Abstract
Virus infection of target cells can result in different biological outcomes: lytic infection, cellular transformation, or cell death by apoptosis. Cells respond to virus infection by the activation of specific transcription factors involved in cytokine gene regulation and cell growth control. The ubiquitously expressed interferon regulatory factor 3 (IRF-3) transcription factor is directly activated following virus infection through posttranslational modification. Phosphorylation of specific C-terminal serine residues results in IRF-3 dimerization, nuclear translocation, and activation of DNA-binding and transactivation potential. Once activated, IRF-3 transcriptionally up regulates alpha/beta interferon genes, the chemokine RANTES, and potentially other genes that inhibit viral infection. We previously generated constitutively active [IRF-3(5D)] and dominant negative (IRF-3 ΔN) forms of IRF-3 that control target gene expression. In an effort to characterize the growth regulatory properties of IRF-3, we observed that IRF-3 is a mediator of paramyxovirus-induced apoptosis. Expression of the constitutively active form of IRF-3 is toxic, preventing the establishment of stably transfected cells. By using a tetracycline-inducible system, we show that induction of IRF-3(5D) alone is sufficient to induce apoptosis in human embryonic kidney 293 and human Jurkat T cells as measured by DNA laddering, terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling assay, and analysis of DNA content by flow cytometry. Wild-type IRF-3 expression augments paramyxovirus-induced apoptosis, while expression of IRF-3 ΔN blocks virus-induced apoptosis. In addition, we demonstrate an important role of caspases 8, 9, and 3 in IRF-3-induced apoptosis. These results suggest that IRF-3, in addition to potently activating cytokine genes, regulates apoptotic signalling following virus infection.
Apoptosis, or programmed cell death, plays a critical role in maintaining the homeostasis of multicellular organisms by specifically removing damaged, spent, or misplaced cells (22). Apoptosis can be induced rapidly through death receptor engagement (3, 41) or in response to a wide variety of cellular stresses, including DNA damage, withdrawal of growth factors, and pathogen assault (51). Following an apoptotic stimulus, cells initiate signalling pathways that lead to the activation of cellular death proteases termed caspases and culminate with the degradation of the cellular machinery, giving rise to the classical apoptotic morphology (11, 50, 51).
Viral replication can induce cells to undergo apoptosis. Cells commit suicide as a host defense mechanism, limiting the spread of progeny virus and preventing oncogenic transformation by oncogenic viruses (42). Induction of apoptosis, however, can also benefit the virus: apoptosis of virus-infected cells can lead to increased viral dissemination while evading immune recognition. Many viruses have evolved strategies to suppress or delay the induction of apoptosis, underscoring the importance of this mechanism in immune control. Epstein-Barr virus encodes a functional homologue of the anti-apoptotic protein Bcl-2 (18), and via its latent membrane protein-1, it up regulates the expression of the anti-apoptotic gene bcl-2 and activates the NF-κB signalling pathway (19, 44), both of which have been implicated in cell survival (1, 6, 41). Other viruses, including human papillomavirus and adenovirus, express proteins which inactivate p53, thereby ablating induction of p53-dependent apoptosis (48). Several other viruses have adopted a common strategy: baculovirus protein p53 and poxvirus CrmA inhibit the activation of caspases (10, 30). Kaposi sarcoma-associated human herpesvirus 8 also specifically encodes an inhibitor of caspase 8, thereby preventing T cells from eliminating infected cells by death receptor induction (49).
One of the immediate cellular responses to viral infection involves the secretion of interferons (IFNs) (52). In addition to their antiviral and immunomodulatory properties, IFNs have recently been shown to be important regulators of virus-induced apoptosis. IFNs elicit an antiviral state in uninfected cells through the transcriptional activation of anti-viral proteins while inducing apoptosis in virus-infected cells (47). IFNs are upregulated through the coordinate activation of transcriptional regulatory proteins NF-κB and IFN regulatory factors (IRFs) (20). Virus-induced posttranslational phosphorylation of IRF-3 is thought to stimulate beta IFN (IFN-β) production (21). Secreted IFN-β (and IFN-α4 in murine cells) then acts through the JAK-STAT pathway to stimulate the production of a distinct member of the IRF family—IRF-7—which in turn contributes to the transcriptional induction of other IFN-α genes (5, 29, 39).
The IRF family of transcription factors includes nine mammalian members; IRF-1 to -7, ICSBP (now designated IRF-8), and p48 (designated IRF-9), as well as several viral homologs (32). IRF-3 is a 55-kDa protein that is expressed constitutively in all tissues (4). In response to virus infection or treatment with double-stranded (ds) RNA, IRF-3 is phosphorylated on specific C-terminal serine and threonine residues (26, 54, 56). This posttranslational modification leads to IRF-3 dimerization and translocation into the nucleus and the transcriptional activation of target genes (26, 27, 53, 54, 56). IRF-3 requires the coactivator CREB-binding protein (CBP)-p300 to mediate transcriptional activation and is important for regulating viral induction of the chemokine RANTES as well as IFN genes (25, 26, 37, 54, 56). Substitution of these serine and threonine residues in the amino acid 396 to 405 region of IRF-3 with aspartic acids creates a constitutively active form of IRF-3 that is able to stimulate transcription in the absence of virus induction (26).
In this study, we demonstrate that IRF-3 is an essential mediator of paramyxovirus-induced apoptosis. Initial experiments demonstrated that expression of the constitutively active form of IRF-3 was toxic in both Jurkat T cells and human embryonic kidney 293 cells, thus preventing the establishment of stably transfected cells. To circumvent this effect, a tetracycline (TET)-inducible system was used to demonstrate that induced expression of the constitutively active form of IRF-3 was sufficient to produce apoptosis in both Jurkat and 293 cells. Wild-type IRF-3 expression augmented paramyxovirus-induced apoptosis in both cell types, whereas expression of a truncated dominant negative form of IRF-3 blocked virus-induced apoptosis. These results suggest that IRF-3, in addition to potently activating cytokine genes, functions to regulate apoptotic signalling in response to virus infection.
MATERIALS AND METHODS
Cell culture and virus infection.
Human embryonic kidney 293 or Jurkat T cells were grown in minimal essential medium alpha (αMEM) (293) or RPMI 1640 (Jurkat) medium (GIBCO-BRL) supplemented with 10% fetal bovine serum (FBS), glutamine, and antibiotics. In experiments where control and IRF-3-expressing cells were virus infected, Sendai virus (80 hemagglutinating units [HAU]/ml) (a kind gift from I. Julkunen) was added to the cells for 1 h in serum-free media. After incubation, cells were washed and placed in growth media containing 10% FBS. For the doxycycline (DOX)-inducible cell lines, cells were pretreated with 1 μg of DOX per ml for 12 h (Jurkat T cells) or for 24 h (293 cells) and were maintained with DOX for the duration of the infection.
Plasmid constructions and mutagenesis.
The wild-type and mutated forms of IRF-3-expressing plasmids were described previously (26).
Generation of IRF-3 and IRF-3(5D) cell lines.
Plasmid CMVt-rtTA (33) was introduced into human embryonic kidney 293 cells by the calcium phosphate method and into human Jurkat T cells by electroporation. Forty-eight hours after transfection, cells were grown in αMEM (GIBCO-BRL) (293 cells) or RPMI 1640 (GIBCO-BRL) (Jurkat cells) medium containing 10% heat-inactivated FBS, glutamine, antibiotics, and 2.5 ng of puromycin (Sigma) per μl. Resistant cells carrying the CMVt-rtTA plasmid (rtTA-293 cells or rtTA-Jurkat cells) were then transfected with the CMVt-IRF-3 and CMVt-IRF-3(5D) plasmids. Cells were placed in selective media (growth media with 2.5 ng of puromycin per μl and 400 μg of G418 per ml) 48 h after transfection and were monitored for approximately 2 weeks. For generation of IRF-3 ΔN cells, IRF-3 ΔN/pEGFPC1 was introduced into 293 cells by the calcium phosphate method, and cells were selected with G418 as described above.
Colony formation assay.
293 cells were transfected with 10 μg each of control pEGFPC1 vector, pEGFPC1-IRF-3, and pEGFPC1-IRF-3(5D). Thirty six hours after transfection, cells were placed in selection media containing 400 μg of G418 (Life Technologies, Inc.) per ml. Two weeks later, cells were fixed with ice-cold methanol and were Giemsa stained (Life Technologies, Inc.). Expression of the green fluorescent protein (GFP) fusion proteins was monitored by fluorescence microscopy.
Western blot analysis.
