Abstract
Clostridioides difficile infection (CDI) is a major cause of healthcare-associated gastrointestinal infections1,2. The exaggerated colonic inflammation caused by C. difficile toxins such as toxin B (TcdB) damages tissues and promotes C. difficile colonization3–6, but how TcdB causes inflammation is unclear. Here we report that TcdB induces neurogenic inflammation by targeting gut-innervating afferent neurons and pericytes, mediated by receptors including Frizzled 1/2/7 (FZD1/2/7) in neurons and chondroitin sulfate proteoglycan 4 (CSPG4) in pericytes. TcdB action stimulates secretion of the neuropeptides substance P (SP) and calcitonin gene-related peptide (CGRP) from neurons and pro-inflammatory cytokines from pericytes. Targeted delivery of the TcdB enzymatic domain, through fusion with a detoxified diphtheria toxin (DT), into peptidergic sensory neurons that express exogeneous DT receptor (an approach we termed toxogenetics) is sufficient to induce neurogenic inflammation and recapitulates major colonic histopathology associated with CDI. Conversely, mice lacking SP, CGRP, or the SP receptor (NK1R) show reduced pathology in both models of cecal TcdB injection and CDI. Blocking SP or CGRP signaling reduces tissue damage and C. difficile burden in mice infected with a standard C. difficile strain or hypervirulent strains expressing the TcdB2 variant. Thus, targeting neurogenic inflammation provides a host-oriented therapeutic approach for treating CDI.
Introduction
CDI is a leading cause of gastrointestinal infections in developed countries, with an estimated half million cases and ~29,000 deaths annually in the United States1,2. C. difficile is a spore-forming opportunistic pathogen that can cause colonic infection when the normal gut microbiota is disrupted1. CDI-associated diseases present with diarrhea and abdominal pain, and can result in pseudomembranous colitis and toxic megacolon in severe cases3. Treatment remains challenging due to frequent recurrent infections, antibiotic resistance, and emerging hypervirulent strains1,2. At tissue levels, CDI is associated with exaggerated inflammatory responses featuring oedema and neutrophil infiltration, leading to colonic epithelial damage and formation of surface plaques filled with neutrophils3. Exaggerated immune responses are associated with worse clinical outcomes4,7–10.
C. difficile causes disease largely through the action of two homologous toxins, toxin A (TcdA) and TcdB. These toxins are large multi-domain proteins that target and enter host cells via receptor-mediated endocytosis, cross the endosomal membrane, and deliver an enzymatic domain into the cytosol of cells11,12. The enzymatic domains of TcdA and TcdB are glucosyltransferases (termed GTD) that modify a key residue on small GTPases and inhibit their activity, leading to disruption of the actin cytoskeleton that eventually results in cell rounding.
Recent studies in mouse models demonstrated that CDI-associated colonic inflammation is toxin-dependent and that toxin-mediated inflammation benefits C. difficile by liberating favorable sources of nutrients and modifying microbiome community structure5,6. However, pathogenic mechanisms that lead to severe colonic inflammation remain unclear.
Neurogenic inflammation is a well-established phenomenon in which sensory neurons release pro-inflammatory neuropeptides such as SP and CGRP, causing rapid vasodilation and increased vascular permeability that results in extravasation of plasma and immune cells into the tissue13,14. SP is well known to act on the endothelium to induce plasma extravasation, and the role of CGRP in vasodilation is also well established. Symptoms consistent with neurogenic inflammation in CDI have been observed, including neutrophilic infiltration, extreme oedema, and increased colonic vascular permeability3,15. However, the role of neurogenic inflammation in CDI pathogenesis has yet to be established.
Previous studies showed that TcdA induces inflammatory responses when injected into ligated small intestinal loops in animal models16–21. These responses could be attenuated by blocking SP or CGRP signaling18,20,21, or by surgical denervation of sensory neurons projecting to the loops22, suggesting a role for neurogenic inflammation induced by TcdA. The relevance of these findings to CDI in vivo, which primarily affects the colon rather than the small intestine, and the underlying molecular mechanisms have not been established.
Recent studies have identified TcdB as the more important toxin associated with pathogenesis and TcdB alone is sufficient to cause the full spectrum of disease23–25. How TcdB induces colonic inflammation and whether SP and CGRP signaling are involved remain unknown. Here, we report that TcdB directly targets gut-innervating peptidergic neurons and perivascular pericytes through distinct receptors, inducing release of neuropeptides from neurons to cause neurogenic inflammation. Genetic or pharmacologic blockade of SP or CGRP signaling reduced pathogenesis and C. difficile colonization in vivo in mouse models of TcdB administration and CDI.
Results
TcdB induces neurogenic inflammation
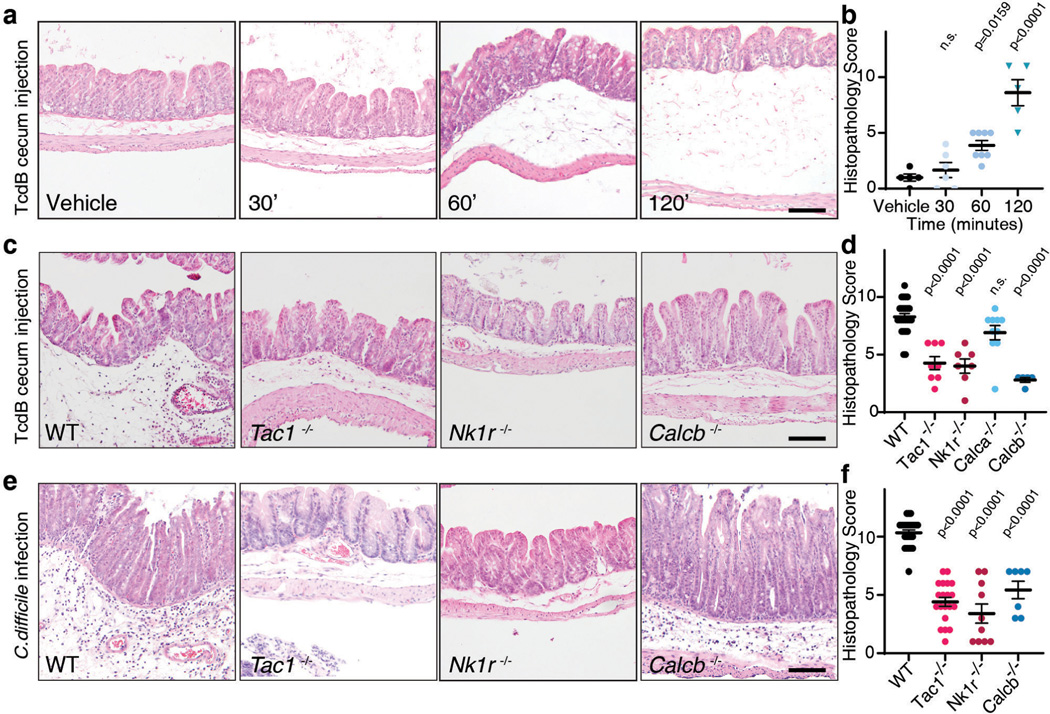
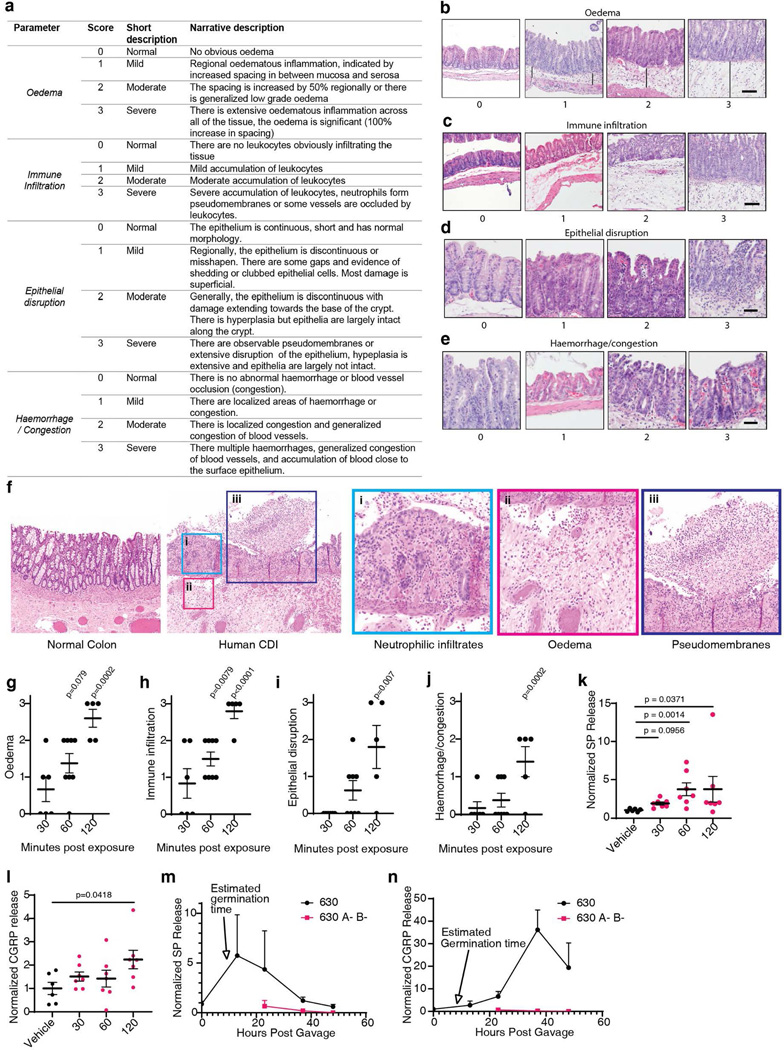
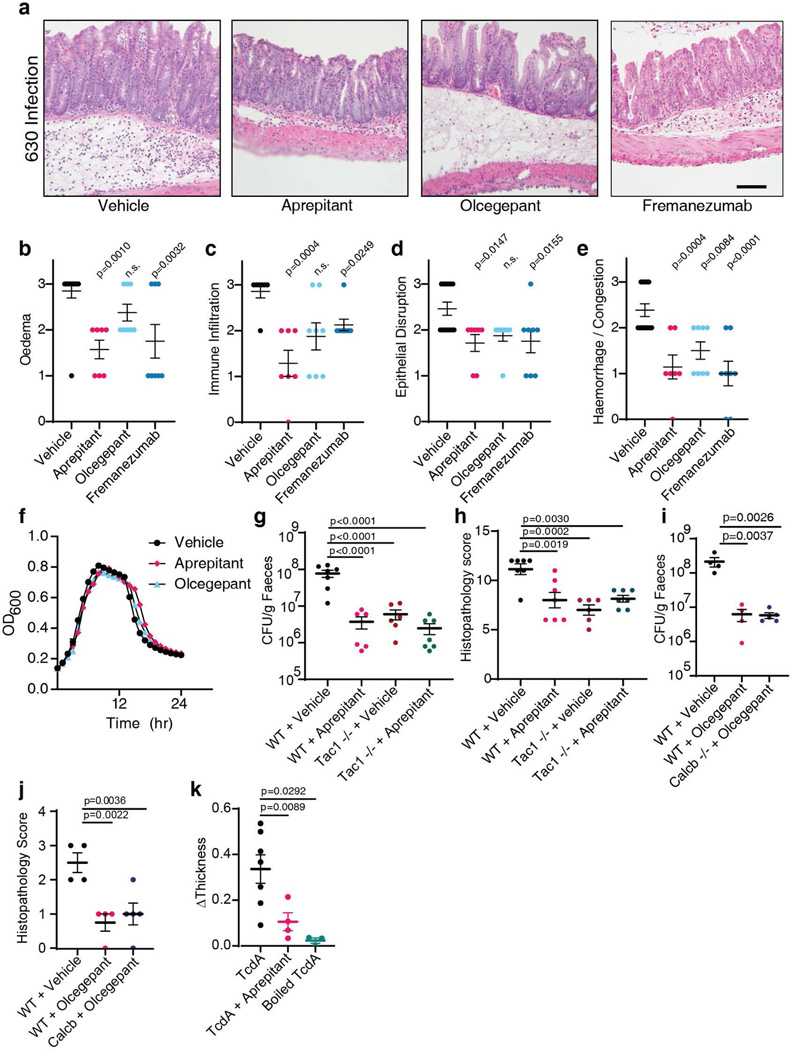
To assess if TcdB induces rapid tissue changes consistent with neurogenic inflammation, we injected TcdB directly into the cecal lumens of wildtype (WT) mice and then isolated tissue for histologic analysis (Fig. 1a). Histopathology was scored based on four criteria: oedema, immune cell (neutrophil) infiltration, epithelium disruption, and tissue haemorrhage (Extended Data Fig. 1a-e), representative of pathologic changes observed in human CDI (Extended Data Fig. 1f)3. TcdB induced oedema and neutrophil infiltration within 1 hour (h) (Fig. 1a-b, Extended Data Fig. 1g-j), reminiscent of classic neurogenic flare responses in the skin13,14.
Fig. 1: SP- and CGRP-mediated neurogenic inflammation in a TcdB cecum injection model and in a CDI mouse model.
a-b, Time course of histopathological changes after injection of TcdB into the cecums of mice. Representative micrographs of Haemotoxylin and Eosin (H&E) stained tissues are shown in a and histopathology scores are shown in b. Scoring criteria and the sub-categories of histopathology scores are shown in Extended Data Fig. 1; n=5, 6, 9, 5 mice.
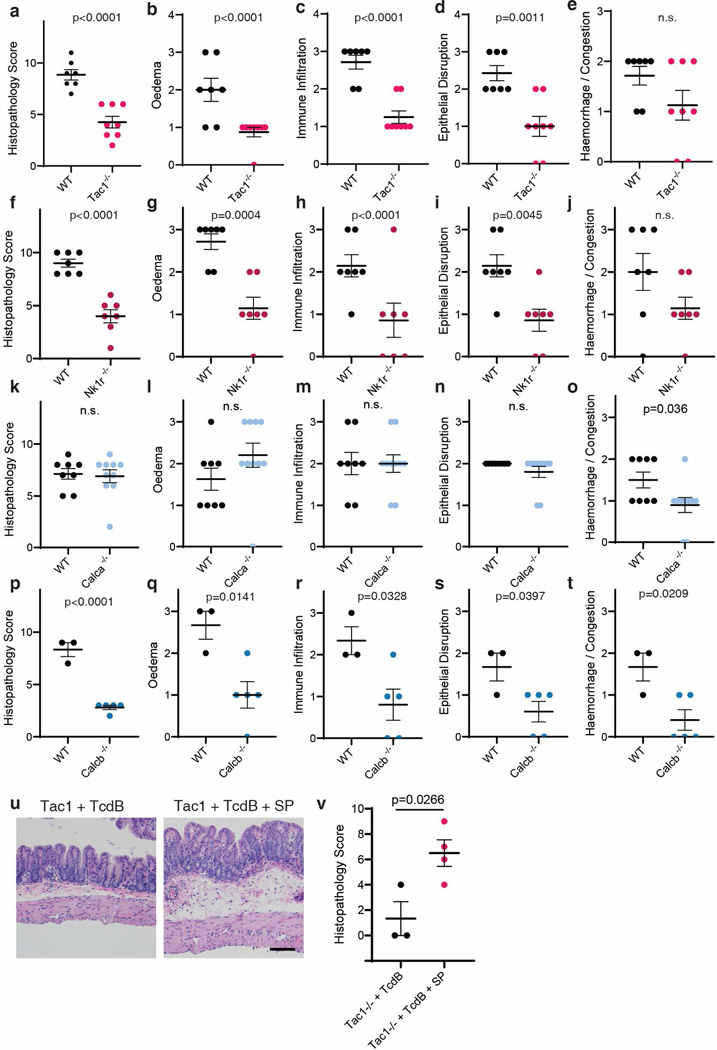
c-d, TcdB was injected into the cecums of wild type (WT) mice or the following KO mice: Tac1 (encoding the precursor for SP), Nk1r (encoding a receptor for SP), Calca (encoding CGRPα), or Calcb (encoding CGRPβ), with representative H&E staining shown in c and histopathology scores shown in d; n=25, 8, 7, 10, 5 mice.
