Abstract
Background: Hepatocellular carcinoma (HCC) is the most common and fatal primary liver cancer. Genetic variants of DNA repair systems can reduce DNA repair capability and increase HCC risk. Objectives: This study aimed to examine, in Egyptian hepatitis C virus (HCV) patients, the relationship between the X-ray repair cross-complementing group 1 (XRCC1) rs1799782 single nucleotide polymorphism (SNP) and HCC susceptibility. Methods: We included 100 adult HCV-positive patients with HCC and 100 adult HCV-positive patients with liver cirrhosis as pathological controls. XRCC1 rs1799782 SNP genotyping was done in both groups using quantitative real-time PCR (qPCR). The distribution of genotypes in patients and controls was compared using several inheritance models. Results: We found that the CT genotype, when analyzed under both the co-dominant (OR (95 % CI): 2.147 (1.184-3.893), p = .012) and the over-dominant (OR (95 % CI): 2.055 (1.153-3.660), p = .015) models, as well as the combined CT and TT genotypes under the dominant model (OR (95 % CI) of 1.991 (1.133-3.497), p = .017), were associated with increased susceptibility to HCC. The frequency of the T allele was higher among HCC participants (32%) compared to those with cirrhosis (23.5%) and carrying the T allele increased the risk of HCC by 1.532 times, however, these associations did not reach statistical significance (p-values >0.05). Moreover, the variant T allele was associated with worse clinical manifestations and laboratory results among the HCC group, but AFP levels were not affected significantly. Conclusions: Egyptians with XRCC1 rs1799782 SNP may have a higher risk of HCV-related HCC. More extensive multi-center prospective investigations must confirm this association.
Keywords: Egyptian, hepatitis C virus, hepatocellular carcinoma, single nucleotide polymorphism, XRCC1
Introduction
One of the worst cancers in humans is hepatocellular carcinoma (HCC), the most prevalent and severe primary liver cancer. 1 It ranks the sixth most frequent type of cancer globally and the fourth most frequent cancer in Egypt. 2 Chronic viral hepatitis is the most common risk factor for HCC globally, with chronic hepatitis C virus (HCV) infection being the leading cause in northern Africa. 3 HCV induces HCC by promoting DNA damage through several processes, including the induction of chronic inflammation and oxidative stress. 4 Therefore, DNA repair machinery is essential for genomic integrity. If the rate of DNA damage exceeds the DNA repair capacity, cells will undergo apoptosis and senescence or acquire chromosomal abnormalities that result in genomic instability and cancers. 5 Multiple DNA repair system genetic variations like single nucleotide polymorphism (SNP) may alter the protein’s structure or function, resulting in decreased DNA repair capacity and increased HCC risk. Understanding these SNPs’ relationship to the risk of HCC may aid in the development of preventative and therapeutic measures for the disease. 6 The gene coding for the X-ray repair cross-complementing group 1 (XRCC1) is located on chromosome 19q13.2 with 17 exons and 32 kb genetic distance. It encodes a critical DNA repair protein for base excision and single-strand break repair. 7 One of the most common XRCC1 gene SNPs is the rs1799782 SNP, also known as Arg194Trp, in which cytosine (C) base is substituted for a thymine (T) base at codon 194 of exon 6 at position 26,304, causing translation into a different amino acid (arginine to tryptophan). 8 Several studies explored the relationship between this SNP and the vulnerability to HCC, but the findings were conflicting.9–11 Thus, the goal of this study was to examine, using various inheritance models, the relationship between the risk of HCC and the XRCC1 rs1799782 gene SNP in Egyptian HCV patients.
Methodology
Study subjects
This pilot case-control study, with ethical approval number FMASU R352/2023, included 100 adult HCV-positive patients with HCC and 100 adult HCV-positive patients with liver cirrhosis as pathological controls. In all participants, HCV infection was confirmed by quantitative real-time PCR (qPCR) using Rotor-Gene Q (QIAGEN, Inc., Hilden, Germany). All participants (n = 200) had active HCV infection without previous successful direct-acting antiviral (DAA) therapy. Patients with positive HBV infection or having metabolic, autoimmune, fatty, or alcoholic liver disease were excluded from the study. Before the study began, all participants provided signed informed consent, and all information was kept private and confidential and used exclusively for research.
Clinical assessment
All participants were subjected to a full clinical examination, especially for features of hepatocellular failure (e.g.: ascites, encephalopathy, jaundice, and edema). HCC was diagnosed by triphasic CT and/or dynamic MRI, and HCC staging was based on The Barcelona Clinic Liver Cancer Staging System (BCLC). Assessment of the extent of liver dysfunction and patients’ prognosis was done using the Child-Pugh scoring system together with Milan criteria to identify patients who were eligible for transplantation. 12
Blood samples and laboratory investigations
Four blood samples were collected from each participant aseptically. The first blood sample (2 mL) was collected into a gel vacutainer tube and was centrifuged at 4000 r/min for 20 min after complete blood clotting. The resultant serum was used for measurement of aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin, and direct bilirubin using the Cobas 6000 analyzer series, c 501 module (Roche Diagnostics, Switzerland), as well as assessment of alpha-fetoprotein (AFP), HCV antibodies and HBsAg using the Cobas e411 autoanalyzer (Roche Diagnostics, Switzerland).
The second blood sample (2 mL) was withdrawn on a 3.2% sodium citrate vacutainer tube and was centrifuged at 4000 r/min for 15 min. The resultant plasma was used to measure prothrombin time (PT), partial thromboplastin time (PTT), and international normalized ratio (INR) using the Sysmex CS-2500 System (Siemens Healthineers, USA).