To screen and characterize the kinetics of IRF-3 and IRF-3(5D) induction, the expressing cells were cultured in the presence of 1 μg of DOX per ml for various times. Cells were washed with phosphate-buffered saline (PBS) and were lysed in 10 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, 0.5% Nonidet P-40, 0.5 mM phenylmethysulfonyl fluoride, 5 μg of leupeptin per ml, 5 μg of pepstatin per ml, and 5 μg of aprotinin per ml. Equivalent amounts of whole-cell extract (20 to 40 μg) were subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) in a 10% (IRF-3) or 15% (CPP-32) polyacrylamide gel. After electrophoresis, the proteins were transferred to Hybond transfer membrane (Amersham) in a buffer containing 30 mM Tris, 200 mM glycine, and 20% methanol for 1 h. The membrane was blocked by incubation in PBS containing 5% milk for 1 h and was then probed with IRF-3 or CPP-32 antibody (kind gifts from P. Pitha and P. R. Sékaly, respectively) in 5% milk-PBS (dilution 1:3,000) at 4°C overnight. After four 10-min washes with PBS, membranes were reacted with a peroxidase-conjugated secondary goat anti-rabbit antibody (Amersham) at a dilution of 1:2,500. The reaction was then visualized with the enhanced chemiluminescence detection system (ECL) as recommended by the manufacturer (Amersham).
Flow cytometry.
For fluorescence-activated cell sorter (FACS) analysis of Jurkat T cells, 5 × 106 cells were washed in PBS and were fixed in 70% ethanol in PBS for 1 h. Cells were then washed three times with PBS and were stained at 4°C overnight in 0.5 mM Tris (pH 8.0), 1.5 mM spermine tetrahydrochloride, 35 μg of RNase A per ml, and 50 μg of propidium iodide per ml. Cells were analyzed by FACStar using the Consort 30 software (Becton Dickinson).
DNA fragmentation.
Following treatments, ∼2 × 106 cells were pelleted, were washed with PBS, were resuspended in 250 μl of lysis buffer (20 mM Tris HCl [pH 7.5], 10 mM borate, 0.25% Nonidet P-40, 0.1 mg of RNase per ml), and were incubated for 1 h at 37°C. Proteinase K was added to a final concentration of 1 mg/ml, and extracts were incubated for an additional 1 h. Samples were separated on a 1.8% agarose gel containing 0.5 μg of ethidium bromide per ml and were visualized by UV illumination.
TUNEL analysis.
Apoptosis was quantified by using an in situ cell death detection kit (Boehringer Mannheim). Approximately 106 cells were centrifuged, washed once with PBS, and resuspended in 20 μl of PBS (on a multichamber slide) or in 100 μl of PBS (FACS analysis). For the multichamber slide, cells were plated, air dried, and fixed with 4% paraformaldehyde for 30 min at room temperature. Slides were rinsed twice with PBS and were incubated for 2 min at 4°C in permeabilization solution (0.1% Triton X-100 and 0.1% sodium citrate), were rinsed with PBS, and were incubated with fluorescein-labelled terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) reaction mixture for 1 h at 37°C in a humid, darkened chamber. Slides were again rinsed with PBS, were incubated with the DNA dye Hoechst 33342 (0.4 ng/ml) to visualize all nuclei, were washed with PBS, and were embedded in mounting solution (10 mM Tris HCL [pH 8.8] and 0.1 M propyl gallate in glycerol). Samples were analyzed by fluorescence microscopy, and the percentage of apoptotic cells was determined by counting a minimum of 350 nuclei (blue filter) and the corresponding TUNEL-positive cells (green filter). For FACS analysis, a similar protocol was used, except that U-bottomed 96-well plates were used instead of multichamber slides. Cells were analyzed by FACStar by using the Consort 30 software (Becton Dickinson).
MTT assay.
293 cells were plated at a concentration of 1,000 cells/well in a 96-well plate. After 24 h, cells were treated with different inducers and were subsequently analyzed by metallothionin (MTT) assay. Briefly, the medium was replaced by fresh medium containing 10 mM of PIPES [4-(2-hydroxyethyl)-1-piperamine-ethanesulfonic acid] buffer (pH 7.4) and 0.5 mg of MTT [3-(4,5-dimethylthiazol-2-γ1)-2,5-diphenyltetrazolium bromide] per ml. The plates were wrapped in aluminum foil and were further incubated for 4 h at 37°C. The medium and MTT were removed, and the crystals that had formed in each well were dissolved with 225 μl of a solution containing 200 μl of dimethylsulfoxide and 25 μl of glycine buffer (0.1 M glycine, 0.1 M NaCl [pH 10.5]). Absorbance was read by using an enzyme-linked immunosorbent assay plate reader (model 450; Bio-Rad Laboratories, Watford, England), interfaced to a Macintosh computer.
RPA.
For ribonuclease protection assay (RPA), total RNA isolated from 293 or Jurkat T cell pellets was prepared using the RNAeasy kit (Qiagen). Total RNA (5 μg) was subjected to RPA by using the hCK-3 chemokine template of the RiboQuant multi-probe RPA kit, following the manufacturer's instructions (Pharmingen, San Diego, Calif.).
Caspase assay.
For quantifying caspase activity. Jurkat T cells (5 × 106 cells) expressing IRF-3(5D) were washed in cold PBS, lyzed, and incubated with fluorogenic substrates for caspase 8 (IETD-AFC; Clontech) or caspase 9 (LEHD-AFC; Enzyme Systems Products) as described in the Apoalert FLICE/Caspase-8 assay kit (Clontech). This kit can also be modified to measure caspase 9 activity (Clontech). AFC fluorescence emission was detected at 505 nm, following excitation at 400 nm with a fluorescence spectrophotometer.
Caspase inhibitor assay.
Noninduced, DOX-induced, and/or Jurkat T cells infected for various times with Sendai virus were aliquoted into the wells of a 96-well plate at a concentration of 50,000 cells/well/100 μl of medium in the presence or absence of a solution containing 200 μM zVAD, zIETH, zLEHD, 4 μg of APO-1-3 per ml (Kamiya Biomedical), and 50 μM Etoposide (Sigma). At 24, 48, or 72 h after treatment, cell viability was determined by Trypan blue exclusion.
RESULTS
Generation of TET-inducible cell lines.
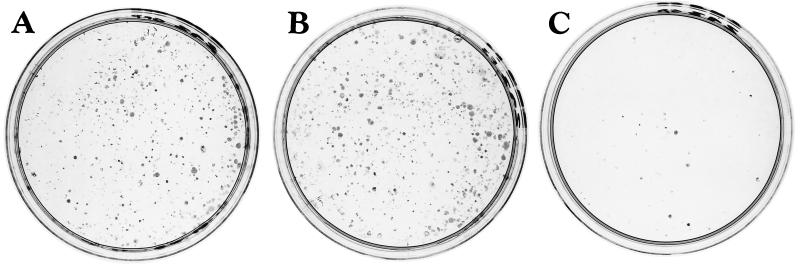
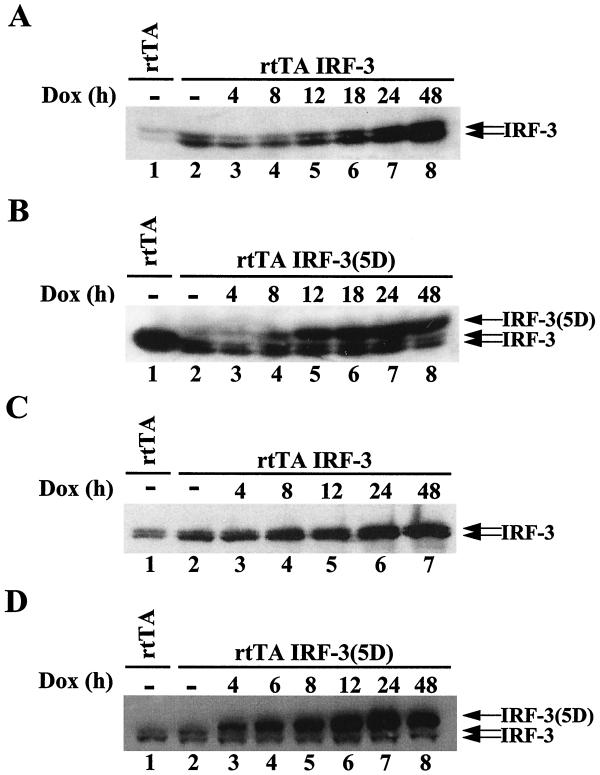
In order to identify genes that may be regulated by the IRF-3 transcription factor, we sought to generate stable cell lines expressing different forms of IRF-3. The pEGFPC1 control vector and vectors encoding GFP-wild-type IRF-3 (wtIRF-3) and a constitutively active form of IRF-3, GFP-IRF-3(5D), were transfected in 293 human embryonic kidney cells. After 2 weeks of selection in G418-containing medium, an equal number of clones (250 to 300 clones per 100-mm-diameter plate) expressing the control vector or wtIRF-3 were obtained (Fig. 1A and B). In contrast, only a few clones (25 to 30 clones per 100-mm-diameter plate) were obtained from cells transfected with IRF-3(5D), revealing a possible toxic effect of the transgene (Fig. 1C). Moreover, clones isolated from the IRF-3(5D)-transfected cells did not express the GFP fusion protein (data not shown). To circumvent this effect, 293 and Jurkat cells that inducibly expressed wtIRF-3 and IRF-3(5D) under the control of a TET-inducible promoter were generated, as previously described (25). Polyclonal populations of 293 and Jurkat cells were screened for DOX-inducible expression of the transgene by immunoblot analysis. wtIRF-3 and IRF-3(5D) were inducible following DOX induction, with high levels of IRF-3 evident at 18 h in rtTA-293 wtIRF-3 cells (Fig. 2A, lane 6), at 8 h in rtTA-293 IRF-3(5D) cells (Fig. 2B, lane 4) and rtTA-Jurkat wtIRF-3 cells (Fig. 2C, lane 4), and as early as at 4 h in rtTA-Jurkat IRF-3(5D) cells (Fig. 2D, lane 3). The IRF-3(5D) protein migrated more slowly on SDS-PAGE than endogenous IRF-3 protein, at a position similar to phosphorylated IRF-3 (25). In contrast to cell lines transfected with wtIRF-3, cell lines transfected with IRF-3(5D) did not exhibit any expression of the transgene in the absence of DOX induction (Fig. 2B and D, lanes 2), further suggesting that leakiness of the IRF-3(5D) transgene under unstimulated conditions prevented the selection of stable cell clones.