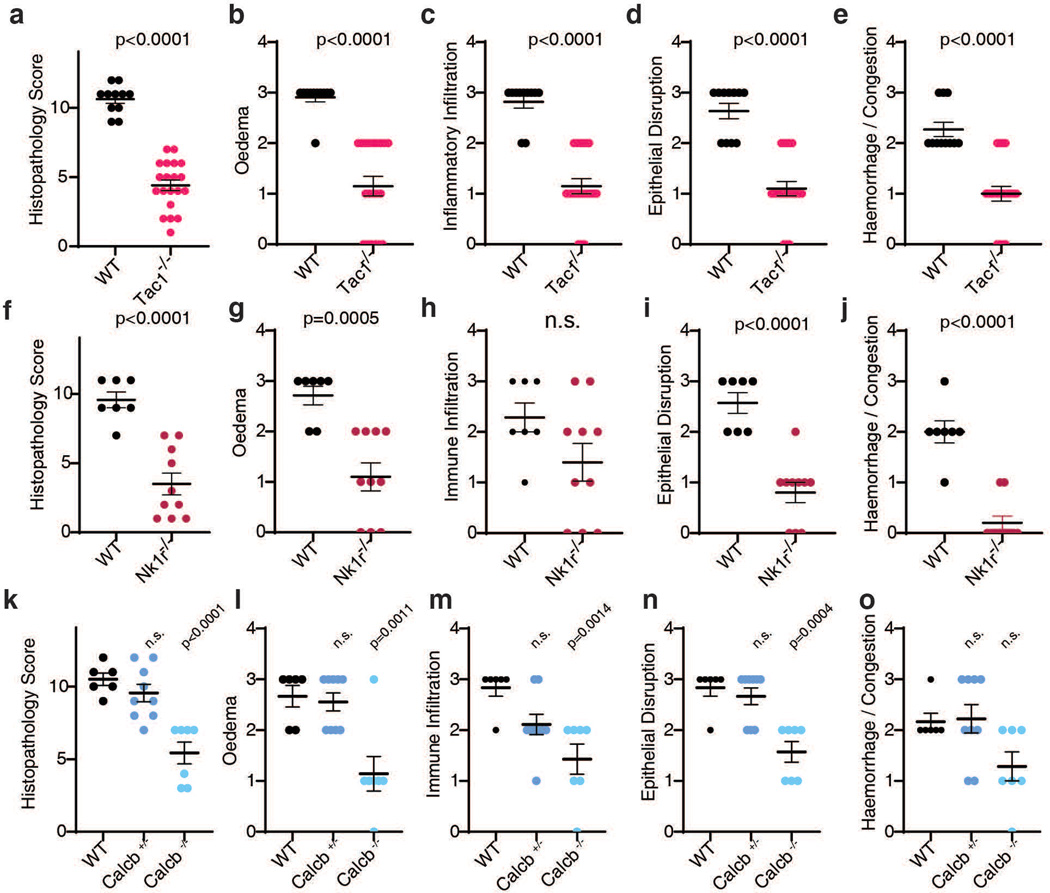
e-f, CDI experiments were carried out on WT, Tac1 KO, Nk1r KO, and Calcb KO mice. Colon tissues were harvested and analyzed 48 h later, with representative H&E staining shown in e and histopathology scores shown in f, n=24, 20, 10, 7 mice.
Statistical analyses in b (p<0.001 overall), d (p<0.001 overall), and f were one-way ANOVAs with post-hoc Dunnett’s test compared with vehicle-treated mice. P values: n.s., not significant, or exact values shown if <0.05. Centre line indicates mean and error bars reflect the standard error of the mean (SEM). Scale bar: 50 μm. “n” is described from left to right for each graph.
SP and CGRP are key drivers of vasodilation and neutrophil infiltration in the skin and other tissues13,14. The levels of both are increased in cecal tissues after exposure to TcdB, with the rapid timescale correlating with histopathological changes (Extended Data Fig. 1k-l). We next examined knockout (KO) mice lacking SP or CGRP signaling. Tac1 (encodes the prepropeptide for SP and neurokinin A) and Nk1r (encodes neurokinin-1 receptor, the high-affinity receptor for SP) KO mice had markedly lower histopathology scores after TcdB injection than WT controls (Fig. 1c-d, Extended Data Fig. 2a-j). CGRP has two closely related forms encoded by distinct genes: Calca (CGRPα) and Calcb (CGRPβ)26. While Calca-null mice had similar histopathology as WT controls, Calcb-null mice showed reduced histopathological scores after TcdB administration (Fig. 1c-d, Extended Data Fig. 2k-t). On the other hand, administering SP together with TcdB into the cecal lumen restored the susceptibility of Tac1 mice to TcdB (Extended Data Fig. 2u-v). These data suggest that SP and CGRPβ are critical to the rapid inflammatory response induced by TcdB.
To validate the role of SP and CGRPβ in CDI, we then utilized an antibiotic treatment mouse model of CDI. Mice pre-treated with antibiotics were infected with the spores of a widely used C. difficile strain (630 Δerm) that produces TcdA and TcdB. Severe pathology including diarrhea, oedema, neutrophil infiltration, and epithelial damage developed in WT mice within 48 h (Fig. 1e-f)1,2. Consistently, SP and CGRP levels in colonic tissues rose during the timeframe of infection, but not in mice infected with a toxin-deficient strain (Extended Data Fig. 1m-n). In contrast to WT mice, Tac1, Nk1r, and Calcb KO mice infected with C. difficile 630 Δerm showed lower histopathological scores (Fig. 1e-f, Extended Data Fig. 3).
TcdB targets sensory neurons via FZD1/2/7
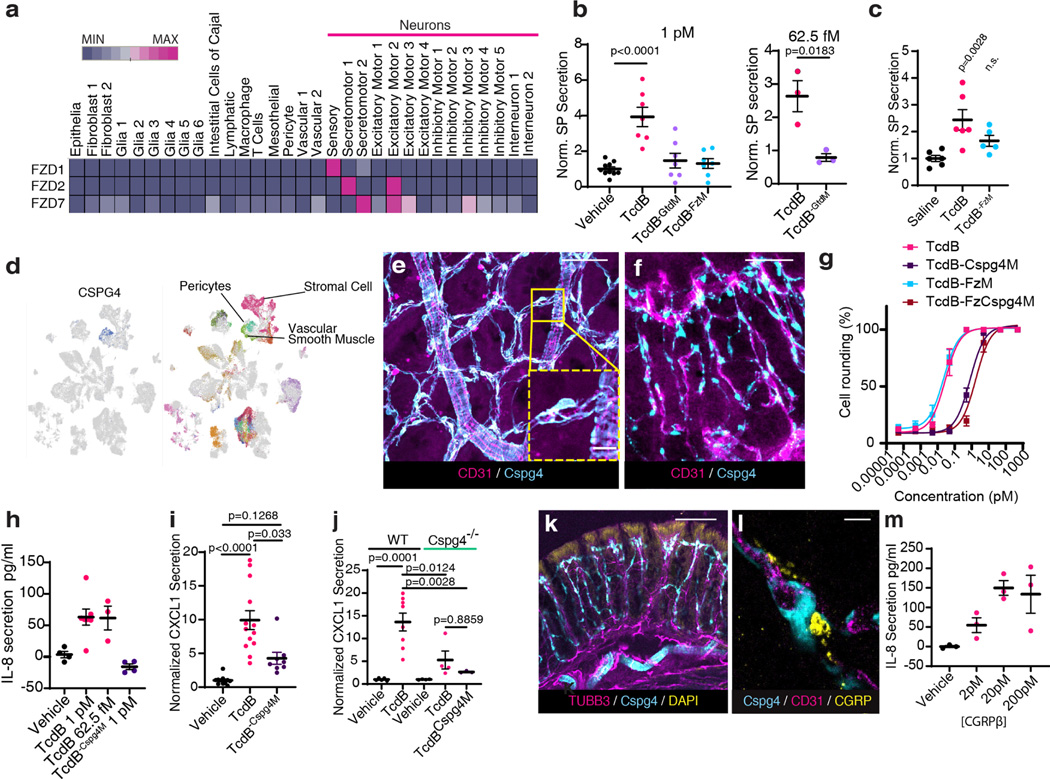
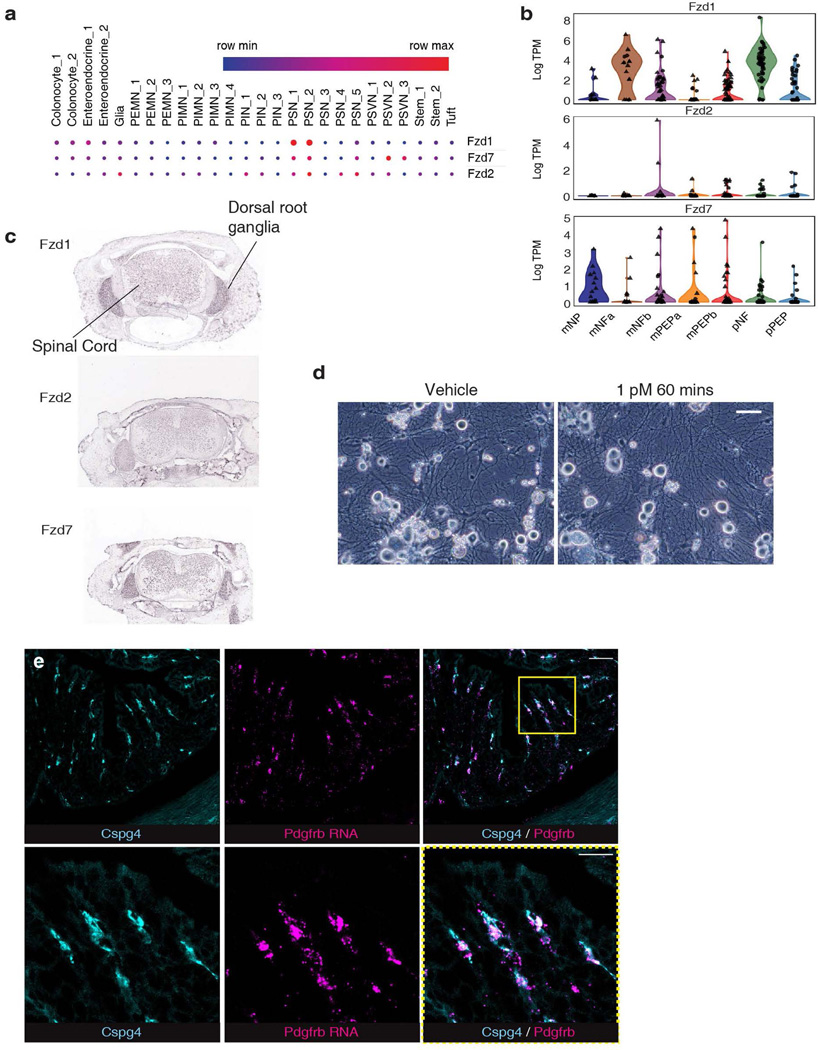
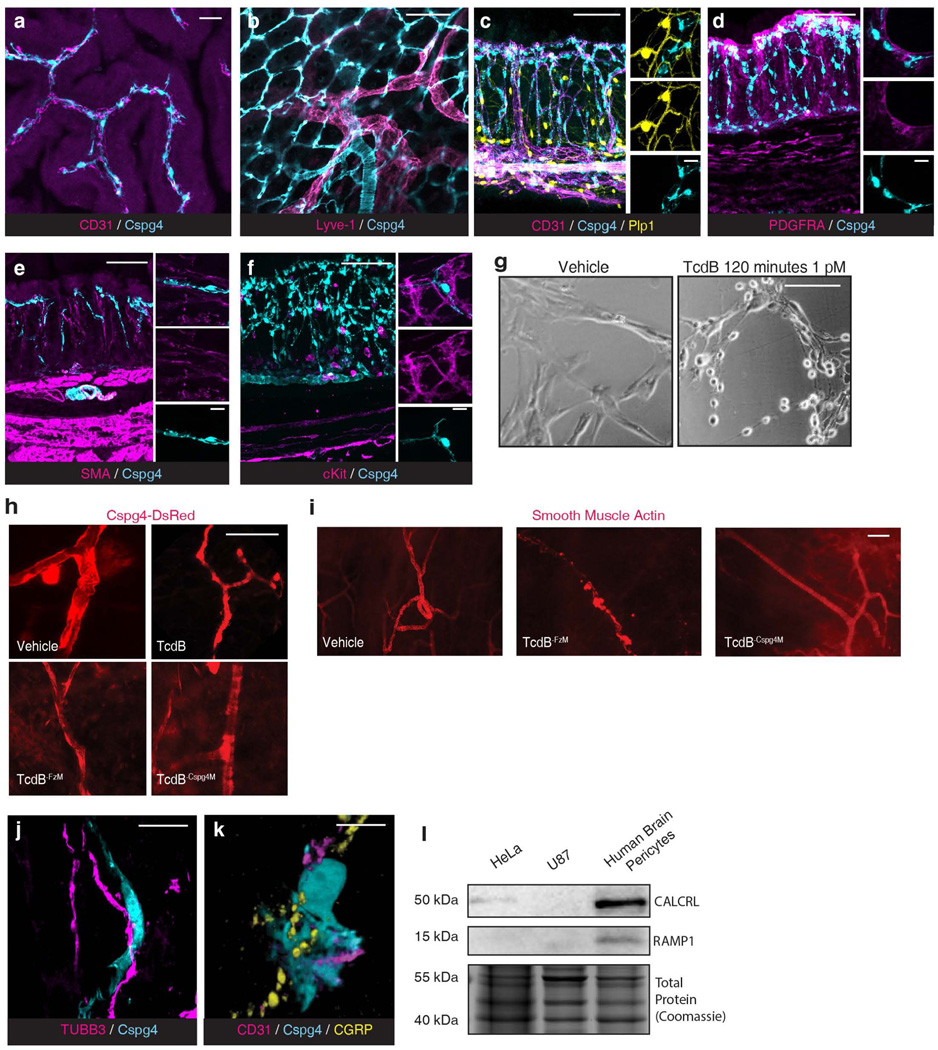
The cecum and colon are innervated by intrinsic neurons of the enteric nervous system (ENS) and projections from extrinsic neurons located in dorsal root ganglia (DRG). FZD1/2/7 and CSPG4 are well-established receptors for TcdB27–30. Analysis of published single cell RNA sequencing (scRNA-seq) data indicated that FZD1/2/7 transcripts are expressed in ENS neurons in mouse and human colon, at levels higher than in many other intestinal cell types including epithelial cells (Fig. 2a, Extended Data Fig. 4a). FZD1/2/7 have been previously reported in DRG neurons and play a role in sensory neuron sensitization31. Single cell RNA sequencing of colon-innervating DRG neurons in mice also showed expression of FZD1/2/7 (Extended Data Fig. 4b). Moreover, in situ hybridization confirmed that FZD1/2/7 are expressed in the DRG (Allen Brain Atlas; Extended Data Fig. 4c).
Fig. 2: TcdB targets neurons via FZD1/2/7 and pericytes via CSPG4.
a, Single cell expression analysis of FZD1, 2 and 7 in human colon tissues71 (red high, blue low).
b, Cultured mouse DRG neurons were exposed to TcdB or TcdB mutants for 1 h. n= 10, 7, 7, 7, 3, 3 culture wells across 3 cultures (each from 5 pooled animals). SP secretion was assessed using ELISA. FzM: TcdB mutant deficient in binding to FZD1/2/7; GtdM: TcdB mutant with mutations in the GTD domain.
c, Mouse cecal explants were exposed to TcdB (6 μg, 6 h), TcdBFzM, or saline, and secretion of SP was measured. n=6,6,5 mice.
d, Single cell analysis of CSPG4 expression in human colonic stroma34.
e-f, Immunostaining of cecum tissues from Cspg4-DsRed mice shows Cspg4+ cells (DsRed, cyan) surrounded by CD31+ vessels (magenta). e: Whole mount muscularis view, f: cross section. Scale bar: 50μm.
g, Cultured human brain pericytes were exposed to TcdB or TcdB mutants for 5 h and cell rounding (%) was assessed. Cspg4M: TcdB mutant deficient in binding to CSPG4; FzCspg4M: a TcdB double mutant deficient in binding to FZD1/2/7 and CSPG4. n= 3 independent cultures.
h, TcdB, but not TcdB-Cspg4M, induced secretion of IL-8 from cultured human pericytes. n=4,7,3,4 from 2 independent cultures.
i-j, TcdB, but not TcdB-Cspg4M, increased CXCL1 in colonic explants after cecal TcdB injections in WT, but not Cspg4 KO mice. In i n= 11, 13, 8 mice, and in j n= 5, 8, 4, 4, 3 mice.
k, Immunostaining of colon tissues from Cspg4-dsRed mice shows extensive overlap and contacts of neurons (Tubb3, magenta) with Cspg4+ cells (cyan). DAPI: yellow. Scale bar: 100 μm.
i, Immunostaining of colon tissues from Cspg4-dsRed mice shows appositions of CGRP (yellow) to Cspg4+ cells (cyan). Blood vessels were labeled with CD31 (magenta). Scale bar: 10 μm.
m, CGRP induced IL-8 release from cultured human pericytes. n=4 culture wells (representative of 3 replicates).