The last two blood samples were collected into two ethylenediaminetetraacetic acid dipotassium salt (K2-EDTA) vacutainer tubes; one blood sample (2 mL) was used for complete blood count (CBC) analysis using the Sysmex XN-1000 six-part differential hematology analyzer (Sysmex, Bornbarch, Germany) and the other sample (2 mL) was stored at −80⸰C until XRCC1 rs1799782 SNP genotyping using qPCR.
PCR and genotyping analysis
DNA extraction
Genomic DNA was extracted from whole blood using the QIAamp DNA blood mini kit (QIAGEN, Inc., Hilden, Germany) in compliance with the manufacturer’s directions. Measurement of extracted DNA concentration was done using Nanodrop One Spectrophotometer (Thermo Fisher Scientific Inc, USA) at 260 nm, while DNA purity was assessed by measurement of the ratio of absorbance at 260 and 280 nm. The accepted ratio was neraly 1.8.
XRCC1 rs1799782 SNP genotyping
TaqMan Universal Master Mix (Applied Biosystems, USA) and TaqMan® SNP genotyping assay kit (cat.no. 4351379, assay ID: C__11463404_10) (Applied Biosystems, USA) were used for XRCC1 rs1799782 SNP genotyping by qPCR. The final reaction volume was twenty μL, set as follows:
• Ten μL of TaqMan Universal Master Mix
• One μL of TaqMan assay (20x)
• Seven μL of nuclease-free water
• Two μL of the extracted DNA
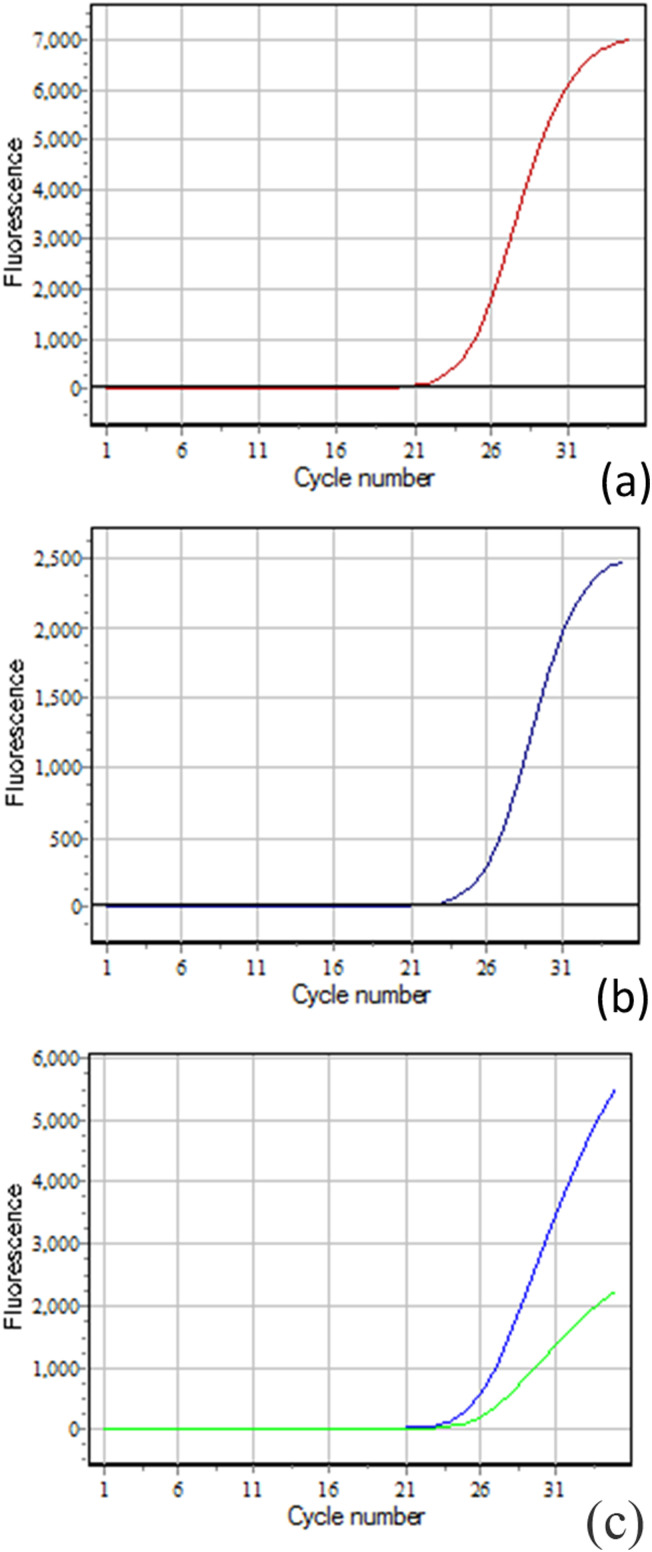
DT-Lite Real-Time PCR system (DNA technology, Russia) was used for amplification according to the following protocol: initial activation at 95°C for 10 min, then 40 cycles of denaturation at 95°C for 15 s and annealing/extension at 60°C for 60 s. Finally, allelic discrimination was done as shown in Figure 1.
Figure 1.
Demonstration of fluorescence curves of the three genotypes. Allele C (wild allele) (FAM dye) and allele T (variant allele) (HEX dye); (a) a significant increase in FAM dye fluorescence indicates the homozygous CC genotype, (b) a significant increase in HEX dye fluorescence indicates the homozygous TT genotype, and (c) a significant increase in both HEX and FAM dye fluorescence indicates the heterozygous CT genotype.
