Abstract
The use of adenovirus vectors for gene therapy has been limited by well-defined cellular and humoral immune responses. We have previously shown that adenovirus vectors rapidly induce the expression of the C-X-C chemokine, interferon-inducible protein 10 (IP-10), in vivo. Various first-generation, type 5 adenovirus vectors, including adCMVβgal and UV-psoralen-inactivated adenovirus, equally induced the expression of IP-10 mRNA as early as 3 h following infection in mouse renal epithelial cells (REC). Luciferase reporter experiments using deletional mutants of the murine IP-10 5′-flanking region revealed that transcriptional activation of the IP-10 promoter by adCMVβgal was dependent on the −161- to −96-bp region upstream of the transcription start site. In electrophoretic mobility shift assays, adCMVβgal, adCMV-GFP, FG140, and transcription-defective adenovirus induced protein binding to oligonucleotides containing a consensus sequence for NF-κB at position −113 of the IP-10 promoter. Supershift assays confirmed an increase in binding activity of NF-κB p65 but not p50 or cRel in REC cells infected with various replication-deficient adenoviruses. Coinfection of REC cells with adCMVβgal and an adenoviral vector expressing IκBα resulted in suppression of adCMVβgal-induced expression of IP-10 at 6 and 16 h, further strengthening the conclusion that adenovirus-induced activation of IP-10 is dependent on NF-κB. The induction of IP-10 appeared to be direct because infection with adenovirus vectors failed to induce the expression of the potent IP-10 stimulators, interferon gamma and tumor necrosis factor alpha. Together, these findings demonstrate that adenovirus vectors directly induce the expression of IP-10 through capsid dependent activation of NF-κB.
Recombinant adenoviruses continue to be used extensively experimentally and clinically due to their safety and their ability to infect a wide range of cells and tissues. Success with adenovirus vectors, however, has been limited by potent host immune responses against viral proteins and the capsid that result in transient gene expression and an inability to readminister vectors of the same serotype to previously immunized subjects (8). Within 1 week of administration, first-generation adenovirus vectors are known to induce major histocompatibility complex class I-restricted CD8+ cytotoxic T lymphocytes (CTLs) directed against adenoviral proteins, a Th1 dominant immune response dependent on gamma interferon (IFN-γ) (28, 29). Ongoing expression of adenoviral proteins has been shown to be a significant factor in the development of the immune response against adenovirus vectors (23). For this reason, newer generations of adenovirus vectors are being developed that drastically reduce the amount of viral DNA packaged within the viral capsid, eliminating the genes encoding viral proteins (19). Unfortunately, the adenoviral capsid is also immunogenic, capable of inducing adenovirus-specific CTLs in the absence of all viral transcription (11). Understanding the biology of the host immune response against replication-deficient adenoviruses is of paramount importance in bringing these very useful vectors closer to effective clinical use.
The chemokines are a superfamily of inducible, proinflammatory proteins (<20 kDa) that contain between two and four highly conserved NH2-terminal cysteine amino acid residues. Chemokines are involved in the recruitment and activation of neutrophils, monocytes/macrophages, and lymphocytes to sites of injury and infection. They are divided into two major subgroups, C-X-C and C-C, based on the presence or absence of an intervening amino acid between the first two conserved NH2-terminal cysteine residues (25). Chemokines exert the majority of their effects through seven transmembrane-spanning, pertussis toxin-sensitive, G-protein-coupled receptors. T lymphocytes express most of the known chemokine receptors and, as a result, T cells, based on their state of differentiation or activation, undergo chemotaxis to many of the known chemokines (25). Recently, there has been significant interest in determining the pattern of chemokine receptor expression on T lymphocytes as markers of their state of differentiation or activation (21, 25). The C-X-C chemokine IP-10 (IFN-inducible protein 10) and its receptor CXCR3 have been associated with Th1 immune responses. CXCR3 is expressed primarily on T lymphocytes of the Th1 phenotype, a finding consistent with the preferential chemotaxis of activated Th1 lymphocytes by IP-10 (1, 4, 25). The association of IP-10 and its receptor CXCR3 with Th1-dependent immunity has been observed in several models of disease, including multiple sclerosis and rheumatoid arthritis (20, 22). These observations raise the possibility that CXCR3 and its ligands, including IP-10, are important mediators of Th1 dominant immune responses.
We have previously demonstrated in an animal model of adenoviral gene therapy, capsid-dependent induction of multiple chemokines occurring within 24 h of infection (15). Of the chemokines identified, IP-10 was found to be highly and rapidly induced in mice as early as 1 h following infection with first-generation and transcription-defective adenovirus vectors. Given the association of IP-10 and its receptor CXCR3 to Th1-dependent immune responses, it is possible that the induction of IP-10 by recombinant adenoviruses represents an important early step in the development of host immunity against these vectors. Therefore, understanding the biology of adenovirus vector-induced expression of IP-10 as a precursor to host Th1 immune responses will have an impact on the development of future generations of adenovirus vectors and identify new targets to reduce the immunogenicity of these vectors. We characterize here the mechanism of adenovirus vector-induced expression of the C-X-C chemokine IP-10 and identify a novel mechanism for the transcriptional activation of this gene.
MATERIALS AND METHODS
Adenovirus vectors.
The type 5, E1-deleted, E3-defective adenoviruses expressing Escherichia coli β-galactosidase (adCMVβgal) (2, 10), green fluorescent protein (GFP) (adCMV-GFP; Quantum, Montreal, Canada), and porcine IκBα (adCMV-IκBα; a generous gift of C. Ferran) (26), all under the control of the cytomegalovirus (CMV) promoter, were propagated in 293 cells and purified as previously described (3). FG140 (Microbix, Toronto, Canada), an E1, E3-defective type 5 adenovirus containing pMX2 plasmid sequences (amp-ori) at 3.8 map units (9) was grown and purified as above. AdCMVβgal was rendered transcription defective based on the protocol by Cotten et al. (7). Briefly, 1 ml of AdCMVβgal was mixed with 1 ml of psoralen (0.50 mg/ml) (Sigma, St. Louis, Mo.) in glycerol and exposed to UV light (365 nm) for 1 h at 4°C. Inactive virus was then dialyzed against 2 liters of 10 mM Tris–1 mM Mg+2–10% glycerol buffer (pH 7.4) for 12 h two times at 4°C and stored at −70°C. Transcriptional activity was tested by plaque assay and determined to be <10−2 PFU/ml. Control vehicle was made by dialyzing 2-ml aliquots of CsCl (1.34 g/ml) without virus against the same dialysis buffer used for the viral preparations (10 mM Tris–1 mM Mg+2–10% glycerol buffer [pH 7.4], 1 liter for 1 h four times at 4°C) and stored at −70°C.