FIG. 1.
Constitutively active IRF-3 is toxic to cells. 293 cells were transfected with 10 μg of control pEGFPC1 vector (A), pEGFPC1-wtIRF-3 (B), or pEGFPC1-IRF-3(5D) (C). Beginning 36 h after transfection, cells were selected in media containing G418 (400 μg/ml); after 2 weeks of selection, cells were fixed in the plate with ice-cold methanol and were stained with Giemsa. Expression of the GFP fusion proteins was monitored by fluorescence microscopy.
FIG. 2.
Inducible expression of IRF-3 and IRF-3(5D). Whole-cell extracts (20 μg) were prepared from rtTA-293 (A and B) and rtTA-Jurkat (C and D) cells. rtTA-293 wtIRF-3 (A), rtTA-293 IRF-3(5D) (B), rtTA-Jurkat wtIRF-3 (C), and rtTA-Jurkat IRF-3(5D) (D) cells were induced with DOX (1 μg/ml) for 0 to 48 h and were analyzed for IRF-3 expression by immunoblot analysis. IRF-3(5D) protein migrated more slowly than endogenous IRF-3 protein on SDS-PAGE at a position similar to phosphorylated IRF-3 protein.
Constitutively active IRF-3 induces apoptosis.
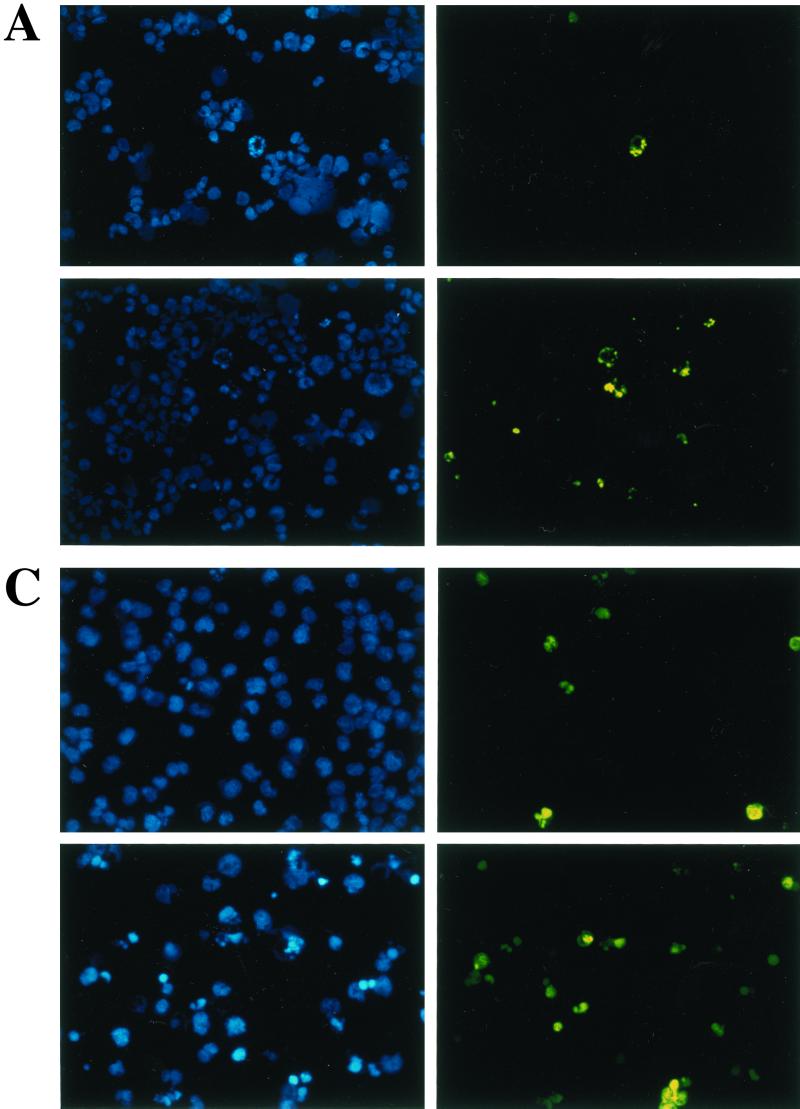
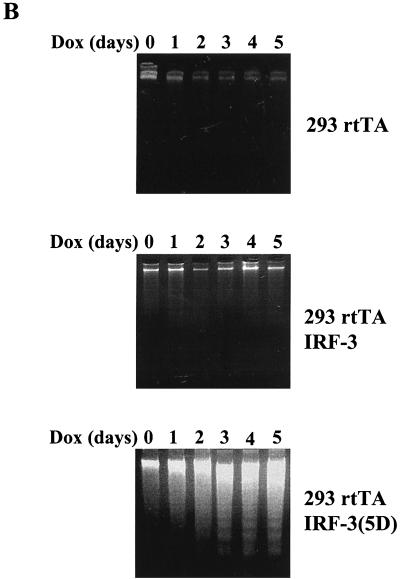
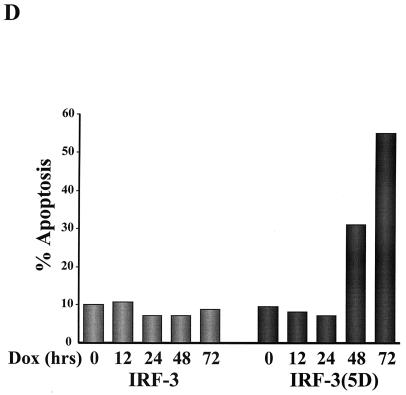
To study the effect of wtIRF-3 and IRF-3(5D) on cell-growth regulation, IRF-3 was expressed following the addition of DOX to the rtTA-293 wtIRF-3- and rtTA-293 IRF-3(5D)-inducible cell lines. The addition of DOX resulted in cell death in IRF-3(5D)-expressing cells beginning at 48 h but did not cause cell death in wtIRF-3-expressing cells. wtIRF-3- and IRF-3(5D)-expressing 293 cells were next assayed for apoptosis by the TUNEL method (Fig. 3A) and by DNA fragmentation (Fig. 3B). DOX-induced control rtTA-293 and rtTA-293 wtIRF-3 cells were TUNEL negative and showed no spontaneous DNA fragmentation (Fig. 3B), while rtTA-293 IRF-3(5D) cells were TUNEL positive (approximately 25%) (Fig. 3A) and exhibited DNA fragmentation starting 2 days after DOX induction with peak levels on days 4 and 5 (Fig. 3B). IRF-3(5D)-induced apoptosis was also examined in rtTA-IRF-3-expressing Jurkat T cells by monitoring for TUNEL-positive staining (Fig. 3C) and DNA content by propidium iodide staining and flow cytometry analysis (Fig. 3D). DOX induction of the IRF-3(5D) resulted in 33% apoptosis by 48 h, and the number of detectable apoptotic cells had increased to 55% by 72 h. Addition of DOX to rtTA-Jurkat control cells and wtIRF-3-expressing Jurkat cells did not increase the low level of apoptosis in these cells (Fig. 3D).
