Abstract
The underlying causes of breast cancer are diverse, however, there is a striking association between type 2 diabetes and poor patient outcomes. Platelet activation is a common feature of both type 2 diabetes and breast cancer and has been implicated in tumourigenesis through a multitude of pathways. Here transcriptomic analysis of type 2 diabetes patient-derived platelet microvesicles revealed an altered miRNA signature compared with normoglycaemic control patients. Interestingly, interrogation of these data identifies a shift towards an oncogenic signature in type 2 diabetes-derived platelet microvesicles, with increased levels of miRNAs implicated in breast cancer progression and poor prognosis. Functional studies demonstrate that platelet microvesicles isolated from type 2 diabetes patient blood are internalised by triple-negative breast cancer cells in vitro, and that co-incubation with type 2 diabetes patient-derived platelet microvesicles led to significantly increased expression of epithelial to mesenchymal transition markers and triple-negative breast cancer cell invasion compared with platelet microvesicles from healthy volunteers. Together, these data suggest that circulating PMVs in type 2 diabetes patients may contribute to the progression of triple-negative breast cancer.
Introduction
There is a profound association between breast cancer (BC) and type 2 diabetes mellitus (T2DM; diabetes), with diabetic women being more likely to both develop BC [1–3] and die from the disease [4, 5]. The mechanisms underpinning this association remain poorly understood. however, platelet activation is a common feature of both diabetes [6] and BC [7, 8] and is driven by metabolic reprogramming, a central hallmark of both pathologies [9]. Indeed, platelet activation is linked to disease progression and poor patient outcomes for a range of cancers [10–14] and has been reported to drive tumourigenesis via a multitude of pathways [15], including enhancing growth and immune invasion [16].
Platelet-derived microvesicles (PMVs) are emerging as key effectors in the aetiology of cardiovascular disease and cancer [17]. PMVs are released from platelets and differ from the widely studied small extracellular vesicles (also known as exosomes) in both their size and their genesis [18]. In addition to their role in thrombosis [19], PMVs also act as reservoirs for bioactive molecules derived from their parent cells [20], with platelet-derived micro-RNAs (miRNAs) attracting significant interest [21, 22]. Underpinning this is a growing body of evidence demonstrating that PMVs deliver miRNAs to a range of cells leading to altered transcriptomes and target cell phenotypes [23–26].
It is becoming increasingly apparent that T2DM induces its own miRNA signature [27–30]. For example, the expression profile of five plasma microRNAs; miR-15a, miR28-3p, miR-126, miR-223, and miR-320, can be used to distinguish T2DM patients from healthy controls [31]. Other groups have highlighted that miR-223 and miR-146a expression is altered in diabetic patient plasma and platelets [27] and there is evidence that altered miRNA expression is associated with poorly controlled T2DM [32]. This raises the intriguing possibility that a T2DM-derived PMV miRNA signature might contribute to the increased risk of breast cancer progression in diabetic patients. Indeed, several studies lend support to this model, including pioneering work establishing that small extracellular vesicles in obesity and metabolic disease are important drivers of cancer progression for several cancer types [33–36]. Moreover, and of particularly relevance to this study, is work reporting that exosomes from diabetic adipocytes and human mesenchymal stem cells drive progression in breast cancer and that plasma exosomes of patients with T2DM carry a distinct profile of miRNAs of known significance for prostate cancer progression that contribute functional effects in a range of cancer models [37].
Here small-RNA sequencing of T2DM- and normoglycaemic-derived PMVs revealed significant differences in the abundance of numerous BC-associated miRNAs. Bioinformatic analysis of these data revealed a shift in T2DM-derived PMVs towards increased expression of pro-tumorigenic miRNA and reduced expression of miRNA reported to function as BC tumour suppressors. To investigate the impact of diabetic PMVs compared with normoglycaemic control PMVs on breast cancer cells, co-incubation experiments were performed. Strikingly, we observed significant increases in cell invasion index and key epithelial-mesenchymal transition (EMT) genes in triple-negative breast cancer (TNBC) cells co-incubated with diabetic PMVs compared with TNBC co-incubated with normoglycaemic PMV controls. Moreover, TNBC cells co-incubated with T2DM-dervied PMVs exhibited increased levels of miRNAs identified as enriched in diabetic samples, suggesting that PMVs may induce these phenotypic changes in BC cells via delivery of differential profiles of miRNAs.
Results
T2DM-derived PMVs harbour a distinct miRNA profile
There is emerging evidence that diabetic patients harbour altered circulating miRNA profiles and that these may have utility as biomarker panels [38]. To investigate if PMVs isolated from poorly controlled T2DM patients possess distinct miRNA cargo human blood was taken by venepuncture from T2DM patients with HbA1C>50mmol/mol and from healthy controls and PMVs isolated by centrifugation. PMVs were subsequently analysed via micro-Bicinchoninic Acid Assay (BCA), NanoSight Tracking Analysis (NTA) (Fig 1A and 1B), and Fluorescence-activated Cell Sorting (FACS) (Fig 1C) to confirm that particles were of the expected size and displayed PMV-associated markers. Total RNA was isolated from purified PMVs and small RNA libraries generated prior to sequencing. Principal component analysis (PCA) distinguished between small RNA sequencing data obtained from T2DM patient derived PMVs and healthy controls (Fig 2A, S1 and S2 Tables). Moreover, subsequent differential gene expression analysis revealed significantly distinct miRNA profiles for T2DM patient-derived PMVs compared with control samples (Fig 2B, S3 Table). Many of the altered miRNAs were down regulated in T2DM (down-regulated = 42) compared up regulated miRNAs (up-regulated = 12). To explore the functions of these altered miRNAs, we performed gene ontology analysis on their experimentally validated mRNA targets (See Methods, S4 Table). We observed that both the up-regulated and down-regulated miRNA-mRNA interactions associate with distinct gene ontologies (Fig 2C, S5 Table). Further, several of the top enriched ontologies were shared between the up and down miRNA-mRNA interactions (Fig 2C, S5 Table), suggesting potential reprogramming within the same core pathways. However, these mRNA targets overlap between the groups (Down: 33% 188 out of 566 and Up: 27% 188 out of 695, S3 Table), which may be related to the fact that a single miRNA can target hundreds of mRNAs. Curation of the miRNAs up-regulated in diabetic PMVs (based on their published functions in cancer [39–42]) revealed an overrepresentation of miRNAs previously reported to act as oncogenes (Fig 2D). Conversely, miRNAs ascribed a tumour-suppressor function were over-represented in genes down-regulated in T2DM-derived PMVs compared with healthy PMV controls (Fig 2D). Collectively, these data show that the miRNA profile of PMVs obtained from T2DM-patients is functionally different to that observed in PMVs obtained from healthy controls and is shifted towards a pro-tumourogenic collection of genes.
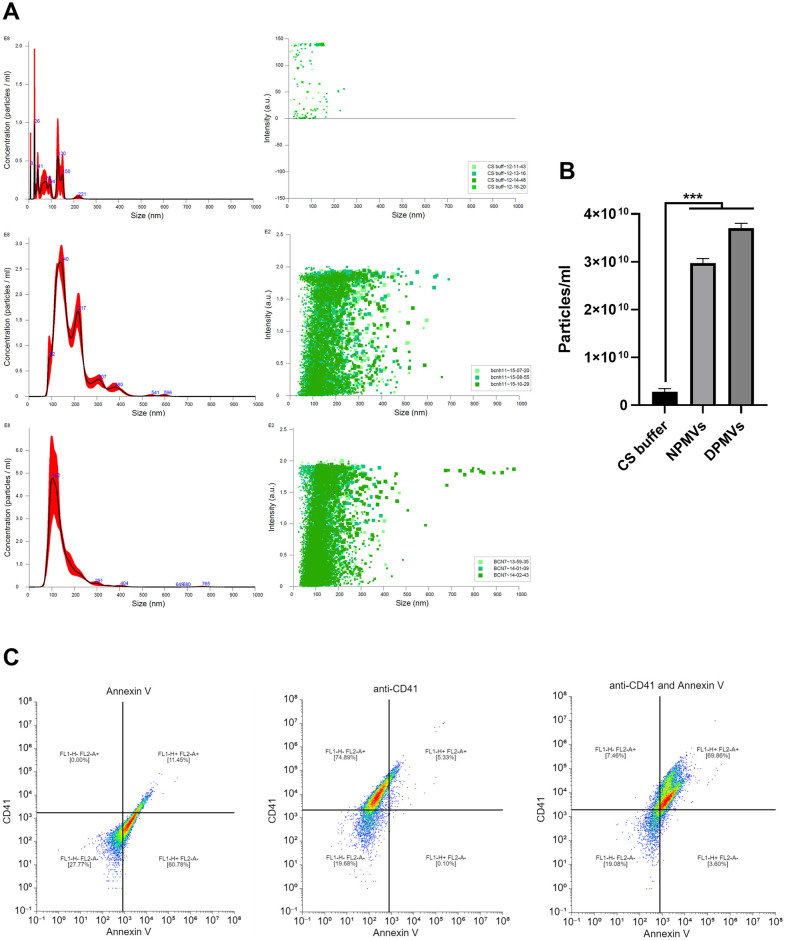
Fig 1. Isolation and characterisation of diabetic PMVs.
