Abstract
Although herpesvirus saimiri-transformed T lymphocytes retain multiple normal T-cell functions, only a few changes have been described. By subtractive hybridization, we have isolated a novel cellular gene, ak155, a sequence homolog of the interleukin-10 gene. Specifically herpesvirus saimiri-transformed T cells overexpress ak155 and secrete the protein into the supernatant. In other T-cell lines and in native peripheral blood cells, but not in B cells, ak155 is transcribed at low levels. AK155 forms homodimers similarly to interleukin-10. As a lymphokine, AK155 may contribute to the transformed phenotype of human T cells after infection by herpesvirus saimiri.
Human T lymphocytes are transformed to stable growth in culture after infection with certain subgroup C strains of herpesvirus saimiri (HVS) (saimiriine herpesvirus type 2), a T-cell tumor virus of New World monkeys (1). The transformed human T cells carry multiple nonintegrated viral episomes; they do not release virions and show only limited virus gene expression (1, 11, 23). In a variety of test systems, HVS-transformed T cells were shown to retain essential functions of their nontransformed parental cells (reviewed in references 4, 13, 30, and 32). In particular, the major histocompatibility complex-restricted antigen-specific reactivity of parental T-cell clones was preserved and resulted in increased proliferation, cytokine release, and cytotoxicity after stimulation (3, 6, 34, 43). In contrast to multiple reports on preserved functions, little is known about cellular features which are clearly changed after transformation. The most pronounced difference is a specific type of hyperreactivity to CD2 stimulation via cell-bound CD58 or cross-linked CD2 antibodies (33). Moreover, unusually high levels of gamma interferon are produced after stimulation, which shifts transformed T helper 2 cells to the T helper 0 phenotype (6). Finally, the nonreceptor tyrosine kinase Lyn is aberrantly expressed and enzymatically active in T cells after HVS transformation (12, 44). Functional consequences of this phenomenon have not yet been defined.
Cloning of ak155 by subtractive hybridization.
In order to describe the phenotypic T-cell alterations after HVS transformation in more detail, we applied the technique of subtractive hybridization for cloning cDNA fragments of transcripts which are specifically present in transformed human T cells and not in their untransformed parental cells (23). Using the acidic phenol extraction method, total cellular RNA was prepared from the phorbol ester-stimulated transformed cell line 3C (CD8+ [11]) and from nontransformed T cells of the same donor. cDNA was generated using purified polyadenylated mRNA and Moloney murine leukemia virus reverse transcriptase (Clontech, Heidelberg, Germany). The second strand was synthesized by a mixture of DNA polymerase I, RNase H, Escherichia coli DNA ligase, and T4 DNA polymerase. Double-stranded cDNA was digested with RsaI to create small fragments. Specific adapters were ligated to the cDNA fragments in order to allow subtraction based on representational difference analysis (PCR-Select; Clontech). Advantage KlenTaq polymerase (Clontech) was applied for PCR. Subtracted PCR products were cloned into pCR2.1 (Invitrogen, Groningen, The Netherlands) and sequenced using M13 reverse and T7 primers with the dye dideoxy terminator method (ABI, Weiterstadt, Germany). The resulting library of 399 sequenced plasmids comprised 280 viral and 119 cellular cDNA clones. Among the cellular cDNAs, 28 clones were not yet represented in the current nucleotide databases (23).
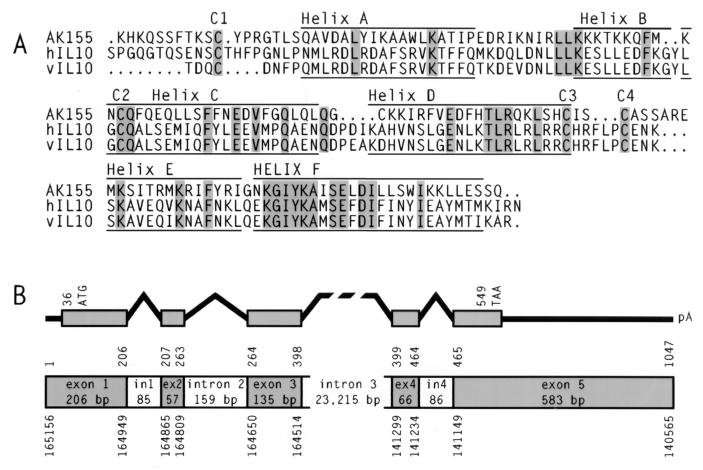
One of these novel cDNA clones, ak155, contained an insert of 506 bp and displayed weak nucleotide homology to the cellular interleukin-10 (IL-10) gene. Subsequently, the cDNA was completed by 5′ and 3′ rapid amplification of cDNA ends (Marathon; Clontech). The resulting cDNA, of 1076 nucleotides (nt), carried 29 nt as a poly(A) tail and a polyadenylation signal at position 1027. The cDNA displayed an open reading frame of 513 nt (position 36 to 549) with coding capacity for a polypeptide of 171 amino acids (aa) and a predicted hydrophobic signal sequence of 21 aa. The isoelectric point was calculated as 10.77. With RNase protection assays, the transcription initiation site was mapped to nt 60 upstream of the ATG, whereas the cDNA clones resulting from 5′ rapid amplification of cDNA ends started 35 nt upstream of the translation initiation site. The predicted AK155 protein showed 24.7% amino acid identity and 47% amino acid similarity to human IL-10 (Fig. 1A). The homology values were similar when AK155 was compared to the human, murine, and bovine IL-10 molecules and to IL-10 of Epstein-Barr virus (EBV). The structural prediction generated with the Genetics Computer Group program package indicated a series of six helices and four highly conserved cysteine residues which are assumed to be relevant for the dimer formation of IL-10. The structural predictions are supported by experimental data on the viral IL-10 variant (45).
