Abstract
HeLa cells expressing the recombinant Marburg virus (MBGV) nucleoprotein (NP) have been studied by immunoelectron microscopy. It was found that MBGV NPs assembled into large aggregates which were in close association with membranes of the rough endoplasmic reticulum. Further analysis of these aggregates revealed that NPs formed tubule-like structures which were arranged in a hexagonal pattern. A similar pattern of preformed nucleocapsids was detected in intracellular inclusions induced by MBGV infection. Our data indicated that MBGV NP is able to form nucleocapsid-like structures in the absence of the authentic viral genome and other nucleocapsid-associated proteins.
Marburg virus (MBGV) and Ebola virus (EBOV) make up the family of Filoviridae, which belongs to the order Mononegavirales. MBGV causes a severe hemorrhagic disease in humans and nonhuman primates (22). The recent outbreak of MBGV hemorrhagic fever in the Democratic Republic of the Congo underlines the emerging potential of this pathogen (25). MBGV is an enveloped virus with a nonsegmented negative-strand RNA genome 19.1 kb in length, which encodes seven structural proteins (5, 8). The nucleocapsid of the virion is composed of the viral RNA and four proteins, the nucleoprotein (NP), P (formerly called VP35), the viral protein VP30, and the catalytic subunit of the polymerase (L). Two putative matrix proteins, VP24 and VP40, are located between the nucleocapsid and the envelope, which is decorated with the surface protein (GP) (2, 3, 7, 13, 15, 20).
Viral reproduction takes place in the cytoplasm, and the infectious particles are released by budding at the cell surface (10, 11, 16–19). MBGV morphogenesis commences with the formation of intracytoplasmic blurred wavy strands which are first arranged in loose accumulations and then scattered (17). More frequently, MBGV inclusions appear as amorphous electron-dense matrices or sheets. It has been observed that MBGV inclusions show the manifestation of nucleocapsid formation only shortly before budding, in contrast to EBOV inclusions, which as a rule contain easily discernible preformed nucleocapsids (11). Sometimes, sections of MBGV inclusions exhibit thin-walled tubule-like structures (TLS) which are organized in a hexagonal pattern (11, 17). However, the exact protein composition of these TLS has not been identified. Immunoelectron microscopic (IEM) analysis revealed that MBGV inclusions contain a large amount of NP, but the internal arrangement of NP has been unknown until now (10, 11).
Encapsidation of the genomic RNA by the major nucleocapsid protein (NP) is presumed to be the first step in the assembly process of MBGV nucleocapsid (1, 19). The mechanism of this initial step is still unknown. Studies with other viruses of the order Mononegavirales showed that nucleoproteins are able to assemble into nucleocapsid-like structures independently of other viral proteins (4, 9, 12, 14, 23). Since the protein composition of the nucleocapsid complexes of Filoviridae is different from that of the other Mononegavirales (four instead of three nucleocapsid proteins [1]), it is unclear whether NP also contains the information for the assembly of nucleocapsid-like structures. Among the proteins of the MBGV nucleocapsid complex, only NP has the potential to self-assemble into large aggregates when it is expressed in the absence of other viral proteins (3). However, features of NP packaging inside the aggregates have not yet been investigated.
To examine whether the NP is able to self-assemble into nucleocapsid-like structures, we have analyzed the ultrastructural organization of aggregates formed by the recombinant NP and compared the results with the ultrastructural organization of viral inclusions in MBGV-infected cells. We report here that recombinant MBGV NP assembles into TLS arranged in a hexagonal pattern. The organization and morphology of TLS in recombinant NP aggregates are very similar to those of the preformed nucleocapsids which we identified in viral inclusions in MBGV-infected cells.
For NP expression, approximately 107 HeLa cells were infected with recombinant vaccinia virus MVA-T7 (24) and subsequently transfected with 1 μg of pT/NP as described previously (15). At 8 h postinfection (hpi), cells were washed with Hanks balanced salt solution (HBS), fixed with HBS containing 2.5% paraformaldehyde and 0.1% glutaraldehyde, dehydrated, and embedded in acrylic resin (LR gold; Sigma). Polymerization was carried out at −20°C by UV irradiation. IEM was performed according to the method of Geisbert and Jahrling (11) by using a monoclonal antibody against NP, diluted 10-fold. The bound antibodies were detected with a goat anti-mouse antiserum conjugated with colloidal gold beads (diameter, 5 nm).
