. Results indicated that factors such as YBX1, SUB1, HMGB1, and CNBP may play specific and critical roles in the UNCs_2 subcluster (Figure 4E, left), while GPBP1, TFDP2, and YBX1 were identified as specific regulators for the UNCs_3 subcluster (Figure 4E, center). Of the three UNCs_2, UNCs_3, and UNCs_5 subcluster-specific regulators, PAGs and PRPs were identified as direct targets of the YBX1 and NPAS2 transcription factors. Immunostaining of early pregnancy placental sections was carried out to determine the roles of YBX1 and NPAS2 in UNCs. The immunofluorescence results revealed that YBX1 and NPAS2 were highly expressed in the placental tissues. Specifically, YBX1 was mainly expressed in the cytoplasm of UNCs and NPAS2 was mainly expressed in the nucleus of UNCs (Figure 4F), suggesting that these transcription factors play key roles in the secretion of pregnancy-associated glycol proteins and PRPs in early pregnancy. Overall, these findings suggest that YBX1 may drive a phenotypic transition between the EG and MG UNC subpopulations.
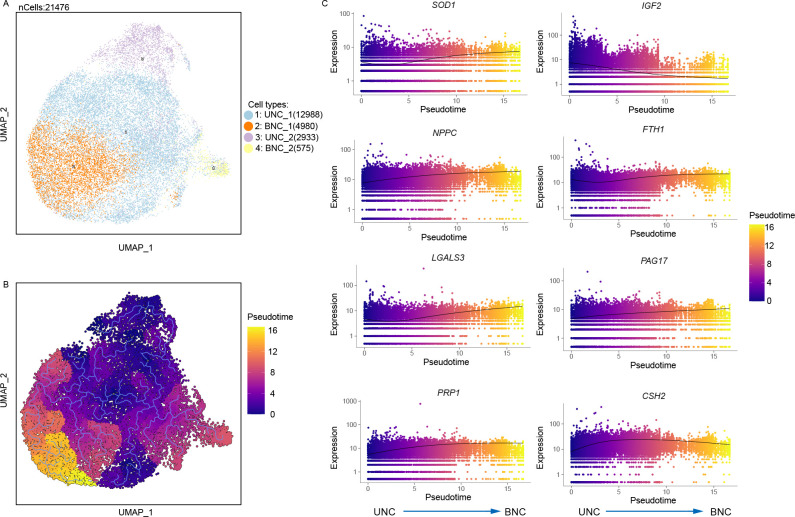
Analysis of differentiation trajectory of trophoblast cells
Next, single-cell trajectory “pseudotime” analysis was conducted using Monocle3 (Figure 5) to better understand how trophoblast cells transit from mononuclear to multinuclear states (Trapnell et al., 2014). In the cow placenta, the trophoblast cell trajectories were anchored in the proliferative UNCs and underwent two distinct transitions before ultimately transitioning into BNCs (Figure 5B). Notably, trajectories became rounded in the BNC trophoblast population, indicating the occurrence of terminal differentiation or differentiation events. A total of 6 414 DEGs with different trajectories were identified by pseudotemporal analysis (Supplementary Table S2). Figure 5C shows the expression patterns of the top eight genes across pseudotime. Notably, results showed increased expression of superoxide dismutase 1 (SOD1), natriuretic peptide C (NPPC), galectin 3 (LGALS3), PRP1, ferritin heavy chain 1 (FTH1), CSH2, and PAG17 and decreased insulin like growth factor 2 (IGF2) expression as the trophoblast cells transited into a terminally differentiated BNC population.
Figure 5.
Pseudotime analysis of trophoblast differentiation
A: Integrated clustering of trophoblast cells at all stages. B: Analyses of pseudotime trajectory of trophoblast differentiation. C: Differentially expressed genes across pseudotime.