The adenovirus vector concentration was determined by measuring the optical density at 260 nm and was expressed as optical particle units (OPU) as described by Mittereder et al. (14).
Animal studies.
DBA/2 (H-2d) mice were obtained from Charles River Laboratories (Wilmington, Mass.) and housed under standard conditions. All animals were used at 10 to 14 weeks of age (28 to 35 g). Under methoxyfluorane general anesthesia, 1011 OPU of adCMVβgal was injected via the femoral vein using a total final volume of 100 μl (virus plus normal saline). Animals were allowed to recover and then were sacrificed at predetermined time points with the livers harvested for analysis. All animal studies were performed in accordance with the Animal Care Committee guidelines at The University of Calgary.
Cell culture.
Primary renal epithelial cells from DBA/2 mice were isolated and grown as previously described (27). An immortalized, nontransformed cell line was derived from these cells through passaging and maintained in Dulbecco modified Eagle medium (DMEM) containing 10% fetal calf serum (FCS) and penicillin-streptomycin (Gibco BRL, Rockville, Md.). Cells were cultured in DMEM containing 1% FCS for 12 h prior to all experiments to slow their proliferation. This maneuver did not affect IP-10 gene expression. Adenoviral infections (except luciferase assays), stimulation with IFN-γ and lipopolysaccharide (LPS) were performed in 60-mm plates with cells at ∼80% confluency (106 cells/plate). For adenoviral infections, media were removed from the plate and replaced with 2 ml of medium containing 5 × 1010 OPU of adenovirus vectors per ml followed by incubation at 37°C in 5% CO2. Adenovirus incubation was terminated at 90 min by removing the medium and replacing it with fresh prewarmed medium followed by incubation at 37°C in 5% CO2. For IFN-γ and LPS stimulation, the medium was removed and replaced with 2 ml of medium containing 0.1, 1, 10, or 100 U of IFN-γ (Pharmingen, San Diego, Calif.) per ml or 0.1, 1, 10, or 100 endotoxin units (EU) of LPS O111:B4 (Sigma) per ml followed by incubation for 6 h at 37°C in 5% CO2. Depending on the experiment, cells were harvested at predetermined time points by scraping or direct lysis.
Endotoxin testing.
Low-endotoxin H2O, buffers, and tissue culture reagents were used for all experiments. Plasmids were grown and purified using the EndoFree Plasmid Kit (Qiagen, Chatsworth, Calif.). Adenovirus vectors and vehicle were routinely tested for the presence of endotoxin using the Limulus Amebocyte Lysate Kit (BioWhittaker, Walkersville, Md.). In the dilutions used, all reagents contained <0.1 EU of endotoxin per ml.
Enzyme-linked immunosorbent assay (ELISA).
Supernatants from renal epithelial cells (REC) cells were harvested 16 h after infection with 5 × 1010 OPU of adCMVβgal per ml or an equivalent volume of vehicle. Then, 100 μl of supernatant was incubated in 96-well plates at 4°C overnight. Wells were washed with phosphate-buffered saline-Tween, followed by the addition of 1 μg of polyclonal goat anti-mouse IP-10 antibody (R&D Systems, Minneapolis, Minn.) per ml, and incubated for 2 h at room temperature. Wells were again washed and incubated for 1 h at room temperature with 1 μg of biotinylated anti-goat immunoglobulin G (IgG) antibody (Santa-Cruz) per ml. After another round of washing, a 1:1,000 dilution of avidin-horseradish peroxidase (Bio-Rad, Hercules, Calif.) was added to each well and incubated for 30 min at room temperature. Wells were washed and incubated with 3,3′,5,5′-tetramethylbenzidine (Sigma) for 8 to 10 min. Samples were analyzed at 600 nm using a microplate reader and Softmax software (Molecular Devices, Menlo Park, Calif.). The protein concentration was measured against a recombinant mouse IP-10 standard (R&D Systems).
RNase protection assays.
Liver tissue and cells were processed for total RNA using RNeasy (Qiagen) according to the manufacturer's protocol. RNase protection assays were performed using the RiboQuant Multi-Probe RNase Protection Assay System (Pharmingen). Briefly, using the multiprobe template set mCK5-mCK3 and a single probe set for IP-10 (Pharmingen), a [32P]UTP-labeled RNA probe was transcribed using T7 polymerase followed by phenol-chloroform extraction and ethanol precipitation. The concentration of the probe was adjusted to 3 × 105 cpm/μl. Then, 5 to 10 μg of RNA per sample was hybridized to 6 × 105 cpm of total probe overnight at 56°C. Samples were then digested with RNase, followed by proteinase K treatment and phenol-chloroform extraction. After ethanol precipitation with 4 M ammonium acetate, protected samples were resuspended in 1× loading buffer and separated on 5.7% acrylamide-bisacrylamide urea gels. After drying, the gels were visualized by autoradiography. Autorads were analyzed by densitometry using Quantity One software (Bio-Rad). Fold induction was determined as the mRNA density ratio of IP-10 to GAPDH (glyceraldehyde-3-phosphate dehydrogenase) within the same sample.
IP-10 promoter constructs.
The 533-bp sequence upstream of the transcription start site of the mouse IP-10 gene (GenBank accession no. L07417) was cloned from genomic DNA derived from the REC cell line using PCR. The PCR product was cloned into pCR 2.1 (Invitrogen) and transformed into competent E. coli. Positive clones were isolated by miniplasmid preparation (Qiagen) and sequenced. The correct 533-bp fragment of the IP-10 promoter was identified, cut with EcoRI, and blunted with large fragment DNA polymerase 1. The resulting DNA fragment was then ligated into the SmaI site of the pGL3-basic vector (Promega, Madison, Wis.) and designated pGL3–IP-10(−533). Deletional mutants at position −161 of the IP-10 5′-flanking region were constructed from pGL3–IP-10(−533) using the unique restriction site BstXI in the promoter. Deletional mutants at positions −237, −190, and −96 of the IP-10 promoter were constructed with PCR using pGL3–IP-10(−533) as a template, sense oligonucleotides containing a HindIII linker corresponding to the desired position, and an antisense oligonucleotide, including the HindIII site of the pGL3 polylinker. PCR products were cloned into pGEM-TEasy vector (Promega) and screened using miniplasmid preparations and restriction enzyme analysis. Positive clones were digested with HindIII, and the DNA fragments were cloned into the HindIII site of pGL3-basic. All pGL3–IP-10 deletional mutants were screened by restriction enzyme digestion.