Statistical analyses in b, c: one way ANOVA (<0.0001 overall) with post-hoc Dunnett’s test compared with vehicle-treated mice, n.s., not significant, p values <0.05 shown. Centre line: mean; error bars: SEM. n is described from left to right.
Consistent with expression of FZD1/2/7, we found that cultured mouse DRG neurons are highly sensitive to TcdB and exposure to TcdB led to SP secretion before visible damage to neurites (Fig. 2b, Extended Data Fig. 4d). A mutant TcdB with mutations that disrupt glucosylation of small GTPases (TcdB-GtdM)32 did not induce SP release (Fig. 2b). We previously generated and validated a TcdB mutant deficient in binding to FZD1/2/7 (TcdB-FzM)28. This mutant toxin did not induce SP release from DRG neurons (Fig. 2b). The effect of TcdB was further confirmed in vivo: TcdB treatment increased SP release within cecal tissue explants following a mouse cecum injection assay while TcdB-FzM showed reduced potency (Fig. 2c). These findings demonstrate that TcdB targets peripheral sensory neurons via FZD1/2/7 and that intoxication induces neuropeptide secretion.
TcdB targets pericytes via CSPG4
Unlike FZD1/2/7, CSPG4 is not expressed by neurons and was previously considered to be expressed by colonic myofibroblasts33. Recent scRNA-seq of human colon tissues, however, suggests that CSPG4 expression is highly enriched in vascular mural cells including pericytes and vascular smooth muscle cells (Fig. 2d)34. Pericytes are contractile cells that wrap around microvascular endothelial cells and are well known for forming and regulating the blood-brain barrier35. To identify Cspg4+ cells in the colon, we utilized Cspg4-DsRed transgenic mice in which the Cspg4 promoter drives fluorescent DsRed reporter expression. Confocal imaging revealed two populations of DsRed+ cells, those that encircled large branching vessels and had morphologies typical of vascular smooth muscle cells (Fig. 2e), and a population with fine processes that wrapped around the subepithelial blood vessels in the colonic mucosa (Fig. 2e-f, Supplementary Video 1). These mucosal Cspg4+ cells always closely apposed vascular endothelial cells marked by CD31, but not lymphatic endothelial cells, and formed a perivascular network consistent with being pericytes (Extended Data Fig. 5a-b. Supplementary Video 2). Mucosal Cspg4+ cells robustly expressed transcripts for pericyte marker platelet derived growth factor receptor beta (PDGFRβ) (Extended Data Fig. 4e)36. They did not colocalize with markers for other intestinal cell types including enteric glia, interstitial cells of Cajal, mast cells, myofibroblasts / colonic smooth muscle cells, and stromal fibroblasts (Extended Data Fig. 5c-f).
To test the necessity of CSPG4 for TcdB to target pericytes, we utilized isolated primary human brain pericytes as we were not able to culture gut pericytes. TcdB was highly potent at inducing cell-rounding of pericytes (Fig. 2g, Extended Data Fig. 5g). A TcdB mutant toxin deficient in binding to CSPG4 (TcdB-Cspg4M) showed markedly reduced potency, unlike TcdB-FzM (Fig. 2g). The double mutant (TcdB-FzCspg4M) showed similar potency as TcdB-Cspg4M (Fig. 2g). To validate whether TcdB targets pericytes via CSPG4 in vivo, we utilized an ear injection model that allows imaging of pericytes surrounding ear arterioles37, which is difficult to do in gut tissues. Injection of TcdB or TcdB-FzM induced shrinkage of pericytes surrounding ear arterioles, whereas injection of TcdB-Cspg4M showed no effect (Extended Data Fig. 5h-i).
Pericytes are known to regulate vascular dilation and immune cell transmigration through changes in cell morphology and secretion of inflammatory cytokines37–39. TcdB, but not TcdB-Cspg4M, induced interleukin 8 (IL-8) secretion from human brain pericytes (Fig. 2h). Consistently, exposure to TcdB in the cecal injection model increased the level of the cytokine CXCL1 (murine IL-8 homolog) in cecum tissues in vivo (Fig. 2i). CXCL1 levels were not increased by TcdB-Cspg4M in WT mice or TcdB in Cspg4 KO mice (Fig. 2j).
Given that both neurons and pericytes are major regulators of vascular function, we assessed their ability to integrate signals. Immunohistochemical staining for the pan-neuronal marker TUBB3 showed an intermingled network of nerves and Cspg4+ pericytes in the mucosa (Fig. 2k). Many of these nerve fibers were immunoreactive for CGRP, and CGRP+ varicosities closely associated with DsRed-labeled pericytes that wrapped around endothelial cells (Fig. 2l, Extended Data Fig. 5j-k, Supplementary Video 3). Cultured human pericytes express the CGRP receptor components CALCRL and RAMP1 (Extended Data Fig. 5l) and adding CGRP to the culture medium induced secretion of IL-8 (Fig. 2m), confirming that pericytes can be regulated by CGRP.
TcdB action in a footpad model
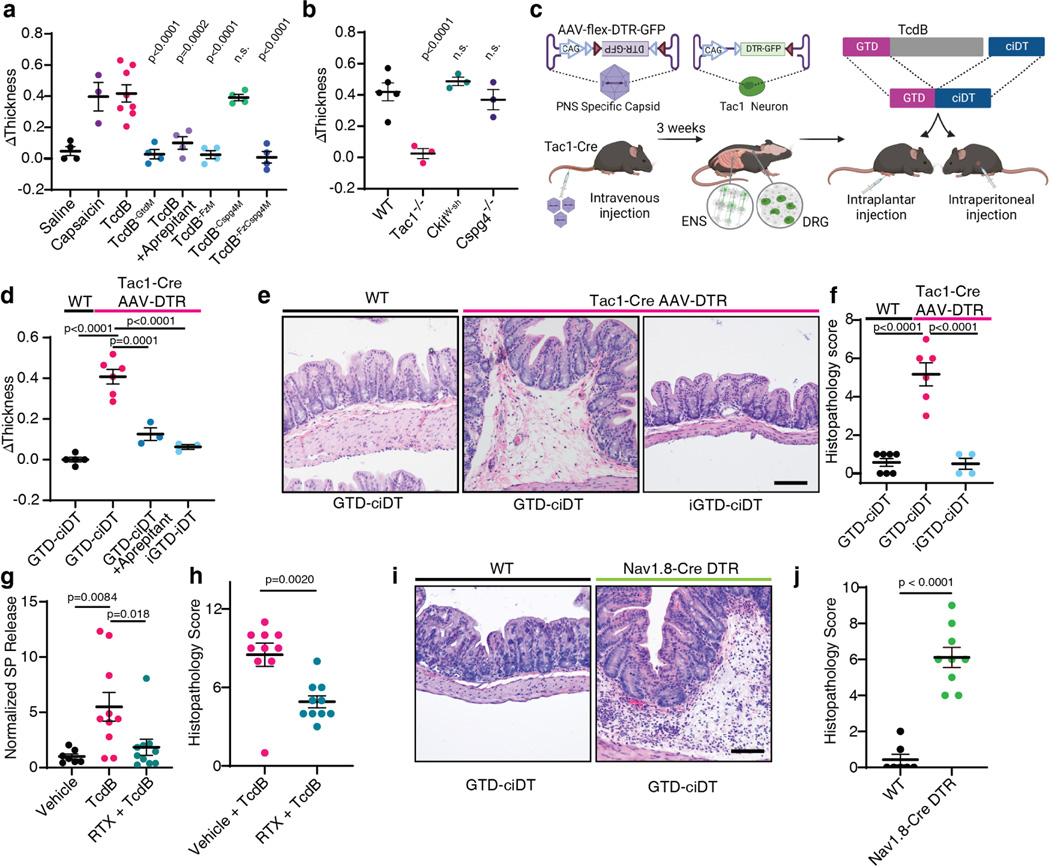
Neurogenic inflammation has been well studied in skin tissues, in which activation of peptidergic nociceptive sensory neurons drives rapid oedema and neutrophil infiltration13. To further assess FZD-mediated targeting of sensory neurons by TcdB, we utilized a subcutaneous footpad injection mouse model, in which the occurrence of neurogenic inflammation can be assessed by measuring oedema (footpad thickness). Injection of TcdB in this model induced rapid development of oedema within 15 min (Fig. 3a), similar to injection of capsaicin, which activates the transient receptor potential cation channel subfamily V member 1 (TRPV1) in nociceptive sensory neurons (Fig. 3a). Blocking SP signaling using an FDA-approved small molecule drug, aprepitant (an NK1R antagonist), prevented oedema induced by TcdB (Fig. 3a). Consistently, TcdB did not induce oedema in Tac1 KO mice (Fig. 3b). Mast-cell-mediated allergic reactions may also induce rapid oedema. TcdB, however, induced oedema in mast-cell-depleted mice (c-KitW-sh mice) similar to WT mice (Fig. 3b). TcdB-FzM did not induce oedema in WT mice whereas TcdB-Cspg4M was as potent as TcdB (Fig. 3a). Furthermore, TcdB induced similar levels of oedema in Cspg4 KO mice and WT mice in this footpad injection model (Fig. 3b). These findings suggest that targeting FZD1/2/7 is sufficient to induce neurogenic inflammation, consistent with a model in which sensory neurons act upstream of pericytes.
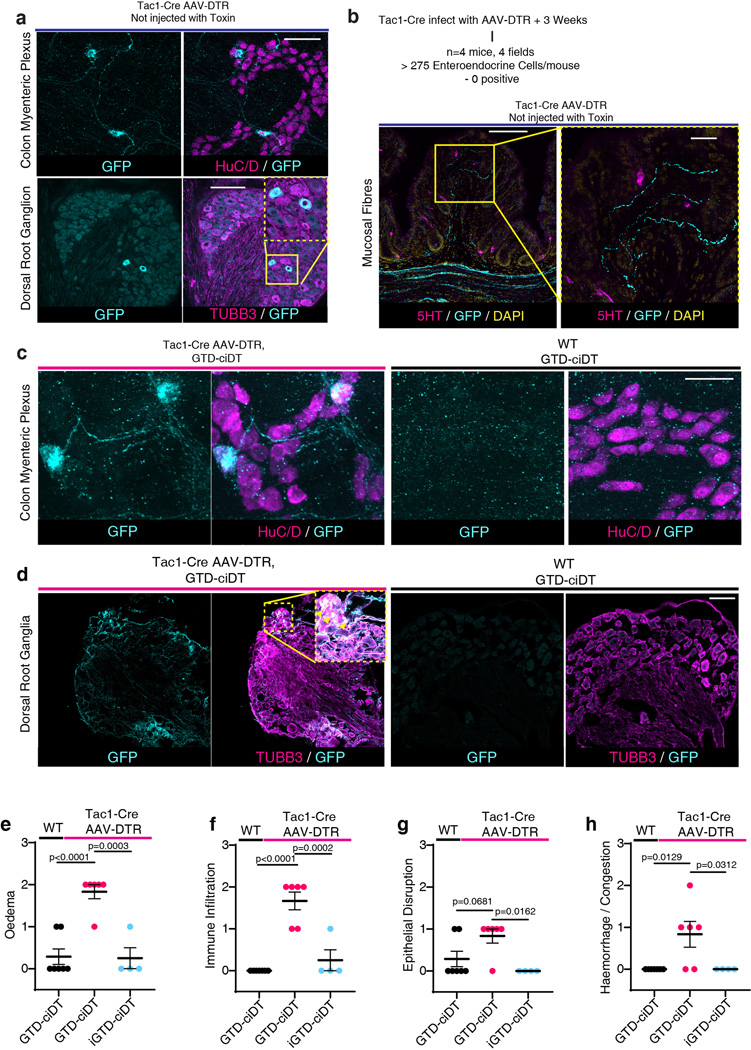
Fig. 3: Delivery of TcdB-GTD into peptidergic sensory neurons using the toxogenetic approach is sufficient to induce neurogenic inflammation.
a, Mouse footpad injection assays were carried out for the indicated toxins or reagents. Footpad oedema was measured and shown as ΔThickness. N=4, 3, 8, 4,4,4,4,4 mice.
b, Footpad injection assays were carried out on WT, Tac1−/−, C-kitW-sh (deficient in mast cells), and Cspg4−/− mice. N=5, 3, 3, 3 mice.
c, Schematic illustration of the toxogenetic approach. PNS: peripheral nervous system. ENS: enteric nervous system. DRG: dorsal root ganglion.
d, DTR was expressed in Tac1+ peptidergic neurons through intravenous injection of AAV.PHP.s.FLEX.DTR:GFP virus into Tac1-Cre mice. Footpad injection assays were carried out. GTD-ciDT: TcdB-GTD fused to a detoxified diphtheria toxin (iDT); iGTD-ciDT: catalytic inactive form of TcdB-GTD fused to ciDT; n=5, 6, 3, 3 mice.
e-f, Intraperitoneal administration of GTD-ciDT induced colonic tissue damage and inflammation in Tac1-Cre mice transduced with AAV.PHP.s.FLEX.DTR:GFP. Representative H&E staining of colonic tissues are shown in e, and histopathology scores are shown in f; n=7, 6, 4 mice.
g-h, Ablating TRPV1+ sensory neurons using RTX reduced the levels of SP and histopathology scores in the TcdB cecal injection model; n=7, 10, 10 mice for g and n=10, 10 mice for h.
i-j Systemic administration of GTD-ciDT induced colonic tissue damage and inflammation in Nav1.8Cre/+ Rosa-LSL-DTR mice; n=7, 9 mice.
Statistical tests in a (p<0.001 overall), b (p<0.001 overall), d (p<0.001), and f (p<0.0001) were one-way ANOVAs. Post-hoc Dunnett’s test compared with TcdB treatment (a) or WT treated with TcdB (b). Post-hoc Tukey’s tests as indicated in d and f. P values are exact or n.s.; Centre line: mean; error bars: SEM. Scale bars are 50 μm. n is described from left to right.
Delivery of TcdB-GTD into neurons induces inflammation
To determine if TcdB-GTD enzymatic activity in neurons is sufficient to induce neurogenic inflammation, we developed a targeted effector delivery approach, termed toxogenetics (Fig. 3c). This approach is similar to widely utilized selective expression of simian diphtheria toxin receptor (DTR) in mouse models for acute ablation of DTR-expressing cells through injection of DT40,. Our approach incorporates two changes: (1) DTR expression (limited by a LoxP-flanked stop cassette) is encoded and delivered using the adeno-associated virus (AAV) serotype PHP.s, which is highly selective toward peripheral neurons41. When injected into Tac1-Cre mice, DTR would be expressed only in Tac1-positive neurons; (2) we utilized a detoxified DT mutant containing three-point mutations that deactivate its enzymatic activity, termed catalytically inactive DT (ciDT), as a carrier to deliver a fused cargo protein into DTR-positive cells in vivo (Fig. 3c).
Injection of AAV-DTR at low titers (~ 1011) into Tac1-Cre mice led to expression of DTR in DRG and ENS neurons, but not non-neuronal Tac1+ cells such as enterochromaffin cells (Extended Data Fig. 6a-d). We constructed a fusion protein composed of the TcdB-GTD domain fused with ciDT (GTD-ciDT). Injection of GTD-ciDT into the foot pads of uninfected control mice did not result in oedema, whereas injection into Tac1-Cre mice infected with AAV-DTR induced rapid development of oedema that was blocked by aprepitant (Fig. 3d). The enzymatic activity of the GTD can be inactivated through a point mutation (D270N, iGTD). Administration of iGTD-ciDT fusion protein did not induce oedema in Tac1-Cre mice infected with AAV-DTR (Fig. 3d).
