Abstract
HLA class II DR is one of the most abundant cell surface proteins incorporated onto human immunodeficiency virus type 1 (HIV-1) during budding. The mechanism for HLA class II protein incorporation is not known and may involve a viral protein. To determine whether Env affects HLA class II protein incorporation, HIV-1 virions, either with or without Env on their surface, were produced from HLA class II-expressing cells and analyzed by whole-virus immunoprecipitation with antisera against HLA class II proteins. HLA class II proteins were detected on virions only when wild-type Env was incorporated, while similar experiments showed that HLA class I proteins were incorporated independent of Env packaging. Therefore, the packaging of HIV-1 Env protein is required for the efficient incorporation of HLA class II but not class I proteins into the virion. Analysis of two Env mutants revealed that the presence of a 43-amino-acid sequence between amino acids 708 and 750 in the gp41TM cytoplasmic tail was required for efficient incorporation of HLA class II proteins. These data show that HIV-1 actively incorporates HLA class II proteins in a process that, either directly or indirectly, requires Env.
Human immunodeficiency virus type 1 (HIV-1) acquires surface glycoproteins, including Env, by budding through the plasma membrane (reviewed in reference 35). Env is a complex made up of the surface glycoprotein gp120SU noncovalently bound to the gp41TM transmembrane (TM) protein. This complex binds to CD4 and various coreceptors and allows for viral entry by mediating virus-cell membrane fusion (reviewed in reference 19). Unlike type C retroviral TM proteins, gp41TM has a relatively long cytoplasmic C terminus (tail) that has been shown to induce cytopathic effects and affect the packaging of Env into the virion (9, 12, 21, 36, 37).
In addition to Env, many different cellular proteins have been detected on the surface of HIV-1 (reviewed in reference 24). Human leukocyte antigen (HLA) class II DR is the most commonly detected and the most abundant host protein on HIV-1 propagated in cell culture and on virus isolated from patient plasma (24, 33). Based on biochemical analysis, HLA class II DR may be present at approximately 1.5 to 2 times the level of gp120SU on the surface of HIV-1 produced from the H9 human T-cell line (2). Although the mechanism by which HIV-1 acquires HLA class II DR is not known, one possibility is that it is merely incorporated by its proximity to the budding virion. However, many host proteins appear to be excluded from enveloped viruses in general (38). Interestingly, only HLA class II DR is appreciably incorporated into virions, even when the other isotypes of HLA class II (DP and DQ) are present on the surface of cells (2, 8, 14, 34). While these results imply that HLA class II DR is specifically incorporated into the virion, this possibility has not been demonstrated.
Expression of Env and HLA class II protein incorporation.
One possible mechanism for HLA class II protein incorporation is that the packaging of Env might cause the placement of HLA class II proteins onto the virion. To test this hypothesis, the presence of HLA class II proteins on virions with and without Env was examined. Since microvesicles containing HLA class II proteins have been shown to contaminate even purified H9 and peripheral blood mononuclear cell (PBMC) virion preparations (4, 14), simply examining virus stocks for HLA class II proteins would detect protein from both virions and microvesicles. Therefore, the presence of HLA class II proteins on the virions was examined by a whole-virus immunoprecipitation with HLA class II antiserum (i.e., without detergent lysis of the particle), followed by detection of precipitated virions by immunoblotting for Gag. To produce Env-deficient virions from HLA class II-expressing cells, these cells were infected with VSV-G pseudotypes of wild-type or Env mutant viruses produced by cotransfection of 293T human kidney cells with a proviral DNA construct (pNL4-3 [1] or its mutants) and a VSV-G expression plasmid, pCMVHg (6), by using a calcium phosphate mammalian cell transfection kit (5 Prime-3 Prime, Inc., Boulder, Colo.). These pseudotyped stocks were then used to infect HLA class II-expressing cells, either H9 cells or phytohemagglutinin-stimulated PBMCs, in the presence of 2 μg of Polybrene per ml. The inoculating virus was removed by washing at 4 h postinfection, and virus-containing clarified supernatants were harvested 48 to 72 h postinfection.