FIG. 3.
Constitutively active IRF-3 induces apoptosis. TUNEL staining of IRF-3(5D)-expressing 293 (A) and Jurkat cells (C). rtTA-293 IRF-3(5D) and rtTA-Jurkat IRF-3(5D) cells were left untreated or induced with DOX for 48 (293) or 72 h (Jurkat). Cells were then stained by the TUNEL method (green filter) and with Hoechst dye to visualize all nuclei (blue filter) as described in Materials and Methods. (B) Kinetics of DNA fragmentation in 293 IRF-3(5D)-expressing cells. Plates of rtTA-, wtIRF-3-, and IRF-3(5D)-expressing 293 cells were induced with DOX (1 μg/ml) for 0 to 5 days. DNA was isolated from each sample and was analyzed by agarose gel electrophoresis as described in Materials and Methods. (D) Kinetics of IRF-3(5D)-induced apoptosis in Jurkat T cells. wtIRF-3- and IRF-3(5D)-expressing Jurkat T cells were induced with DOX (1 μg/ml) for 0 to 72 h as indicated. Cells (5 × 106) were fixed and stained with propidium iodide. Cellular DNA content was analyzed by flow cytometry.
IRF-3 mediates virus-induced apoptosis.
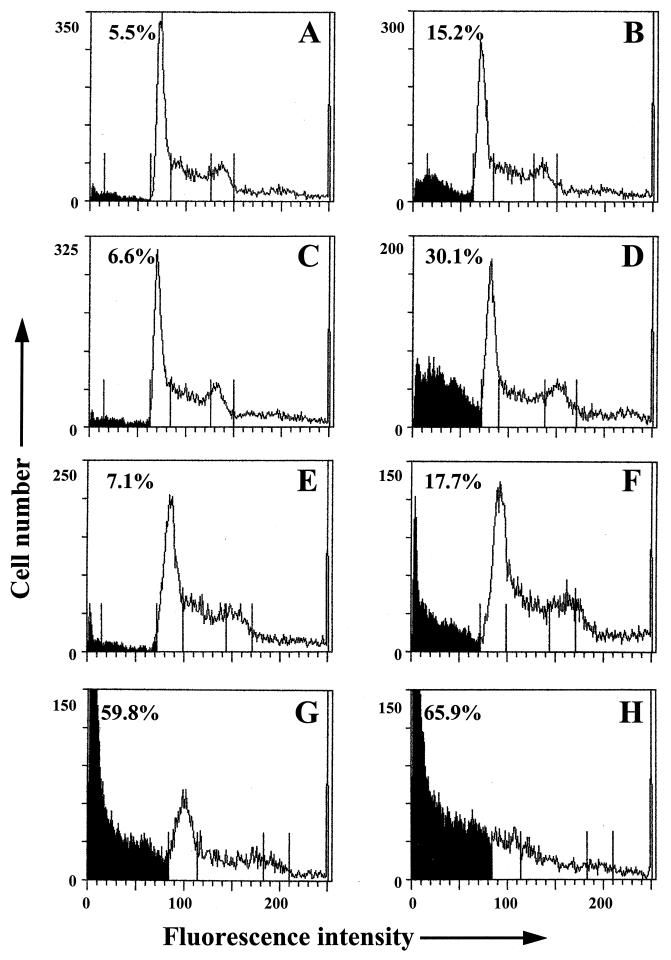
Since IRF-3 is directly activated following virus infection, we sought to examine whether IRF-3 was involved in virus-induced apoptosis. Control and Jurkat cells overexpressing wtIRF-3 or IRF-3(5D) were infected with Sendai virus and were subsequently stained with propidium iodide for FACS analysis (Fig. 4). Sendai virus infection of rtTA-Jurkat cells resulted in approximately 15% apoptosis (Fig. 4B), while overexpression of wtIRF-3 increased the number of apoptotic cells to 30% (Fig. 4D). The response to virus infection in uninduced IRF-3(5D)-expressing Jurkat cells was similar to control cells (Fig. 4F) while overexpression of IRF-3(5D)-induced apoptosis in 60% of the population (Fig. 4G); the combination of virus infection and overexpression of IRF-3(5D) resulted in almost 66% cell death (Fig. 4H).
FIG. 4.
IRF-3 potentiates virus-induced apoptosis. rtTA-Jurkat (A and B), rtTA-Jurkat wtIRF-3 (C and D), and rtTA-Jurkat IRF-3(5D) (E to H) were cultured in the presence (A to D, G, and H) or absence of DOX (1 μg/ml) (E and F). After 12 h, cells were either left untreated (A, C, E, and G) or were infected with Sendai virus (80 HAU/ml) for 72 h (B, D, F, and H). Cells (5 × 106) were fixed and stained with propidium iodide. Cellular DNA content was analyzed by flow cytometry.
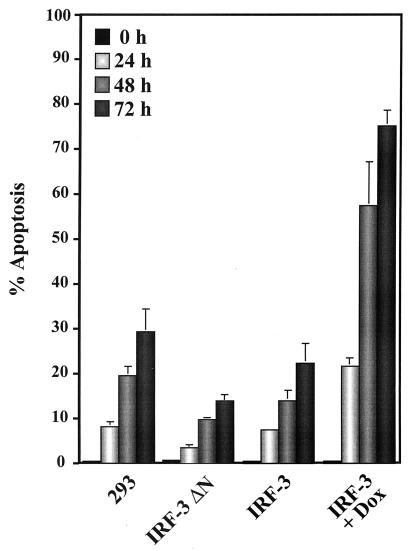
Next, a dominant negative mutant of IRF-3 lacking most of the DNA binding domain (IRF-3 ΔN) was used to examine the involvement of IRF-3 in virus-mediated apoptosis. This dominant negative mutant of IRF-3 has been shown to block downstream target gene activation (25). 293 cells overexpressing wtIRF-3 or IRF-3 ΔN were infected with Sendai virus and were analyzed for virus induced apoptosis by TUNEL analysis. Virus infection of control 293 cells resulted in 25 to 30% apoptosis by 72 h after infection, whereas the level of apoptosis in wtIRF-3-expressing cells was more pronounced. Nonstimulated rtTA-293 wtIRF-3 cells showed approximately the same amount of apoptosis as control 293 cells (25% at 72 h postinfection), whereas the same cells exhibited a threefold increase in TUNEL positive cells following DOX induction. By 24 h postinfection, 20% of the cells were apoptotic; the level of apoptosis increased to approximately 60% after 48 h, and by 72 h after virus infection about 75% of the cell population was apoptotic (Fig. 5). The essential role of IRF-3 in mediating virus-induced apoptosis was highlighted by the observation that expression of the dominant negative form of IRF-3 blocked the induction of apoptosis by viral infection; less than 15% of the cells were apoptotic at 72 h postinfection (Fig. 5).
FIG. 5.
Inhibition of virus-induced apoptosis. Control 293 and 293 IRF-3 ΔN-expressing cells were left untreated or were infected with Sendai virus (80 HAU/ml) for 24, 48, and 72 h. rtTA-293 wtIRF-3 were cultured in the presence or absence of DOX (1 μg/ml) as indicated. After 24 h, cells were either left untreated or were infected with Sendai virus as described above. The number of apoptotic cells was determined by TUNEL staining as described in Materials and Methods.
IFN release is not implicated in IRF-3-induced apoptosis.