Systemic administration of TcdB via intraperitoneal (IP) injection can cause death of mice. In contrast, injection of GTD-ciDT did not affect viability, likely because GTD-ciDT targets only Tac1+ cells. This allowed us to administer GTD-ciDT systemically, which caused diarrhea within 3 h in Tac1-Cre mice infected with AAV-DTR. Histopathological analysis revealed severe oedema as well as mild neutrophil infiltration and epithelium disruption in colonic tissues (Fig. 3e-f, Extended Data Fig. 6e-h). In contrast, neither administration of GTD-ciDT into control mice uninfected with AAV-DTR nor administration of inactive iGTD-ciDT into Tac1-Cre mice infected with AAV-DTR resulted in any diarrhea or damage to colonic tissues (Fig. 3e-f). These findings demonstrate that targeted delivery of the GTD domain into peripheral Tac1+ neurons alone is sufficient to replicate major histopathological changes associated with CDI.
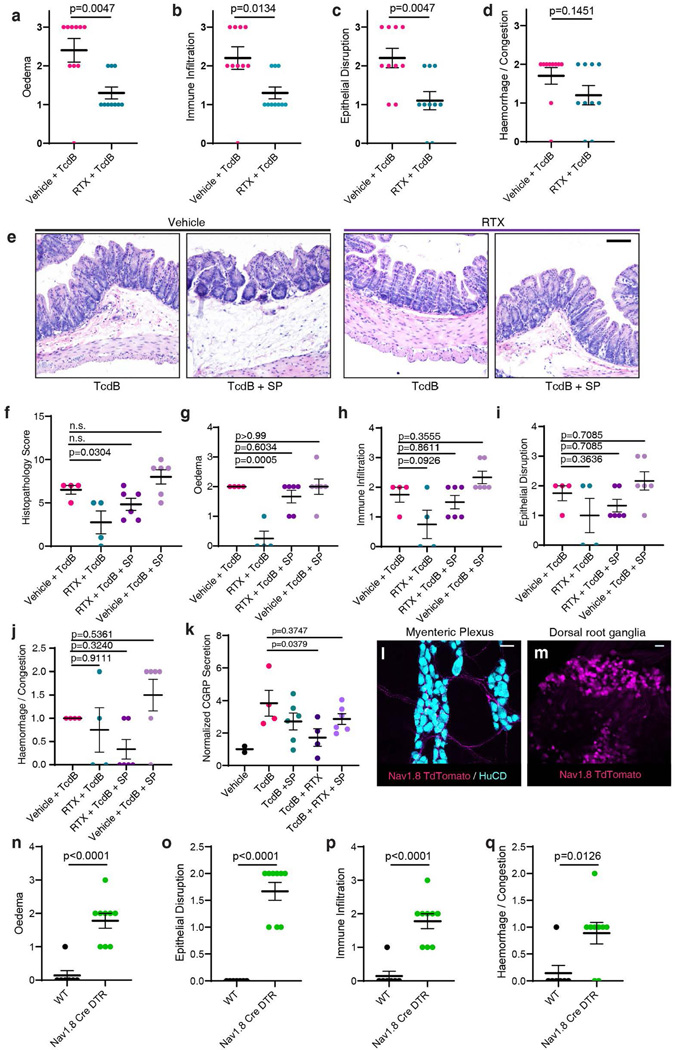
To determine the relative contributions of intrinsic and extrinsic Tac1+ neurons, we took two complementary approaches. First, we performed cecal TcdB injections in mice pre-treated mice with resiniferatoxin (RTX), a strong TRPV1 agonist that causes depletion of TRPV1+ sensory nerve terminals42. RTX-treated mice showed reduced SP levels as well as less severe histopathology upon TcdB administration compared to vehicle-treated controls (Fig. 3g-h, Extended Data Fig. 7a-d). Co-administration of SP with TcdB to RTX-treated mice restored histopathological scores (Extended Data Fig. 7e-k).
Second, we have previously shown that sodium channel Nav1.8 (Scn10a) is selectively expressed in gut-innervating extrinsic sensory neurons, but not enteric neurons in the small intestine42. We confirmed this to be the case in the colon using Scn10aCre/+ Rosa25LSL-tdTomato/+ mice (Extended Data Fig. 7i-m). By crossing Scn10aCre/+ mice with Rosa-LSL-DTR mice, we obtained mice with DTR selectively expressed in extrinsic sensory neurons but not enteric neurons. Injection of GTD-ciDT into Nav1.8-Cre-DTR mice induced histopathological changes typical of CDI (Fig. 3i-j, Extended Data Fig. 7n-q). These data suggest that targeting extrinsic sensory neurons is sufficient to elicit neurogenic inflammation in the colon and recapitulates major elements of CDI pathogenesis.
Blocking inflammation reduces C. difficile colonization
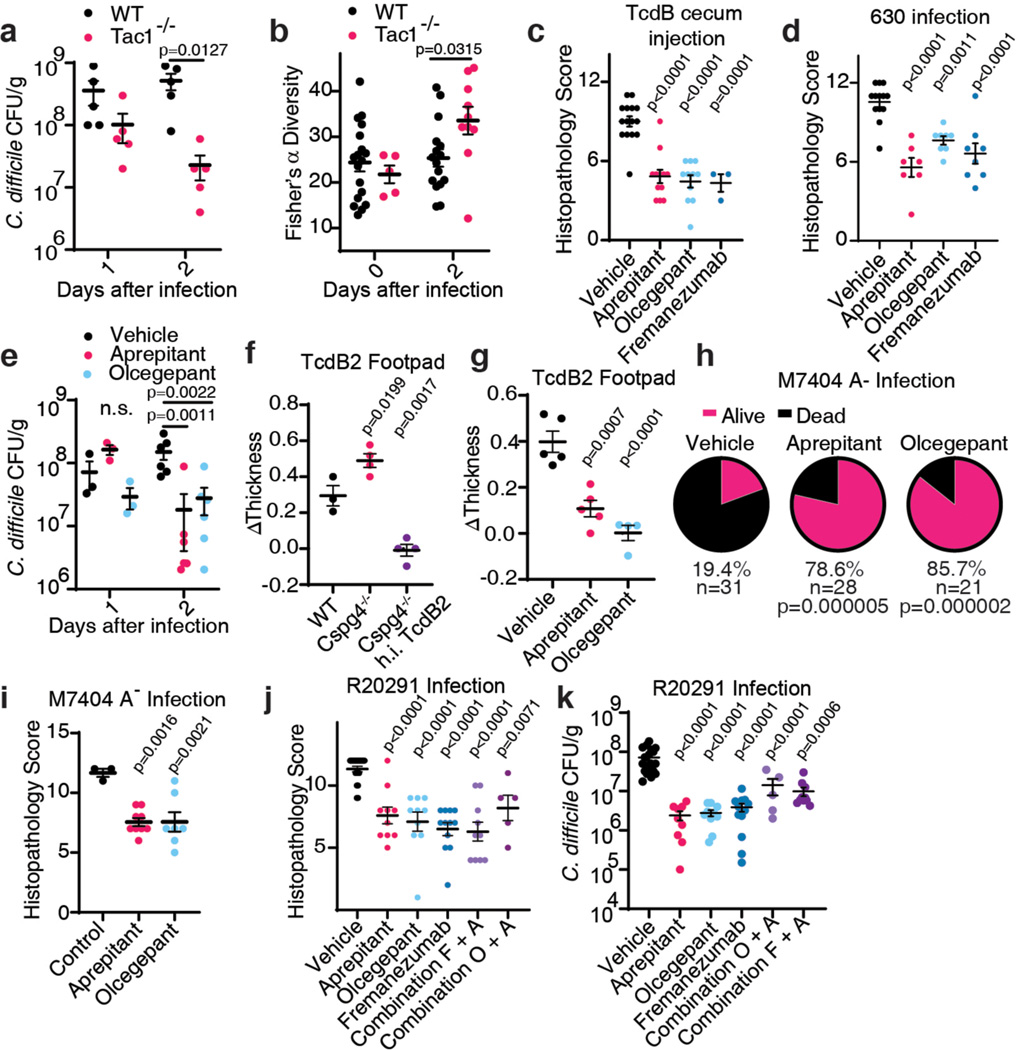
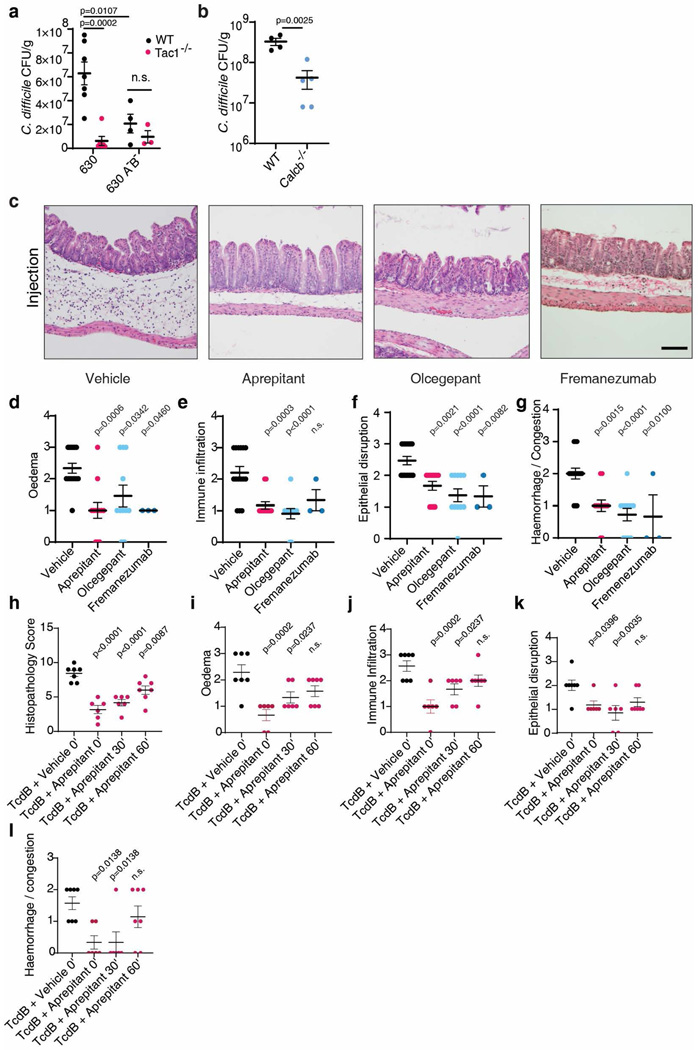
Recent studies showed that toxin-induced inflammation promotes C. difficile colonization5,6. Consistently, although C. difficile burden measured by colony-forming units (CFUs) on the first day of infection was at similar levels between WT and Tac1 KO mice, Tac1 KO mice showed a greatly reduced C. difficile burden by day 2 (Fig. 4a). This reduced pathogen load was at similar levels to a mutant C. difficile lacking TcdA and TcdB administered to WT and Tac1 KO mice (Extended Data Fig. 8a). Similarly, C. difficile burden was reduced on day 2 in Calcb KO mice compared with WT mice (Extended Data Fig. 8b).
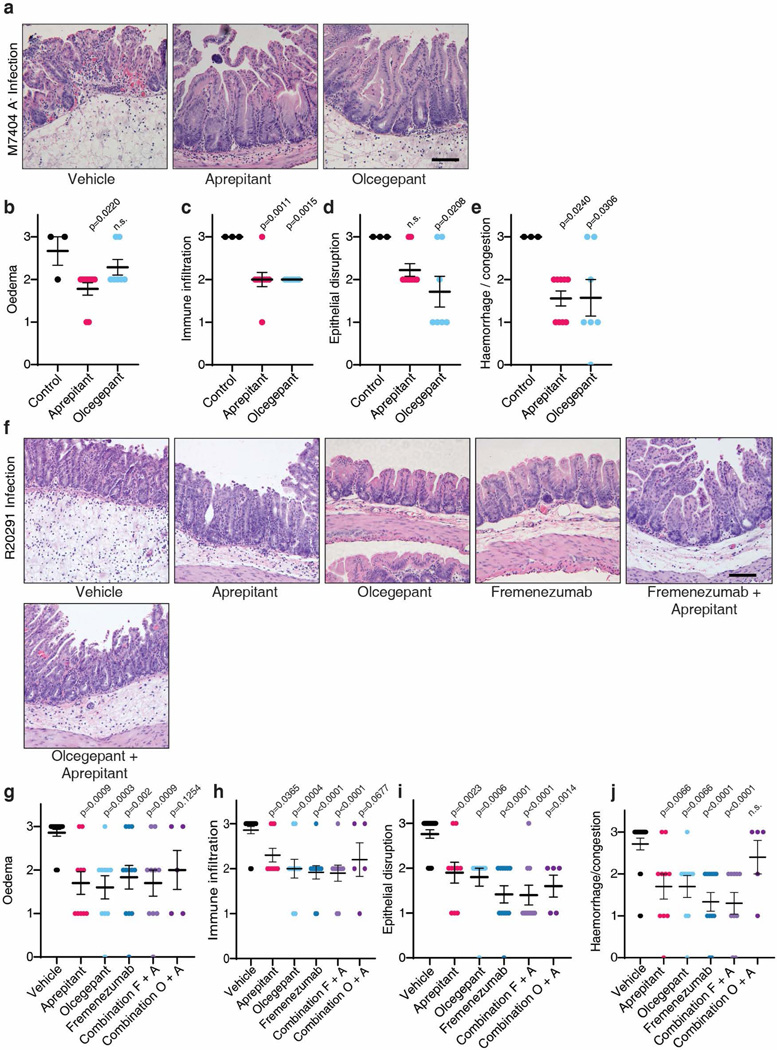
Fig. 4: Inhibiting SP or CGRP signaling alleviates inflammation and reduces C. difficile colonization for endemic and hypervirulent epidemic strains.
a-b, CDI were carried out (630 Δerm strain), and colony forming units (CFUs) of C. difficile in faeces were assessed and plotted in a; n=5, 5 mice. Microbiome diversity was assessed and presented as Fisher’s alpha diversity in b; n=18, 16, 5, 10 mice.
c, Cecum injection assays were carried out with TcdB on mice pretreated with vehicle, aprepitant, olcegepant, or fremanezumab; n=15, 12, 11, 3.
d-e, CDI were carried out (630 Δerm) on mice pretreated with vehicle, aprepitant, olcegepant, or fremanezumab; n=13, 7, 8, 8 mice in d; n=3 (d1) and n=6 (d2) in e.
f-g, TcdB2 induces oedema in WT and Cspg4−/− mice in the footpad injection assay; n=3, 4, 4 mice. Aprepitant and olcegepant can prevent oedema induced by TcdB2. h.i. TcdB2: heat inactivated TcdB2, n=5, 5, 4 mice.
h-i, CDI with the strain M7404 (TcdA-/TcdB2) of mice pre-treated with vehicle, aprepitant, or olcegepant. Survival is plotted in h, n=31, 28, 21. Histological scores are plotted in i, n=3, 9, 7 mice.
j-k, CDI with the hypervirulent strain R20291 of mice pre-treated with the indicated inhibitor (Combination F+A: fremanezumab plus aprepitant; Combination O+A: olcegepant plus aprepitant). Histopathological scores are shown in j, n= 21, 10, 10, 12, 10, 5 mice; CFUs are shown in k, n= 21, 9, 10, 12, 10, 5 mice.
Statistical analyses for a, b, and e: two-way ANOVA, variables are time and genotype/drug treatment, post-hoc Sidak’s test with comparisons by day. For c, d, f, g, i, j and k: one-way ANOVA (<0.0001 overall) with post-hoc Dunnett’s test compared with vehicle (c, d, g, i, j and k). For h, drugs were compared by Fisher’s test to vehicle controls. Centre line: mean; error bars: SEM; n.s., not significant; p values are exact. n is described from left to right.
It has been reported that toxin-mediated inflammation suppresses the recovery of microbiome diversity, thus maintaining a non-normative state favorable for CDI5. Consistently, recovery of microbiome community diversity, measured by 16S ribosomal RNA gene sequencing and quantified by Fisher’s α diversity index, is suppressed in WT mice between days 1 and 2 of CDI, whereas microbiome diversity begins to increase on day 2 in Tac1 KO mice (Fig. 4b). Thus, depletion of inflammatory neuropeptides not only directly reduces damage to colonic tissues during CDI, but also decreases C. difficile colonization and promotes recovery of microbiome diversity.