Spatial gene modules uncovered the interaction of different spatial regions
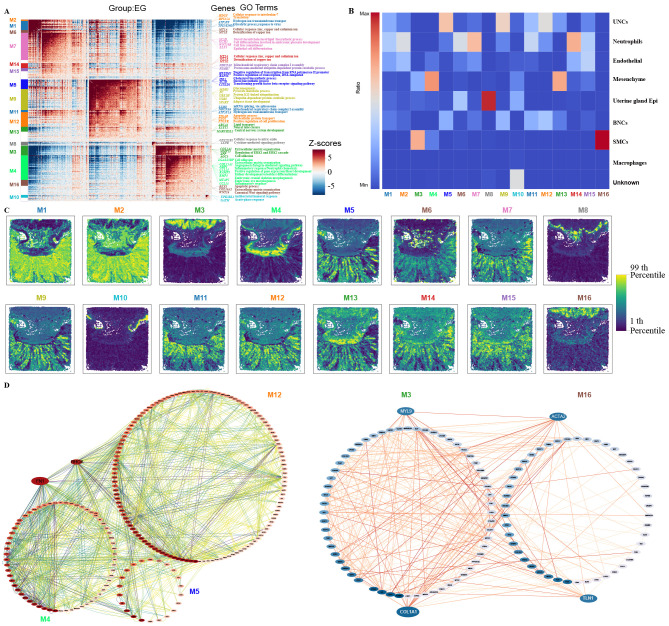
The spatial patterns of gene expression depicted diverse functional regions in the placental slices. To identify co-variant genes with similar spatial distribution, Hotspot (an algorithm that measures nonrandom variation to recognize informative gene programs) (Detomaso & Yosef, 2021) was employed to identify gene modules from our placental Stereo-seq dataset. In total, 16 spatial modules were identified for the EG placental sections (Figure 6A, C). GO enrichment analysis of spatially related genes of each module was conducted to explore their potential biological functions, which showed consistency with the spatial clusters. For example, module 1 (M1) genes were involved in glycolysis, response to viruses, hydrogen ion transmembrane transport, and response to nutrient levels, while module 3 (M3) genes were related to signal transduction, extracellular matrix organization, collagen fibril organization, and positive regulation of ERK1 and ERK2 cascades. In addition, almost all module genes were involved in cell division, cycle, and proliferation. At the MG stage, 15 spatial modules were identified (Supplementary Figure S11), with GO functional enrichment results reasonably matching the spatial location. Interestingly, certain clusters contained several gene expression modules. For instance, in the EG placenta, UNCs were divided into M1, M5, M9, and M12 subspatial modules, while SMCs were divided into M3 and M16 subspatial modules. Single gene modules also corresponded to multiple subregions. For example, M13 in the EG placenta was distributed across various clusters, including BNCs, mesenchymal cells, and UNCs (Figure 6B). To explore the connections among these four cell clusters, the search tool for retrieval of interacting genes (STRING) was used to identify potential gene interactions, followed by visualization with Cytoscape (Shannon et al., 2003) (Figure 6D). Among the identified genes, fibronectin 1 (FN1) and Ras homolog family member A (RHOA) were highlighted within the network. FN1, found in module M4 interacted with ACTR3 and CENNA1 in M5 and CD44 and PRKAR1AR in M13. Among the potential interacting genes revealed by STRING, COL1A1 and MYL9 in M3 interacted with ACTA2 in M16. Hence, spatial modules, by delineating functional regions or subregions within an organism, provide a powerful tool for exploring gene interactions between different spatial regions, serving as a reliable resource for discovering novel functions and unknown gene interactions.
Figure 6.
Spatial modules identified by Hotspot showing interactions among spatial regions in EG placenta
A: Heatmap showing genes with significant spatial autocorrelation (12 750 genes, false discovery rate (FDR)<0.05) grouped into 16 gene modules based on pairwise spatial correlations of gene expression in multiple sections of EG placenta. Selected genes and GO terms related to representative modules are highlighted on the right side. B: Heatmap showing ratio of Bins with high module scores (>3) in each module distributed in different spatial clusters. Spatial clusters in EG placenta come from Stereo-seq dataset in Figure 1C. C: Spatial visualization of module scores for modules in EG placental sections. D: Protein-protein interaction (PPI) network of genes in M12, M4, M5 (left), M3, and M16 (right) modules. Network was visualized using Cytoscape.
DISCUSSION
Exploring organ development can offer profound insights into its cellular and biological foundations (Yuan et al., 2022). However, a comprehensive understanding of the biological functions, networks, and interactions among the various cell types underlying organ and disease development requires both cellular information and a spatial context (Asp et al., 2019). Here, we utilized Stereo-seq to generate an unbiased spatial transcriptome profile of cow placental development at near single-cell resolution and analytically resolved specific cell-type populations and their transcriptome profiles to advance our understanding of the ruminant placenta (Chen et al., 2022; Liu et al., 2022). Based on Stereo-seq analysis, 10 major cell populations were identified at the maternal-fetal interface, effectively distinguishing between UNC from BNC trophoblasts within the placenta. Many of these cell subsets are known to play crucial roles in cow physiology and pregnancy (Palmieri et al., 2008; Sağsöz et al., 2022; Wooding et al., 2005). Furthermore, the relative abundance of most cell clusters differed significantly between the EG and MG placentas. Thus, these findings provide a framework to assess the spatial dynamics of gene expression and cellular structure at the maternal-fetal interface of the cow placenta.