Statistical analysis
Version 26.0 of the Statistical Package for Social Science (IBM Corp., Armonk, NY, USA) was used to analyze the data. For non-parametric numerical data, medians and interquartile ranges (IQR) were employed in descriptive statistics; for non-numerical data, frequencies and percentages were used. To determine the difference between non-parametric variables in two groups, the Mann-Whitney Test (U test) was employed. The difference between more than two groups was assessed using the Kruskal-Wallis test. 13 The association between two qualitative variables was investigated using the Chi-Square test. The relationship between the XRCC1 rs1799782 gene SNP and the risk of HCC was evaluated using odds ratios (OR). To evaluate the accuracy of the estimations, the allowable margin of error was set to 5%, while the confidence interval (CI) was set to 95%. Consequently, the p-value was considered statistically significant if < 0.05.
Results
Features of the study groups
This case-control study included 100 adult HCV-positive patients with HCC and 100 matching adult HCV-positive patients with liver cirrhosis as pathological controls. The mean (±SD) age of the included participants was 56.25 (±7.80) years, and the male-to-female ratio was 2:1 in each group. The study groups’ clinical features are compared in Table 1.
Table 1.
Comparison of the clinical characteristics of the study groups.
| Group | Chi-square | ||||||
|---|---|---|---|---|---|---|---|
| HCC n = 100 | Cirrhosis n = 100 | ||||||
| n | % | n | % | X2 | p-value | ||
| Ascites | None | 36 | 36.00 | 23 | 23.00 | 11.333 | 0.010* |
| Mild | 10 | 10.00 | 24 | 24.00 | |||
| Moderate | 22 | 22.00 | 30 | 30.00 | |||
| Massive | 32 | 32.00 | 23 | 23.00 | |||
| Encephalopathy | None | 82 | 82.00 | 71 | 71.00 | 12.408 | 0.015* |
| Grade I | 3 | 3.00 | 16 | 16.00 | |||
| Grade II | 3 | 3.00 | 5 | 5.00 | |||
| Grade I-II | 10 | 10.00 | 8 | 8.00 | |||
| Grade III | 2 | 2.00 | 0 | 0.00 | |||
| Child-Pugh score | Child A | 21 | 21.00 | 13 | 13.00 | 7.348 | 0.025* |
| Child B | 51 | 51.00 | 41 | 41.00 | |||
| Child C | 28 | 28.00 | 46 | 46.00 | |||
| Jaundice | Positive | 59 | 59.00 | 59 | 59.00 | 0.000 | 1.000 |
| Negative | 41 | 41.00 | 41 | 41.00 | |||
| Edema | Positive | 64 | 64.00 | 75 | 75.00 | 2.854 | 0.091 |
| Negative | 36 | 36.00 | 25 | 25.00 | |||
| Milan criteria | Within Milan | 38 | 38.00 | - | - | - | - |
| Beyond Milan | 62 | 62.00 | - | - | |||
| BCLC staging | Stage 0 | 5 | 5.00 | - | - | - | - |
| Stage A | 16 | 16.00 | - | - | |||
| Stage B | 30 | 30.00 | - | - | |||
| Stage C | 21 | 21.00 | - | - | |||
| Stage D | 28 | 28.00 | - | - | |||
* p-value <0.05 indicates statistical significance.
Compared to the cirrhosis group as regards the laboratory data, the HCC group showed significantly higher median (IQR) values of AFP (p < .001), ALT (p = .002), AST (p = .007), platelets (p = .009), and albumin (p = .002) and significantly lower values of PT, PTT, and INR (p < .001). There were no significant differences between the study groups as regards the median (IQR) values of hemoglobin (p = .981), total leukocytic count (p = .250), total protein (p = .593), total bilirubin (p = .604) and direct bilirubin (p = .406). Table 2.
Table 2.
Comparison of the laboratory data of the study groups.
| Group | Mann-Whitney test | |||||
|---|---|---|---|---|---|---|
| HCC n = 100 | Cirrhosis n = 100 | |||||
| Median | IQR | Median | IQR | Z | p-value | |
| AFP (IU/ml) | 20.10 | 13.20-83.00 | 1.60 | 1.31-2.20 | 12.154 | <0.001* |
| PTT (seconds) | 42.00 | 39.00-46.00 | 46.00 | 40.00-54.00 | 3.759 | <0.001* |
| PT (seconds) | 18.88 | 15.30-21.59 | 20.10 | 17.70-31.40 | 4.102 | <0.001* |
| INR | 1.46 | 1.28-1.70 | 1.68 | 1.40-2.50 | 5.192 | <0.001* |
| Hemoglobin (gm/dl) | 10.40 | 9.30-11.80 | 10.50 | 9.80-11.50 | 0.023 | 0.981 |
| Platelets (×103/µl) | 141.00 | 115.50-226.00 | 135.00 | 72.00-162.50 | 2.606 | 0.009* |
| Total leukocytic count (×103/µl) | 8.50 | 6.15-13.00 | 7.20 | 5.50-11.80 | 1.150 | 0.250 |
| Total protein (g/dl) | 6.30 | 5.60-6.80 | 6.40 | 5.70-6.90 | 0.535 | 0.593 |
| Albumin (g/dl) | 2.90 | 2.40-3.40 | 2.60 | 2.40-2.90 | 3.079 | 0.002* |
| ALT (IU/L) | 25.00 | 17.50-46.00 | 21.00 | 15.00-27.00 | 3.086 | 0.002* |
| AST (IU/L) | 41.00 | 29.00-89.00 | 36.00 | 26.00-42.00 | 2.674 | 0.007* |
| Total bilirubin (mg/dl) | 2.20 | 1.25-4.90 | 2.70 | 1.50-4.10 | 0.518 | 0.604 |
| Direct bilirubin (mg/dl) | 1.60 | 0.50-3.20 | 1.80 | 0.80-2.30 | 0.832 | 0.406 |
* p-value <0.05 indicates statistical significance.
