Abstract
Objectives
Anaphylaxis is a severe life-threatening allergic reaction, and its accurate identification in healthcare databases can harness the potential of “Big Data” for healthcare or public health purposes.
Materials and methods
This study used claims data obtained between October 1, 2015 and February 28, 2019 from the CMS database to examine the utility of machine learning in identifying incident anaphylaxis cases. We created a feature selection pipeline to identify critical features between different datasets. Then a variety of unsupervised and supervised methods were used (eg, Sammon mapping and eXtreme Gradient Boosting) to train models on datasets of differing data quality, which reflects the varying availability and potential rarity of ground truth data in medical databases.
Results
Resulting machine learning model accuracies ranged from 47.7% to 94.4% when tested on ground truth data. Finally, we found new features to help experts enhance existing case-finding algorithms.
Discussion
Developing precise algorithms to detect medical outcomes in claims can be a laborious and expensive process, particularly for conditions presented and coded diversely. We found it beneficial to filter out highly potent codes used for data curation to identify underlying patterns and features. To improve rule-based algorithms where necessary, researchers could use model explainers to determine noteworthy features, which could then be shared with experts and included in the algorithm.
Conclusion
Our work suggests machine learning models can perform at similar levels as a previously published expert case-finding algorithm, while also having the potential to improve performance or streamline algorithm construction processes by identifying new relevant features for algorithm construction.
Keywords: anaphylaxis, machine learning, public health, allergy, electronic health records, Centers for Medicare & Medicaid Services
Introduction
The term anaphylaxis was first introduced in 1901 by Charles Richet and Paul Portier while trying to “immunize” dogs to the venom of the sea anemone. They identified that some dogs developed an increased sensitivity rather than prophylaxis, which they named anaphylaxis.1–3 Today, anaphylaxis is recognized as a severe life-threatening allergic reaction that must be differentiated from milder allergic reactions. Since anaphylaxis shares many signs and symptoms with allergic reactions, identifying these cases can be difficult. In addition, anaphylaxis cases can be underdiagnosed and underreported in the United States and remain a burden in the healthcare system.4,5
Accurate detection of anaphylaxis cases in healthcare databases can help to harness the potential of “Big Data” in healthcare and public health through the use of available data for surveillance or research purposes. Retrospective identification of anaphylaxis in administrative claims data is not meant to immediately impact patient care, rather it can help with general surveillance to gauge the incidence of anaphylaxis in the population, or to assess the rate of anaphylaxis after exposure to a specific medical product.6,7 This can help assess if a specific intervention may be associated with a higher rate of anaphylaxis than expected, or to compare to other similar products, which can be useful for benefit-risk assessment.8,9 In addition, identification and surveillance of vaccine-induced anaphylaxis cases, especially for vaccines developed in response to pandemics, can be useful for public health and safety.10
Identifying anaphylaxis outcomes within healthcare databases, particularly in administrative claims data, can be a challenging task. To accomplish this, healthcare professionals have relied on the use of ICD-9 and ICD-10 codes for the detection of anaphylaxis cases.11,12 A rule-based algorithm, devised by experts for anaphylaxis identification was created analyzing exposure-agnostic anaphylaxis patterns in claims data. The algorithm was validated post-vaccination, and yielded a 58% positive predictive value (PPV, also known as Precision) and 100% sensitivity (also known as Recall), with potential PPV increase at the cost of diminished sensitivity.7 This instance underscores the difficulty of crafting accurate algorithms for medical outcome identification, especially for conditions like anaphylaxis with varied presentations and coding. Our proposal suggests machine learning (ML) methods as a promising approach to enhance anaphylaxis case detection. To demonstrate the usefulness of the proposed method, we compared performance metrics of ML algorithms against the rule-based algorithms created by experts.
To explore the effectiveness of ML in detecting anaphylaxis cases, we used CMS (Centers for Medicare & Medicaid Services) Fee For Service (FFS) claims data. We deviated from the classical supervised ML paradigm, as “ground truth” or chart-confirmed results are frequently rare or unavailable in healthcare databases. Instead, we used a minimally curated dataset created with simple, well-recognized aspects of anaphylaxis, such as specific treatments and relevant symptoms, to divide CMS claims into subsets with varying likelihoods of containing real anaphylaxis cases.
We compared the ML model predictions to those identified through a rule-based anaphylaxis algorithm, previously developed by subject matter experts, to find potential cases within CMS claims data, along with a subset featuring chart-confirmed outcomes.7 This evaluation allowed us to gauge the ML model’s capacity to mimic human approaches and assess its accuracy. Our findings indicate that ML can efficiently select pertinent claim codes, expediting algorithm creation through collaboration with human experts who can then investigate the ML-identified features. This approach holds promise for enhancing accuracy by optimizing available data utilization. Furthermore, we compared features identified by human algorithms and those identified by ML models. This comparison aimed to highlight areas of concordance in processes and measure accuracy indicators such as PPV.
Methods
Study data
This project utilized CMS FFS claims data from October 1, 2015 to February 28, 2019, which contains codes from ICD-10-Clinical Modification (ICD-10-CM), the ICD-10 Procedure Coding System (ICD-10-PCS), Healthcare Common Procedure Coding System (HCPCS), and Current Procedural Terminology (CPT). These codes are used to describe patient interactions with the healthcare system and are submitted by providers to CMS for reimbursement. Prior experience demonstrated that the presence or absence of an ICD anaphylaxis code on its own did not definitively identify chart-confirmed anaphylaxis, so combinations of codes were utilized to identify claims that were possible or probable cases of chart-confirmed anaphylaxis.7,11
Claims data used in this study consists of 3 cohorts. Cohort 1 is the Minimally Curated Dataset (MCD) that was constructed using simple, easily identifiable rules that generate four groups of interest: the allergy claims (unlikely anaphylaxis), the possible anaphylaxis (also named AR group), probable anaphylaxis (index group), and random (control) background claims. (See File S1 Sheet: “Rules for Minimal Data Curation” for the list of specific codes for each subset). The allergy claims subset consists of non-anaphylaxis allergy codes without anaphylaxis-specific treatments and explicitly excludes possible anaphylaxis (AR) cases. The AR subset contains claims with initial visit anaphylaxis codes, while the probable anaphylaxis group contains claims with the same anaphylaxis codes as in the possible anaphylaxis group combined with an anaphylaxis treatment, such as epinephrine. The background subset contains a random selection of claims from the entire CMS dataset. The allergy, possible anaphylaxis, and probable anaphylaxis subsets were constructed to be mutually exclusive. The probable anaphylaxis, possible anaphylaxis, and allergy subsets have 500 episodes each, and the background subset has 1000 episodes. The total number of codes present in at least one episode of the MCD is 4313.