Pregnancy is an intricate process associated with various biological changes in the maternal body. Fetal implantation and pregnancy success rely on the creation of an immune tolerant environment, achieved primarily through two mechanisms: the presence of non-functional immune cells, including essential decidual natural killer (dNK) cells, and the action of synergistic molecules at the maternal-fetal interface, including secretory leukocyte protease inhibitor (SLPI), SERPINA14, and GRP, particularly in ruminants (Bauersachs et al., 2006; Budipitojo et al., 2003; Davoodi et al., 2016; Dráb et al., 2014; McNeel et al., 2017). Previous studies have shown that uterine serpins (SERPINA14) are abundantly expressed in ruminants during pregnancy, while in vitro experiments have demonstrated that SERPINA14 inhibits lymphocyte proliferation and uterine natural killer-like cell activity (Ulbrich et al., 2009). Ovine SERPINA14 is also known to form complexes with immunoglobulin A (IgA) and IgM, preventing antibody-mediated actions against the fetus (Ulbrich et al., 2009). These genes were detected in the current study, with SERPINA14 expression mostly restricted to the endometrial gland of the placentas. Recent studies on buffalo (Bubalus bubalis) (Kandasamy et al., 2010; Padua & Hansen, 2010), sheep (Ovine) (Song et al., 2008), sika deer (Cervus nippon) (Ulbrich et al., 2009), and mouse deer (Tragulus nigricans) (Kimura et al., 2006) have revealed comparable trends, suggesting the conservation of mechanisms for placental development. However, these molecular features have not been characterized in recent single-cell transcriptome data from human and mouse placentas, suggesting that they may serve as unique placental maternal-fetal immune tolerance traits in ruminants. In addition, SLPI, SERPINA14, and GRP were found to have more positive sites in early pregnancy, suggesting they may play an important role in pro-/anti-inflammatory balance of the local immune microenvironment in the uterus during pregnancy, important for a healthy pregnancy due to maternal-fetal immune tolerance regulation in cows. The identification of these molecules is crucial for advancing our understanding of the immune tolerance mechanisms in cow pregnancy and for informing livestock production strategies.
Trophoblast cells characteristically emerge in early gestation in the cow placenta. They secrete pivotal hormones and cytokines for fetal-maternal communication, including placental lactogens (CSH1), PRP1, and PAG1, which are necessary for pregnancy establishment in cows (Zhou et al., 2012). In this study, the UNC and BNC clusters showed the strongest signaling output capacity among the 10 identified clusters (Supplementary Figure S12), indicating their importance in microenvironment homeostasis regulation. Cell trajectory analysis of trophoblasts predicted a framework for UNC differentiation to BNCs (Figure 5). In this framework, UNCs_1 and UNCs_2 differentiated into two distinct types of BNCs, marked by up-regulation of BNC-specific genes (CSH2, PAG17, and PRP1), as reported in previous research (Davenport et al., 2023b). Despite this, our results also showed that the expression levels of SOD1, NPPC, LGALS3, and FTH1 increased during the differentiation of UNCs into BNCs, and interestingly, that IGF2 expression decreased during this process, different from that observed in humans and mice (Pringle & Roberts, 2007). In the microenvironment of the maternal-fetal interface in cow placentas, we identified several key signaling pathways involved in cell adhesion, migration, epithelial cell proliferation, differentiation, and apoptosis, specifically IGF, CDH, PROS. These pathways demonstrated pronounced signaling activity in UNCs and BNCs, with gene expression density primarily observed in these cells, a discovery not previously reported. Given that ruminant placentas feature BNCs that fuse with the uterine epithelium and release their granules into the maternal tissue beneath the uterine epithelium (Klisch & Schraner, 2020), these signaling pathways may play a role in the regulation of signals involved in this process, which warrants further experimental validation. Notably, we identified seven cell-type subpopulations within the UNCs at both developmental stages. However, analysis revealed four significant UNC subclusters that were more abundant in MG than in EG, and two UNC subclusters that were more abundant in EG than in MG. These findings coincide with the notable increase in the UNC cell population at MG, supporting our analytical findings. Among these, subgroups 2, 3, 5, and 7 were spatially localized in the cotyledon section, while subpopulations 1, 2, and 4 were localized in the cotyledon, caruncle, and inter-cotyledonary regions (Supplementary Figure S9). By isolating the spatial localization and gene expression patterns of UNC subpopulations without considering the BNC clusters, we were able to map the PAG and PRP expression regions more precisely. For example, PAG11 was localized in UNCs_6 of the inter-cotyledonary trophoblast and the chorionic plate of the cotyledon. These results showed a high specificity in the expression and cellular localization of PAGs, providing evidence for their multiple biological functions in placental tissue. Trophoblast cell invasion and migration are strongly influenced by cell-cell and cell-matrix interactions. Notably, the CellChat results demonstrated strong signaling output of the CDH signaling pathway in the UNCs_3 and UNCs_5 subpopulations, with stronger signaling in EG than in MG. Significant differences in ECM composition were observed among UNCs_1, UNCs_2, UNCs_3, UNCs_4, UNCs_5, UNCs_6, and UNCs_7, with enhanced anchoring of UNCs_2 and UNCs_5 in the surrounding matrix, which may be associated with their greater migratory capacity and invasive phenotype at MG. Overall, our results indicated that COL18A1, COL1A2, and COL3A1 may play important roles in the differentiation, migration, interaction, and adhesion of trophoblast cells to the uterine epithelium. Furthermore, the UNCs_2 and UNCs_5 subpopulations may be key cells for establishing interactions with the maternal uterus in EG.