Luciferase assays.
REC cells were plated in six-well plates at 4 × 105 cells/well and incubated overnight. Then, 2 μg of plasmid DNA was transfected into each well using 6 μl of Lipofectamine reagent (Gibco BRL), followed by incubation for 5 h in serum-free DMEM at 37°C. Fetal bovine serum was added to each well to give a final concentration of 1%, and the cells were incubated for 16 h at 37°C. A 1-ml portion of medium containing 2.5 × 1010 OPU of adCMVβgal or an equivalent volume of vehicle was added to the cells and incubated at 37°C for 90 min. The medium was removed and replaced with fresh prewarmed DMEM containing 1% FBS, and the cells were incubated for 24 h. Cells were washed with phosphate-buffered saline, harvested by scraping, and centrifuged into a pellet, followed by resuspension in lysis buffer (1% Triton X-100; 25 mM glycylglycine, pH 7.8; 15 mM MgSO4; 4 mM EGTA; 1 mM dithiothreitol [DTT]). Samples were centrifuged at 14,000 rpm for 10 min, and 150 μl of supernatant was added to 300 μl of assay buffer (25 mM glycylglycine, pH 7.8; 15 mM K2PO4, pH 7.8; 15 mM MgSO4; 4 mM EGTA; 1 mM DTT; 2 mM ATP) in polystyrene tubes. The luciferase activity of each sample was measured in a luminometer following the addition of 100 μl of luciferin (0.3 mg/ml) (Promega).
Nuclear extracts.
REC cells were grown to 80% confluency in 60-mm plates. A total of 5 × 1010 OPU of adenovirus vector per ml or an equivalent volume of vehicle in 2 ml of DMEM containing 1% FBS was added to the plates and then incubated for 6 or 12 h. Medium was removed and replaced with lysis buffer (10 mM HEPES [1 ml], 10 mM KCl, 0.1 M EDTA, 0.5% NP-40, 3 mM DTT, 1 mM phenylmethylsulfonyl fluoride [PMSF], and 2 μg of leupeptin and 20 μg of aprotinin per ml) and then incubated on ice for 15 min. Nuclei were scraped and briefly centrifuged at 14,000 rpm. Supernatant was removed, and cells were resuspended in extraction buffer (20 mM HEPES, 420 mM NaCl, 5 mM EDTA, 10% glycerol, 5 mM DTT, 1 mM PMSF) and gently rocked for 30 min at 4°C. Samples were centrifuged at 14,000 rpm for 10 min at 4°C, and the supernatant was removed and stored at −70°C. The protein concentration was determined by Bradford assay (Bio-Rad).
Electrophoretic mobility shift assays (EMSAs).
Oligonucleotides were synthesized and annealed to obtain double-stranded DNA fragments. The oligonucleotide sequences were as follows: IP-10(−124/−94), 5′-TCGGTTTACAGGGGACTTCCCTCGGGTTGCG-3′; Oligo 1, 5′-CACTTATGATACCGGCCAATGCTTGGT-3′; and Oligo 2, 5′-ACTAACCTTAGGGGATGCCCCTCAACTGGC-3′.
DNA binding reactions were performed in 20-μl samples containing 4 μg of nuclear extract, 2 μg of d(I-C) (Pharmacia, Piscataway, N.J.), 2 μg of bovine serum albumin, 20 mM HEPES, 50 mM NaCl, 0.1 mM EDTA, 1 mM DTT, and 10% glycerol at room temperature for 20 min. For supershift assays, 1 μl of polyclonal goat antibodies directed against NF-κB proteins cRel, p50, and p65 (Santa Cruz, Santa Cruz, Calif.) was also added. For competitor EMSA, 0.001, 0.005, or 0.01 pmol of unlabeled oligonucleotides was added 10 min into the first incubation, prior to the addition of labeled probe. After 20 min, 0.5 ng of 32P-labeled IP-10(−124/−94) (30,000 cpm) was added to the samples and incubated for 20 min at room temperature. Samples were loaded and run on 4% polyacrylamide gels with 1× TGE buffer (25 mM Tris, pH 8.3; 190 mM glycine; 1 mM EDTA). Gels were dried and visualized using autoradiography.
Statistical analysis.
All experiments were performed at least three times. Values are expressed as the mean ± the standard deviation (SD) and compared using the Student's t test.
RESULTS
Adenovirus vector-induced expression of IP-10.
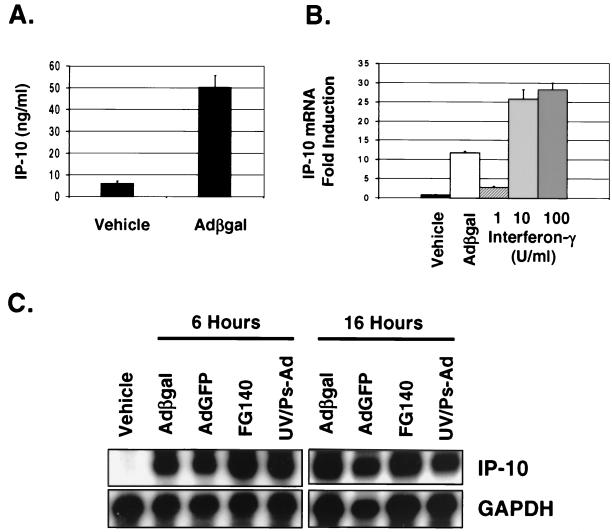
We have previously demonstrated that various adenovirus vectors induce the expression of IP-10 in DBA/2 mice in vivo (15). Using REC cells derived from DBA/2 mice, we tested the ability of adenovirus vectors to induce the expression of IP-10 in vitro. REC cells were found to constitutively express low levels of IP-10. Infection of REC cells with 5 × 1010 OPU of adCMVβgal per ml resulted in significant production of IP-10 protein at 16 h compared to vehicle-treated cells as detected by ELISA using an anti-mouse IP-10 antibody (50.3 ± 5.7 versus 6.1 + 1.1 ng/ml, P < 0.0002) (Fig. 1). AdCMVβgal also increased the expression of IP-10 mRNA in REC cells as early as 3 h and continued 6 and 16 h postinfection (Fig. 1). Expression of IP-10 mRNA was not increased following treatment with vehicle. The induction of IP-10 in REC cells by 5 × 1010 OPU of adCMVβgal per ml was comparable to stimulation using moderate doses of IFN-γ (Fig. 1). At 6 h, adCMVβgal induced the expression of IP-10 mRNA more than stimulation with 1 U/ml, but the level was approximately 50% less than stimulation with 10 or 100 U of IFN-γ per ml. In contrast, LPS did not induce the expression of IP-10 in these cells (data not shown).