FIG. 1.
Amino acid sequence alignment and genomic structure of ak155. (A) The amino acid sequences of AK155, human IL-10 (hIL10), and EBV IL-10 (vIL10) were aligned. Identical amino acids are shaded. Cysteine residues C1 to C4 are conserved. Six predicted helical areas (helices A to F) for the three proteins are marked. (B) The genomic exon-intron structure and the ak155 coding region are depicted. The nucleotide positions above the structure refer to ak155 cDNA. The nucleotide positions below the structure correspond to the genomic sequence of the human chromosome 12q15 region.
Chromosomal localization and genomic structure.
Upon further database searches, we detected a local nucleotide sequence identity of ak155 to chromosome 12q15 at a genomic sequence-tagged site (accession no. U29151) used for mapping the genomic region in a 6-Mb yeast artificial chromosome contig (15, 38). Similarly to the gamma interferon gene, downstream at a distance of 41 kb, ak155 is oriented towards the centromere. We obtained the respective genomic cosmids and plasmid subclones from E. Schoenmakers (Louvain, Belgium) and determined the exon-intron structure of the gene (Fig. 1B). The five ak155 exons of 206, 57, 135, 66, and 583 bp are disrupted by three small introns (85, 159, and 86 bp) and one large intron of more than 23 kb. We sequenced the 5′ and 3′ flanking regions, the exons, and the small introns from the cosmid clones. Recently, the genomic sequence of the respective region of human chromosome 12q15 has become available in GenBank (accession no. AC007458; 191,111 bp; BAC RPCI11-444B24). Within this entry of high-throughput genome sequence data, our genomic sequences correspond to nt 140063 to 141674 (1,612 nt comprising exons 4 and 5 and the 3′ region) and 159771 to 166615 (6,839 nt comprising the promoter region, exons 1 to 3, and a part of intron 3) with a gap in intron 3. Whereas the exons are strictly conserved in both genomic sequences, some allelic divergence was observed in the 5′ upstream region (four point mutations and deletion of 3 nt within a region of 395 nt) and within intron 3 (nine point mutations, one insertion of 3 nt, and one deletion of 6 nt within a stretch of 3,635 nt).
Overexpression of ak155 in HVS-transformed lymphocytes.
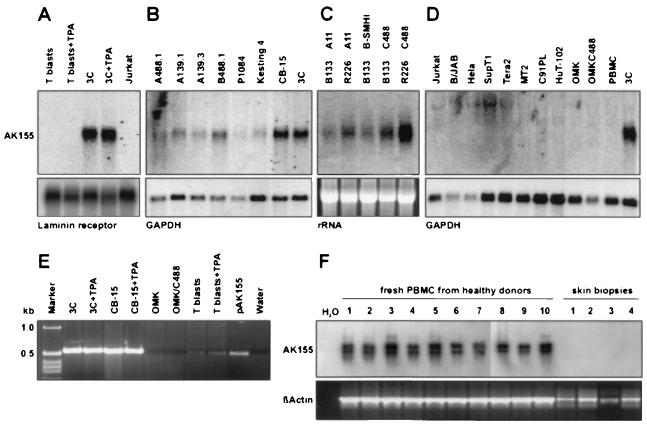
In the next step, the expression pattern was studied. The ak155 cDNA had been cloned from an HVS-transformed human CD8+ T-cell line (3C, transformed by virus strain C488 [11, 23]). First, we analyzed ak155 transcription by Northern blotting utilizing total cellular RNA and the coding region as probe DNA. Whereas strong ak155 signals at a position corresponding to 1.3 kb were readily detectable in T-cell line 3C, there was no hybridization found with mRNA from the human T-cell leukemia line Jurkat or from primary T cells after mitogen stimulation and cultivation in the presence of IL-2. Additional stimulation with the phorbol ester tetradecanoyl phorbol acetate (TPA) (2 ng/ml for 6 h) did not induce ak155 transcription (Fig. 2A). A series of other HVS-transformed T-cell lines contained ak155 transcripts as well (Fig. 2B): CB-15, Kesting, and A488.1 (CD4+; transformed by C488 [1, 11, 12]), P1084 and B488.1 (CD8+; transformed by C488 [1, 12]), and the C139-transformed T-cell lines A139.1 (γδ T-cell receptor) and A139.3 (αβ, CD4+ [12]). Moreover, we were able to demonstrate ak155 transcripts in transformed T cells from New World monkeys (Saguinus oedipus) (T cells from donors B133 and R226 [24]). Remarkably, ak155 expression was not specific for the virus subgroup used for transformation and was similarly detectable in T cells transformed by the virus strains A11, B-SMHI, and C488 (Fig. 2C). Additionally, various other cell types were tested for ak155 transcripts by Northern blotting (Jurkat, SupT1, MT2, C91PL, HuT-102, B/JAB, HeLa, and Tera2 [Fig. 2D]). With this method, we were unable to identify additional ak155-positive cell types. Infection of the permissive epithelial cell line OMK with HVS C488 did not induce ak155 transcription.
