Abstract
Scaffolding proteins play a critical role in the assembly of certain viruses by directing the formation and maturation of a precursor capsid. Using electron cryomicroscopy difference mapping, we have identified an altered arrangement of a mutant scaffolding within the bacteriophage P22 procapsid. This mutant scaffolding allows us to directly visualize scaffolding density within the P22 procapsid. Based on these observations we propose a model for why the mutant prevents scaffolding release and capsid maturation.
Critical stages in many viral life cycles involve large-scale conformational transitions. In certain classes of viruses, including the herpesviruses, adenoviruses, and double-stranded-DNA bacteriophages, the regulation of these transitions requires scaffolding proteins, molecules not found in the mature virion but essential for assembly (2, 4, 17). The assembly pathway for these viruses is epitomized by the Salmonella phage P22, a T=7 icosahedral phage. Assembly of P22 requires approximately 300 scaffolding subunits in addition to 420 coat subunits (3). These coassemble into a precursor structure, the procapsid, that contains scaffolding subunits within the capsid instead of DNA and is smaller and rounder than the mature virion. Upon the commencement of DNA packaging, all of the scaffolding molecules exit the procapsid intact, probably through channels present at the centers of the hexameric capsomeres, and are recycled in further rounds of procapsid assembly (9). The DNA is packaged into the capsid and the capsid undergoes conformational transitions resulting in expansion, angularization, and closure of the hexon channels (16). The scaffolding protein plays a critical role in assembly, as in the absence of scaffolding protein only aberrant, incorrectly sized, or otherwise nonproductive capsids are formed (5).
An additional role for scaffolding in the maturation transition is suggested by the phenotype of a temperature-sensitive scaffolding mutant, R74C/L177I. The mutant scaffolding protein assembles into procapsids, but the procapsids fail to package DNA (6). The R74C/L177I scaffolding protein appears to be defective in the release step, because experiments in vitro demonstrate that the mutant scaffolding is difficult to extract from the procapsids (6). Although the mutant scaffolding proteins can form disulfide-linked dimers (15), the mutant scaffolding shows altered in vitro extraction kinetics even under reducing conditions (B. Greene, unpublished data), suggesting that the cross-link itself is not the cause of the mutant phenotype. The site of the mutation is not within the scaffolding domain required for binding to the coat protein but in a region involved in mediating scaffolding-scaffolding interactions (8, 14). This suggests that the arrangement of scaffolding subunits might be altered within the mutant procapsids. In order to determine if this is the case, we used electron cryomicroscopy difference mapping to localize the binding sites of the mutant scaffolding protein within the procapsid. Although the scaffolding location was previously inferred using difference mapping (19), the scaffolding density was not visible. This mutant permitted us to directly observe for the first time density representing the scaffolding protein itself.
Localization of scaffolding in R74C/L177I procapsids reveals coat-scaffolding interactions different from wild-type scaffolding.
The three-dimensional structures of the R74C/L177I scaffolding-containing procapsids and procapsids assembled in the absence of scaffolding (20) were compared to determine the locations of the R74C/L77I scaffolding-coat interaction. Figure 1 shows the identified differences. Specifically, significant differences are present at the trimer tips of the b, c, d, e, f, and g subunits within the procapsid (Fig. 1). This localization of R74C/L177I scaffolding differs from that of wild-type scaffolding, in which coat-scaffolding interactions are present only at the trimer tips of the b, c, f, and g subunits (19). As with the wild-type scaffolding localization, the relatively small size of the densities is believed to be the result of a highly disordered region immediately preceding the C-terminal 30 amino acids that interact with the coat protein (18).
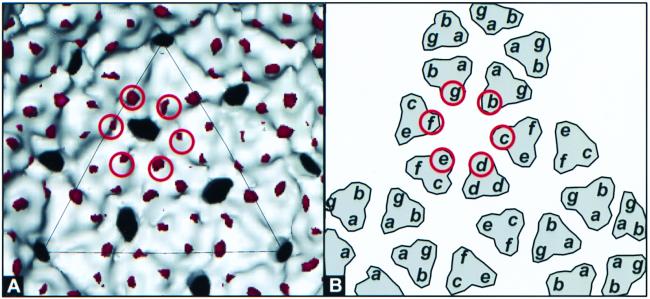
FIG. 1.