FIG. 1.
IP-10 expression in REC cells following infection with adenovirus vectors. (A) REC cells infected with 5 × 1010 OPU of adCMVβgal per ml express IP-10 protein at 16 h as detected by ELISA (6.1 ± 1.1 versus 50.3 ± 5.7 ng/ml; P < 0.0002). (B) IP-10 mRNA expression detected by RNase protection assay in REC cells following stimulation with IFN-γ. At 6 h, adCMVβgal induces the expression of IP-10 more than stimulation with 1 U/ml but less than stimulation with 10 U of IFN-γ per ml. Bars represent the SD. (C) AdCMVβgal, adCMV-GFP, FG140, and UV-psoralen-inactivated adenovirus (UV/Ps-Ad) induce the expression of IP-10 mRNA in REC cells 6 and 16 h following infection as seen by RNase protection. Vehicle-treated cells do not induce the expression of IP-10.
The induction of IP-10 in REC cells occurred independent of the transgene since 5 × 1010 OPU of adCMV-GFP or FG140 (9), a replication-deficient adenovirus lacking a transgene, per ml induced a similar pattern of IP-10 mRNA expression (Fig. 1). The early induction of IP-10 and lack of a transgene effect suggested that this event occurs independent of adenovirus vector and transgene transcription. Therefore, we tested the ability of transcription-defective adenovirus to induce the expression of IP-10. AdCMVβgal was rendered transcription defective by UV-psoralen inactivation as described by Cotten et al. (7). These adenovirus particles maintain the ability to bind to and be internalized by a host cell. The presence of active virus following inactivation was <10−2 PFU/ml by standard plaque assay in 293 cells. In addition, infection of REC cells with 5 × 1010 OPU of psoralen-UV-inactivated adCMVβgal per ml yielded no β-galactosidase activity at 24 h, whereas a similar titer of adCMVβgal resulted in nearly 75% of cells expressing β-galactosidase (data not shown). At 6 and 16 h, UV-psoralen-inactivated adenovirus also induced IP-10 mRNA expression comparable to the other adenovirus vectors (Fig. 1). The ability of UV-psoralen-inactivated adenovirus to induce the expression of IP-10 confirmed that this response occurs independent of viral transcription and is likely dependent on the viral capsid.
Adenovirus vector activation of the IP-10 promoter.
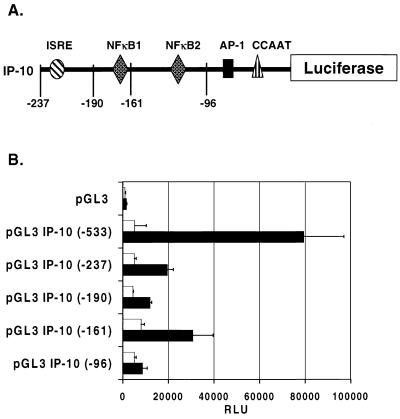
The rapid and large induction of IP-10 mRNA by various adenovirus vectors suggested that this process was mediated by transcription. This suggestion is supported by previous studies demonstrating that the expression of IP-10 in response to various stimuli also occurs through increased transcription (16–18). Therefore, we employed luciferase reporter assays to examine the transcriptional activation of the IP-10 promoter in response to infection with adenovirus vectors. The 533-bp region upstream of the transcription start site of the murine IP-10 gene was cloned from DBA/2 genomic DNA using PCR and inserted into the luciferase reporter vector pGL3 to give pGL3–IP-10(−533) (Fig. 2).
FIG. 2.
Adenovirus vector-induced activation of the IP-10 promoter in REC cells. (A) Organization of the 237-bp fragment upstream of the transcription start site of the mouse IP-10 gene. Positions of the deletional mutants are noted in relation to the potential cis elements involved. (B) Luciferase reporter assay. AdCMVβgal (■) activates the 533-bp fragment of the IP-10 promoter >10-fold more than the vehicle (□)-treated cells. AdCMVβgal-induced luciferase activity is diminished but still significantly greater than with the vehicle in REC cells transfected with IP-10 promoter deletion constructs pGL3–IP-10(−237), pGL3–IP-10(−190), and pGL3–IP-10(−161). Luciferase activity falls to baseline levels in REC cells transfected with the deletional mutant pGL3–IP-10(−96), thus confirming that the minimal elements responsible for IP-10 promoter activation are located between positions −161 and −96 of the IP-10 5′-flanking region. Bars represent the SD.
In REC cells transiently transfected with pGL3 basic vector, infection with 2.5 × 1010 OPU of adCMVβgal per ml resulted in low-level induction of luciferase activity relative to vehicle (1,583 ± 165 versus 842 ± 332 relative light units [RLU]; P was not significant) likely due to nonspecific vector-protein interactions. In REC cells transfected with pGL3–IP-10(−533), infection with 2.5 × 1010 OPU of adCMVβgal per ml induced >10-fold expression of luciferase compared to vehicle-treated cells (79,295 ± 17,758 versus 5,246 ± 5,192 RLU; P < 0.02) (Fig. 2). Deletional mutants of the IP-10 promoter were constructed to further identify the cis elements involved in adenovirus vector-induced transcription of IP-10. The fold induction of luciferase activity by adCMVβgal diminished substantially in REC cells transfected with the deletional mutant pGL3–IP-10(−237) but was still threefold higher than in vehicle-treated cells. (19,411 ± 1,538 versus 5,064 ± 452 RLU; P < 0.01). Compared to vehicle, adCMVβgal also significantly induced the expression of luciferase in REC cells transfected with the deletional mutants pGL3–IP-10(−190) and pGL3–IP-10(−161). AdCMVβgal-induced luciferase activity returned to baseline in REC cells transfected with the deletional mutant pGL3–IP-10(−96) (8,576 ± 12,487 versus 5,089 ± 495 RLU; P was not significant) (Fig. 2). These results demonstrated that the minimal elements required for activation of the IP-10 promoter by adCMVβgal are contained within positions −161 to −96 of the IP-10 5′-flanking region.