FIG. 2.
ak155 transcription pattern. The transcription of ak155 was analyzed by Northern blotting (A to D) and RT-PCR (E and F). (A) Strong ak155 transcript bands were demonstrated for the HVS-transformed CD8+ human T-cell line 3C, but not for either nontransformed T cells or Jurkat cells. Phorbol ester stimulation (TPA; 2 ng/ml for 6 h) did not affect ak155 transcription. Laminin receptor transcripts are shown as a control. (B) A series of additional HVS-transformed T-cell lines also transcribed ak155: CB-15, Kesting, and A488.1 (CD4+; transformed by C488), P1084 and B488.1 (CD8+; transformed by C488), and the C139-transformed T-cell lines A139.1 (γδ T-cell receptor) and A139.3 (αβ, CD4+). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) transcripts are shown as a control. (C) ak155 transcripts were also demonstrated in transformed T cells from New World monkeys (S. oedipus) (T cells from donors B133 and R226 [24]). ak155 expression was not specific for the virus subgroup used for transformation and was similarly detectable in T cells transformed by the virus strains A11, B-SMHI, and C488. rRNA bands are shown as a transfer control. (D) Various other cell types were tested by Northern blotting for ak155 transcripts (Jurkat, SupT1, MT2, C91PL, HuT-102, B/JAB, HeLa, and Tera2). No additional ak155-positive lines were identified. Infection of the permissive epithelial cell line OMK with HVS C488 did not induce ak155 transcription. GAPDH transcripts are shown as a control. (E) By using RT-PCR we confirmed that HVS-transformed human T-cell lines (CB-15 and 3C) transcribed ak155 at a high level (540-bp fragment). Transcripts were also detected in T blasts but at low levels. Additional phorbol ester stimulation did not change the signal intensity. The cDNA plasmid pAK155 served as a positive control. (F) Weak RT-PCR signals for ak155 transcripts were detected from unstimulated fresh peripheral blood mononuclear cells (PBMC) of 10 healthy blood donors by ethidium bromide staining and confirmed by Southern blot hybridization. β-Actin transcripts are shown as a positive control.
By using reverse transcription (RT)-PCR with random hexamer primers, reverse transcriptase (Superscript; Gibco), and the specific primers HF123 (GTG-AAC-GGA-AAT-GCT-GGT-G) and HF126 (GGC-TTT-GGT-TTA-CTG-ACT-G), we confirmed that a large number of HVS-transformed human T-cell lines transcribed ak155 at high levels (540-bp fragment [example shown in Fig. 2E]). With a RT-PCR protocol with increased sensitivity (Superscript II; Gibco), we screened various laboratory cell lines, mainly of hematopoietic lineages (Table 1) (31). In addition, we tested the same RNA samples for IL-10 and for β-actin transcripts as a positive control. The primers HF360 (TCT-CAA-GGG-GCT-GGG-TCA-GCT-ATC-CCA) and HF361 (ATG-CCC-CAA-GCT-GAG-AAC-CAA-GAC-CCA-GAC) were used for demonstrating IL-10 transcripts, and HF291 (CGG-GAA-ATC-GTG-CGT-GAC-AT) and HF292 (GAA-CTT-TGG-GGG-ATG-CTC-GC) were used for demonstrating β-actin transcripts. The results (Table 1) were monitored in a simple semiquantitative way. Strong signals were easily detectable in ethidium bromide-stained agarose gels (+++); weak signals were still detected by simple ethidium bromide staining (++ [example in Fig. 2F]), whereas in some cases, faint signals were detectable only after Southern blot hybridization of the same gels (+). In order to confirm specificity, all IL-10 gene and ak155 RT-PCR gels were analyzed by Southern blot hybridization. Additionally, selected PCR product samples were tested for specificity by direct sequencing. Whereas the IL-10 gene was transcribed in most cell lines of the T or B lineage, ak155 transcription was rather specific for T cells. A series of leukemia T-cell lines and human T-cell leukemia virus (HTLV)-transformed T-cell lines, as well as primary mitogen-stimulated T cells, showed ak155 transcripts. In contrast, most other cell lines tested were negative for ak155 transcripts. The human herpesvirus 8 (HHV-8)-containing cell line BCBL-1 and the Hodgkin's lymphoma line L428 harbored small amounts of transcripts (+). The ak155 transcript amounts did not seem to depend on the level of T-cell activity: phorbol ester stimulation or inhibitory treatment with cyclosporine did not change the transcript levels observed. Moreover, unstimulated fresh peripheral blood cells of 10 healthy blood donors were positive for ak155 mRNA (++ [Fig. 2F and Table 1]). Thus, we conclude that ak155 is normally expressed by certain T cells at low levels and specifically overexpressed by T cells after HVS transformation.
TABLE 1.