For comparative electron microscopical studies, Vero cells were infected with MBGV strain Musoke at a multiplicity of infection of 1 PFU per cell and harvested at 43 hpi. Cells were fixed with HBS containing 4% paraformaldehyde, postfixed with HBS containing 1% osmium tetraoxide, dehydrated, and embedded in Epon and Araldite. Ultrathin sections were stained with uranyl acetate and lead citrate (19). The gold particles and structures of infected cells were visualized using a Hitachi 600 electron microscope. Measurements were made from prints at a final magnification of 200,000. Numerical values are presented as means ± standard deviations (n = 20). Student's t test was used for statistical analysis (P < 0.01).
IEM analysis of HeLa cells expressing NP revealed electron-dense inclusions within the cytoplasms, which were heavily labeled with gold particles (Fig. 1). Areas of the cytoplasm which were devoid of NP inclusions did not show gold labeling. As a consequence of the vaccinia virus infection, viral factories, immature viral particles, and mature viral particles were observed. The used antibody did not cross-react with vaccinia virus-specific structures. Furthermore, background staining of uninfected cells was not detectable (data not shown).
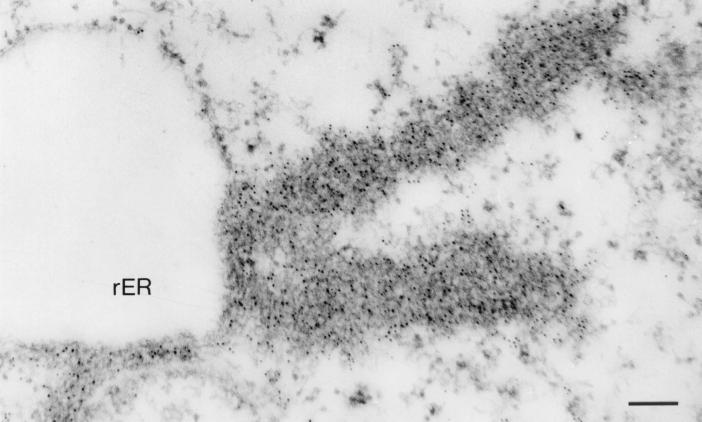
FIG. 1.
IEM analysis of ultrathin section of MVA-T7-infected HeLa cells expressing recombinant NP. Immunogold labeling was carried out using a monoclonal antibody directed against MBGV NP (dilution, 1:10) and a goat anti-mouse antibody conjugated with colloidal gold (bead diameter, 5 nm) (dilution, 1:50). NP aggregates appeared as two long sheets of a reticular network of electron-dense material heavily labeled with gold particles. The NP aggregates are in close association with the membrane of the rER. Bar, 150 nm.
The NP aggregates appeared as a reticular network of electron-dense material interspersed with electron-lucent material. The NP aggregates were detected mostly in close association with rough endoplasmic reticulum (rER) membranes and sometimes with the outer membrane of the nuclear envelope. The NP aggregates formed long sheets with one end in close association with rER channels or were localized between rER channels (Fig. 1). It is assumed that the NP-induced sheets were formed because nascent NP peptide chains were attached to the preexisting NP molecules, thus preventing diffuse spreading throughout the cytoplasm.
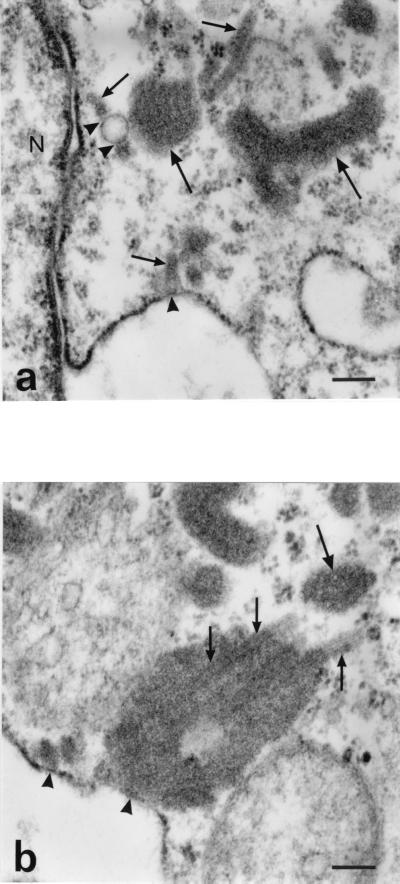
To investigate whether intracytoplasmic MBGV-specific inclusions are also associated with the membrane of the rER, we have studied ultrathin sections of MBGV-infected Vero cells harvested at 43 hpi. Association of intracytoplasmic MBGV inclusions with rER membranes did not appear as frequently as with aggregates of recombinant NP. However, in several cases, an association between rER membranes and both small wavy strands (Fig. 2a) and large sheets of intracytoplasmic MBGV inclusions was observed (Fig. 2b). The differences between MBGV inclusions and NP aggregates with regard to their rER associations have yet to be explained.