SP or CGRP blockade alleviates infection
SP signaling can be blocked using aprepitant, an FDA-approved agent for treating nausea and vomiting. CGRP signaling can be inhibited with either small molecule CGRP receptor antagonists, such as olcegepant, or FDA-approved monoclonal antibodies that bind CGRP, such as fremanezumab, which are used for treating migraine headaches. Administration of aprepitant, olcegepant, or fremanezumab reduced histopathological scores in both cecum injection assays with TcdB and in CDI mouse models using C. difficile 630 Δerm (Fig. 4c-d, Extended Data Fig. 8c-l and 9a-e). In mice with CDI, both aprepitant and olcegepant reduced pathogen burden on day 2 after infection (Fig. 4e). Neither drug influenced C. difficile growth rate in vitro nor had an effect in Tac1 or Calcb KO mice, respectively (Extended Data Fig. 9f-j), suggesting that reduced pathogen colonization in CDI is not due to a direct effect on C. difficile.
A major challenge for treating CDI is the emergence of hypervirulent strains such as ribotype 02743. These strains often encode a major variant of TcdB classified as TcdB2, which contains ~8% sequence variation from the standard TcdB in C. difficile 630 Δerm and does not bind to FZD1/2/744–48. We found that injection of TcdB2 into mouse footpads induced oedema, which was prevented by aprepitant or olcegepant (Fig. 4f), indicating that TcdB2 also induces neurogenic inflammation. The effect is independent of Cspg4, as TcdB2 induced footpad oedema in Cspg4 KO mice (Fig. 4g). Furthermore, TcdA also induced oedema in footpad injection assays, which was blocked by aprepitant (Extended Data Fig. 9k).
We then utilized a genetically modified C. difficile strain that expresses TcdB2, but not TcdA (M7404 A-/B+)25, and is known to be hypervirulent, causing death in the majority of mice in a CDI model (Fig. 4h). Treatment with aprepitant or olcegepant markedly reduced fatality as well as histopathological scores of colitis in surviving mice (Fig. 4h-i, Extended Data Fig. 10a-e). We further examined therapeutic effects against a WT hypervirulent strain R20291 that expresses both TcdA and TcdB2. Treatment with aprepitant, olcegepant, or fremanezumab all reduced histopathological scores in colonic tissues in CDI models (Fig. 4j, Extended Data Fig. 10f-j), and reduced colonization of R20291 (Fig. 4k). Combinations of aprepitant and olcegepant or aprepitant and fremanezumab did not show additive effects over monotherapy (Fig. 4j-k, Extended Data Fig. 10f-j).
Discussion
Here we established that TcdB induces neurogenic inflammation by targeting gut-innervating peptidergic neurons and pericytes in mouse models. The specificity of TcdB is mediated by its receptors FZD1/2/7, which are enriched in ENS and DRG neurons, and CSPG4, which is a marker for pericytes. The ability to induce neurogenic inflammation is shared by TcdB2 and TcdA, reflecting a central role of neurogenic inflammation in CDI. TcdB2 and TcdA do not utilize FZD1/2/7 as receptors and alternative potential receptors have been suggested49–51, although the molecular basis for their targeting of neurons remains to be established.
Using our toxogenetic approach, we demonstrated that TcdB in Tac1+ neurons alone was sufficient to induce neurogenic inflammation and recapitulate major histopathologic changes associated with CDI. We note that expression of FZD1/2/7 is not restricted to neurons and the pro-inflammatory effects of TcdB are likely multifaceted including damage to colonic epithelium, stem cells, and immune cells27,52. Additionally, tissue damage may generate an inflammatory environment that activates sensory neurons and further promotes neurogenic inflammation.
How exactly TcdB induces vesicle secretion remains to be established. It has been reported that the cortical actin network underneath the plasma membrane forms a physical barrier preventing exocytosis of vesicles and that depolymerization of the cortical actin network, for instance through latrunculin treatment or deactivation of small GTPases, promotes exocytosis of large dense core vesicles and secretory granules53. Consistently, it has been shown that TcdB can induce degranulation and secretion of hexosaminidase from the human mast cell line HMC-154. Interestingly, TcdB-GTD contains a membrane localization region that localizes it to the plasma membrane after entering the cytosol55, suggesting that TcdB-GTD may preferentially disrupt the cortical actin network underneath the plasma membrane.
A limitation of our studies was that the relative contribution of intrinsic enteric neurons remains to be established. Additionally, the role of pericytes remains underexplored. We found that nearly all micro-vessels in the colonic mucosa are wrapped with Cspg4+ pericytes, suggesting that pericytes are an important part of the gut vascular barrier (GVB)56,57. The close contact between neurons and pericytes suggests that they may form an integral part of a neurovascular structure that dynamically regulates the GVB. Although direct targeting of pericytes by toxins was dispensable for inducing neurogenic inflammation in the acute footpad injection model, it is possible that simultaneous targeting of both neurons and pericytes by TcdB is needed to have a maximal effect during infection. Pericytes are a major source of cytokines such as CXCL1 and could play an important immunomodulatory role in CDI29, although the exact contribution of pericytes on inflammatory kinetics during CDI remains to be established.
Lastly, we have shown that targeting neurogenic inflammation with agents blocking SP and CGRP is an effective, host-oriented therapeutic approach in rodent models of CDI. These agents have already proven safe and effective in other clinical contexts. Clinical trials are needed to determine if these agents might have efficacy in treating human CDI.
Methods
Mouse models
All experiments were performed in accordance with animal protocols approved by the at Boston Children’s Hospital Institutional Animal Care and Use Committee. Animals were housed in a specific pathogen free facility, except for infection experiments which were conducted in a facility that contains specific pathogens (ABSL2+). Animals were housed under standard conditions with ad libitum food and water, 22˚C+/− 2C, 35–70% humidity, and 12-hour light-dark cycle. WT mice were from Envigo (CD1, Female 18–20g), Jackson Lab (C57/BL6J, JAX: 000664), or bred in our colony (C57/BL6N, JAX: 005304). CalcbeGFP/+ (kindly provided by Ron Emeson, Vanderbilt University)58, CalcaCreERT2/+ (kindly provided by Pao-Tien Chuang, UCSF)59, Cspg4-DsRed (JAX: 008241)60, Tac1-Cre (JAX: 021877)61, Tac1−/− (JAX: 004103)62, Cspg4−/− (kindly provided by William Stallcup)63, Nk1rCreERT2 (JAX: 035046)64, Nav1.8-cre (JAX: 036564)65, Rosa-LSL-DTR (JAX: 007900)66, Rosa-LSL-TdTomato (JAX: 007909)67, c-kitW-sh (JAX:030764)68, and PLP1-EGFP (JAX033357) mice, have all been previously described. CalcbeGFP/eGFP mice are referred to as Calcb KO, CalcaCreERT2/CreERT2 as Calca KO, and NK-1rCreERT2/CreERT2 as Nk1r KO. Tac1 KO, c-kitW-sh, and Cspg4 KO were backcrossed extensively, maintained as homozygous colonies, and compared to C57/BL6J mice raised at the same time. Tac1-Cre and Nav1.8-Cre DTR were compared to C57/BL6J. Calca KO and Calcb KO were compared to littermate controls. Nk1r KO mice were compared to C57/Bl6NJ mice given that they were raised on this background. Mice were co-housed where littermate controls were used and across all pharmacological experiments. Tac1 KO and NK1R KO mice were not cohoused routinely, except that Tac1 KO mice for the experiments described in Extended Data Fig. 9g-h were cohoused with their WT control mice from weaning.
Cecum Injection Assays
Cecum injection assays followed a previously established protocol69. Mice were fasted 12–16 h prior to experiments and anaesthetized by isoflurane (2–4%) with oxygen (0.8 L/min). The cecum was exposed using a midline laparotomy and 6 μg of toxins (TcdB, TcdB-FzM, TcdB-Cspg4M or vehicle control) was injected into the lumen in 100 μl of normal saline. Internal fascia were then closed with absorbable sutures (Johnson and Johnson, Vicryl 4/0 RC) and overlying skin was closed using wound clips. Following surgery, all mice were given buprenorphine (Ethiqa XR, Fidelis) analgesia (1 mg/kg) by subcutaneous administration then allowed to recover. After the indicated incubation periods (6 h unless otherwise stated in the figure legend), mice were euthanized by CO2 inhalation, and the colon was dissected, flushed with PBS, and processed for downstream experiments. In experiments involving transgenic mice, both male and female mice were used with their respective matched genetic controls, which did not display sexually dimorphic responses. For pharmacological assays, only female CD1 mice were used for cecum injection assays. CD1 mice were injected with pharmacological agents 30 min to 1 h prior to cecum injection. Aprepitant (100 mg/kg, Tocris, 6486) and olcegepant (5 mg/kg, Medchemexpress, HY-10095) were dissolved in DMSO and diluted into 2% Tween-80 in normal saline and administered to mice by intraperitoneal (IP) injection. Fremanezumab (100 mg/kg, Ajovy, 225 mg One-Prefilled Syringe, obtained via Boston Children’s Hospital Central Pharmacy (BCHP), NDC 51759–204-10) was dissolved in normal saline. For the rescue experiments TcdB was co-administered with 10 nmoles of SP, and mice were euthanized 2 h later.
CDI Infection model
We utilized an antibiotic-treatment-induced CDI mouse model previously developed70. These experiments were performed in an A-BSL2 facility with autoclaved food (Prolab RMH 3500, Autoclavable), cages, and water. C difficile strains 630 Δerm, 630 Δerm (TcdA-/TcdB-)23, M7404 (TcdA-)25, and R20291 were used. A mixture of antibiotics including vancomycin (0.4 mg/mL, Sigma-Aldrich, A94747) colistin (850 U/mL, Sigma-Aldrich, C4461), metronidazole (0.215 mg/mL, Sigma-Aldrich, M1547), gentamicin (0.035 mg/mL, Research Products International, G38000), and kanamycin (0.045 mg/mL, Sigma-Aldrich, K1377) were diluted in drinking water and used to feed mice for 3 days, followed by regular water for 2 more days, then mice were injected with clindamycin 10 mg/kg, IP, Mylan pharmaceuticals, obtained from BCHP, NDC 67457–816-00) in normal saline. Twenty-four h later, mice were gavaged with 104 spores diluted in PBS. Mice were monitored twice daily and euthanized if they became moribund (trouble breathing, >15% body weight loss).
For experiments utilizing transgenic mice: 8–15-week-old mice were infected with C. difficile 630 Δerm, their tissues were collected two days after infection and analyzed for histopathology. Both male and female mice were used. For simplicity, the results of WT mice that reflect multiple genetic backgrounds are combined in main figures (they are presented separately in Extended Data Figures). Pharmacological assays for CDI infection were performed in WT C57/BL6J mice. Mice were injected daily with the small molecule antagonists. Fremanezumab was injected at the time of spore gavage. In experiments involving R20291, mice were injected with fremanezumab prior to the experiments at the same time clindamycin was injected.
Histology analysis and scoring
For histological analyses, tissues were trimmed, placed in neutral buffered formalin (10%), and then transferred to PBS at 4oC prior to embedding by the Beth Israel Deaconess Medical Center (BIDMC) core facility. Blocks were cut at 6 μm and subjected to haemotoxylin and eosin (H&E) staining. Briefly, slides were baked, deparaffinized in xylene, re-hydrated with serial ethanol washes, immersed in Gill’s haemotoxylin (Number 3, Thermo Fisher, Shandon), washed, and then blued with Scott’s Tap Water (0.1% W/V sodium bicarbonate in water) prior to immersion in acidic eosin. Samples were dehydrated through serial changes of ethanol and then xylene prior to mounting in DPX. Slides were then left to dry overnight in a fume hood. Sections were imaged and scored blindly according to standardized criteria (oedema, epithelial disruption, immune infiltration, and haemorrhage or vascular congestion), scored from 0–3 (0 – Normal, 1- Mild, 2- Moderate or 3 – Severe, see Extended Data Fig. 1a-e). Only animals that were euthanized at the indicated endpoint were included in scoring. De-identified images of sample histopathology from human CDI compared to healthy colon were provided by Dr. Jeffrey Goldsmith. Sample images were from cases that had been previously collected, processed, sectioned and H&E stained as part of routine patient care at Boston Children’s Hospital and Brigham & Women’s Hospital.
Single cell data analysis
Single cell data were extracted from published datasets. For analysis of human and mouse Frizzled expression in colonic cells, we extracted the data from a comprehensive atlas of mouse/human enteric neurons using the Broad institute single cell expression platform71, normalized the data in Excel (data were normalized to the maximum expression), and generated plots shown in Tableau software (v2022.2). For analysis of DRG neuron expression of Frizzled in colonic innervating DRG neurons, we extracted data from the author’s analysis tool72. For the in situ hybridization dataset, we extracted spinal cord images from P4 mice from the Allen Brain atlas73. For analysis of CSPG4 expression in human colon, we used the EMBL single cell analysis platform to visualize expression in a published dataset34.
Immunohistochemistry
For analyses of Cspg4-dsRed, Nav1.8-TdTomato, and AAV-injected mice, intestines and/or DRG were isolated, and drop fixed in 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS) pH 7.4. For AAV-injected and DT-treated mice, mice were euthanized with CO2 inhalation, a thoracotomy was performed to expose the heart, and then 15 ml of PBS was perfused followed by PFA through a 21G winged catheter. Following tissue isolation and fixation, intestines or DRG were processed for whole mount staining or cryoprotected using 30% sucrose. DRG were exposed by spinal laminectomy and isolated from mice by microdissection.
Whole mount staining was performed as previously described74. Briefly, tissues were rinsed six times with PBS + 0.5% Triton X-100 followed by incubation in blocking solution (PBS + 5% normal goat or donkey serum + 0.5% Triton X-100 + 20% DMSO) with diluted primary antibody (Supplementary Table 1) for 48–72 h at room temperature (RT). Tissues were then rinsed and incubated overnight in blocking solution with secondary antibody at RT. Tissues were rinsed again with PBS + 0.5% Triton X-100 six times and then mounted with Vectashield mounting media. For some samples, tissues were subjected to optical tissue clearing (BABB solution protocol as described previously)74. As controls, for analyses involving Cspg4-DsRed we used littermate non-carrier mice not expressing DsRed; for analyses involving GFP we used mice that were not injected with virus. For all other routine staining, initial experiments used a section stained with only the secondary antibodies. Antibodies utilized are listed in Supplementary Table 1.
For cryosections, intestinal segments and DRG were equilibrated in 30% sucrose/PBS overnight at 4°C and embedded in optimal cutting medium (OCT, Sakura). For the intestine, cross-sections of 40, 70, and 100 μm were collected onto slides. DRG were sectioned at 10–12 μm. Staining was performed as previously described74. Briefly, slides were air dried for >20 minutes, equilibrated with PBS, and then incubated in blocking solution (PBS + 5% normal goat or donkey serum + 0.1% Triton X-100 +/− 5% BSA for intestinal segments, PBS + 2% BSA or 5% normal donkey serum + 0.1% Triton X-100 for DRG) for one hour at RT, followed by incubation with primary antibody in blocking solution (Supplementary Table 1) overnight at 4°C. Slides were rinsed three times in PBS followed by incubation in secondary antibody diluted in blocking solution for 60 min. Slides were then rinsed three times in PBS and mounted in Vectashield media for intestinal segments, and in Vectashield or fluoromount media for DRG. For nuclear staining, DAPI (1:500 from 200 μg/ml concentrate) was either diluted in blocking solution during the secondary antibody incubation step or Vectashield/Fluoromount containing DAPI was utilized during tissue mounting. As controls, for analyses involving Cspg4-dsRed we used wild-type mice not expressing DsRed and stained them in the same way; for analyses involving GFP we used mice not injected with virus. For all other routine staining initial experiments, we used a section stained with only the secondary antibodies.