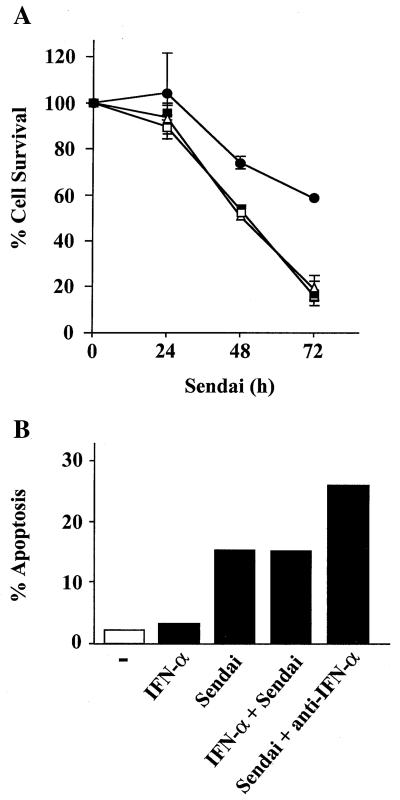
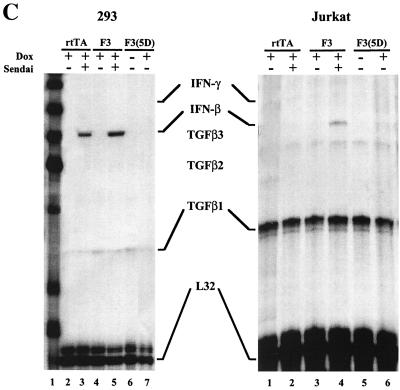
Since constitutively active IRF-3 has been shown to be a strong activator of interferon and cytokine gene expression (25, 26), we investigated the possibility that IRF-3 may stimulate IFN production which in turn may induce apoptosis, since IFN has been shown to induce apoptosis in virus-infected cells (47). IFN treatment alone did not induce apoptosis in Jurkat cells. As shown in Fig. 6A and B, the addition of a neutralizing antibody against IFN-α did not protect 293 or Jurkat cells from Sendai virus-induced apoptosis, as measured by MTT or TUNEL assays, respectively. The MTT assay, which reflects mitochondrial activity, was used as a marker of cell death in Fig. 6A. Moreover, cotreatment of both cell types with Sendai virus and IFN-α did not increase the level of cell death beyond that induced by virus alone (Fig. 6A and B). Expression of IRF-3 ΔN abrogated 50% of Sendai virus-induced apoptosis (Fig. 6A); this data was similar to that by TUNEL assay presented in Fig. 5. The efficacy of the neutralizing antibody was demonstrated by the inhibition of IFN-α stimulation of an IFN-stimulated response element (ISRE)-containing reporter gene construct (ISG-15-CAT) in transient transfections (data not shown). RNase protection analysis further demonstrated that while Sendai virus infection induced IFN-β mRNA in 293 and Jurkat cells, IRF-3(5D) overexpression alone was not sufficient to induce IFN-β mRNA in either cell type (Fig. 6C). Similarly, no IFN-γ mRNA was detected (Fig. 6C). Finally, Jurkat T cells grown in the conditioned media derived from rtTA-Jurkat or rtTA-Jurkat IRF-3(5D) cells induced with DOX for 72 h did not undergo apoptosis (data not shown). Altogether, these data indicated that IFN production was not involved in the induction of apoptosis by IRF-3.
FIG. 6.
IFN release is not implicated in IRF-3-induced apoptosis. (A) Control 293 and 293 IRF-3 ΔN-expressing cells were left untreated or were infected with Sendai virus (80 HAU/ml) for 24, 48, and 72 h in the presence or absence of IFN-α (400 IU/ml) or neutralizing antibody for alpha/beta interferon (1/100) (Sigma) as indicated. Viability was measured by using an MTT assay as described in Materials and Methods. Symbols: ■, 293; □, 293 plus IFN-α; ▵, 293 plus anti-IFN-α; ●, 293 IRF-3 ΔN. (B) TUNEL staining of Jurkat cells. The rtTA-Jurkat cells were either left untreated, were infected with Sendai virus (80 HAU/ml), or were treated with IFN-α (400 IU/ml) for 72 h; anti-IFN-α antibody was added with Sendai virus. The number of apoptotic cells was determined by TUNEL as described in Materials and Methods. (C) RPA of IFN-β and IFN-γ mRNA production. The rtTA-, wtIRF-3-, and IRF-3(5D)-expressing 293 and Jurkat cells were cultured in the presence or absence of DOX, as indicated, for 24 h. Cells were then either left untreated or were infected with Sendai virus for 72 h. Total RNA was isolated from each sample and was analyzed by RNase protection analysis by using the human CK-3 RPA kit (Pharmingen), according to manufacturer's instructions.
Activation of CPP-32/caspase 3.
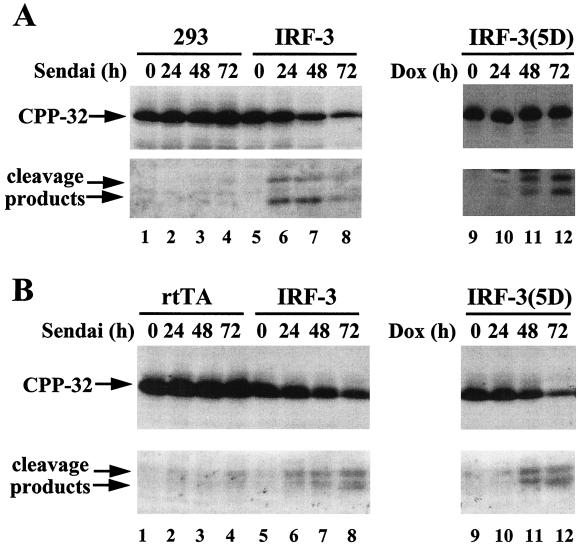
To examine which members of the caspase family were involved in these apoptotic processes, CPP-32/caspase 3 activation was monitored by immunoblot analysis (Fig. 7). Following an apoptotic stimulus, CPP-32 proenzyme is processed into two subunits, the active subunit (p17) and a smaller subunit (p12) (35). Whereas only low levels of the active p17 subunit of caspase 3 could be detected in control 293 and rtTA-Jurkat cells after Sendai virus infection (Fig. 7A and B, lanes 1 to 4), both Jurkat and 293 cell lines overexpressing wtIRF-3 showed a progressive decrease in the amount of full-length caspase 3 and an increase in the levels of active p17 caspase 3 subunit after virus infection (Fig. 7A and B, lanes 5 to 8). Induction of the constitutively active IRF-3(5D) with DOX had a similar effect: caspase 3 was activated 48 h after DOX induction (Fig. 7A and B, lanes 11), in agreement with the kinetics of induction of apoptosis.
FIG. 7.
CPP-32 activation in virus-infected and IRF-3(5D)-expressing cells. (A) Whole-cell extracts from 293 and DOX-induced rtTA-wtIRF-3-, and IRF-3(5D)-expressing 293 cells infected with Sendai virus (80 HAU/ml) or treated with DOX for the times indicated were subjected to SDS-PAGE and were transferred to nitrocellulose membrane. (B) Whole-cell extracts from untreated or DOX-induced rtTA-, wtIRF-3-, or IRF-3(5D)-expressing Jurkat cells infected with Sendai virus (80 HAU/ml) or induced with DOX for the times indicated were subjected to SDS-PAGE and were transferred to nitrocellulose membrane. CPP-32 and its cleavage products were detected by immunoblot analysis by using a polyclonal CPP-32 antibody (a gift from P. R. Sekaly).
Caspase 8 and caspase 9 are involved in IRF-3-dependent activation of CPP-32/caspase 3.
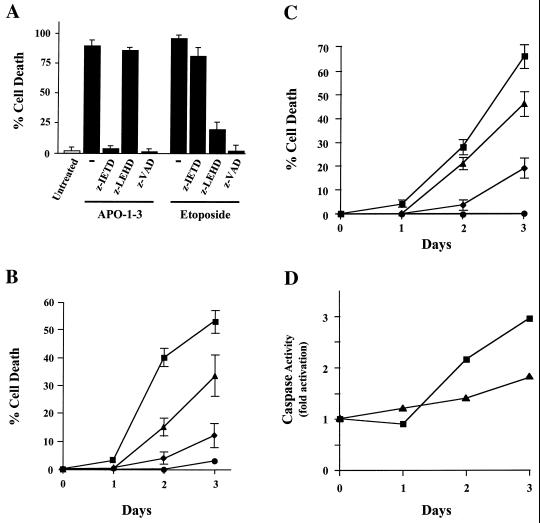
We next examined which apoptotic pathways led to caspase 3 activation in Sendai virus-infected or IRF-3(5D)-overexpressing cells. Several studies have reported that at least two caspases (caspases 8 and 9) can activate CPP-32/caspase 3 following death receptor ligation or exposure to cytotoxic agents (8, 15, 24, 31, 43). To verify if these caspases are involved in Sendai virus- and IRF-3(5D)-induced apoptosis, highly specific inhibitors of the caspases were used in a pharmacological approach, and the effects of these inhibitors in IRF-3-expressing Jurkat cells are shown in Fig. 8A. The use of a broad-spectrum caspase inhibitor, z-VAD, completely abrogated anti-Fas antibody (APO-1-3)- and etoposide-induced apoptosis in rtTA-Jurkat IRF-3(5D). Under the same conditions, z-IETD—which is selective for caspase 8—inhibited the effect of anti-Fas antibody which stimulates FADD-dependent cell death though caspase 8 activation, but had no effect on etoposide-induced apoptosis. In contrast, the caspase 9 specific inhibitor z-LEHD blocked the apoptotic effect of etoposide which bypasses caspase 8 and induces apoptosis by triggering cytochrome c release and caspase 9 activation.
FIG. 8.