FIG. 2.
Electron microscopical analysis of ultrathin sections of MBGV-infected Vero cells at 43 hpi. (a) Morphological variability of MBGV inclusions. MBGV inclusions appeared as thin strands (small arrows) or as thick sheets with longitudinal striation (large arrows). The thin strands of MBGV inclusions are in close association with the outer membrane of the nuclear envelope and with a smooth membrane structure (arrowheads). N, nucleus. Bar, 200 nm. (b) Large sheets of MBGV inclusions. Cross-sectioned TLS in MBGV inclusions, which represent preformed nucleocapsids, display circles in a regular arrangement (large arrow). Longitudinally sectioned thin-walled TLS surrounded by more-electron-dense matrices (mature nucleocapsids) are seen inside and outside the large MBGV inclusion (small arrows). The association of MBGV inclusions with an rER membrane is indicated by arrowheads. Bar, 150 nm.
Detailed analysis of aggregates formed by recombinant NP in HeLa cells revealed in some sections gold-labeled regular striation or regularly arranged circles (Fig. 3). The periodicity within the striated patterns was approximately 25 nm. In some areas of longitudinal sections the gold particles were arranged in rows (Fig. 3a, inset). The circular domains were hexagonally organized (Fig. 3b). The inner diameters of the circles were 18.5 ± 0.4 nm, and the outer diameters were 27.9 ± 0.4 nm. The distances between the circles were 16.1 ± 0.6 nm, and the distances between the centers of the circles were 44.0 ± 0.5 nm. As individual circular domains were labeled with up to six gold particles delineating their perimeters (Fig. 3b, inset) it became clear that these circular domains are comprised of NP molecules.
FIG. 3.
IEM analysis of ultrathin sections of aggregates formed by recombinant NP. Immunogold labeling was performed as described in the legend to Fig. 1. (a) NP aggregate labeled with gold showing striated matrices with a periodicity of approximately 25 nm. The inset shows a row of gold particles which occurred in a longitudinal section. Bar, 100 nm. (b) NP aggregates labeled with gold showing a hexagonal pattern (hexagons are indicated by squares and crosses). Bar, 100 nm. The inset shows one circular domain labeled by five gold particles, one of which is slightly dislocated.
Based on the maximum number of antibodies binding to one of the ring-like structures, it is presumed that these structures are formed by at least six subunits which are composed of NP. However, it cannot be ruled out that the subunits are composed of a multitude of NP molecules, e.g., six dimers, trimers, or even tetramers. Peters et al. (17) described in a study performed with transmission electron microscopy that one cross-sectioned nucleocapsid in MBGV inclusions is composed of 24 grains with diameters of less than 5 nm. The helical nucleocapsid of Sendai virus contains 13 NP subunits in each turn (6).
The varied patterns of NP aggregates in ultrathin sections were related to different planes of the sections. Thus, the presence of striated or hexagonal patterns or a reticular network suggested that NP forms TLS which are connected to each other. The regularly arranged TLS in NP aggregates that have a striated appearance in a longitudinal section appear as hexagons in cross-sections and exhibit a reticular network in a skewed section. It is noteworthy that in some sections of NP aggregates all three kinds of appearances of TLS could be observed, indicating that these aggregates are curved (not shown). This result might reflect a certain flexibility of TLS.
It has been shown previously that recombinant nucleoproteins of Paramyxoviridae form helices (4, 9, 14, 23) and that recombinant nucleoproteins of rabies virus can form both helices and rings, depending on conditions of purification (12). Analysis of postnuclear supernatant of HeLa cells expressing MBGV NP, fractionated on sucrose gradients (5 to 30%), revealed filamentous structures or extremely elongated helices. The rings detected with rabies virus were not detected (data not shown). This result could be explained by the extreme fragility of MBGV nucleocapsids, which might prevent preservation of the original structure during the isolation procedure (1). Although our results rather suggested a helical arrangement of TLS, detailed analysis has to be carried out before definite statements on the three-dimensional packaging of NP inside the TLS can be made.