(A) Analysis of Tyrode’s buffer, healthy control PMVs (NPMV) and diabetic-patient derived PMVs (DPMV) by NTA. Left panels display particle concentrations (1x108) and size, right panels display individual particle size verses intensity (a.u.), n = 3. (B) PMV particle concentration for Tyrode’s buffer, NPMV and DPMV, n = 3. (C) PMVs were labelled with Annexin V-FITC and/or CD41a-PE, gated around singlets and 10,000 events collected. Example dot plot for single and dual labelled PMVs with quadrant analysis, n = 3. Data are representative of all PMV samples, *** = p<0.001.
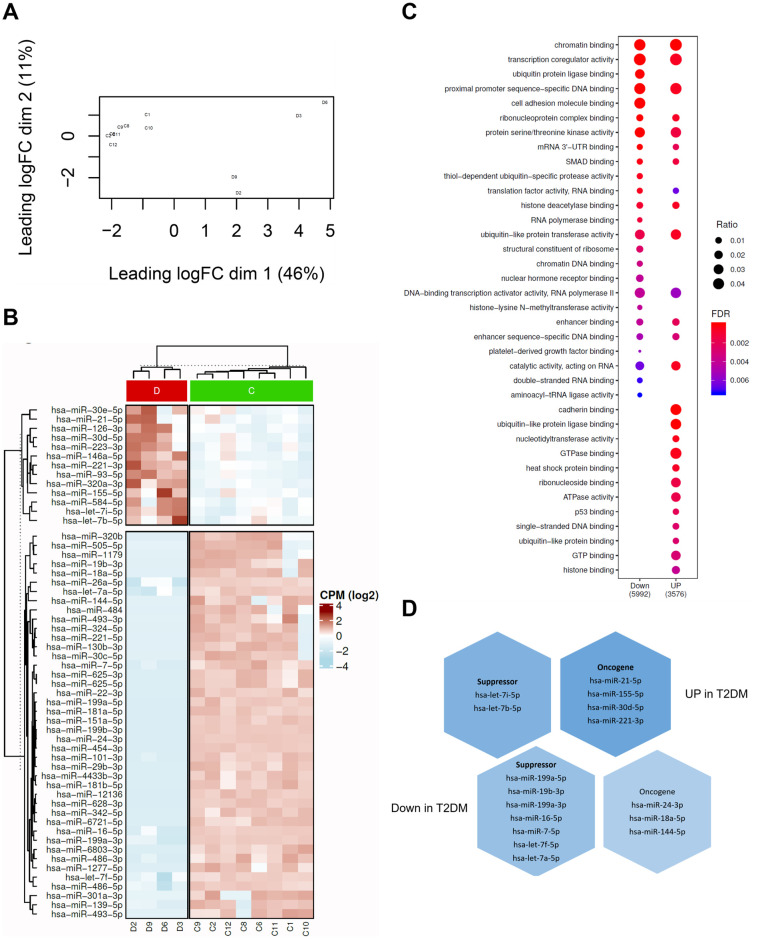
Fig 2. PMVs isolated from poorly controlled diabetic patients harbour an oncogene-polarised miRNA profile compared with PMVs derived from healthy volunteers.
(A) Multi-dimensional scaling (MDS) plot of miRNA profiles in diabetic PMV samples (D; n = 4) and healthy controls (C; n = 8). (B) Heatmap of expression profiles of all differentially expressed miRNAs in diabetic PMVs compared with normal PMVs. Within the plot, red colour shows diabetic PMVs and green normal. (C) Cluster plot of enriched biological processes associated with mRNA targets of the down regulated (Down) or up regulated (Up) miRNAs. Gene ratio—genes in ontology/total gene set and false discovery rate (FDR)-. mRNA transcripts represented in the plot are experimentally validated miRNA-mRNA interactions from the miRTarBase database. (D) Distribution of known tumour suppressor and oncogenic miRNAs in the set of differentially expressed miRNAs in diabetic PMVs compared with normal PMVs.
T2DM-derived PMVs demonstrate tropism for TNBC cell uptake
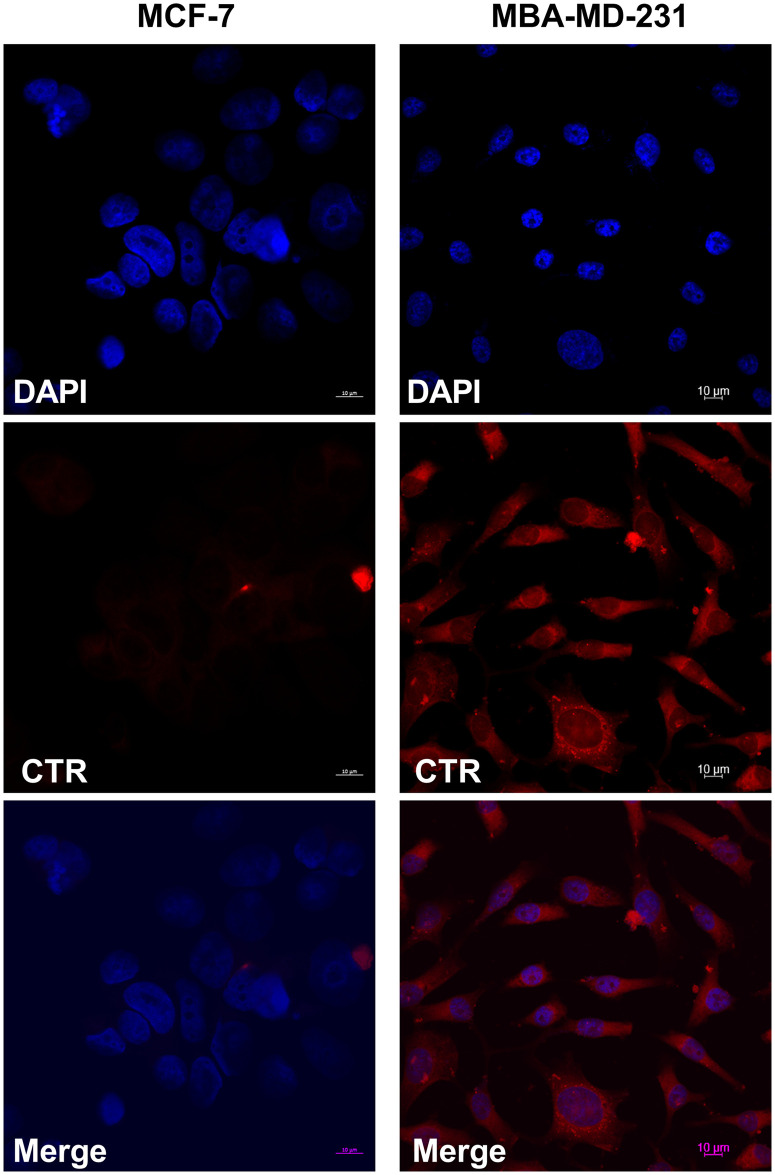
Recently it was reported that PMVs exhibited efficient uptake when co-cultured with the TNBC cell line, MDA-MB-231, but not the luminal ER+ Her2- MCF-7 cell line [43]. To determine if PMVs derived from T2DM patient blood exhibited similar preferential uptake, co-culture experiments were carried out with T2DM-derived PMVs labelled with the membrane dye, CellTracker™ Red (CTR). Labelled PMVs were cultured with MDA-MB-231 or MCF-7 cells for 30 minutes, prior to fixation and counterstaining with DAPI. As shown in Fig 3, we observed clear uptake of labelled PMVs in MDA-MB-231 cells but only nominal CTR signal in MCF-7 cells, consistent with previously published work38. Together these data demonstrate that PMVs from T2DM patients are internalised by TNBC cells, in vitro.
Fig 3. PMVs isolated from poorly controlled diabetic volunteers are internalised by TNBC cells, in vitro.
PMVs were stained with 1mM CellTracker™ Red CMTPX, free-dye removed and labelled PMVs cocultured with MCF-7 or MDA-MB-231 cells for 4h prior to analysis by confocal microscope.