Transcription factors, together with their downstream gene networks, are key drivers of cell-fate transition during placental development. Here, we identified several transcription factors potentially involved in the phenotypic transformation of different cell types, including YBX1, SUB1, HMGB1, CNBP, and NPAS2. The YBX1 gene encodes a highly conserved cold shock domain protein with broad nucleic acid binding properties (Suresh et al., 2018). Our data suggested potential new roles for several transcription factors. For example, YBX1 and NPAS2 may regulate the secretion of trophoblast pregnancy proteins PAGs and PRPs, which has not been reported previously. Based on the Stereo-seq data, spatial modules at different gestation stages were identified and integrated with spatial regions to investigate key co-varying genes. However, the interaction patterns among spatially correlated gene sets, both within and across modules forming a regulatory network, remain unclear. Unraveling these interactions should improve our understanding of the molecular mechanisms underlying mammalian pregnancy. For instance, studies have explored the roles of fibronectin, a major extracellular glycoprotein influencing cell proliferation and differentiation, and laminin, a principal constituent of the basement membrane, in cow placentas during early pregnancy (Sağsöz et al., 2022). Our findings indicated that endosomal stroma/floor plate provides directional signals to the surrounding tissue during pregnancy, with the FN1 signal potentially playing an important role. Developing an in vitro culture system to derive mature BNCs from UNCs presents a promising avenue for future research, offering insights into the critical cellular and molecular processes governing trophoblast differentiation in the cow placenta.
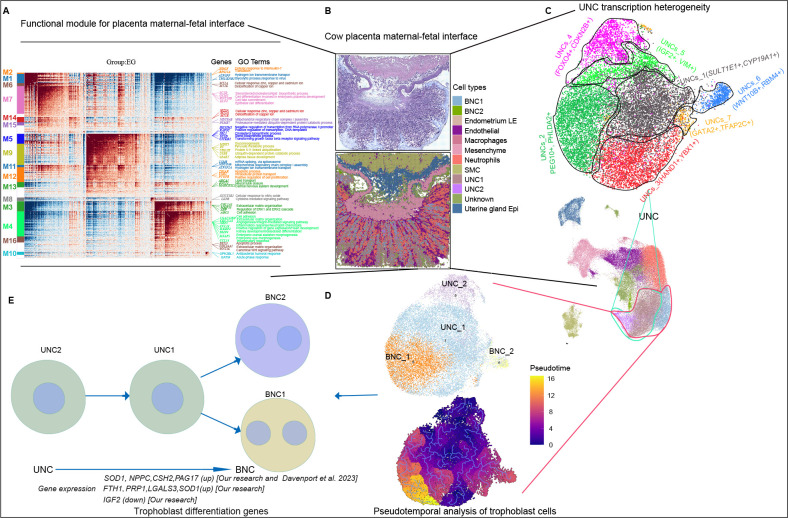
In conclusion, Stereo-seq was used to generate unbiased spatial transcriptome profiles of cow placental development at near single-cell resolution, enabling accurate assignment of cell type and stage information with high precision. Detailed mining of trophoblast cell heterogeneity and differentiation was also performed, highlighting the importance of these cells in the chorionic villi for the successful establishment of pregnancy and formation of the placenta to maintain gestation and support fetal growth to term in ruminants. As outlined in Figure 7, this study provides a valuable framework and resource for advancing our understanding of ruminant placental development, with potential long-term benefits for cow breeding and reproductive strategies.
Figure 7.
Overview of interface between cow placenta and fetus
A: Functional module of maternal-fetal interface. B: Molecular characterization of maternal-fetal interface. C: UNC heterogeneity. D, E: Trophoblastic cell differentiation (UNCs differentiate into BNCs). Compared with previous research (Davenport et al., 2023b), our study showed up-regulated FTH1, PRP1, LGALS3, and SOD1 expression and down-regulated IGF2 expression during differentiation process.
SUPPLEMENTARY DATA
Supplementary data to this article can be found online.
Acknowledgments
COMPETING INTERESTS
The authors declare that they have no competing interests.