The second cohort is the Expert Driven Dataset (EDD), consisting of claims from vaccinated individuals discovered by the rule-based human expert algorithm. In addition, this dataset contained some claims not satisfying the algorithm, which helped human experts evaluate decisions made during algorithm construction. In EDD, a suspected anaphylaxis case is a claim that satisfies either the extended or core expert algorithm’s criteria to be an anaphylaxis case, while likely non-anaphylaxis cases are claims that do not satisfy the expert algorithm criteria.7 The anaphylaxis cases included those due to the vaccine and those due to other triggers. This dataset size is 363 episodes and 1252 codes.
The third cohort is the Chart Confirmed Dataset (CCD), which contains Ground Truth for this study. The dataset is the subset of EDD cases for which charts were requested, received, and chart-confirmed to verify anaphylaxis status. In CCD, we used the Brighton Collaboration Anaphylaxis case definition which utilizes medical chart data to validate if a chart satisfies clinical criteria to be considered a case of anaphylaxis or not.13 The Brighton case definition is comparable and informed by the NIAID/FAAN anaphylaxis case definition, which is used for non-vaccine exposures.14 The CCD sample size is 174 episodes and contains 1252 CMS codes.
When training and testing ML models in this study, we used cohort-specific anaphylaxis and non-anaphylaxis claims designations. For example, in the MCD, cases in the high-likelihood subset are considered anaphylaxis for model training, while chart-confirmed cases in the CCD are considered reference anaphylaxis cases for model testing.
Manual case finding algorithm
For comparison against ML approaches, this study utilized a rule-based case definition algorithm which was constructed to differentiate incident anaphylaxis cases from prevalent, historical cases, and less-severe allergy cases in claims data.7 Experts built the rules and selected codes by utilizing their clinical knowledge, an understanding of Medicare claims, and the expected acuity patterns in the office (PB), inpatient (IP), and outpatient (OP) claims settings in the United States (Supplementary Information: Description of Cohorts).
Data preprocessing and harmonization
Medical claims were converted into tables, where codes were arranged as columns and episodes as rows. If a code was present for a case, the table value assigned would be 1; otherwise, it was 0. Episodes were allocated to the corresponding subsets for each cohort, creating mutually exclusive datasets. Since CMS codes assigned to a claim reflected the specific clinical situation at that time, the three cohorts did not contain the same codes. However, to enable analysis across the datasets, they were merged, and the missing features were filled with zeros. This approach accounted for variation among patients, medical history, clinical processes, providers, and institutions while permitting further analysis.
Experimental design
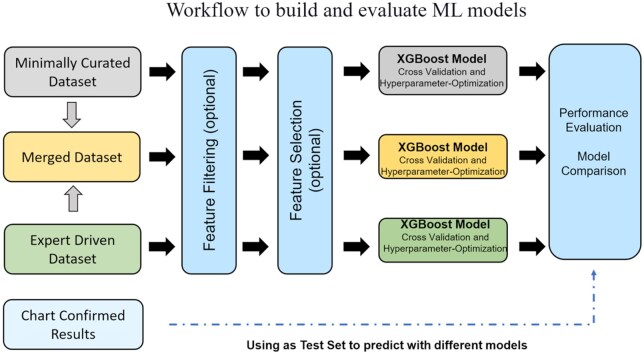
To train ML classifiers for anaphylaxis identification, we utilized various configurations for model training inputs. These include (1) minimally curated dataset (MCD), (2) expert-driven dataset (EDD), and a merged dataset (3) that combines MCD and EDD, and (4) chart confirmed data (CCD) (Figure 1). For each dataset, we applied three different configurations: (1) no feature filtering or selection, (2) feature filtering, and (3) both feature filtering and selection. All model predictions were tested on CCD, and a separate experiment was conducted to train models on the ground truth data using a 20% held-out portion of CCD for testing.
Figure 1.
Workflow to build and evaluate ML models.
Feature filtering
ML model training aimed to accurately identify both “strong” and “weak” influencers within the data, and to prevent data leakage,15,16 by removing the codes that were part of the case definitions used to construct datasets. These codes included word stems such as “allerg,” “epinephrine,” “endotracheal,” and “anaph.” Additionally, removing case-defining codes tested whether enough structure remained in the data to build predictive models. Filtering these codes reduced the number of features to 4228 in MCD (from 4313), and 1197 in EDD and CCD (from 1252). Without code filtering, the total number of codes across all datasets was 4539, and after filtering, it was 4450.
Feature selection
The selection process seeks to identify key features across datasets that effectively detect anaphylaxis events. We achieve this by training models with each dataset separately, using repeated stratified K-fold cross-validation. We employ multiple feature selection methods that are not bound by specific algorithms or statistical assumptions. By doing so, we increase the likelihood of identifying features that are relevant and generalize well across datasets.
Our study utilizes 5 different statistical tests and ML algorithms to find salient features.17 These tests include:
Chi-square analysis.18
Recursive feature elimination using Logistic Regression.19,20
Hyperparameter tuned Logistic regression classifier (High error rate).20,21
Hyperparameter tuned Random Forest classifier.22
Hyperparameter tuned Light Gradient Boosting classifier.23
We used Sklearn Python 3 implementation for Methods 1-4 with default parameters and the Python package lightgbm for the Light Gradient Boosting classifier. The top 200 features chosen by each method were combined, and only the codes selected by at least one feature selector were retained in the claims dataset. After applying this feature selection method to each dataset separately, the workflow identified 352 codes in MCD and 338 codes in EDD as relevant for classification. We also created a feature set of 131 codes by taking the intersection of the selected features from MCD and EDD. Additional information on the selected features for individual cohorts and the common features can be found in File S1, sheets: “MCD Selected Features,” “EDD Selected Features,” and “Common Features.”
