Abstract
BACKGROUND:
Hypertrophic cardiomyopathy (HCM) is caused by sarcomere gene mutations (genotype-positive HCM) in ≈50% of patients and occurs in the absence of mutations (genotype-negative HCM) in the other half of patients. We explored how alterations in the metabolomic and lipidomic landscape are involved in cardiac remodeling in both patient groups.
METHODS:
We performed proteomics, metabolomics, and lipidomics on myectomy samples (genotype-positive N=19; genotype-negative N=22; and genotype unknown N=6) from clinically well-phenotyped patients with HCM and on cardiac tissue samples from sex- and age-matched and body mass index–matched nonfailing donors (N=20). These data sets were integrated to comprehensively map changes in lipid-handling and energy metabolism pathways. By linking metabolomic and lipidomic data to variability in clinical data, we explored patient group–specific associations between cardiac and metabolic remodeling.
RESULTS:
HCM myectomy samples exhibited (1) increased glucose and glycogen metabolism, (2) downregulation of fatty acid oxidation, and (3) reduced ceramide formation and lipid storage. In genotype-negative patients, septal hypertrophy and diastolic dysfunction correlated with lowering of acylcarnitines, redox metabolites, amino acids, pentose phosphate pathway intermediates, purines, and pyrimidines. In contrast, redox metabolites, amino acids, pentose phosphate pathway intermediates, purines, and pyrimidines were positively associated with septal hypertrophy and diastolic impairment in genotype-positive patients.
CONCLUSIONS:
We provide novel insights into both general and genotype-specific metabolic changes in HCM. Distinct metabolic alterations underlie cardiac disease progression in genotype-negative and genotype-positive patients with HCM.
Keywords: cardiomyopathies, genotype, hypertrophy, lipidomics, metabolism, metabolomics
Hypertrophic cardiomyopathy (HCM) is the most common inherited cardiomyopathy.1 Clinical hallmarks of HCM include diastolic impairment and asymmetrical left ventricular hypertrophy, often giving rise to left ventricle outflow tract obstruction.2,3 Approximately 50% of all patients harbor a pathogenic heterozygous variant (mutation) in genes encoding sarcomere proteins (ie, genotype-positive [Gpositive]). Despite the expansion of the number of genes for diagnostic screening, no causative mutation is identified in the other half of patients.4–6 As a result, it remains unknown what drives disease in these genotype-negative (Gnegative) individuals. Nowadays, the majority of newly diagnosed patients with HCM are Gnegative5; hence, insight into disease mechanisms in Gnegative HCM is warranted.
Impairment of cardiac bioenergetics is a well-established feature in all patients with HCM. This is evident from imaging studies showing lower phosphocreatine/adenosine triphosphate ratios and reduced myocardial external efficiency7,8 and from proteomic studies in myectomy tissue samples demonstrating widespread downregulation of proteins involved in mitochondrial energy metabolism.9,10 Reports from the past few years have aimed to define the metabolomic and lipidomic signatures of HCM to gain insight into, for example, substrate utilization, formation of toxic lipids, and alterations in biosynthetic pathways. While these studies consistently show downregulation in the oxidation of fatty acids, findings on lipid handling and storage, phospholipids, glycolytic intermediates, amino acids, redox metabolites, and purine/pyrimidine metabolism remain inconclusive.11–13 Possible explanations for these reported discrepancies may include methodological differences, intercohort dissimilarities (eg, demographics), and suboptimal matching to scarcely available tissue samples from nonfailing donor hearts.
Whereas the underlying cause of the disease is different in Gpositive and Gnegative patients, studies leveraging multiomics analyses of myectomy tissue samples uniformly agree on the absence of differences at the group level between these patient populations.10–14 However, we recently demonstrated that mitochondrial dysfunction was tightly linked to the degree of septal hypertrophy in Gnegative patients, while, in Gpositive patients, mitochondrial function was not associated with hypertrophy.15 This challenges the idea that similar metabolic remodeling underlies disease in Gpositive and Gnegative patients, and it remains to be determined whether the metabolome and lipidome correlate differently with cardiac phenotype in Gpositive and Gnegative patients.
To address these unresolved matters, we here performed proteomics, metabolomics, and lipidomics on a large number of myectomy samples (N=47) from a clinically well-phenotyped cohort of patients with HCM and cardiac tissue samples from sex- and age-matched and body mass index–matched nonfailing donors (N=20). We provide a comprehensive overview of alterations in lipid-handling and energy metabolism pathways, integrating 3 complementary omics analyses. By linking metabolomic and lipidomic profiles to cardiac phenotype, we unveil that pathological cardiac remodeling and fatty acid oxidation impairment in HCM were not associated with increased fatty acid storage or ceramide-induced lipotoxicity. Moreover, in Gnegative patients, we observed striking negative associations between cardiac remodeling and abundance of acylcarnitines, redox metabolites, amino acids, pentose phosphate pathway (PPP) intermediates, purines, and pyrimidines. In contrast, in Gpositive patients, cardiac remodeling was positively associated with levels of redox metabolites, amino acids, PPP intermediates, purines, and pyrimidines. Combined, the present study provides novel insights into general and genotype-specific metabolic alterations in relation to cardiac phenotype in HCM and indicates that differential and even opposite metabolic changes underlie cardiac disease in Gpositive and Gnegative patients.
METHODS
An extended methods section and an overview of the study design (Figure S1) are provided in the Supplemental Material.
Metabolomics and Lipidomics
Metabolomics and lipidomics analyses were performed on snap-frozen cardiac tissue as previously described.16,17 A detailed description is provided in the Supplemental Material. Metabolic pathway enrichment analysis was performed using MetaboAnalyst 5.0 (www.MetaboAnalyst.ca).18
Proteomics
Proteomics analyses were performed on cardiac tissue samples that were fractionated according to the IN-Seq method as described previously,19 followed by liquid chromatography-tandem mass spectrometry according to a previously published workflow.20 A detailed description is provided in the Supplemental Material.
Respirometry
Mitochondrial respiration data used in this study were previously generated.15 Here, we analyzed these data in relation to metabolite and lipid levels in corresponding samples.
Statistical Analyses
Statistical analyses were performed using GraphPad Prism, v9, software. Metabolomics and lipidomics data were log2 transformed and analyzed by the Welch t test. P values were corrected for multiple testing via the Benjamini-Hochberg false discovery rate procedure. A corrected P value (ie, Q-value) <0.05 was considered significant. Correlations were tested using linear regression and are described by the Pearson R coefficient. Because the aim of this study was to explore associations between metabolites/lipids and clinical phenotype/mitochondrial function, we did not correct significance levels of correlations for multiple testing. Data on glycogen levels were analyzed via the Mann-Whitney U test when 2 groups were compared or 1-way ANOVA with the Tukey multiple comparisons test when 3 groups were compared; blood pressure data were analyzed via the Student t test or the Welch t test. These data are displayed as mean±SEM. Categorical distributions are presented as percentages and were analyzed using the Fisher exact test.
Study Approval
Study protocols were approved by local medical ethics review committees. Before surgery, written informed consent was obtained from all patients.
Data Availability Statement
Metabolomics and lipidomics data have been submitted to the MetaboLights database under accession number MTBLS8695.21 Proteomics data are available via the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD046187.22 Processed metabolomics, lipidomics, and proteomics data are furthermore available as a Supplemental Material (Multi-Omics Data Sets). The remaining data underlying this article will be shared on reasonable request to the corresponding author.