Adenovirus vector activation of NF-κB.
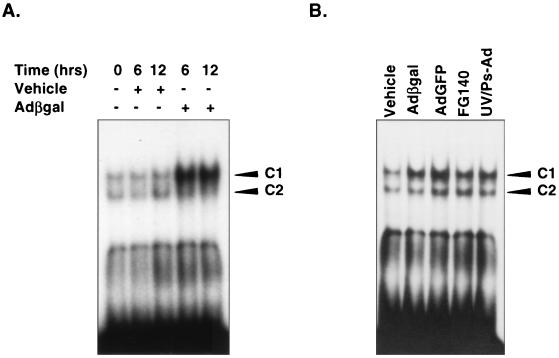
To determine the nuclear factors responsible for the expression of IP-10 by adenovirus vectors, EMSAs were employed. Nuclear extracts were prepared from untreated, vehicle-treated, or adenovirus vector-infected REC cells at 6 and 12 h, time points at which IP-10 mRNA is highly expressed following infection with adenovirus vectors. The IP-10 promoter fragment at positions −161 to −96 contains a consensus sequence for NF-κB, GGGACTTCC, at position −113, an element which has been demonstrated to be instrumental in the transcriptional activation of IP-10 (16–18). Therefore, a DNA probe spanning positions −124 to −94 of the IP-10 5′-flanking region, IP-10(−124/−94), was synthesized and used in the EMSA. Nuclear extracts from untreated REC cells display constitutive binding to radiolabeled IP-10(−124/−90) probe in two complexes (designated C1 and C2) (Fig. 3). At 6 and 12 h, infection with 5 × 1010 OPU of adCMVβgal per ml increased the level of C1 complex in EMSA but had minimal effect on the C2 complex (Fig. 3). Nuclear extracts from vehicle-treated REC cells demonstrate no increase in DNA-protein interactions compared to untreated cells.
FIG. 3.
EMSA of REC cell nuclear extracts. (A) Nuclear extracts from untreated and vehicle-treated REC cells constitutively form DNA-protein complexes C1 and C2 when incubated with the radiolabeled oligonucleotide, IP-10(−124/−94). IP-10(−124/−94) corresponds to base pairs −124 to −94 of the IP-10 5′-flanking region and contains the NF-κB sequence GGGACTTCC at position −113. AdCMVβgal significantly increases C1 complex 6 and 12 h following infection in REC cells. (B) EMSA using radiolabeled DNA fragment IP-10(−124/−94) and REC nuclear extracts following infection with 5 × 1010 OPU of various adenovirus vectors per ml. C1 complex is increased by adCMV-GFP, FG140, and UV-psoralen-inactivated adenovirus (UV/Ps-Ad) 6 h following infection, a result similar to the pattern induced by adCMVβgal. C2 complex is minimally or not increased significantly over baseline levels following infection with different adenovirus vectors.
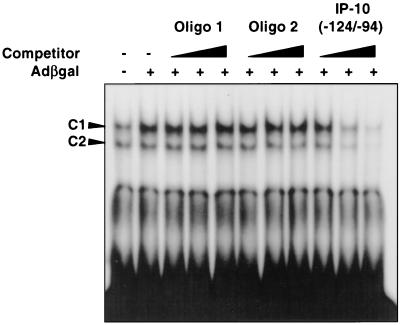
Since we have demonstrated that various adenovirus vectors are capable of inducing the expression of IP-10 in REC cells, we performed EMSA using nuclear extracts from REC cells infected with 5 × 1010 OPU of adCMV-GFP, FG140, and UV-psoralen-inactivated adenovirus per ml. All vectors were capable of inducing the C1 complex as measured 6 h after infection, confirming that this response is not specific to adCMVβgal and occurs independent of viral transcription (Fig. 3). Again, as with adCMVβgal, the C2 complex was minimally or not significantly induced over the baseline level. Competitor studies confirm the specificity of binding to IP-10(−124/−90). C1 complex is unaffected by increasing concentrations of cold nonspecific sequence oligonucleotide (Oligo 1). Competition with an oligonucleotide encoding an alternate NF-κB consensus motif GGGATGCCC (Oligo 2) also had little effect on the C1 complex; however, the protein-DNA interaction was competed for successfully by unlabeled IP-10(−124/−90) (Fig. 4).
FIG. 4.
Competitor EMSA using nuclear extracts from adCMVβgal-infected REC cells and radiolabeled probe IP-10(−124/−94). C1 complex is not competed away by the nonspecific DNA fragment (Oligo 1) or by a DNA fragment containing an alternate NF-κB sequence GGGATGCCC (Oligo 2). DNA-protein interaction is competed for effectively with unlabeled IP-10(−124/−94), confirming the specificity of this interaction.
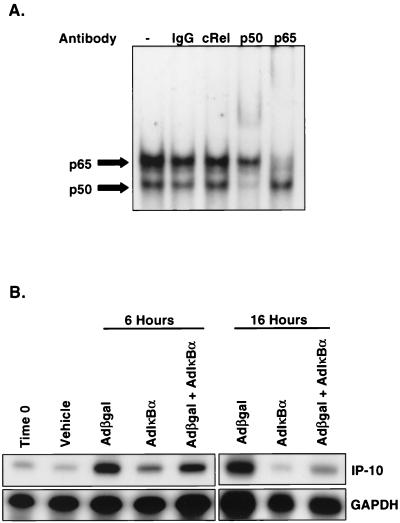
Supershift assays were performed on nuclear extracts from adCMVβgal-infected REC cells at 6 h to further characterize the DNA-protein complexes. Anti-NF-κB p65 supershifted the C1 complex, whereas anti-NF-κB p50 supershifted C2. Anti-cRel and control IgG had no effect on the complexes (Fig. 5). These results demonstrated that adenovirus vectors primarily activate NF-κB p65 following infection in REC cells, a process that occurs independent of viral or transgene transcription.
FIG. 5.