Dimensionality reduction and Sammon maps
Our study used t-SNE,24 PCA,25 and Multi-Dimensional Scaling26 (MDS or Sammon Mapping) algorithms to observe topological patterns in the claims data and to assess the distinctiveness of compared classes. Hyperparameters for each unsupervised method used to create the plots can be found in File S1, sheet name: “Hyperparameters.”
Supervised ML analysis
In our study, we employed various ML algorithms to train classifiers and assess model performance metrics. Our ML algorithm selection included SVM,27 Random Forests,22 and XGBoost.28 To implement SVM and Random Forests, we used the Sklearn Python 3 library with the default algorithm parameters. We used the Python implementation of XGBoost Classifier for XGBoost, with the parameters tested within a range of 100-500 for the number of estimators, 6-20 for the maximum tree depth, and 0.001-0.1 for min_child_weight. However, we observed no substantial differences in model performance within this parameter range. The Hyperparameters sheet in File S1 contains the exact hyperparameters we used for training the ML models. We obtained feature importance for XGBoost models using the importance type “weight.”
To measure model performance, we calculated the number of false positives, false negatives, true negatives, and true positives, from which we derived sensitivity, specificity, accuracy, PPV, and NPV using standard definitions.
We also evaluated the impact of feature removal on model performance beyond XGBoost. For this, we trained 27 supervised algorithms using the lazypredict package,29 recording accuracy, balanced accuracy, F1 score, and ROC score for each model, as well as the mean values for individual datasets. These results are available in the Effect of Feature Removal sheet of File S1.
Results
Dimensionality reduction results
We utilized the multi-dimensional scaling algorithm to explore the underlying patterns in the data. The three maps in Figure 2 are a result of the same Sammon mapping that used the combined dataset before feature removal and selection, but are color and shape-coded based on different properties. Color coding in Figure 2A by cohort demonstrates that chart-confirmed results tend to be placed within the EDD, which in turn are clustered within the MCD. Likewise, Figure 2B illustrates clustering by clinical setting. Finally, Figure 2C puts anaphylaxis designations within each dataset and the clinical setting together to show that anaphylaxis cases cluster in three areas by setting, and that these tend to be more proximally surrounded by cases that were thought more likely to be true anaphylaxis (eg, satisfying the human algorithm), while cases less likely to be anaphylaxis (eg, allergy and random cases) are located more distally to the chart confirmed anaphylaxis cases.
Figure 2.
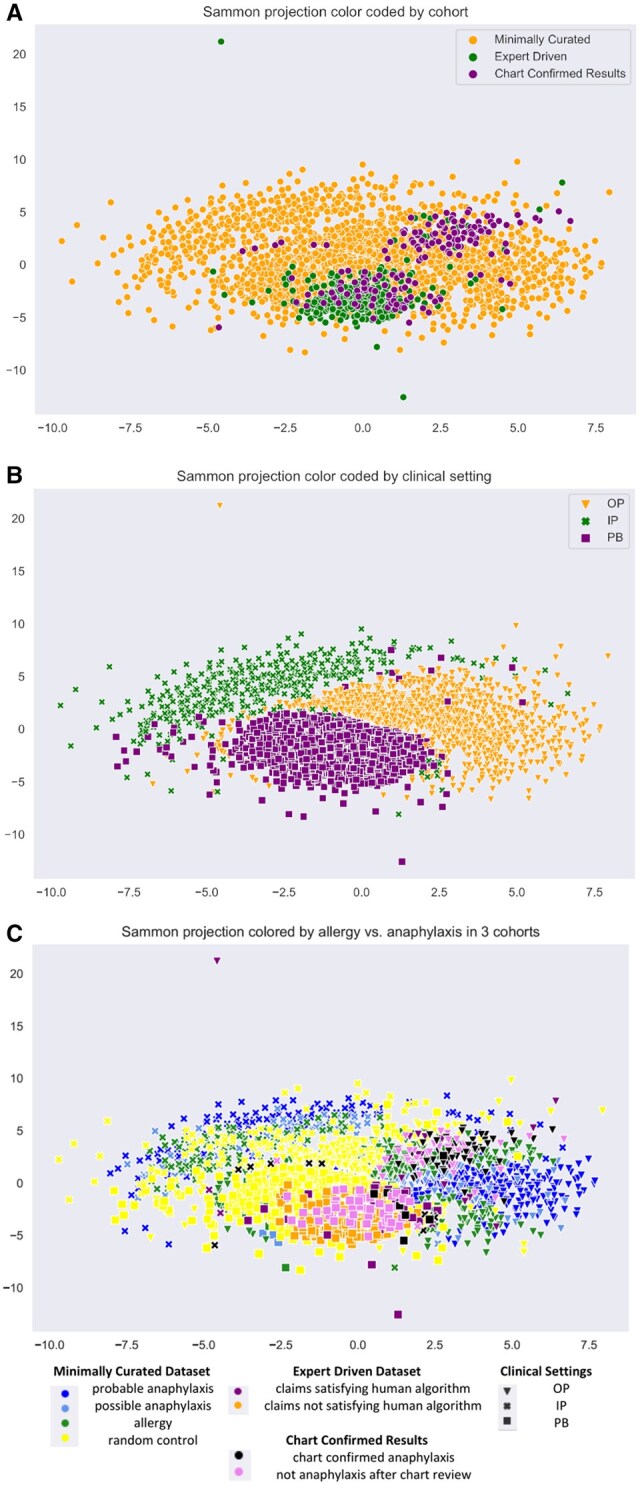
(A) Sammon projection color coded by cohort. (B) Sammon projection of datasets by color coded clinical setting. OP is outpatient, IP is inpatient, and PB is ambulatory (office) setting. (C) Sammon projection. Results for allergy versus anaphylaxis. Comparison for each cohort MCD, EDD, and CCD in order of appearance before feature selection. The graph includes the clinical setting information and separation by case definitions for distinct datasets.