RESULTS
We performed multiomic profiling of myectomy tissue from clinically well-phenotyped patients with HCM and cardiac tissue from nonfailing donors to comprehensively map metabolic alterations in HCM. Echocardiographic data were acquired as described previously23 and are summarized in Table S1. Detailed individual information of each patient and donor is available in Table S2. Patients with obstructive HCM and nonfailing donors were comparable in terms of sex distribution, age, and body mass index. Gpositive and Gnegative patients did not differ demographically and displayed similar indexed septal thickness and diastolic function. Left ventricle outflow tract pressure gradients were significantly higher in Gnegative versus Gpositive patients, in line with observations in the NHLBI HCM Registry.5,24
Metabolomics Reveals Impaired Fatty Acid Oxidation and Alterations in Glycolytic and Biosynthetic Pathways in HCM
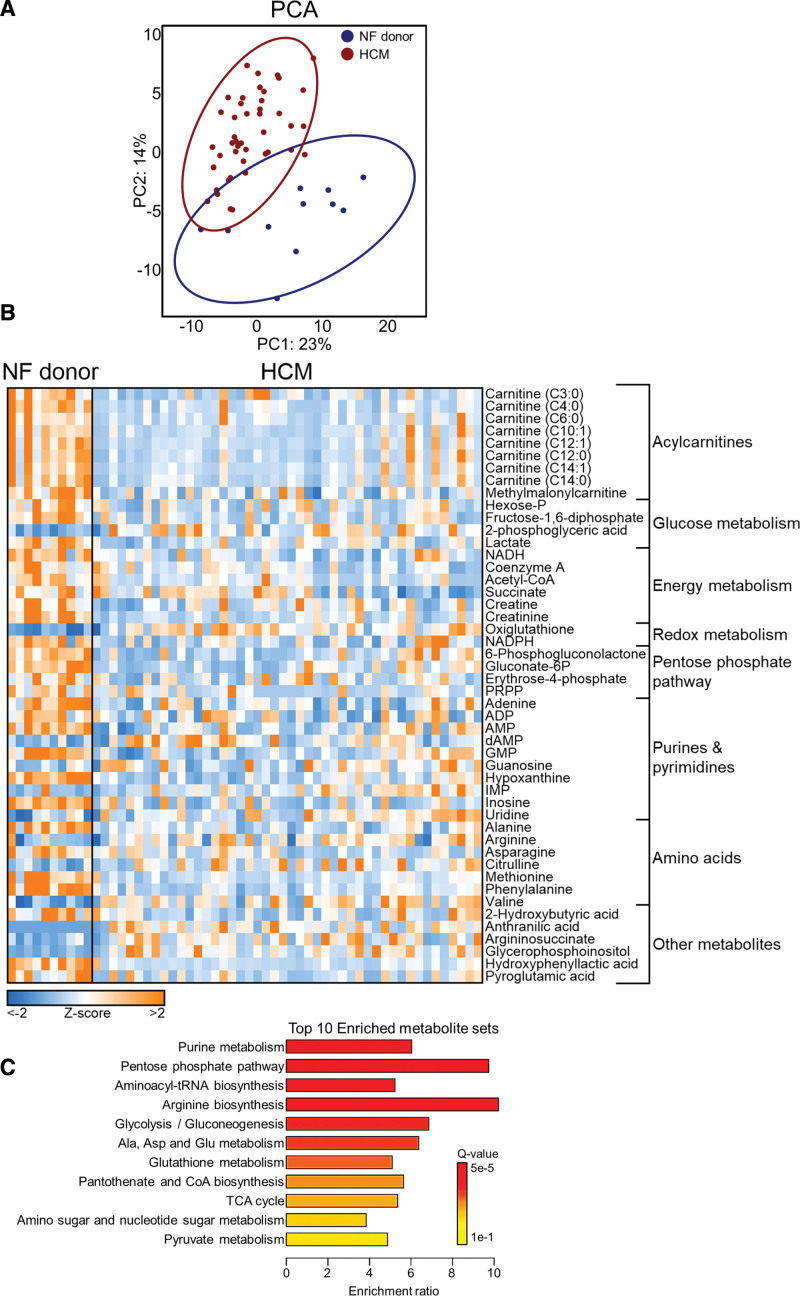
We performed metabolomics on myectomy samples from patients with HCM (N=47) and nonfailing donor cardiac samples (N=12) to characterize the metabolite profile in HCM. Metabolomic differences between nonfailing donors and patients with HCM are shown in Figure 1. In total, 141 metabolites were detected, 51 of which were differently abundant in HCM versus nonfailing donors (Q<0.05). HCM and nonfailing donor samples displayed separate clustering on principal component analysis (Figure 1A), indicating clearly different metabolite profiles. Gpositive and Gnegative HCM samples did not cluster separately (Figure S2A), and no metabolites were significantly different between these groups. A heatmap of all statistically significant different metabolites in HCM versus nonfailing donors is shown in Figure 1B. Multiple acylcarnitines were less abundant in HCM versus nonfailing donors. We additionally observed altered levels of several glycolytic intermediates and a reduced abundance of metabolites involved in energy metabolism. Oxidized glutathione was elevated, while nicotinamide adenine dinucleotide phosphate was lowered in HCM versus nonfailing donors, indicative of oxidative stress. Major changes were also seen in metabolites linked to biosynthetic pathways such as the pentose phosphate pathway and purine/pyrimidine metabolism. Numerous amino acids were furthermore altered in HCM versus nonfailing donors. Accordingly, the most significantly enriched metabolic pathways were purine metabolism, the pentose phosphate pathway, and biosynthesis of tRNAs and arginine (Figure 1C). Altogether, metabolomic profiling in HCM yields a marked downregulation of fatty acid oxidation, while changes in glycolytic and biosynthetic pathways were rather bidirectional.
Figure 1.
Overview of metabolomic differences between hypertrophic cardiomyopathy (HCM) myectomy tissue and cardiac tissue from nonfailing (NF) donors. A, Principal component analysis (PCA) showing distinct clustering of patients with HCM and NF donors. B, Heatmap showing z scores of all significantly different metabolites in HCM vs NF donor hearts (Q<0.05). C, Top 10 enriched metabolite sets in HCM vs NF donors analyzed via MetaboAnalyst 5.0.18 dAMP indicates deoxyadenosine monophosphate; Gluconate-6P, gluconate-6-phosphate; GMP, guanosine monophosphate; Hexose-P, hexose-phosphate; IMP, inosine monophosphate; NADH, reduced nicotinamide adenine dinucleotide; NADPH, reduced nicotinamide adenine dinucleotide phosphate; PRPP, phosphoribosyl pyrophosphate; and TCA, tricarboxylic acid.
Decreased Fatty Acid Storage and No Ceramide-Induced Lipotoxicity in HCM Despite Downregulation of Fatty Acid Oxidation
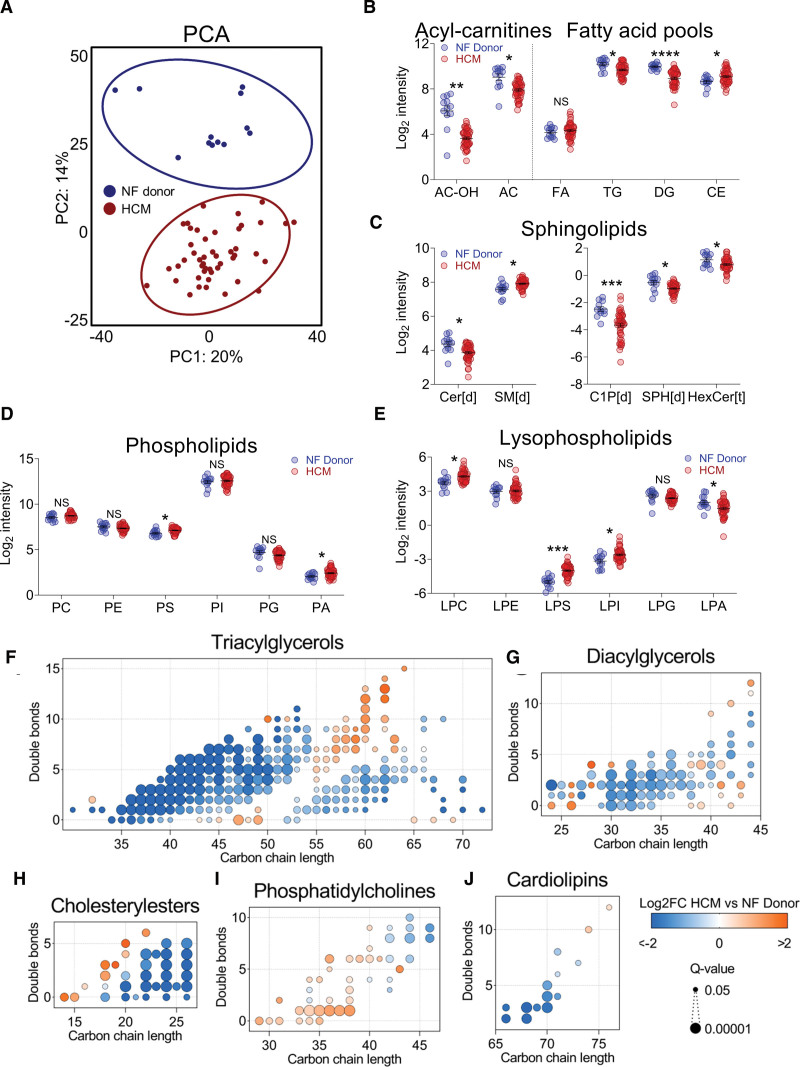
To identify changes in lipid biology in HCM, we analyzed lipid profiles in HCM myectomy samples (N=47) and nonfailing donor cardiac samples (N=12) via lipidomics. Lipidomic observations are displayed in Figure 2. A total of 2139 lipids were detected, divided over 55 lipid classes (see Table S3 for an overview of abbreviations). Principal component analysis revealed markedly distinct clustering of HCM and nonfailing donor samples (Figure 2A). No differences were observed between Gpositive and Gnegative patients (Figure S2B). Consistent with metabolomic data, we observed markedly lowered levels of acylcarnitines in HCM versus nonfailing donors, which is in good agreement with previous work reporting downregulation of fatty acid oxidation in HCM10–13,15 (Figure 2B). Interestingly, despite defective fatty acid oxidation capacity, we observed unchanged levels of free fatty acids and less storage of fatty acids in major storage depots (ie, diacylglycerol and triacylglycerol). Levels of lipotoxic ceramides and associated sphingolipids ceramide-1-phosphate, sphingosine, and hexosyl-ceramide were furthermore reduced in HCM versus nonfailing donors, suggesting that fatty acids in HCM hearts were not directed more toward ceramide formation despite diminished oxidation and storage. Sphingomyelins were however more abundant in HCM versus nonfailing donors, indicating that excess fatty acids may be preferably routed toward this class of sphingolipids in HCM hearts (Figure 2C). Changes in terms of phospholipids included elevation of both phosphatidylserine and lysophosphatidylserine, higher levels of phosphatidic acid but lower levels of lysophosphatidic acid, and increased abundance of lysophosphatidylcholine and lysophosphatidylinositol in HCM versus nonfailing donors (Figure 2D and 2E). Whereas total triacylglycerol levels were reduced in HCM versus nonfailing donors, we observed an elevation of triacylglycerol species with relatively long and less saturated acyl chains (Figure 2F). Regarding the diacylglycerol pool, particularly medium-length species were depleted (Figure 2G). While the total pool of cholesteryl esters was increased, the majority of these species were actually less abundant, and a shift toward shorter chain cholesteryl esters was observed in HCM versus nonfailing donors (Figure 2H). Phosphatidylcholines were in addition relatively shorter and more saturated in HCM versus nonfailing donor hearts (Figure 2I). Total levels of the mitochondrial membrane–specific phospholipid cardiolipin were unchanged; however, we did observe an apparent shift from short, saturated cardiolipin toward longer, more unsaturated cardiolipin species in HCM (Figure 2J).
Figure 2.
Overview of lipidomic differences between hypertrophic cardiomyopathy (HCM) myectomy tissue and cardiac tissue from nonfailing (NF) donors. A, Principal component analysis (PCA) showing distinct clustering of patients with HCM and NF donors. B through E, log2-transformed data for total amounts of lipids within a class, grouped per theme. Abbreviations of lipid classes are detailed in Table S3. F through J, Major alterations in individual lipid species abundance within a lipid class as dot plots, in which each dot represents a significantly altered lipid species. Vertical axes indicate double bond number, and horizontal axes indicate the carbon chain length of lipid species. The color and size of dots indicate fold change (FC) between groups and the significance of these changes, respectively.