AUTHORS’ CONTRIBUTIONS
G.H.T., S.J.L., Y. Zheng, and F.W.: conceptualization, methodology, data curation, writing original draft preparation. D.F.Z., G.H.T., F.N.L., and J.Y.L.: programming, software development; designing computer programs; implementation of the computer code and supporting algorithms; testing of existing code components. G.H.T., T.S., H.K.L., S.J.L., M.L.D., H.P.W., A.Z., and J.Y.L.: visualization, investigation. Y. Zhang, Y.J., Y. Zheng, and F.W. worked on the final approval of the version to be submitted and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors read and approved the final version of the manuscript.
ACKNOWLEDGMENTS
We thank the High-Performance Computing Platform of Northwest A&F University and the National Supercomputing Center in Xi’an and Hefei for providing the computing resources for the bioinformatic analyses.
Funding Statement
This work was supported by the National Key R&D Program of China (2022YFF1000100), Technology Application and Development Program for Rapid Propagation of Cow Breeding (20211117000005), Basic Science (Agricultural Biology) Research Center of Shaanxi (K3030922016), Ningxia Hui Autonomous Region Key R&D Projects (2021BEF01001), and Natural Science Basic Research Program of Shaanxi (2022JQ-171)
Contributor Information
Yi Zheng, Email: y.zheng@nwafu.edu.cn.
Fei Wang, Email: wangfei2840@nwafu.edu.cn.
DATA AVAILABILITY
Raw data were deposited in the National Center for Biotechnology Information database (NCBI) under BioProjectID PRJNA1093563, Genome Sequence Archive (GSA) under accession number PRJCA023784, and Science Data Bank (DOI: 10.57760/sciencedb.16428). The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.
References
- Abdisa T Mechanism of retained placenta and its treatment by plant medicine in ruminant animals in Oromia, Ethiopia. Journal of Veterinary Medicine and Animal Health. 2018;10(6):135–147. doi: 10.5897/JVMAH2018.0672. [DOI] [Google Scholar]
- Anbo H, Sato M, Okoshi A, et al Functional segments on intrinsically disordered regions in disease-related proteins. Biomolecules. 2019;9(3):88. doi: 10.3390/biom9030088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asp M, Giacomello S, Larsson L, et al. 2019. A spatiotemporal organ-wide gene expression and cell atlas of the developing human heart. Cell, 179 (7): 1647–1660. e19.
- Averill MM, Kerkhoff C, Bornfeldt KE S100A8 and S100A9 in cardiovascular biology and disease. Arteriosclerosis, Thrombosis, and Vascular Biology. 2012;32(2):223–229. doi: 10.1161/ATVBAHA.111.236927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awad M, Kizaki K, Takahashi T, et al Dynamic expression of SOLD1 in bovine uteroplacental tissues during gestation. Placenta. 2013;34(8):635–641. doi: 10.1016/j.placenta.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Bauersachs S, Ulbrich SE, Gross K, et al Embryo-induced transcriptome changes in bovine endometrium reveal species-specific and common molecular markers of uterine receptivity. Reproduction. 2006;132(2):319–331. doi: 10.1530/rep.1.00996. [DOI] [PubMed] [Google Scholar]
- Budipitojo T, Sasaki M, Matsuzaki S, et al Expression of gastrin-releasing peptide (GRP) in the bovine uterus during the estrous cycle. Archives of Histology and Cytology. 2003;66(4):337–346. doi: 10.1679/aohc.66.337. [DOI] [PubMed] [Google Scholar]
- Chatuphonprasert W, Jarukamjorn K, Ellinger I Physiology and pathophysiology of steroid biosynthesis, transport and metabolism in the human placenta. Frontiers in Pharmacology. 2018;9:1027. doi: 10.3389/fphar.2018.01027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A, Liao S, Cheng MN, et al. 2022. Spatiotemporal transcriptomic atlas of mouse organogenesis using DNA nanoball-patterned arrays. Cell, 185 (10): 1777–1792. e21.
- Dai H, Jin QQ, Li L, et al Reconstructing gene regulatory networks in single-cell transcriptomic data analysis. Zoological Research. 2020;41(6):599–604. doi: 10.24272/j.issn.2095-8137.2020.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport KM, Ortega MS, Johnson GA, et al. 2023a. Review: implantation and placentation in ruminants. Animal, 17 Suppl 1 : 100796.
- Davenport KM, Ortega MS, Liu HY, et al Single-nuclei RNA sequencing (snRNA-seq) uncovers trophoblast cell types and lineages in the mature bovine placenta. Proceedings of the National Academy of Sciences of the United States of America. 2023b;120(12):e2221526120. doi: 10.1073/pnas.2221526120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoodi S, Cooke RF, Fernandes ACC, et al Expression of estrus modifies the gene expression profile in reproductive tissues on Day 19 of gestation in beef cows. Theriogenology. 2016;85(4):645–655. doi: 10.1016/j.theriogenology.2015.10.002. [DOI] [PubMed] [Google Scholar]
- DeTomaso D, Yosef N. 2021. Hotspot identifies informative gene modules across modalities of single-cell genomics. Cell Systems, 12 (5): 446–456. e9.