Caspase 8 and caspase 9 are involved in IRF-3-dependent activation of CPP-32/caspase-3. (A) rtTA-Jurkat IRF-3(5D) cells were cultured in the presence of 4 μg of APO-1-3 per ml or 50 μM Etoposide for 48 h in the continuous presence of caspase blockers zVAD, zIETH, and zLEHD (200 μM), as indicated. Viability was evaluated by using trypan blue exclusion. (B) wtIRF-3-expressing Jurkat cells were treated for 48 h with DOX (5 μg/ml) to stimulate IRF-3 production and were then infected with Sendai virus (400 HAU/ml/106 cells) for 24, 48, or 72 h in the continuous presence of caspase blockers zVAD, zIETH, and zLEHD (200 μM). Viability was evaluated by using trypan blue exclusion. (C) IRF-3(5D)-expressing Jurkat cells were treated with DOX (5 μg/ml) for 24, 48, or 72 h to stimulate IRF-3(5D) production. DOX treatment was accomplished in the continuous presence of caspase blockers zVAD, zIETH, and zLEHD (200 μM). Viability was evaluated by using trypan blue exclusion. Symbols in B and C: ■, Sendai virus; ▴, Sendai virus plus zLEHD; ⧫, Sendai virus plus zIETD; ●, Sendai virus plus zVAD. (D) IRF-3(5D)-expressing Jurkat cells were treated with DOX (5 μg/ml) for 24, 48, and 72 h; whole-cell extracts were prepared at different times after treatment with DOX and were incubated with fluorogenic substrates for caspase 8 (■) and caspase 9 (▴) as described in Materials and Methods. Caspase activity is represented by the fluorescence ratio between DOX-induced versus noninduced cells.
By using a similar approach, we next verified the pathway by which Sendai virus and IRF-3(5D) induce apoptosis. In Fig. 8B, treatment of wtIRF-3 Jurkat cells with the caspase inhibitor z-IETD abrogated almost 70% of the apoptotic response observed at 72 h after Sendai virus infection. The use of z-LEHD partially impaired Sendai virus-induced cell death, suggesting that caspase 9 may also be involved in the pathway leading to apoptosis. These caspase inhibitors produced similar apoptotic inhibitor effects in IRF-3(5D)-expressing cells (Fig. 8C). These data illustrate the high potency and selectivity of these apoptosis inhibitors and, more importantly, demonstrate that caspase 8 and to a lesser extent caspase 9 are involved in IRF-3(5D)- and Sendai virus-induced apoptosis.
To demonstrate that IRF-3(5D) is sufficient to induce the activation of caspases 8 and 9, lysates from DOX-induced rtTA-Jurkat IRF-3(5D)-expressing cells were used in combination with fluorogenic substrates for both caspases to quantify their respective activities. As shown in Fig. 8D, the activity of both caspases was significantly increased 48 to 72 h after DOX induction, a time interval that corresponds with the activation of CPP-32/caspase 3 (Fig. 7B). Similar results were also obtained when cleavage of proapoptotic caspase 8 was monitored by immunoblot analysis (data not shown). Caspase 9 was not as strongly activated as caspase 8; however, as shown in Fig. 8B and C, IRF-3(5D) induction of apoptosis may involve multiple pathways, concomitantly requiring both caspase 8 and 9.
DISCUSSION
In this study, the potential growth modulatory properties of the IRF-3 transcription factor were analyzed in cell lines constitutively or inducibly expressing IRF-3 transgenes. The constitutively active form of IRF-3 [IRF-3(5D)] was shown to induce apoptosis in human embryonic kidney 293 and Jurkat T cells, while wtIRF-3 had no effect on its own. Viral infection of cells overexpressing wtIRF-3 enhanced the number of cells undergoing apoptosis by two to threefold, depending on the cell type, while a dominant negative form of IRF-3 (IRF-3 ΔN) interfered with virus-induced apoptosis in 293 cells, thus demonstrating that IRF-3 activation may initiate apoptotic signalling. The ability of IRF-3 to signal apoptosis is likely limited to virus-infected cells, since virus infection is the only mechanism known to activate IRF-3 (21); furthermore, dominant negative IRF-3 did not inhibit apoptosis induced by osmotic stress (data not shown). Activation of caspase 8, and to a lesser extent caspase 9 and CPP-32/caspase 3, were involved in both Sendai virus- and IRF-3(5D)-induced apoptosis. However, it is not clear whether IRF-3 is able to independently regulate both the mitochondrial apoptotic pathway as well as the death-induced signaling complex (DISC) that comprises caspase 8. These effects may depend on the cell type. For example, in type II cells, including Jurkat cells, it has been reported that caspase 8 and caspase 3 are activated downstream of the mitochondria (40). In contrast, in type I cells, apoptosis is relatively mitochondrion independent, and caspase 8 is rapidly activated at the DISC prior to caspase 3 activation (40). However, inhibition of caspase 8, rather than mitochondrion-related caspase 9, in IRF-3(5D)-expressing cells profoundly repressed apoptosis. It is plausible that IRF-3 may induce an unknown gene(s) that contributes to apoptosis by regulating the DISC to convert Jurkat cells into type I cells. Therefore, inhibiting caspase 9 would not dramatically affect IRF-3-mediated apoptosis, as described in this study. Nevertheless, some caspase 9 activity was observed and may be explained by recent data demonstrating that activated caspase 8 can recruit mitochondrial input into the caspase-9-mediated activation of caspase 3 via cleavage of Bid-2 (28). It remains to be determined what proapoptotic genes may be induced by IRF-3; interestingly, recent studies using DNA array technology revealed that IFN induces a number of proteins known to be involved in the regulation of apoptosis that were not previously considered to harbor ISREs in their promoter regions (14).
In uninfected cells, IRF-3 resides in the cytoplasm in a closed conformation that maintains it in a latent inactive state (21, 27). Hence, overexpression of wtIRF-3 per se does not induce transcriptional up regulation of target genes. Virus-induced phosphorylation of IRF-3 leads to a conformational change that relieves autoinhibition and permits its dimerization and translocation to the nucleus and transcriptional activation of target genes. The constitutively active form of IRF-3—IRF-3(5D)—does not require this posttranslational modification; it is able to dimerize, interact with the CBP/p300 coactivator, and localize to the nucleus where it is a potent transactivator (21, 26, 27). These results are consistent with the observation that wtIRF-3 overexpression alone does not induce apoptosis without activation by virus infection, whereas IRF-3(5D) is sufficient to induce apoptosis.
Other members of the IRF family of transcription factors have been implicated in growth control and apoptosis. IRF-1 functions as a tumor suppressor, while overexpression of IRF-2 results in cell transformation of NIH 3T3 cells and tumor formation in nude mice (17, 34). IRF-1 mediates oncogene- and DNA damage-induced apoptosis by transcriptionally activating target genes such as caspase 1 and by cooperating with p53 to induce p21/WAF expression (36, 45, 46). Chromosomal deletion at the IRF-1 locus has been associated with leukemia and preleukemic myelodysplasia, further highlighting the important functions of IRF proteins in growth control (55). Although IRF-1 is activated upon viral infection, Tanaka et al. using IRF-1 knockout cells demonstrated that IRF-1 is not necessary for virus-induced apoptosis (47). This study further demonstrated that alpha/beta IFN was essential for virus-induced apoptosis and neutralization of IFN reduced viral induced death. Furthermore, IFNAR and Stat1−/− cells, were also deficient for virus-induced apoptosis (47). It is possible that IRF-3 may subvert the function of IRF-1 since both proteins bind to the same DNA sequence element (GAAANNGAAANN); alternatively, IRF-3 may in some circumstances induce IRF-1 expression through the ISRE in the IRF-1 promoter. In our experiments, both Sendai virus- and IRF-3(5D)-induced apoptosis were not diminished by the use of antibody capable of neutralizing alpha/beta IFNs. Furthermore, RNase protection analysis showed that in 293 and Jurkat cells, overexpression of IRF-3(5D) alone was not sufficient to induce mRNA production for either IFN-β or IFN-γ. These results imply that IFN secretion is not essential for Sendai virus or IRF-3 activation of apoptosis, but rather indicate that several mechanisms of virus-induced apoptosis may exist. This hypothesis is supported by the observation that Sendai virus can induce apoptosis through the activation of FLICE/caspase 8 and CPP-32/caspase 3 by a mechanism independent of tumor necrosis factor and Fas receptor ligand binding (8, 9).