Still, the class separation is incomplete as some nodes blend across data classes. In addition, the mapping identifies outliers, which could be investigated further. Similar observations were made with alternative dimensionality reduction methods PCA and t-SNE.
Feature filtering and feature selection results
Data preprocessing and filtering of core case definition codes (eg, code descriptions which contain “allerg,” “epinephrine,” “endotracheal,” and “anaph”) removed 4228 features in the MCD, and 1197 features in the EDD. In total, there were 4539 features before feature filtering and 4450 after the filtering. About 131 codes were consistent between MCD and EDD.
We have performed the feature selection process for each dataset separately, as described in M&) (Figure 1).
Supervised machine learning model results
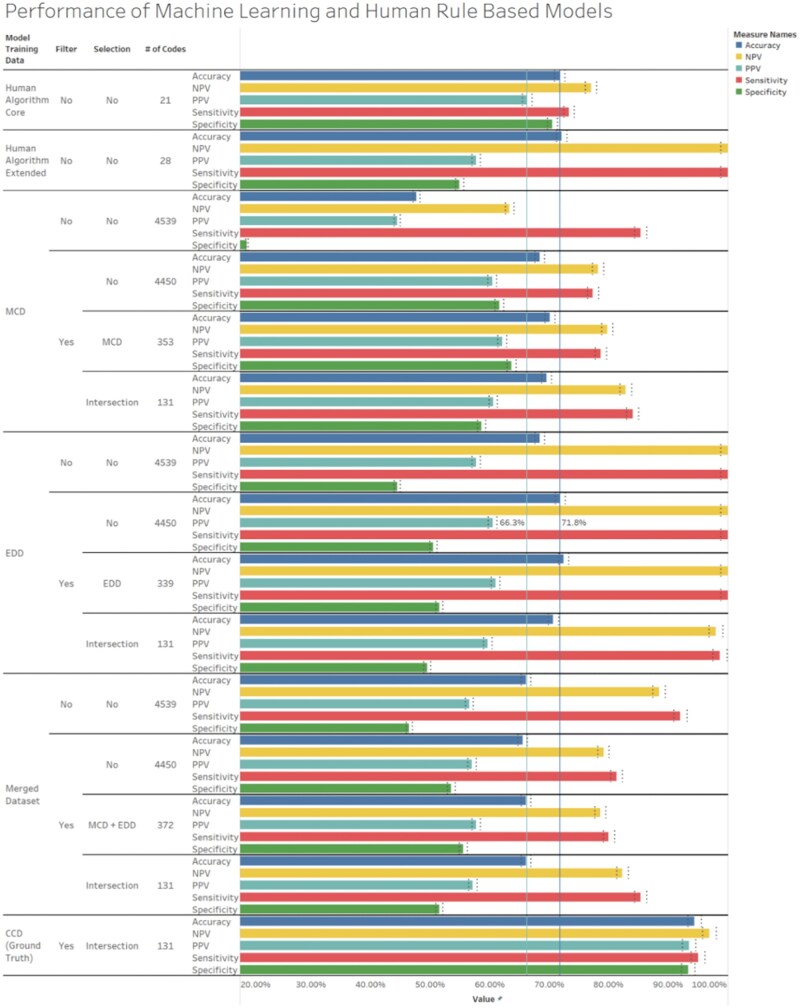
The results of model performance tested against ground truth data are reported as accuracy, sensitivity, specificity, PPV, and NPV (Figure 3). The first two rows present the results of these metrics for the human expert rule-based algorithms that were previously constructed. The core human algorithm achieves a balanced sensitivity, specificity, and accuracy of 73.3%, 70.5%, 71.8%, while the extended human algorithm has a much higher sensitivity (100%), but lower specificity (54.9%), and an accuracy of 72.1%. This contrasts with the various ML algorithms, for which accuracy ranged from ∼47.7% to ∼72.4% in the generated models, and PPV was between 44.4% to 62.1%, with the highest values being better than results from the human expert algorithm, that is, 62.1% versus ∼57.7%. Model performance metrics improved as data quality increased, progressing from models using MCD (using simple-rules-based probable anaphylaxis as a proxy for ground truth) to EDD (using expert algorithm-identified anaphylaxis cases as ground truth), and finally, those using CCD (Ground Truth). The extended human expert model, however, had the highest sensitivity at 100%. The model constructed using MCD performed comparably with the core human expert algorithm after feature removal and dataset-specific feature selection. In Figure 3, the EDD model had a slightly higher accuracy and PPV, 72.4% and ∼61%, respectively, compared to MCD model (70.1% and 62.1%). In fact, the model’s accuracy and PPV values were relatively stable and only markedly improved when data quality improved significantly with models constructed using chart confirmed or ground truth data. For the CCD/ground truth model, all performance metrics were above 93.5% after feature removal and selection (131 common features identified by the workflow) and indicated the importance of data quality as the main factor of model performance.
Figure 3.
Model performance shows a progression in accuracy, sensitivity, and PPV when models are trained on datasets with an increasingly reliable estimate of ground truth (MCD), then, EDD, and last Ground Truth using the common features obtained by feature selection on MCD and EDD. Model performance is increased with core case definition code removal in models trained using MCD and EDD when tested against ground truth data. All models use chart-confirmed data to assess model performance, except the model generated by chart-confirmed data, which uses 20% of a held-out portion of itself. The dark blue and light blue reference lines represent the accuracy and PPV baseline for the Human Core Algorithm. The light gray reference bands specify the confidence intervals for the generated models. Finally, the feature content of trained models suggests using a lower number of potent features common between datasets can be as effective as a high number of dataset-specific features.