- Diskin MG, Waters SM, Parr MH, et al Pregnancy losses in cattle: potential for improvement. Reproduction, Fertility and Development. 2016;28(1-2):83–93. doi: 10.1071/RD15366. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, et al STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dráb T, Kračmerová J, Hanzlíková E, et al The antimicrobial action of histones in the reproductive tract of cow. Biochemical and Biophysical Research Communications. 2014;443(3):987–990. doi: 10.1016/j.bbrc.2013.12.077. [DOI] [PubMed] [Google Scholar]
- Ge W, Tan SJ, Wang SH, et al Single-cell transcriptome profiling reveals dermal and epithelial cell fate decisions during embryonic hair follicle development. Theranostics. 2020;10(17):7581–7598. doi: 10.7150/thno.44306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green JA, Geisert RD, Johnson GA, et al. 2021. Implantation and placentation in ruminants. In: Geisert RD, Spencer T. Placentation in Mammals. Cham: Springer, 129–154.
- Green JA, Xie SC, Quan X, et al Pregnancy-associated bovine and ovine glycoproteins exhibit spatially and temporally distinct expression patterns during pregnancy. Biology of Reproduction. 2000;62(6):1624–1631. doi: 10.1095/biolreprod62.6.1624. [DOI] [PubMed] [Google Scholar]
- Hashizume K, Ushizawa K, Patel OV, et al Gene expression and maintenance of pregnancy in bovine: roles of trophoblastic binucleate cell-specific molecules. Reproduction, Fertility and Development. 2007;19(1):79–90. doi: 10.1071/RD06118. [DOI] [PubMed] [Google Scholar]
- Hayashi KG, Hosoe M, Sakumoto R, et al Temporo-spatial expression of adrenomedullin and its receptors in the bovine placenta. Reproductive Biology and Endocrinology. 2013;11:62. doi: 10.1186/1477-7827-11-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heaton H, Talman AM, Knights A, et al Souporcell: robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nature Methods. 2020;17(6):615–620. doi: 10.1038/s41592-020-0820-1. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Green JA, Garbayo JM, et al Adaptive diversification within a large family of recently duplicated, placentally expressed genes. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(7):3319–3323. doi: 10.1073/pnas.050002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin SQ, Guerrero-Juarez CF, Zhang L, et al Inference and analysis of cell-cell communication using CellChat. Nature Communications. 2021;12(1):1088. doi: 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Bazer FW, Burghardt RC, et al Cellular events during ovine implantation and impact for gestation. Animal Reproduction. 2018;15(S1):843–855. doi: 10.21451/1984-3143-AR2018-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandasamy S, Jain A, Kumar R, et al Molecular characterization and expression profile of uterine serpin (SERPINA14) during different reproductive phases in water buffalo (Bubalus bubalis) Animal Reproduction Science. 2010;122(1-2):133–141. doi: 10.1016/j.anireprosci.2010.08.005. [DOI] [PubMed] [Google Scholar]
- Kimura J, Budipitojo T, Sasaki M, et al Immunolocalization of gastrin-releasing peptide (GRP) in the uteroplacenta of the mouse deer. Anatomia, Histologia, Embryologia. 2006;35(4):217–220. doi: 10.1111/j.1439-0264.2005.00662.x. [DOI] [PubMed] [Google Scholar]
- King GJ Comparative placentation in ungulates. Journal of Experimental Zoology. 1993;266(6):588–602. doi: 10.1002/jez.1402660609. [DOI] [PubMed] [Google Scholar]
- Klisch K, Schraner EM Intraluminal vesicles of binucleate trophoblast cell granules are a possible source of placental exosomes in ruminants. Placenta. 2020;90:58–61. doi: 10.1016/j.placenta.2019.12.006. [DOI] [PubMed] [Google Scholar]
- Li H, Huang QH, Liu Y, et al Single cell transcriptome research in human placenta. Reproduction. 2020;160(6):R155–R167. doi: 10.1530/REP-20-0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, Li R, Li Y, et al. 2022. Spatiotemporal mapping of gene expression landscapes and developmental trajectories during zebrafish embryogenesis. Developmental Cell, 57 (10): 1284–1298. e5.