Expression of IL-10 and AK155 in cell lines
| Cell type | Cell linea | Features | RT-PCR signal intensity
|
||
|---|---|---|---|---|---|
| IL-10 | AK155 | β-actin | |||
| T cells | PHA T blasts | T, primary | ++ | +++ | +++ |
| PHA T blasts, TPA (6 h) | T, primary | ++ | +++ | +++ | |
| CB-15 | T, HVS transformed, CD4 | +++ | +++ | +++ | |
| CB-15, TPA (6 h) | T, HVS transformed, CD4 | +++ | +++ | +++ | |
| 3C | T, HVS transformed, CD8 | ++ | +++ | +++ | |
| 3C, TPA (6 h) | T, HVS transformed, CD8 | ++ | +++ | +++ | |
| Jurkat | T, mature | +++ | +++ | +++ | |
| Karpas-45 | T, Pro-ALLb | + | +++ | +++ | |
| Molt-15 | T, Pro-ALL | + | +++ | +++ | |
| Molt-3 | T, cortical | +++ | + | +++ | |
| Molt-16 | T, mature | + | + | +++ | |
| SupT1 | T, mature | + | +++ | +++ | |
| MT2 | T, HTLV-1 transformed | +++ | + | +++ | |
| MT2, TPA (6 h) | T, HTLV-1 transformed | +++ | + | +++ | |
| C91PL | T, HTLV-1 transformed | +++ | + | +++ | |
| C91PL, TPA (6 h) | T, HTLV-1 transformed | +++ | ++ | +++ | |
| HUT102 | T, HTLV-1 transformed | + | ++ | +++ | |
| HUT102, TPA (6 h) | T, HTLV-1 transformed | ++ | +++ | +++ | |
| B cells | B/JAB | B, Burkitt | +++ | − | +++ |
| Raji | B, Burkitt | +++ | − | +++ | |
| Daudi | B, Burkitt | + | − | +++ | |
| Jijoye | B, Burkitt | +++ | − | +++ | |
| SS-EBV | B-LCL, EBV transformed | +++ | − | +++ | |
| AC-EBV | B-LCL, EBV transformed | +++ | − | +++ | |
| ES-EBV | B-LCL, EBV transformed | +++ | − | +++ | |
| RPMI8226 | Plasma cell | ++ | − | +++ | |
| BCBL-1 | B, HHV-8 containing | +++ | + | +++ | |
| BCBL-1, TPA (6 h) | B, HHV-8 containing | +++ | + | +++ | |
| BCBL-1, TPA (24 h) | B, HHV-8 containing | +++ | + | +++ | |
| Other hematological cell lines | THP-1 | Monocytes | + | − | +++ |
| U937 | Monocytes | + | − | +++ | |
| K562 | Erythroleukemia | − | − | +++ | |
| L428 | Hodgkin's disease | + | + | +++ | |
| Fibroblasts | HFF | Fibroblasts | − | − | +++ |
| Carcinoma cell lines | Pancl | Pancreas carcinoma | − | − | +++ |
| HeLa | Cervix carcinoma | − | − | +++ | |
| Tera2 | Teratocarcinoma | ++ | − | +++ | |
| Skin biopsy cells | Skin | Donor 1 | ++ | − | +++ |
| Skin | Donor 2 | ++ | − | +++ | |
| Skin | Donor 3 | ++ | − | +++ | |
| Skin | Donor 4 | ++ | − | +++ | |
| Blood cells | Fresh PBMC | Donor 1 | ++ | ++ | +++ |
| Fresh PBMC | Donor 2 | ++ | ++ | +++ | |
| Fresh PBMC | Donor 3 | ++ | ++ | +++ | |
| Fresh PBMC | Donor 4 | ++ | ++ | +++ | |
| Fresh PBMC | Donor 5 | ++ | ++ | +++ | |
| Fresh PBMC | Donor 6 | ++ | ++ | +++ | |
| Fresh PBMC | Donor 7 | ++ | ++ | +++ | |
| Fresh PBMC | Donor 8 | ++ | ++ | +++ | |
| Fresh PBMC | Donor 9 | ++ | ++ | +++ | |
| Fresh PBMC | Donor 10 | ++ | ++ | +++ | |
Details on the individual cell lines are given in reference 31. PBMC, peripheral blood mononuclear cells.
Pro-ALL, acute lymphocytic leukemia.
Dimer formation and secretion of AK155 from human T cells.