T2DM-derived PMVs potentiate TNBC cell invasion
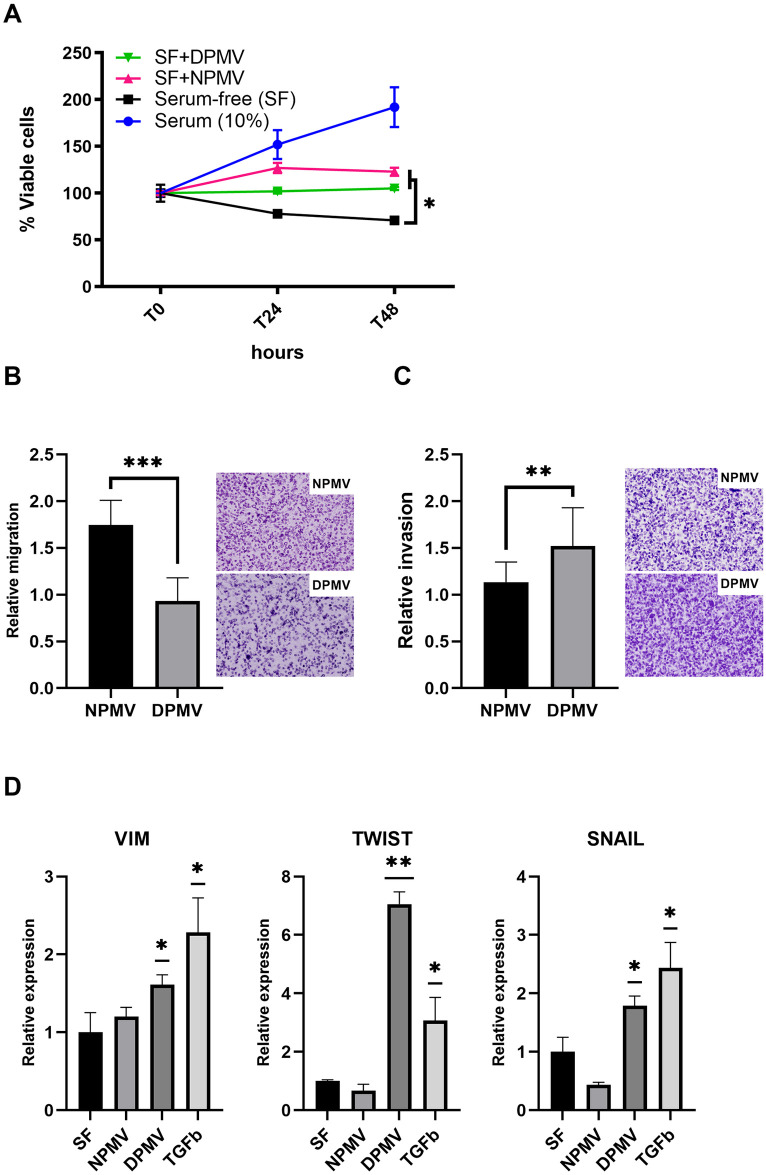
PMVs were previously reported to increase MDA-MB-231 cell migration [43]. Given the bias towards oncogenic miRNA cargo in T2DM-derived PMVs we were interested in assessing how T2DM-derived PMVs impact TNBC cells compared with PMVs isolated from healthy volunteers. To investigate this, MDA-MB-231 cells were serum starved prior to co-culture with T2DM-derived PMVs or control PMVs and cell viability, cell migration, and cell invasion assays carried out. As shown in Fig 4A, while co-culture of MDA-MB-231 cells with control and T2DM-derived PMVs led to a nominal increase in total viable cell number 48h post co-incubation compared with control, we observed no significant difference between diabetic and healthy PMVs. Next, we assessed if T2DM-derived PMVs induce cell migration more avidly than control PMVs. MDA-MB-231 cells were cultured in serum-free media prior to seeding in transwell inserts and co-incubation with either vehicle, T2DM-derived PMVs or PMVs isolated from healthy controls. As shown in Fig 4B TNBC cells co-incubated with T2DM-derived PMVs exhibited a significant increase in migration compared with control, however, to our surprise this was not as pronounced as the impact of PMVs from healthy volunteers. Cancer cell invasion is one of the first steps in the metastatic cascade and so we sought to investigate the impact of T2DM-derived PMV co-incubation with TNBC cells on invasion index. MDA-MB-231 cells were cultured in serum-free media prior to seeding in transwell inserts coated with basement membrane matrix (Geltrex®) and co-incubated with either vehicle, T2DM-derived PMVs or PMVs isolated from healthy controls. As shown in Fig 4C, PMVs from healthy volunteers did not increase TNBC cell invasion compared with control, however, we observed a significant increase in cell invasion index for TNBC cells co-incubated with PMVs from diabetic patients. Cancer cell invasion is often associated with the expression of drivers of mesenchymal cell state. To determine if T2DM-derived PMVs were increasing the expression of key mesenchymal genes, we analysed the relative expression of TWIST, SNAIL, and VIM (vimentin) via reverse-transcriptase quantitative PCR (RT-qPCR) in TNBC cells co-incubated with T2DM-derived PMVs, PMVs isolated from healthy controls or TGFβ, which served as a positive control for the induction of mesenchymal gene expression. PMVs isolated from healthy controls failed to induce significant increases in mesenchymal genes, however, we observed significant increases in TWIST, SNAIL, and vimentin transcripts levels in cells co-incubated with T2DM-derived PMVs (Fig 4D). Together, these data demonstrate that diabetic PMVs drive a significant increase in TNBC cell invasion, possibly via the induction of EMT-associated genes.
Fig 4. PMVs isolated from poorly controlled diabetic volunteers potentiate TNBC invasion, in vitro.
(A) MDA-MB-231 cells were cultured for 48h in complete media plus vehicle (PBS), serum-free (SF) media plus vehicle (PBS), SF media with NPMV, or SF media with DPMV and total viable cells assessed via CellTiter-Glo®, n = 3. (B and C) MDA-MB-231 cells were cultured in SF media or co-incubated with either healthy control PMVs (NPMV), or diabetic-patient derived PMVs (DPMV) in either uncoated or extra-cellular matrix (ECM)-coated transwell inserts to determine migration (B) and invasion (C), respectively. Migration and invasion rates are relative to cells cultured in SF media, n = 6. (D) MDA-MB-231 cells were cultured in SF media or co-incubated in SF media with either healthy control PMVs (NPMV), diabetic-patient derived PMVs (DPMV), or TGFβ (10ng/ml) for 24h prior to isolation of total RNA and analysis of target gene expression by RT-qPCR, n = 3. * = p>0.05; ** = p<0.01; *** = p<0.001.
TNBC cells co-incubated with T2DM-dervied PMV contain increased amount of diabetic PMV-enriched miRNAs
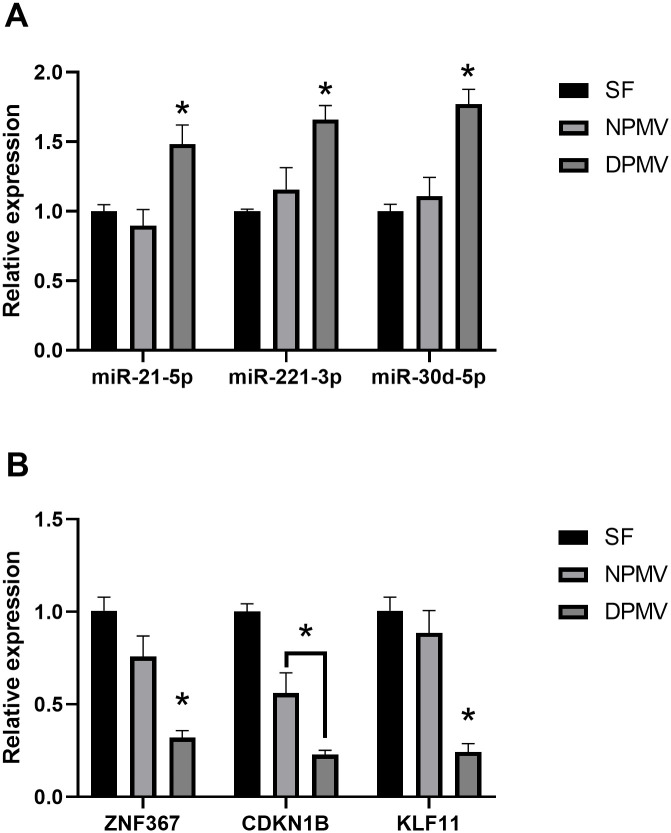
Given the uptake of PMVs by TNBC cells and the enrichment of several protumourigenic miRNAs in our T2DM-derived PMV RNA-seq data, we set out to determine if co-incubation of TNBC cells with T2DM-derived PMVs resulted in increased levels of these miRNAs, when compared with TNBC cells co-incubated with vehicle alone or PMVs isolated from healthy controls. To control for changes in miRNA target levels being driven endogenously via altered expression of cellular pre-miRNAs, TNBC cells were treated with actinomycin D prior to co-incubation with PMVs or control for 48h. Total RNA was isolated, and miRNA transcript levels determined via miRCURY LNA miRNA PCR. As shown in Fig 5A, we observed increased amounts of mir-21-5p, miR-221-3p, and miR-30d-5p in MDA-MB-231 cells co-incubated with diabetic PMVs compared with both control PMVs and vehicle alone. To investigate if these increases corresponded with a functional impact, we assessed the transcript levels of Zinc Finger Protein 367 (ZNF367), Cyclin Dependent Kinase Inhibitor 1B (CDKN1B), and KLF Transcription Factor 11 (KLF11). These three genes have previously been validated, in a breast cancer cell context, to be direct targets of mir-21-5p [44], miR-221-3p [45], and miR-30d-5p [46], respectively. As shown in Fig 5B, we observed a significant decrease in transcript levels in cells treated with PMVs from both healthy and T2DM volunteers, however, this decrease was significantly greater in cells co-incubated with T2DM-derived PMVs, suggesting that elevated levels of mir-21-5p, miR-221-3p, and miR-30d-5p in DPMVs may exert a functional impact on target gene expression in TNBC cells.