Our study further clarifies the molecular pathway used by paramyxoviruses to induce apoptosis. Notably, the fact that IRF-3(5D) overexpression per se is sufficient for caspase 8 and caspase 9 activation and induction of apoptosis implies that phosphorylation of IRF-3 and subsequent nuclear translocation of IRF-3 is part of the general pathway involved in the regulation of cell death by paramyxoviruses. It is not possible to rule out the parallel activation of other apoptotic pathways since overexpression of a dominant negative mutant of IRF-3 (IRF-3 ΔN) decreased Sendai virus-induced apoptosis by 50% but did not completely block virus-induced apoptosis (Fig. 5). Likewise, it is not possible to rule out the synthesis of a low level of endogenous IFN that may escape detection but nonetheless contribute to IRF-3-mediated apoptosis (8). The recent observation that the dsRNA protein kinase PKR regulates apoptosis through the involvement of the death receptors represents another pathway used by viruses to induce apoptosis in target cells (7).
A role for IRF-3 in mediating virus-induced apoptosis is supported by studies with the human papilloma virus E6 protein. E6, which induces cellular transformation via p53-dependent and independent mechanisms, was found to interact with IRF-3 and block its transactivation potential (38). Since p53 gene deletion cannot account for the impaired differentiation seen in E6-expressing transgenic mice, E6 may induce oncogenesis by modulating IRF-3-regulated cell proliferation or apoptosis (38). Our group and others have also demonstrated that virus infection leads to NF-κB activation in target cells (2, 13, 16). Furthermore, it is well established that NF-κB provides a protective antiapoptotic function in most cell types (12) and therefore could act as a physiological antagonist of IRF-3 with regard to the induction of apoptosis. In support of this contention, the apoptotic effect of IRF-3 is partially masked by NF-κB activation, since expression of a super repressor form of IκB (2NΔ4) (23) increased Sendai virus-induced apoptosis by 70% in 293 cells (data not shown).
Activation of the IRF-3 transcription factor by viral infection may thus serve several functions. A role for IRF-3 in immediate-early activation of immunomodulatory RANTES and IFN gene expression has recently been demonstrated (21, 25). These proteins are necessary for the host to mount an effective immune response. Our results illustrate that IRF-3 plays an additional role in mediating Sendai virus-induced apoptosis. Effective viral containment requires that infected cells are quickly eliminated. IRF-3 is an attractive candidate for mediating virus-induced apoptosis since IRF-3 is only activated upon viral infection and its activation—as demonstrated by transfection studies with IRF-3(5D)—induces up to a 200-fold increase in the transcriptional activity of responsive genes (25, 26). Activation of IRF-3 would therefore allow an immediate-early response to virus infection that involves both the stimulation of the antiviral response and the elimination of virus-infected cells by apoptosis.
ACKNOWLEDGMENTS
We thank Pierre Rafick Sékaly, Illka Julkunen, and Paula Pitha-Rowe for providing reagents used in this study.
This research was supported by operating grants from the Medical Research Council of Canada (MRC) (J.H.) and the National Institutes of Health (G.B.). C.H. was supported by an MRC studentship, M.J.S. was supported by an MRC postdoctoral Fellowship, C.D. was supported by a McGill Major studentship, R.L. was supported by a Fraser Monat McPherson Fellowship from McGill University, and J.H. was supported by an MRC Senior Scientist award.
REFERENCES
- 1.Adams J M, Cory S. The Bcl-2 protein family: arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 2.Algarte M, Kwon H, Genin P, Hiscott J. Identification by in vivo genomic footprinting of a transcriptional switch containing NF-κB and Sp1 that regulates the IκBα promoter. Mol Cell Biol. 1999;19:6140–6153. doi: 10.1128/mcb.19.9.6140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ashkenazi A, Dixit V M. Death receptors: signaling and modulation. Science. 1998;281:1305–1308. doi: 10.1126/science.281.5381.1305. [DOI] [PubMed] [Google Scholar]
- 4.Au W-C, Moore P A, Lowther W, Juang Y-T, Pitha P M. Identification of a member of the interferon regulatory factor family that binds to the interferon-stimulated response element and activates expression of interferon-induced genes. Proc Natl Acad Sci USA. 1995;92:11657–11661. doi: 10.1073/pnas.92.25.11657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Au W-C, Moore P A, LaFleur D W, Tombal B, Pitha P M. Characterization of the interferon regulatory factor-7 and its potential role in the transcription activation of interferon A genes. J Biol Chem. 1998;273:29210–29217. doi: 10.1074/jbc.273.44.29210. [DOI] [PubMed] [Google Scholar]
- 6.Baichwal V R, Baeuerle P A. Apoptosis: activate NF-κB or die? Curr Biol. 1997;7:R94–R96. doi: 10.1016/s0960-9822(06)00046-7. [DOI] [PubMed] [Google Scholar]
- 7.Balachandran S, Kim C N, Yeh W C, Mak T W, Bhalla K, Barber G N. Activation of the dsRNA-dependent protein kinase, PKR, induces apoptosis through FADD-mediated death signaling. EMBO J. 1998;17:6888–6902. doi: 10.1093/emboj/17.23.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balachandran S, Roberts C, Kipperman T, Bhalla K, Compans R, Archer D, Barber G. Alpha/beta interferons potentiate virus-induced apoptosis through activation of the FADD/Caspase-8 death signaling pathway. J Virol. 2000;74:1513–1523. doi: 10.1128/jvi.74.3.1513-1523.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bitzer M, Prinz F, Bauer M, Spiegel M, Neubert W J, Gregor M, Schulze-Osthoff K, Lauer U. Sendai virus infection induces apoptosis through activation of caspase-8 (FLICE) and caspase-3 (CPP32) J Virol. 1999;73:702–708. doi: 10.1128/jvi.73.1.702-708.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bump N J, Hackett M, Hagunin M, Seshagiri S, Brady K, Chen P, Ferenz C, Franklin S, Ghayur T, Li P, Lican P, Mankovich J, Shi L, Greenberg A H, Miller L K, Wong W W. Inhibition of ICE family proteases by baculovirus antiapoptotic protein p35. Science. 1995;269:1885–1888. doi: 10.1126/science.7569933. [DOI] [PubMed] [Google Scholar]
- 11.Cryns V, Yuan J. Proteases to die for. Genes Dev. 1998;12:1551–1570. doi: 10.1101/gad.12.11.1551. [DOI] [PubMed] [Google Scholar]
- 12.DeLuca C, Kwon H, Lin R, Wainberg M, Hiscott J. NF-κB activation and HIV-1 induced apoptosis. Cytokine Growth Factor Rev. 1999;10:235–253. doi: 10.1016/s1359-6101(99)00015-5. [DOI] [PubMed] [Google Scholar]
- 13.DeLuca C, Petropoulos L, Zmeureanu D, Hiscott J. Nuclear IκBβ maintains persistent NF-κB activation in HIV-1-infected myeloid cells. J Biol Chem. 1999;274:13010–13016. doi: 10.1074/jbc.274.19.13010. [DOI] [PubMed] [Google Scholar]
- 14.Der S D, Zhou A, Williams B R G, Silverman R H. Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Faleiro L, Kobayashi R, Fearnhead H, Lazebnik Y. Multiple species of CPP32 and Mch2 are the major active caspases present in apoptotic cells. EMBO J. 1997;16:2271–2281. doi: 10.1093/emboj/16.9.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garoufalis E, Kwan I, Lin R, Mustafa A, Pepin N, Roulston A, Lacoste J, Hiscott J. Viral induction of the human interferon beta promoter: modulation of transcription by NF-κB/rel proteins and interferon regulatory factors. J Virol. 1994;68:4707–4715. doi: 10.1128/jvi.68.8.4707-4715.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harada H, Kitagawa M, Tanaka N, Yamamoto H, Harada K, Ishihara M, Taniguchi T. Anti-oncogenic and oncogenic potentials of interferon regulatory factors-1 and -2. Science. 1993;259:971–974. doi: 10.1126/science.8438157. [DOI] [PubMed] [Google Scholar]
- 18.Henderson S, Huen D, Rowe M, Dawson C, Johnson G, Rickinson A. Epstein-Barr virus-coded BHRF1 protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc Natl Acad Sci USA. 1993;15:8479–8483. doi: 10.1073/pnas.90.18.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson S, Rowe M, Gregory C, Croom-Carter D, Wang F, Longnecker R, Kieff E, Rickinson A. Induction of bcl-2 expression by Epstein-Barr virus latent membrane protein-1 protects infected B-cells from programmed cell death. Cell. 1991;65:1107–1115. doi: 10.1016/0092-8674(91)90007-l. [DOI] [PubMed] [Google Scholar]