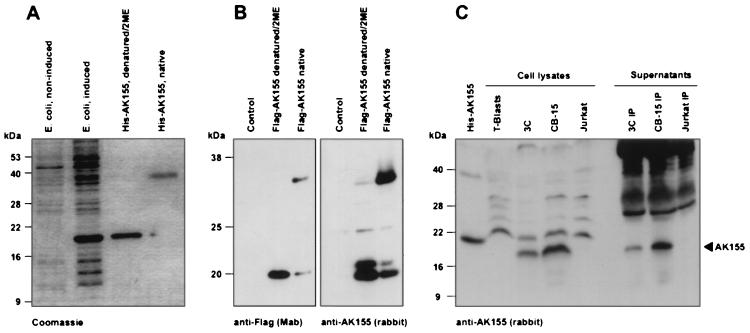
We cloned the ak155 open reading frame without the N-terminal 21-aa signal peptide into the bacterial expression vector pQE30 (Qiagen, Hilden, Germany). After isopropyl-β-d-thiogalactopyranoside (IPTG) induction in E. coli K-12/M15/pRep4, the recombinant N-terminal histidine-tagged protein was purified under denaturing conditions on nickel-nitrilotriacetic acid-agarose columns and renatured by dialysis (Fig. 3A). The denatured recombinant protein migrated as a 19-kDa band in sodium dodecyl sulfate (SDS) gel electrophoresis. When the protein was loaded in the absence of β-mercaptoethanol and without heat denaturation, the 19-kDa band shifted to the 36-kDa position. This is an indication of spontaneous dimer formation and functional protein folding after renaturation. The recombinant protein was used to raise polyclonal rabbit antisera. Moreover, the predicted mature protein coding sequence was fused to a CD8 leader sequence and N-terminal Flag epitope tag as described for IL-10 (18, 26). This construct was cloned into the eukaryotic expression vector pME18S under the control of the SRα hybrid promoter (28, 41). After transfection of COS-7 cells, the recombinant protein was easily detectable by Western blotting either with an anti-Flag monoclonal antibody (Integra, Fernwald, Germany) or with rabbit antiserum. The eukaryotically expressed protein efficiently formed dimers when tested under nondenaturing conditions with either of the two antibodies (Fig. 3B). Finally, the endogenous AK155 protein of HVS-transformed human T cells was demonstrated by Western blotting utilizing rabbit antiserum. AK155 protein was detected in lysates of the transformed T-cell lines 3C (CD8+) and CB-15 (CD4+) without previous immunoprecipitation and in culture supernatants after immunoprecipitation with rabbit antiserum and protein G-agarose (Roche) (Fig. 3C).
FIG. 3.
AK155 dimerization and production by HVS-transformed human T cells. (A) Recombinant amino-terminally histidine-tagged AK155 protein was expressed after induction in E. coli. The protein was purified by nickel-nitrilotriacetic acid-agarose chromatography. The denatured recombinant protein was demonstrated as a 19-kDa band in SDS gels. In absence of 2-mercaptoethanol (2ME) and without heat denaturation, the 19-kDa band shifted to the 36-kDa position. A Coomassie-stained SDS gel is shown. (B) Recombinant amino-terminally Flag-tagged AK155 protein was expressed in COS-7 cells after transfection. The recombinant protein was easily detectable by Western blotting either with the anti-Flag monoclonal antibody or with rabbit antiserum. In both cases, the eukaryotically expressed protein efficiently formed dimers when tested under nondenaturing conditions. (C) The endogenous AK155 protein from HVS-transformed human T cells (3C and CB-15) was demonstrated by Western blotting with rabbit antiserum. Protein was detected in lysates of the transformed T-cell lines 3C and CB-15 without previous immunoprecipitation and in their supernatants after immunoprecipitation with rabbit antiserum and protein G-agarose. As a control, bacterially expressed His-AK155 is shown in the first lane. Due to the histidine tag, this protein appears at a slightly larger size than the endogenous protein from 3C and CB-15 cells. AK155 was detectable neither in Jurkat cells nor in their supernatant. The Western blots were developed with chemiluminescence reactions.
Although many functional features of parental T-cell clones are maintained after transformation by HVS, little is known about functional changes besides CD2 hyperreactivity, aberrant Lyn expression, and the tendency towards the T helper 1 phenotype due to high levels of gamma interferon (6, 12, 33, 44). Although the viral genes stpC and tip, which are essential for transformation, have been identified and functionally characterized, it is still unclear by which mechanism they finally do cause the transformed phenotype of T cells (2, 10, 20, 23; reviewed in references 4 and 21). Using the nonbiased approach of subtractive hybridization and representational difference analysis, we have isolated a novel cellular gene, ak155, which is strongly expressed in HVS-transformed T cells. Northern blotting analysis indicated that ak155 overexpression is highly specific for this cell type.
ak155 has been mapped to the human chromosome 12q15. This locus is in the vicinity of a chromosomal breakpoint region called the multiple-aberration region in benign tumors, such as leiomyomas of the uterus, lipomas, and pleomorphic adenomas of the salivary gland (38, 42). A salivary gland adenoma cell line carried a complex genomic rearrangement in which the high-mobility-group protein HMGIC gene from the 12q15 multiple-aberration region was inserted into the large intron of ak155 (15). The gamma interferon gene is situated downstream of ak155 at a distance of approximately 41 kb. Both the gamma interferon gene and ak155 are overexpressed in HVS-transformed T cells. Thus, a common regulatory mechanism for the two genes is conceivable. In contrast, the human IL-10 gene is localized to human chromosome 1 (22). Simple gene duplication is unlikely, since the sequence homology is rather low. Although the overall intron-exon structure is relatively similar, a large intron of 23 kb is not found in the IL-10 genes of various species.