Understanding and explaining the machine learning model
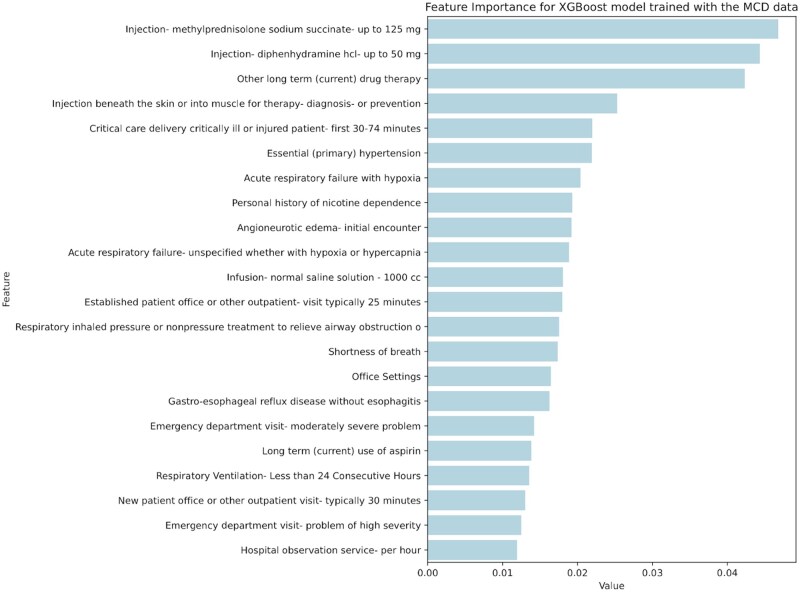
All models were investigated further with feature importance ranking to understand which features were the most useful for the ML model decision process. Figure 4 shows the most important codes found. Results demonstrate the ML process replicated aspects of the human algorithm creation process. This is clear in the MCD model’s identification of injections of diphenhydramine, methylprednisolone, and the angioneurotic edema codes as helpful when identifying anaphylaxis. Similarly, the MCD model identified the healthcare setting as important when identifying anaphylaxis (ER as emergency, PB as the office setting), reflecting the human expert’s decision to use varying decision rules according to clinical setting.7 All these features were not part of the codes used for minimal curation and MCD generation, but were rather identified through ML.
Figure 4.
Feature importance for eXtreme gradient boosting model trained with the MCD data. The top 22 features are shown. PB is Ambulatory (Office) setting.
In addition, the high-ranking features were consistent with symptoms, signs, tests, or interventions naturally occurring in the setting of anaphylaxis cases. These include respiratory compromise resulting in shortness of breath, and the ordering of a chest x-ray to identify potential non-anaphylactic causes in complex older patients that may have multiple comorbidities. Similarly, ML identified the code for normal saline infusion as important; this could represent the need for fluid resuscitation that can occur in a patient with cardiovascular compromise. All codes identified by ML, however, may not be useful, and potentially due to noise or an artifact in the data; this is likely the case with codes for conditions such as diabetes, or nicotine dependence, which are not readily associated with anaphylaxis.
Lastly, ML identified salient codes not found through the human expert process, and that were useful in identifying cases of anaphylaxis. ML specifically identified “Injection beneath the skin or into muscle for therapy, diagnosis, or prevention” as valuable, and inspection of this code determined that it was useful in the PB, or office setting, where an anaphylaxis code is combined with a treatment code to differentiate acute cases of anaphylaxis from individuals receiving follow-up care. In the office setting, this injection code had a higher PPV (100%) than the epinephrine injection code (PPV = 37.5%), which is the first-line treatment for anaphylaxis and was used to identify probable anaphylaxis cases in the MCD. A detailed description of how a decision tree-based model uses these features to classify anaphylaxis cases can be seen in Figure S1.
Discussion
This study demonstrated the applicability of ML methods for anaphylaxis identification in large administrative medical databases, even when ground truth data may or may not be available, with ML models performing comparably or better than an expert rule-based algorithm for identifying anaphylaxis. Our findings suggest ML models can be leveraged to enhance the manual process of building new rule-based case finding algorithms.5 Namely, the determination of feature importance metrics through ML model training permits the identification of codes associated with anaphylaxis outcomes. As these codes were found meaningful by the subject matter experts, the algorithm construction process can be automated and streamlined with ML once a dataset with case/control pairs is formed with either minimal curation, or with ground truth data, if it is available. In addition, we showed ranks of feature importance between important variables, which facilitates understanding of the generated models, ensuring their explainability and credibility in a decision-making process to human experts and healthcare practitioners.
We found filtering out codes used in the minimal data curation process (case-defining codes) improved the performance of the ML models. This observation was verified by training classifiers with filtered (without the case defining codes) vs. unfiltered data using 27 supervised ML algorithms in the sklearn library and demonstrating model performance gains on ground truth data after the feature removal process (File S1, sheet “Effect of Feature Removal”). The observation suggests that the presence of case-determining codes is reducing the ML models’ ability to generalize well, causing lower performance on new datasets. This observation is similar to the phenomenon of “data leakage” (more specifically, feature leakage) known in ML literature.15,16 An example of data leakage would be a model that uses the target variable as a predictor, for example, concluding that “It will snow on snowy days.” A remedy to this problem would be to remove predictor codes from model training. Similarly, in our case, removing codes that effectively define the anaphylaxis cases when building the ML model has improved ML performance. In summary, our analysis showed that removing certain case-defining codes during the curation process of the minimally curated dataset, and to a lesser extent, the EDD, improved the ML model’s robustness.
Our analyses demonstrated that if strong predictors are selected across cohorts, the ML model predictions are more reliable and well generalized but, as a downside, become less accurate (Figure 3). In theory, identifying common aspects of two datasets could reduce noise introduced in a single dataset by lowering any mislabeled episodes’ contribution to the final model.30 We found that feature removal reduces the ML model bias and error,30,31 while cohort-specific feature selection followed by common feature identification guarantees the recognition of well-defined properties and reduces variance once a model is trained using these features. The benefit of such a workflow is the identification of well-defined patterns with a small number of features. The experiments on MCD and EDD confirm this hypothesis with incremental increases in model performance to a certain point (Figure S2), depending on the strictness of the feature selection workflow and the amount of agreement between feature selection algorithms. Feature selection is helpful in both settings when we use single or multiple datasets to identify essential features and train models with the inclusion of these identified features.