In-Sequence Proteomics Yields Downregulation of Mitochondrial Translation and Increased Expression of Proteasome Proteins as Novel HCM Disease Features
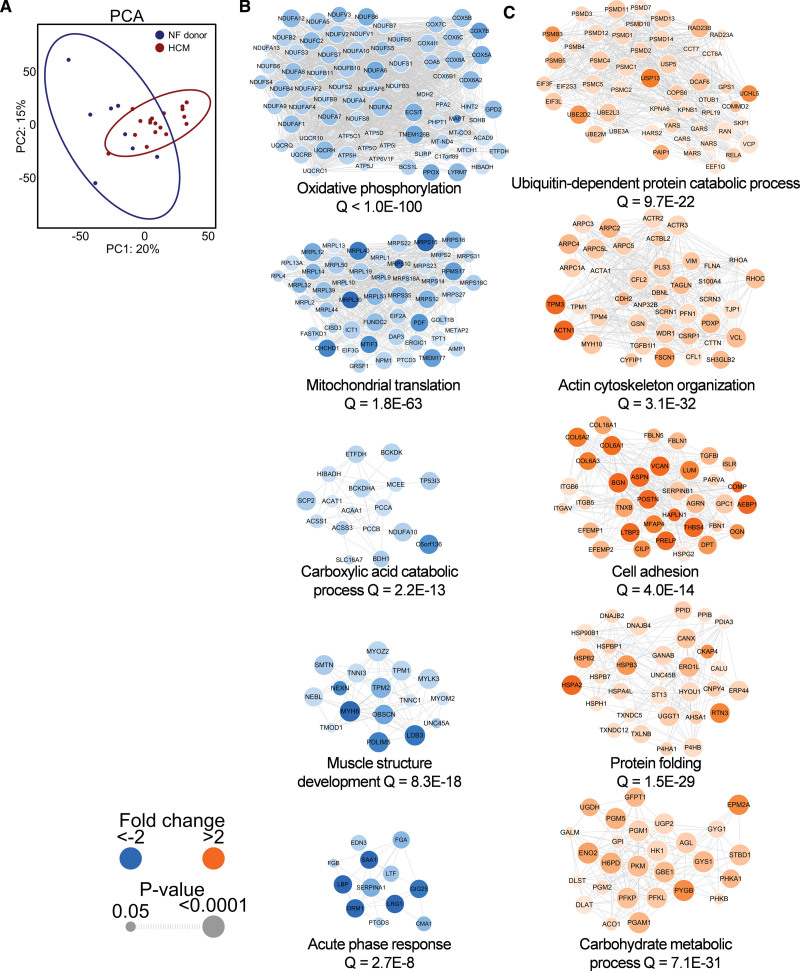
Proteomics was performed on HCM myectomy samples (N=18) and nonfailing donor cardiac samples (N=8). An overview of proteomic findings is shown in Figure 3. To achieve maximum coverage of myocardial proteins, we performed in-sequence mass spectrometry on cytosolic-, myofilament-, and membrane-enriched fractions. In total, 3258 proteins were detected. HCM samples clustered separately from nonfailing donor samples (Figure 3A); Gpositive and Gnegative HCM samples did not differ from one another (Figure S2C). To identify downregulated and upregulated cellular processes in HCM versus nonfailing donor hearts, we performed gene ontology analysis on clusters of interacting proteins, as per the STRING database.25 Observations consistent with previous reports included lower levels of oxidative phosphorylation proteins, upregulation of proteins associated with the actin- and tubulin cytoskeleton (Figure S3), elevated deposition of extracellular matrix (ie, cell adhesion) proteins, and activation of protein quality control (ie, protein folding) in HCM versus nonfailing donor hearts (Figure 3B and 3C).9,10,12,13,26 We also report a marked reduction of proteins involved in mitochondrial translation in HCM versus nonfailing donors, many of which are components of the mitochondrial ribosomal machinery (Figure 3B), suggesting an overall impairment of mitochondrial protein synthesis in HCM. Additionally, we observed a higher abundance of proteins involved in ubiquitin-dependent proteasomal protein degradation (Figure 3C) in HCM versus nonfailing donors. Taken together, using high-coverage proteomics, we confirm key cellular hallmarks of HCM and identify downregulation of mitochondrial translation and upregulation of proteasome proteins as novel factors implicated in HCM pathophysiology.
Figure 3.
Summary of proteomic differences between hypertrophic cardiomyopathy (HCM) and nonfailing (NF) donors. A, Principal component analysis (PCA) showing distinct clustering of patients with HCM and NF donors. Top 5 downregulated clusters of interacting proteins (B) and the top 5 upregulated clusters of interacting proteins (C) according to cluster size. The color and size of protein nodes indicate fold change between groups and the significance of these changes, respectively.
Integration of Multiomics Data Unveils Complex Alterations in Energy Metabolism Pathways
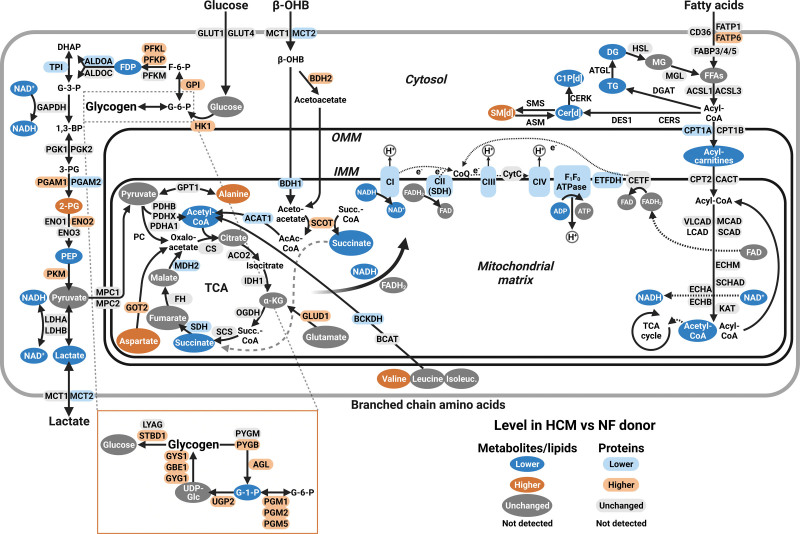
To comprehensively map alterations in lipid-handling and energy metabolism pathways, we integrated proteomic, metabolomic, and lipidomic data (Figure 4). Surprisingly, despite the downregulation of fatty acid oxidation in HCM, the vast majority of proteins involved in fatty acid uptake and subsequent shuttling to the mitochondria and β-oxidation were unchanged in HCM versus nonfailing donors. Only 1 protein of the carnitine shuttle (CPT1A [carnitine palmitoyltransferase I]) and ETFDH (electron transferring flavoprotein dehydrogenase) that feeds electrons from β-oxidation-derived FADH2 into the electron transfer system were less abundant in HCM versus nonfailing donors. This suggests that impaired fatty acid oxidation capacity in HCM is not primarily regulated by protein abundance. In terms of glucose metabolism, we found that the majority of proteins involved in glycolysis were more abundant in HCM versus nonfailing donors. This was accompanied by decreased levels of fructose-1,6-diphosphate and phosphoenolpyruvate and higher levels of 2-phosphoglycerate. Interestingly, we also found a global upregulation of proteins involved in glycogen metabolism (eg, glycogen synthetase, glycogen phosphorylase), but it was unclear whether these data would predict accumulation or depletion of myocardial glycogen levels. We measured glycogen amounts in cardiac tissue from patients with HCM collected at the time of myectomy and explanted tissue from patients with HCM with end-stage heart failure to assess disease stage–specific levels of glycogen. Compared with nonfailing donors, we observed no myocardial accumulation of glycogen in patients with HCM at any disease stage (Figure S4A). There were also no differences in myocardial glycogen levels between Gnegative and Gpositive patients at the time of myectomy (Figure S4B). Thus, higher levels of proteins involved in glycogen metabolism without altered glycogen levels may suggest increased glycogen synthesis and breakdown in HCM. Lower lactate abundance suggests that anaerobic glycolysis was not increased in HCM versus nonfailing donors and may possibly indicate increased usage of lactate to fuel pyruvate production, which has been observed in failing hearts.27 Acetyl-CoA levels were lower in HCM versus nonfailing donors; however, we found no evidence of downregulation of pyruvate to acetyl-CoA conversion. Consequently, lower acetyl-CoA levels may particularly result from impaired acetyl-CoA input from mitochondrial fatty acid oxidation. While most tricarboxylic acid (TCA) cycle enzymes and intermediates were unchanged, it appears that acetyl-CoA conversion to citrate is maintained to prevent TCA intermediate depletion. Additionally, we observed an increased abundance of several proteins that participate in anaplerosis, replenishing TCA intermediates via transamination of amino acids (ie, GOT2 [aspartate aminotransferase], GLUD1 [glutamate dehydrogenase 1]), raising the possibility that amino acids may be alternative substrates to fuel the TCA cycle. Catabolism of branched-chain amino acids, however, appeared to be blunted, which has also been reported in other settings of heart failure.28 We found conflicting observations with respect to ketone oxidation. Cytosolic BDH (3-hydroxybutyrate dehydrogenase) 2 and SCOT (succinyl-CoA-3-oxaloacid CoA transferase) were elevated, while mitochondrial BDH1 and ACAT1 (acetyl-CoA acetyltransferase 1) were less abundant in HCM. Thus, it remains unclear whether ketones are differentially used in HCM hearts. In summary, altered energy metabolism in HCM is typified by (1) downregulation of fatty acid oxidation without overtly lowered levels of proteins involved in this pathway; (2) generally higher abundance of glycolysis and glycogen turnover–associated proteins, which may suggest increased usage of glucose as energy fuel; and (3) no indications of impaired pyruvate entry into the TCA cycle and no hints toward increased anaerobic conversion of pyruvate to lactate.
Figure 4.
Comprehensive overview of alterations in energy metabolism and lipid-handling pathways in patients with hypertrophic cardiomyopathy (HCM) vs nonfailing (NF) donors integrating metabolomics, lipidomics, and proteomics data. Metabolomic, lipidomic, and proteomic data were integrated to provide an overview of changes in major energy metabolism and lipid-handling pathways. Metabolites and lipids are indicated by objects with a dark fill and white text; proteins are indicated by objects with a light fill and black text. Blue, orange, and gray indicate lower, higher, and unchanged abundances in HCM vs NF donors, respectively. Created with BioRender.com.