IL-10 is a multifunctional, pleiotropic cytokine with stimulatory and suppressive effects on B and T cells (reviewed in references 9, 17, and 35). HVS-transformed human T-cell lines are able to produce IL-10 (29) (Table 1). IL-10 is the relevant growth factor for suppressive regulatory T cells (16). The IL-10 receptor consists of two chains (18, 25, 26). The distant homology of AK155 to IL-10 suggests that use of the IL-10 receptor by AK155 is unlikely. Several viruses carry their own variants of the IL-10 gene. The IL-10 homologs of EBV (19) and equine herpesvirus type 2 (37) are much more homologous (approximately 70% amino acid identity) to IL-10 than to AK155 (approximately 25% amino acid identity). EBV IL-10 does engage the IL-10 receptor (27). In mice, EBV IL-10 was shown to inhibit the rejection of transplanted organs and of allogeneic and syngeneic tumors (36, 39). IL-10 of the orf parapoxvirus also exhibits some inhibitory effect on T-cell proliferation (14). IL-10 of EBV has been studied in detail. Although there are subtle functional differences between viral and cellular IL-10 (27), cellular IL-10 seems to be functionally dominant in EBV-transformed B cells (5). However, the viral IL-10 gene is dispensable for virus replication and B-cell transformation by EBV (40). Latently infected EBV-transformed B cells were shown to express a novel IL-12 p40-related cytokine, called EBV-induced gene 3 (7, 8). The situation seems to be analogous for HVS, in which the overexpression of ak155 is one of rare changes between native and transformed T cells. AK155 is a good candidate to play a role in the autocrine growth stimulation leading to spontaneous proliferation of T cells after HVS infection.
Nucleotide sequence accession numbers.
EMBL accession no. AJ251549 to AJ251551 have been assigned to the cDNA and the genomic sequences of ak155.
Acknowledgments
A. Knappe and S. Hör contributed equally to this work.
We are grateful to K. Moore and Y. Liu (Palo Alto, Calif.) for valuable experimental advice and for providing reagents, including the eukaryotic expression vector. We thank E. Schoenmakers (Louvain, Belgium) for genomic cosmids, M. Gramatzki (Erlangen, Germany) for several hematological tumor cell lines, P. von den Driesch (Erlangen, Germany) for skin biopsy samples, F. Brière (Dardilly, France) for stimulating discussions, and B. Fleckenstein (Erlangen, Germany) for continuous support.
This project was funded by the Deutsche Forschungsgemeinschaft, Bonn, Germany (Sonderforschungsbereich 466).
REFERENCES
- 1.Biesinger B, Müller-Fleckenstein I, Simmer B, Lang G, Wittmann S, Platzer E, Desrosiers R C, Fleckenstein B. Stable growth transformation of human T lymphocytes by herpesvirus saimiri. Proc Natl Acad Sci USA. 1992;89:3116–3119. doi: 10.1073/pnas.89.7.3116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Biesinger B, Tsygankov A Y, Fickenscher H, Emmrich F, Fleckenstein B, Bolen J B, Bröker B M. The product of the herpesvirus saimiri open reading frame 1 (tip) interacts with T cell-specific kinase p56lck in transformed cells. J Biol Chem. 1995;270:4729–4734. doi: 10.1074/jbc.270.9.4729. [DOI] [PubMed] [Google Scholar]
- 3.Bröker B M, Tsygankov A Y, Müller-Fleckenstein I, Guse A H, Chitaev N A, Biesinger B, Fleckenstein B, Emmrich F. Immortalization of human T cell clones by herpesvirus saimiri. Signal transduction analysis reveals functional CD3, CD4 and IL-2 receptors. J Immunol. 1993;151:1184–1192. [PubMed] [Google Scholar]
- 4.Bröker B M, Fickenscher H. Herpesvirus saimiri strategies for T cell stimulation and transformation. Med Microbiol Immunol. 1999;187:127–136. doi: 10.1007/s004300050084. [DOI] [PubMed] [Google Scholar]
- 5.Burdin N, Peronne C, Banchereau J, Rousset F. Epstein-Barr virus transformation induces B lymphocytes to produce human interleukin 10. J Exp Med. 1993;177:295–304. doi: 10.1084/jem.177.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Carli M, Berthold S, Fickenscher H, Müller-Fleckenstein I, D'Elios M, Gao Q, Biagiotti R, Giudizi M, Kalden J, Fleckenstein B, Romagnani S, Del Prete G. Immortalization with herpesvirus saimiri modulates the cytokine secretion profile of established Th1 and Th2 human T cell clones. J Immunol. 1993;151:5022–5030. [PubMed] [Google Scholar]
- 7.Devergne O, Hummel M, Koeppen H, Le Beau M M, Nathanson E C, Kieff E, Birkenbach M. A novel interleukin-12 p40-related protein induced by latent Epstein-Barr virus infection in B lymphocytes. J Virol. 1996;70:1143–1153. doi: 10.1128/jvi.70.2.1143-1153.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devergne O, Birkenbach M, Kieff E. Epstein-Barr virus-induced gene 3 and the p35 subunit of interleukin 12 form a novel heterodimeric hematopoietin. Proc Natl Acad Sci USA. 1997;94:12041–12046. doi: 10.1073/pnas.94.22.12041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Waal Malefyt R, Moore K W. Interleukin-10. In: Thomson A, editor. The cytokine handbook. San Diego, Calif: Academic Press; 1998. pp. 333–364. [Google Scholar]