RNAscope fluorescent in situ hybridization was carried out via the vendor-provided protocol for fixed-frozen tissue sample and preparation (Advanced Cell Diagnostics) with Pdgfrb probe (Cat. No. 411381) with the following modifications: Activated ribonucleoside vanadyl complexes (New England Biolabs Cat. No. S1402S) were included in PBS at 10 mM during tissue collection. The tissue was fixed in EM grade 4% PFA at 4°C for 24 h, equilibrated in 30% sucrose solution in PBS, and embedded in OCT media. 8 μm cryo-sections of colon were then collected and used. Opal 690 dye (Akoya Biosciences, FP1497001KT) was used at 1:1500. DapB (bacterial transcript) probe (Cat.No. 200470) was used as a negative control. Immunohistochemistry and tissue mounting were then performed as outlined above.
Footpad injection assay
The footpad toxin injection assay was performed as we previously described75. Briefly, mice were restrained in an incontinence pad and the footpad was held and subsequently injected using a 29.5G syringe into the plantar surface. Mice were observed for 15 minutes, and measurements of paw diameter were performed by digital caliper measurement from the plantar surface to the top of the foot. Paw diameter changes were assessed by calculating a ratio relative to the un-injected foot. For each test toxin, 500 ng were administered in a volume of 20 μl of normal saline. Pharmacological assays were performed in female CD1 mice, with pre-treatment with drugs 30 min to 1 h prior to toxin injection. For experiments involving transgenic mice, experiments were performed in male and female mice (age matched, 8–15 weeks). Capsaicin was dissolved in ethanol and diluted into a 10% Tween solution; 1 μg of capsaicin was injected into the footpad.
Ear toxin injection assay
CD1 mice were anesthetized with isoflurane (2–3%), injections were made intradermally into the ear using a 29.5G syringe. Toxins (1 μg) were diluted in normal saline. Mice were monitored and then euthanized after two hours. Their ears were dissected, split down the midline using No.5 style forceps and then fixed with acetone for 1 min prior to being washed in PBS, blocked in 2% BSA + 0.3% Triton X-100, and stained for 2 h in blocking buffer at RT with anti-smooth muscle actin (Sigma Aldrich, 1A4, 1:200) and then secondary antibody staining for 1 h at RT. Experiments were replicated in Cspg4-DsRed mice, for those experiments we used 4% paraformaldehyde in PBS (pH 7.4) as fixative in replication experiments because acetone disrupted the signal of the DsRed.
DRG neuron culture
DRG cultures were based on a protocol previously established with minor modifications76. DRG were isolated into ice-cold Neurobasal A media (Gibco, 21103049) by laminectomy from exsanguinated mice following euthanasia by CO2 inhalation. DRG were dissociated in collagenase (5 mg/mL, Sigma, 10103578001) plus dispase (1 mg/mL, Sigma, 4942078001) in Hank’s Buffered Salt Saline containing calcium (HBSS, Gibco) for 70 min. The enzymatic digest was neutralized by 10% Fetal Bovine Serum (FBS) and then centrifuged to pellet the DRG cells. DRG cells were resuspended into 2 mL of complete neurobasal media A (Gibco,10888022), with nerve growth factor complex 2.5s 50 ng/mL (Sigma Aldrich, N6009), B27 plus 1X (Thermo Fisher Scientfic, A3582801), glial-derived neurotrophic factor (2 ng/mL, Sigma Aldrich, SRP3239), penicillin/streptomycin 1X (Gibco, 21103049), and Glutamax (1 mM, Gibco, 35050061). Neurons were triturated 20 times with a wide bore flame polished glass pipette, then 20 times with a narrow bore glass pipette. The cells underwent gradient purification through a 10% Bovine serum albumin (BSA) gradient (in PBS) and were overlaid on top of the BSA solution using a glass pipette. Gradient purification was performed at 400 g for 12 min. Following gradient purification, the pellet was retained and resuspended in 1 mL complete media. The cells were examined by microscopy and then plated at a density of ~2000 neurons per well in a 96 well or scaled accordingly for larger wells. Plates were coated in Poly-D-Lysine (0.1 mg/mL, Sigma, P6407) for 2 h, washed three times with water, then incubated with Laminin (10 μg/mL in PBS, Sigma, 11243217001) for 60 min. Plates were washed with PBS and dried before plating. Cells were allowed to grow for 3–5 days prior to experiments.
Confocal images
Confocal z-stack images for figures and supplementary videos were captured using the Zeiss LSM 880 using super resolution deconvolution laser scanning confocal microscopy mode using objectives ranging from 10X to 63X prior to Airy deconvolution using Zeiss Zen Black software. In some cases, microscope images were captured using a Zeiss LSM 770. Routine fluorescent images were captured using an Olympus IX83 using a Lambda DG4 light source (Sutter Instruments) connected to a Hammatsu Orca Fusion camera and an Olympus BX53 using a LED light source and collected using Olympus Cellsens. Following deconvolution, z-stack images were rendered into maximum intensity projections and images were adjusted for brightness and contrast in ImageJ or Zeiss Zen Black software. For supplementary videos, z-stack images were processed and rendered into 3D image videos using Aivia image analysis software (Aivia, Inc., Leica Microsystems.) Videos were edited and converted to .mov format using Wondershare Uniconverter™ 14 software.
Measuring release of SP and cytokines
SP in cell culture medium for DRG neurons was measured using the well-established enzyme-linked immunosorbent assay (ELISA) with a kit following the manufacturer’s instruction (Cayman chemical, 583751). CGRP (Cayman, 589001) and CXCL1 (Biolegend, 22141) in cecal samples were measured using established kits and following manufacturer’s instructions. SP/CGRP/CXCL1 release from cecum samples was measured after cecum injection assays (6 μg toxins, 6 h incubation). Briefly, cecum tissues were dissected and flushed with PBS, a section of cecum was taken and weighed, release was measured for 60 min by incubation with agitation (400 rpm), at 37°C in DMEM (Gibco, 11965084) without supplements. Media were collected and release assays for SP were performed using the ELISA assay. Data were normalized internally to the controls collected in parallel. For the infection and time course experiments mice were treated as normal for infection and collected at the indicated time points before SP and CGRP release was measured from explants.
Primary human cultured brain vascular pericytes (ScienCell, #1200) were routinely cultured in special medium with supplement, FBS and penicillin streptomycin provided by the cell’s vendor (ScienCell, #1201). Cells were subcultured when confluent and used up to passage 10. For secretion assays these were performed on early passage cells (passages 3–8). Cytokine secretion in cell culture medium was determined using ELISA based assays against IL-8 (Biolegend, 431504). The effect of CGRP on cultured pericytes was determined by incubating pericytes for 1 h with human CGRPβ (Cayman, 24725).
Cell rounding assay
Cells were seeded on 96 well plates, serially diluted toxins were applied in their respective medias and incubated for five hours. Images were obtained on an Olympus IX83, and an Olympus IX71. Round cells were determined by eye, and the proportions were plotted in Graphpad Prism.
Immunoblot
Cultured pericytes, U87 and HeLa cells were collected using trypsin-EDTA (0.05%, ThermoFisher scientific). Cells were collected by centrifugation, the pellet was washed in PBS. Cells were lysed in Radio-Immunoprecipitation Assay Buffer (50 mM Tris 8.0, 0.1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate, 150 mM NaCl) with protease inhibitor cocktail, then cell lysates were clarified by centrifugation at 14,000 g for 10 min. Subsequently, 10 μL of sample were diluted in Laemmli buffer containing DTT, then boiled for 10 min at 95°C. Samples were loaded into a hand-cast, 12% acrylamide gel (Bio-Rad), proteins were separated by SDS-PAGE at 125V prior to transfer to a nitrocellulose membrane using a semi-dry transfer system (Bio-Rad, Trans-blot Turbo). Membranes were blocked in blocking buffer (3% BSA in Tris Buffered Saline, 0.1% Tween-20, TBS-T), then incubated overnight at 4°C with anti-RAMP1 (1:500, Alomone; ARR-021), anti-CALCRL (1:1000, Sigma-Aldrich, HPA008070), or anti-CALCR (1:1000, Bioss; bs-1860R) diluted in blocking buffer, the membrane was washed three times in TBS-T, then incubated in secondary antibody for 60 min (Goat anti-Rabbit HRP, BioRad, 1:2000). Blots were then washed three times in TBS-T prior to development using SuperSignal Chemiluminescent Substrate (ThermoFisher Scientific, #34080) and imaging on a BioRad GelDoc. See supplementary information figure 1 for blots.
Protein purification
BL21 were transformed with expression plasmids and plated on kanamycin LB plates. Single clones were selected, sub-cultured, and transferred to larger volume flasks in autoinduction media with agitation, on an incubated shaking platform (at 37oC) until they reached OD600 of 0.8. The temperature was then dropped to 18oC for 48 h. Bacteria were collected by centrifugation at 4000 g for 15 minutes. For His tagged proteins, bacteria were lysed in TBS (50 mM Tris-HCl, pH7.5, 150 mM NaCl, 0.05% Tween-20, and 1 mM phenylmethylsulfonyl fluoride (PMSF)), sonicated (3 sec/8 sec time of on/off for 5 min total sonication time), supernatants were collected after centrifugation and then loaded onto prewashed Ni-NTA resin containing column, washed with 70 column volumes of wash buffer (TBS, 0.05% Tween-20, 10 mM imidazole), then eluted with TBS with 200 mM imidazole, buffer exchanged into TBS.
Recombinant TcdB refers to TcdB1 sequence from VPI10463, codon optimized from VPI10463 and purified from E. coli as previously described 29. Recombinant TcdB2 refers to TcdB2 sequence from R20291, purified using B. megaterium. Recombinant TcdA refers to TcdA sequence from VPI10463, purified from B. megaterium. Recombinant TcdB mutants were all purified in E. coli. TcdB-FzM (also known as TcdB-GFE), bears mutations disrupting Frizzled binding (V1595G/N1596F/F1597E, plus deletion of L1598, Q1599, and S1600)28. TcdB-Cspg4M bears mutations disrupting Cspg4 binding (S567E/Y603G/D1812G)29. TcdB-FzCspg4M is a combinational mutant containing both the mutations disrupting Frizzled binding and those disrupting Cspg4 binding. TcdB-GtdM contains mutations disrupting Rho GTPase binding wherein residues G444–K452 were replaced with the corresponding residues in TcdB from C. difficile strain M68, I445–S45332.
Plasmids
GTD-ciDT was constructed by fusion of the TcdB-GTD (TcdB 1–543, based on the sequence in VPI10463, codon optimized for E. coli expression) in frame with ciDT containing three-point mutations (K51E/G52E/E148K, inactivating the enzyme domain) into pET28a vector (Novagen, with a C-terminal His-tag) through Gibson Assembly (NEB, E2621). A linker region (EFGSGSGSGA) was added between the GTD and ciDT. iGTD-ciDT was generated by site directed mutagenesis introducing a D270N mutation in GTD, which disrupts manganese co-ordination as previously described for full length TcdB77. All constructs were confirmed by Sanger sequencing (Azenta).
Toxogenetic experiments
AAV-PHP.s Flex DTR:GFP viral particles were prepared by the Boston Children’s Hospital virus core facility, with titer information provided. Viral particles (1×1011) were diluted in normal saline. Mice (4–8 weeks) were transduced through intravenous (IV) injection, into the tail vein, using a 29.5G syringe with a volume of 100 μL under BSL2 conditions in a biosafety cabinet. Mice were tested/examined 3 weeks after virus transduction. For IP injection of GTD-ciDT and iGTD-ciDT, 500 ng of protein was diluted in normal saline and injected into mice. Mice were observed for 3 h, followed by euthanization. Cecum tissues were collected, processed, and analyzed. For germline expression of DTR in extrinsic afferent neurons, Nav1.8-cre mice were crossed with Rosa-LSL-DTR mice to generate Nav1.8 Cre/+: Rosa-LSL-DTR/+ mice and analyzed at 8 weeks of age. Parallel injections of toxins into wild-type C57/BL6J mice were used as controls.
RTX-induced sensory neuron denervation
For denervation of TRPV1+ cells, resiniferatoxin (Adipogen) or vehicle (0.2% Tween / 2% DMSO in normal saline) was administered to 4-week-old mice by IP injection at escalating doses of 30, 70, and 100 μg/kg on 3 consecutive days. Mice were used for experiments at 8 weeks of age.
C. difficile strains and spore preparation
C. difficile 630 Δerm and 630 Δerm (A-/B-) strains were kindly provided by Nigel Minton (University of Nottingham, Nottingham, United Kingdom). C. difficile C. difficile R20291 and M7404 (A-/B+) were kindly provided by Dena Lyras (Monash University, Melbourne, Australia)25. Cultures were grown in Sporulation broth medium (400 ml) for 5 days78. Cells were centrifuged and washed twice in sterile PBS (20 ml), before being shocked with 50% v/v ethanol at room temperature for 1 h. The suspension was then washed five times in PBS, before collecting the spores. For 630 Δerm strains, cultures of C difficile were grown in Brain heart infusion (BD, Difco) overnight before spreading on 70:30 plates. Bacteria were cultivated for 10 days, collected by cotton swab into PBS and then ethanol shocked. Spores were enumerated on ChromID plates and stored at −80.
C. difficile CFUs
Faeces were collected as raw stool from mice one day and two days post gavaging C. difficile spores. Faeces were immediately weighed and resuspended to 50 mg/mL in pre-reduced PBS (allowed to reduce under anaerobic conditions for 24 hours). Live bacteria were enumerated by plating multiple dilutions on pre-reduced commercial ChromID C. difficile plates (Biomerieux, #43871) after incubation for 24 h (37°C) in an anerobic chamber (Coy Labs). For experiments involving R20291, cecal content rather than faeces was used for CFU enumeration as it was difficult to collect enough faeces from infected mice. CFUs were blindly counted.
16S rDNA phylotyping
Faecal samples were collected and stored at −80oC. Genomic DNA for downstream 16S rDNA amplicon next generational sequencing was isolated using the ZymoBIOMICSTM – 96 DNA Kit (Zymo Research, D4309). The 16S amplicon library was prepared in a 96-well format using dual-index barcodes as we previously described79. Each library was cleaned with the DNA Clean and Concentrator TM – 5 Kit (Zymo Research, D4014) and then quantified by qPCR (NEBNext Library Quant Kit, NEB, E7630). 20 pM of DNA were loaded onto an Illumina MiSeq and sequenced (v3, 600-cycle). Primers used for amplification are: Uni16S_V3: TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-CCTACGGGNGGCWGCAG (1st PCR primer for universal 16S V3V4; “-” stands for 4~11nt spacer sequences); Uni16S-V4R: GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-GGACTACNVGGGTWTCTAAT (1st PCR primer for universal 16S V4); Nextera_i5: AATGATACGGCGACCACCGAGATCTACAC-[i5]-TCGTCGGCAGCGTC (2nd PCR primers, shared between different rDNA amplicons; i5/i7 refers to Nextera indexes); Nextera_i7: CAAGCAGAAGACGGCATACGAGAT-[i7]-GTCTCGTGGGCTCGG.
To generate the Operational Taxonomic Unit (OTU) table for downstream analyses of gut microbiome composition and diversity, Illumina raw reads were de-multiplexed, paired end joined (PEAR v0.9.6)80, adapter trimmed (Cutadapt v3.1), quality filtered, dereplicated, and denoised (vsearch v2.23.0) and mapped into an OTU table (FAST v1.2.2) Sequences were mapped against the publicly available 16S rDNA databases SILVA (v132)81 using QIME2 (2022.11)82and clustered into OTUs >= 97% nucleotide sequence identity. OTU-based microbial community diversity was estimated by calculating the Fisher’s alpha diversity index in phyloseq79.