Virions were produced from H9 cells by using wild-type DNA and two NL4-3 mutant constructs that do not incorporate Env (25): Env− that produces no Env and the p6Gag mutant Y36S-L41P that expresses but does not incorporate Env. Immunoprecipitation was carried out with cell-free supernatants containing equal amounts of p24CA (typically 10 to 30 ng, as determined by a p24CA enzyme-linked immunosorbent assay (ELISA) kit [New England Nuclear, Boston, Mass.]) with 1% (vol/vol) rabbit anti-HLA class II β-chain serum (DJ-31681; AIDS Vaccine Program, National Cancer Institute-Frederick Cancer Research and Development Center [NCI-FCRDC]) and 2 μg (dry weight) of hydrated protein A-Sepharose beads (Amersham Pharmacia Biotech, Inc., Piscataway, N.J.). After the antibody-bead complexes were washed three times with phosphate-buffered saline (Life Technologies Inc., Gaithersburg, Md.), the amount of virus precipitated was visualized by immunoblot analysis (essentially as described in reference 17) with a mouse monoclonal antibody against p24CA (AIDS Vaccine Program, NCI-FCRDC) and the Enhanced ChemiLuminescence procedure (Amersham Pharmacia Biotech). To eliminate most of the signals produced by the secondary antibody binding to the light and heavy chains of the precipitating antibody, the gels for these immunoblots were run under nonreducing conditions to keep the antibody molecules intact. Under these conditions, p24CA and Pr55Gag migrated similarly under reducing conditions (data not shown), while the majority of the background from the precipitating antibodies migrated at a higher molecular weight (∼180 kDa).
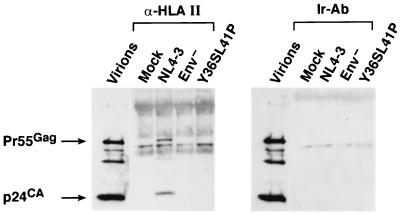
Wild-type and mutant virions produced from H9 cells were analyzed by using this HLA class II immunoprecipitation assay. The p24CA immunoblot of the samples showed that only the lane containing the wild-type immunoprecipitate sample contained bands corresponding to p24CA and Pr55Gag, indicating that the wild-type HIV-1 was precipitated by the anti-HLA class II serum (Fig. 1). A blot of a precipitation experiment with an irrelevant antibody (rabbit anti-horse immunoglobulin G [IgG]; Sigma Chemical Co., St. Louis, Mo.) did not produce any Gag signal, demonstrating that precipitation was specific for the HLA class II antibody. Virions produced from cells that do not express HLA class II proteins did not precipitate in this assay (data not shown). These data indicate that HLA class II proteins are present on the surface of the wild-type virion, as expected. In addition to the viral bands in the immunoblot, other bands were present that did not appear to be viral and were present in the mock control, possibly associated with the HLA class II-precipitating antibody. In contrast to the wild-type result, samples of mutant virions that either fail to express the Env complex, Env−, or express Env on the cell surface but fail to incorporate Env, Y36S-L41P, did not produce any detectable Gag signal in this assay. The inability of these mutant virions to be precipitated indicates that HLA class II proteins were not incorporated onto the virion surface in the absence of Env packaging.
FIG. 1.
Immunoblots of whole-virus immunoprecipitates. The p24CA blots of virions precipitated with either HLA class II or an irrelevant antibody (rabbit anti-horse IgG [Ir]) are presented. The precipitated samples are identified above their respective lanes. A sample of wild-type virions is included for reference.
HLA class II protein incorporation and gp41TM.