The mixture of feature selection and common feature identification using multiple datasets can improve the robustness of ML models. This conclusion was reached by comparing selected MCD and EDD features. Although common feature identification can reduce the accuracy of the model due to the reduction of relevant information content, it is beneficial in determining salient codes. By using ML models and feature selection workflows, we improved the accuracy of existing rule-based classification algorithms by identifying additional codes and patterns that can be useful for identifying anaphylaxis cases. Identification of the injection code is an example that enabled the generated models to outperform the epinephrine code in an office setting. This finding suggests that combining expert knowledge with salient codes identified by ML methods and feature selection workflows can enhance existing case-finding algorithms.
One of the strengths of this study is the use of various levels of data quality and different ML methods to construct successful classifiers. This allowed us to demonstrate the potential utility of ML in healthcare claims data analysis, even when ground truth data is not available, as is the case with the MCD model which performed at levels comparable to the human expert rule-based models. Additionally, by comparing ML models to a human expert process, we were able to show that ML models can replicate aspects of expert rule-based approaches. Finally, the use of transparent ML model construction methods and model explainers helps healthcare professionals to comprehend decision-making by the ML model better.
One limitation of our study is the need for an assessment regarding whether the ML models can uncover new cases in claims data, a process demanding resource-intensive chart reviews that exceeds the scope of our study. Moreover, our method’s adaptability to other outcomes of interest remains to be determined. Instead of arduous chart-review, expert review of ML identified features could be used to qualitatively assess the validity of newly constructed algorithms. By incorporating a broader range of features using ML compared to existing algorithms, our approach aims to uncover more edge cases, thereby enhancing the development of a resilient case definition and reinforcing the overall method’s robustness.
Clinical variability in drug usage, as with drugs like diphenhydramine, methylprednisolone, and epinephrine, which extend beyond anaphylaxis treatment can complicate accurate case identification solely through drug codes. Our study acknowledges the intricacy of crafting a manual case definition due to numerous variables, highlighting the value of ML models. These models unveil latent patterns and validate trends that may be too complicated for manual methods. By identifying connections across variables like drug usage, signs, and symptoms, these models facilitate anaphylaxis case identification, underscoring ML's pivotal role in overcoming challenges tied to utilizing claims data for this purpose.
Many studies have used the medical claims database and EHR to create ML classifiers for case/control pairs, including diabetes mellitus, systemic sclerosis, pulmonary hypertension, rheumatoid arthritis, and many others.32–34 A common trend in these studies is the use of chart-confirmed data for training and validation of ML models. Few studies focus on the feature selection and removal process or identifying important variables for classification purposes without the readiness of ground truth data. Even fewer studies are investigating ML methods for enhancing the existing rule-based classification methods with features identified by ML, or a workflow that could be successful without chart-confirmed data. Currently, we have not observed any studies that investigate possible data leakage problems with generated ML models using a claims database.
The challenge of isolating anaphylaxis using claims codes is documented, with anaphylaxis codes proving inadequate for the precise identification of cases and associated mortality.11 Earlier rule-based algorithms crafted by experts and utilizing claims data have exhibited variable PPVs ranging from 42% to 66%.6,7,35,36 These algorithms can be adjusted to enhance specificity, albeit at the expense of diminished sensitivity.6 Attempts to automate anaphylaxis detection using diverse data sources (eg, VAERS reports, electronic medical records) and methods like natural language processing have shown similar complexity, yielding PPVs around 60% and trading off sensitivity for higher specificity.37,38
Our study underscores the potency of ML) for the intricate task of accurately identifying anaphylaxis in claims data. Our model utilizing ground truth data boasts sensitivity, specificity, and PPV exceeding 90%. Moreover, models like the MCD models, absent ground truth data, achieved PPVs around 60%, akin to expert rule-based approaches. This suggests that ML methods can construct highly accurate algorithms with ample ground truth and, when such data is lacking, can swiftly develop algorithms comparable in performance to expert rule-based methods with minimal data curation.
Conclusion
Based on our experience, we propose the following workflow for researchers working with claims data. If chart-confirmed data is unavailable, a minimally curated dataset can be generated with simple rules that contain a data subset with a high density of cases and a negative control data subset with a very low likelihood of being the outcome of interest. The generated data pairs could be used with unsupervised techniques and investigated for underlying patterns or outliers. If the data can be represented with clear clusters, simple supervised ML techniques could be used for case classification due to the underlying properties of the data. If there are any specific, highly potent codes used to tag cases, filtering them out would be beneficial for identifying underlying patterns and features. In addition, with explainable algorithms such as decision trees, one could use model explainers to identify salient codes, which could be presented to experts and incorporated into a rule-based algorithm, for tasks where rule-based algorithms are required. Alternatively, if ample ground truth data is available, researchers can utilize supervised ML methods using ground truth data for algorithm construction and validation. The ability to use ML methods with varying levels of starting data quality, and the potential to combine with expert opinion, could make algorithm construction more efficient and accurate, which could help make claims data more practical for research and surveillance purposes.
Supplementary Material
Contributor Information
Kamil Can Kural, Center for Biologics Evaluation and Research (CBER), Food and Drug Administration, Silver Spring, MD 20993, United States; School of Systems Biology, George Mason University, Manassas, VA 20110, United States.
Ilya Mazo, Center for Biologics Evaluation and Research (CBER), Food and Drug Administration, Silver Spring, MD 20993, United States.
Mark Walderhaug, Center for Biologics Evaluation and Research (CBER), Food and Drug Administration, Silver Spring, MD 20993, United States.
Luis Santana-Quintero, Center for Biologics Evaluation and Research (CBER), Food and Drug Administration, Silver Spring, MD 20993, United States.
Konstantinos Karagiannis, Center for Biologics Evaluation and Research (CBER), Food and Drug Administration, Silver Spring, MD 20993, United States.
Elaine E Thompson, Center for Biologics Evaluation and Research (CBER), Food and Drug Administration, Silver Spring, MD 20993, United States.