Adenovirus vector-induced activation of NF-κB. (A) EMSA using nuclear extracts from adCMVβgal-infected REC cells and radiolabeled probe IP-10(−124/−94). Anti-NF-κB p65 supershifts C1 complex, whereas anti-NF-κB p50 supershifts C2 complex, confirming the activation of p65 primarily by adenovirus vectors. NF-κB p50 is present in the nuclear extracts but is minimally increased by adenovirus vectors compared to untreated or vehicle-treated cells. (B) RNase protection assay of REC cell RNA following infection with 5 × 1010 OPU of adCMVβgal or adCMV-IκBα per ml. Unlike other adenovirus vectors, adCMV-IκBα does not significantly induce the expression of IP-10 mRNA at 6 and 16 h following infection. Coinfection of adCMVβgal and adCMV-IκBα results in suppression of adCMVβgal-induced expression of IP-10 mRNA at 6 and 16 h, confirming the importance of NF-κB in mediating the transcription of IP-10 after infection with adenovirus vectors.
Although we have shown binding and the requirement of the proximal NF-κB site for IP-10 promoter activation, this observation does not confirm the functional importance of NF-κB in the adenovirus vector-induced transcription of IP-10. Therefore, we employed an adenovirus expressing the NF-κB inhibitor, IκBα (26), to confirm that NF-κB was involved in the adenovirus-induced transcription of IP-10. Infection of REC cells with 5 × 1010 OPU of adCMV-IκBα per ml resulted in reduced or absent expression of IP-10 mRNA at 6 and 16 h compared to the same titer of adCMVβgal. Furthermore, adCMV-IκBα was able to suppress the expression of IP-10 by adCMVβgal when coinfected in REC cells at 6 and 16 h; however, the suppression of IP-10 mRNA at 16 h was greater than at 6 h (Fig. 5). The latter finding is likely due to a delay in IκBα expression by adCMV-IκBα, which is expected to be much higher at 16 h versus 6 h. Therefore, based on the above findings, nuclear translocation of NF-κB is a necessary component of the adenovirus vector-induced transcription of IP-10.
Adenovirus vector-induced expression of IP-10 occurs in the absence of TNF-α.
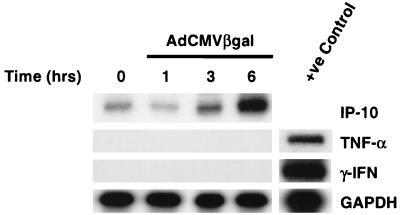
The activation of the IP-10 promoter and transcriptional regulation of IP-10 has been studied in response to LPS, tumor necrosis factor alpha (TNF-α), IFN-γ, and measles virus (16–18). In the case of IFN-γ and TNF-α, synergistic induction of IP-10 was seen with IFN-γ acting through the ISRE at position −224 and with TNF-α acting on either of the two proximal NF-κB sites on the promoter through the activation of p65-p50 heterodimers (17). The transcription of IP-10 induced by our adenovirus vectors depends primarily on p65, a pattern different than those previously described, suggesting a mechanism independent of TNF-α and IFN-γ. Furthermore, in the promoter studies, the expression of IP-10 was not shown to be dependent on the ISRE. However, since TNF-α and IFN-γ are both induced by adenovirus vectors in vivo (Fig. 6), we screened the REC cells for the presence of these cytokines. Infection of REC cells with 5 × 1010 OPU of adCMVβgal per ml did not induce the expression of mRNA for IFN-γ or TNF-α at 1, 3, or 6 h, whereas the expression of IP-10 has increased as early as 3 h following infection (Fig. 6). Cells were also tested for IFN-β and TNF-β mRNA, both of which were not induced by adCMVβgal (data not shown). These results support the hypothesis that adenovirus vectors are capable of directly inducing the expression of IP-10 independent of cytokines such as IFN-γ and TNF-α.
FIG. 6.
RNase protection assay of REC cells infected with adCMVβgal. IP-10 mRNA is induced by adCMVβgal as early as 3 h following infection in the absence of TNF-α and IFN-γ. RNA from DBA/2 mouse liver harvested 16 h following infection with 2 × 1011 OPU of adCMVβgal is used as a positive control for TNF-α and IFN-γ expression.
DISCUSSION
We have previously demonstrated significant expression of IP-10 and other chemokines in an in vivo model of gene therapy (15). Up to this point, the molecular mechanisms underlying the expression of IP-10 and other chemokines by adenovirus vectors were unknown. In the present study, we demonstrate that adenovirus vectors induce the expression of IP-10 independent of viral and transgene transcription. Furthermore, we show that adenovirus vector-induced transcription of IP-10 is mediated by NF-κB and occurs in the absence of TNF-α and IFN-γ. Adenovirus vectors for gene therapy elicit potent Th1 immune responses consisting of CD8+-restricted CTLs directed against adenoviral proteins that continue to be expressed at low levels despite the absence of E1 (28, 29). Kafri et al. have also demonstrated that replication-deficient adenoviruses are capable of inducing adenovirus-specific CTLs in the absence of viral gene transcription, confirming the immunogenic properties of the adenoviral capsid (11). The induction of IP-10 by adenovirus vectors, including transcription-defective adenoviruses, is consistent with the Th1 dominant host immune response described above. IP-10 is a potent chemoattractant for activated T lymphocytes of the Th1 phenotype and shares the receptor CXCR3 with related chemokines, Mig and I-TAC (6, 13, 24). IP-10 and its receptor CXCR3 have been implicated as important players in the development of host Th1 immune responses in experimental and clinical models of disease, including models of multiple sclerosis and rheumatoid arthritis (21, 22). Thus, the induction of IP-10 by adenovirus vectors may represent a critical step in the development of host antiadenoviral immunity.
The activation of NF-κB by adenovirus vectors in our studies is consistent with the in vivo findings by Lieber et al. who demonstrated NF-κB activation in the livers of mice infected with adenovirus vectors (12). Our results show that NF-κB is necessary for the transcription of IP-10 induced by adenoviral vectors, since the expression of IP-10 mRNA was effectively suppressed by IκBα. Based on our promoter studies, the proximal NF-κB site at position −113 is the minimal element required for transcriptional activation of IP-10 by adenovirus vectors. However, optimal transcriptional activation of the IP-10 gene also involves elements located upstream of the NF-κB sites identified. The region of the IP-10 promoter from positions −533 to −237 includes, among others, two AP-1 sites, an ets binding domain, and a c/EBP motif. Loss of this region of the promoter leads to a reduction in the fold induction of luciferase activity by adCMVβgal in our reporter experiments. The exact role of this promoter region in the transcriptional activation of IP-10 by adenovirus vectors is not known and will be the focus of future studies.