Fig 5. PMVs isolated from poorly controlled diabetic volunteers deliver oncogenic miRNA to TNBC cells, in vitro.
(A) MDA-MB-231 cells were cultured in the presence of actinomycin D (1 μg/ml) for 24h cultured prior to culturing for a further 24h in serum-free (SF) media, SF media with healthy control PMVs (NPMV), or SF media with diabetic-patient derived PMVs (DPMV). Total RNA was isolated from cells and RT-qPCR performed against target miRNAs, n = 3. (B) Total RNA from (A) was utilised to assess via RT-qPCR the transcript levels of validated cognate targets of each of the miRNA shown in panel A, n = 3. * = p<0.05.
Discussion
Increased platelet activation is common to both T2DM and BC patients, and is a key driver of the hypercoagulable state found in both sets of patients, while also impacting directly on cancer metastasis across a range of neoplasms, including breast cancer [47]. Platelet activation also results in increased release of PMVs and there is compelling evidence that demonstrates a potent correlation between high PMV concentration in the blood and breast cancer progression [48], however, the mechanisms by which PMVs support tumour growth and dissemination remain to be fully elucidated. While a T2DM-induced increase in circulating PMV concentration may contribute to elevated risk of BC, qualitative differences in the miRNA cargo of T2DM patient PMVs may also be influential. This is particularly relevant given the body of research demonstrating that diabetic plasma harbours significantly altered miRNA profiles and the mooted utility of this observation in terms of diagnosis and prognosis of comorbidities [38, 49]. Here we performed small RNA sequencing on PMV total RNA isolated from the blood of poorly controlled T2DM patients and healthy volunteers. We observed a clear differentiation of small RNA transcripts between the two groups via PCA (Fig 2A), suggesting that reported differences in the miRNA profile of platelets in diabetic patients also translates to PMVs being released by these cells. Several studies have reported platelet-specific miRNA expression profiles [29, 50, 51] with a recent study elegantly demonstrating, via subtractive approaches, a panel of platelet-enriched miRNA genes [52]. Analysis of our PMV sequencing data reveals the presence of the widely reported platelet-enriched (PE) miRNAs miR-223-3p and miR-21-5p, moreover, we identified all the PE miRNA genes described by Krammer et al [52] in both control and T2DM data, suggesting that platelet-specific miRNA profiles are mirrored in released PMVs. Moreover, DE analysis revealed significant changes across T2DM patient PMV miRNA cargo compared with healthy control PMVs (Fig 2B). In many cases this equated to an alternation in the transcript abundance of PE transcripts, for example miR-223-3p and miR-21-5p were significantly higher in T2DM PMVs, whereas PE miRNA miR-151-5p and miR-148b-3p were significantly decreased.
There is great interest in diagnostic, prognostic, and on-treatment miRNA biomarkers for T2DM, however, findings often lack consensus, likely due to inherent variability between patients and issues with circulating transcript ‘noise’. To assess if observed differences in PMV miRNA levels in our two groups reflected published work reporting on circulating biomarkers for T2DM we cross-referenced our findings with a curated list of circulating T2DM miRNA biomarkers which have appeared and been validated by at least two independent experimental studies [53]. The picture here was mixed with some of our differential targets (miR-24-3p↓, miR-146a-5p↑, miR-30d-5p↑) mirroring abundance changes reported in diabetic patient plasma, however, both miR-126-3p and miR-223-3p are decreased in T2DM patient plasma but increased in our T2DM PMVs. One explanation for this could be the small sample size used in this study, but it is also likely that changes to circulating miRNA levels in diabetic patients arise via several routes and that the input from platelet-derived miRNA is just one of many contributing factors.
The focus of this study was the impact of diabetic PMVs on BC. Inspection of our T2DM PMV miRNA data in terms of changes to genes that have been shown to contribute to pro- and anti-tumourogenic progression in breast cancer [39–42], revealed a skewed pattern of changes in T2DM PMVs that favoured increased miRNA transcript levels for genes that have been previously reported to function in a pro-tumourogenic capacity in breast cancer (Fig 2D), including miR21-5p, miR-221-3p, and miR-30d-5p [46, 54–57]. Understanding how observed DE in miRNA genes between T2DM and control PMV relates to both the diabetic state of the donor and wider biology, specifically breast cancer cell growth and progression is to an extent a Gordian knot. To begin unravelling this, one approach is to focus on those differentially expressed PMV miRNAs upregulated in T2DM plasma that also have reported roles in BC progression. Two genes that meet these criteria in our data are miR-21-5p and miR-30d-5p, indeed a single study reported that miR-30d-5 expression correlates with both insulin resistance and BC development, however, it is important to note that this was generally in the context of ER+ BC and so if this also applies to a TNBC context is unclear [58]. To investigate the impact of increased pro-tumourogenic miRNA transcripts in T2DM PMVs on BC cells a series of in vitro co-incubation experiments were performed. Previously, PMVs have been reported to exhibit efficient internalisation by the TNBC cell line, MDA-MB-231, and in contrast poor uptake by the ER+ BC cell line, MCF-7 [43], an observation recently confirmed via a second study [59]. We observed similar results using T2DM PMVs, with only nominal uptake in MCF-7 cells compared with extensive uptake in MDA-MB-231 cells (Fig 3). The mechanisms governing PMV internalisation are poorly understood. There is some evidence that in endothelial cells uptake occurs via active endocytosis [60], and that in breast cancer cells PMV binding is integrin αIIbβ3-independent, however, αIIbβ3 antagonists reported to be ineffective at blocking PMV uptake into MDA-MB-231 cells [43]. Limited amounts of clinical sample prevented more widespread analysis of PMV uptake across additional BC cells lines in this study, however, more research is needed, both in terms of PMV cell surface proteins and their contribution to target cell tropism and to the precise mechanisms governing uptake and if these are uniform across different target cell types.
Co-incubation of MDA-MB-231 cells with PMVs resulted in increased cell growth and migration, as previously reported [43], however, there was no significant T2DM-specific impact on cell growth and surprisingly while migration was increased in MBA-MD-231 cells co-incubated with T2DM-derived PMVs this was at a significantly lower level than PMVs isolated from healthy controls (Fig 4A and 4B). In contrast, co-incubation with T2DM- derived PMVs, resulted in a significant increase in MBA-MD-231 cell invasion and expression of EMT-associated genes compared with PMV controls (Fig 4C and 4D). As PMV concentration (mg/mL) used in in vitro assays was the same for control and diabetic samples, the invasion-specific effect of T2DM PMVs on TNBC cells is presumably due to qualitative differences in these PMVs in terms of either their uptake efficiency, the cargo they deliver to cells, or both. We observed no obvious difference in MBA-MD-231 PMV uptake between diabetic and control, however, it is important to note that the method employed here does not facilitate true quantitative analysis of PMV uptake and therefore we cannot exclude the possibility that the phenotypic and biochemical differences we observed in MBA-MD-231 cells are due to a general increase in diabetic PMV internalisation, and that our analysis of EMT gene expression was limited to analysis of RNA transcript levels. Interestingly, we did observe an increase T2DM-enriched PMV miRNA in MBA-MD-231 cells co-incubated with diabetic PMVs compared with control, suggesting that the increased abundance of these miRNAs in diabetic PMVs results in elevated levels of these miRNAs in target cells. Importantly, this increase was observed in cells pre-treated with the RNA polymerase II inhibitor, actinomycin D, suggesting that elevated levels were due to transfer of miRNA from PMVs, rather than indirect effects of PMV co-incubation on miRNA transcription (Fig 5A). Furthermore, quantitative analysis of reported cognate mRNA targets of miRNAs elevated in diabetic PMVs revealed a significant decrease in transcript abundance compared with the same targets in MBA-MD-231 cells co-incubated with control PMVs, suggesting that the increased levels of some miRNA in T2DM-derived PMVs manifested a tangible and significant impact on the levels of target messenger RNA in BC cells (Fig 5B). Several studies have demonstrated that high levels of miR-21-5p positively correlate with poor outcome and increased invasive index in TNBC [61–63], with one study reporting that increased levels of miR-21-5p in serum is associated with TNBC metastasis and prognosis [63]. While we observed changes in miRNA levels of TNBC cells co-incubated with PMVs, it is likely that PMV protein cargo and PMV-associated receptors also exert an influence or target cells, further investigation is required to better understand how these biomolecules differ between healthy and diabetic PMVs.