Jeffrey A Kelman, Centers for Medicare & Medicaid Services, Washington, DC 20001, United States.
Ravi Goud, Center for Biologics Evaluation and Research (CBER), Food and Drug Administration, Silver Spring, MD 20993, United States.
Author contributions
K.C.K. had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. K.C.K. is responsible for the concept and design, acquisition, analysis, or interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, statistical analysis, and Administrative, technical, or material support. I.M. is responsible for the concept and design, acquisition, analysis, or interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, and Administrative, technical, or material support. E.E.T. is responsible for the concept and design. R.G. is responsible for the concept and design, acquisition, analysis, or interpretation of data, drafting of the manuscript, critical revision of the manuscript for important intellectual content, and Administrative, technical, or material support. M.W. is responsible for the concept and design, acquisition, analysis, or interpretation of data, critical revision of the manuscript for important intellectual content, and Administrative, technical, or material support. L.S.Q. and K.K. are responsible for the acquisition, analysis, or interpretation of data, critical revision of the manuscript for important intellectual content, and Administrative, technical, or material support. J.A.K. is responsible for the acquisition, analysis, or interpretation of data, and critical revision of the manuscript for important intellectual content. All the authors read and approved the final version of the manuscript.
Supplementary material
Supplementary material is available at JAMIA Open online.
Funding
This project was funded by internal FDA resources.
Conflicts of interests
None declared.
Data availability
The data were collected by the Centers for Medicare & Medicaid Services (CMS) and shared with the United States Food and Drug Administration (FDA) and is not available to share due to confidentiality and potential leakage of healthcare and patient data.
Ethical declarations
Disclaimer: The authors are employees or contractors of the U.S. Food and Drug Administration or the Centers for Medicare & Medicaid Services; however, other officials at the U.S. Food and Drug Administration and the Centers for Medicare & Medicaid Services had no role in the design and conduct of the study; collection, analysis, and interpretation of the data; or in the preparation, review, or approval of the manuscript.
The manuscript was subject to administrative review before submission, but this review did not alter the content of the manuscript. The views expressed are those of the authors and not necessarily those of the Department of Health and Human Services, the U.S. Food and Drug Administration, or the Centers for Medicare & Medicaid Services.
Ethics statement
The study utilized deidentified retrospective data and did not require IRB (Institutional Review Board) review as determined by human subject protection at FDA. To ensure patient privacy and confidentiality, all patient data used in this study were deidentified. Personally identifiable information such as names, addresses, and other identifying details were removed from the dataset prior to any analysis or processing. The deidentified data were securely stored and accessed only by authorized researchers involved in the study.
The deidentification process helped mitigate potential risks associated with the disclosure of sensitive patient information. Strict data access controls and security measures were implemented to safeguard the privacy and confidentiality of the deidentified patient data throughout the study.
References
- 1. Lieberman P. Anaphylaxis and anaphylactoid reactions. In: Middleton E, ed. Allergy: principles and Practice. 5th ed. Mosby; 1998:1079-1089. [Google Scholar]
- 2. Samter M, Cohen SG. Excerpts from classics in allergy; edited for the 25th anniversary committee of the American Academy of Allergy by Max Samter. Ross Laboratories; 1969.
- 3. Lieberman P, Nicklas RA, Randolph C, et al. Anaphylaxis—a practice parameter update 2015. Ann Allergy Asthma Immunol. 2015;115(5):341-384. 10.1016/j.anai.2015.07.019 [DOI] [PubMed] [Google Scholar]
- 4. Sclar DA, Lieberman PL. Anaphylaxis: underdiagnosed, underreported, and undertreated. Am J Med. 2014;127(1 Suppl):S1-S5. 10.1016/j.amjmed.2013.09.007 [DOI] [PubMed] [Google Scholar]
- 5. Li X, Ma Q, Yin J, et al. A clinical practice guideline for the emergency management of anaphylaxis (2020). Front Pharmacol. 2022;13:845689. 10.3389/fphar.2022.845689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Walsh KE, Cutrona SL, Foy S, et al. Validation of anaphylaxis in the Food and Drug Administration’s mini-sentinel. Pharmacoepidemiol Drug. 2013;22(11):1205-1213. 10.1002/pds.3505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Goud R, Thompson D, Welsh K, et al. ICD-10 anaphylaxis algorithm and the estimate of vaccine-attributable anaphylaxis incidence in Medicare. Vaccine. 2021;39(38):5368-5375. 10.1016/j.vaccine.2021.08.004 [DOI] [PubMed] [Google Scholar]
- 8. Wang C, Graham DJ, Kane RC, et al. Comparative risk of anaphylactic reactions associated with intravenous iron products. JAMA. 2015;314(19):2062-2068. 10.1001/jama.2015.15572 [DOI] [PubMed] [Google Scholar]
- 9. Bennett CL, Jacob S, Hymes J, et al. Anaphylaxis and hypotension after administration of peginesatide. N Engl J Med. 2014;370(21):2055-2056. 10.1056/nejmc1400883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Turner PJ, Campbell DE, Motosue MS, et al. Global trends in anaphylaxis epidemiology and clinical implications. J Allergy Clin Immunol Pract. 2020;8(4):1169-1176. 10.1016/j.jaip.2019.11.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tuttle KL, Wickner P. Capturing anaphylaxis through medical records. Ann Allergy Asthma Immunol. 2020;124(2):150-155. 10.1016/j.anai.2019.11.026 [DOI] [PubMed] [Google Scholar]