- 10.Duboise S M, Guo J, Czajak S, Desrosiers R C, Jung J U. STP and Tip are essential for herpesvirus saimiri oncogenicity. J Virol. 1998;72:1308–1313. doi: 10.1128/jvi.72.2.1308-1313.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fickenscher H, Biesinger B, Knappe A, Wittmann S, Fleckenstein B. Regulation of the herpesvirus saimiri oncogene stpC, similar to that of T-cell activation genes, in growth-transformed human T lymphocytes. J Virol. 1996;70:6012–6019. doi: 10.1128/jvi.70.9.6012-6019.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fickenscher H, Bökel C, Knappe A, Biesinger B, Meinl E, Fleischer B, Fleckenstein B, Bröker B M. Functional phenotype of transformed human αβ and γδ T cells determined by different subgroup C strains of herpesvirus saimiri. J Virol. 1997;71:2252–2263. doi: 10.1128/jvi.71.3.2252-2263.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fickenscher H, Fleckenstein B. Growth transformation of human T cells. Methods Microbiol. 1998;25:573–602. [Google Scholar]
- 14.Fleming S B, McCaughan C A, Andrews A E, Nash A D, Mercer A A. A homolog of interleukin-10 is encoded by the poxvirus orf virus. J Virol. 1997;71:4857–4861. doi: 10.1128/jvi.71.6.4857-4861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Geurts J M, Schoenmakers E F, van de Ven W J. Molecular characterization of a complex chromosomal rearrangement in a pleomorphic salivary gland adenoma involving the 3′-UTR of HMGIC. Cancer Genet Cytogenet. 1997;95:198–205. doi: 10.1016/s0165-4608(96)00411-6. [DOI] [PubMed] [Google Scholar]
- 16.Groux H, O'Garra A, Bigler M, Rouleazu M, Antonenko S, de Vries J E, Roncarolo M G. A CD4+ T-cell subset inhibits antigen-specific T-cell responses and prevents colitis. Nature. 1997;389:737–742. doi: 10.1038/39614. [DOI] [PubMed] [Google Scholar]
- 17.Groux H, Bigler M, de Vries J E, Roncarolo M G. Inhibitory and stimulatory effects of IL-10 on human CD8+ T cells. J Immunol. 1998;160:3188–3193. [PubMed] [Google Scholar]
- 18.Ho A S, Liu Y, Khan T A, Hsu D H, Bazan J F, Moore K W. A receptor for interleukin 10 is related to interferon receptors. Proc Natl Acad Sci USA. 1993;90:11267–11271. doi: 10.1073/pnas.90.23.11267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsu D H, de Waal Malefyt R, Fiorentino D F, Dang M N, Vieira P, de Vries J, Spits H, Mosmann T R, Moore K W. Expression of interleukin-10 activity by Epstein-Barr virus protein BCRF1. Science. 1990;250:830–832. doi: 10.1126/science.2173142. [DOI] [PubMed] [Google Scholar]
- 20.Jung J U, Desrosiers R C. Association of the viral oncoprotein STP-C488 with cellular ras. Mol Cell Biol. 1995;15:6506–6512. doi: 10.1128/mcb.15.12.6506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung J U, Choi J K, Ensser A, Biesinger B. Herpesvirus saimiri as a model for gammaherpesvirus oncogenesis. Semin Cancer Biol. 1999;9:231–239. doi: 10.1006/scbi.1998.0115. [DOI] [PubMed] [Google Scholar]
- 22.Kim J M, Brannan C I, Copeland N G, Jenkins N A, Khan T A, Moore K W. Structure of the mouse IL-10 gene and chromosomal localization of the mouse and human genes. J Immunol. 1992;148:3618–3623. [PubMed] [Google Scholar]
- 23.Knappe A, Hiller C, Thurau M, Wittmann S, Hofmann H, Fleckenstein B, Fickenscher H. The superantigen-homologous viral immediate-early gene ie14/vsag in herpesvirus saimiri-transformed human T cells. J Virol. 1997;71:9124–9133. doi: 10.1128/jvi.71.12.9124-9133.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Knappe A, Hiller C, Niphuis H, Fossiez F, Thurau M, Wittmann S, Kuhn E-M, Lebecque S, Banchereau J, Rosenwirth B, Fleckenstein B, Heeney J, Fickenscher H. The interleukin-17 gene of herpesvirus saimiri. J Virol. 1998;72:5797–5801. doi: 10.1128/jvi.72.7.5797-5801.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kotenko S V, Krause C D, Izotova L S, Pollak B P, Wu W, Pestka S. Identification and functional characterization of a second chain of the interleukin-10 receptor complex. EMBO J. 1997;16:5894–5903. doi: 10.1093/emboj/16.19.5894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Y, Wei S H, Ho A S, de Waal Malefyt R, Moore K W. Expression cloning and characterization of a human IL-10 receptor. J Immunol. 1994;152:1821–1829. [PubMed] [Google Scholar]
- 27.Liu Y, de Waal Malefyt R, Briere F, Parham C, Bridon J M, Banchereau J, Moore K W, Xu J. The EBV IL-10 homologue is a selective agonist with impaired binding to the IL-10 receptor. J Immunol. 1997;158:604–613. [PubMed] [Google Scholar]