A limitation of this study is that our phenotypic assessment of patient derived PMVs occurs only through in vitro based assays. Conceptually, the likely impact of a differential PMV miRNA signature for T2DM patients who develop BC is via altered concentrations and cargo of circulating PMVs in the blood. PMVs represent the overwhelming majority (up to 90%) of all cell-derived vesicles in blood [17, 64] and as such it is conceivable that changes to platelet biology and by extension the microvesicles that they shed in disease like T2DM might have a measurable impact on systemic biology, including development and progression of cancer. Moreover, evidence demonstrating that PMVs increase endothelial cell permeability suggest a manner by which these vesicles might access tumour cells [65]. Clearly, clinical and in vivo studies are sorely needed, however, attempting to disentangle PMV-induced effects from circulating extracellular vesicles is an ongoing challenge in the field [66]. With regards to the T2DM PMV miRNA profile described in this study, while we observed a pro-tumourogenic consensus across increased miRNAs, it is important that we do not discount the fact that numerous changes to the DE of miRNAs in T2DM-derived PMV compared with healthy control are represented by significant decreases in miRNAs reported to function in an anti-tumourogenic capacity (Fig 2D). It seems possible that a dramatic decrease in a PMV-associated circulating pool of anti-tumourogenic miRNA, due to T2DM, may propagate an environment that is more receptive of both tumourigenesis and cancer metastasis. This model is also consistent with studies that have reported PMVs to function in a predominantly protective manner [67]. Interestingly, decreased miRNAs in our T2DM-derived PMV data include miR-199a-5p and miR-221-5p, which have previously been linked to tumour-suppressive roles in TNBC [68, 69].
In summary, we have shown that PMVs isolated from poorly controlled T2DM patients harbour a distinct onco-miR-enriched miRNA profile, are taken up by TNBC cells and potentiate cell invasion (Fig 6). In addition, with the utility of extracellular vesicle biomarker signatures for BC becoming increasingly clear [70–72], future studies on a larger patient cohort will be essential to determine if a suitable miRNA signature can be identified within diabetic PMVs that might be used to identify diabetic patients at increased risk of developing breast cancer or indeed diabetic breast cancer patients at risk of developing metastatic disease.
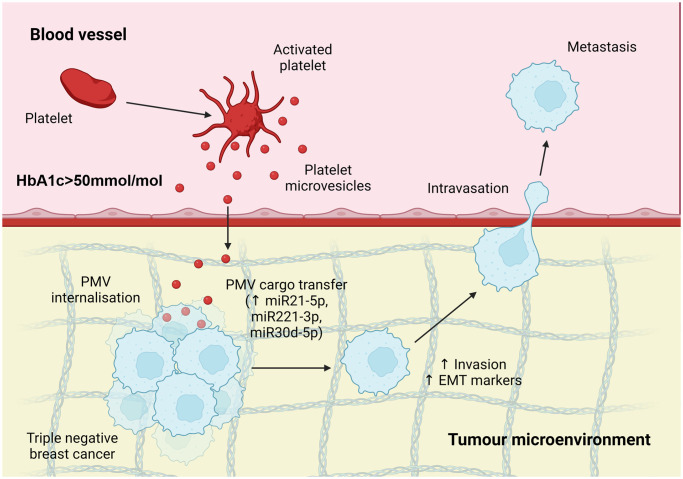
Fig 6. Model for how DPMV might impact on TNBC progression.
Circulating PMVs harbour an altered and pro-oncogenic miRNA profile in T2DM patients (indicated by HbA1c>50mmol/mol) that impact on TNBC invasion, in vitro, and might similarly drive tumourigenesis in diabetic patients with breast cancer.
Methods
PMV isolation and analysis
Ethical approval was obtained prior to the start of this study (IRAS project ID 235632). The recruitment period for the study was 31st May 2018 to 31st May 2020. Written informed consent, witnessed, and documented by a research nurse (JT), was obtained from participants prior to the collection of samples. Samples were stored in a fully anonymised form. PMV isolation was carried out as previously described [23]. Briefly, human blood was taken by venepuncture from healthy adult volunteers and T2DM patients with HbA1C>50 mmol/mol, anonymised demographic data for consented patients can be seen in S6 Table. Blood was added to acid citrate dextrose (ACD; 29.9mM sodium citrate, 113.8mM glucose, 72.6mM sodium chloride and 2.9mM citric acid, pH 6.4) and platelet-rich plasma (PRP) was obtained by centrifugation of whole blood at 200g at 20°C, 20 minutes. Washed platelets were isolated from the PRP by centrifugation at 800g at 20°C, 12 minutes in the presence of prostaglandin E1 (PGE1; 50 ng/ml). Platelets were resuspended in Tyrode’s buffer (150mM NaCl, 5mM HEPES, 0.55mM NaH2PO4, 7mM NaHCO3, 2.7mM KCl, 0.5mM MgCl2, 5.6mM glucose). PMV were isolated as reported [23] and suspensions activated with thrombin (0.1 U/ml) for 60 minutes. Platelet activation was stopped by EDTA (20 mM) and platelets pelleted by centrifugation (3200g, 10 minutes). PMVs were isolated by centrifugation at 20,000g for 90 minutes at 18°C, then resuspended in Tyrode’s buffer. Protein content was measured using micro-BCA assay (Thermofisher Scientific, UK), and 100mg/ml of pooled PMV suspension used in all assays. PMVs were diluted 100-fold in PBS and analysed by flow cytometry as previously reported [21, 31] for phosphatidylserine exposure detected by Fluorescein (FITC)-Annexin-V (BD Biosciences) binding or platelet specific marker CD41a expression detected with APC-conjugated anti human CD41a antibody or APC-conjugated mouse IgG isotope control (BD Biosciences). Following incubation for 30 minutes, samples were diluted 5-fold in PBS and FACS analysis was completed using the BD Accuri™ C6 Flow Cytometer (BD Biosciences, UK), making sure to QC instrument before each use using the BD CS&T RUO Beads (BD Biosciences, UK). For tracking cellular uptake PMVs were stained with 1mM CellTracker™ Red CMTPX Dye (ThermoFisher, UK) for 30 minutes at 37°C and incorporated dye removed via centrifugation in Exosome Spin Columns (ThermoFisher, UK) prior to co-incubation with cells for 4 hours, followed by fixation in 4% paraformaldehyde for 10 minutes, staining with DAPI and visualization via LSM 880 with Airyscan (Zeiss).
Cell culture
Breast cancer cell lines were obtained from European Collection of Authenticated Cell Cultures and certified mycoplasma-free. MDA-MB-231 and MCF-7 cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM; ThermoFisher, UK), supplemented with 10% foetal bovine serum (FBS), 100 U/ml of penicillin and 100 μg/ml of streptomycin. All cells were cultured at 37°C and 5% CO2. LNA inhibitors were transfected into cells using HiPerfect (QIAgen, UK), as previously described [23]. To assess PMV transfer of miRNA, MDA-MB-231 cells were treated with actinomycin D (1mg/mL), as previously described [73]. For RT-qPCR analysis of EMT markers, cells were co-incubated with 10ng/ml of TGFβ as a positive control for induction of mesenchymal gene expression.
Viable cell growth
Cells were seeded at a density of 1x104 cells/well in white-walled 96-well plates (ThermoFisher, UK). After 24 hours, media was replaced with 100μl of serum-free media (SFM) and cells incubated at 37°C for a further 24, 48 and 72 hours in the presence of either vehicle alone (PBS), NPMV, DPMV or 10% FBS. Viable cell growth was assessed at each time point using CellTiter-Glo® Luminescent Cell Viability Assay (Promega, UK) and luminescence recorded 10 minutes after reagent addition using the GloMax® Explorer system (Promega, UK).
Transwell assays
Cells were grown for at least 24 hours in serum media, then serum starved for 24 hours prior to detachment and resuspension in the indicated media at a concentration of 1x105 cell/ml. Depending on the experiment, 1ml of serum-free or complete media was added to the lower chambers of a 24 well plate and 500μL of the cell suspension added to an 8μm tissue culture (TC)-insert (Sarstedt, UK). For invasion assays, TC-inserts were coated with 50μL Geltrex™ LDEV-Free Reduced Growth Factor Basement Membrane Matrix (Gibco) and incubated at 37°C for 1 hour. To assess migration and invasion index, cells were incubated for 24 hours with vehicle or PMVs at 37°C and a cotton bud used to gently remove any non-migrated cells from the apical side of the transwell insert membrane. Prior to being fixed with ice cold 70% ethanol for 30 minutes. Insert membranes were stained in 0.2% crystal violet (Fisher Science, UK) for 10 minutes, then mounted onto slides. 5 random high-power images were captured (EVOS XL Core Cell Imaging System, 4x objective) and the number of migrated cells calculated, relative to serum-free control.