Localization of the scaffolding protein within the R74C/L177I procapsid by difference maps computed between procapsids assembled in the absence of the scaffolding protein (20) and the 22-Å structure of the R74C/L177I capsids. The R74C/L177I three-dimensional structure was determined using 161 particle images obtained from purified R74C/L177I (6) capsids imaged at ∼1.0 μm underfocus with 100-kV flood beam imaging. Prior to calculation of the difference maps, radial scaling and density scaling were performed (1, 10, 22). Both algebraic difference maps (11) and the Student t test (12, 13, 21) were used to identify and confirm statistically significant structural differences (19). Shown in white is the three-dimensional structure of the procapsid assembled in the absence of the scaffolding protein (20). Shown in red is the density corresponding to the differences attributable to the scaffolding protein. No significant differences were observed on the outer procapsid surface. (A) Inner surface, with differences at the trimer tips of the b, c, d, e, f, and g subunits circled. (B) Schematic diagram depicting the procapsid icosahedral lattice inner surface view. The subunits with R74C/L177I scaffolding bound are identified by red circles.
Visualization of R74C/L177I scaffolding within the procapsid.
Members of our group have also determined the 15-Å structure of the R74C/L177I procapsid (23), which depicts the point of interaction between the R74C/L177I scaffolding and coat proteins. Figure 2 shows an enlarged view of this 15-Å procapsid structure inner surface contoured at 118% molecular volume (contour chosen based on the absence of floating noise). At this contour the inner procapsid surface contains knob-like densities at the trimer tips of the b, c, d, e, f, and g subunits (Fig. 2, red density). Because this contour does not result in the addition of floating noise but does reveal densities consistent with our computed difference maps, we believe that the additional densities present at this contour are significant. Furthermore, the observed density corresponds to an approximately 15- by 20- by 25-Å region with an approximate molecular mass of 8 kDa that is consistent with the expected size of the C-terminal 30-amino-acid scaffolding protein coat binding domain (18). Consequently, we attribute the observed densities at the trimer tips of the b, c, d, e, f, and g subunits to this ordered region of the scaffolding protein.
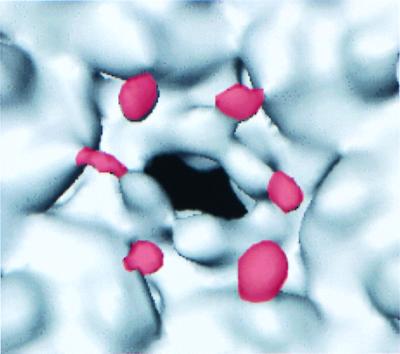
FIG. 2.
View of the 15-Å-resolution R74C/L177I procapsid three-dimensional structure inner surface contoured at 118% molecular volume. The three-dimensional structure was determined using 697 400-kV flood beam particle images with defocus values between 0.9 and 1.4 μm underfocus (23). Shown is the region surrounding one hexon hole, with the density attributed to the R74C/L177I scaffolding protein colored in red. The boundaries for the red density were determined by visual comparison of the difference map shown in Fig. 1A and the densities present at the trimer tips of the 15-Å-resolution R74C/L177I procapsid structure.
Effects of the R74C/L177I mutation on scaffolding binding.
In the wild-type scaffolding localization (19), we proposed that the scaffolding was bound to the coat lattice as dimers spanning approximately 40 Å across the coat trimer tips within a single trimer cluster (Fig. 3A, b-g and c-f subunits). In this mutant scaffolding localization, we observed these coat-scaffolding interactions and two additional interactions at the e-d subunits. To account for these additional observed densities, we propose that the R74C/L177I mutation allows the scaffolding dimer to alter its conformation such that the dimer is capable of spanning a greater distance. Thus, this altered scaffolding dimer would allow stable binding of a scaffolding dimer between neighboring trimer clusters spanning a distance of 50 Å (Fig. 3B, d-e subunits) while also allowing the wild-type dimeric scaffolding interactions to remain. The temperature sensitivity of this mutant could be explained if the scaffolding subunits must partially unfold in order to form this interaction. This possibility is consistent with observations that a domain of the mutant scaffolding protein is significantly destabilized with respect to the wild-type protein (8).