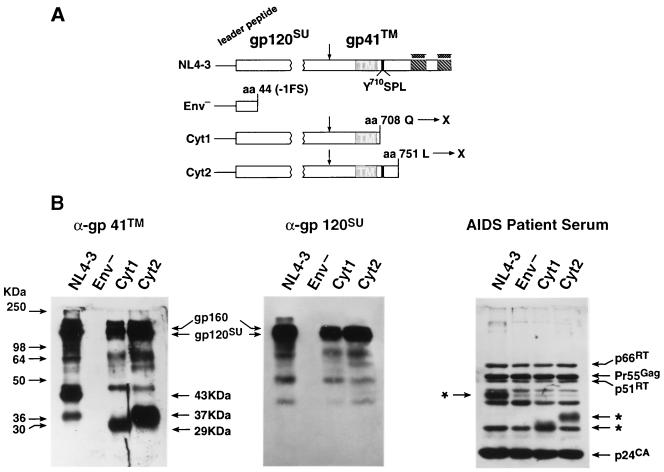
Since the HIV-1 Env complex contains a long cytoplasmic tail that could interact with HLA class II proteins, two mutants were constructed that truncated the C terminus of gp41TM (summarized in Fig. 2A) by the insertion of a nonsense codon at either amino acid (aa) 708 (C to T at nucleotide 8342 [1]), Cyt1, or aa 751, Cyt2 (T to G at nucleotide 8472). The Cyt2 mutation did not alter the overlapping rev reading frames. DNA fragments that contained the mutation were produced by the PCR overlap extension procedure (18) and then reinserted into the pNL4-3 molecular clone. All mutants were confirmed by dideoxy sequencing. Following transfection, the two mutant DNA constructs produced wild-type levels of particles, as measured by reverse transcriptase and p24CA ELISA (Table 1). Virions were centrifuged through a 20% sucrose cushion (as previously described [25]), and equal amounts of particles (100 ng by p24CA) were analyzed by immunoblot to examine the Env proteins in the virions. Probing a blot with a gp41TM antiserum (Fitzgerald International, Inc., Concord, Mass.) revealed that the wild-type samples contained a band at 43 kDa, corresponding to gp41TM, while those from the Cyt1 and Cyt2 mutant virions contained bands at 29 and 37 kDa, respectively (Fig. 2B). These sizes were close to those predicted for the nonglycosylated sizes of the three species: 39.7 kDa for gp41TM, 22.8 kDa for Cyt1, and 27.5 kDa for Cyt2 (Fig. 2B). Since this antiserum also reacts with gp120SU, other higher-migrating bands were present in the immunoblot in addition to the gp41TM bands (Fig. 2B). These same larger bands, corresponding to gp160Env and gp120SU, were present in an immunoblot using a gp120SU monoclonal antibody (AIDS Vaccine Program), confirming that these mutants incorporated Env (Fig. 2B). In contrast, results of blots with either gp120SU or gp41TM antiserum showed that the Env− mutant did not contain either protein as expected (Fig. 2B). Blotting with the AIDS patient serum (AIDS Vaccine Program) also detected the full-length and the truncated gp41TM proteins in the wild-type and mutant samples, respectively. Similar amounts of the other viral proteins (e.g., Gag and Pol) were present in both the wild-type and the mutant samples (Fig. 2B).
FIG. 2.
Immunoblot analysis of gp41TM mutants. (A) Env truncation mutants are presented with mutations indicated above the altered regions. Vertical arrow indicates the Env cellular protease-processing site. (B) The gp41TM, gp120SU, and AIDS patient serum immunoblots of virions (100 ng of p24CA in each lane) produced from 293T cells are presented. Virion samples are labeled above their respective lanes. Positions of the molecular weight standards are presented at left, and those of the viral proteins are identified at the margins of the blots. ∗, gp41TM bands in the AIDS patient serum blot.
TABLE 1.
Infectivity of Env mutants
| Virus | Concn of p24 (ng/ml)a | RT activity (cpm/ml)b | No. of BCFUc (U/ml) |
|---|---|---|---|
| Mock | <0.008 | 1.3 × 103 | <1 |
| NL4-3 | 28.7 | 8.1 × 106 | 1 × 104 |
| Env− | 13.9 | 5.4 × 106 | <1 |
| Cyt1 | 25.3 | 7.4 × 106 | 1 × 102 |
| Cyt2 | 27.4 | 6.8 × 106 | 3 × 102 |
p24CA content determined by enzyme-linked immunosorbent assay kit (Retro-Tek, Buffalo, N.Y.).
RT, reverse transcriptase. RT activity assay was performed as previously described (16).
BCFU, blue cell-forming units.
To test for mutant Env function, the Cyt1 and Cyt2 mutants were tested in an HCLZ single-round infectivity assay (long terminal repeat-driven LacZ complementation assay as previously described [25]). Supernatants containing equivalent p24CA levels derived from two independent transfections were tested. The results showed that both the Cyt1 and Cyt2 mutants were infectious and that the truncated Env proteins were functional, though their infectivity was 100-fold lower than the wild-type virus (Table 1). As expected, the Env− mutant was not significantly infectious since it lacks Env.