C. difficile growth in vitro
Frozen stock cultures of bacteria were stored at −80oC in cryogenic vials. C. difficile isolates were grown on brain-heart-infusion supplemented with hemin (50 mg/L) and vitamin K1 (0.25 mg/L) (BHIS) agar plates or in basal media (BSG; proteose peptone (20 g/L), yeast (5 g/L), NaCl (5 g/L), glucose (5 g/L), potassium phosphate dibasic (5 g/L), cysteine (0.5 g/L), hemin (50 mg/L), and vitamin K1 (0.25 mg/L). All isolates were recovered on and grown in pre-reduced media in a Coy anaerobic chamber (Coy labs). To determine the fitness effects of olcegepant or aprepitant on C. difficile, C. difficile 630 Δerm was recovered on a BHIS agar plate and incubated anaerobically at 37oC for 48 h. A starter culture (1 mL) of BSG was inoculated with C. difficile and grown for 15 h. This starter culture was then used to inoculate a BSG sub-culture (1 mL) at a final dilution of 1:10. This sub-culture was grown for 3 h to an OD600~0.5 before being used to inoculate experimental cultures at a final dilution of 1:50. The experimental media consisted of a modified YCFA media (mYCFA; casitone (2.5 g/L), yeast extract (0.625 g/L), cysteine (0.5 g/L), magnesium sulfate heptahydrate (f.c. 365 μM), calcium chloride dihydrate (f.c. 610 μM), sodium bicarbonate (4 g/L), dipotassium phosphate (0.45 g/L), monopotassium phosphate (0.45 g/L), sodium chloride (0.9 g/L), sodium acetate (2.71 g/L), Hemin (50 mg/L), Vitamin K1 (0.25 mg/L), ferrous sulfate (4 μg/mL), ATCC vitamin mix (1% v/v) supplemented with 0.25% glucose and 20 μM olcegepant, 20 μM aprepitant, or vehicle (0.04% DMSO). Bacterial growth was monitored by measuring the optical density at 600 nm (OD600) of each culture in a 384 well plate with an Epoch2 spectrophotometer (BioTek Instruments) for 24 h.
Statistical analysis and reproducibility
Data were plotted and all statistical analysis was performed using Graphpad Prism. No statistical methods were used to pre-determine sample sizes, rather the experience of the investigators and pilot data determined subsequent sample sizes. Statistical tests and post hoc tests are reported in the experiment’s figure legend. Our threshold for statistical significance is p <0.05, exact p values are reported for the experiments in each figure and the accompanying source data. For comparisons of multiple inhibitors, comparisons were made to the vehicle group using Dunnett’s multiple comparison test. For multiple comparisons between several groups, a Tukey’s post hoc test was utilized. Where data showed a non-parametric distribution, a Kruskal-Wallis test was used followed by post-hoc’s test. For tests involving two groups, a Student’s Two-tailed T-test was used and for categorical data, Fisher’s exact test was used.
For qualitative microscopy experiments, we utilized a minimum of N=3 independent animals per antibody or genetic reporter and the core observations were independently assessed by 3 independent investigators (MR, MM, JM or AS). For qualitative western blots, these were repeated twice, plus an independent antibody was assessed for Calcrl. For in vitro bacterial growth experiments, these were performed twice with 3 wells per condition.
Extended Data
Extended Data Fig. 1: Histopathology score analysis and time course of TcdB-induced inflammation in a mouse cecum injection model.
a, A list of criteria for assessing histopathology scores of colonic tissues.
b-e, H&E micrographs illustrating oedema (b, black line indicates oedema extent); epithelial disruption (c); haemorrhage or congestion (d); immune cell infiltration (e).
f, Representative histopathology of human CDI colon tissues compared to a normal control, illustrating neutrophilia, oedematous inflammation, and pseudomembranes.
g-j, The subcategories for the histopathology scoring in Fig. 1b: oedema (g), immune cell infiltration (h), epithelial disruption (i), and haemorrhage or congestion (j). n = 5, 6, 9, 5 mice.
k-l, SP (k) and CGRP (l) levels in colonic explants from the mouse cecal TcdB injection model at the indicated time (min) following TcdB injection. Vehicle was at 120 min. n = 6, 7, 7, 7, 7 mice.
m-n, The levels of SP (m) and CGRP (n) in colonic explants from CDI mouse models infected with either C. difficile 630 or a toxin-null strain (630 A- B-) at 23 h (n = 5), 37 h (n= 5), and 48 h (n = 3), normalized to mice treated with antibiotics but gavaged with saline (time point 0, n= 5).
g-j, assessed by one-way ANOVA with post hoc Dunnett’s test. k-l, assessed by Kruskall-Wallis test with post-hoc Dunn’s test. Centre line, mean; error bars reflect the standard error of the mean (SEM); n.s., not significant, p values are exact. Scale bar is 50 μm in b, c and 20 μm in d and e. n is described from left to right.
Extended Data Fig. 2: Histological scoring for cecum injections of TcdB in the indicated mouse models as described in Fig. 1c-d.
a-e, Histopathology total scores and subscores of TcdB injected Tac1 KO mice, n = 7, 8 mice.
f-j, Histopathology total scores and subscores of TcdB injected Nk1r KO mice, n = 7, 7 mice.
k-o, Histopathology total scores and subscores of TcdB injected Calca KO mice, n = 8, 10 mice.
p-t, Histopathology total scores and subscores of TcdB injected Calcb KO mice, n = 3, 5 mice.
u, Representative histopathology of Tac1 KO mice injected with TcdB compared to Tac1 KO mice injected with TcdB and SP in the cecal TcdB injection model (120 min incubation).
v, Histopathology scores of experiments described in u, n = 3, 4 mice.
Assessed by Student’s T-test, two tailed, relative to WT control mice. Centre line: mean; error bars: SEM; n.s., not significant, p values are exact. n is described from left to right.
Extended Data Fig. 3: Histological scoring for CDI in the indicated KO mouse models as described in Fig. 1e-f.
a-e, Histopathology total scores and subscores of CDI in Tac1 KO mice, n = 11, 20 mice.
f-j, Histopathology total scores and subscores of CDI in Nk1r KO mice, n = 7, 10 mice.
k-o, Histopathology total scores and subscores of CDI in Calcb KO mice, n = 6, 9, 7 mice.
a-j, assessed by Student’s T-test, two tailed, relative to WT control mice. k-o, assessed by one-way ANOVA with post hoc Dunnett’s test. Centre line: mean; error bars: SEM; n.s., not significant, p values are exact. n is described from left to right.
Extended Data Fig. 4: FZD1/2/7 are expressed in DRG and enteric neurons.
a, Single cell RNA-seq analysis of FZD1, 2, and 7 expression in different cell types within colonic tissues in mice71. Size of dot indicates proportion expressing whereas the colour indicates expression level (red high, blue low). Putative excitatory motor neuron (PEMN), putative inhibitory motor neuron (PIMN), putative sensory neuron (PSN), putative sudomotor and vasodilator neurons (PSVN), putative interneuron (PIN).
b, FZD1, 2, and 7 expression in DRG neurons that innervate colonic tissues are displayed as violin plots72.
c, In situ hybridization of spinal cord sections shows enriched expression of FZD1/2/7 in dorsal root ganglia areas. Data are from Allen Brain Atlas73.
d, Phase-contrast images show that DRG neurons exposed to TcdB (1 pM, 60 min) exhibit no morphological changes. Scale bar is 50 μm. n=3 independent replicates.
e, RNAscope in situ hybridization showing localization of Pdgfrb expression (magenta) within Cspg4+ cells (cyan, Cspg4-dsRed mice) in colonic tissues. Panels in bottom row show regions in yellow boxed area at higher magnification. Scale bar: 50 μm and lower panel is 20 μm.
Extended Data Fig. 5: Cspg4+ pericytes wrap around blood vessels and form close contact with CGRP-containing afferent neurons.
a, Colon tissues from Cspg4-DsRed mice were harvested and subjected to whole mount immunofluorescence analysis, using an antibody against CD31 (a marker for blood vessel endothelial cells; magenta). DsRed (cyan) signals were detected directly. DsRed-positive cells surround CD31-positive capillaries in colonic crypts. Maximum intensity projection of the whole stack is presented in Figure 2e. Scale bar is 50 μm.
b, Experiments were carried out as in panel a, except that lymphatic endothelial cells were labeled with an antibody against Lyve-1 (magenta) and DsRed signals were detected using an antibody against DsRed. DsRed-positive cells are clearly separate from lymphatic endothelial cells. Scale bar is 50 μm.
c-f, Experiments were carried out as described in panel a. Enteric glial cells were visualized with GFP fluorescence in examining proteolipid protein 1 (PLP1)-GFP/-Cspg4-dsRed dual reporter mice (panel c). Fibroblasts were labeled using an antibody against platelet-derived growth factor receptor alpha (PGDFRA, magenta, panel d). Myofibroblasts were labeled using an antibody against smooth muscle actin (SMA, magenta, panel e). Mast cells and interstitial cells of Cajal were labeled using an antibody against cKit (magenta, panel f). Scale bar: 10 μm.
g, TcdB induced cell-rounding of cultured human brain vascular pericytes. Scale bar: 25 μm
h, The indicated TcdB mutants were injected into mouse ears via intradermal injection37. Pericytes surrounding ear arterioles were labeled and visualized through DsRed. TcdB and TcdB-FzM induced morphological changes of Cspg4+ pericytes surrounding ear arterioles, whereas TcdB-Cspg4M showed no effect. Scale bar, 50 μm. n= 3 mice/group.
i, Experiments were carried out as described in panel h, except that WT mice were utilized and pericytes were detected by immunostaining using an antibody against smooth muscle actin (labeling pericytes). TcdB-FzM disrupted pericytes around ear arterioles, whereas TcdB-Cspg4M has no effect. Scale bar: 50 μm. n= 3 mice/group.
j, Experiments were carried out as in panel a, except that neuronal processes were labeled using an antibody against Tubb3 (magenta, a marker for neuronal processes), showing that neuronal processes are extending alongside DsRed-positive pericytes. Scale bar: 10 μm.
k, Experiments were carried out as in panel a, except that an antibody against CGRP was added to detect CGRP-positive nerve terminals. 3-dimensional reconstruction of images showed that DsRed+ pericytes (cyan) surround the vasculature (CD31 endothelial marker; magenta) and contact CGRP-expressing nerve terminals (CGRP; yellow). Scale bar: 10 μm.
l, Immunoblot analysis of cell lysates showed expression of CGRP receptors (CALCRL: calcitonin receptor like receptor; and RAMP1: receptor activity modifying protein 1) in primary cultured human brain vascular pericytes. Two human cell lines, HeLa and U87, were analyzed in parallel as controls, which do not express detectable levels of CGRP receptors. Total protein staining with Coomassie blue was used as a control for protein loading. For gel source data, see Supplementary Figure 1. N = 2 replicates.
Extended Data Fig. 6: AAV-DTR mediates selective expression of DTR in ENS and DRG neurons in Tac1-Cre mice.
a, AAV-DTR was administered to Tac1-Cre mice via tail vein injection. Colon tissues were harvested 3-weeks later and subjected to whole mount immunofluorescence analysis. DRG were harvested and their cryosections were analyzed. HuC/D (magenta) marks neuron soma in colon tissues. An antibody against β3-tubulin (TUBB3, magenta) was utilized to label DRG neurons. DTR-GFP was labeled using an antibody against GFP (cyan). Scale for DRG: 100 μm; for myenteric plexus: 50 μm. Images were collected using a confocal microscope and maximal intensity projection is presented. GFP is directly fused to the C-terminus of DTR, thus cyan color represents DTR expression. n = 4 mice.
b, Experiments were carried out as described in upper panel. 5-HT (magenta) labels enterochromaffin cells in colonic tissues, showing no co-expression of DTR-GFP (cyan). Scale bars represent 100 μm (50 μm for right panel, which shows boxed region at higher magnification).
c-d, Experiments were carried out as described in Fig. 3e. Colonic tissues (panel c) and DRG (panel d) were isolated and subjected to analysis as described in panel a. WT mice not exposed to AAV-DTR were analyzed in parallel as a negative control (right panels). HuC/D labels myenteric plexus neurons within the proximal colon and TUBB3 labels DRG neurons of a wildtype mouse injected with AAV-DTR. No GFP positive cell bodies or fibers were detected in WT mice. Scale bar is 50 μm.
e-h, Sub-scoring of histopathology, related to Fig. 3f, n = 7, 6, 4 mice.
One-way ANOVAs were applied with post hoc Tukey’s test as indicated. Centre line: mean; error bars: SEM; n.s., not significant, p values are exact. n is described from left to right.
Extended Data Fig. 7: Extrinsic afferent neuron-dependent secretion of SP is necessary and sufficient for TcdB mediated pathology.
a-d; Histopathology subscoring of experiments following TcdB administration to RTX treated mice. Related to Fig. 3h, n = 10, 10 mice.
e-j, Histopathology of RTX treated/vehicle mice treated with TcdB alone or TcdB and SP for 120 min, demonstrating that SP can restore the effects of TcdB in RTX-treated mice. Representative histopathology in e, and histopathology scores f-j, n = 4, 4, 6, and 6 mice. The scale bar is 50 μm.
k, CGRP secretion from RTX treated mice is reduced in the cecal TcdB injection model, and is restored by co-administration of SP with TcdB. n = 2, 4, 6, 4, and 6 mice.
l-m, Nav1.8Cre/+ Rosa-LSL-Tdtomato mice have no evident Tomato expression (magenta) in the ENS (myenteric neurons marked with HuC/D, cyan) with only sparse Tomato+ nerve fibers from extrinsic neurons labeled in the colon (l). In contrast, there are many Tomato+ neurons in DRG (m). Scale bar: 50 μm.
n-q, Histopathology subscores of Nav1.8Cre/+, Rosa-LSL-DTR mice treated with GTD-iDT related to Fig 3i-j; n = 7, 9 mice.
For a-d and n-q, significance was assessed by student’s two-tailed t-test. For f-k, significance was assessed by one-way ANOVA with post hoc Dunnett’s test. Centre line: mean; error bars: SEM. p values are exact. n is described from left to right.
Extended Data Fig. 8: Inhibiting SP or CGRP signaling reduces severity of C. difficile colitis.
a, CDI models were carried out on WT versus Tac1 KO mice, using either C. difficile 630 Δerm or a toxin-null mutant strain (A-/B-). C. difficile CFUs from faeces were analyzed at 2 days post infection. The toxin-null strain showed reduced colonization on WT mice compared with the standard 630 Δerm, suggesting that the presence of toxins facilitates C. difficile colonization. In contrast, 630 Δerm showed similar levels of CFUs as the toxin-null strain in Tac1 KO mice, suggesting that the toxin’s contribution to C. difficile colonization relies on intact SP signaling. n= 7, 6, 4, 3 mice.
b, CDI models were carried out on WT versus Calcb KO mice (using C. difficile 630 Δerm). C. difficile CFUs from faeces showed reduced colonization in Calcb KO mice compared with WT mice, n= 4, 5 mice.
c, Representative H&E images for the experiments described in Fig. 4c. Scale bar is 50 μm.
d-h, Sub-categories of histopathological scoring of Fig. 4c; n = 15, 12, 11, 3.
h-l, Histopathology scoring of mice administered vehicle or aprepitant at the indicated times (0, 30 or 60 min following cecum injection and wound closure); n = 7, 6, 6, 7 mice.
Significance was assessed for a – Two-Way ANOVA with post hoc Sidak’s test and by T-test for b. One Way ANOVA with post hoc Dunnett’s test was used for d-l. Centre line: mean; error bars: SEM. p values are exact. n is described from left to right.
Extended Data Fig. 9: Inhibiting SP or CGRP reduces histopathological scores in cecum injection assays and in CDI models.
a, Representative H&E images for the experiment described in Fig. 4d. Scale bar is 50 μm.
b-e, Sub-categories of histopathological scoring of Fig. 4d, n = 13,7, 8, 8 mice.
f, C. difficile 630 Δerm fitness (measured by OD600) was not affected by treatment with aprepitant or olcegepant (20 μM) in vitro. Representative of 2 independent experiments (3 wells/group).
g-h, CDI experiments (with C. difficile 630) were carried out on WT and Tac1 KO mice (co-housed from weaning), followed by treatment with vehicle or aprepitant. C. difficile colonization (panel g) and histopathological scores (panel h) were shown, n = 7, 7, 6, 7 mice.
i-j, CDI experiments (with C. difficile 630) were carried out on WT and Calcb KO mice (littermates, co-housed from birth), followed by treatment with vehicle or olcegepant. C. difficile colonization (panel i) and histopathological scores (panel j) were shown. n = 4, 4, 5, 7 mice.
k, TcdA induced oedema in footpad injection assays in mice, which is prevented by pre-treatment with aprepitant. Boiled TcdA solution did not induce any oedema. n = 7,4, 3 mice.
Significance was assessed by one-way ANOVA with post hoc Dunnett’s test. Centre line: mean; error bars: SEM. n.s., not significant, p values are exact. n is described from left to right.