RNA extraction and RT-qPCR
Total RNA was extracted from PMVs, or PMV treated cells using the Aurum total RNA Mini kit (BIO-RAD, UK) according to the manufacturer’s instructions. To assess PMV transfer of miRNA, cells were treated with actinomycin D (1mg/mL), as previously described [73]. For RT-qPCR analysis of EMT markers, cells were co-incubated with 10ng/ml of TGFβ as a positive control. 500ng of total RNA was used to generate cDNA using SsoAdvanced SYBR master mix (Bio-Rad, UK), and RT-qPCR analysis carried out using the CFX96 system (Bio-Rad, UK). Micro-RNA expression was analysed using miRCURY LNA miRNA PCR assays (Qiagen, UK) and data normalised against UniSp6 spike RNA. A list of oligonucleotides used during this study can be found in S7 Table.
RNA sequencing analysis
Small RNA-Seq libraries were prepared using NEBNext® Ultra™ DNA Library Prep Kit for Illumina® (NEB, UK) and analysed via NextSeq 500 (Illumina, USA) and data deposited at GEO (TBC). Raw reads were subjected to adaptor trimming (Illumina sequencing adapters) using Cutadapt v2.5 [74]. Trimmed reads were aligned to the GRCh38/hg38 assembly of the human genome using Bowtie2 v2.3.5.1 [75]. HTSeq-count v0.11.1 [76] was used to quantify the miRNA abundance based on the human miRNA annotation (miRBase Release 22.1) with parameters (-s no -a 10 -m union—nonunique none). Expression levels were normalised by “counts per million” (CPM). Differential expression (DE) analyses between diabetic and non-diabetic samples were performed using the edgeR R package [77]. The DE microRNAs were defined at adjusted p-value < 0.05.
Gene ontology analysis
The mRNA targets of the DE miRNAs were extracted from the miRTarBase database [78]. To reduce the rate of false-positive enrichments, only experimentally validated miRNA-mRNA pairs were extracted for further analysis. Biological processes associated with the mRNA targets were determined using R ClusterProfiler and the human Bioconductor annotation database (org.Hs.eg.db) to compare the enriched biological processes between diabetic and non-diabetic targets. All enrichment analyses were performed using p-value and q-value <0.01 cut-off with reduced redundancies by semantic similarity analysis [79].
Statistical analysis
Except stated otherwise, graphical data shown represent mean ± standard deviation of mean (SD) from at least three independent experiments. Differences between means was analysed by Student’s t-test, verified by Bonferroni, Mann Whitney Wilcoxon test, or ANOVA. Statistics were considered significant at p<0.05.
Supporting information
Raw count of miRNA transcript across all samples.
(XLSX)
Le CPM normalised expression levels of miRNA transcripts across all samples.
(XLSX)
Differential expression results for miRNAs passing minimum expression threshold (1 CPM in 3 samples).
(XLSX)
Functionally validated miRNA-mRNA interactions for the differentially expressed miRNA in S3 Table found miRTarBase.
(XLSX)
Complete gene ontology analysis of molecular functions associated the target mRNA between each group.
(XLSX)
Anonymised data for T2DM patients recruited to the study.
(XLSX)
Details of oligonucleotides used in this study.
(XLSX)
Acknowledgments
We would like to thank all the patients and volunteers who agreed to take part in this study.
Data Availability
The datasets generated during and/or analysed during the current study are available in the GEO repository under accession GSE249475 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE249475).
Funding Statement
This work was funded by a Breast Cancer NOW studentship award to JRB (BCN-2016NovPhD851). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Boyle P, Boniol M, Koechlin A, Robertson C, Valentini F, Coppens K, et al. Diabetes and breast cancer risk: a meta-analysis. British journal of cancer. 2012;107(9):1608–17. Epub 2012/09/22. doi: 10.1038/bjc.2012.414 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Larsson SC, Mantzoros CS, Wolk A. Diabetes mellitus and risk of breast cancer: a meta-analysis. International journal of cancer. 2007;121(4):856–62. Epub 2007/04/03. doi: 10.1002/ijc.22717 . [DOI] [PubMed] [Google Scholar]
- 3.Liao S, Li J, Wei W, Wang L, Zhang Y, Li J, et al. Association between diabetes mellitus and breast cancer risk: a meta-analysis of the literature. Asian Pac J Cancer Prev. 2011;12(4):1061–5. Epub 2011/07/28. . [PubMed] [Google Scholar]
- 4.Peairs KS, Barone BB, Snyder CF, Yeh HC, Stein KB, Derr RL, et al. Diabetes mellitus and breast cancer outcomes: a systematic review and meta-analysis. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(1):40–6. Epub 2010/12/01. doi: 10.1200/JCO.2009.27.3011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao XB, Ren GS. Diabetes mellitus and prognosis in women with breast cancer: A systematic review and meta-analysis. Medicine (Baltimore). 2016;95(49):e5602. Epub 2016/12/09. doi: 10.1097/MD.0000000000005602 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pretorius L, Thomson GJA, Adams RCM, Nell TA, Laubscher WA, Pretorius E. Platelet activity and hypercoagulation in type 2 diabetes. Cardiovasc Diabetol. 2018;17(1):141. Epub 2018/11/06. doi: 10.1186/s12933-018-0783-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiesner T, Bugl S, Mayer F, Hartmann JT, Kopp HG. Differential changes in platelet VEGF, Tsp, CXCL12, and CXCL4 in patients with metastatic cancer. Clin Exp Metastasis. 2010;27(3):141–9. Epub 2010/02/26. doi: 10.1007/s10585-010-9311-6 . [DOI] [PubMed] [Google Scholar]
- 8.Zara M, Canobbio I, Visconte C, Canino J, Torti M, Guidetti GF. Molecular mechanisms of platelet activation and aggregation induced by breast cancer cells. Cell Signal. 2018;48:45–53. Epub 2018/05/01. doi: 10.1016/j.cellsig.2018.04.008 . [DOI] [PubMed] [Google Scholar]
- 9.Vander Heiden MG, DeBerardinis RJ. Understanding the Intersections between Metabolism and Cancer Biology. Cell. 2017;168(4):657–69. Epub 2017/02/12. doi: 10.1016/j.cell.2016.12.039 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buergy D, Wenz F, Groden C, Brockmann MA. Tumor-platelet interaction in solid tumors. International journal of cancer. 2012;130(12):2747–60. Epub 2012/01/21. doi: 10.1002/ijc.27441 . [DOI] [PubMed] [Google Scholar]
- 11.Chadha AS, Kocak-Uzel E, Das P, Minsky BD, Delclos ME, Mahmood U, et al. Paraneoplastic thrombocytosis independently predicts poor prognosis in patients with locally advanced pancreatic cancer. Acta Oncol. 2015;54(7):971–8. Epub 2015/01/23. doi: 10.3109/0284186X.2014.1000466 . [DOI] [PubMed] [Google Scholar]
- 12.Holmes CE, Levis JE, Schneider DJ, Bambace NM, Sharma D, Lal I, et al. Platelet phenotype changes associated with breast cancer and its treatment. Platelets. 2016;27(7):703–11. Epub 2016/11/05. doi: 10.3109/09537104.2016.1171302 . [DOI] [PubMed] [Google Scholar]
- 13.Jefferson K, Persad R. Poor prognosis associated with thrombocytosis in patients with renal cell carcinoma. BJU Int. 2001;87(7):715–6. Epub 2001/05/15. doi: 10.1046/j.1464-410x.2001.02169.x . [DOI] [PubMed] [Google Scholar]
- 14.Monreal M, Fernandez-Llamazares J, Pinol M, Julian JF, Broggi M, Escola D, et al. Platelet count and survival in patients with colorectal cancer—a preliminary study. Thromb Haemost. 1998;79(5):916–8. Epub 1998/06/03. . [PubMed] [Google Scholar]
- 15.Chaudhary PK, Kim S, Kim S. An Insight into Recent Advances on Platelet Function in Health and Disease. Int J Mol Sci. 2022;23(11). Epub 2022/06/11. doi: 10.3390/ijms23116022 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang L, Wang X, Guo E, Mao X, Miao S. Emerging roles of platelets in cancer biology and their potential as therapeutic targets. Frontiers in oncology. 2022;12:939089. Epub 2022/08/09. doi: 10.3389/fonc.2022.939089 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lazar S, Goldfinger LE. Platelets and extracellular vesicles and their cross talk with cancer. Blood. 2021;137(23):3192–200. Epub 2021/05/04. doi: 10.1182/blood.2019004119 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morel O, Jesel L, Freyssinet JM, Toti F. Cellular mechanisms underlying the formation of circulating microparticles. Arterioscler Thromb Vasc Biol. 2011;31(1):15–26. Epub 2010/12/17. doi: 10.1161/ATVBAHA.109.200956 . [DOI] [PubMed] [Google Scholar]