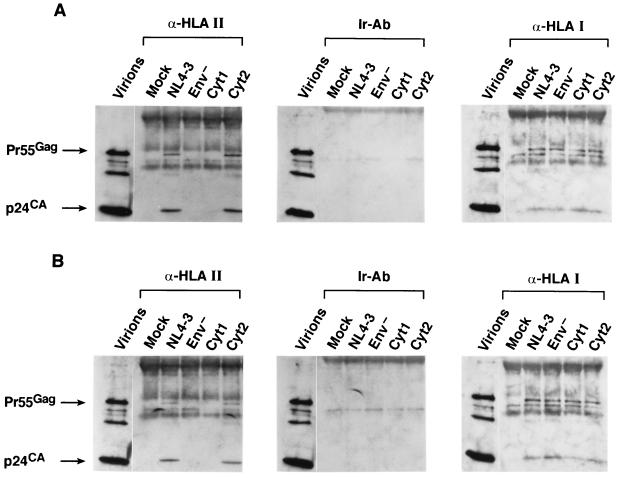
To determine if the truncations in the cytoplasmic tail of gp41TM had an effect on HLA class II protein incorporation, wild-type and mutant viruses were produced from H9 cells or PBMCs and examined by the whole-virus anti-HLA class II immunoprecipitation-immunoblot protocol. The blots of the wild-type and the Cyt2 virion precipitates produced from either H9 cells (Fig. 3A) or from PBMCs (Fig. 3B) readily detected both p24CA and Pr55Gag. In contrast, the blots of the Env− and Cyt1 samples did not detect any Gag proteins (Fig. 3A and B). In addition, experiments with irrelevant antibody also failed to recover Gag signal from any of the samples (Fig. 3). Therefore, the Cyt2 mutant was precipitated by anti-HLA class II serum, indicating that this mutant incorporates HLA class II proteins. In contrast, the Cyt1 mutant was not precipitated, indicating that there were no detectable HLA class II proteins on the virion surface of the Cyt1 mutant, even though they were present in the pelleted supernatant, most likely associated with microvesicles (data not shown). Therefore, the presence of sequences between aa 708 and 750 within gp41TM are required for the efficient incorporation of HLA class II proteins.
FIG. 3.
HLA incorporation of gp41TM mutants. The p24CA immunoblots of whole-virus precipitates of virions produced from H9 cells (A) or PBMCs (B) by using rabbit anti-HLA class II antibodies, anti-horse IgG (Ir), or anti-HLA class I antibodies are presented. The precipitated samples are identified above their respective lanes. A sample of wild-type virions is included for reference.
HLA class I protein incorporation is Env independent.
HLA class I proteins are also present on HIV-1 particles but at lower levels than HLA class II proteins (reviewed in reference 24). Based on our results, we examined whether Env influences HLA class I protein incorporation onto the surface of the virions. Supernatants containing virions (with equivalent p24CA amounts) produced from H9 cells or PBMCs were subjected to the whole-virus immunoprecipitation procedure with antiserum to HLA class I proteins (DJ-31679; AIDS Vaccine Program, NCI-FCRDC). Immunoblotting of the HLA class I precipitations detected p24CA and Pr55Gag in all of the virus-containing samples from H9 and PBMCs (Fig. 3). The samples from the irrelevant antibody precipitation control contained no Gag proteins, showing that the results were specific for HLA class I proteins. The ability to precipitate virions with anti-HLA class I serum shows that HLA class I proteins were present on the wild-type virus as well as the Env mutant viruses. Thus, incorporation of HLA class I proteins onto the surface of HIV-1 is independent of the presence of Env.
Tyrosine endocytosis signal.
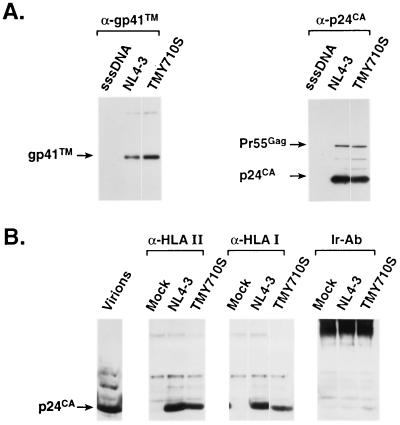
Recent studies have shown that a YXXL endocytosis signal in gp41TM, YSPL at aa 710 to 713 of NL4-3, interacts with the clathrin-mediated endocytosis system to internalize HIV-1 Env (3, 5, 13, 32). Additionally, this signal is a determinant for polar budding of HIV-1 in lymphocytes (11). Since the YSPL sequence is within the aa 708 to 750 region identified above, the tyrosine at aa 710 of pNL4-3 was changed to a serine (TMY710S) and mutant virions were analyzed. Consistent with the report of a similar mutation made in the HXB2 DNA clone (11), this virus incorporated its Env (Fig. 4A) and was infectious (data not shown). Both of the HLA class II and class I whole-virus immunoprecipitation experiments could readily detect p24CA in the blots from both the wild-type and TMY710S virion samples produced from H9 cells (Fig. 4B). Slight differences in signal intensities among the blots were not reproducible: in total, all of the samples produced similar results. Therefore, this mutant contains HLA class II proteins on the virion surface in the absence of the intact YSPL signal, showing that this motif is not required for efficient HLA class II protein incorporation.