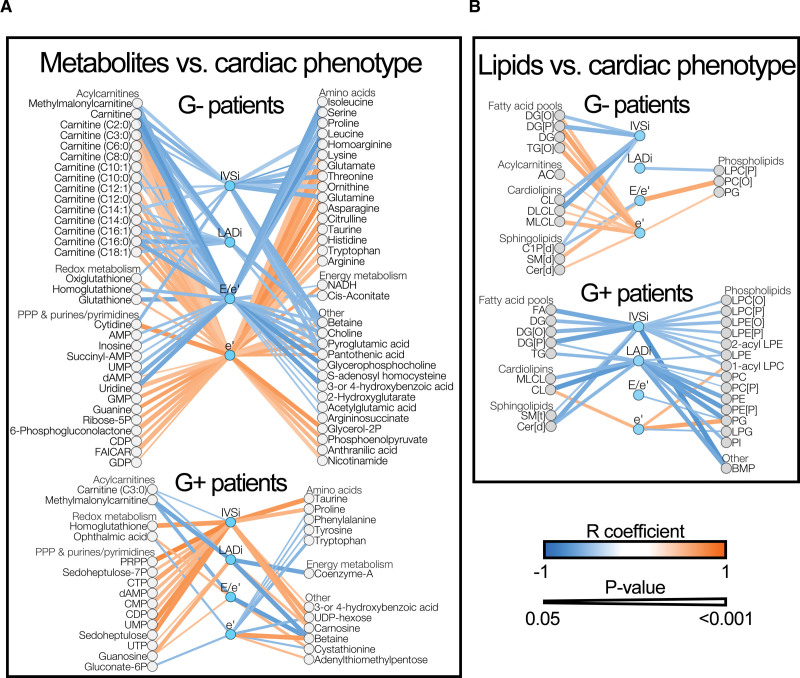
Cardiac Remodeling Is Differentially Associated With Metabolite Levels in Gnegative and Gpositive Patients
We previously observed that mitochondrial dysfunction was tightly linked to septal hypertrophy in Gnegative HCM but not in Gpositive HCM,15 which suggests that also metabolic alterations may correlate to cardiac remodeling differentially in these patient groups. Therefore, we explored how metabolites and lipids were associated with parameters of cardiac function in Gnegative and Gpositive patients with HCM (Figure 5). Strikingly, in Gnegative patients, we observed that numerous acylcarnitines were negatively associated with septal hypertrophy, atrial dilatation, and diastolic dysfunction. In contrast, we did not observe such a clear pattern in Gpositive patients, in whom only 2 acylcarnitines negatively correlated with pathological cardiac remodeling. These findings are in line with the notion that particularly in Gnegative HCM, cardiac remodeling is intertwined with impaired bioenergetic homeostasis.15 Another remarkable discrepancy was seen with respect to metabolites involved in the pentose phosphate pathway and downstream synthesis of purines and pyrimidines, which constitute the building blocks of RNA and DNA. In Gnegative patients, these metabolites were negatively correlated to diastolic dysfunction, whereas in Gpositive patients, these metabolites displayed strong positive associations with septal hypertrophy and modest positive correlations with diastolic impairment. Similarly, in Gnegative samples, a vast array of amino acids showed negative associations with septal hypertrophy and diastolic dysfunction, whereas in Gpositive patients, several amino acids were positively associated with septal hypertrophy and negatively with diastolic function. These findings suggest that in Gnegative HCM, the progression of cardiac disease is associated with the depletion of metabolites that are necessary for the synthesis of proteins and nucleosides, while in Gpositive HCM, septal hypertrophy is accompanied by elevated levels of such metabolites. Finally, several metabolites involved in reactive oxygen species defense (ie, redox metabolites) were negatively correlated with septal hypertrophy and diastolic dysfunction in Gnegative patients, while in Gpositive patients, such metabolites were positively associated with adverse cardiac changes. This may suggest that oxidative stress is differently involved in disease progression in Gnegative and Gpositive HCMs.
Figure 5.
Correlations between cardiac phenotype and metabolites and lipids in patients with hypertrophic cardiomyopathy (HCM). Correlations were tested via linear regression and are described by the Pearson R coefficient. A, All significant correlations between cardiac parameters and metabolites in genotype-negative (Gnegative; G−) and genotype-positive (Gpositive; G+) patients. B, All significant correlations between cardiac parameters and lipids classes in Gnegative and Gpositive patients. Blue and orange lines indicate negative and positive correlations, respectively. Line thickness corresponds with the significance level of the correlation. Exact R coefficients and P values, as well as group comparison statistics between nonfailing donors and G− or G+ patients of metabolites and lipids in these correlations, are available as Supplemental Material (Correlation statistics). IVSi indicates septal thickness indexed to body surface area; and LADi, left atrial diameter indexed to body surface area.
Regarding associations between lipids and cardiac phenotype, we did not observe drastic differences between Gnegative and Gpositive patients with HCM (Figure 5B). Remarkably, in both patient groups, we nearly exclusively observed negative associations between lipid amounts and pathological cardiac alterations. In Gpositive patients, many lipid levels were inversely linked to septal hypertrophy and atrial dilatation. This included free fatty acids and fatty acid storage depots (ie, diacylglycerol, triacylglycerol) and lipotoxic ceramides. Gnegative patients displayed similar albeit less pronounced patterns, characterized by diastolic compliance being positively linked to diacylglycerol and ceramide levels. Thus, in both patient groups, cardiac hypertrophy and functional impairment were accompanied by reduced fatty acid storage and ceramide levels.
In previous work, we measured mitochondrial respiratory parameters in fresh myectomy samples from the same patients whose myectomy samples were analyzed in the present study.15 We, therefore, also explored correlations between mitochondrial respiratory function and metabolites and lipids (Figure S5). In Gnegative patients, we observed numerous positive associations between mitochondrial respiratory parameters on the one hand and acylcarnitines, redox metabolites, PPP and purine/pyrimidine metabolites, amino acids, and metabolites involved in energy metabolism on the other hand (Figure S5A). These metabolites were negatively associated with septal hypertrophy and diastolic impairment in Gnegative patients (Figure 5), thus suggesting that in Gnegative HCM, pathological cardiac remodeling and functional impairment are interwoven with fatty acid oxidation impairment, mitochondrial respiratory dysfunction, impaired antioxidant defense, and reduced levels of biosynthetic metabolites. In contrast, Gpositive samples displayed only few associations between metabolites and mitochondrial function (Figure S5B), which is consistent with the observation that mitochondrial impairment in Gpositive HCM is not linked to cardiac remodeling.15 By extension, in samples from Gnegative patients, but not Gpositive patients, mitochondrial respiration was overall positively associated with lipids that displayed negative associations with cardiac hypertrophy and diastolic dysfunction.
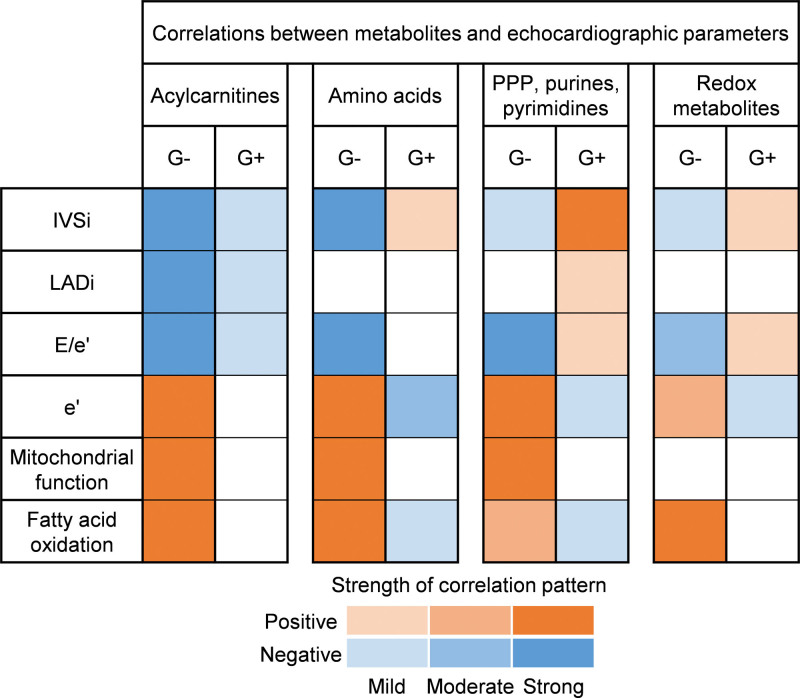
Combined, by linking echocardiographic and mitochondrial respiration data to metabolomic and lipidomic data, we observed markedly different correlation patterns in Gnegative and Gpositive patients with HCM, as summarized in Figure 6. Gnegative patients displayed an overall connectedness between cardiac remodeling, mitochondrial dysfunction, fatty acid oxidation impairment, and lowering of acylcarnitines, biosynthetic metabolites, and redox metabolites. Conversely, in Gpositive patients, both biosynthetic metabolites and redox metabolites were positively associated with cardiac remodeling. Collectively, these observations imply major pathophysiological differences between Gnegative and Gpositive patients with HCM.
Figure 6.