XRCC1 rs1799782 genotype distribution
The co-dominant model showed a significant difference (p = .041) in the distribution of XRCC1 rs1799782 genotypes between the study groups, with the CT (48%) genotype being the most frequent among the HCC group and the CC (61%) genotype among the cirrhosis group. As regards the association with the risk of HCC, the CT genotype was associated with a significant (p = .012) OR (95 % CI) of 2.147 (1.184-3.893), while the TT genotype was associated with a non-significant (p = .544) OR (95 % CI) of 1.386 (0.483–3.977).
When the CT and TT genotypes were added together in the dominant model, they showed a significant (p = .017) association with the risk of HCC with an OR (95 % CI) of 1.991 (1.133–3.497), the HCC group showed a significant (p = .016) higher frequency of CT and TT genotypes grouped together (56%) compared to the cirrhosis group (39%).
When using the recessive model, the TT genotype showed no significant difference in the distribution between the HCC and cirrhosis groups (p = 1.000) and a non-significant (p = 1.000) risk for HCC with an OR (95 % CI) of 1.000 (0.360-2.778).
On the other hand, when using the over-dominant model, the heterozygous CT genotype showed a significantly (p = .014) higher frequency among the HCC group (48%) compared to the cirrhosis group (31%) and showed a significant (p = .015) OR (95 % CI) of 2.055 (1.153-3.660). Table 3.
Table 3.
Comparison of XRCC1 rs1799782 genotype frequency of the study groups.
| Model | Group | Chi-square | OR (95 % CI) | p-value | |||||
|---|---|---|---|---|---|---|---|---|---|
| HCC n = 100 | Cirrhosis n = 100 | ||||||||
| n | % | n | % | X2 | p-value | ||||
| Co-dominant | CC | 44 | 44.00 | 61 | 61.00 | 6.411 | 0.041* | - | |
| CT | 48 | 48.00 | 31 | 31.00 | 2.147 (1.184-3.893) | 0.012* | |||
| TT | 8 | 8.00 | 8 | 8.00 | 1.386 (0.483-3.977) | 0.544 | |||
| Dominant | CC | 44 | 44.00 | 61 | 61.00 | 5.794 | 0.016* | - | |
| CT + TT | 56 | 56.00 | 39 | 39.00 | 1.991 (1.133-3.497) | 0.017* | |||
| Recessive | CC + CT | 92 | 92.00 | 92 | 92.00 | 0.000 | 1.000 | - | |
| TT | 8 | 8.00 | 8 | 8.00 | 1.000 (0.360-2.778) | 1.000 | |||
| Over-dominant | CC + TT | 52 | 52.00 | 69 | 69.00 | 6.047 | 0.014* | - | |
| CT | 48 | 48.00 | 31 | 31.00 | 2.055 (1.153-3.660) | 0.015* | |||
* p-value <0.05 indicates statistical significance.
XRCC1 rs1799782 allele distribution
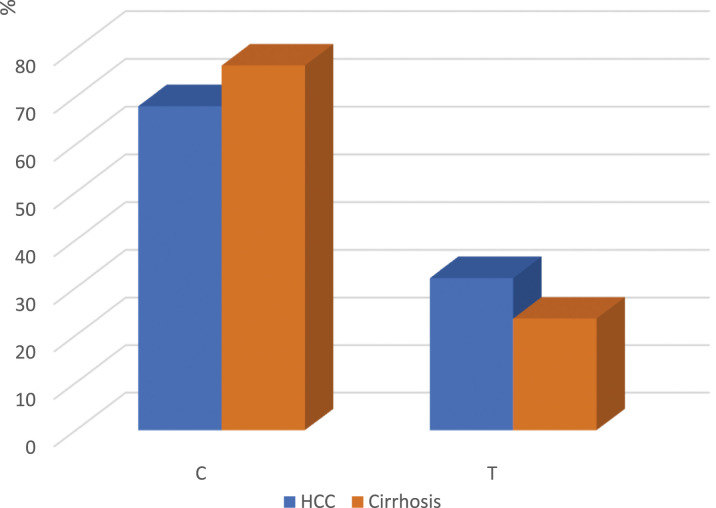
The XRCC1 rs1799782 allele frequency distribution showed no significant differences (p = .074) between the HCC and cirrhosis groups. The C allele frequency was 68% (136/200) and 76.5% (153/200), while the T allele frequency was 32% (64/200) and 23.5% (47/200) in the HCC and cirrhosis groups, respectively with a non-significant OR (95 % CI) of 1.532 (0.985-2.383) (p = .058). Figure 2.
Figure 2.
Bar chart showing XRCC1 rs1799782 allele frequency in hepatocellular carcinoma (HCC) group and liver cirrhosis (pathological control) group (p = .074).
Clinical characteristics of the XRCC1 rs1799782 genotypes among HCC participants
The clinical characteristics of the HCC participants were compared according to the XRCC1 rs1799782 genotypes and showed significant differences.
Regarding the Milan criteria, beyond Milan criteria was found in 52.27% of HCC participants with the CC genotype, 64.58% of HCC participants with the CT genotype, and 100% of HCC participants with the TT genotype (p = .033).
Regarding ascites, most of the HCC participants with the CC genotype had no (65.91%) or mild (15.91%) ascites. In comparison, most of the HCC participants with the CT genotype had massive ascites (50%) or moderate (29.17%) ascites. On the other hand, HCC participants with the TT genotype only had massive (62.50%) or moderate (7.50%) ascites (p < .001).