FIG. 4.
Endocytosis signal mutant. (A) Immunoblot analysis of the TMY710S mutant by using gp41TM antibody and p24CA anti-serum. Each blot is labeled with the immune reagent used, and the virion samples are identified above their respective lanes. Lane sssDNA, the sample of a negative control transfected with sheared salmon sperm DNA. (B) The p24CA immunoblots of whole virus precipitates of viruses produced from H9 cells by using rabbit anti-HLA class II antibodies, anti-horse IgG, or anti-HLA class I antibodies are presented. The precipitated samples are identified above their respective lanes. A sample of wild-type virions is included for reference.
Incorporation of HLA class II proteins and Env.
The results presented above show that the packaging of Env is required for efficient incorporation of HLA class II proteins onto HIV-1 in both cultured and primary cells. The phenomenon is specific for this major histocompatibility complex protein as the incorporation of HLA class I proteins was not appreciably affected by Env packaging. Incorporation, not just expression of Env, is required for this effect as the Y36S-L41P Gag mutant did not incorporate detectable amounts of HLA class II proteins. Together, these data show that efficient HLA class II protein incorporation requires the packaging of Env. However, HLA class II proteins are not required for Env packaging since Env incorporation is not affected when produced from cells that lack HLA class II proteins, such as 293T cells (M. T. Esser, NCI-FCRDC, personal communication).
The requirement for Env-mediated HLA class II protein incorporation suggests that Env brings HLA class II proteins into the virion. Our mutagenic analysis of gp41TM showed that sequences in its cytoplasmic tail, between Env aa 708 and 750, were important for this effect. Our data also show that the YXXL endocytosis signal within this 43-aa region is not required for efficient HLA class II protein incorporation. Additionally, this region is upstream from the two amphipathic helices present in the gp41TM cytoplasmic region (Fig. 2A) that are thought to be involved in the cytopathic effect of Env (21). Currently, it is not obvious what sequence or sequences in this region are required for HLA class II protein incorporation and whether the absence of the 43-aa sequence disrupts the conformation of other regions of Env. Additional experiments are being carried out to define the sequences within this 43-aa region important for efficient HLA class II protein incorporation as well as the portion of Env that is sufficient for this effect.
The results presented here demonstrate that HLA class II proteins are actively incorporated into HIV-1 particles and not by random association with the budding virion. The finding that a 43-aa stretch of gp41TM close to the inner leaflet of the plasma membrane is required for HLA class II protein incorporation suggests that this occurs either by a direct or a third-party interaction between the molecules. However, a direct interaction between HLA class II proteins and Env has not been demonstrated. It has been observed that HLA class II proteins and Env share some antigenic properties (30), and these could be a basis for interaction. However, at least some of these homologous regions are present in the portion of the C-terminal tail (15) that is dispensable for HLA class II protein incorporation.
An alternative mechanism, that Env directs the budding of HIV-1 to regions of the plasma membrane that contain HLA class II proteins and thereby indirectly cause HLA class II protein incorporation, cannot be formally ruled out. HIV-1 has been observed specifically budding from pseudopodia of activated T cells (27). It has been shown that Env can cause HIV-1 to bud basolaterally in polarized epithelium (26) and that this effect requires the same sequences in the cytoplasmic tail of gp41TM that are deleted in our Cyt2 mutant (22). Since HLA class II proteins were present on the Cyt2 mutant, it is unlikely that specific HLA class II protein incorporation is due to this type of polarized budding. Likewise, the YSPL signal in gp41TM that can also produce polarized budding of HIV-1 (11) is dispensable for HLA class II protein incorporation. However, the presence of the first 10 membrane-proximal amino acids (including YSPL) of the gp41TM cytoplasmic tail can redirect the localization of chimeric VSV-G proteins on the cell surface, suggesting that this sequence might also influence Env localization (20). However, it has not been determined if sequences in this region other than YSPL also influence HIV-1 budding; therefore, its relevance for HLA class II protein incorporation is unclear. One study has shown that redirection of Env does not necessarily affect the site of Gag budding (29). Based on the current understanding of Env and viral budding, HLA class II protein incorporation does not appear to be due to Env redirecting the assembling virus to an HLA class II-rich site on the cell.