Numerous groups have studied the transcriptional regulation of IP-10. A variety of hematopoietic and nonhematopoietic cell types, including macrophages, epithelial cells, and neutrophils, can be induced to express IP-10 in response to IFN-γ, TNF-α, or LPS. Induction of IP-10 in response to these stimuli is dependent primarily on transcription (5, 17, 18). In response to IFN-γ or TNF-α, Ohimori and Hamilton have demonstrated that optimal IP-10 promoter activation in NIH 3T3 cells requires the synergistic action of both cytokines acting on the ISRE plus one or both of the two downstream NF-κB sites on the promoter (17). Activation of the IP-10 promoter by IFN-γ occurred through STAT-1, while TNF-α acted through the nuclear translocation of NF-κB p65 and p50 heterodimers. Interestingly, activation of the IP-10 promoter by measles virus, a negative-strand RNA paramyxovirus, was also found to be dependent on the ISRE plus a downstream NF-κB site, events mediated by viral gene transcription in human glioma cell lines (16). The transcriptional regulation of IP-10 following infection with adenovirus vectors differs significantly from the transcriptional mechanisms activated in response to TNF-α and IFN-γ. Adenovirus vectors are capable of inducing IP-10 gene transcription independent of the ISRE element located upstream on the promoter. These findings plus the activation of p65 in excess of p50 following infection with adenovirus vectors is consistent with the absence of TNF-α and IFN-γ in our system and represent a novel mechanism for the activation and expression of IP-10. We are not, however, suggesting that the direct induction of IP-10 by adenovirus vectors is the only mechanism driving the expression of this chemokine in vivo. Rather, this process represents another mechanism by which adenovirus vectors can activate this gene. TNF-α and IFN-γ are known to be upregulated early following infection with replication-deficient adenoviral vectors (12) and almost certainly contribute to the expression of IP-10 in vivo.
The mechanisms by which the adenoviral capsid is capable of inducing the transcription of IP-10 and the signaling pathways involved are unknown. Several possibilities exist, including the interaction of the adenoviral fiber protein with the coxsackievirus-adenovirus receptor, interactions between the penton base and surface integrins required for viral internalization, or a relationship to endosome formation following internalization. Future studies will clarify the role of virus-cell interactions and viral entry in the biology of adenovirus-induced inflammation.
The implications of the findings described here are numerous. First, these data increase our understanding of the host immune response to replication-deficient adenoviruses, the biggest obstacle to successful implementation of these vectors in humans. Identifying and characterizing the molecular mechanism of adenovirus vector-induced expression of IP-10, a potentially critical immunoregulatory chemokine, will permit the development of strategies to target the expression of this chemokine and/or its receptor to modulate antiadenoviral host immunity at a very early stage. These results also have implications for newer generations of adenovirus vectors, particularly third-generation vectors that have the majority of the adenovirus genome deleted. Although these modifications reduce the host immune response stemming from low-level adenoviral gene expression, they do not address the immunogenic properties of the adenoviral capsid. Understanding the mechanism by which the adenoviral capsid triggers host antiviral immunity will allow modifications or interventions to further reduce the immunogenicity of these potentially useful third-generation adenovirus vectors. Targeting IP-10, its receptor CXCR3, or events leading to their expression may prove to be an effective approach to overcome the major obstacle for the application of adenoviral gene therapy in humans.
ACKNOWLEDGMENTS
This study was funded by the Alberta Heritage Foundation for Medical Research and the Banting Research Foundation.
We thank B. Winston and M. Hollenberg for advice and reviews of the manuscript.
REFERENCES
- 1.Annunziato F, Cosmi L, Galli G, Beltrame C, Romagnani P, Manetti R, Romagnani S, Maggi E. Assessment of chemokine receptor expression by human Th1 and Th2 cells in vitro and in vivo. J Leukoc Biol. 1999;65:691–699. doi: 10.1002/jlb.65.5.691. [DOI] [PubMed] [Google Scholar]
- 2.Becker T C, BeltrandelRio H, Noel R J, Johnson J H, Newgard C B. Overexpression of hexokinase I in isolated islets of Langerhans via recombinant adenovirus. Enhancement of glucose metabolism and insulin secretion at basal but not stimulatory glucose levels. J Biol Chem. 1994;269:21234–21238. [PubMed] [Google Scholar]
- 3.Becker T C, Noel R J, Coats W S, Gomez-Foix A M, Alam T, Gerard D R, Newgard C B. Use of recombinant adenovirus for metabolic engineering of mammalian cells. Methods Cell Biol. 1994;43(Pt. A):161–189. doi: 10.1016/s0091-679x(08)60603-2. [DOI] [PubMed] [Google Scholar]
- 4.Bonecchi R, Bianchi G, Bordignon P P, D'Ambrosio D, Lang R, Borsatti A, Sozzani S, Allavena P, Gray P A, Mantovani A, Sinigaglia F. Differential expression of chemokine receptors and chemotactic responsiveness of type 1 helper cells (Th1) and Th2s. J Exp Med. 1998;187:129–134. doi: 10.1084/jem.187.1.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cassatella M A, Gasperini S, Calzetti F, Bertagnin A, Luster A, McDonald P P. Regulated production of the interferon-γ-inducible protein-10 (IP-10) chemokine by human neutrophils. Eur J Immunol. 1997;27:111–115. doi: 10.1002/eji.1830270117. [DOI] [PubMed] [Google Scholar]
- 6.Cole K E, Strick C A, Paradis T J, Ogborne K T, Loetscher M, Gladue R P, Lin W, Boyd J G, Moser B, Wood D E, Sahagan B G, Neote K. Interferon-inducible T cell alpha chemoattractant (I-TAC): a novel non-ELR CXC chemokine with potent activity on activated T cells through selective high-affinity binding to CXCR3. J Exp Med. 1998;187:2009–2021. doi: 10.1084/jem.187.12.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cotten M, Saltik M, Kursa M, Wagner E, Maass G, Birnstiel M L. Psoralen treatment of adenovirus particles eliminates virus replication and transcription while maintaining the endosomolytic activity of the virus capsid. Virology. 1994;205:254–261. doi: 10.1006/viro.1994.1641. [DOI] [PubMed] [Google Scholar]