- McNeel AK, Ondrak JD, Amundson OL, et al Timing of transcriptomic and proteomic changes in the bovine placentome after parturition. Theriogenology. 2017;100:1–7. doi: 10.1016/j.theriogenology.2017.05.020. [DOI] [PubMed] [Google Scholar]
- Moraes JGN, Behura SK, Geary TW, et al Uterine influences on conceptus development in fertility-classified animals. Proceedings of the National Academy of Sciences of the United States of America. 2018;115(8):E1749–E1758. doi: 10.1073/pnas.1721191115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaya-C M, Fritsch M, Bernal JE Immunohistochemical protein expression profiling of growth- and apoptotic-related factors in relation to umbilical cord length. Early Human Development. 2015;91(5):291–297. doi: 10.1016/j.earlhumdev.2015.03.001. [DOI] [PubMed] [Google Scholar]
- Padua MB, Hansen PJ Evolution and function of the uterine serpins (SERPINA14) American Journal of Reproductive Immunology. 2010;64(4):265–274. doi: 10.1111/j.1600-0897.2010.00901.x. [DOI] [PubMed] [Google Scholar]
- Palmieri C, Loi P, Ptak G, et al Review paper: a review of the pathology of abnormal placentae of somatic cell nuclear transfer clone pregnancies in cattle, sheep, and mice. Veterinary Pathology. 2008;45(6):865–880. doi: 10.1354/vp.45-6-865. [DOI] [PubMed] [Google Scholar]
- Polei M, Günther J, Koczan D, et al Gene expression profiles of bovine uninucleate trophoblast cells and trophoblast giant cells: a data note. BMC Research Notes. 2020a;13(1):115. doi: 10.1186/s13104-020-04964-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polei M, Günther J, Koczan D, et al Trophoblast cell differentiation in the bovine placenta: differentially expressed genes between uninucleate trophoblast cells and trophoblast giant cells are involved in the composition and remodeling of the extracellular matrix and O-glycan biosynthesis. BMC Molecular and Cell Biology. 2020b;21(1):1. doi: 10.1186/s12860-020-0246-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pringle KG, Roberts CT New light on early post-implantation pregnancy in the mouse: roles for insulin-like growth factor-II (IGF-II)? Placenta. 2007;28(4):286–297. doi: 10.1016/j.placenta.2006.04.006. [DOI] [PubMed] [Google Scholar]
- Ramilowski JA, Goldberg T, Harshbarger J, et al A draft network of ligand–receptor-mediated multicellular signalling in human. Nature Communications. 2015;6(1):7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryckman C, Vandal K, Rouleau P, et al Proinflammatory activities of S100: proteins S100A8, S100A9, and S100A8/A9 induce neutrophil chemotaxis and adhesion. The Journal of Immunology. 2003;170(6):3233–3242. doi: 10.4049/jimmunol.170.6.3233. [DOI] [PubMed] [Google Scholar]
- Sağsöz H, Liman N, Akbalık ME, et al Expression of cadherins and some connective tissue components in cow uterus and placenta during pregnancy. Research in Veterinary Science. 2022;151:64–79. doi: 10.1016/j.rvsc.2022.07.006. [DOI] [PubMed] [Google Scholar]
- Sánchez JM, Mathew DJ, Passaro C, et al Embryonic maternal interaction in cattle and its relationship with fertility. Reproduction in Domestic Animals. 2018;53(S2):20–27. doi: 10.1111/rda.13297. [DOI] [PubMed] [Google Scholar]
- Sartori R, Bastos MR, Wiltbank MC Factors affecting fertilisation and early embryo quality in single- and superovulated dairy cattle. Reproduction, Fertility and Development. 2010;22(1):151–158. doi: 10.1071/RD09221. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Gerber D, Soley JT, et al Histo-morphology of the uterus and early placenta of the African buffalo (Syncerus caffer) and comparative placentome morphology of the African buffalo and cattle (Bos taurus) Placenta. 2006;27(8):899–911. doi: 10.1016/j.placenta.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, et al Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G, Satterfield MC, Kim J, et al Gastrin-releasing peptide (GRP) in the ovine uterus: regulation by interferon tau and progesterone. Biology of Reproduction. 2008;79(2):376–386. doi: 10.1095/biolreprod.108.068403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer TE, Hansen TR. 2015. Implantation and establishment of pregnancy in ruminants. In: Geisert RD, Bazer FW. Regulation of Implantation and Establishment of Pregnancy in Mammals. Cham: Springer, 105–135.