Regarding encephalopathy, all HCC participants with the CC genotype (100%) and most HCC participants with the CT genotype (72.92%) had no encephalopathy. Of the HCC participants with TT genotype, 37.50% had no encephalopathy, 37.50% had grade I-II, and 25% had grade III (p < .001).
Regarding the Child-Pugh scores, none of the HCC participants with the CC genotype had Child C, and most of them were Child B (61.36%). Of the HCC participants with the CT genotype, 50% were Child B, 41.67% were Child C, and only 8.33% were Child A. On the other hand, all HCC participants with the TT genotype were Child C (p < .001).
Regarding the BCLC staging, most of the HCC participants with the CC genotype had stage B (52.27%) or stage A (36.36%), while most of the HCC participants with the CT genotype had stage C (43.75%) or stage D (41.67%). All HCC participants with the TT genotype had stage D (p < .001).
Regarding jaundice and edema, most of the HCC participants with the CC genotype had no jaundice (56.80%) or edema (72.23%). On the other hand, most of the HCC participants with the CT genotype and all HCC participants with the TT genotype had jaundice and edema (66.70%, 100% and 91.67%, 100%, respectively) (p = .004, <0.001, respectively). Table 4.
Table 4.
Comparison of the clinical characteristics of the HCC group (n = 100) according to XRCC1 rs1799782 genotypes (co-dominant model).
| HCC | XRCC1 rs1799782 Co-dominant | Chi-square | |||||||
|---|---|---|---|---|---|---|---|---|---|
| CC n = 44 | CT n = 48 | TT n = 8 | |||||||
| n | % | n | % | n | % | X2 | p-value | ||
| Milan criteria | Within Milan | 21 | 47.73 | 17 | 35.42 | 0 | 0.00 | 6.806 | 0.033* |
| Beyond Milan | 23 | 52.27 | 31 | 64.58 | 8 | 100.00 | |||
| Ascites | None | 29 | 65.91 | 7 | 14.58 | 0 | 0.00 | 43.102 | <0.001* |
| Mild | 7 | 15.91 | 3 | 6.25 | 0 | 0.00 | |||
| Moderate | 5 | 11.36 | 14 | 29.17 | 3 | 37.50 | |||
| Massive | 3 | 6.82 | 24 | 50.00 | 5 | 62.50 | |||
| Encephalopathy | None | 44 | 100.00 | 35 | 72.92 | 3 | 37.50 | 45.112 | <0.001* |
| Grade I | 0 | 0.00 | 3 | 6.25 | 0 | 0.00 | |||
| Grade II | 0 | 0.00 | 3 | 6.25 | 0 | 0.00 | |||
| Grade I-II | 0 | 0.00 | 7 | 14.58 | 3 | 37.50 | |||
| Grade III | 0 | 0.00 | 0 | 0.00 | 2 | 25.00 | |||
| Child-Pugh score | Child A | 17 | 38.64 | 4 | 8.33 | 0 | 0.00 | 47.214 | <0.001* |
| Child B | 27 | 61.36 | 24 | 50.00 | 0 | 0.00 | |||
| Child C | 0 | 0.00 | 20 | 41.67 | 8 | 100.00 | |||
| BCLC staging | Stage 0 | 5 | 11.36 | 0 | 0.00 | 0 | 0.00 | 93.289 | <0.001* |
| Stage A | 16 | 36.36 | 0 | 0.00 | 0 | 0.00 | |||
| Stage B | 23 | 52.27 | 7 | 14.58 | 0 | 0.00 | |||
| Stage C | 0 | 0.00 | 21 | 43.75 | 0 | 0.00 | |||
| Stage D | 0 | 0.00 | 20 | 41.67 | 8 | 100.00 | |||
| Jaundice | Positive | 19 | 43.20 | 32 | 66.70 | 8 | 100.00 | 11.277 | 0.004* |
| Negative | 25 | 56.80 | 16 | 33.30 | 0 | 0.00 | |||
| Edema | Positive | 12 | 27.27 | 44 | 91.67 | 8 | 100.00 | 46.207 | <0.001* |
| Negative | 32 | 72.73 | 4 | 8.33 | 0 | 0.00 | |||
* p-value <0.05 indicates statistical significance.
Laboratory characteristics of the XRCC1 rs1799782 genotypes among HCC participants
The laboratory data of the HCC participants were compared according to XRCC1 rs1799782 genotypes. No significant differences between genotypes were found as regards median (IQR) values of AFP (p = .446), PTT (p = .472), PT (p = .078), hemoglobin (p = .963), TLC (p = .145), ALT (p = .277), and AST (p = .473). On the other hand, INR values showed a significant increase (p = .022) in HCC participants with the CT genotype compared to those with the CC genotype. In addition, compared to HCC participants with the CC genotype, platelet values showed a significant decrease in HCC participants with the CT (p = .001) and TT (p = .021) genotypes. Regarding total protein values, they showed a significant decrease in HCC participants with the CT (p = .049) and TT (p = .001) genotypes compared to those with the CC genotype. Also, total protein values showed a significant decrease (p = .040) in HCC participants with the TT genotype compared to those with the CT genotype. Similarly, albumin values showed a significant decrease in HCC participants with the CT (p < .001) and TT (p = .006) genotypes compared to those with the CC genotype. Finally, total and direct bilirubin values showed a significant increase in HCC participants with the TT genotype compared to those with the CC (p = .002 and .021, respectively) and CT (p < .001 and = 0.002, respectively) genotypes. Table 5.d
Table 5.