Contrasting metabolic alterations underlie cardiac remodeling in genotype-negative (G−) and genotype-positive (G+) patients with hypertrophic cardiomyopathy (HCM). Matrix summarizing correlation patterns between metabolite classes and (functional) cardiac parameters in G− and G+ patients with HCM. Negative and positive correlation patterns are colored blue and orange, respectively. The color intensity reflects the strength of correlation patterns. IVSi indicates interventricular septum thickness indexed to body surface area; LADi, left atrial diameter indexed to body surface area; and PPP, pentose phosphate pathway;
DISCUSSION
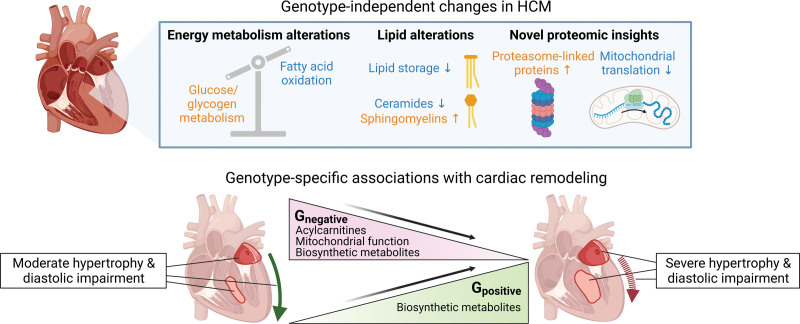
To identify general and genotype-specific metabolic alterations in HCM, we conducted multiomics analyses on myectomy samples from a well-phenotyped cohort of patients with HCM and cardiac tissue from age- and sex-matched and body mass index–matched nonfailing donors. The main findings of our study are summarized in Figure 7. Our data suggest that common features of HCM are (1) a shift away from fatty acid oxidation toward glucose metabolism and glycogen turnover; (2) reduced lipid storage and ceramide formation, but elevated sphingomyelin levels; and (3) upregulation of proteins involved in proteasomal protein degradation and downregulation of mitochondrial translation machinery. In Gnegative patients with HCM, cardiac remodeling was associated with impaired fatty acid oxidation, mitochondrial dysfunction, and lower abundance of biosynthetic metabolites (ie, PPP intermediates, purines, pyrimidines, and amino acids) and redox metabolites. In Gpositive patients with HCM, biosynthetic metabolites and redox metabolites were positively associated with cardiac remodeling. These findings indicate contrasting metabolic alterations underlie cardiac disease in Gpositive and Gnegative patients.
Figure 7.
Summary of the main findings of this study. Genotype-independent changes in hypertrophic cardiomyopathy (HCM) included downregulation of fatty acid oxidation and increased glucose metabolism and glycogen turnover; decreased lipid storage and ceramide formation and increased sphingomyelin abundance; and proteasome upregulation and downregulation of mitochondrial translation proteins. Gnegative patients with HCM displayed negative associations between cardiac remodeling and fatty acid oxidation, mitochondrial function, and levels of biosynthetic metabolites (ie, pentose phosphate pathway [PPP] intermediates, purines, pyrimidines, and amino acids). Conversely, Gpositive patients with HCM displayed positive associations between biosynthetic metabolites and cardiac remodeling. Thus, opposite metabolic changes are linked to cardiac disease in Gpositive and Gnegative patients. Created with BioRender.com.
Proteasome Upregulation and Reduced Mitochondrial Translation as Novel Features of HCM
Using high-coverage in-sequence proteomics, we here identified upregulation of proteasome-linked proteins and downregulation of mitochondrial translation proteins as novel hallmarks in HCM, irrespective of genotype. Impairment of the ubiquitin-dependent proteasome system has been observed in HCM mouse models and human HCM myocardium,29,30 which has been linked to oxidative damage of proteasomal subunits.30,31 Our analyses indicate that proteasome malfunctioning is unlikely to stem from the depletion of proteasomal machinery and raise the question of whether the observed upregulation of proteasomal subunits might reflect a compensatory response to functional impairment. We observed a marked reduction of proteins involved in mitochondrial translation, particularly mitochondrial ribosomal subunits, suggesting reduced mitochondrial protein synthesis in HCM. The involvement of altered mitochondrial translation in heart disease has not been extensively studied. Several studies report a link between mitochondrial ribosome protein gene mutations and cardiomyopathy.32,33 Gao et al34 recently demonstrated that knockout of MRPS5 (mitochondrial ribosomal protein S5) resulted in cardiac hypertrophy and heart failure, which was accompanied by mitochondrial dysfunction and metabolic remodeling. In contrast, mrps-5 suppression extended lifespan in Caenorhabditis elegans,35,36 and reduction of mitochondrial protein synthesis in heterozygous Mrpl54 mutant mice did not affect cardiac health.37 These studies show that manipulating mitochondrial ribosome function can incite both beneficial and detrimental effects, most likely related to dosage effects, and further study is warranted to elucidate the functional consequences of reduced mitochondrial ribosomal protein levels in HCM.
Energy Metabolism and Lipid Alterations in HCM
By integrating multiomics data, we comprehensively mapped alterations in energy metabolism pathways. Consistent with elevated myocardial glucose uptake in patients with HCM,38 we found that the majority of proteins involved in glycolysis were increased in HCM versus nonfailing donor samples. This did not seem to be accompanied by increased anaerobic glycolysis, as lactate levels were lower in HCM samples. Glucose did also not seem to be stored excessively as glycogen, which even appeared to be reduced at end-stage HCM. Thus, although glycogen storage diseases may manifest as HCM phenocopy,39 glycogen storage is not a general feature of HCM. These observations are discordant with previous work reporting elevated glycogen deposition in myocardium of patients with HCM displaying impaired contractile reserve.40 Importantly, that study did not differentiate patients with HCM based on genotype; thus, it is unknown whether that study cohort included patients with glycogen storage diseases.
Pyruvate levels and pyruvate-derived TCA input did not seem abnormal in HCM versus nonfailing donors, suggesting that oxidative glucose metabolism was sustained in HCM. Surprisingly, despite the reduced use of fatty acids as energy substrate in HCM, the only proteins involved in fatty acid oxidation that were less abundant were CPT1A and ETFDH. While downregulation of these proteins may impair acylcarnitine shuttling and electron input into the electron transfer system, respectively, it is likely that other factors than enzyme abundance (eg, posttranslational modifications of fatty acid oxidation enzymes41) contribute substantially to the downregulation of mitochondrial fatty acid oxidation in HCM. In other heart failure etiologies, impaired fatty acid oxidation capacity is typically met with a mismatch in lipid uptake, causing accumulation of free fatty acids and lipotoxic ceramides and increased lipid storage.42,43 We did not observe this and even found that fatty acids, diacylglycerols, triacylglycerols, and ceramides were negatively associated with cardiac remodeling, which is consistent with previous work showing a global reduction of lipid levels in advanced heart failure tissue.44 Notably, in Gnegative patients, ceramide and ceramide-1-phosphate levels were positively correlated with fatty acid oxidation capacity. Altogether, these observations suggest that during cardiac remodeling in HCM, no overt mismatch between fatty acid uptake and oxidation occurs. Instead, we put forward that reduced use of fatty acids as fuel is orchestrated in concert with reduced lipid uptake and storage. This would be in line with our observation of numerous phospholipids being negatively associated with cardiac remodeling, as phospholipid synthesis requires fatty acids. Previous work provided indications of reduced fatty acid uptake and decreased CD36 expression in nongenotyped HCM.45 However, in our hands, the levels of fatty acid transport proteins were unchanged; thus, it would be pertinent to study whether the expression and activity of CD36 in HCM with well-characterized genotype are altered due to posttranslational modifications and changes in localization.46
Divergent Associations Between Cardiac and Metabolic Remodelings in Gpositive and Gnegative HCMs
We observed contrasting patterns of correlations between cardiac remodeling and metabolite levels in Gnegative and Gpositive patients. In Gnegative patients, cardiac remodeling was associated with mitochondrial dysfunction and lowering of acylcarnitines and biosynthetic metabolites (ie, PPP intermediates, purines, pyrimidines, and amino acids). The energetic cost of production of purines and pyrimidines via the PPP and protein synthesis from amino acids is substantial47,48; thus, reductions in the levels of metabolites related to these processes are to be expected when bioenergetic homeostasis is compromised as a consequence of mitochondrial dysfunction. The added hemodynamic burden from left ventricle outflow tract obstruction further deteriorates cardiac function in obstructive HCM49 and is likely to exacerbate the already high myocardial energy consumption.7,8 Consistent with previous studies, the obstruction of the left ventricle outflow tract is more pronounced in Gnegative patients (Table S1).5,24 Our observations reinforce the notion that Gnegative HCM is a condition driven by energetic and metabolic impairment15 although the primary cause of disease remains unknown. Further study is warranted to assess how the downregulation of biosynthetic processes may contribute to disease progression in Gnegative HCM. In contrast to Gnegative patients, Gpositive patients displayed positive associations between cardiac remodeling and biosynthetic metabolites. These observations may suggest that in Gpositive HCM, biosynthesis of nucleotides and proteins is upregulated to facilitate cardiac hypertrophy.50 Wang et al13 proposed that interfering with the PPP might represent a therapeutic strategy in HCM. Based on our observations, such a strategy may indeed theoretically inhibit hypertrophy in Gpositive HCM by reducing the availability of nucleotides needed to promote cardiac growth but would be inadvisable in Gnegative HCM, as, in these patients, loss of PPP, purine, and pyrimidine intermediates was intertwined with cardiac remodeling. Taken together, our findings clearly suggest that different patterns of metabolic alterations underpin cardiac remodeling in Gnegative and Gpositive HCM, and future research is needed to gain mechanistic insight.