Viral inclusions in MBGV-infected cells were analyzed to compare the organization of these structures with that of aggregates of recombinant NP. A remarkable morphological variability of intracytoplasmic MBGV inclusions was detected, as has also been shown in previous reports (10, 11, 16–19) (Fig. 2). Depending on the plane of the section, MBGV inclusions exhibited either hexagonally arranged thin-walled circles (Fig. 4) or a striated pattern (Fig. 2), suggesting the presence of TLS as in NP aggregates. In some sections of MBGV inclusions, both striation and circles could be observed, suggesting that TLS in MBGV inclusions, like the TLS formed by recombinant NP, were rather curved (not shown). The inner diameters of the thin-walled tubules in MBGV inclusions were 17.6 ± 0.3 nm, and the outer diameters were 26.3 ± 0.4 nm. The distances between the thin-walled tubules were 18.2 ± 0.5 nm, and the distances between the centers of TLS in MBGV inclusions were 44.5 ± 0.6 nm. Thus, thin-walled TLS in MGBV inclusions and in NP-expressing HeLa cells were highly similar. The slight differences in the dimensions were not statistically significant and could be related to the different processing of samples for IEM and conventional EM.
FIG. 4.
Comparative analysis of ultrathin sections of viral inclusions in MBGV-infected cells and aggregates formed by recombinant NP. (a) Cross-section of viral inclusion in MBGV-infected Vero cells at 43 hpi. Preformed nucleocapsids are arranged in a hexagonal pattern (white squares) and embedded in electron-dense matrices. Some of the thin-walled TLS in the periphery of the inclusion are surrounded by more-electron-dense matrices and represent mature nucleocapsids (arrows). Inset 1 (fragment of the hexagonal pattern which is marked by white squares) shows thin-walled TLS surrounded by a narrow electron-translucent region (arrow). Inset 2 (fragment of the viral inclusion in panel a [one of the marked mature nucleocapsids]) represents thin-walled TLS surrounded by a region of high electron density. Inset 3 shows a cross-section of an MBGV particle. The thicknesses and electron densities of the regions surrounding the mature nucleocapsid within the MBGV inclusion and within the viral particle are almost identical. Bar, 25 nm. Inset 4 shows a cross-sectioned TLS formed by recombinant NP (the fragment of Fig. 3b which is marked by black squares). The photo of inset 4 is printed at the same magnification as the photo of the MBGV inclusion. Bar, 70 nm. (b and c) Schematic illustrations of cross-sectioned inclusions induced either by recombinant NP infection (b) or by MBGV infection (c). Hexagonally arranged black circles represent TLS inside an NP aggregate (b) and preformed nucleocapsids inside an MBGV inclusion (c). MBGV inclusions (c) contain preformed nucleocapsids embedded in electron-dense matrices (shown by shading) and mature nucleocapsids which are characterized by their highly electron-dense appearance (shown by superimposed circular shading).
In contrast to their appearance in NP aggregates, the thin-walled TLS in viral inclusions were embedded in electron-dense matrices. Besides containing thin-walled TLS which were surrounded by a narrow electron-translucent region (Fig. 4a, inset 1), inclusions contained another type of TLS surrounded by a region of high electron density (Fig. 4a, inset 2). These TLS were morphologically identical to single nucleocapsids outside the inclusions and to nucleocapsids in mature virions. Both were surrounded by an electron-dense region (Fig. 4a, inset 3) which had a thickness of 13 to 20 nm. The variation in the thickness of the region might be induced by the stepwise incorporation of viral matrix proteins. These results suggested that MBGV inclusions contain preformed nucleocapsids at different steps of maturation. The exact protein compositions of the nucleocapsids, which differ in electron density, have yet to be determined.