Comparison of the laboratory data of the HCC group (n = 100) according to XRCC1 rs1799782 genotypes (co-dominant model).
| HCC | XRCC1 rs1799782 Co-dominant | Kruskal-Wallis test | Mann-Whitney test | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| CC | CT | TT | |||||||||
| Median | IQR | Median | IQR | Median | IQR | X2 | p-value | CC&CT | CC&TT | CT&TT | |
| AFP (IU/ml) | 18.10 | 14.00-29.00 | 35.00 | 13.10-213.00 | 15.00 | 13.20-50.00 | 1.615 | 0.446 | |||
| PTT (seconds) | 42.50 | 40.00-46.00 | 42.00 | 40.00-47.00 | 38.00 | 35.00-55.00 | 1.503 | 0.472 | |||
| PT (seconds) | 18.20 | 15.75-20.66 | 20.30 | 15.25-23.79 | 21.59 | 15.24-22.85 | 5.092 | 0.078 | |||
| INR | 1.40 | 1.27-1.55 | 1.56 | 1.29-1.90 | 1.90 | 1.20-2.10 | 6.149 | 0.046* | 0.022* | 0.153 | 0.466 |
| Hemoglobin (gm/dl) | 10.40 | 9.30-11.80 | 10.15 | 9.70-11.90 | 10.40 | 9.75-11.10 | 0.076 | 0.963 | |||
| Platelets (×103/µl) | 172.50 | 138.50-255.00 | 121.00 | 79.00-189.00 | 135.00 | 130.50-142.00 | 13.004 | 0.002* | 0.001* | 0.021* | 0.639 |
| Total leukocytic count (×103/µl) | 8.15 | 6.65-9.95 | 9.30 | 5.85-13.30 | 11.20 | 8.85-13.90 | 3.860 | 0.145 | |||
| Total protein (g/dl) | 6.50 | 6.15-6.90 | 6.00 | 5.40-6.70 | 5.50 | 5.00-6.10 | 11.373 | 0.003* | 0.049* | 0.001* | 0.040* |
| Albumin (g/dl) | 3.10 | 2.95-3.60 | 2.50 | 2.20-3.00 | 2.70 | 2.65-2.90 | 21.845 | <0.001* | <0.001* | 0.006* | 0.204 |
| ALT (IU/L) | 25.00 | 21.00-55.00 | 25.00 | 16.00-42.00 | 35.00 | 29.50-38.00 | 2.568 | 0.277 | |||
| AST (IU/L) | 55.00 | 31.00-89.00 | 38.50 | 25.00-76.50 | 33.00 | 32.00-98.00 | 1.498 | 0.473 | |||
| Total bilirubin (mg/dl) | 1.80 | 0.90-5.15 | 2.60 | 1.40-4.00 | 7.20 | 6.85-8.00 | 13.645 | 0.001* | 0.511 | 0.002* | <0.001* |
| Direct bilirubin (mg/dl) | 0.90 | 0.30-4.40 | 1.70 | 0.50-2.60 | 4.50 | 2.40-6.60 | 8.755 | 0.013* | 0.301 | 0.021* | 0.002* |
* p-value <0.05 indicates statistical significance.
Discussion
The pathogenesis of HCC is a multifactorial and complex process influenced by both environmental and genetic variables. Exploring those variables may help clarify the several factors that contribute to the development of liver cancer, enhancing screening practices for individuals at high risk and developing targeted therapy. 14 Oxidative stress, in which elevated amounts of reactive oxygen species destroy cell nucleic acid, is the primary factor influencing HCV-induced HCC. A malfunction in the DNA repair machinery can lead to unrepaired DNA damage, resulting in genomic instability and making liver cells malignant, predisposing for HCC. 15 XRCC1 is a major DNA repair gene involved in the base excision repair (BER) pathway, which is the primary defense against endogenous agents that damage DNA, such as viruses. 16
In the current study, we aimed to investigate the link between XRCC1 rs1799782 SNP and the risk of HCC. We identified an association between the CT genotype in the co-dominant and over-dominant models and the CT and TT genotypes grouped together in the dominant model with the susceptibility to HCC. Although the T allele frequency was higher in HCC participants compared to those with cirrhosis, and carrying the T allele increased the risk of HCC by 1.532 times, neither reached statistical significance, which could be attributed to the relatively small sample size in the current study. Moreover, in the current study, harboring the variant T allele was associated with worse clinical manifestations of HCC. In addition, the laboratory parameters were affected by the presence of the variant T allele in HCC participants, e.g., a decrease in platelets, total protein, and albumin and an increase in INR, total bilirubin, and direct bilirubin values; AFP levels, on the other hand, did not differ significantly.