- 8.Crystal R G. Transfer of genes to humans: early lessons and obstacles to success. Science. 1995;270:404–410. doi: 10.1126/science.270.5235.404. [DOI] [PubMed] [Google Scholar]
- 9.Graham F L. Covalently closed circles of human adenovirus DNA are infectious. EMBO J. 1984;3:2917–2922. doi: 10.1002/j.1460-2075.1984.tb02232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herz J, Gerard R D. Adenovirus-mediated transfer of low density lipoprotein receptor gene acutely accelerates cholesterol clearance in normal mice. Proc Natl Acad Sci USA. 1993;90:2812–2816. doi: 10.1073/pnas.90.7.2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kafri T, Morgan D, Krahl T, Sarvetnick N, Verma I. Cellular immune response to adenoviral vector infected cells does not require de novo viral gene expression: implications for gene therapy. Proc Natl Acad Sci USA. 1998;95:11377–11382. doi: 10.1073/pnas.95.19.11377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lieber A, He C Y, Meuse L, Schowalter D, Kirillova I, Winther B, Kay M A. The role of Kupffer cell activation and viral gene expression in early liver toxicity after infusion of recombinant adenovirus vectors. J Virol. 1997;71:8798–8807. doi: 10.1128/jvi.71.11.8798-8807.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loetscher M, Gerber B, Loetscher P, Jones S A, Piali L, Clark-Lewis I, Baggiolini M, Moser B. Chemokine receptor specific for IP-10 and Mig: structure, function, and expression in activated T-lymphocytes. J Exp Med. 1996;184:963–969. doi: 10.1084/jem.184.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mittereder N, March K L, Trapnell B C. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muruve D A, Barnes M J, Stillman I E, Libermann T A. Rapid induction of multiple chemokines by replication-deficient adenoviral vectors results in acute neutrophil-dependent hepatic toxicity in vivo. Hum Gene Ther. 1999;10:965–976. doi: 10.1089/10430349950018364. [DOI] [PubMed] [Google Scholar]
- 16.Nazar A S M I, Cheng G, Shin H S, Brothers P N, Dhib-Jalbut S, Shin M L, Vanguri P. Induction of IP-10 chemokine promoter by measles virus: comparison with interferon-γ shows the use of the same response element but with differential DNA-protein binding profiles. J Neuroimmunol. 1997;77:116–127. doi: 10.1016/s0165-5728(97)00070-2. [DOI] [PubMed] [Google Scholar]
- 17.Ohimori Y, Hamilton T A. The interferon-stimulated response element and a κB site mediate synergistic induction of murine IP-10 gene transcription by IFN-γ and TNF-α. J Immunol. 1995;154:5235–5244. [PubMed] [Google Scholar]
- 18.Ohimori Y, Hamilton T A. Cooperative interaction between interferon stimulus response element and κB sequence motifs controls IFN-γ and lipopolysaccharide-stimulated transcription from the murine IP-10 promoter. J Biol Chem. 1993;268:6677–6688. [PubMed] [Google Scholar]
- 19.Parks R J, Chen L, Anton M, Sankar U, Rudnicki M A, Graham F L. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc Natl Acad Sci USA. 1996;93:13565–13570. doi: 10.1073/pnas.93.24.13565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qin S, Rottman J B, Myers P, Kassam N, Weinblatt M, Loetscher M, Koch A E, Moser B, MacKay C R. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J Clin Investig. 1998;101:746–754. doi: 10.1172/JCI1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sallusto F, MacKay C R, Lanzavecchia A. Selective expression of the eotaxin receptor CCR3 by human T-helper cells. Science. 1997;277:2005–2007. doi: 10.1126/science.277.5334.2005. [DOI] [PubMed] [Google Scholar]
- 22.Sorensen T L, Tani M, Jensen J, Pierce V, Lucchinetti C, Folcik V A, Qin S, Rottman J, Sellebjerg F, Streiter R M, Frederiksen J L, Ransohoff R M. Expression of specific chemokines and chemokine receptors in the central nervous system of multiple sclerosis patients. J Clin Investig. 1999;103:807–815. doi: 10.1172/JCI5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Spergel J M, Hsu W, Akira S, Thimmappaya B, Kishimoto T, Chen-Kiang S. NF-IL6, a member of the C/EBP family regulates E1A-responsive promoters in the absence of E1A. J Virol. 1992;66:1021–1030. doi: 10.1128/jvi.66.2.1021-1030.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taub D D, Lloyd A R, Conlon K, Wang J M, Ortaldo J R, Harada A, Matsushima K, Kelvin D J, Oppenheim J J. Recombinant human interferon-inducible protein 10 is a chemoattractant for human monocytes and T lymphocytes and promotes T cell adhesion to endothelial cells. J Exp Med. 1993;177:1809–1814. doi: 10.1084/jem.177.6.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward S G, Bacon K, Westwick J. Chemokines and T-lymphocytes: more than just an attraction. Immunity. 1998;9:1–11. doi: 10.1016/s1074-7613(00)80583-x. [DOI] [PubMed] [Google Scholar]
- 26.Wrighton C J, Hofer-Warbinek R, Moll T, Eytner R, Bach F H, de Martin R. Inhibition of endothelial cell activation by adenovirus-mediated expression of IκBα, an inhibitor of the transcription factor NFκB. J Exp Med. 1996;183:1013–1022. doi: 10.1084/jem.183.3.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wuthrich R P, Glimcher L H, Yui M A, Jevnikar A M, Dumas S E, Kelley V E. MHC class II, antigen presentation and tumor necrosis factor in renal tubular epithelial cells. Kidney Int. 1990;37:783–792. doi: 10.1038/ki.1990.46. [DOI] [PubMed] [Google Scholar]
- 28.Yang Y, Ertl H C J, Wilson J M. MHC class 1-restricted cytotoxic T lymphocytes to viral antigens destroy hepatocytes in mice infected with E1-deleted recombinant adenoviruses. Immunity. 1994;1:433–442. doi: 10.1016/1074-7613(94)90074-4. [DOI] [PubMed] [Google Scholar]
- 29.Yang Y, Xiang Z, Ertl H C J, Wilson J M. Upregulation of class I major histocompatibility complex antigens by interferon-γ is necessary for T-cell-mediated elimination of recombinant-infected hepatocytes in vivo. Proc Natl Acad Sci USA. 1995;92:7257–7261. doi: 10.1073/pnas.92.16.7257. [DOI] [PMC free article] [PubMed] [Google Scholar]