Lack of Consensus Among Studies Into Metabolomic and Lipidomic Landscapes in HCM
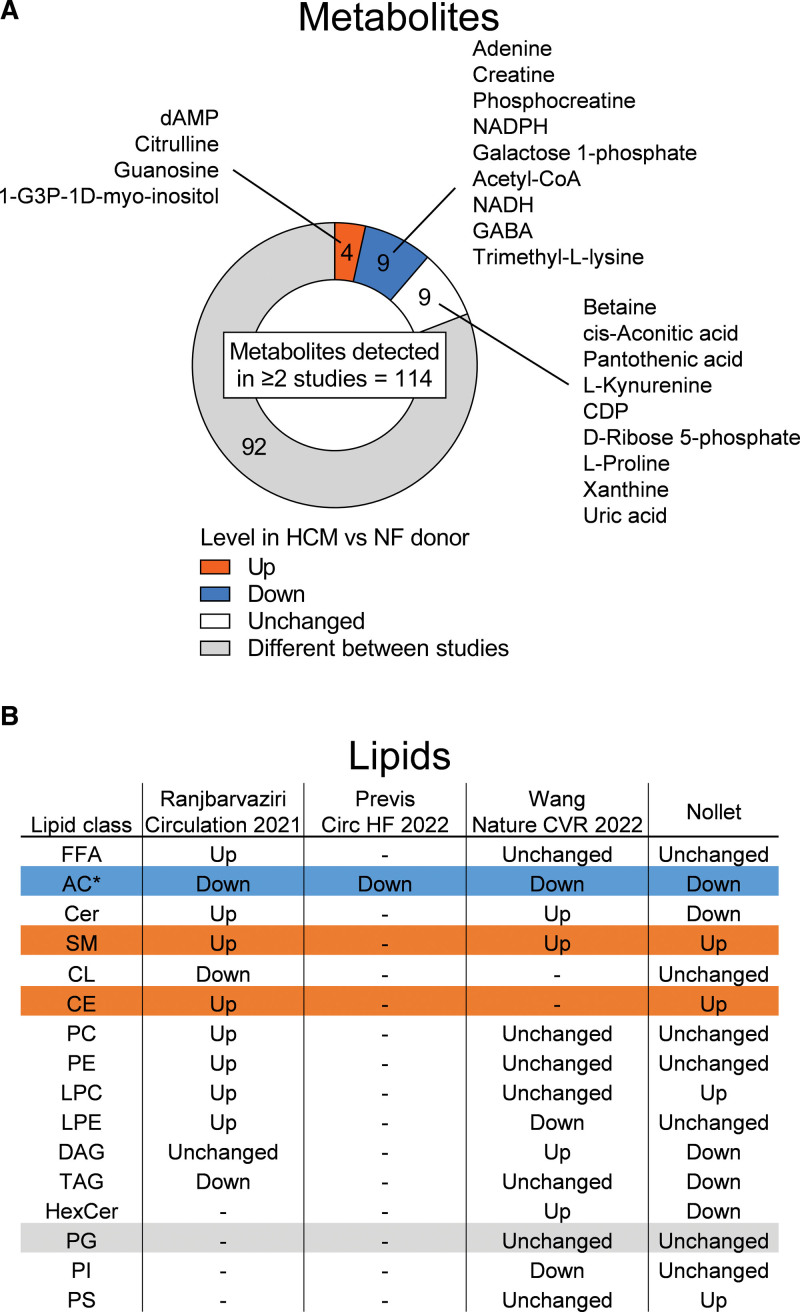
Three other recent studies have aimed to define the lipidome and metabolome in cardiac tissue of patients with obstructive HCM relative to nonfailing donor tissue.11–13 An overview of patient and nonfailing donor demographics in these studies and the present study is shown in Table S4. To map the degree of agreement between studies, we visualized the extent to which metabolomic and lipidomic data were consistent (Figure 8). In total, 114 metabolites were detected in at least 2 studies (Figure 8A; Table S5). Twenty-two of these metabolites displayed similar behavior in HCM versus nonfailing donor tissue. Depletion of acetyl-CoA, phosphocreatine, nicotinamide adenine dinucleotide, and nicotinamide adenine dinucleotide phosphate was consistently observed, suggesting that energetic impairment is a robust feature of HCM. However, the vast majority of metabolomic findings differed between studies. Similarly, lipidomic findings were mostly different between studies, only showing consistent reductions in acylcarnitines and elevations in sphingomyelins and cholesteryl esters in HCM versus nonfailing donors (Figure 8B). These discrepancies suggest that the external validity of group differences observed between nonfailing donors and patients with HCM is relatively limited. Possible explanations underlying this poor generalizability may include cohort differences, such as the degree of cardiac remodeling at the time of myectomy, the demographic and genetic makeup of the HCM patient study population, and the presence of comorbidities and medication use. Regarding comorbidities, average body mass index differed substantially between patient cohorts (Table S4), and it is unclear to what extent this might have influenced results in previous studies.11–13 With respect to medication usage, in our study, a substantial number of patients received antihypertensive drugs, suggesting the presence of hypertension in these patients, potentially modifying our findings. Usage of antihypertensive drugs was, however, not associated with overt hypertension (Figure S6). Moreover, omitting samples from patients on antihypertensive drugs from our correlation analyses (Figure S7) yielded correlation patterns consistent with Figure 5. Thus, the usage of antihypertensive drugs in our patient cohort did not drastically influence our observations. In conclusion, we argue that the involvement of metabolic and lipid alterations in HCM pathophysiology is more optimally identified by linking metabolomic and lipidomic data to variability in clinical patient data. This approach enabled us to identify Gnegative and Gpositive patient–specific associations between metabolic and cardiac remodeling and may serve as a valuable approach to unveil mutation type–specific associations as well when applied to larger cohorts.
Figure 8.
Limited agreement among studies into metabolomic and lipidomic changes in tissue of patients with hypertrophic cardiomyopathy (HCM) compared with nonfailing (NF) donor tissue. A, Pie chart visualizing agreement in metabolite findings in HCM vs NF donor cardiac tissue samples; 114 metabolites were detected in at least 2 studies. Of these metabolites, 92 displayed inconsistent outcomes in HCM vs NF donor tissue. B, Table displaying consistency in lipidomic findings in HCM vs NF donor tissue. Blue and orange indicate consistently lowered and elevated levels, respectively, in HCM vs NF donors. *Derived from both metabolomics and lipidomics.
Study Limitations and Future Implications
We acknowledge several limitations of the study. First, the hearts of nonfailing donors were subjected to cold cardioplegia solution for a longer period of time than the hearts of patients, which may have masked or exacerbated some metabolic changes. Second, all patient samples were collected in the Netherlands, whereas nonfailing donor samples originated from Australia and the United States, which may potentially have introduced bias into our data. Third, given the observational nature of our study, we cannot infer causality of correlations. Nonetheless, our findings underscore that an integrated approach, where we combine clinical and experimental data, is a valuable approach to explore patient group–specific disease mechanisms. Establishing causality should be reserved for further research, utilizing mouse models and human stem cell–derived models of HCM. Our integrated multiomics data suggest an increase in glucose metabolism along with a downturn in fatty acid uptake and catabolism in HCM although our data do not allow for direct evaluation of substrate flux. Impaired fatty acid catabolism is in line with findings in a novel 2-hit mouse model of HCM, in which the onset of cardiomyopathy was associated with depressed fatty acid oxidation.51 A recent study found that failing human hearts with reduced ejection fraction surprisingly displayed retained capability of shifting energy metabolism toward fatty acid oxidation during intralipid infusion. This shift was accompanied by improved cardiac energetics and cardiac output,52 suggesting therapeutic benefit of improved myocardial delivery of fatty acids. Such a strategy could be equally effective as well in patients with HCM, given that cardiac remodeling appeared to be coupled to decreased fatty acid uptake.
Conclusions
Metabolic remodeling in HCM was characterized by a shift away from fatty acid oxidation toward increased glucose and glycogen metabolism. Cardiac remodeling was associated with reduced ceramide formation and lipid storage. In Gnegative patients, cardiac remodeling was linked to mitochondrial dysfunction and decreased levels of acylcarnitines and biosynthetic metabolites. Conversely, in Gpositive patients, we observed positive associations between the abundance of biosynthetic metabolites and cardiac remodeling. Combined, our findings suggest that distinct metabolic alterations contribute to cardiac disease in Gpositive and Gnegative HCM.
ARTICLE INFORMATION
Acknowledgments
The authors are grateful to Georges Janssens (Amsterdam UMC, the Netherlands) for insightful discussions about the contents of this study.
Sources of Funding
This work was supported by the Netherlands Organization for Scientific Research (NWO VICI; grant 91818602) and the Leducq Foundation (grant 20CVD01).
Disclosures
Dr Margulies is a consultant for Bristol Myers Squibb and receives research support from Amgen.
Supplemental Material
Supplemental Methods
Figures S1–S7
Tables S1–S5
Data Set Quality Data
Data Set Multi-Omics Data Sets
Data Set Correlation Statistics
Supplementary Material
Nonstandard abbreviations and acronyms
- ACAT1
- acetyl-CoA acetyltransferase
- BDH
- 3-hydroxybutyrate dehydrogenase
- CPT1A
- carnitine palmitoyltransferase I
- ETFDH
- electron transferring flavoprotein dehydrogenase
- GLUD1
- glutamate dehydrogenase 1
- Gnegative
- genotype-negative
- GOT2
- aspartate aminotransferase
- Gpositive
- genotype-positive
- HCM
- hypertrophic cardiomyopathy
- MRPS5
- mitochondrial ribosomal protein S5
- PPP
- pentose phosphate pathway
- SCOT
- succinyl-CoA-3-oxaloacid CoA transferase
- TCA
- tricarboxylic acid cycle
For Sources of Funding and Disclosures, see page 252.
Supplemental Material is available at https://www.ahajournals.org/doi/suppl/10.1161/CIRCGEN.123.004369.
Contributor Information
Edgar E. Nollet, Email: e.e.nollet@amsterdamumc.nl.
Maike Schuldt, Email: maike.schuldt@gmail.com.
Vasco Sequeira, Email: Sequeira_V@ukw.de.
Aleksandra Binek, Email: Aleksandra.Binek@cshs.org.
Mark Jansen, Email: Mark_jansen@live.nl.
Bauke V. Schomakers, Email: b.schomakers@amsterdamumc.nl.
Michel van Weeghel, Email: m.vanweeghel@amsterdamumc.nl.
Riekelt H. Houtkooper, Email: r.h.houtkooper@amsterdamumc.nl.
Jennifer E. Van Eyk, Email: Jennifer.Vaneyk@cshs.org.
Connie R. Jimenez, Email: c.jimenez@amsterdamumc.nl.
Michelle Michels, Email: m.michels@erasmusmc.nl.
Kenneth B. Margulies, Email: ken.margulies@uphs.upenn.edu.
Cristobal G. dos Remedios, Email: crisdosremedios@gmail.com.
Diederik W.D. Kuster, Email: d.kuster@amsterdamumc.nl.