The association of XRCC1 rs1799782 SNP and the risk of HCC could be explained by the fact that DNA ligase III, poly (ADP-ribose) polymerase (PARP), and DNA polymeraseβ (polyβ) form DNA repair complexes with the XRCC1 wild-type protein. When XRCC1 rs1799782 SNP occurs, it modifies the wild-type protein structure and function, thus altering XRCC1 DNA repair capacity and putting one at risk for metastasis or recurrence, shortened survival, resistance to chemotherapy, and carcinogenesis. 17
Several studies have assessed the link between XRCC1 rs1799782 SNP and the risk of HCC; however, conflicting findings were found across populations. A meta-analysis by Merchant et al., 2023, found a statistically significant association between XRCC1 rs1799782 SNP and the risk of HCC in both Asian and Caucasian populations, according to subgroup analysis based on ethnic backgrounds. 11 Similarly, Mattar et al., 2018, reported that XRCC1 rs1799782 SNP was significantly associated with a 2.14-fold increased risk of HCC development among the Egyptian population compared to healthy controls but did not find a significant difference in AFP levels according to XRCC1 rs1799782 genotypes. 4 Moreover, Kiran et al., 2009, reported that XRCC1 rs1799782 SNP was significantly associated with a 2.27-fold increased risk of HCC in Indian patients with hepatitis. They also discovered that the combined heterozygous genotypes Arg194Trp + Arg280His among the normal controls showed a significant cumulative positive correlation for developing HCC. Still, they did not find a similar correlation in chronic viral hepatitis participants, as chronic viral hepatitis infection could allow cells to evade some of the typical multisteps that genetic changes cause, leading to hepatocarcinogenesis. 10 Also, Guo et al., 2012, reported that the XRCC1 rs1799782 TT genotype was significantly associated with the risk of HCC compared to the CC genotype in the Chinese population. 18
According to different genetic models, Ghaderi-Zefrehi et al., 2021, reported the overall analysis for XRCC1 rs1799782 SNP revealed a significant association with increased HCC susceptibility under the allelic, dominant, heterozygote, and homozygote genetic models. 6 Another study by Mandal and Mittal, 2021, demonstrated that only the variant T allele was associated with an increased risk of overall cancer by 1.301. Still, the heterozygous, homozygous, dominant, and recessive genetic models did not indicate any statistical association. 19 In addition, in a meta-analysis of 13 studies evaluating the XRCC1 rs1799782 SNP in HCC, the homozygous, heterozygous, dominant, and recessive genetic models showed no significant association with the susceptibility to HCC. 9
Contrasting our results, Zeng et al., 2010, suggested that XRCC1 rs1799782 SNP does not predispose to HCC but interacts with HCC risk factors like smoking and chronic HBV infection, 20 suggesting that gene-environment interactions can influence the effect of XRCC1 rs1799782 SNP on HCC risk. Similarly, Yang and Zhao, 2015, reported that XRCC1 rs1799782 genotypes did not show any significant differences in distribution between HCC and controls and were not associated with the risk of HCC. Instead, they reported that the XRCC1 rs1799782 CT and TT genotypes were associated with the risk of chemotherapy resistance. 21 Several other studies and meta-analyses suggested no association between XRCC1 rs1799782 SNP and the risk of HCC in different populations.22–25
The heterogeneity between studies could be attributed to sample selection bias due to small sample sizes in most studies and the differences in the selected control, ethnicity of the included participants, family genetic background, and the molecular technique used for XRCC1 rs1799782 genotyping. Moreover, the T allele of XRCC1 rs1799782 SNP may be in linkage disequilibrium with other polymorphisms to predispose to HCC.
This study had several limitations; it was a single-center pilot study with a limited sample size and lacked patient follow-up, particularly regarding their medication regimens. Additionally, the study did not account for other potential confounding factors that could have influenced the outcomes. Therefore, to draw definitive conclusions, further prospective multicenter studies with adequate sample size determination, extended patient follow-up, and consideration of confounding factors are recommended. Additionally, validation experiments using DNA sequencing are necessary to confirm the findings.
Conclusions
In conclusion, the XRCC1 rs1799782 SNP may be linked to an increased risk of developing HCV-related HCC in the Egyptian population, particularly with a more severe clinical presentation. However, the definitive confirmation of this association requires validation through large-scale, multi-center prospective studies. Establishing this association could aid in the early identification of HCV patients at risk of HCC and pave the way for the development of novel targeted therapies.
Footnotes
Author contributions: The study design was done by all authors. R.A.G. did the investigations and the statistics and reviewed the manuscript. S.H.F. shared in clinical evaluation and reviewed the manuscript. Y.E. shared in clinical evaluation and reviewed the manuscript. M.H. shared in clinical evaluation and sample collection and reviewed the manuscript. S.I.T shared in clinical evaluation and sample collection and wrote the manuscript.
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
Ethical statement
Ethical approval
Ethical approval for this study was obtained from The Research Ethics Committee of the Faculty of Medicine, Ain Shams University, Cairo, Egypt with ethical approval number FMASU R352/2023.
Informed consent
Written informed consent was obtained from all subjects before the study.
Availability of data and materials: All the data needed to support the current findings will be available upon request.