While HIV-1 Env is important for HLA class II protein incorporation, it does not seem to affect HLA class I protein incorporation. HLA class I and class II proteins follow different biosynthetic and transport pathways (23, 28): class I molecules traffic through compartments of the secretory pathway, whereas class II molecules interact with the endocytic pathway. Even though both HLA class II molecules and HIV-1 Env are endocytosed, our finding that a gp41TM endocytosis signal is not required for HLA class II protein uptake suggests that this process is not important for its incorporation. However, Env peptides are presented by HLA class II proteins in the absence of endocytosis (32), possibly suggesting that Env and HLA class II proteins could associate during transit to the cell surface.
The presence of HLA class II proteins on the virion surface might also influence HIV-1 pathogenesis. It has been shown that the presence of HLA class II proteins on HIV-1 modestly increased viral infectivity in vitro by a factor of 1.6 to 2.3 (7). Given the number of rounds of replication in an infected individual, even this modest increase could be compounded into a very large difference in viral load over time (10). It has also been proposed that antigen-bearing HLA class II proteins on HIV-1 could induce anergy and possibly apoptosis in the CD4+ T cells (2, 31). HIV-1 might specifically incorporate HLA class II proteins to selectively eliminate virus-specific CD4 helper T cells that could mount immune responses in an infected individual. This might explain why HIV-1 has a mechanism, either direct or indirect, to actively incorporate HLA class II proteins. Further study of the mechanism of HLA class II incorporation might further our understanding of HIV-1 pathology and disease.
Acknowledgments
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-56000.
We thank Jane Burns, University of California, San Diego Medical Center, for the kind gift of pCMVHg; David Waters, NCI-FCRDC, for the kind gift of the HCLZ cells; Ray Sowder for help with computer molecular weight predictions; Mark Esser for sharing 293T data before publication; and Lou Henderson, Jeffrey Rossio, and Mark Esser for helpful suggestions on the manuscript.
REFERENCES
- 1.Adachi A, Koenig S, Gendelman H E, Daugherty D, Gattoni-Celli S, Fauci A S, Martin M A. Productive, persistent infection of human colorectal cell lines with human immunodeficiency virus. J Virol. 1987;61:209–213. doi: 10.1128/jvi.61.1.209-213.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arthur L O, Bess J W, Jr, Sowder II R C, Benveniste R E, Mann L D, Chermann J-C, Henderson L E. Cellular proteins bound to immunodeficiency viruses: implications for pathogenesis and vaccines. Science. 1992;258:1935–1938. doi: 10.1126/science.1470916. [DOI] [PubMed] [Google Scholar]
- 3.Berlioz-Torrent C, Shacklett B L, Erdtmann L, Delamarre L, Bouchaert I, Sonigo P, Dokhelar M C, Benarous R. Interactions of the cytoplasmic domains of human and simian retroviral transmembrane proteins with components of the clathrin adaptor complexes modulate intracellular and cell surface expression of envelope glycoproteins. J Virol. 1999;73:1350–1361. doi: 10.1128/jvi.73.2.1350-1361.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bess J W, Jr, Gorelick R J, Bosche W J, Henderson L E, Arthur L O. Microvesicles are a source of contaminating cellular proteins found in purified HIV-1 preparations. Virology. 1997;230:134–144. doi: 10.1006/viro.1997.8499. [DOI] [PubMed] [Google Scholar]
- 5.Boge M, Wyss S, Bonifacino J S, Thali M. A membrane-proximal tyrosine-based signal mediates internalization of the HIV-1 envelope glycoprotein via interaction with the AP-2 clathrin adaptor. J Biol Chem. 1998;273:15773–15778. doi: 10.1074/jbc.273.25.15773. [DOI] [PubMed] [Google Scholar]
- 6.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cantin R, Fortin J-F, Lamontagne G, Tremblay M. The presence of host-derived HLA-DR1 on human immunodeficiency virus type 1 increases viral infectivity. J Virol. 1997;71:1922–1930. doi: 10.1128/jvi.71.3.1922-1930.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cantin R, Fortin J-F, Tremblay M. The amount of host HLA-DR proteins acquired by HIV-1 is virus strain- and cell type-specific. Virology. 1996;218:372–381. doi: 10.1006/viro.1996.0206. [DOI] [PubMed] [Google Scholar]