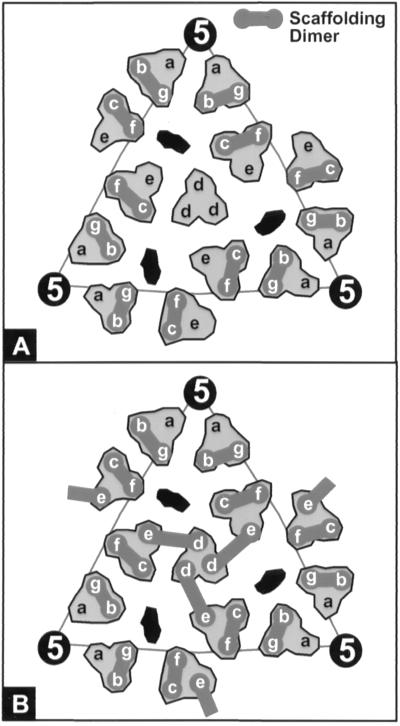
FIG. 3.
Schematic of proposed scaffolding dimer organization within the P22 procapsid. Shown is the icosahedral lattice of the inner surface unit triangle with the seven quasi-equivalent coat subunits labeled a to g. The dark gray dumbbells connecting pairs of scaffolding-containing subunits indicate the positions of scaffolding dimers. The elongated black regions indicate the locations of the hexon holes. (A) Dimeric organization of wild-type scaffolding is within trimer clusters between the b-g and c-f subunits, as previously proposed (19). (B) Proposed dimeric organization of R74C/L177I scaffolding. Dimers form both within trimer clusters (b-g and c-f subunits) and across trimer clusters (e-d subunits).
This model suggests that scaffolding monomers are able to bind to any trimer cluster tip but that only those scaffolding molecules that have formed dimers are stably bound. Interestingly, the mutant procapsids do not appear to contain more molecules of scaffolding protein than wild-type procapsids (6), suggesting that wild-type procapsids contain a class of scaffolding molecules that do not bind the coat lattice in an ordered fashion consistent with this model.
Implications for scaffolding release.
This altered R74C/L177I scaffolding dimer may explain the difficulty in release of the mutant scaffolding proteins through either steric hindrance or extremely tight coat-scaffolding interactions. Specifically, the presence of the additional scaffolding dimers, which are presumably monomers in the wild-type procapsid, may sterically hinder exit of scaffolding through the approximately 35- by 40-Å hexon hole. Alternatively, if the coat-scaffolding interaction at the d-e coat subunits is extremely tight, due to the altered conformation of the scaffolding dimer at these sites, then this scaffolding dimer may never release. At concentrations of denaturant twice that sufficient to extract all the wild-type scaffolding in vitro, approximately a third of the mutant scaffolding subunits remain bound to the coat lattice (6), so this population may represent the subunits bound as altered dimers.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health (AI43656), the National Center for Research Resources of the National Institutes of Health (P41RR02250), the National Institute of General Medical Sciences of the National Institutes of Health (GM47980 to P.E.P.), and the Robert A. Welch Foundation.
REFERENCES
- 1.Booy F P, Trus B L, Newcomb W W, Brown J C, Conway J F, Steven A C. Finding a needle in a haystack: detection of a small protein (the 12-kDa VP26) in a large complex (the 200-MDa capsid of herpes simplex virus) Proc Natl Acad Sci USA. 1994;91:5652–5656. doi: 10.1073/pnas.91.12.5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calender R, editor. The bacteriophages. New York, N.Y: Plenum Publishing; 1988. pp. 15–91. [Google Scholar]
- 3.Casjens S, King J. P22 morphogenesis. I. Catalytic scaffolding protein in capsid assembly. J Supramol Struct. 1974;2:202–224. doi: 10.1002/jss.400020215. [DOI] [PubMed] [Google Scholar]
- 4.D'Halluin J-C M, Martin G R, Torpier G, Boulanger P. Adenovirus type 2 assembly analyzed by reversible cross-linking of labile intermediates. J Virol. 1978;26:357–363. doi: 10.1128/jvi.26.2.357-363.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Earnshaw W, King J. Structure of phage P22 coat protein aggregates formed in the absence of the scaffolding protein. J Mol Biol. 1978;126:721–747. doi: 10.1016/0022-2836(78)90017-7. [DOI] [PubMed] [Google Scholar]
- 6.Greene B, King J. Scaffolding mutants identifying domains required for P22 procapsid assembly and maturation. Virology. 1996;225:82–96. doi: 10.1006/viro.1996.0577. [DOI] [PubMed] [Google Scholar]