- 19.Sinauridze EI, Kireev DA, Popenko NY, Pichugin AV, Panteleev MA, Krymskaya OV, et al. Platelet microparticle membranes have 50- to 100-fold higher specific procoagulant activity than activated platelets. Thromb Haemost. 2007;97(3):425–34. Epub 2007/03/06. . [PubMed] [Google Scholar]
- 20.Edelstein LC. The role of platelet microvesicles in intercellular communication. Platelets. 2017;28(3):222–7. Epub 2016/12/09. doi: 10.1080/09537104.2016.1257114 . [DOI] [PubMed] [Google Scholar]
- 21.Laffont B, Corduan A, Ple H, Duchez AC, Cloutier N, Boilard E, et al. Activated platelets can deliver mRNA regulatory Ago2*microRNA complexes to endothelial cells via microparticles. Blood. 2013;122(2):253–61. Epub 2013/05/09. doi: 10.1182/blood-2013-03-492801 . [DOI] [PubMed] [Google Scholar]
- 22.Xia L, Zeng Z, Tang WH. The Role of Platelet Microparticle Associated microRNAs in Cellular Crosstalk. Frontiers in cardiovascular medicine. 2018;5:29. Epub 2018/04/20. doi: 10.3389/fcvm.2018.00029 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anene C, Graham AM, Boyne J, Roberts W. Platelet microparticle delivered microRNA-Let-7a promotes the angiogenic switch. Biochimica et biophysica acta Molecular basis of disease. 2018;1864(8):2633–43. Epub 20180421. doi: 10.1016/j.bbadis.2018.04.013 . [DOI] [PubMed] [Google Scholar]
- 24.Laffont B, Corduan A, Rousseau M, Duchez AC, Lee CH, Boilard E, et al. Platelet microparticles reprogram macrophage gene expression and function. Thromb Haemost. 2016;115(2):311–23. Epub 2015/09/04. doi: 10.1160/TH15-05-0389 . [DOI] [PubMed] [Google Scholar]
- 25.Liang H, Yan X, Pan Y, Wang Y, Wang N, Li L, et al. MicroRNA-223 delivered by platelet-derived microvesicles promotes lung cancer cell invasion via targeting tumor suppressor EPB41L3. Molecular cancer. 2015;14:58. Epub 20150311. doi: 10.1186/s12943-015-0327-z . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Semple JW. Platelets deliver small packages of genetic function. Blood. 2013;122(2):155–6. Epub 2013/07/13. doi: 10.1182/blood-2013-05-502609 . [DOI] [PubMed] [Google Scholar]
- 27.Duan X, Zhan Q, Song B, Zeng S, Zhou J, Long Y, et al. Detection of platelet microRNA expression in patients with diabetes mellitus with or without ischemic stroke. Journal of diabetes and its complications. 2014;28(5):705–10. Epub 2014/06/09. doi: 10.1016/j.jdiacomp.2014.04.012 . [DOI] [PubMed] [Google Scholar]
- 28.Elgheznawy A, Shi L, Hu J, Wittig I, Laban H, Pircher J, et al. Dicer cleavage by calpain determines platelet microRNA levels and function in diabetes. Circulation research. 2015;117(2):157–65. Epub 2015/05/07. doi: 10.1161/CIRCRESAHA.117.305784 . [DOI] [PubMed] [Google Scholar]
- 29.Nagalla S, Shaw C, Kong X, Kondkar AA, Edelstein LC, Ma L, et al. Platelet microRNA-mRNA coexpression profiles correlate with platelet reactivity. Blood. 2011;117(19):5189–97. Epub 2011/03/19. doi: 10.1182/blood-2010-09-299719 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willeit P, Zampetaki A, Dudek K, Kaudewitz D, King A, Kirkby NS, et al. Circulating microRNAs as novel biomarkers for platelet activation. Circulation research. 2013;112(4):595–600. Epub 2013/01/04. doi: 10.1161/CIRCRESAHA.111.300539 . [DOI] [PubMed] [Google Scholar]
- 31.Zampetaki A, Kiechl S, Drozdov I, Willeit P, Mayr U, Prokopi M, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circulation research. 2010;107(6):810–7. doi: 10.1161/CIRCRESAHA.110.226357 . [DOI] [PubMed] [Google Scholar]
- 32.Stratz C, Nuhrenberg T, Fiebich BL, Amann M, Kumar A, Binder H, et al. Controlled type II diabetes mellitus has no major influence on platelet micro-RNA expression. Results from micro-array profiling in a cohort of 60 patients. Thromb Haemost. 2014;111(5):902–11. Epub 2013/12/20. doi: 10.1160/TH13-06-0476 . [DOI] [PubMed] [Google Scholar]
- 33.Clement E, Lazar I, Attané C, Carrié L, Dauvillier S, Ducoux-Petit M, et al. Adipocyte extracellular vesicles carry enzymes and fatty acids that stimulate mitochondrial metabolism and remodeling in tumor cells. The EMBO journal. 2020;39(3):e102525. Epub 20200110. doi: 10.15252/embj.2019102525 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Clement E, Lazar I, Muller C, Nieto L. Obesity and melanoma: could fat be fueling malignancy? Pigment Cell Melanoma Res. 2017;30(3):294–306. Epub 20170420. doi: 10.1111/pcmr.12584 . [DOI] [PubMed] [Google Scholar]
- 35.Lazar I, Clement E, Attane C, Muller C, Nieto L. A new role for extracellular vesicles: how small vesicles can feed tumors’ big appetite. J Lipid Res. 2018;59(10):1793–804. Epub 20180420. doi: 10.1194/jlr.R083725 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lazar I, Clement E, Dauvillier S, Milhas D, Ducoux-Petit M, LeGonidec S, et al. Adipocyte Exosomes Promote Melanoma Aggressiveness through Fatty Acid Oxidation: A Novel Mechanism Linking Obesity and Cancer. Cancer Res. 2016;76(14):4051–7. Epub 20160523. doi: 10.1158/0008-5472.CAN-16-0651 . [DOI] [PubMed] [Google Scholar]
- 37.Jafari N, Chen A, Kolla M, Pompa IR, Qiu Y, Yu R, et al. Novel plasma exosome biomarkers for prostate cancer progression in co-morbid metabolic disease. Adv Cancer Biol Metastasis. 2022;6. Epub 20221104. doi: 10.1016/j.adcanc.2022.100073 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angelescu MA, Andronic O, Dima SO, Popescu I, Meivar-Levy I, Ferber S, et al. miRNAs as Biomarkers in Diabetes: Moving towards Precision Medicine. Int J Mol Sci. 2022;23(21). Epub 20221025. doi: 10.3390/ijms232112843 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frixa T, Donzelli S, Blandino G. Oncogenic MicroRNAs: Key Players in Malignant Transformation. Cancers (Basel). 2015;7(4):2466–85. Epub 20151218. doi: 10.3390/cancers7040904 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Otmani K, Lewalle P. Tumor Suppressor miRNA in Cancer Cells and the Tumor Microenvironment: Mechanism of Deregulation and Clinical Implications. Frontiers in oncology. 2021;11:708765. Epub 20211015. doi: 10.3389/fonc.2021.708765 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. Epub 20160128. doi: 10.1038/sigtrans.2015.4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W, Liu J, Wang G. The role of microRNAs in human breast cancer progression. Tumour Biol. 2014;35(7):6235–44. Epub 20140618. doi: 10.1007/s13277-014-2202-8 . [DOI] [PubMed] [Google Scholar]
- 43.Vismara M, Zara M, Negri S, Canino J, Canobbio I, Barbieri SS, et al. Platelet-derived extracellular vesicles regulate cell cycle progression and cell migration in breast cancer cells. Biochim Biophys Acta Mol Cell Res. 2021;1868(1):118886. Epub 2020/10/12. doi: 10.1016/j.bbamcr.2020.118886 . [DOI] [PubMed] [Google Scholar]
- 44.Du L, Tao X, Shen X. Human umbilical cord mesenchymal stem cell-derived exosomes inhibit migration and invasion of breast cancer cells via miR-21-5p/ZNF367 pathway. Breast Cancer. 2021;28(4):829–37. Epub 20210326. doi: 10.1007/s12282-021-01218-z . [DOI] [PubMed] [Google Scholar]
- 45.Li Y, Liang C, Ma H, Zhao Q, Lu Y, Xiang Z, et al. miR-221/222 promotes S-phase entry and cellular migration in control of basal-like breast cancer. Molecules. 2014;19(6):7122–37. Epub 20140530. doi: 10.3390/molecules19067122 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han M, Wang Y, Guo G, Li L, Dou D, Ge X, et al. microRNA-30d mediated breast cancer invasion, migration, and EMT by targeting KLF11 and activating STAT3 pathway. J Cell Biochem. 2018;119(10):8138–45. Epub 20180619. doi: 10.1002/jcb.26767 . [DOI] [PubMed] [Google Scholar]