- Suresh PS, Tsutsumi R, Venkatesh T YBX1 at the crossroads of non-coding transcriptome, exosomal, and cytoplasmic granular signaling. European Journal of Cell Biology. 2018;97(3):163–167. doi: 10.1016/j.ejcb.2018.02.003. [DOI] [PubMed] [Google Scholar]
- Szafranska B, Miura R, Ghosh D, et al Gene for porcine pregnancy-associated glycoprotein 2 (poPAG2): its structural organization and analysis of its promoter. Molecular Reproduction and Development. 2001;60(2):137–146. doi: 10.1002/mrd.1070. [DOI] [PubMed] [Google Scholar]
- Touzard E, Reinaud P, Dubois O, et al Specific expression patterns and cell distribution of ancient and modern PAG in bovine placenta during pregnancy. Reproduction. 2013;146(4):347–362. doi: 10.1530/REP-13-0143. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Cacchiarelli D, Grimsby J, et al The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nature Biotechnology. 2014;32(4):381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulbrich SE, Frohlich T, Schulke K, et al Evidence for estrogen-dependent uterine serpin (SERPINA14) expression during estrus in the bovine endometrial glandular epithelium and lumen. Biology of Reproduction. 2009;81(4):795–805. doi: 10.1095/biolreprod.108.075184. [DOI] [PubMed] [Google Scholar]
- Vento-Tormo R, Efremova M, Botting RA, et al Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563(7731):347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QL, Li JL, Wang SP, et al Single-cell transcriptional profiling reveals cellular and molecular divergence in human maternal-fetal interface. Scientific Reports. 2022;12(1):10892. doi: 10.1038/s41598-022-14516-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Song R, Wang ZY, et al S100A8/A9 in inflammation. Frontiers in Immunology. 2018;9:1298. doi: 10.3389/fimmu.2018.01298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedemann I, Krebs T, Momberg N, et al mRNA expression profiling in cotyledons reveals significant up-regulation of the two bovine pregnancy-associated glycoprotein genes boPAG-8 and boPAG-11 in early gestation. Veterinary Medicine and Science. 2018;4(4):341–350. doi: 10.1002/vms3.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiltbank MC, Baez GM, Garcia-Guerra A, et al Pivotal periods for pregnancy loss during the first trimester of gestation in lactating dairy cows. Theriogenology. 2016;86(1):239–253. doi: 10.1016/j.theriogenology.2016.04.037. [DOI] [PubMed] [Google Scholar]
- Wooding FBP The ruminant placental trophoblast binucleate cell: an evolutionary breakthrough. Biology of Reproduction. 2022;107(3):705–716. doi: 10.1093/biolre/ioac107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wooding FBP, Fowden AL, Bell AW, et al Localisation of glucose transport in the ruminant placenta: implications for sequential use of transporter isoforms. Placenta. 2005;26(8-9):626–640. doi: 10.1016/j.placenta.2004.09.013. [DOI] [PubMed] [Google Scholar]
- Wooding FBP, Wathes DC Binucleate cell migration in the bovine placentome. Journal of Reproduction and Fertility. 1980;59(2):425–430. doi: 10.1530/jrf.0.0590425. [DOI] [PubMed] [Google Scholar]
- Xie SC, Green J, Beckers JF, et al The gene encoding bovine pregnancy-associated glycoprotein-1, an inactive member of the aspartic proteinase family. Gene. 1995;159(2):193–197. doi: 10.1016/0378-1119(94)00928-L. [DOI] [PubMed] [Google Scholar]
- Yang Y, Zhu QY, Liu JL Deciphering mouse uterine receptivity for embryo implantation at single-cell resolution. Cell Proliferation. 2021;54(11):e13128. doi: 10.1111/cpr.13128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu N, Wu JL, Xiao J, et al HIF-1α regulates angiogenesis via Notch1/STAT3/ETBR pathway in trophoblastic cells. Cell Cycle. 2019;18(24):3502–3512. doi: 10.1080/15384101.2019.1689481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Sun DM, Qin T, et al Single-cell transcriptomic landscape of the sheep rumen provides insights into physiological programming development and adaptation of digestive strategies. Zoological Research. 2022;43(4):634–647. doi: 10.24272/j.issn.2095-8137.2022.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng ZM, Xu P, He YQ, et al Acetylation of atp5f1c mediates cardiomyocyte senescence via metabolic dysfunction in radiation-induced heart damage. Oxidative Medicine and Cellular Longevity. 2022;2022:4155565. doi: 10.1155/2022/4155565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Wang Z, Zhang Z, et al Delivery of AP-2α siRNA into cultured bovine trophoblast cells by electroporation repressed key placenta-specific gene expression. Gene. 2012;499(1):169–175. doi: 10.1016/j.gene.2012.03.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data to this article can be found online.
Data Availability Statement
Raw data were deposited in the National Center for Biotechnology Information database (NCBI) under BioProjectID PRJNA1093563, Genome Sequence Archive (GSA) under accession number PRJCA023784, and Science Data Bank (DOI: 10.57760/sciencedb.16428). The datasets generated and/or analyzed during the current study are available from the corresponding author upon reasonable request.