- 9.Chen S S, Ferrante A A, Terwilliger E F. Characterization of an envelope mutant of HIV-1 that interferes with viral infectivity. Virology. 1996;226:260–268. doi: 10.1006/viro.1996.0654. [DOI] [PubMed] [Google Scholar]
- 10.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 11.Deschambeault J, Lalonde J P, Cervantes-Acosta G, Lodge R, Cohen E A, Lemay G. Polarized human immunodeficiency virus budding in lymphocytes involves a tyrosine-based signal and favors cell-to-cell viral transmission. J Virol. 1999;73:5010–5017. doi: 10.1128/jvi.73.6.5010-5017.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dubay J W, Roberts S J, Hahn B H, Hunter E. Truncation of the human immunodeficiency virus type 1 transmembrane glycoprotein cytoplasmic domain blocks virus infectivity. J Virol. 1992;66:6616–6625. doi: 10.1128/jvi.66.11.6616-6625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Egan M A, Carruth L M, Rowell J F, Yu X, Siliciano R F. Human immunodeficiency virus type 1 envelope protein endocytosis mediated by a highly conserved intrinsic internalization signal in the cytoplasmic domain of gp41 is suppressed in the presence of the Pr55gag precursor protein. J Virol. 1996;70:6547–6556. doi: 10.1128/jvi.70.10.6547-6556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gluschankof P, Mondor I, Gelderblom H R, Sattentau Q J. Cell membrane vesicles are a major contaminant of gradient-enriched human immunodeficiency virus type-1 preparations. Virology. 1997;230:125–133. doi: 10.1006/viro.1997.8453. [DOI] [PubMed] [Google Scholar]
- 15.Golding H, Robey F A, Gates III F T, Linder W, Beining P R, Hoffman T, Golding B. Identification of homologous regions in human immunodeficiency virus I gp41 and human MHC class II beta 1 domain. I. Monoclonal antibodies against the gp41-derived peptide and patients' sera react with native HLA class II antigens, suggesting a role for autoimmunity in the pathogenesis of acquired immune deficiency syndrome. J Exp Med. 1988;167:914–923. doi: 10.1084/jem.167.3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gorelick R J, Nigida S M, Bess J W, Jr, Henderson L E, Arthur L O, Rein A. Noninfectious human immunodeficiency virus type 1 mutants deficient in genomic RNA. J Virol. 1990;64:3207–3211. doi: 10.1128/jvi.64.7.3207-3211.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henderson L E, Sowder II R C, Smythers G, Oroszlan S. Chemical and immunological characterizations of equine infectious anemia virus gag-encoded proteins. J Virol. 1988;61:2587–2595. doi: 10.1128/jvi.61.4.1116-1124.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horton R M, Cai Z, Ho S N, Pease L R. Gene splicing by overlap extension: tailor-made genes using polymerase chain reaction. BioTechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 19.Hunter E. Synthesis, assembly, and processing of viral proteins. In: Coffin J, Hughes S, Varmus H, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Press; 1997. pp. 71–120. [PubMed] [Google Scholar]
- 20.Johnson J E, Rodgers W, Rose J K. A plasma membrane localization signal in the HIV-1 envelope cytoplasmic domain prevents localization at sites of vesicular stomatitis virus budding and incorporation into VSV virions. Virology. 1998;251:244–252. doi: 10.1006/viro.1998.9429. [DOI] [PubMed] [Google Scholar]
- 21.Kliger Y, Shai Y. A leucine zipper-like sequence from the cytoplasmic tail of the HIV-1 envelope glycoprotein binds and perturbs lipid bilayers. Biochemistry. 1997;36:5157–5169. doi: 10.1021/bi962935r. [DOI] [PubMed] [Google Scholar]
- 22.Lodge R, Gottlinger H, Gabuzda D, Cohen E A, Lemay G. The intracytoplasmic domain of gp41 mediates polarized budding of human immunodeficiency virus type 1 in MDCK cells. J Virol. 1994;68:4857–4861. doi: 10.1128/jvi.68.8.4857-4861.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neefjes J J, Stollorz V, Peters P J, Geuze H J, Ploegh H L. The biosynthetic pathway of MHC class II but not class I molecules intersects the endocytic route. Cell. 1990;61:171–183. doi: 10.1016/0092-8674(90)90224-3. [DOI] [PubMed] [Google Scholar]