- 47.Roweth HG, Battinelli EM. Lessons to learn from tumor-educated platelets. Blood. 2021;137(23):3174–80. doi: 10.1182/blood.2019003976 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chaari M, Ayadi I, Rousseau A, Lefkou E, Van Dreden P, Sidibe F, et al. Impact of breast cancer stage, time from diagnosis and chemotherapy on plasma and cellular biomarkers of hypercoagulability. BMC Cancer. 2014;14:991. Epub 20141222. doi: 10.1186/1471-2407-14-991 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Al-Mahayni S, Ali M, Khan M, Jamsheer F, Moin ASM, Butler AE. Glycemia-Induced miRNA Changes: A Review. Int J Mol Sci. 2023;24(8). Epub 20230419. doi: 10.3390/ijms24087488 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Landry P, Plante I, Ouellet DL, Perron MP, Rousseau G, Provost P. Existence of a microRNA pathway in anucleate platelets. Nat Struct Mol Biol. 2009;16(9):961–6. Epub 20090809. doi: 10.1038/nsmb.1651 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Plé H, Landry P, Benham A, Coarfa C, Gunaratne PH, Provost P. The repertoire and features of human platelet microRNAs. PloS one. 2012;7(12):e50746. Epub 20121204. doi: 10.1371/journal.pone.0050746 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krammer TL, Zeibig S, Schrottmaier WC, Pirabe A, Goebel S, Diendorfer AB, et al. Comprehensive Characterization of Platelet-Enriched MicroRNAs as Biomarkers of Platelet Activation. Cells. 2022;11(8). Epub 20220407. doi: 10.3390/cells11081254 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grieco GE, Besharat ZM, Licata G, Fignani D, Brusco N, Nigi L, et al. Circulating microRNAs as clinically useful biomarkers for Type 2 Diabetes Mellitus: miRNomics from bench to bedside. Transl Res. 2022;247:137–57. Epub 20220327. doi: 10.1016/j.trsl.2022.03.008 . [DOI] [PubMed] [Google Scholar]
- 54.Pan Y, Li J, Zhang Y, Wang N, Liang H, Liu Y, et al. Slug-upregulated miR-221 promotes breast cancer progression through suppressing E-cadherin expression. Scientific reports. 2016;6:25798. Epub 20160513. doi: 10.1038/srep25798 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Roscigno G, Quintavalle C, Donnarumma E, Puoti I, Diaz-Lagares A, Iaboni M, et al. MiR-221 promotes stemness of breast cancer cells by targeting DNMT3b. Oncotarget. 2016;7(1):580–92. doi: 10.18632/oncotarget.5979 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26(19):2799–803. Epub 20061030. doi: 10.1038/sj.onc.1210083 . [DOI] [PubMed] [Google Scholar]
- 57.Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ, Hou JH, et al. Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast cancer research: BCR. 2011;13(1):R2. Epub 20110110. doi: 10.1186/bcr2803 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang S, Guo LJ, Zhang G, Wang LL, Hao S, Gao B, et al. Roles of microRNA-124a and microRNA-30d in breast cancer patients with type 2 diabetes mellitus. Tumour Biol. 2016;37(8):11057–63. Epub 20160220. doi: 10.1007/s13277-016-4981-6 . [DOI] [PubMed] [Google Scholar]
- 59.Veilleux V, Pichaud N, Boudreau LH, Robichaud GA. Mitochondria Transfer by Platelet-Derived Microparticles Regulates Breast Cancer Bioenergetic States and Malignant Features. Mol Cancer Res. 2024;22(3):268–81. doi: 10.1158/1541-7786.MCR-23-0329 . [DOI] [PubMed] [Google Scholar]
- 60.Faille D, El-Assaad F, Mitchell AJ, Alessi MC, Chimini G, Fusai T, et al. Endocytosis and intracellular processing of platelet microparticles by brain endothelial cells. J Cell Mol Med. 2012;16(8):1731–8. doi: 10.1111/j.1582-4934.2011.01434.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dong G, Liang X, Wang D, Gao H, Wang L, Wang L, et al. High expression of miR-21 in triple-negative breast cancers was correlated with a poor prognosis and promoted tumor cell in vitro proliferation. Med Oncol. 2014;31(7):57. Epub 20140615. doi: 10.1007/s12032-014-0057-x . [DOI] [PubMed] [Google Scholar]
- 62.Fang H, Xie J, Zhang M, Zhao Z, Wan Y, Yao Y. miRNA-21 promotes proliferation and invasion of triple-negative breast cancer cells through targeting PTEN. Am J Transl Res. 2017;9(3):953–61. Epub 20170315. . [PMC free article] [PubMed] [Google Scholar]
- 63.Yang L, Feng Y, Qi P, Xu S, Zhou Y. Mechanism of serum miR-21 in the pathogenesis of familial and triple negative breast cancer. J Biol Regul Homeost Agents. 2016;30(4):1041–5. . [PubMed] [Google Scholar]
- 64.Berckmans RJ, Nieuwland R, Böing AN, Romijn FP, Hack CE, Sturk A. Cell-derived microparticles circulate in healthy humans and support low grade thrombin generation. Thromb Haemost. 2001;85(4):639–46. . [PubMed] [Google Scholar]
- 65.Miyazawa B, Trivedi A, Togarrati PP, Potter D, Baimukanova G, Vivona L, et al. Regulation of endothelial cell permeability by platelet-derived extracellular vesicles. J Trauma Acute Care Surg. 2019;86(6):931–42. doi: 10.1097/TA.0000000000002230 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alberro A, Iparraguirre L, Fernandes A, Otaegui D. Extracellular Vesicles in Blood: Sources, Effects, and Applications. Int J Mol Sci. 2021;22(15). Epub 20210729. doi: 10.3390/ijms22158163 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Michael JV, Wurtzel JGT, Mao GF, Rao AK, Kolpakov MA, Sabri A, et al. Platelet microparticles infiltrating solid tumors transfer miRNAs that suppress tumor growth. Blood. 2017;130(5):567–80. Epub 20170512. doi: 10.1182/blood-2016-11-751099 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen J, Shin VY, Siu MT, Ho JC, Cheuk I, Kwong A. miR-199a-5p confers tumor-suppressive role in triple-negative breast cancer. BMC Cancer. 2016;16(1):887. Epub 20161114. doi: 10.1186/s12885-016-2916-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Das K, Paul S, Singh A, Ghosh A, Roy A, Ansari SA, et al. Triple-negative breast cancer-derived microvesicles transfer microRNA221 to the recipient cells and thereby promote epithelial-to-mesenchymal transition. The Journal of biological chemistry. 2019;294(37):13681–96. Epub 20190724. doi: 10.1074/jbc.RA119.008619 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guo Q, Pan K, Qiu P, Liu Z, Chen J, Lin J. Identification of an exosome-related signature associated with prognosis and immune infiltration in breast cancer. Scientific reports. 2023;13(1):18198. Epub 20231024. doi: 10.1038/s41598-023-45325-7 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Qiu P, Guo Q, Lin J, Pan K, Chen J, Ding M. An exosome-related long non-coding RNAs risk model could predict survival outcomes in patients with breast cancer. Scientific reports. 2022;12(1):22322. Epub 20221224. doi: 10.1038/s41598-022-26894-5 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang H, Wang R, Luo L, Hong J, Chen X, Shen K, et al. An exosome-based specific transcriptomic signature for profiling regulation patterns and modifying tumor immune microenvironment infiltration in triple-negative breast cancer. Front Immunol. 2023;14:1295558. Epub 20231206. doi: 10.3389/fimmu.2023.1295558 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bi O, Anene CA, Nsengimana J, Shelton M, Roberts W, Newton-Bishop J, et al. SFPQ promotes an oncogenic transcriptomic state in melanoma. Oncogene. 2021;40(33):5192–203. Epub 20210703. doi: 10.1038/s41388-021-01912-4 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. 2011. 2011;17(1):3. Epub 2011-08-02. doi: 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- 75.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9. Epub 20120304. doi: 10.1038/nmeth.1923 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Anders S, Pyl PT, Huber W. HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–9. Epub 20140925. doi: 10.1093/bioinformatics/btu638 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40. Epub 20091111. doi: 10.1093/bioinformatics/btp616 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic acids research. 2011;39(Database issue):D163–9. Epub 20101110. doi: 10.1093/nar/gkq1107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang JZ, Du Z, Payattakool R, Yu PS, Chen CF. A new method to measure the semantic similarity of GO terms. Bioinformatics. 2007;23(10):1274–81. Epub 20070307. doi: 10.1093/bioinformatics/btm087 . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Raw count of miRNA transcript across all samples.
(XLSX)
Le CPM normalised expression levels of miRNA transcripts across all samples.
(XLSX)
Differential expression results for miRNAs passing minimum expression threshold (1 CPM in 3 samples).
(XLSX)
Functionally validated miRNA-mRNA interactions for the differentially expressed miRNA in S3 Table found miRTarBase.
(XLSX)
Complete gene ontology analysis of molecular functions associated the target mRNA between each group.
(XLSX)
Anonymised data for T2DM patients recruited to the study.
(XLSX)
Details of oligonucleotides used in this study.
(XLSX)
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in the GEO repository under accession GSE249475 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE249475).