REFERENCES
- 1.Semsarian C, Ingles J, Maron MS, Maron BJ. New perspectives on the prevalence of hypertrophic cardiomyopathy. J Am Coll Cardiol. 2015;65:1249–1254. doi: 10.1016/j.jacc.2015.01.019 [DOI] [PubMed] [Google Scholar]
- 2.Ommen SR, Mital S, Burke MA, Day SM, Deswal A, Elliott P, Evanovich LL, Hung J, Joglar JA, Kantor P, et al. 2020 AHA/ACC guideline for the diagnosis and treatment of patients with hypertrophic cardiomyopathy: a report of the American College of Cardiology/American Heart Association Joint Committee on Clinical Practice Guidelines. J Am Coll Cardiol. 2020;76:e159–e240. doi: 10.1016/j.jacc.2020.08.045 [DOI] [PubMed] [Google Scholar]
- 3.Maron BJ. Clinical course and management of hypertrophic cardiomyopathy. N Engl J Med. 2018;379:1977. doi: 10.1056/NEJMc1812159 [DOI] [PubMed] [Google Scholar]
- 4.Ho CY, Charron P, Richard P, Girolami F, Van Spaendonck-Zwarts KY, Pinto Y. Genetic advances in sarcomeric cardiomyopathies: state of the art. Cardiovasc Res. 2015;105:397–408. doi: 10.1093/cvr/cvv025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neubauer S, Kolm P, Ho CY, Kwong RY, Desai MY, Dolman SF, Appelbaum E, Desvigne-Nickens P, DiMarco JP, Friedrich MG, et al. ; HCMR Investigators. Distinct subgroups in hypertrophic cardiomyopathy in the NHLBI HCM registry. J Am Coll Cardiol. 2019;74:2333–2345. doi: 10.1016/j.jacc.2019.08.1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Watkins H. Time to think differently about sarcomere-negative hypertrophic cardiomyopathy. Circulation. 2021;143:2415–2417. doi: 10.1161/CIRCULATIONAHA.121.053527 [DOI] [PubMed] [Google Scholar]
- 7.Crilley JG, Boehm EA, Blair E, Rajagopalan B, Blamire AM, Styles P, McKenna WJ, Ostman-Smith I, Clarke K, Watkins H. Hypertrophic cardiomyopathy due to sarcomeric gene mutations is characterized by impaired energy metabolism irrespective of the degree of hypertrophy. J Am Coll Cardiol. 2003;41:1776–1782. doi: 10.1016/s0735-1097(02)03009-7 [DOI] [PubMed] [Google Scholar]
- 8.Guclu A, Knaapen P, Harms HJ, Parbhudayal RY, Michels M, Lammertsma AA, van Rossum AC, Germans T, van der Velden J. Disease stage-dependent changes in cardiac contractile performance and oxygen utilization underlie reduced myocardial efficiency in human inherited hypertrophic cardiomyopathy. Circ Cardiovasc Imaging. 2017;10:e005604. doi: 10.1161/CIRCIMAGING.116.005604 [DOI] [PubMed] [Google Scholar]
- 9.Coats CJ, Heywood WE, Virasami A, Ashrafi N, Syrris P, Dos Remedios C, Treibel TA, Moon JC, Lopes LR, McGregor CGA, et al. Proteomic analysis of the myocardium in hypertrophic obstructive cardiomyopathy. Circ Genom Precis Med. 2018;11:e001974. doi: 10.1161/CIRCGEN.117.001974 [DOI] [PubMed] [Google Scholar]
- 10.Schuldt M, Pei J, Harakalova M, Dorsch LM, Schlossarek S, Mokry M, Knol JC, Pham TV, Schelfhorst T, Piersma SR, et al. Proteomic and functional studies reveal detyrosinated tubulin as treatment target in sarcomere mutation-induced hypertrophic cardiomyopathy. Circ Heart Fail. 2021;14:e007022. doi: 10.1161/CIRCHEARTFAILURE.120.007022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ranjbarvaziri S, Kooiker KB, Ellenberger M, Fajardo G, Zhao M, Vander Roest AS, Woldeyes RA, Koyano TT, Fong R, Ma N, et al. Altered cardiac energetics and mitochondrial dysfunction in hypertrophic cardiomyopathy. Circulation. 2021;144:1714–1731. doi: 10.1161/CIRCULATIONAHA.121.053575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Previs MJ, O’Leary TS, Morley MP, Palmer BM, LeWinter M, Yob JM, Pagani FD, Petucci C, Kim MS, Margulies KB, et al. Defects in the proteome and metabolome in human hypertrophic cardiomyopathy. Circ Heart Fail. 2022;15:e009521. doi: 10.1161/CIRCHEARTFAILURE.121.009521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Wang J, Yao K, Wang S, Nie M, Zhao Y, Wang B, Pang H, Xu J, Wu G, et al. Metabolic characterization of hypertrophic cardiomyopathy in human heart. Nat Cardiovasc Res. 2022;1:445–461. doi: 10.1038/s44161-022-00057-1 [Google Scholar]
- 14.Garmany R, Bos JM, Tester DJ, Giudicessi JR, Dos Remedios CG, Dasari S, Nagaraj NK, Nair AA, Johnson KL, Ryan ZC, et al. Multi-omic architecture of obstructive hypertrophic cardiomyopathy. Circ Genom Precis Med. 2023;16:e003756. doi: 10.1161/CIRCGEN.122.003756 [DOI] [PubMed] [Google Scholar]
- 15.Nollet EE, Duursma I, Rozenbaum A, Eggelbusch M, Wust RCI, Schoonvelde SAC, Michels M, Jansen M, van der Wel NN, Bedi KC, et al. Mitochondrial dysfunction in human hypertrophic cardiomyopathy is linked to cardiomyocyte architecture disruption and corrected by improving NADH-driven mitochondrial respiration. Eur Heart J. 2023;44:1170–1185. doi: 10.1093/eurheartj/ehad028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Molenaars M, Schomakers BV, Elfrink HL, Gao AW, Vervaart MAT, Pras-Raves ML, Luyf AC, Smith RL, Sterken MG, Kammenga JE, et al. Metabolomics and lipidomics in Caenorhabditis elegans using a single-sample preparation. Dis Model Mech. 2021;14:dmm047746. doi: 10.1242/dmm.047746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schomakers BV, Hermans J, Jaspers YRJ, Salomons G, Vaz FM, van Weeghel M, Houtkooper RH. Polar metabolomics in human muscle biopsies using a liquid-liquid extraction and full-scan LC-MS. STAR Protoc. 2022;3:101302. doi: 10.1016/j.xpro.2022.101302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques PE, Li S, Xia J. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:W388–W396. doi: 10.1093/nar/gkab382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kane LA, Neverova I, Van Eyk JE. Subfractionation of heart tissue: the “in sequence” myofilament protein extraction of myocardial tissue. Methods Mol Biol. 2007;357:87–90. doi: 10.1385/1-59745-214-9:87 [DOI] [PubMed] [Google Scholar]
- 20.Mc Ardle A, Binek A, Moradian A, Chazarin Orgel B, Rivas A, Washington KE, Phebus C, Manalo DM, Go J, Venkatraman V, et al. Standardized workflow for precise mid- and high-throughput proteomics of blood biofluids. Clin Chem. 2022;68:450–460. doi: 10.1093/clinchem/hvab202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haug K, Cochrane K, Nainala VC, Williams M, Chang J, Jayaseelan KV, O’Donovan C. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020;48:D440–D444. doi: 10.1093/nar/gkz1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perez-Riverol Y, Bai J, Bandla C, Garcia-Seisdedos D, Hewapathirana S, Kamatchinathan S, Kundu DJ, Prakash A, Frericks-Zipper A, Eisenacher M, et al. The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences. Nucleic Acids Res. 2022;50:D543–D552. doi: 10.1093/nar/gkab1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nijenkamp L, Bollen IAE, van Velzen HG, Regan JA, van Slegtenhorst M, Niessen HWM, Schinkel AFL, Kruger M, Poggesi C, Ho CY, et al. Sex differences at the time of myectomy in hypertrophic cardiomyopathy. Circ Heart Fail. 2018;11:e004133. doi: 10.1161/CIRCHEARTFAILURE.117.004133 [DOI] [PubMed] [Google Scholar]
- 24.Hermida U, Stojanovski D, Raman B, Ariga R, Young AA, Carapella V, Carr-White G, Lukaschuk E, Piechnik SK, Kramer CM, et al. Left ventricular anatomy in obstructive hypertrophic cardiomyopathy: beyond basal septal hypertrophy. Eur Heart J Cardiovasc Imaging. 2023;24:807–818. doi: 10.1093/ehjci/jeac233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D612. doi: 10.1093/nar/gkaa1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dorsch LM, Schuldt M, dos Remedios CG, Schinkel AFL, de Jong PL, Michels M, Kuster DWD, Brundel B, van der Velden J. Protein quality control activation and microtubule remodeling in hypertrophic cardiomyopathy. Cells. 2019;8:741. doi: 10.3390/cells8070741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murashige D, Jang C, Neinast M, Edwards JJ, Cowan A, Hyman MC, Rabinowitz JD, Frankel DS, Arany Z. Comprehensive quantification of fuel use by the failing and nonfailing human heart. Science. 2020;370:364–368. doi: 10.1126/science.abc8861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn VS, Petucci C, Kim MS, Bedi KC, Jr, Wang H, Mishra S, Koleini N, Yoo EJ, Margulies KB, Arany Z, et al. Myocardial metabolomics of human heart failure with preserved ejection fraction. Circulation. 2023;147:1147–1161. doi: 10.1161/CIRCULATIONAHA.122.061846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schlossarek S, Englmann DR, Sultan KR, Sauer M, Eschenhagen T, Carrier L. Defective proteolytic systems in Mybpc3-targeted mice with cardiac hypertrophy. Basic Res Cardiol. 2012;107:235. doi: 10.1007/s00395-011-0235-3 [DOI] [PubMed] [Google Scholar]