- 7.Greene B, King J. Folding and stability of mutant scaffolding proteins defective in P22 capsid assembly. J Biol Chem. 1999;274:16141–16146. doi: 10.1074/jbc.274.23.16141. [DOI] [PubMed] [Google Scholar]
- 8.Greene B, King J. In vitro unfolding/refolding of wild-type phage P22 scaffolding protein reveals capsid-binding domain. J Biol Chem. 1999;274:16135–16140. doi: 10.1074/jbc.274.23.16135. [DOI] [PubMed] [Google Scholar]
- 9.King J, Casjens S. Catalytic head assembling protein in virus morphogenesis. Nature. 1974;251:112–119. doi: 10.1038/251112a0. [DOI] [PubMed] [Google Scholar]
- 10.Lawton J A, Prasad B V V. Automated software package for icosahedral virus reconstruction. J Struct Biol. 1996;116:209–215. doi: 10.1006/jsbi.1996.0032. [DOI] [PubMed] [Google Scholar]
- 11.Marvik O J, Dokland T, Nokling R H, Jacobsen E, Larsen T, Lindqvist B H. The capsid size-determining protein Sid forms an external scaffold on phage P4 procapsids. J Mol Biol. 1995;251:59–75. doi: 10.1006/jmbi.1995.0416. [DOI] [PubMed] [Google Scholar]
- 12.McGough A, Way M, DeRosier D. Determination of the alpha-actinin binding site on actin filaments by cryoelectron microscopy and image analysis. J Cell Biol. 1994;126:433–443. doi: 10.1083/jcb.126.2.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milligan R A, Flicker P F. Structural relationships of actin, myosin, and tropomyosin revealed by cryo-electron microscopy. J Cell Biol. 1987;105:29–39. doi: 10.1083/jcb.105.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker M, Casjens S, Prevelige P E., Jr Functional domains of bacteriophage P22 scaffolding protein. J Mol Biol. 1998;281:69–79. doi: 10.1006/jmbi.1998.1917. [DOI] [PubMed] [Google Scholar]
- 15.Parker M H, Stafford III W F, Prevelige P E., Jr Bacteriophage P22 scaffolding protein forms oligomers in solution. J Mol Biol. 1997;268:655–665. doi: 10.1006/jmbi.1997.0995. [DOI] [PubMed] [Google Scholar]
- 16.Prasad B V V, Prevelige P E, Marietta E, Chen R O, Thomas D, King J, Chiu W. Three-dimensional transformation of capsids associated with genome packaging in a bacterial virus. J Mol Biol. 1993;231:65–74. doi: 10.1006/jmbi.1993.1257. [DOI] [PubMed] [Google Scholar]
- 17.Rixon F J. Structure and assembly of herpesviruses. Semin Virol. 1993;4:135–144. [Google Scholar]
- 18.Sun, Y., M. H. Parker, P. Weigele, S. Casjens, P. E. Prevelige, Jr., and N. R. Krishna. Structure of the coat protein-binding domain of the scaffolding protein from a double-stranded DNA virus. J. Mol. Biol., in press. [DOI] [PubMed]
- 19.Thuman-Commike P, Greene B, Malinski J, McGough A, Burbea M, Chiu W, Prevelige P E., Jr Mechanism of scaffolding-directed virus assembly suggested by comparison of scaffolding-containing and scaffolding-lacking P22 procapsids. Biophys J. 1999;76:3267–3277. doi: 10.1016/S0006-3495(99)77479-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thuman-Commike P, Greene B, Malinski J A, King J, Chiu W. Role of the scaffolding protein in P22 procapsid size determination suggested by T=4 and T=7 procapsid structures. Biophys J. 1998;74:559–568. doi: 10.1016/S0006-3495(98)77814-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Trachtenberg S, DeRosier D. Three-dimensional structure of the frozen, hydrated flagellar filament. The left-handed filament of Salmonella typhimurium. J Mol Biol. 1987;195:581–601. doi: 10.1016/0022-2836(87)90184-7. [DOI] [PubMed] [Google Scholar]
- 22.Venien-Bryan C, Fuller S D. The organization of the spike complex of Semliki Forest virus. J Mol Biol. 1994;236:572–583. doi: 10.1006/jmbi.1994.1166. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, Z., B. Greene, P. A. Thuman-Commike, J. Jakana, P. E. Prevelige Jr., J. King, and W. Chiu. Visualization of the maturation transition in bacteriophage P22 by electron cryomicroscopy. J. Mol. Biol., in press. [DOI] [PubMed]