ORCID iDs
Shaimaa H. Fouad https://orcid.org/0000-0003-4052-9454
Sara I. Taha https://orcid.org/0000-0001-8224-8701
References
- 1.Verma S, Sahu BD, Mugale MN. (2023) Role of lncRNAs in hepatocellular carcinoma. Life Sci 325: 121751. DOI: 10.1016/j.lfs.2023.121751. [DOI] [PubMed] [Google Scholar]
- 2.Rashed WM, Kandeil MAM, Mahmoud MO, et al. (2020) Hepatocellular Carcinoma (HCC) in Egypt: a comprehensive overview. J Egypt Natl Cancer Inst 32(1): 5. DOI: 10.1186/s43046-020-0016-x. [DOI] [PubMed] [Google Scholar]
- 3.Yang JD, Hainaut P, Gores GJ, et al. (2019) A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol 16(10): 589–604. DOI: 10.1038/s41575-019-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mattar MM, Zekri AN, Hussein N, et al. (2018) Polymorphisms of base-excision repair genes and the hepatocarcinogenesis. Gene 675: 62–68. DOI: 10.1016/j.gene.2018.06.056. [DOI] [PubMed] [Google Scholar]
- 5.Kumari R, Jat P. (2021) Mechanisms of cellular senescence: cell cycle arrest and senescence associated secretory phenotype. Front Cell Dev Biol 9: 645593. DOI: 10.3389/fcell.2021.645593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghaderi-Zefrehi H, Rezaei M, Sadeghi F, et al. (2021) Genetic polymorphisms in DNA repair genes and hepatocellular carcinoma risk. DNA Repair 107: 103196. DOI: 10.1016/j.dnarep.2021.103196. [DOI] [PubMed] [Google Scholar]
- 7.Li Q, Ma R, Zhang M. (2018) XRCC1 rs1799782 (C194T) polymorphism correlated with tumor metastasis and molecular subtypes in breast cancer. OncoTargets Ther 11: 8435–8444. DOI: 10.2147/OTT.S154746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai W, Liu X, Li Y, et al. (2020) New sights on the associations between the XRCC1 gene polymorphisms and hepatocellular carcinoma susceptibility. J Cell Biochem 121(2): 1005–1022. DOI: 10.1002/jcb.29335. [DOI] [PubMed] [Google Scholar]
- 9.Xiong Y, Zhang Q, Ye J, et al. (2018) Associations between three XRCC1 polymorphisms and hepatocellular carcinoma risk: a meta-analysis of case-control studies. PLoS One 13(11): e0206853. DOI: 10.1371/journal.pone.0206853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kiran M, Saxena R, Chawla YK, et al. (2009) Polymorphism of DNA repair gene XRCC1 and hepatitis-related hepatocellular carcinoma risk in Indian population. Mol Cell Biochem 327(1-2): 7–13. DOI: 10.1007/s11010-009-0035-3. [DOI] [PubMed] [Google Scholar]
- 11.Merchant N, Alam A, Bhaskar LV. (2023) The correlation between hepatocellular carcinoma susceptibility and XRCC1 polymorphisms Arg194Trp, Arg280His, and Arg399Gln–A meta-analysis. Hum Genet: 201165. [Google Scholar]
- 12.Ferrante ND, Pillai A, Singal AG. (2020) Update on the diagnosis and treatment of hepatocellular carcinoma. Gastroenterol Hepatol 16(10): 506–516. [PMC free article] [PubMed] [Google Scholar]
- 13.Nahm FS. (2016) Nonparametric statistical tests for the continuous data: the basic concept and the practical use. Korean J Anesthesiol 69(1): 8–14. DOI: 10.4097/kjae.2016.69.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naguib M, Helwa MM, Soliman MM, et al. (2020) XRCC1 gene polymorphism increases the risk of hepatocellular carcinoma in Egyptian population. Asian Pac J Cancer Prev APJCP 21(4): 1031–1037. DOI: 10.31557/APJCP.2020.21.4.1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krupa R, Czarny P, Wigner P, et al. (2017) The relationship between single-nucleotide polymorphisms, the expression of DNA damage response genes, and hepatocellular carcinoma in a polish population. DNA Cell Biol 36(8): 693–708. DOI: 10.1089/dna.2017.3664. [DOI] [PubMed] [Google Scholar]
- 16.Seeberg E, Eide L, Bjørås M. (1995) The base excision repair pathway. Trends Biochem Sci 20(10): 391–397. DOI: 10.1016/s0968-0004(00)89086-6. [DOI] [PubMed] [Google Scholar]
- 17.Dai Q, Luo H, Li XP, et al. (2015) XRCC1 and ERCC1 polymorphisms are related to susceptibility and survival of colorectal cancer in the Chinese population. Mutagenesis 30(3): 441–449. DOI: 10.1093/mutage/geu088. [DOI] [PubMed] [Google Scholar]
- 18.Guo LY, Jin XP, Niu W, et al. (2012) Association of XPD and XRCC1 genetic polymorphisms with hepatocellular carcinoma risk. Asian Pac J Cancer Prev APJCP 13(9): 4423–4426. DOI: 10.7314/apjcp.2012.13.9.4423. [DOI] [PubMed] [Google Scholar]
- 19.Mandal RK, Mittal RD. (2021) Genetic variant XRCC1 rs1799782 (C194T) and risk of cancer susceptibility in Indian population: a meta-analysis of case-control studies. Indian J Clin Biochem 36(2): 175–184. DOI: 10.1007/s12291-020-00877-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng XY, Yu HP, Qiu XQ, et al. (2010) A case-control study of polymorphism of XRCC1 gene and the risk of hepatocellular carcinoma. Zhongguo Jibing Kongzhi Zazhi 14: 760–763. [Google Scholar]
- 21.Yang Z, Zhao J. (2015) Effect of APE1 and XRCC1 gene polymorphism on susceptibility to hepatocellular carcinoma and sensitivity to cisplatin. Int J Clin Exp Med 8(6): 9931–9936. [PMC free article] [PubMed] [Google Scholar]
- 22.Li W, Yang F, Gui Y, et al. (2014) DNA repair gene XRCC1 Arg194Trp polymorphism and susceptibility to hepatocellular carcinoma: a meta-analysis. Oncol Lett 8(4): 1725–1730. DOI: 10.3892/ol.2014.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Li Z, Feng L, et al. (2013) Polymorphisms of DNA repair gene XRCC1 and hepatocellular carcinoma risk among East Asians: a meta-analysis. Tumour Biol 34(1): 261–269. DOI: 10.1007/s13277-012-0546-5. [DOI] [PubMed] [Google Scholar]
- 24.Su HY. (2008) A case-control study on association between genetic polymorphisms of DNA repair and hepatic cell cancer susceptibility. Taichung, Taiwan: China Medical University. https://dwanfangdatacomcn/Thesis_Y1300729aspx (article in Chinese). [Google Scholar]
- 25.Jinlong Y. (2004) Influence of human XRCC1-399 single nucleotide polymorphism on primary hepatocytic carcinoma. Tumor 24(4): 322–324. [Google Scholar]