Comparison of inclusions in MBGV-infected cells and aggregates composed of recombinant NP further revealed that the hexagonal organization of the nucleocapsids is related to that of NP. For illustration, the arrangement of the nucleocapsid-like structures inside the inclusions, either induced by recombinant NP or by MBGV infection, is schematically drawn in Fig. 4b and c. It is shown that the overall organization of thin-walled TLS formed by recombinant NP (Fig. 4a, inset 4, and b) is conserved in MBGV-induced inclusions (Fig. 4a and c), with the difference that viral inclusions additionally contained electron-dense material inside and surrounding the TLS that were not detected in aggregates of recombinant NP. It cannot yet be ruled out that the interaction between the TLS is made by cellular proteins. In any case, it seems very likely that NP is a scaffold for viral assembly. It is interesting that the hexagonal arrangement of preformed nucleocapsids is also observed in EBOV inclusions. Other families of the order Mononegavirales, however, display viral inclusions with irregularly arranged nucleocapsids (21; our unpublished observations). Self-assembly of nucleoproteins into nucleocapsid-like structures has been detected with the families Paramyxoviridae (4, 9, 14, 23) and Rhabdoviridae (12). Thus, the ability of nucleoproteins to determine the nucleocapsid structure appears to be a general pattern among the order Mononegavirales.
In this report we demonstrated for the first time that (i) MBGV NP assembles into TLS arranged in a hexagonal pattern in the absence of the authentic viral genome and the other nucleocapsid-associated proteins, (ii) TLS in NP aggregates are curved, (iii) there is a close association between NP aggregates and membranes of the rER, and (iv) the organization and morphology of TLS in recombinant NP aggregates are very similar to those of the preformed nucleocapsids which were identified in inclusions of MBGV-infected cells.
Acknowledgments
We thank Andreas Holzenburg, University of Leeds, for helpful discussion, critical reading of the manuscript, and encouragement. We also thank Brigitte Agricola and Claudia Schmidt (Institute of Cell Biology, Marburg, Germany), Sonja Heck (Institute of Virology, Marburg, Germany), and Marina Vorobjova (SRC VB Vector Novosibirsk, Russia) for assisting with ultrastructural analysis. In addition, we thank Michael Weik for preparing the artwork.
This work was supported by the European Union (grant INTAS 96-1361) and by the Deutsche Forschungsgemeinschaft (grants SFB 286, TPA6).
REFERENCES
- 1.Becker S, Rinne C, Hofsäß U, Klenk H-D, Mühlberger E. Interaction of Marburg virus nucleocapsid proteins. Virology. 1998;249:406–417. doi: 10.1006/viro.1998.9328. [DOI] [PubMed] [Google Scholar]
- 2.Becker S, Klenk H-D, Mühlberger E. Intracellular transport and processing of the Marburg virus surface protein in vertebrate and insect cells. Virology. 1996;225:145–155. doi: 10.1006/viro.1996.0582. [DOI] [PubMed] [Google Scholar]
- 3.Becker S, Huppertz S, Klenk H D, Feldmann H. The nucleoprotein of Marburg virus is phosphorylated. J Gen Virol. 1994;75:809–818. doi: 10.1099/0022-1317-75-4-809. [DOI] [PubMed] [Google Scholar]
- 4.Buchholz C J, Spehner D, Drillien R, Neubert W J, Homann H E. The conserved N terminal region of Sendai virus nucleocapsid protein NP is required for nucleocapsid assembly. J Virol. 1993;67:5803–5812. doi: 10.1128/jvi.67.10.5803-5812.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bukreyev A A, Volchkov V E, Blinov V M, Dryga S A, Netesov S V. The complete nucleotide sequence of the Popp (1967) strain of Marburg virus: a comparison with the Musoke (1980) strain. Arch Virol. 1995;140:1589–1600. doi: 10.1007/BF01322532. [DOI] [PubMed] [Google Scholar]
- 6.Egelman E H, Wu S S, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldmann H, Will C, Schikore M, Slenczka W, Klenk H-D. Glycosylation and oligomerization of the spike protein of Marburg virus. Virology. 1991;182:353–356. doi: 10.1016/0042-6822(91)90680-A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feldmann H, Mühlberger E, Randolf A, Will C, Kiley M P, Sanchez A, Klenk H-D. Marburg virus, a filovirus: messenger RNAs, gene order, and regulatory elements of the replication cycle. Virus Res. 1992;24:1–19. doi: 10.1016/0168-1702(92)90027-7. [DOI] [PubMed] [Google Scholar]