Extended Data Fig. 10: Inhibiting SP or CGRP reduces histopathological scores and colonization of hypervirulent strains that express TcdB2.
a, Representative H&E images for the experiments described in Fig. 4i. Scale bar is 50 μm.
b-e, Sub-categories of histopathological scoring of Fig. 4i, n = 3, 9, 7 mice.
f, Representative H&E images for the experiment described in Fig. 4j. Scale bar is 50 μm.
g-j, Sub-categories of histopathological scoring of Fig. 4j, n = 21, 10, 10, 12, 10, 5 mice.
Significance was assessed by one-way ANOVA with post hoc Dunnett’s test. Centre line: mean; error bars: SEM; n.s., not significant; p values are exact. n is described from left to right.
Supplementary Material
Acknowledgements
We thank members of the Dong and Rao labs for technical assistance and suggestions. This study was partially supported by grants from the National Institute of Health (NIH) (R01NS080833, R01NS117626, R01AI132387, and R01AI139087 to M.D.; K08DK110532, R01DK135707 and R01DK130836 to M.R; T32DK007477 for supporting M.A.M.; and R01 1R01HL150106 for K.Y.). G.A.K. was supported by an NDSEG Fellowship and A.S. by a Schmidt Science Fellowship.
We acknowledge support from the NIH-funded Harvard Digestive Disease Center (P30DK034854) and Boston Children’s Hospital Intellectual and Developmental Disabilities Research Center (P30HD18655). M.R. was supported by the Odyssey Award from the Richard and Susan Smith Family Foundation. M.D. holds the Investigator in the Pathogenesis of Infectious Disease award from the Burroughs Wellcome Fund. Figure 3c was created with BioRender.
Footnotes
Competing Interests
The authors declare no conflicts of interest.
Additional Information
Supplementary Information is available for this paper. Correspondence and requests for materials should be addressed to M.D.: min.dong@childrens.harvard.edu or M.R.: meenakshi.rao@childrens.harvard.edu. Reprints and permissions information is available at www.nature.com/reprints.
Data and material availability statement
All data generated or analyzed during this study are included in this published article (and its supplementary data and source data files). All biological materials are available upon request from the co-corresponding authors. The following datasets were utilized for this work:
Allen Brain Atlas (https://mousespinal.brain-map.org/)
SILVA https://www.arb-silva.de/no_cache/download/archive/release_132/
Mouse and human intestinal cell data from Broad Institute Single Cell portal accession: SCP1038
NCBI – For stromal data: GSE132465, GSE132257 and GSE144735
Colon innervating DRG neurons (https://hockley.shinyapps.io/ColonicRNAseq/) NCBI: GSE102962
References
- 1.Smits WK, Lyras D, Lacy DB, Wilcox MH & Kuijper EJ Clostridium difficile infection. Nat Rev Dis Primers 2, 16020 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guh AY et al. Trends in U.S. Burden of Clostridioides difficile Infection and Outcomes. N Engl J Med 382, 1320–1330 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kelly CP, Pothoulakis C. & LaMont JT Clostridium difficile colitis. N Engl J Med 330, 257–262 (1994). [DOI] [PubMed] [Google Scholar]
- 4.El Feghaly RE et al. Markers of intestinal inflammation, not bacterial burden, correlate with clinical outcomes in Clostridium difficile infection. Clin Infect Dis 56, 1713–1721 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fletcher JR et al. Clostridioides difficile exploits toxin-mediated inflammation to alter the host nutritional landscape and exclude competitors from the gut microbiota. Nat Commun 12, 462 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pruss KM & Sonnenburg JLC difficile exploits a host metabolite produced during toxin-mediated disease. Nature 593, 261–265 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yu H. et al. Cytokines Are Markers of the Clostridium difficile-Induced Inflammatory Response and Predict Disease Severity. Clin Vaccine Immunol 24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Solomon K. et al. Mortality in patients with Clostridium difficile infection correlates with host pro-inflammatory and humoral immune responses. J Med Microbiol 62, 1453–1460 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Kelly CP et al. Host Immune Markers Distinguish Clostridioides difficile Infection From Asymptomatic Carriage and Non-C. difficile Diarrhea. Clin Infect Dis 70, 1083–1093 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abhyankar MM et al. Immune Profiling To Predict Outcome of Clostridioides difficile Infection. mBio 11 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aktories K, Schwan C. & Jank T. Clostridium difficile Toxin Biology. Annu Rev Microbiol 71, 281–307 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Kordus SL, Thomas AK & Lacy DB Clostridioides difficile toxins: mechanisms of action and antitoxin therapeutics. Nat Rev Microbiol (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choi JE & Di Nardo A. Skin neurogenic inflammation. Semin Immunopathol 40, 249–259 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chiu IM, von Hehn CA & Woolf CJ Neurogenic inflammation and the peripheral nervous system in host defense and immunopathology. Nat Neurosci 15, 1063–1067 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang J. et al. Clostridium difficile toxins induce VEGF-A and vascular permeability to promote disease pathogenesis. Nat Microbiol 4, 269–279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castagliuolo I. et al. Neuronal involvement in the intestinal effects of Clostridium difficile toxin A and Vibrio cholerae enterotoxin in rat ileum. Gastroenterology 107, 657–665 (1994). [DOI] [PubMed] [Google Scholar]
- 17.Kelly CP et al. Neutrophil recruitment in Clostridium difficile toxin A enteritis in the rabbit. J Clin Invest 93, 1257–1265 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pothoulakis C. et al. CP-96,345, a substance P antagonist, inhibits rat intestinal responses to Clostridium difficile toxin A but not cholera toxin. Proc Natl Acad Sci U S A 91, 947–951 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Castagliuolo I. et al. Increased substance P responses in dorsal root ganglia and intestinal macrophages during Clostridium difficile toxin A enteritis in rats. Proc Natl Acad Sci U S A 94, 4788–4793 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Castagliuolo I. et al. Neurokinin-1 (NK-1) receptor is required in Clostridium difficile- induced enteritis. J Clin Invest 101, 1547–1550 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keates AC et al. CGRP upregulation in dorsal root ganglia and ileal mucosa during Clostridium difficile toxin A-induced enteritis. Am J Physiol 274, G196–202 (1998). [DOI] [PubMed] [Google Scholar]
- 22.Mantyh CR, McVey DC & Vigna SR Extrinsic surgical denervation inhibits Clostridium difficile toxin A-induced enteritis in rats. Neurosci Lett 292, 95–98 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Lyras D. et al. Toxin B is essential for virulence of Clostridium difficile. Nature 458, 1176–1179 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuehne SA et al. The role of toxin A and toxin B in Clostridium difficile infection. Nature 467, 711–713 (2010). [DOI] [PubMed] [Google Scholar]
- 25.Carter GP et al. Defining the Roles of TcdA and TcdB in Localized Gastrointestinal Disease, Systemic Organ Damage, and the Host Response during Clostridium difficile Infections. MBio 6, e00551 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell FA, King R, Smillie SJ, Kodji X. & Brain SD Calcitonin gene-related peptide: physiology and pathophysiology. Physiol Rev 94, 1099–1142 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tao L. et al. Frizzled proteins are colonic epithelial receptors for C. difficile toxin B. Nature 538, 350–355 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen P. et al. Structural basis for recognition of frizzled proteins by Clostridium difficile toxin B. Science 360, 664–669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen P. et al. Structural basis for CSPG4 as a receptor for TcdB and a therapeutic target in Clostridioides difficile infection. Nat Commun 12, 3748 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan P. et al. Chondroitin sulfate proteoglycan 4 functions as the cellular receptor for Clostridium difficile toxin B. Cell Res 25, 157–168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simonetti M. et al. Wnt-Fzd signaling sensitizes peripheral sensory neurons via distinct noncanonical pathways. Neuron 83, 104–121 (2014). [DOI] [PubMed] [Google Scholar]
- 32.Liu Z. et al. Structural basis for selective modification of Rho and Ras GTPases by Clostridioides difficile toxin B. Sci Adv 7, eabi4582 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Terada N. et al. Immunohistochemical study of NG2 chondroitin sulfate proteoglycan expression in the small and large intestines. Histochem Cell Biol 126, 483–490 (2006). [DOI] [PubMed] [Google Scholar]
- 34.Lee HO et al. Lineage-dependent gene expression programs influence the immune landscape of colorectal cancer. Nat Genet 52, 594–603 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Sweeney MD, Ayyadurai S. & Zlokovic BV Pericytes of the neurovascular unit: key functions and signaling pathways. Nat Neurosci 19, 771–783 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muhl L. et al. Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat Commun 11, 3953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Proebstl D. et al. Pericytes support neutrophil subendothelial cell crawling and breaching of venular walls in vivo. J Exp Med 209, 1219–1234 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hartmann DA et al. Brain capillary pericytes exert a substantial but slow influence on blood flow. Nat Neurosci 24, 633–645 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stark K. et al. Capillary and arteriolar pericytes attract innate leukocytes exiting through venules and ‘instruct’ them with pattern-recognition and motility programs. Nat Immunol 14, 41–51 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Saito M. et al. Diphtheria toxin receptor-mediated conditional and targeted cell ablation in transgenic mice. Nat Biotechnol 19, 746–750 (2001). [DOI] [PubMed] [Google Scholar]
- 41.Chan KY et al. Engineered AAVs for efficient noninvasive gene delivery to the central and peripheral nervous systems. Nat Neurosci 20, 1172–1179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai NY et al. Gut-Innervating Nociceptor Neurons Regulate Peyer’s Patch Microfold Cells and SFB Levels to Mediate Salmonella Host Defense. Cell 180, 33–49 e22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He M. et al. Emergence and global spread of epidemic healthcare-associated Clostridium difficile. Nat Genet 45, 109–113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lanis JM, Heinlen LD, James JA & Ballard JD Clostridium difficile 027/BI/NAP1 encodes a hypertoxic and antigenically variable form of TcdB. PLoS Pathog 9, e1003523 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mansfield MJ et al. Phylogenomics of 8,839 Clostridioides difficile genomes reveals recombination-driven evolution and diversification of toxin A and B. PLoS Pathog 16, e1009181 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shen E. et al. Subtyping analysis reveals new variants and accelerated evolution of Clostridioides difficile toxin B. Commun Biol 3, 347 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez-Urena D. et al. Toxin B Variants from Clostridium difficile Strains VPI 10463 and NAP1/027 Share Similar Substrate Profile and Cellular Intoxication Kinetics but Use Different Host Cell Entry Factors. Toxins (Basel) 11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chung SY et al. The Conserved Cys-2232 in Clostridioides difficile Toxin B Modulates Receptor Binding. Front Microbiol 9, 2314 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo J. et al. TFPI is a colonic crypt receptor for TcdB from hypervirulent clade 2 C. difficile. Cell 185, 980–994 e915 (2022). [DOI] [PubMed] [Google Scholar]
- 50.Schottelndreier D, Langejurgen A, Lindner R. & Genth H. Low Density Lipoprotein Receptor-Related Protein-1 (LRP1) Is Involved in the Uptake of Clostridioides difficile Toxin A and Serves as an Internalizing Receptor. Front Cell Infect Microbiol 10, 565465 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tao L. et al. Sulfated glycosaminoglycans and low-density lipoprotein receptor contribute to Clostridium difficile toxin A entry into cells. Nat Microbiol 4, 1760–1769 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mileto SJ et al. Clostridioides difficile infection damages colonic stem cells via TcdB, impairing epithelial repair and recovery from disease. Proc Natl Acad Sci U S A 117, 8064–8073 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Porat-Shliom N, Milberg O, Masedunskas A. & Weigert R. Multiple roles for the actin cytoskeleton during regulated exocytosis. Cell Mol Life Sci 70, 2099–2121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Meyer GK et al. Clostridium difficile toxins A and B directly stimulate human mast cells. Infect Immun 75, 3868–3876 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Geissler B, Tungekar R. & Satchell KJ Identification of a conserved membrane localization domain within numerous large bacterial protein toxins. Proc Natl Acad Sci U S A 107, 5581–5586 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brescia P. & Rescigno M. The gut vascular barrier: a new player in the gut-liver-brain axis. Trends Mol Med 27, 844–855 (2021). [DOI] [PubMed] [Google Scholar]
- 57.Spadoni I. et al. A gut-vascular barrier controls the systemic dissemination of bacteria. Science 350, 830–834 (2015). [DOI] [PubMed] [Google Scholar]
Additional References
- 58.Thompson BJ et al. Protective roles of alpha-calcitonin and beta-calcitonin gene-related peptide in spontaneous and experimentally induced colitis. Dig Dis Sci 53, 229–241 (2008). [DOI] [PubMed] [Google Scholar]
- 59.Song H. et al. Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc Natl Acad Sci U S A 109, 17531–17536 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu X, Bergles DE & Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development 135, 145–157 (2008). [DOI] [PubMed] [Google Scholar]
- 61.Harris JA et al. Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front Neural Circuits 8, 76 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cao YQ et al. Primary afferent tachykinins are required to experience moderate to intense pain. Nature 392, 390–394 (1998). [DOI] [PubMed] [Google Scholar]
- 63.Grako KA, Ochiya T, Barritt D, Nishiyama A. & Stallcup WB PDGF (alpha)-receptor is unresponsive to PDGF-AA in aortic smooth muscle cells from the NG2 knockout mouse. J Cell Sci 112 ( Pt 6), 905–915 (1999). [DOI] [PubMed] [Google Scholar]
- 64.Huang H. et al. Generation of a NK1R-CreER knockin mouse strain to study cells involved in Neurokinin 1 Receptor signaling. Genesis 54, 593–601 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nassar MA et al. Nociceptor-specific gene deletion reveals a major role for Nav1.7 (PN1) in acute and inflammatory pain. Proc Natl Acad Sci U S A 101, 12706–12711 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Buch T. et al. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods 2, 419–426 (2005). [DOI] [PubMed] [Google Scholar]
- 67.Madisen L. et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13, 133–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lyon MF & Glenister PH A new allele sash (Wsh) at the W-locus and a spontaneous recessive lethal in mice. Genet Res 39, 315–322 (1982). [DOI] [PubMed] [Google Scholar]
- 69.Zhang Y. et al. The role of purified Clostridium difficile glucosylating toxins in disease pathogenesis utilizing a murine cecum injection model. Anaerobe 48, 249–256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen X. et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology 135, 1984–1992 (2008). [DOI] [PubMed] [Google Scholar]
- 71.Drokhlyansky E. et al. The Human and Mouse Enteric Nervous System at Single-Cell Resolution. Cell 182, 1606–1622 e1623 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hockley JRF et al. Single-cell RNAseq reveals seven classes of colonic sensory neuron. Gut 68, 633–644 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lein ES et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 (2007). [DOI] [PubMed] [Google Scholar]
- 74.Rao M. et al. Enteric glia express proteolipid protein 1 and are a transcriptionally unique population of glia in the mammalian nervous system. Glia 63, 2040–2057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lau MT et al. Molecular dissection of box jellyfish venom cytotoxicity highlights an effective venom antidote. Nat Commun 10, 1655 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davies AJ et al. Natural Killer Cells Degenerate Intact Sensory Afferents following Nerve Injury. Cell 176, 716–728 e718 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Jank T, Giesemann T. & Aktories K. Clostridium difficile glucosyltransferase toxin B-essential amino acids for substrate binding. J Biol Chem 282, 35222–35231 (2007). [DOI] [PubMed] [Google Scholar]
- 78.Wilson KH, Kennedy MJ & Fekety FR Use of sodium taurocholate to enhance spore recovery on a medium selective for Clostridium difficile. J Clin Microbiol 15, 443–446 (1982). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rao C. et al. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature 591, 633–638 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhang J, Kobert K, Flouri T. & Stamatakis A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Quast C. et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41, D590–596 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bolyen E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37, 852–857 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article (and its supplementary data and source data files). All biological materials are available upon request from the co-corresponding authors. The following datasets were utilized for this work:
Allen Brain Atlas (https://mousespinal.brain-map.org/)
SILVA https://www.arb-silva.de/no_cache/download/archive/release_132/
Mouse and human intestinal cell data from Broad Institute Single Cell portal accession: SCP1038
NCBI – For stromal data: GSE132465, GSE132257 and GSE144735
Colon innervating DRG neurons (https://hockley.shinyapps.io/ColonicRNAseq/) NCBI: GSE102962