- 30.Predmore JM, Wang P, Davis F, Bartolone S, Westfall MV, Dyke DB, Pagani F, Powell SR, Day SM. Ubiquitin proteasome dysfunction in human hypertrophic and dilated cardiomyopathies. Circulation. 2010;121:997–1004. doi: 10.1161/CIRCULATIONAHA.109.904557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Day SM, Divald A, Wang P, Davis F, Bartolone S, Jones R, Powell SR. Impaired assembly and post-translational regulation of 26S proteasome in human end-stage heart failure. Circ Heart Fail. 2013;6:544–549. doi: 10.1161/CIRCHEARTFAILURE.112.000119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smits P, Saada A, Wortmann SB, Heister AJ, Brink M, Pfundt R, Miller C, Haas D, Hantschmann R, Rodenburg RJ, et al. Mutation in mitochondrial ribosomal protein MRPS22 leads to Cornelia de Lange-like phenotype, brain abnormalities and hypertrophic cardiomyopathy. Eur J Hum Genet. 2011;19:394–399. doi: 10.1038/ejhg.2010.214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carroll CJ, Isohanni P, Poyhonen R, Euro L, Richter U, Brilhante V, Gotz A, Lahtinen T, Paetau A, Pihko H, et al. Whole-exome sequencing identifies a mutation in the mitochondrial ribosome protein MRPL44 to underlie mitochondrial infantile cardiomyopathy. J Med Genet. 2013;50:151–159. doi: 10.1136/jmedgenet-2012-101375 [DOI] [PubMed] [Google Scholar]
- 34.Gao F, Liang T, Lu YW, Fu X, Dong X, Pu L, Hong T, Zhou Y, Zhang Y, Liu N, et al. A defect in mitochondrial protein translation influences mitonuclear communication in the heart. Nat Commun. 2023;14:1595. doi: 10.1038/s41467-023-37291-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Molenaars M, Janssens GE, Williams EG, Jongejan A, Lan J, Rabot S, Joly F, Moerland PD, Schomakers BV, Lezzerini M, et al. A conserved mito-cytosolic translational balance links two longevity pathways. Cell Metab. 2020;31:549–563.e7. doi: 10.1016/j.cmet.2020.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature. 2013;497:451–457. doi: 10.1038/nature12188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reid K, Daniels EG, Vasam G, Kamble R, Janssens GE, Hu IM, Green AE, Houtkooper RH, Menzies KJ. Reducing mitochondrial ribosomal gene expression does not alter metabolic health or lifespan in mice. Sci Rep. 2023;13:8391. doi: 10.1038/s41598-023-35196-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Aoyama R, Takano H, Kobayashi Y, Kitamura M, Asai K, Amano Y, Kumita SI, Shimizu W. Evaluation of myocardial glucose metabolism in hypertrophic cardiomyopathy using 18F-fluorodeoxyglucose positron emission tomography. PLoS One. 2017;12:e0188479. doi: 10.1371/journal.pone.0188479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Arad M, Maron BJ, Gorham JM, Johnson WH, Jr, Saul JP, Perez-Atayde AR, Spirito P, Wright GB, Kanter RJ, Seidman CE, et al. Glycogen storage diseases presenting as hypertrophic cardiomyopathy. N Engl J Med. 2005;352:362–372. doi: 10.1056/NEJMoa033349 [DOI] [PubMed] [Google Scholar]
- 40.Unno K, Isobe S, Izawa H, Cheng XW, Kobayashi M, Hirashiki A, Yamada T, Harada K, Ohshima S, Noda A, et al. Relation of functional and morphological changes in mitochondria to myocardial contractile and relaxation reserves in asymptomatic to mildly symptomatic patients with hypertrophic cardiomyopathy. Eur Heart J. 2009;30:1853–1862. doi: 10.1093/eurheartj/ehp184 [DOI] [PubMed] [Google Scholar]
- 41.Stram AR, Payne RM. Post-translational modifications in mitochondria: protein signaling in the powerhouse. Cell Mol Life Sci. 2016;73:4063–4073. doi: 10.1007/s00018-016-2280-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bertero E, Maack C. Metabolic remodelling in heart failure. Nat Rev Cardiol. 2018;15:457–470. doi: 10.1038/s41569-018-0044-6 [DOI] [PubMed] [Google Scholar]
- 43.Sharma S, Adrogue JV, Golfman L, Uray I, Lemm J, Youker K, Noon GP, Frazier OH, Taegtmeyer H. Intramyocardial lipid accumulation in the failing human heart resembles the lipotoxic rat heart. FASEB J. 2004;18:1692–1700. doi: 10.1096/fj.04-2263com [DOI] [PubMed] [Google Scholar]
- 44.Bedi KC, Jr, Snyder NW, Brandimarto J, Aziz M, Mesaros C, Worth AJ, Wang LL, Javaheri A, Blair IA, Margulies KB, et al. Evidence for intramyocardial disruption of lipid metabolism and increased myocardial ketone utilization in advanced human heart failure. Circulation. 2016;133:706–716. doi: 10.1161/CIRCULATIONAHA.115.017545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tanaka T, Sohmiya K, Kawamura K. Is CD36 deficiency an etiology of hereditary hypertrophic cardiomyopathy? J Mol Cell Cardiol. 1997;29:121–127. doi: 10.1006/jmcc.1996.0257 [DOI] [PubMed] [Google Scholar]
- 46.Luiken JJ, Chanda D, Nabben M, Neumann D, Glatz JF. Post-translational modifications of CD36 (SR-B2): implications for regulation of myocellular fatty acid uptake. Biochim Biophys Acta. 2016;1862:2253–2258. doi: 10.1016/j.bbadis.2016.09.004 [DOI] [PubMed] [Google Scholar]
- 47.Pedley AM, Benkovic SJ. A new view into the regulation of purine metabolism: the purinosome. Trends Biochem Sci. 2017;42:141–154. doi: 10.1016/j.tibs.2016.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, Buttgereit F, Bennink JR, Yewdell JW. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18:343–354. doi: 10.1016/s1074-7613(03)00051-7 [DOI] [PubMed] [Google Scholar]
- 49.Steendijk P, Meliga E, Valgimigli M, Ten Cate FJ, Serruys PW. Acute effects of alcohol septal ablation on systolic and diastolic left ventricular function in patients with hypertrophic obstructive cardiomyopathy. Heart. 2008;94:1318–1322. doi: 10.1136/hrt.2007.139535 [DOI] [PubMed] [Google Scholar]
- 50.Ritterhoff J, Tian R. Metabolic mechanisms in physiological and pathological cardiac hypertrophy: new paradigms and challenges. Nat Rev Cardiol. 2023;20:812–829. doi: 10.1038/s41569-023-00887-x [DOI] [PubMed] [Google Scholar]
- 51.Nollet EE, Algül S, Goebel M, Schlossarek S, van der Wel NN, Jans JJ, van de Wiel MA, Knol JC, Pham TV, Piersma SR, et al. Western diet triggers cardiac dysfunction in heterozygous Mybpc3-targeted knock-in mice: a two-hit model of hypertrophic cardiomyopathy. J Mol Cell Cardiol Plus. 2023;6:100050. doi: 10.1016/j.jmccpl.2023.100050 [Google Scholar]
- 52.Watson WD, Green PG, Lewis AJM, Arvidsson P, De Maria GL, Arheden H, Heiberg E, Clarke WT, Rodgers CT, Valkovic L, et al. Retained metabolic flexibility of the failing human heart. Circulation. 2023;148:109–123. doi: 10.1161/CIRCULATIONAHA.122.062166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.van Velzen HG, Schinkel AFL, Baart SJ, Huurman R, van Slegtenhorst MA, Kardys I, Michels M. Effect of gender and genetic mutations on outcomes in patients with hypertrophic cardiomyopathy. Am J Cardiol. 2018;122:1947–1954. doi: 10.1016/j.amjcard.2018.08.040 [DOI] [PubMed] [Google Scholar]
- 54.Chen CY, Caporizzo MA, Bedi K, Vite A, Bogush AI, Robison P, Heffler JG, Salomon AK, Kelly NA, Babu A, et al. Suppression of detyrosinated microtubules improves cardiomyocyte function in human heart failure. Nat Med. 2018;24:1225–1233. doi: 10.1038/s41591-018-0046-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dos Remedios CG, Lal SP, Li A, McNamara J, Keogh A, Macdonald PS, Cooke R, Ehler E, Knoll R, Marston SB, et al. The Sydney Heart Bank: improving translational research while eliminating or reducing the use of animal models of human heart disease. Biophys Rev. 2017;9:431–441. doi: 10.1007/s12551-017-0305-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Soetkamp D, Gallet R, Parker SJ, Holewinski R, Venkatraman V, Peck K, Goldhaber JI, Marban E, Van Eyk JE. Myofilament phosphorylation in stem cell treated diastolic heart failure. Circ Res. 2021;129:1125–1140. doi: 10.1161/CIRCRESAHA.119.316311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Demichev V, Messner CB, Vernardis SI, Lilley KS, Ralser M. DIA-NN: neural networks and interference correction enable deep proteome coverage in high throughput. Nat Methods. 2020;17:41–44. doi: 10.1038/s41592-019-0638-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pham TV, Henneman AA, Jimenez CR. iq: an R package to estimate relative protein abundances from ion quantification in DIA-MS-based proteomics. Bioinformatics. 2020;36:2611–2613. doi: 10.1093/bioinformatics/btz961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3:Article3. doi: 10.2202/1544-6115.1027 [DOI] [PubMed] [Google Scholar]
- 60.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551 [DOI] [PubMed] [Google Scholar]
- 62.Nepusz T, Yu H, Paccanaro A. Detecting overlapping protein complexes in protein-protein interaction networks. Nat Methods. 2012;9:471–472. doi: 10.1038/nmeth.1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rohart F, Gautier B, Singh A, Le Cao KA. mixOmics: an R package for ‘omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13:e1005752. doi: 10.1371/journal.pcbi.1005752 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Metabolomics and lipidomics data have been submitted to the MetaboLights database under accession number MTBLS8695.21 Proteomics data are available via the ProteomeXchange Consortium via the PRIDE partner repository with the data set identifier PXD046187.22 Processed metabolomics, lipidomics, and proteomics data are furthermore available as a Supplemental Material (Multi-Omics Data Sets). The remaining data underlying this article will be shared on reasonable request to the corresponding author.