- 28.Liu Y C, Kawagishi M, Mikayama T, Inagaki Y, Takeuchi T, Ohashi H. Processing of a fusion protein by endoprotease in COS-1 cells for secretion of mature peptide by using a chimeric expression vector. Proc Natl Acad Sci USA. 1993;90:8957–8961. doi: 10.1073/pnas.90.19.8957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mackewicz C E, Orque R, Jung J, Levy J A. Derivation of herpesvirus saimiri-transformed CD8+ T cell lines with noncytotoxic anti-HIV activity. Clin Immunol Immunopathol. 1997;82:274–281. doi: 10.1006/clin.1996.4292. [DOI] [PubMed] [Google Scholar]
- 30.Meinl E, Hohlfeld R, Wekerle H, Fleckenstein B. Immortalization of human T cells by herpesvirus saimiri. Immunol Today. 1995;16:55–58. doi: 10.1016/0167-5699(95)80087-5. [DOI] [PubMed] [Google Scholar]
- 31.Meinl E, Fickenscher H. Commonly used human lymphoma lines. In: Rowland-Jones S, McMichael A, editors. Lymphocytes—a practical approach. Oxford, United Kingdom: IRL/OUP; 2000. pp. 337–341. [Google Scholar]
- 32.Meinl E, Fickenscher H. Viral transformation of lymphocytes. In: Rowland-Jones S, McMichael A, editors. Lymphocytes—a practical approach. Oxford, United Kingdom: IRL/OUP; 2000. pp. 55–74. [Google Scholar]
- 33.Mittrücker H, Müller-Fleckenstein I, Fleckenstein B, Fleischer B. CD2-mediated mutual stimulation of herpesvirus saimiri-transformed human T lymphocytes. J Exp Med. 1992;176:909–913. doi: 10.1084/jem.176.3.909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mittrücker H, Müller-Fleckenstein I, Fleckenstein B, Fleischer B. Herpesvirus saimiri-transformed human T lymphocytes: normal functional phenotype and preserved T cell receptor signalling. Int Immunol. 1993;5:985–990. doi: 10.1093/intimm/5.8.985. [DOI] [PubMed] [Google Scholar]
- 35.Moore K W, O'Garra A, de Waal Malefyt R, Vieira P, Mosmann T R. Interleukin-10. Annu Rev Immunol. 1993;11:165–190. doi: 10.1146/annurev.iy.11.040193.001121. [DOI] [PubMed] [Google Scholar]
- 36.Qin L, Chavin K D, Ding Y, Tahara H, Favaro J P, Woodward J E, Suzuki T, Robbins P D, Lotze M T, Bromberg J S. Retrovirus-mediated transfer of viral IL-10 gene prolongs murine cardiac allograft survival. J Immunol. 1996;156:2316–2323. [PubMed] [Google Scholar]
- 37.Rode H J, Janssen W, Rösen-Wolff A, Bugert J J, Thein P, Becker Y, Darai G. The genome of equine herpesvirus type 2 harbors an interleukin 10 (IL10)-like gene. Virus Genes. 1993;7:111–116. doi: 10.1007/BF01702353. [DOI] [PubMed] [Google Scholar]
- 38.Schoenmakers E F, Geurts J M, Kools P F, Mols R, Huysmans C, Bullerdiek J, van den Berghe H, van de Ven W J. A 6-Mb yeast artificial chromosome contig and long-range physical map encompassing the region on chromosome 12q15 frequently rearranged in a variety of benign solid tumors. Genomics. 1995;29:665–678. doi: 10.1006/geno.1995.9952. [DOI] [PubMed] [Google Scholar]
- 39.Suzuki T, Tahara H, Narula S, Moore K W, Robbins P D, Lotze M T. Viral interleukin 10 (IL-10), the human herpes virus 4 cellular IL-10 homologue, induces local anergy to allogeneic and syngeneic tumors. J Exp Med. 1995;182:477–486. doi: 10.1084/jem.182.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swaminathan S, Hesselton R, Sullivan J, Kieff E. Epstein-Barr virus recombinants with specifically mutated BCRF1 genes. J Virol. 1993;67:7406–7413. doi: 10.1128/jvi.67.12.7406-7413.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Takebe Y, Seiki M, Fujisawa J-I, Hoy P, Yokota K, Arai K-I, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wanschura S, Kazmierczak B, Schoenmakers E, Meyen E, Bartnitzke S, van de Ven W, Bullerdiek J, Schloot W. Regional fine mapping of the multiple-aberration region involved in uterine leiomyoma, lipoma, and pleomorphic adenoma of the salivary gland to 12q15. Genes Chromosomes Cancer. 1995;14:68–70. doi: 10.1002/gcc.2870140112. [DOI] [PubMed] [Google Scholar]
- 43.Weber F, Meinl E, Drexler K, Czlonkowska A, Huber S, Fickenscher H, Müller-Fleckenstein I, Fleckenstein B, Wekerle H, Hohlfeld R. Herpesvirus saimiri-transformed human T cell lines expressing functional receptor for myelin basic protein. Proc Natl Acad Sci USA. 1993;90:11049–11054. doi: 10.1073/pnas.90.23.11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wiese N, Tsygankov A, Klauenberg U, Bolen J, Fleischer B, Bröker B. Selective activation of T cell kinase p56lck by herpesvirus saimiri protein Tip. J Biol Chem. 1996;271:847–852. doi: 10.1074/jbc.271.2.847. [DOI] [PubMed] [Google Scholar]
- 45.Zdanov A, Schalk-Hihi C, Menon S, Moore K W, Wlodawer A. Crystal structure of Epstein-Barr virus protein BCRF1, a homolog of cellular interleukin-10. J Mol Biol. 1997;268:460–467. doi: 10.1006/jmbi.1997.0990. [DOI] [PubMed] [Google Scholar]