- 9.Fooks A R, Stephenson J R, Warnes A, Dowsett A B, Rima B K, Wilkinson G W G. Measles virus nucleocapsid protein expressed in insect cells assembles into nucleocapsid-like structures. J Gen Virol. 1993;74:1439–1444. doi: 10.1099/0022-1317-74-7-1439. [DOI] [PubMed] [Google Scholar]
- 10.Geisbert T W, Jaax N K. Marburg hemorrhagic fever: report of a case studied by immunohistochemistry and electron microscopy. Ultrastruct Pathol. 1998;22:3–17. doi: 10.3109/01913129809032253. [DOI] [PubMed] [Google Scholar]
- 11.Geisbert T W, Jahrling P B. Differentiation of filoviruses by electron microscopy. Virus Res. 1995;39:129–150. doi: 10.1016/0168-1702(95)00080-1. [DOI] [PubMed] [Google Scholar]
- 12.Iseni F, Barge A, Baudin F, Blondel D, Ruigrok R W. Characterization of rabies virus nucleocapsids and recombinant nucleocapsid-like structures. J Gen Virol. 1998;79:2909–2919. doi: 10.1099/0022-1317-79-12-2909. [DOI] [PubMed] [Google Scholar]
- 13.Lötfering B, Mühlberger E, Tamura T, Klenk H-D, Becker S. The nucleoprotein of Marburg virus is target for multiple cellular kinases. Virology. 1999;255:50–62. doi: 10.1006/viro.1998.9577. [DOI] [PubMed] [Google Scholar]
- 14.Meric C, Spehner D, Mazarin V. Respiratory syncytial virus nucleocapsid protein (N) expressed in insect cells forms nucleocapsid-like structures. Virus Res. 1994;31:187–201. doi: 10.1016/0168-1702(94)90003-5. [DOI] [PubMed] [Google Scholar]
- 15.Mühlberger E, Lötfering B, Klenk H-D, Becker S. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J Virol. 1998;72:8756–8764. doi: 10.1128/jvi.72.11.8756-8764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy F A, van der Groen G, Whitfield S G, Lange J V. Ebola and Marburg virus morphology and taxonomy. In: Murphy F A, editor. Ebola virus haemorrhagic fever. Amsterdam, The Netherlands: Elsevier/North-Holland; 1978. pp. 61–82. [Google Scholar]
- 17.Peters D, Müller G, Slenczka W G. Morphology, development, and classification of the Marburg virus. In: Martini G A, Siegert R, editors. Marburg virus disease. Berlin, Germany: Springer-Verlag; 1971. pp. 68–83. [Google Scholar]
- 18.Ryabchikova E, Vorontsova L A, Skripchenko A A, Shestopalov A M, Sandakhchiev L S. The peculiarities of internal organ damage in experimental animals infected with Marburg virus. Bull Exp Biol Med. 1994;4:430–434. [PubMed] [Google Scholar]
- 19.Ryabchikova E, Strelets L, Kolesnikova L, Pyankov O, Sergeev A. Respiratory Marburg virus infection in guinea pig. Arch Virol. 1996;141:2177–2190. doi: 10.1007/BF01718224. [DOI] [PubMed] [Google Scholar]
- 20.Sanchez A, Kiley M P, Klenk H-D, Feldmann H. Sequence analysis of the Marburg virus nucleoprotein gene: comparison to Ebola virus and other non-segmented negative-strand RNA viruses. J Gen Virol. 1992;73:347–357. doi: 10.1099/0022-1317-73-2-347. [DOI] [PubMed] [Google Scholar]
- 21.Scheid A. Paramyxoviridae. In: Nermut M V, Steven A C, editors. Animal virus structure. Amsterdam, The Netherlands: Elsevier; 1987. pp. 233–252. [Google Scholar]
- 22.Siegert R, Shu H-L, Slenczka W, Peters D, Müller G. Zur Aetiologie einer unbekannten, von Affen ausgegangenen menschlichen Infektioskrankheit. Dtsch Med Wochenschr. 1967;51:2341–2343. doi: 10.1055/s-0028-1106144. [DOI] [PubMed] [Google Scholar]
- 23.Spehner D, Kirn A, Drillien R. Assembly of nucleocapsid-like structures in animal cells infected with a vaccinia virus recombinant encoding the measles virus nucleoprotein. J Virol. 1991;65:6296–6300. doi: 10.1128/jvi.65.11.6296-6300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sutter G, Ohlmann M, Erfle V. Non-replicating vaccinia vector efficiently expresses bacteriophage T7 RNA polymerase. FEBS Lett. 1995;371:9–12. doi: 10.1016/0014-5793(95)00843-x. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization. Marburg fever, Democratic Republic of the Congo. WER. 1999;74:145. [PubMed] [Google Scholar]