- 24.Ott D E. Cellular proteins in HIV. Rev Med Virol. 1997;7:167–180. doi: 10.1002/(sici)1099-1654(199709)7:3<167::aid-rmv199>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 25.Ott D E, Chertova E N, Busch L K, Coren L V, Gagliardi T D, Johnson D G. Mutational analysis of the hydrophobic tail of the human immunodeficiency virus type 1 p6(Gag) protein produces a mutant that fails to package its envelope protein. J Virol. 1999;73:19–28. doi: 10.1128/jvi.73.1.19-28.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Owens R J, Dubay J W, Hunter E, Compans R W. Human immunodeficiency virus envelope protein determines the site of virus release in polarized epithelial cells. Proc Natl Acad Sci USA. 1991;88:3987–3991. doi: 10.1073/pnas.88.9.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearce-Pratt R, Malamud D, Phillips D M. Role of the cytoskeleton in cell-to-cell transmission of human immunodeficiency virus. J Virol. 1994;68:2898–2905. doi: 10.1128/jvi.68.5.2898-2905.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peters P J, Neefjes J J, Oorschot V, Ploegh H L, Geuze H J. Segregation of MHC class II molecules from MHC class I molecules in the Golgi complex for transport to lysosomal compartments. Nature. 1991;349:669–676. doi: 10.1038/349669a0. [DOI] [PubMed] [Google Scholar]
- 29.Pfeiffer T, Zentgraf H, Freyaldenhoven B, Bosch V. Transfer of endoplasmic reticulum and Golgi retention signals to human immunodeficiency virus type 1 gp160 inhibits intracellular transport and proteolytic processing of viral glycoprotein but does not influence the cellular site of virus particle budding. J Gen Virol. 1997;78:1745–1753. doi: 10.1099/0022-1317-78-7-1745. [DOI] [PubMed] [Google Scholar]
- 30.Puppo F, Ruzzenenti R, Brenci S, Lanza L, Scudeletti M, Indiveri F. Major histocompatibility gene products and human immunodeficiency virus infection. J Lab Clin Med. 1991;117:91–100. [PubMed] [Google Scholar]
- 31.Rossio J L, Bess J W, Jr, Henderson L E, Cresswell P, Arthur L O. HLA class II on HIV-1 particles is functional in superantigen presentation to human T cells: implications for HIV pathogenesis. AIDS Res Hum Retrovir. 1995;11:1433–1439. doi: 10.1089/aid.1995.11.1433. [DOI] [PubMed] [Google Scholar]
- 32.Rowell J F, Stanhope P E, Siliciano R F. Endocytosis of endogenously synthesized HIV-1 envelope protein. Mechanism and role in processing for association with class II MHC. J Immunol. 1995;155:473–488. [PubMed] [Google Scholar]
- 33.Saarloos M-N, Sullivan B L, Czerniewski M A, Parameswar K D, Spear G T. Detection of HLA-DR associated with monocytropic, primary, and plasma isolates of human immunodeficiency virus type 1. J Virol. 1997;71:1640–1643. doi: 10.1128/jvi.71.2.1640-1643.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schols D, Pauwels R, Desmyter J, De Clercq E. Presence of class II histocompatibility DR proteins on the envelope of human immunodeficiency virus demonstrated by FACS analysis. Virology. 1992;189:374–376. doi: 10.1016/0042-6822(92)90719-6. [DOI] [PubMed] [Google Scholar]
- 35.Swanstrom R, Wills J. Synthesis, assembly, and processing of viral proteins. In: Coffin J, Hughes S, Varmus H, editors. Retroviruses. Plainview, N.Y: Cold Spring Harbor Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 36.Wilk T, Pfeiffer T, Bosch V. Retained in vitro infectivity and cytopathogenicity of HIV-1 despite truncation of the C-terminal tail of the env gene product. Virology. 1992;189:167–177. doi: 10.1016/0042-6822(92)90692-i. [DOI] [PubMed] [Google Scholar]
- 37.Yu X, Yuan X, McLane M F, Lee T H, Essex M. Mutations in the cytoplasmic domain of human immunodeficiency virus type 1 transmembrane protein impair the incorporation of Env proteins into mature virions. J Virol. 1993;67:213–221. doi: 10.1128/jvi.67.1.213-221.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zavada J. The pseudotypic paradox. J Gen Virol. 1982;63:15–24. doi: 10.1099/0022-1317-63-1-15. [DOI] [PubMed] [Google Scholar]