- 20.Hiscott J, Nguyen H, Lin R. Molecular mechanisms of interferon beta gene induction. Semin Virol. 1995;6:161–173. [Google Scholar]
- 21.Hiscott J, Pitha P, Génin P, Nguyen H, Heylbroeck C, Mamane Y, Algarté M, Lin R. Triggering the interferon response: the role of IRF-3 transcription factor. J Interferon Cytokine Res. 1999;19:1–13. doi: 10.1089/107999099314360. [DOI] [PubMed] [Google Scholar]
- 22.Jacobson M D, Weil M, Raff M C. Programmed cell death in animal development. Cell. 1997;88:347–354. doi: 10.1016/s0092-8674(00)81873-5. [DOI] [PubMed] [Google Scholar]
- 23.Kwon H, Pelletier N, DeLuca C, Genin P, Cisternas S, Lin R, Wainberg M A, Hiscott J. Inducible expression of IκBα repressor mutants interferes with NF-κB activity and HIV-1 replication in Jurkat T cells. J Biol Chem. 1998;273:7431–7440. doi: 10.1074/jbc.273.13.7431. [DOI] [PubMed] [Google Scholar]
- 24.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 25.Lin R, Heylbroeck C, Genin P, Pitha P, Hiscott J. Essential role of IRF-3 in direct activation of RANTES gene transcription. Mol Cell Biol. 1999;19:959–966. doi: 10.1128/mcb.19.2.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential and proteasome mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin R, Mamane Y, Hiscott J. Structural and functional analysis of interferon regulatory factor 3: localization of the transactivation and autoinhibitory domains. Mol Cell Biol. 1999;19:2465–2474. doi: 10.1128/mcb.19.4.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 29.Marie I, Durbin J E, Levy D E. Differential viral induction of distinct interferon-α genes by positive feedback through interferon regulatory factor-7. EMBO J. 1998;17:6660–6669. doi: 10.1093/emboj/17.22.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miura M, Zhu H, Rotello R, Hartwieg E A, Yuan J. Induction of apoptosis in fibroblasts by IL-1 beta-converting enzyme, a mammalian homolog of the C. elegans cell death gene ced-3. Cell. 1993;19:653–660. doi: 10.1016/0092-8674(93)90486-a. [DOI] [PubMed] [Google Scholar]
- 31.Muzio M, Salvesen G S, Dixit V M. FLICE induced apoptosis in a cell-free system. Cleavage of caspase zymogens. J Biol Chem. 1997;272:2952–2956. doi: 10.1074/jbc.272.5.2952. [DOI] [PubMed] [Google Scholar]
- 32.Nguyen H, Hiscott J, Pitha P M. The growing family of IRF transcription factors. Cytokine Growth Factor Rev. 1997;8:293–312. doi: 10.1016/s1359-6101(97)00019-1. [DOI] [PubMed] [Google Scholar]
- 33.Nguyen H, Lin R, Hiscott J. Activation of multiple growth regulatory genes following inducible expression of IRF-1 or IRF/RelA fusion proteins. Oncogene. 1997;15:1425–1435. doi: 10.1038/sj.onc.1201318. [DOI] [PubMed] [Google Scholar]
- 34.Nguyen H, Mustafa A, Hiscott J, Lin R. Transcription factor IRF-2 exerts its oncogenic phenotype through the DNA binding/transcription repression domain. Oncogene. 1995;11:537–544. [PubMed] [Google Scholar]
- 35.Nicholson D W, Ali A, Thornberry N A, Vaillancourt J P, Ding C K, Gallant M, Gareau Y, Griffin P R, Labelle M, Lazebnik Y A, Munday N A, Raju S M, Smulson M E, Yamin T-T, Yu V L, Miller D K. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature. 1995;376:37–43. doi: 10.1038/376037a0. [DOI] [PubMed] [Google Scholar]
- 36.Ozawa H, Matsuyama T, Mak T W, Aizawa S, Tokino T, Oren M, Taniguchi T. Cooperation of the tumour suppressors IRF-1 and p53 in response to DNA damage. Nature. 1996;382:816–818. doi: 10.1038/382816a0. [DOI] [PubMed] [Google Scholar]
- 37.Parekh B S, Maniatis T. Virus infection leads to localized hyperacetylation of histones H3 and H4 at the IFN-β promoter. Mol Cell. 1999;3:125–129. doi: 10.1016/s1097-2765(00)80181-1. [DOI] [PubMed] [Google Scholar]
- 38.Ronco L, Karpova A, Vidal M, Howley P. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sato M, Hata N, Asagiri M, Nakaya T, Taniguchi T, Tanaka N. Positive feedback regulation of type I IFN genes by the IFN-inducible transcription factor IRF-7. FEBS Lett. 1998;441:106–110. doi: 10.1016/s0014-5793(98)01514-2. [DOI] [PubMed] [Google Scholar]
- 40.Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli K, Debatin K, Krammer P, Peter M. Two CD95 (APO-1/Fas) signaling pathways. EMBO J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schulze-Osthoff K, Ferrari D, Los M, Wesselborg S, Peter M E. Apoptosis signaling by death receptors. Eur J Biochem. 1998;254:439–459. doi: 10.1046/j.1432-1327.1998.2540439.x. [DOI] [PubMed] [Google Scholar]
- 42.Shen Y, Shenk T E. Viruses and apoptosis. Curr Opin Gen Dev. 1995;5:105–111. doi: 10.1016/s0959-437x(95)90061-6. [DOI] [PubMed] [Google Scholar]
- 43.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Litwack G, Alnemri E S. Molecular ordering of the Fas-apoptotic pathway: the Fas/Apo-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple ced-3/ICE-like cysteine proteases. Proc Natl Acad Sci USA. 1996;93:14486–14491. doi: 10.1073/pnas.93.25.14486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sylla B S, Hung S C, Davidson D M, Hatzivassiliou E, Malinin N L, Wallach D, Gilmore T, Kieff E, Mosialos G. Epstein-Barr virus-transforming protein latent infection membrane protein 1 activates transcription factor NF-κB through a pathway that includes the NF-κB-inducing kinase and the IκB kinases IKKα and IKKβ. Proc Natl Acad Sci USA. 1998;95:10106–10111. doi: 10.1073/pnas.95.17.10106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura T, Ishihara M, Lamphier M S, Tanaka N, Oishi I, Aizawa S, Matsuyama T, Mak T W, Taki S, Taniguchi T. An IRF-1-dependent pathway of DNA damage-induced apoptosis in mitogen-activated T-lymphocytes. Nature. 1995;376:596–599. doi: 10.1038/376596a0. [DOI] [PubMed] [Google Scholar]
- 46.Tanaka N, Ishihara M, Kitagawa M, Harada H, Kimura T, Matsuyama T, Lamphier M S, Aizawa S, Mak T W, Taniguchi T. Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell. 1994;77:829–839. doi: 10.1016/0092-8674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka N, Sato M, Lamphier M S, Nozawa H, Oda E, Noguchi S, Schreiber R D, Tsujimoto Y, Taniguchi T. Type I interferons are essential mediators of apoptotic death in virally infected cells. Genes Cells. 1998;3:29–37. doi: 10.1046/j.1365-2443.1998.00164.x. [DOI] [PubMed] [Google Scholar]
- 48.Teodoro J G, Branton P E. Regulation of apoptosis by viral gene products. J Virol. 1997;71:1739–1746. doi: 10.1128/jvi.71.3.1739-1746.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thome M, Schneider P, Hofmann K, Kickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer J-L, Schröter M, Scaffidi C, Krammer P H, Peter M E, Tschopp J. Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature. 1997;386:517–521. doi: 10.1038/386517a0. [DOI] [PubMed] [Google Scholar]
- 50.Thornberry N A, Lazebnik Y. Caspases: enemies within. Science. 1998;281:1312–1316. doi: 10.1126/science.281.5381.1312. [DOI] [PubMed] [Google Scholar]
- 51.Vaux D L, Strasser A. The molecular biology of apoptosis. Proc Natl Acad Sci USA. 1996;93:2239–2244. doi: 10.1073/pnas.93.6.2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vilcek J, Sen G. Interferons and other cytokines. In: Fields B, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 375–399. [Google Scholar]
- 53.Wathelet M G, Lin C H, Parakh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 54.Weaver B K, Kumar K P, Reich N C. Interferon regulatory factor 3 and CREB-binding protein/p300 are subunits of double-stranded RNA-activated transcription factor DRAF1. Mol Cell Biol. 1998;18:1359–1368. doi: 10.1128/mcb.18.3.1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Willman C L, Sever C E, Pallavicini M G, Harada H, Tanaka N, Slovak M L, Yamamoto H, Harada K, Meeker T C, List A F, Taniguchi T. Deletion of IRF-1, mapping to chromosome 5q31.1, in human leukemia and preleukemic myelodysplasias. Science. 1993;259:968–971. doi: 10.1126/science.8438156. [DOI] [PubMed] [Google Scholar]
- 56.Yoneyama M, Suhara W, Fukuhara Y, Fukada M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]