- 12. Eldredge CE, Pracht E, Gallagher J, et al. Direct versus indirect query performance of ICD-9/-10 coding to identify anaphylaxis. J Allergy Clin Immunol Pract. 2023;11(4):1190-1197.e2. 10.1016/j.jaip.2022.12.034 [DOI] [PubMed] [Google Scholar]
- 13. Kohl KS, Bonhoeffer J, Braun MM, et al. ; The Brighton Collaboration. The Brighton Collaboration: creating a global standard for case definitions (and guidelines) for adverse events following immunization. In: Henriksen K, Battles JB, Marks ES, et al. , eds. Advances in Patient Safety: From Research to Implementation (Volume 2: Concepts and Methodology). Agency for Healthcare Research and Quality; 2005. [PubMed] [Google Scholar]
- 14. Sampson HA, Muñoz-Furlong A, Campbell RL, et al. Second symposium on the definition and management of anaphylaxis: summary report—Second National Institute of Allergy and Infectious Disease/Food Allergy and Anaphylaxis Network symposium. J Allergy Clin Immunol. 2006;117(2):391-397. 10.1016/j.jaci.2005.12.1303 [DOI] [PubMed] [Google Scholar]
- 15. Kaufman S, Rosset S, Perlich C, et al. Leakage in data mining: formulation, detection, and avoidance. ACM Trans Knowl Discovery Data. 2012;6(4):1-21. 10.1145/2382577.2382579 [DOI] [Google Scholar]
- 16. Kapoor S, Narayanan A. Leakage and the reproducibility crisis in machine-learning-based science. Patterns (N Y). 2023;4(9):100804. 10.1016/j.patter.2023.100804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Shardlow M. An analysis of feature selection techniques. Univ Manchester. 2016;1(2016):1-7. [Google Scholar]
- 18. McHugh ML. The chi-square test of independence. Biochem Med. 2013;23(2):143-149. 10.11613/bm.2013.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Su R, Liu X, Wei L. Mine-RFE: determine the optimal subset from RFE by minimizing the subset-accuracy–defined energy. Brief Bioinformatics. 2019;21(2):687-698. 10.1093/bib/bbz021 [DOI] [PubMed] [Google Scholar]
- 20. Peng C-YJ, Lee KL, Ingersoll GM. An introduction to logistic regression analysis and reporting. J Edu Res. 2002;96(1):3-14. 10.1080/00220670209598786 [DOI] [Google Scholar]
- 21. Kohavi R, John GH. Wrappers for feature subset selection. Artif. Intell. 1997;97(1-2):273-324. 10.1016/s0004-3702(97)00043-x [DOI] [Google Scholar]
- 22. Breiman L. Random forests. Mach Learn. 2001;45(1):5-32. 10.1023/a:1010933404324 [DOI] [Google Scholar]
- 23. Ke G, Meng Q, Finley T, et al. LightGBM: a highly efficient gradient boosting decision tree. In: Proceedings of the 31st International Conference on Neural Information Processing Systems (NIPS'17), Long Beach, CA, USA; 2017.
- 24. Van der Maaten L, Hinton G. Visualizing data using t-SNE. J Mach Learn Res. 2008;9(86):2579-2605. [Google Scholar]
- 25. Wold S, Esbensen K, Geladi P. Principal component analysis. Chemomet Intell Lab Syst. 1987;2(1-3):37-52. 10.1016/0169-7439(87)80084-9 [DOI] [Google Scholar]
- 26. Sammon JW. A nonlinear mapping for data structure analysis. IEEE Trans Comput. 1969;C-18(5):401-409. 10.1109/T-C.1969.222678 [DOI] [Google Scholar]
- 27. Cortes C, Vapnik V. Support-vector networks. Mach Learn. 1995;20(3):273-297. 10.1007/bf00994018 [DOI] [Google Scholar]
- 28. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Association for Computing Machinery Published Online First; 2016. 10.1145/2939672.2939785 [DOI]
- 29. Pandala S. Lazypredict. PyPI. 2019. Accessed April 25, 2023. https://pypi.org/project/lazypredict/
- 30. Domingos P. A unified bias-variance decomposition and its applications. In: Proceedings of the 17th International Conference on Machine Learning, Stanford, CA. Morgan Kaufmann; 2000: 231–238.
- 31. Belkin M, Hsu D, Ma S, et al. Reconciling modern machine-learning practice and the classical bias–variance trade-off. Proc Natl Acad Sci USA. 2019;116(32):15849-15854. 10.1073/pnas.1903070116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bolón-Canedo V, Sánchez-Maroño N, Alonso-Betanzos A. Feature selection and classification in multiple class datasets: an application to KDD Cup 99 dataset. Expert Syst Appl. 2011;38(5):5947-5957. 10.1016/j.eswa.2010.11.028 [DOI] [Google Scholar]
- 33. Kopitar L, Kocbek P, Cilar L, et al. Early detection of type 2 diabetes mellitus using machine learning-based prediction models. Sci Rep. 2020;10(1):11981. 10.1038/s41598-020-68771-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ong MS, Klann JG, Lin KJ, et al. Claims‐based algorithms for identifying patients with pulmonary hypertension: a comparison of decision rules and machine‐learning approaches. J Am Heart Associat. 2020;9(19):e016648. 10.1161/jaha.120.016648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bann MA, Carrell DS, Gruber S, et al. Identification and validation of anaphylaxis using electronic health data in a population-based setting. Epidemiology. 2021;32(3):439-443. 10.1097/ede.0000000000001330 [DOI] [PubMed] [Google Scholar]
- 36. Mesfin YM, Cheng AC, Tran AHL, et al. Positive predictive value of ICD‐10 codes to detect anaphylaxis due to vaccination: a validation study. Pharmacoepidemiol Drug Saf. 2019;28(10):1353-1360. 10.1002/pds.4877 [DOI] [PubMed] [Google Scholar]
- 37. Botsis T, Woo EJ, Ball R, et al. Application of information retrieval approaches to case classification in the vaccine adverse event reporting system. Drug Saf. 2013;36(7):573-582. 10.1007/s40264-013-0064-4 [DOI] [PubMed] [Google Scholar]
- 38. Ball R, Toh S, Nolan J, et al. Evaluating automated approaches to anaphylaxis case classification using unstructured data from the FDA Sentinel System. Pharmacoepidemiol Drug Saf. 2018;27(10):1077-1084. 10.1002/pds.4645 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data were collected by the Centers for Medicare & Medicaid Services (CMS) and shared with the United States Food and Drug Administration (FDA) and is not available to share due to confidentiality and potential leakage of healthcare and patient data.